Abstract
In the social amoeba Dictyostelium, a terminal step in development is regulated by environmental O2. Prolyl 4-hydroxylase-1 (P4H1) was previously implicated in mediating the O2 signal, and P4H1-null cells require elevated O2 to culminate. The E3-ubiquitin ligase adaptor Skp1 is a P4H1 substrate, and here we investigate the function of PgtA, a dual function β3-galactosyltransferase/α2-fucosyltransferase that contributes the 2nd and 3rd sugars of the pentasaccharide cap formed on Skp1 hydroxyproline. Although pgtA-null cells, whose Skp1 contains only a single sugar (N-acetylglucosamine or GlcNAc), show wild-type O2 dependence of culmination, cells lacking AgtA, an α3-galactosyltransferase required to extend the trisaccharide, require elevated O2 as for P4H1-null cells. Skp1 is the only detectable protein modified by purified PgtA added to pgtA-null extracts. The basis for specificity of PgtA was investigated using native Skp1 acceptor glycoforms and a novel synthetic peptide containing GlcNAcα1,4-hydroxy(trans)proline. Cysteine-alkylation of Skp1 strongly inhibited modification by the PgtA galactosyltransferase but not the fucosyltransferase. Furthermore, native and synthetic Skp1 glycopeptides were poorly galactosylated, not processively fucosylated, and negligibly inhibitory, whereas the fucosyltransferase was active toward small substrates. In addition, the galactosyltransferase exhibited an atypical concentration dependence on UDP-galactose. The results provide the first evidence that Skp1 is the functional target of P4H1 in O2 regulation, indicate a gatekeeper function for the β3-galactosyltransferase in the PgtA dual reaction, and identify an unexpected P4H1-dependent yet antagonistic function for PgtA that is reversed by AgtA.
Eukaryotic organisms have evolved multiple mechanisms to sense O2 availability and accordingly regulate their functions. A major pathway in animals is the O2-dependent prolyl 4-hydroxylation of the transcriptional factor subunit hypoxia-inducible factor-1α (HIFα),2 which renders it a target of the VHL (von-Hippel Lindau)-type ubiquitin ligase leading to subsequent degradation by the 26S proteasome (1). In the mycetozoan Dictyostelium, the ortholog of animal HIFα-type prolyl 4-hydroxylases, P4H1, is also involved in O2 sensing (2). Genetic disruption of P4H1 increases the O2 requirement for culmination, the developmental process in which the multicellular slug converts to a fruiting body. In contrast, overexpression of P4H1 reduces the O2 requirement. These and other results suggest a model in which environmental O2 is a cue that influences the decision of the motile slug to differentiate, after migration to the soil surface, into a sessile aerial fruiting body best poised for dispersal of spores to more favorable environments.
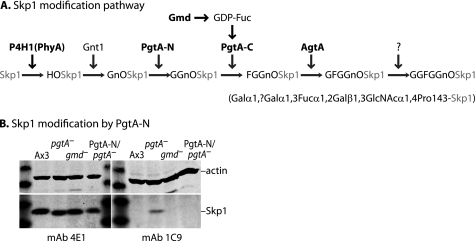
Dictyostelium lacks an obvious ortholog of HIFα but a known substrate for Dictyostelium P4H1 is Skp1, an adaptor in SCF (Skp1/Cullin 1/F-box protein)-type Ub ligases (3). E3SCFUb ligases and E3VHLUb ligases are evolutionarily related, and it is interesting that a subunit of the latter, VHL protein, recognizes a hydroxyproline target whereas in Dictyostelium, a subunit of the former type, Skp1, is prolyl 4-hydroxylated. Subsequent to hydroxylation, the Skp1 HyPro (HyPro143) is sequentially modified by five glycosylation reactions resulting in a pentasaccharide cap (Ref. 4, see Fig. 2A). A previous study showed that the O2 requirement for a mutant disrupted in pgtA, required for addition of the second and subsequent sugars, was normal (2). This suggested that, except possibly for the first sugar GlcNAc whose addition is mediated by Gnt1, the remainder of the glycosylation pathway fulfills an unrelated function, and raised the possibility that an alternative unknown protein actually mediates P4H1-dependent O2 signaling.
FIGURE 2.
Diagram of the Skp1 glycosylation pathway and glycoforms examined. A, glycoforms accumulating as a result of disruption of modification genes are indicated. Knock-out strains are available for all modification genes (in bold) except Gnt1 and the second αGalT activity. gmd encodes GDP-mannose 4,6-dehydratase involved in synthesis of GDP-Fuc from GDP-Man. B, glycosylation status of PgtA-N/pgtA-null strain. Whole cell extracts (100 μg of protein) were subjected to SDS-PAGE, Western-blotted, and probed with anti-actin (upper half of each blot), or mAb 4E1 for total Skp1, or mAb 1C9 for Gn-O-Skp1.
Addition of the second and third sugars of the Skp1 glycan is mediated by PgtA, a dual function β3GalT/α2FucT that results in formation of the blood group H type 1 antigen Fucα1,2Galβ1,3GlcNAcα1- on HyPro143 (5). Although the separate domains exhibit partial activity in vitro, the 2-domain architecture of PgtA supports processive addition of both sugars (6). Addition of the fourth sugar is mediated by AgtA, resulting in modification of Fuc by Gal in an α1,3-linkage (7). We report here that agtA-null cells, like P4H1-null cells, also require elevated O2 to culminate. Thus PgtA has an unexpected function that was masked by the next processing enzyme in the pathway. This implicates Skp1 as the functional target of P4H1 because PgtA negates the action of P4H1/Gnt1 and, as also shown here, Skp1 is the only acceptor substrate detected in crude extracts by biochemical complementation. Specificity for Skp1 is supported by in vitro studies of native Skp1 and a synthetic analog of the acceptor site prepared using a novel synthetic approach. PgtA might be subject to novel regulation as it exhibits unusual dependence on UDP-Gal concentration compared with AgtA. These properties offer clues for the mechanism of a proposed hierarchical regulation of culmination by additional cues whose effects are contingent upon initial O2-dependent prolyl-hydroxylation of Skp1. This novel pathway, distinct from other known cytoplasmic glycosylation pathways (8), may be important for the diverse aerobic protists whose genomes harbor potential coding sequences related to Skp1 modification genes in Dictyostelium (4), including pathogenic organisms such as the agent for human toxoplasmosis, Toxoplasma gondii (9).
EXPERIMENTAL PROCEDURES
Growth and Development
Cells were grown axenically, and tested for O2 dependence of development as described (2). Briefly, cells were deposited on filters in PDF buffer and allowed to develop for 42 h in the presence of the indicated concentration of O2 with the balance made up with N2. Development was evaluated morphologically and by counting spores in a hemacytometer.
Synthesis of the Skp1 Glycopeptide
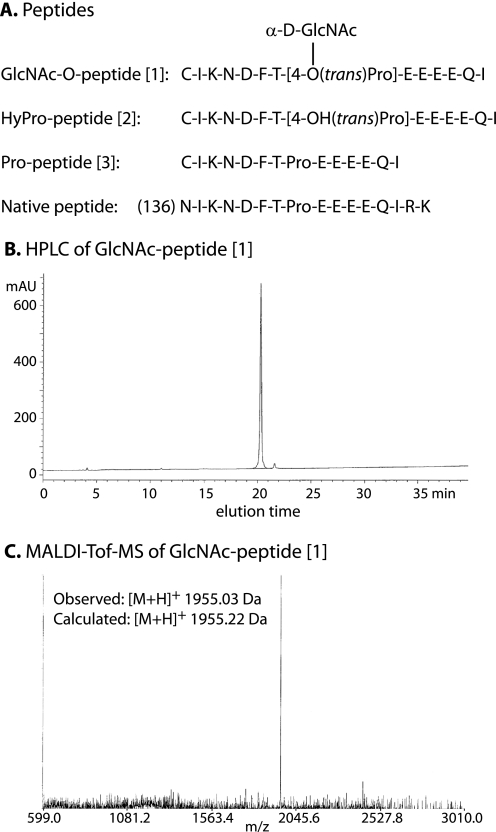
The full method and documentation are described under supplemental data. Briefly, an efficient approach was developed for the chemical synthesis of the GlcNAcα1,4-HyPro-containing glycopeptide [1] (Fig. 1A). First, peracetylated Fmoc-Hyp(α-d-GlcNAc)-OH was synthesized, which was employed in combination with other Fmoc-protected amino acids, for stepwise solid phase glycopeptide synthesis using a Rink Amide AM resin. The polymer-bound glycopeptide was liberated from the resin with concurrent cleavage of the acid-labile side-chain protecting groups, and the acetyl esters of the GlcNAc moiety of the glycopeptide were removed using 5% aqueous hydrazine hydrate in the presence of dithiothreitol. The resulting crude glycopeptide was purified by reverse-phase HPLC on a C18 column (Fig. 1B). The structural identity of the compound was confirmed by MALDI-TOF-MS (Fig. 1C) and NMR (see supplemental data). The corresponding HyPro-peptide [2] and Pro-peptide [3] (Fig. 1A) were prepared similarly. In analytical scale separations, all three peptides resolved as two closely spaced peaks with indistinguishable m/z values (data not shown). Re-chromatography of the separated fractions regenerated the same pair of peaks, indicative of potential interconversion of cis-trans isomers of the Thr-Pro peptide bond.
FIGURE 1.
Peptides synthesized. A, sequences of the 3 Skp1 peptide isoforms synthesized, and the corresponding Endo-Lys C-generated peptide from Skp1. B, reverse-phase (C18) HPLC analysis of GlcNAc-O-peptide [1] in an ascending gradient of acetonitrile in 0.1% trifluoroacetic acid, monitored at A215. C, MALDI-TOF-MS analysis of GlcNAc-O-peptide [1].
Antibodies
The Gn-O-peptide was coupled to keyhole limpet hemocyanin and bovine serum albumin (see supplemental data). The former was used to immunize BALB/c female mice, in a mixture with Freund's complete adjuvant for the primary injection, with incomplete adjuvant for boosting, and finally alone as an intravenous booster 3 days before euthanasia, as described (10). Hybridomas were prepared from spleen cells, and culture supernatants were screened for selective reactivity against the Gn-O-peptide relative to the unmodified peptide in ELISA assays. Positive supernatants were subjected to secondary screening by Western blot analysis against a panel of whole cell extracts, from mutant strains expressing discrete glycoforms of Skp1 (Fig. 2A). mAbs 1C9 (Fig. 2B) and 2F8 (not shown) were specific for the GlcNAc isoform.
Mass Spectrometry of Skp1
Skp1 was purified by C8-reversed phase HPLC (5) and analyzed by MALDI-TOF-TOF-MS using sinapinic acid on a Bruker Ultraflex II operated in positive ion mode. An aliquot was digested with endo-Lys C, fractionated on a C2/C18 reversed phase column (Amersham Biosciences), and analyzed similarly using α-hydroxy-4-cyano-cinnamic acid as the matrix (5).
PgtA/FT85
PgtA/FT85 was purified to near homogeneity from Dictyostelium as described (11).
Skp1
GlcNAc-O-Skp1 (Gn-O-Skp1) was purified to near homogeneity from strain HW260 (5), in which the pgtA gene is disrupted leading to accumulation of the monosaccharide form. GlcNAc-O-Skp1 was subjected to heating at 60 °C for 15 min, at 100 °C for 3 min, or to 6 m urea at 60 °C for 15 min. An aliquot of the urea-treated sample was treated with 7.5 mm DTT, slightly above the concentration (5 mm) in the activity assay, and alkylated with 17.5 mm iodoacetamide as described (12). The preparations were centrifuged at 15,000 g × 10 min after treatment to remove any insoluble material. The concentrations of urea, DTT, and iodoacetamide were lowered by multiple cycles of concentration/dilution in a centrifugal ultrafiltration device to levels that did not inhibit enzyme activity (data not shown). Gn-O-Skp1 was cleaved by incubation in a 1:50 (w/w) preparation of trypsin (Promega, mass spectrometry grade) in 0.08 m NH4CO3 (pH 8.0), 20% acetonitrile, at 37 °C for 20 h. GGn-O-Skp1 was purified to near homogeneity from strain HL250, which is mutationally disabled in its ability to form GDP-Fuc (13, 14).
Glycosyltransferase Assays
PgtA (FT85; β3GalT/α2FucT) was incubated with acceptor substrate, in the presence of 2 μm UDP-[3H]Gal or GDP-[3H]Fuc as indicated, for 2 h at 29 °C as described (5). Incorporation into Gn-O-Skp1 or GGn-O-Skp1 was assayed using the SDS-PAGE assay (15). Briefly, the reaction mixture was supplemented with 2 μg of soybean trypsin inhibitor (Sigma) as a marker that comigrates with Skp1 and electrophoresed on a 15–20% gradient SDS-PAGE gel. After fixation and minimal staining with Coomassie Blue, the gel was equilibrated in water, and 5 slices surrounding the trypsin inhibitor band were excised and analyzed in a liquid scintillation counter. Incorporation into peptide acceptor substrates was assayed by reversed phase HPLC on a C18 column, using an ascending gradient of acetonitrile in water with 0.1% trifluoroacetic acid. Peptide fractions were detected based on A215 and counted in a liquid scintillation counter. In the combined β3GalT/α2FucT reaction with Gn-O-peptide, which contained GDP-[3H]Fuc, dpm detected in the absence of UDP-Gal were subtracted as background. Incorporation into pNP or benzyl conjugates of sugars were assayed by adsorption of the reaction product to a C18-SepPak, and elution with methanol and counting by liquid scintillography (6).
UDP-Gal Hydrolysis Assay
The first two water flow-through fractions from reaction mixtures applied to a C18-SepPak for the glycosyltransferase reaction (see above) were pooled and applied to 0.8 ml Dowex-1 (HCl-form) packed in a column. The flow-through and wash fractions were pooled and quantitated in a liquid scintillation counter.
RESULTS
Roles of PgtA and AgtA in Development
Skp1 is sequentially modified by six enzyme activities encoded by at least four genes. Strains carrying disruptions of phyA/P4H1 (3), pgtA (5), or agtA (7) have been described, and these strains accumulate unmodified Skp1 or Skp1 bearing a glycan consisting of one or three sugars, respectively, compared with the pentasaccharide that accumulates in the parental normal strain (Fig. 2A). In addition, a gmd− strain (HL250) with a deficiency in conversion of GDP-Man to GDP-Fuc accumulates Skp1 with a disaccharide (13, 14, 16). Because this strain is defective in all fucosylation, we reexamined a previously constructed strain in which the N-terminal domain of the PgtA diglycosyltransferase (PgtA-N) is constitutively expressed in a pgtA-null background, and whose extracts exhibit β3-GalT but not α2FucT activity as expected (5). To determine if Skp1 is modified by Gal in vivo, a mAb was isolated that could differentiate between Gn-O-Skp1 and GGn-O-Skp1. To serve as an immogen (and later as a potential substrate for PgtA), a homolog of the Skp1 glycopeptide was chemically synthesized as described under “Experimental Procedures,” Fig. 1 and supplemental data. A new synthetic route was developed to reach the glycosylated HyPro building block, whose structure was validated by 1H- and 13C-NMR, for stepwise solid phase peptide synthesis. The purity and identity of the peptide were confirmed by HPLC and MALDI-TOF-MS (Fig. 1 and supplemental data). The Gn-O-Skp1 glycopeptide was coupled to KLH and used to generate independent mAbs 1C9 and 2F8 that are specific for Gn-O-Skp1 compared with unmodified Skp1 or Skp1 expressing 2, 3, or 5 sugars (Fig. 2B and data not shown). Skp1 accumulated normally in the PgtA-N/pgtA− strain, based on probing of cell extracts with the pan-specific mAb 4E1. As observed for the GDP-Fuc-negative strain HL250, Skp1 in this strain was not detected by mAb 1C9 (Fig. 2B), indicating processing to the disaccharide glycoform.
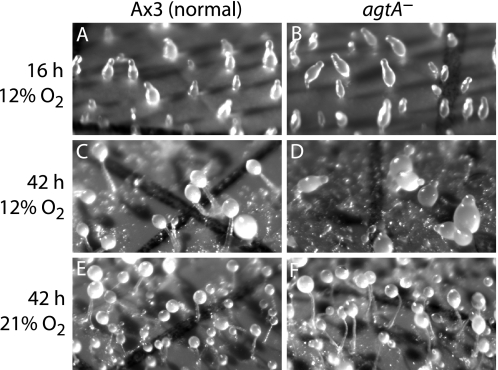
Each of the pathway mutants formed normal appearing fruiting bodies when developed on filters under standard conditions including overhead room fluorescent lighting and 21% (normal atmospheric) O2, as reported previously (2, 7). However, striking differences were observed when cells were developed in mild hypoxia, 12% O2. Whereas Ax3 (normal) and pgtA-null cells (not shown, Ref. 2) formed normal appearing fruiting bodies within 21 h, P4H1-null cells (not shown; 2) and, unexpectedly, agtA-null cells did not progress past the slug stage even after 42 h (Fig. 3, C and D). Accumulation of slime trails and larger slug size suggested merging of slugs during extensive migration. Slug formation in hypoxia was normal (Fig. 3, A and B), and culmination was normal at 21% O2 (Fig. 3, E and F). This suggests that PgtA has a cellular function that negates the P4H1-dependent checkpoint for the slug-to-fruit switch. In turn, AgtA appears to act on the product of the PgtA reaction to neutralize the negative PgtA signal, thereby restoring the initial action of P4H1.
FIGURE 3.
PgtA induces developmental arrest in mild hypoxia in the absence of subsequent processing. The action of PgtA was examined using a strain disrupted in the subsequent pathway gene, agtA. The normal strain Ax3 and the agtA-null strain were developed at 12% (hypoxic) or 21% (ambient) O2. A and B, both strains formed normal upright slugs at 16 h in hypoxia or normoxia (not shown). C and D, in hypoxia, Ax3 (normal) cells culminated into fruiting bodies by 22 h (42 h shown), whereas agtA-null cells remained as slugs. E and F, both strains culminated by 22 h in normoxia. Fruiting bodies consist of aerial, spherical spore clusters supported by vertical stalks anchored to the filter surface. The grid lines are 150-μm wide.
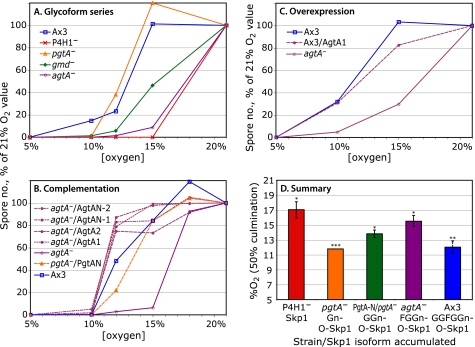
P4H1-null cell culmination is rescued at higher O2 levels, by an unknown mechanism that does not involve Skp1 modification (2). Therefore, O2 thresholds for the slug-to-fruit switch were determined for mutant strains incubated under a range of defined O2 levels. Culmination was quantified by counting the number of spores formed, which in all cases correlated with the extent of fruiting body formation as visualized microscopically (data not shown). In the trial shown in Fig. 4A, normal and pgtA-null cells required 15% O2 for all slugs to culminate, in contrast to P4H1- and agtA-null cells, which required 21% O2. As observed previously (2), the exact O2 threshold for maximal development varied from trial to trial, and is influenced by light, NH3, and other factors. The relative thresholds shown in Fig. 4 were conserved in all trials. Average 50% values, calculated from the levels of O2 required for 50% spore formation relative to the value required for 50% spore formation of the normal strain Ax3 in the same trial, are shown in Fig. 4D.
FIGURE 4.
Effects of pathway mutations on O2 requirement for culmination. Development was quantitated by counting the number of spores produced after 42–45 h, and plotting relative to the value at 21% O2 (100%). A, strains expressing Skp1 (phyA−), Gn-O-Skp1 (pgtA−), GGn-O-Skp1 (gmd−), FGGn-O-Skp1 (agtA−), and GGFGGn-O-Skp1 (normal Ax3) were examined. 100% values were 1.6–2.3 × 107 spores. B, GGn-Skp1 accumulation was recapitulated by expressing Pgt-N, which encodes the β3GalT domain of PgtA, in a pgtA− strain (see Fig. 2B). agtA-null cells were complemented by overexpression of either full-length AgtA (2 clones) or AgtA-N (2 clones) under control of the discoidin promoter. Ax3 and agtA-null cells were included for reference. 100% values were 1.1–3.8 × 107 spores. C, a strain overexpressing AgtA is compared with Ax3 and the agtA-null strain. 100% values were 3.0–4.4 × 107 spores. Values at 2.5% O2 were the same as at 5%, and values at 40 and 70% O2 were very similar to those at 21% (data not shown). Trends were identical in all trials; and each result was confirmed in one or more additional trials. D, summary of data from these, and replicate trials are not shown. The average value of O2 required for 50% maximal spore formation of each strain was obtained by averaging the differences from the 50% value for Ax3 in each same trial, and adding those values (±S.E.,*) to the average value for Ax3 across all trials. Colors correspond to traces in panels A–C. **, value for Ax3 includes its own S.E. ***, only a single trial confirming previous results (2) was performed for pgtA− cells in this series.
Strain HL250 (gmd−), which accumulates the disaccharide form of Skp1 (GGn-O-Skp1) because of inability to form GDP-Fuc, showed an intermediate requirement for O2 to culminate (Fig. 4A). Because HL250 cells are globally deficient in fucosylation and therefore the effect might be indirect, the PgtA-N/pgtA− strain described in Fig. 2, which also accumulates the disaccharide form, was examined. This strain similarly showed a modestly elevated requirement for O2 relative to that of the pgtA-null and Ax3 strains in this (Fig. 4B) and other trials, as summarized in Fig. 4D.
To verify that the agtA-null phenotype was specific to the absence of AgtA, full-length His6-AgtA was expressed under control of the semi-constitutive discoidin promoter, which was shown previously to highly overexpress the enzyme activity in a normal genetic background (7). The O2 threshold was restored to slightly lower than the wild-type level in each of two clones (Fig. 4B). However, overexpression in the normal background did not decrease the O2 threshold (Fig. 4C). The results suggest AgtA expression negates the action of PgtA by completing the glycan chain.
AgtA consists of an N-terminal CAZy family GT77 domain, which alone catalyzes the attachment of αGal-1 to Fucα1-benzyl, and C-terminal repeats that may fold as a β-propeller (7). To test whether the N-terminal domain is sufficient for rescuing the high O2 requirement for agtA− culmination, AgtA-N was expressed in the agtA− background. These cells exhibited a reduced O2 requirement indistinguishable from that of cells rescued with full-length AgtA (Fig. 4B). To determine if AgtA-N rescued Skp1 galactosylation as expected, Skp1 was purified from the AgtA-N/agtA− strain. MALDI-TOF-MS analysis showed an m/z value for the full-length protein (Fig. 5A) of 52 Da above the expected value considering expected processing events (5), which is most consistent with the presence of 5 sugars. This was confirmed by analysis of the glycopeptide after endo-Lys C digestion (Fig. 5B), suggesting that complementation by AgtA was due to rescued glycosylation per se rather than an effect of its C-terminal domain.
FIGURE 5.
Skp1 α-galactosylation is rescued by overexpressed AgTA-N. A, MALDI-TOF-MS analysis of Skp1 purified from AgtA-N/agtA− cells. Note that Skp1 consists of two genetic isoforms (12, 18), which differ by a Ser or Ala at codon 39 (difference of 16 Da) which broadens the peak. B, similar analysis of glycopeptide purified by RP-HPLC from endo-Lys C-digested Skp1.
PgtA Acceptor Substrates in Cells
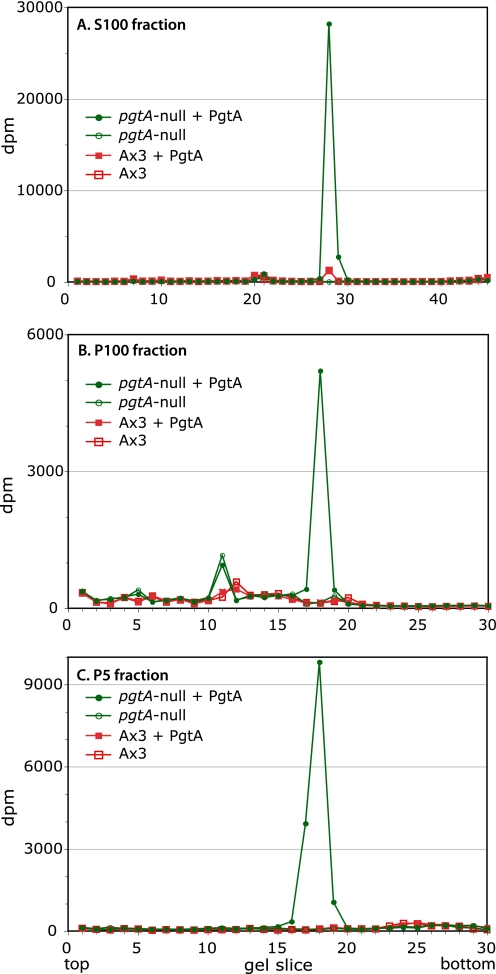
To identify potential substrates that might mediate PgtA function in regulating the O2 threshold, a biochemical complementation approach was implemented by incubating crude extracts of pgtA-null cells with recombinant PgtA in the presence of UDP-[3H]Gal. Analysis of the desalted cytosol (S100) reaction by SDS-PAGE showed that a protein at the position of Skp1 was the major species that incorporated 3H (Fig. 6A). Less than 0.1% of the incorporation at this position was observed in the absence of PgtA. On the order of 5% incorporation was observed in parental Ax3 extracts, indicating that a fraction of Skp1 normally exists in the monosaccharide form. No other gel slices were detectably labeled over the PgtA-minus controls. Two particulate fractions from mutant cells, P100 (mitochondria, lysosomes, and microsomes) and P5 (nuclear), also showed substantial and specific labeling of Skp1 that was not observed in normal cell extracts (Fig. 6, B and C), confirming previous studies that Skp1 is widely distributed across cell fractions (10, 18). The minimal PgtA-independent incorporation of label seen at other Mr positions in the particulate extracts probably reflects the activity of endogenous enzymes. Thus, no other PgtA targets could be detected in the extracts, suggesting that Skp1 may be the only acceptor substrate for the β3GalT activity of PgtA, as observed previously for later enzymes in the pathway (11, 7).
FIGURE 6.
Specificity of PgtA in extracts. Extracts were prepared from stationary phase pgtA-null and parental Ax3 cells and incubated with purified PgtA and UDP-[3H]Gal. The reaction mixture was subjected to SDS-PAGE. After fixation, the gel lanes were cut into 30–45 slices, and incorporated radioactivity was determined by liquid scintillation counting. A, S100 fractions were incubated in the presence or absence of PgtA. B, P100 fractions were analyzed. C, P5 fractions were analyzed.
Parameters of PgtA Recognition of Skp1 in Vitro
The above studies indicate that PgtA contributes a critical function to O2-dependent culmination, and that Skp1 may be the functional target of the enzyme in cells. The activity of highly purified PgtA was investigated to address the basis for its high degree of specificity toward Skp1. Gn-O-Skp1 was isolated from pgtA-null cells and purified to near homogeneity. Using a highly purified preparation of native PgtA from Dictyostelium, and the donor UDP- [3H]Gal, assay conditions were developed in which transfer of Gal to Gn-O-Skp1, which becomes β3-linked to the GlcNAc (5), was dependent on the concentration of enzyme and substrate, and time. <10% of substrate was consumed, and product greatly exceeded the molar level of enzyme (data not shown). Incorporation into Skp1 was determined by excising the Skp1 band after SDS-PAGE, which allowed high sensitivity due to low backgrounds (typically <40 dpm).
To assess the importance of the folded state for acceptor activity, Gn-O-Skp1 was subjected to denaturing conditions and recovered. Western blotting was employed to correct for protein recovery (Fig. 7A, inset). Skp1 remained soluble after heating to 100 °C, treatment with urea, or alkylation with iodoacetamide in the presence of urea. Gn-O-Skp1 acceptor activity was 70% stable to heating or urea-treatment (Fig. 7A). In contrast, alkylation of the urea-treated sample reduced acceptor activity to 20%, or less than 30% of the activity of urea-treated Skp1. The limited availability of Gn-O-Skp1 precluded verification of alkylation, but parallel treatment of native unmodified Skp1 purified under non-denaturing conditions from Escherichia coli and MALDI-TOF-TOF-MS analysis of the intact and endo-Lys C-digested protein indicated that the majority of each of the 5 Cys residues was carboxamidomethylated3 as observed previously (16, 5). Although Skp1 was treated with 7.5 mm DTT prior to alkylation in both studies, this is similar to the concentration of DTT used in the assay (5 mm) and found not to affect Skp1 substrate activity. In addition, similar analysis of an endo-Lys C-digest of untreated Skp1 from E. coli failed to reveal evidence for preformed disulfide bonds,3 indicating that the alkylation protocol did not inhibit Skp1 due to reduction of disulfide bonds. Since Cys residues are not adjacent to the Pro-143 linkage site either in the primary sequence or in crystal structures of Skp1 complexed to F-box proteins, optimal activity of β3GalT may depend on a distal determinant affected by alkylation.
FIGURE 7.
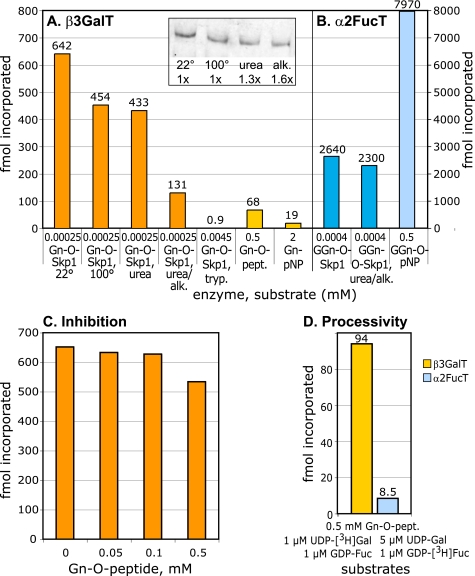
Acceptor substrate specificity of PgtA. A, β3GalT activity. Purified PgtA was incubated with the indicated acceptor in the presence of 1 μm UDP-[3H]Gal for 2 h at 29 °C. Gn-O-Skp1 was isolated from pgtA− cells, and pretreated as indicated. Gn-O-Skp1 was repurified after treatment with 8 m urea or urea and iodoacetamide (alk.), and the volume assayed was corrected by a factor based on recovery as determined by Western blotting of identical volumes (inset). Assays using Skp1 were quantitated by the SDS-PAGE method and background values (<40 dpm) were subtracted. Trypsinized Skp1, which was not detectable by Western blotting (data not shown), was treated with soybean trypsin inhibitor, aprotinin and phenylmethylsulfonyl fluoride, which do not inhibit PgtA. Incorporation into glycopeptides was assayed using the HPLC method, and incorporation into GlcNAcα1-pNP was assayed using the C18-SepPak method. B, α2FucT activity. Purified PgtA was assayed similarly in the presence of 1 μm GDP-[3H]Fuc. GGn-O-Skp1 was isolated from strain HL250, and pretreated as indicated. C, sensitivity of the GalT activity to the Skp1 glycopeptide. The GalT reaction using Gn-O-Skp1 described in panel A was conducted in the presence of the indicated concentration of synthetic glycopeptide. D, reaction using Gn-O-peptide was tested for processivity by conducting the reaction in the presence of the indicated concentrations of UDP-Gal and GDP-Fuc. The GalT and FucT reactions were monitored in parallel reactions using UDP-[3H]Gal and GDP-[3H]Fuc, respectively. Incorporation of [3H]Fuc in absence of UDP-Gal was subtracted as background. The FucT reaction was much slower despite the 5× higher level of UDP-Gal. 1 fmol is 44 dpm in these reactions. All results were confirmed in independent trials.
In a second approach, the sufficiency of the acceptor GlcNAc, in the context of nearby amino acids, to serve as an acceptor was tested. Digestion of Skp1 with trypsin dramatically reduced the acceptor activity. A ∼20-fold higher concentration of trypsinized Skp1 yielded about 500-fold less incorporation of Gal in the GalT assay (Fig. 7A), suggesting that the glycopeptide was 10,000-fold less active. To test the acceptor glycopeptide at higher concentration and in the absence of potential interference from other Skp1 peptides, the synthetic glycopeptide [1] described above (Fig. 1), similar though not identical to the version expected after trypsin digestion, was employed. The corresponding HyPro- and Pro-peptides were also synthesized and used as negative controls. At the highest concentration of synthetic Gn-O-peptide [1] tested, 0.5 mm, PgtA exhibited only 10% of the activity relative to 0.25 μm Skp1 (Fig. 7A). Although by this measure the glycopeptide was 20,000-fold less active than Skp1, similar to the 10,000-fold difference based on analyzing the tryptic glycopeptide, incorporation was specific based on inactivity of the corresponding HyPro- or Pro-peptides (data not shown). In addition, the Gn-O-peptide [1] was more active than GlcNAcα1-pNP, exhibiting 3-fold higher activity at 4-fold lower concentration, or about a 12-fold difference. The ability of the glycopeptide to bind PgtA was tested by measuring its effect on the reaction with native Skp1. Even at a 2000-fold concentration excess relative to Skp1, the glycopeptide inhibited the reaction <15% (Fig. 7C). Thus the GlcNAc-O-peptide may not adopt a conformation preferred by the PgtA active site. Altogether, the results indicate that the acceptor activity requires presentation of the modification site in the context of the full-length protein, and is consistent with the high degree of specificity of PgtA for Skp1 in cell extracts.
PgtA processively catalyzes the transfer of Fuc from GDP-Fuc to the β3Gal in an α2-linkage (5). In contrast to the GalT acceptor activity, the FucT acceptor activity of GGn-O-Skp1 (isolated from strain HL250) was unaffected by treatment with urea and alkylation (Fig. 7B). Furthermore, the α2FucT activity exhibited robust activity toward a small molecule form of its acceptor substrate. Although GGn-O-pNP exhibited only a 3-fold higher activity than native Skp1 when tested at a 1000-fold higher concentration (Fig. 6B), the 330-fold reduced activity of the disaccharide acceptor by this measure is 30–60-fold better than the relative activity of the glycopeptide analog for the GalT reaction. Therefore the FucT has different requirements for processing Skp1 and is less dependent on determinants distal to the glycosylation site.
Finally, when sugar nucleotide donor substrates for both reactions were included in the glycopeptide reaction, the FucT activity did not keep pace with the GalT activity (Fig. 7D), in contrast to reactions with full-length Skp1 (5). Therefore, though the glycan modification site of Skp1 has weak activity in isolation, other parts of Skp1 are required for processive processing.
Unusual Donor Substrate Dependence of PgtA in Vitro
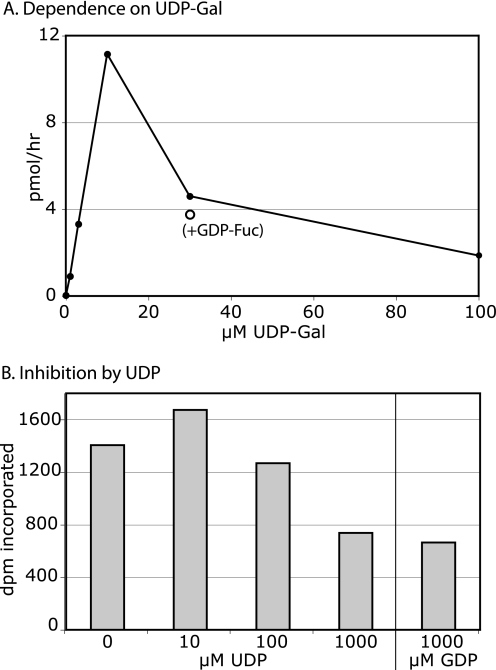
The GalT activity was approximately linearly dependent on UDP-Gal concentration up to 10 μm but reduced at higher concentrations (Fig. 8A). Addition of GDP-Fuc to the reaction did not relieve inhibition. PgtA exhibited negligible UDP-Gal hydrolysis activity (data not shown), indicating that reduced activity was not the result of UDP-Gal depletion. To determine if inhibition might be caused by accumulation of UDP from the transferase reaction, the assay was conducted in the presence of increasing amounts of UDP (Fig. 8B). No inhibition was observed except at the highest level tested, 1 mm, which was also observed using 1 mm GDP and was far above the concentration of UDP-Gal in the reactions. In contrast, the FucT exhibits Michaelis-Menten-type dependence on its sugar nucleotide donor, GDP-Fuc, and is strongly inhibited by the product GDP (11).
FIGURE 8.
Donor substrate specificity of β3GalT activity of PgtA. A, GalT assay was conducted in the presence of 1.25 μm Gn-O-Skp1 and the indicated concentration of UDP-[3H]Gal for 10 min at 29 °C. (○) marks a data point for a reaction that included 5 μm GDP-Fuc. Incorporation into Skp1 was measured using the SDS-PAGE method. B, GalT assay was conducted in the presence of 7 μm GlcNAc-peptide-BSA conjugate, 10 μm UDP-[3H]Gal, and the indicated concentration of nucleotide diphosphate, for 2 h at 29 °C. Results are representative of two independent trials.
DISCUSSION
Function of Cytoplasmic Glycosylation in O2 Regulation
In the absence of all pathway activity (P4H1−), slugs required 17% O2 for half-maximal culmination (Fig. 4D). The mechanism by which 17% O2 is sensed in P4H1− cells is unknown (2). With full pathway activity (wild-type), only 12% O2 is required. We assume that the pathway contributes a positive signal to allow culmination at as low as 12% O2, which is still higher than the 2.5% O2 level required for proliferation and slug formation (2). As shown here, partial pathway activity as determined by genetic perturbations results in intermediate O2 requirements. Whereas genetic elimination of PgtA results in a 12% requirement similar to when the pathway has full activity, restoration of only the β3GalT activity of PgtA raises the requirement to 14%, an effect that was confirmed in a global GDP-Fuc− strain indicating that the action of PgtA involves its enzyme activity. Restoration of both PgtA activities, as determined using agtA− cells, raised the O2 requirement to almost 16%, close to the value when the pathway is inactive. Because overexpression of the catalytic domain of AgtA alone restored both glycosylation (Fig. 5) and the normal O2 requirement, it is likely that the effect of AgtA depletion is due to the glycosylation deficiency per se. Although the effects on culmination occurred over a rather narrow range of O2 levels, the consequences of hypoxia on the population of organisms in the culture dish can be dramatic as shown in Fig. 3, where at 12% O2, agtA− cells do not culminate but normal cells culminate normally. The up-down-up-down excursions of the O2 requirement revealed by genetic analysis of the pathway (Fig. 4D) suggest the existence of a hierarchy of dependent control steps that are brought to bear on the slug to ultimately permit execution of the slug-to-fruit switch in its normal soil environment.
Potential Involvement of Skp1 as the Functional PgtA Acceptor Substrate in O2 Regulation
Previous studies suggested that Skp1 is the only protein modified by the FucT activity of PgtA in cells, based on analysis of cytoplasmic proteins that are metabolically labeled using [3H]Fuc, or that can be labeled by GDP-[3H]Fuc in the presence of PgtA in a cytosolic extract of gmd− cells (19). However, the possible occurrence of an acceptor substrate on cytoplasmically exposed surfaces of structures in the particulate fraction, could not be easily addressed because of microsomal FucT activities. Because the pgtA knock-out only affects galactosylation mediated by PgtA, it was possible to search for potential substrates in cytosolic, microsomal, nuclear and other particulate fractions of pgtA− cells. Only evidence for Skp1 labeling was obtained (Fig. 6), suggesting that Skp1 may be the only protein modified by PgtA in cells. This strongly implies that Skp1 is the functional target of PgtA in its effects on O2 regulation. Previous evidence that Skp1 is the only detectable substrate in the cytosol of AgtA (7), which acts on the product of PgtA and reverses its effect on P4H1, is consistent with this conclusion.
Potentially PgtA could be specific for GlcNAc, which itself might be selectively applied to Skp1 based on specificity of P4H1 or Gnt1. However, the specificities of these enzymes are not known. There is no straightforward method for determining acceptor specificity of prolyl hydroxylases, and a Gnt1 mutant strain is not available to test the specificity of the Skp1 αGlcNAcT (Gnt1). Although no genes that might encode a cytoplasmic protein related to Gnt1 are evident, a Spy-like sequence, related to genes that encode O-β-GlcNAc-transferase in plants and certain protists, is found in the genome (20). Considering the possible presence of β-GlcNAc on proteins, and occurrence of β-GlcNAc on a cytoplasmic precursor for N-glycan synthesis and α-GlcNAc on UDP-GlcNAc, there may be a need for additional determinants for the specificity observed for Skp1.
Whereas subjecting Gn-O-Skp1 to denaturing conditions, including boiling or 6 m urea, does not markedly reduce its GalT acceptor activity, alkylation with iodoacetamide in the presence of urea reduces activity by 80% (Fig. 7A). Dd-Skp1 has five Cys residues, which are each distant from the P143 modification site in both primary sequence and in crystal structures of Skp1 with F-box proteins (e.g. 21). All but one shows some degree of conservation throughout phylogeny, though none are absolutely conserved in other protists with pgtA-like genes,4 suggesting that alkylation may not be acting by inhibiting a critical Cys-function per se. In contrast, both the denaturing treatments and alkylation abolish acceptor activity toward the next enzyme AgtA (17). Therefore, these enzymes appear to depend on distinct determinants of Skp1 separate from the region of the Pro-143 modification site. This is consistent with the weak activities of PgtA GalT (Fig. 7) and AgtA (17) toward small molecule mimics of the Skp1 acceptor site. Skp1 glycopeptides (native or synthetic) were about 4 orders of magnitude less active than full-length Skp1 as β3GalT substrates (Fig. 7A), though still an order of magnitude better than simple Gn-pNP. The negligible inhibitor activity of the Gn-O-peptide (Fig. 7C), which suggests that it is not recognized by the enzyme, is consistent with a model in which appropriate presentation of the GlcNAc acceptor site to the β3GalT depends on other parts of the Skp1 protein. Whether alkylation disturbs this presentation, or directly affects recognition by PgtA remains to be determined. These aspects of Skp1 folding may contribute to the specificity of PgtA for Skp1 in extracts (Fig. 6) and cells.
In contrast, the acceptor activity of Skp1 toward the FucT activity was not affected by denaturing treatments or alkylation (Fig. 7B), and the GGn acceptor disaccharide-linked to pNP or benzyl is a relatively good acceptor albeit not as nearly as effective as full-length Skp1 (11). Interestingly, the free FucT domain can modify the disaccharide conjugate but not full-length Skp1 in vitro (6). This suggests that the GalT domain assists in presenting Skp1 to the FucT consistent with the processive nature of the in vitro reaction. However, presentation to the FucT is not affected by alkylation, indicating that the affected determinants are important for galactosylation per se. Thus the FucT may unconditionally extend the glycan once modified by the GalT, contingent on GDP-Fuc availability.
The GalT activity of PgtA exhibits an unusual dependence on UDP-Gal concentration, with activity peaking at 10 μm and declining at higher concentrations (Fig. 8). Inhibition is not due to inability to complete the double reaction, not due to accumulation of UDP, as the enzyme is not inhibited by this nucleotide, or to introduced defects as native untagged proteins were used. Since the donor substrate typically binds first in superfamily A GTs (22), UDP-Gal may have novel allosteric activity. In comparison, the FucT activity of PgtA (11), and the subsequent AgtA activity (17), exhibit normal Michaelis-Menten dependence on their sugar nucleotide donor substrates with sub-μm and μm Km values, respectively. The divergent dependences on UDP-Gal concentration by sequential pathway enzymes suggests that UDP-Gal dynamics, potentially coupled to strong up-regulation of UDP-Glc epimerase activity during development (23) and consumption of UDP-Gal by spore coat polysaccharide synthesis (24), may influence β3-galactosylation of Skp1 in cells.
Role of Skp1 in O2 Signaling
As discussed above, the specificity of PgtA for Skp1, based on the biochemical complementation data and the dependence of PgtA on critical structural features of Skp1, indicate that PgtA signals via Skp1 in cells. The ability of PgtA to reverse the effect of P4H1 action strongly implicates Skp1 as the mediator of P4H1 activity in O2 signaling as well. The serial modifications of Skp1 represent a potential hierarchy of stepwise control points for culmination that are contingent upon O2. Light, NH3 and other environmental cues also influence the slug-to-fruit switch (2), and their effects are potentially integrated by means of this hierarchical pathway, possibly via the state of Skp1 or internal metabolic signals such as UDP-Gal or GDP-Fuc. This model raises the question of how Skp1 functions to regulate the slug-to-fruit switch. The animal model shows that hydroxylation by PHD2 controls the stability of HIFα, which in turn controls the proteome by a transcriptional mechanism (1). In contrast, the hydroxylation target in Dictyostelium appears to be a subunit of the large family of E3SCFUb-ligases evolutionarily related to the E3 that degrades animal HIFα (25). This suggests that hydroxylation/glycosylation in Dictyostelium more directly modulates the proteome, not transcriptionally, but by controlling its stability via polyubiquitylation as a signal for proteasomal degradation. This concept is in accord with many examples of involvement of E3SCFUb-ligases in cellular regulation (25), but represents a highly novel and sophisticated form of SCF-regulation. Further studies are required to determine if modification of Skp1, which involves >90% of the steady state cellular pool, is contingent during development, and if SCF-associated Skp1 is affected similarly.
Supplementary Material
Acknowledgments
We thank Scherwin Henry (University of Florida ICBR Hybridoma Core Laboratory) for preparing the monoclonal antibodies and Wendy Ives for the HPLC analysis of synthetic peptides.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM84383 and 2R01-GM37539 (to C. M. W.) and 2R01-CA088986 (to G. J. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental data, Schemes S1–S3, and Figs. S1–S3.
H. van der Wel, J. M. Johnson, and C. M. West, unpublished data.
C. M. West and H. van der Wel, unpublished studies.
- HIF
- hypoxia-inducible factor
- AgtA
- Dictyostelium α3-galactosyltransferase (α3GalT) encoded by agtA
- Fuc
- l-fucose (pyranose form)
- Gal
- d-galactose (pyranose form)
- GlcNAc
- N-acetyl-d-glucosamine (pyranose form)
- Gmd
- GDP-mannose 4,6-dehydratase
- GT
- glycosyltransferase
- HyPro
- hydroxyproline
- mAb
- monoclonal antibody
- Man
- d-mannose (pyranose)
- P4H1
- Dictyostelium prolyl 4-hydroxylase-1 encoded by phyA locus
- PgtA
- dual-function Dictyostelium β3-galactosyltransferase/α2-fucosyltransferase (β3GalT/α2FucT) encoded by pgtA
- pNP
- paranitrophenyl
- SCF
- complex consisting of Skp1, cullin 1, and an F-box protein
- VHL
- von-Hippel Lindau protein
- Ub
- ubiquitin
- DTT
- dithiothreitol
- MALDI-TOF-MS
- matrix-assisted laser desorption/ionization-time of flight-mass spectrometry.
REFERENCES
- 1.Kaelin W. G., Jr., Ratcliffe P. J. (2008) Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 2.West C. M., van der Wel H., Wang Z. A. (2007) Development 134, 3349–3358 [DOI] [PubMed] [Google Scholar]
- 3.van der Wel H., Ercan A., West C. M. (2005) J. Biol. Chem. 280, 14645–14655 [DOI] [PubMed] [Google Scholar]
- 4.West C. M., van der Wel H., Sassi S., Gaucher E. A. (2004) Biochim. Biophys. Acta. 1673, 29–44 [DOI] [PubMed] [Google Scholar]
- 5.van der Wel H., Fisher S. Z., West C. M. (2002) J. Biol. Chem. 277, 46527–46534 [DOI] [PubMed] [Google Scholar]
- 6.van der Wel H., Morris H. R., Panico M., Paxton T., North S. J., Dell A., Thomson J. M., West C. M. (2001) J. Biol. Chem. 276, 33952–33963 [DOI] [PubMed] [Google Scholar]
- 7.Ercan A., Panico M., Sutton-Smith M., Dell A., Morris H. R., Matta K. L., Gay D. F., West C. M. (2006) J. Biol. Chem. 281, 12713–12721 [DOI] [PubMed] [Google Scholar]
- 8.Funakoshi Y., Suzuki T. (2009) Biochim. Biophys. Acta. 1790, 81–94 [DOI] [PubMed] [Google Scholar]
- 9.West C. M., van der Wel H., Blader I. J. (2006) Methods Enzymol. 417, 389–404 [DOI] [PubMed] [Google Scholar]
- 10.Kozarov E., van der Wel H., Field M., Gritzali M., Brown R. D., Jr., West C. M. (1995) J. Biol. Chem. 270, 3022–3030 [DOI] [PubMed] [Google Scholar]
- 11.West C. M., Scott-Ward T., Teng-umnuay P., van der Wel H., Kozarov E., Huynh A. (1996) J. Biol. Chem. 271, 12024–12035 [DOI] [PubMed] [Google Scholar]
- 12.West C. M., Kozarov E., Teng-umnuay P. (1997) Gene 200, 1–10 [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Yanes B., Mandell R. B., Girard M., Henry S., Aparicio O., Gritzali M., Brown R. D., Jr., Erdos G. W., West C. M. (1989) Dev. Biol. 133, 576–587 [DOI] [PubMed] [Google Scholar]
- 14.Schiller B., Hykollari A., Voglmeir J., Pöltl G., Hummel K., Razzazi-Fazeli E., Geyer R., Wilson I. B. H. (2009) Biochem. J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng-Umnuay P., van der Wel H., West C. M. (1999) J. Biol. Chem. 274, 36392–36402 [DOI] [PubMed] [Google Scholar]
- 16.Teng-umnuay P., Morris H. R., Dell A., Panico M., Paxton T., West C. M. (1998) J. Biol. Chem. 273, 18242–18249 [DOI] [PubMed] [Google Scholar]
- 17.Ketcham C., Wang F., Fisher S. Z., Ercan A., van der Wel H., Locke R. D., Sirajud-Doulah K., Matta K. L., West C. M. (2004) J. Biol. Chem. 279, 29050–29059 [DOI] [PubMed] [Google Scholar]
- 18.Sassi S., Sweetinburgh M., Erogul J., Zhang P., Teng-Umnuay P., West C. M. (2001) Glycobiology 11, 283–295 [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Yanes B., Cicero J. M., Brown R. D., Jr., West C. M. (1992) J. Biol. Chem. 267, 9595–9605 [PubMed] [Google Scholar]
- 20.Banerjee S., Robbins P. W., Samuelson J. (2009) Glycobiology 19, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. (2000) Nature 408, 381–386 [DOI] [PubMed] [Google Scholar]
- 22.Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 23.Telser A., Sussman M. (1971) J. Biol. Chem. 246, 2252–2257 [PubMed] [Google Scholar]
- 24.West C. M., Nguyen P., van der Wel H., Metcalf T., Sweeney K. R., Blader I. J., Erdos G. W. (2009) Eukaryot. Cell 8, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willems A. R., Schwab M., Tyers M. (2004) Biochim. Biophys. Acta. 1695, 133–170 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.