Abstract
Group I metabotropic glutamate receptors (mGluRs) are coupled via phospholipase Cβ to the hydrolysis of phosphoinositides and function to modulate neuronal excitability and synaptic transmission at glutamatergic synapses. The desensitization of Group I mGluR signaling is thought to be mediated primarily via second messenger-dependent protein kinases and G protein-coupled receptor kinases. We show here that both mGluR1 and mGluR5 interact with the calcineurin inhibitor protein (CAIN). CAIN is co-immunoprecipitated in a complex with Group I mGluRs from both HEK 293 cells and mouse cortical brain lysates. Purified CAIN and its C-terminal domain specifically interact with glutathione S-transferase fusion proteins corresponding to the second intracellular loop and the distal C-terminal tail domains of mGluR1. The interaction of CAIN with mGluR1 could also be blocked using a Tat-tagged peptide corresponding to the mGluR1 second intracellular loop domain. Overexpression of full-length CAIN attenuates the agonist-stimulated endocytosis of both mGluR1a and mGluR5a in HEK 293 cells, but expression of the CAIN C-terminal domain does not alter mGluR5a internalization. In contrast, overexpression of either full-length CAIN or the CAIN C-terminal domain impairs agonist-stimulated inositol phosphate formation in HEK 293 cells expressing mGluR1a. This CAIN-mediated antagonism of mGluR1a signaling appears to involve the disruption of receptor-Gαq/11 complexes. Taken together, these observations suggest that the association of CAIN with intracellular domains involved in mGluR/G protein coupling provides an additional mechanism by which Group I mGluR endocytosis and signaling are regulated.
Metabotropic glutamate receptors (mGluRs)2 play an essential role in regulating neuronal plasticity, development, and neurotoxicity and belong to the G protein-coupled receptor superfamily of integral membrane proteins (1–4). The mGluR family can be subclassified into three groups based on sequence homology, G protein specificity, and pharmacology. Group I mGluRs (mGluR1 and mGluR5) couple via the heterotrimeric Gαq/11 proteins to the activation of phospholipase Cβ, resulting in the formation of inositol 1,4,5-triphosphate and diacylglycerol, the release of Ca2+ from intracellular stores, and the activation of protein kinase C (PKC) (4–6).
The regulation of mGluR signal transduction involves numerous proteins that function to regulate signaling at both the level of the heterotrimeric G protein and the receptor (6–8). At the level of the receptor, Group I mGluR activity is regulated by a process termed desensitization, which protects against both acute and chronic receptor overstimulation (9, 10). The attenuation of Group I mGluR signaling can be mediated by both phosphorylation-dependent and phosphorylation-independent processes (11). The phosphorylation-independent attenuation of Group I mGluR signaling is mediated by GRK2 (G protein-coupled receptor kinase 2), which is composed of three functional domains: an N-terminal RGS (regulator of G protein signaling) homology domain, a central catalytic domain, and a C-terminal Gβγ-binding pleckstrin homology domain (12). GRK2-mediated desensitization of Group I mGluRs does not require catalytic activity but rather requires the interaction of the GRK2 RGS homology domain with both the second intracellular loop domain of mGluR1 and the α-subunit of Gαq/11, thereby attenuating heterotrimeric G protein coupling (13–15). Phosphorylation-independent desensitization of mGluR1 signaling is also mediated by optineurin, an effect that is enhanced by the expression of mutant huntingtin (16). Phosphorylation-dependent desensitization of Group I mGluR responsiveness involves the phosphorylation of PKC consensus sequence localized within the intracellular loop and C-terminal tail domains of mGluR1 and mGluR5 by PKC (17, 18). It is proposed that calcineurin and mGluR5 may exist in a signaling complex in the brain and that calcineurin may function to modulate mGluR5 signaling by directly dephosphorylating the receptor at a PKC consensus site that contributes to mGluR5 desensitization (19). Calcineurin is also linked to the regulation of endocytosis via its interaction with dynamin-1 (20).
On the basis of the observation that calcineurin may form a complex with Group I mGluRs, we hypothesized that CAIN (calcineurin inhibitor protein) might also interact with Group I mGluRs and modulate their endocytosis and signaling. CAIN, also known as Cabin1 (calcineurin-binding protein), was first identified as a protein that binds to calcineurin and was shown to inhibit calcineurin catalytic activity (21–23). Previous studies also demonstrated that CAIN may interact with amphiphysin-1, dynamin-1, and α-adaptin and led to the suggestion that CAIN functions as a component of synaptic endocytic complexes (24). Consistent with this hypothesis, the overexpression of CAIN in human embryonic kidney (HEK 293) cells resulted in attenuated transferrin receptor endocytosis.
We show here that CAIN interacts with the second intracellular loop and C-terminal tail domains of Group I mGluRs, inhibits Group I mGluR internalization, and attenuates mGluR1a signaling by disrupting receptor-Gαq/11 complexes. Taken together, these results describe an additional mechanism by which Group I mGluR activity may be regulated.
EXPERIMENTAL PROCEDURES
Materials
HEK 293 cells were obtained from the American Type Culture Collection (Manassas, VA). Tissue culture reagents were purchased from Invitrogen. Quisqualate was purchased from Tocris Cookson Inc. (Ellisville, MO). myo-[3H]Inositol was acquired from PerkinElmer Life Sciences. The Dowex 1-X8 resin (formate form) with 200–400 mesh was purchased from Bio-Rad. EZ-Link sulfo-NHS-SS-biotin and immobilized NeutrAvidin were purchased from Pierce. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG secondary antibodies, protein G-Sepharose beads, enhanced chemiluminescence (ECL) Western blotting detection reagents, and mouse anti-hemagglutinin (HA) monoclonal antibody (12CA5) were purchased from GE Healthcare. Mouse anti-glutathione S-transferase (GST) monoclonal antibody and rabbit anti-Gαq/11 polyclonal antibody were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Rabbit anti-mGluR5 and anti-mGluR1 polyclonal antibodies were obtained from Upstate (Lake Placid, NY). Rabbit anti-CAIN polyclonal antibody was obtained from Calbiochem. Alexa Fluor 488-conjugated goat anti-mouse IgG and Alexa Fluor 568-conjugated goat anti-rabbit IgG were purchased from Invitrogen. All other biochemical reagents were purchased from Sigma, Fisher, and VWR Scientific (Mississauga, Ontario, Canada).
Plasmid Construction
All recombinant cDNA procedures were carried out in accordance with standard protocols. The HA-CAIN construct was generously provided by Dr. Solomon H. Snyder (Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore). This construct contained the entire rat CAIN open reading frame (ORF) (nucleotides 1–6549) and its 3′-untranslated region. The HA-CAIN and its 3′-untranslated region were subcloned into the SalI-XbaI sites of the Escherichia coli expression vector pPROEX-HT to create His6-CAIN. A His6-tagged C-terminal domain CAIN construct (nucleotides 5643–6519, amino acid residues 1519–2182) was generated by PCR and cloned into the Bg1II-XbaI sites of pPROEX-HT to create His6-CAIN-CTer. This construct encodes both the amphiphysin- and calcineurin-binding domains of CAIN (24). The GST-tagged rat mGluR1a constructs were generated by PCR and cloned into the EcoRI-XhoI sites of the E. coli expression vector pGEX-4T (Amersham Biosciences). The PCR-generated sequences encode mGluR1a intracellular loop 1 (GST-IL1, nucleotides 1777–1950 of the ORF, amino acid residues 593–650), loop 2 (GST-IL2, nucleotides 1969–2181 of the ORF, amino acid residues 657–727), loop 3 (GST-IL3, nucleotides 2251–2421 of the ORF, amino acid residues 751–807), the membrane-proximal portion of the C-terminal tail (CT-N, nucleotides 2446–2580 of the ORF, amino acid residues 816–860), and the rest of the C-terminal tail (CT-C, nucleotides 2581–3597 of the ORF, amino acid residues 861–1199). pPROEX-HT- and pGEX-4T-based constructs were transformed into E. coli BL21. Full-length CAIN was cloned into the SalI and XbaI sites of pEGFP-C3, and the C-terminal portion of the CAIN ORF was cloned into the HindIII and SacII sites of pEGFP-C3. All constructs were subjected to automated DNA sequencing to confirm sequence integrity.
Cell Culture and Transfection
HEK 293 cells were maintained in Eagle's minimal essential medium supplemented with 10% (v/v) fetal bovine serum (Invitrogen) and 50 μg/ml gentamycin. Cells were transfected using a modified calcium phosphate method as described previously (25). Following transfection (18 h), the cells were incubated with fresh medium and allowed to recover for 24 h for co-immunoprecipitation studies. Otherwise, they were allowed to recover for 6–8 h and reseeded either into 12-well dishes coated with collagen or containing collagen-coated coverslips or into 24-well dishes and then grown for an additional 18 h prior to experimentation.
Primary neuronal cultures were prepared from the cortexes of embryonic day 15 CD1 mouse embryos. Animal procedures were approved by the University of Western Ontario Animal Care Committee. Cells were plated on either 100-mm dishes or 15-mm glass coverslips coated with poly-l-ornithine (Invitrogen) in Neurobasal medium with B-27 and N-2 supplements, 2 mm GlutaMAX, 50 μg/ml penicillin, and 50 mg/ml streptomycin. Cells were incubated at 37 °C and 5% CO2 in a humidified incubator and cultured for up to 21 days in vitro with media replenishment every 3 days.
His6-CAIN, His6-CAIN-CTer, and GST-mGluR1a Fusion Protein Purification and Pulldown Assays
GST-mGluR1a peptides were generated by growing recombinant E. coli BL21 at 37 °C to an A600 of 06–1.0. His6-CAIN and His6-CAIN-CTer (amino acid residues 1519–2182) cultures were induced for 3 h with 0.6 mm isopropyl β-d-thiogalactopyranoside, whereas GST-mGluR1a cultures were induced for 3 h with 1 mm isopropyl β-d-thiogalactopyranoside. After induction, cultures were pelleted, resuspended in phosphate-buffered saline (PBS) containing protease inhibitors, and lysed by mild sonication. The bacterial lysates were cleared of cellular debris by centrifugation, and the His6-CAIN and His6-CAIN-CTer lysates were then applied to Talon metal affinity resin. GST-mGluR1a lysates were applied to glutathione-Sepharose 4B and incubated on an orbital shaker overnight at 4 °C. Matrix-bound His6-CAIN, His6-CAIN-CTer, and GST-mGluR1a fusion proteins were washed extensively with PBS-containing 0.3% Triton X-100 (PBS-T) and then eluted off the resin in phosphate-buffered 500 mm imidazole. The imidazole was subsequently removed by chromatography using NAP-10 columns, and the proteins were recovered in phosphate buffer. One μg of His6-CAIN, His6-CAIN-CTer, and GST-mGluR1a peptides was used in the pulldown assays. Glutathione-Sepharose 4B-bound GST fusion proteins and either purified His6-CAIN or His6-CAIN-CTer were incubated together and mixed overnight on an orbital shaker at 4 °C. The Sepharose-bound protein complexes were washed extensively with PBS-T and then eluted off the Sepharose in SDS-PAGE loading buffer. Eluted samples were analyzed by immunoblotting with anti-CAIN antibody (1:1000 dilution).
Co-immunoprecipitation
Co-immunoprecipitation experiments were performed using 500–1000 μg of total cell lysate protein solubilized from HEK 293 cells transiently transfected with the cDNA constructs as described in the figure legends. Cells were solubilized in lysis buffer (25 mm HEPES, pH 7.5, 300 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, and 0.1% Triton X-100) containing protease inhibitors. FLAG-mGluR1a and FLAG-mGluR5a were immunoprecipitated with anti-FLAG monoclonal antibody M2 affinity Sepharose beads by 2 h of rotation at 4 °C. In experiments utilizing a peptide corresponding to the second intracellular loop domain of mGluR1a (IARILAGSKKKICTRKPRFMS) fused to the human immunodeficiency virus Tat protein membrane transducing domain (YGRKKRRQRRR), HEK 293 cells were treated with peptide for 2 h prior to co-immunoprecipitation with FLAG-mGluR1a. For the co-immunoprecipitation of endogenous proteins from cortical extracts, adult mouse brains were employed. Tissue was dissected and homogenized on ice in lysis buffer containing protease inhibitors. The particulate fraction was removed by centrifugation, and 2 mg of supernatant protein was incubated with rabbit anti-CAIN polyclonal antibody and protein G-Sepharose beads by 2 h of rotation at 4 °C. Afterward, the beads were washed twice with lysis buffer and once with PBS, and proteins were eluted in SDS-PAGE loading buffer by warming the samples at 55 °C for 5 min. Eluted samples were subjected to SDS-PAGE, followed by electroblotting onto nitrocellulose membranes for immunoblotting with antibodies described in the figure legends. Immunoblots were visualized by chemiluminescence using an ECL kit. Intensities of immunoblot signals were determined with the 4.0.2 version of the data acquisition and analysis software from Scion Corp. (Frederick, MD).
Confocal Microscopy and Immunostaining
Confocal microscopy was performed using a Zeiss LSM-510 META laser scanning confocal microscope equipped with a Zeiss 63×, 1.4 numerical aperture, oil immersion lens. HEK 293 cells expressing either green fluorescent protein (GFP)-CAIN alone or coexpressing GFP-CAIN and FLAG-mGluR1a or neuronal primary culture cells were seeded on 15-mm glass coverslips. Cells were washed with HEPES-buffered saline solution (HBSS; 116 mm NaCl, 20 mm HEPES, 11 mm glucose, 5 mm NaHCO3, 4.7 mm KCl, 2.5 mm CaCl2, 1.2 mm MgSO4, and 1.2 mm KH2PO4, pH 7.4; Molecular Probes, Eugene, OR) and fixed in 4% paraformaldehyde. Afterward, HEK 293 cells were permeabilized with 0.05% Triton X-100 and incubated with rabbit anti-FLAG and mouse anti-HA primary antibodies for 1 h at room temperature. Cells were washed and labeled with Alexa Fluor 568-conjugated anti-rabbit and Alexa Fluor 488-conjugated anti-mouse secondary antibodies. Primary neuronal cultures were permeabilized with 0.05% Triton X-100, stained for endogenous mGluR5a with rabbit anti-mGluR5a antibody (1:2000) conjugated to Alexa Fluor 568 following the manufacturer's specifications using a Zenon IgG labeling kit, and washed three times with HBSS. Cells were washed again with HBSS and labeled for 20 min with rabbit anti-CAIN antibody (1:500) conjugated to Alexa Fluor 488. Cells were washed three times with HBSS, fixed in 4% paraformaldehyde, washed again with HBSS, mounted in Immu-Mount (Thermo Shandon, Pittsburgh, PA) onto glass slides, and allowed to air dry before viewing. Co-localization studies were performed using dual excitation (488 and 543 nm) and emission (505–530 nm band pass and 560 nm long pass for Alexa Fluor 488 and 568, respectively) filter sets. The specificity of labeling and the absence of signal crossover were established by examination of single-labeled samples.
Inositol Phosphate Formation
HEK 293 cells transiently transfected with 3 μg of FLAG-mGluR1a construct and 3 μg of either empty plasmid vector or plasmid vector containing GFP-CAIN or GFP-CAIN-CTer were seeded into 24-well dishes. Inositol lipids were radiolabeled by incubating the cells overnight with 1 μCi/ml myo-[3H]inositol in inositol-free Dulbecco's modified Eagle's medium. Unincorporated myo-[3H]inositol was removed by washing the cells with HBSS. Cells were preincubated for 1 h in HBSS at 37 °C and then preincubated in 500 μl of the same buffer containing 10 mm LiCl for an additional 10 min at 37 °C. Next, cells were incubated in the absence or presence of quisqualate for 30 min at 37 °C. The reaction was stopped on ice by the addition of 500 μ1 of 0.8 m perchloric acid and then neutralized with 400 μ1 of 0.72 m KOH and 0.6 m KHCO3. The total myo-[3H]inositol incorporated into the cells was determined by counting the radioactivity present in 50 μ1 of the cell lysate. Total inositol phosphate was purified from the cell extracts by anion exchange chromatography using Dowex 1-X8 200–400 mesh anion exchange resin (formate form). [3H]Inositol phosphate formation was determined by liquid scintillation using a Beckman LS 6500 scintillation system.
Receptor Internalization
HEK 293 cells transiently coexpressing either FLAG-mGluR1 and HA-CAIN or FLAG-mGluR5 and either GFP-CAIN or GFP-CAIN-CTer were washed and incubated on ice in HBSS. Plasma membrane proteins were then biotinylated with EZ-Link sulfo-NHS-SS-biotin for 1 h on ice. To quench the biotinylation reaction, cells were washed and incubated for 30 min with cold 100 mm glycine in HBSS, followed by three washes with cold HBSS. Subsequently, cells were incubated for varying times with 30 μm quisqualate in HBSS at 37 °C to allow receptor internalization. Cells were then transferred to ice to stop internalization and washed once with ice-cold HBSS. Biotin remaining at the cell surface was stripped by incubating the cells with freshly prepared 150 mm mercaptoethanesulfonic acid sodium salt in HBSS at 4 °C for 45 min. To assess the amount of receptor initially in the plasma membrane, a control sample was kept on ice without mercaptoethanesulfonic acid sodium salt stripping. Cells were then lysed in the same lysis buffer described above. Biotinylated proteins were pulled down with NeutrAvidin-agarose resin, eluted from the beads with 3× SDS loading buffer, subjected to SDS-PAGE, and immunoblotted for the receptors. Internalization of the receptors at the various time points was expressed as a percentage of biotinylated receptor on the surface of the non-stimulated and non-stripped control.
Data Analysis
The means ± S.E. are shown for values obtained for the number of independent experiments indicated in the figure legends. GraphPad Prism software was used to analyze data for statistical significance as well as to analyze and fit dose-response and time-course data. The statistical significance was determined by t test.
RESULTS
Interaction between CAIN and Group I mGluRs
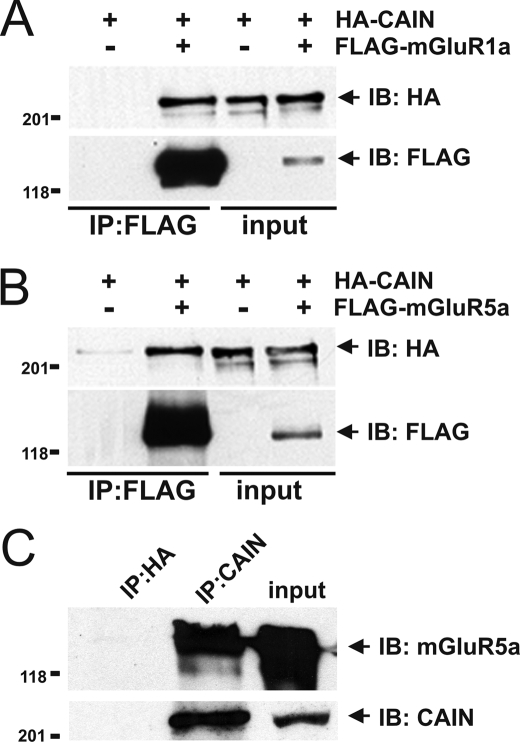
Previous studies demonstrated that calcineurin interacts with Group I mGluRs and that CAIN negatively regulates both calcineurin function and transferrin receptor endocytosis (19, 21, 24). Therefore, in initial experiments, we tested whether CAIN associates with either mGluR1a or mGluR5a by co-immunoprecipitation. We found that HA epitope-tagged CAIN was co-immunoprecipitated from HEK 293 cells with both FLAG-tagged mGluR1a and mGluR5a (Fig. 1, A and B). We also found that endogenous mGluR5 could be co-immunoprecipitated with endogenous CAIN from mouse cortical brain lysates using a rabbit CAIN-specific polyclonal antibody (Fig. 1C). Taken together, these data suggested that CAIN was associated in a protein complex with Group I mGluRs.
FIGURE 1.
CAIN co-immunoprecipitation with Group I mGluRs. The representative immunoblots show the co-immunoprecipitation (IP) of HA-CAIN from HEK 293 cells coexpressing either FLAG-mGluR1a (A) or FLAG-mGluR5 (B). Lysate from cells expressing only HA-CAIN was used as a control. The immunoblot (IB) is representative of three independent experiments, and HEK 293 cells were transfected with 3 μg of pcDNA3 plasmid cDNA encoding either FLAG-mGluR1a or FLAG-mGluR5 and 3 μg of either empty pcDNA3 plasmid cDNA or pcDNA3 plasmid cDNA encoding HA-CAIN. FLAG-mGluR1a was immunoprecipitated from 500–1000 μg of protein lysate. The expression of mGluR1a, mGluR5, and CAIN shown in input lanes corresponds to 100 μg of protein lysate. C, co-immunoprecipitation of endogenous mGluR5 with endogenous CAIN from mouse cortical lysates (2 mg of total protein) using anti-CAIN polyclonal antibody. Anti-HA polyclonal antibody was used as a negative control. Endogenous expression of both mGluR5 and CAIN shown in input lanes corresponds to 100 μg of protein lysate. The immunoblot is representative of three independent experiments.
Co-localization of CAIN and Group I mGluRs
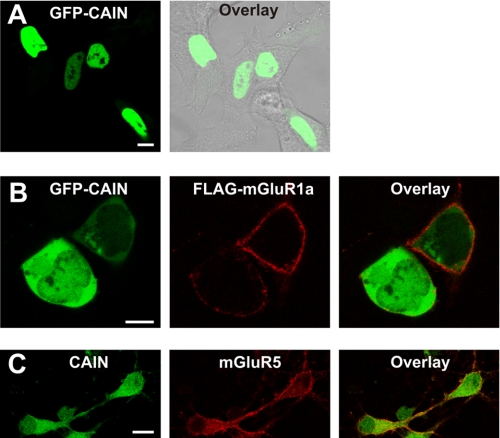
Previously, Snyder and co-workers (21) reported that CAIN is predominantly a cytosolic protein, whereas in lymphocytes, CAIN is localized to the nucleus, where it negatively regulates MEF2 transcription factor activity (22, 23, 26). Therefore, because the current information in the literature regarding the subcellular distribution of CAIN is contradictory, we examined the subcellular localization of GFP-CAIN in HEK 293 cells in the absence and presence of mGluR1a expression. In HEK 293 cells lacking FLAG-mGluR1a expression, GFP-CAIN localization was restricted to the nucleus (Fig. 2A), consistent with the report indicating that CAIN is a nuclear protein. However, in cells expressing both FLAG-mGluR1a and GFP-CAIN, the subcellular distribution of GFP-CAIN was altered such that CAIN was localized to the cytoplasm and, to a lesser extent, the nucleus (Fig. 2B). In addition, primary mouse cortical neurons that stained positive for endogenous mGluR5 were positive for the expression of CAIN in both the nucleus and the cytoplasm (Fig. 3C). Taken together, the data indicated that CAIN was appropriately localized to the cytoplasm in both HEK 293 cells and neurons, which was consistent with the observation that CAIN forms a complex with Group I mGluRs.
FIGURE 2.
CAIN co-localizes with Group I mGluRs in HEK 293 cells and in primary mouse cortical neurons. A, shown is a representative micrographic image of a field of HEK 293 cells transfected with 1 μg of pEGFP-C1 plasmid cDNA encoding CAIN. GFP-CAIN is shown in green and is overlaid with a phase-contrast image of the cells in gray. The micrograph is representative of six independent experiments. B, shown is a representative micrographic image of a field of HEK 293 cells transfected with 1 μg of pEGFP-C1 plasmid cDNA encoding CAIN and 3 μg of pcDNA3 plasmid cDNA encoding FLAG-mGluR1a. GFP-CAIN is shown in green, and FLAG-mGluR1a is labeled with rabbit anti-FLAG polyclonal antibody conjugated to Alexa Fluor 568 (shown in red). The micrograph is representative of four independent experiments. C, shown is a representative micrograph of primary mouse cortical neurons stained with rabbit anti-mGluR5a polyclonal antibody conjugated to Alexa Fluor 568 and rabbit anti-CAIN polyclonal antibody conjugated to Alexa Fluor 488. The micrograph is representative of three independent experiments. Scale bars = 10 μm.
FIGURE 3.
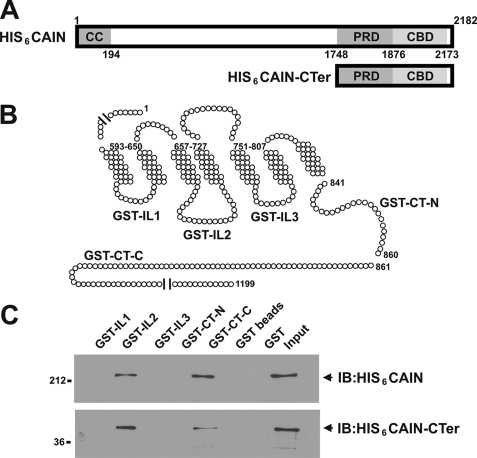
Purified His6-CAIN binds to GST fusion proteins corresponding to the mGluR1a second intracellular loop and distal C-terminal tail domains. A, shown are the full-length CAIN and CAIN C-terminal domains purified from E. coli as His6 fusion proteins and the localization of the coil-coiled (CC) domain, the proline-rich domain (PRD) that binds amphiphysin, and the calcineurin-binding domain (CBD). B, the schematic representation of mGluR1a illustrates the regions of mGluR1a that were prepared as GST fusion proteins. The GST fusion proteins correspond to the first (GST-IL1), second (GST-IL2), and third (GST-IL3) intracellular loops as well as the proximal (GST-CT-N) and distal (GST-CT-C) C-terminal tails. C, 1 μg of purified GST-IL1, GST-IL2, GST-IL3, GST-CT-N, and GST-CT-C was incubated with 1 μg of either full-length His6-tagged CAIN (HIS6CAIN) or the His6-tagged CAIN C-terminal domain (HIS6CAIN-CTer). CAIN eluted with the GST fusion proteins was determined by immunoblotting (IB) with rabbit anti-CAIN polyclonal antibody. Data are representative of three independent experiments.
CAIN Interacts Directly with Group I mGluRs
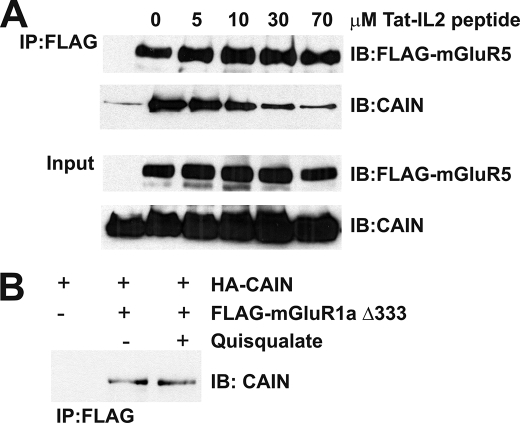
To determine whether CAIN interacts directly with Group I mGluRs and to ascertain which intracellular domains bind to CAIN, both full-length CAIN and the C terminus of CAIN were expressed in and purified from E. coli as His6 fusion proteins (Fig. 3A). The purified CAIN proteins were subsequently tested for their ability to bind in vitro to GST fusion proteins corresponding to the mGluR1a intracellular loops (IL1, IL2, and IL3) as well as the proximal (GST-CT-C) and distal (GST-CT-N) portions of the mGluR1a C-terminal tail (Fig. 3B). We found that CAIN specifically bound to GST fusion proteins corresponding to the IL2 and CT-N domains of mGluR1a (Fig. 3C). In addition, the CAIN-CTer construct, which encodes the CAIN calcineurin- and amphiphysin-binding domains, also bound to the IL2 and CT-N domains of mGluR1a (Fig. 3C). Treatment of the cells with increasing concentrations of Tat-tagged peptide corresponding to the second intracellular loop domain of mGluR1a resulted in a dose-dependent attenuation of HA-CAIN co-immunoprecipitation with FLAG-mGluR1a (Fig. 4A). HA-CAIN co-immunoprecipitated with FLAG-mGluR1a was reduced by 78 ± 9% of the control in the presence of 70 μm Tat-tagged peptide. CAIN was also co-immunoprecipitated from HEK 293 cells with a FLAG-mGluR1a construct lacking the last 333 amino acids of the C-terminal tail, and the association of CAIN with the truncated receptor was not affected by agonist stimulation (Fig. 4B). Thus, IL2, which is essentially identical in amino acid composition between mGluR1a and mGluR5a, represents the primary site of interaction for CAIN and Group I mGluRs.
FIGURE 4.
mGluR1a intracellular loop 2 but not the C-terminal tail is essential for CAIN interactions. A, shown is a representative immunoblot (IB) of HA-CAIN co-immunoprecipitated (IP) with FLAG-mGluR1a from HEK 293 cells in the presence of increasing concentrations of Tat-tagged peptide corresponding to the second intracellular loop domain of mGluR1a (0, 5, 10, 30, and 70 μm peptide). HEK 293 cells were transfected with 3 μg of pcDNA3 plasmid encoding FLAG-mGluR1a and 3 μg of pcDNA3 plasmid encoding HA-CAIN. FLAG-mGluR1a was immunoprecipitated from 500 μg of protein lysate. The blot is representative of three independent experiments. B, shown is a representative immunoblot of HA-CAIN co-immunoprecipitated with FLAG-mGluR1a-(1–866) (FLAG-mGluR1aΔ333), lacking the last 333 amino acids of the receptor C-terminal tail, from HEK 293 cells. HEK 293 cells were transfected with 3 μg of pcDNA3 plasmid cDNA encoding FLAG-mGluR1aΔ333 and either 3 μg of empty pcDNA3 plasmid cDNA or pcDNA3 plasmid cDNA encoding CAIN. HEK 293 cells were treated with or without 100 μm quisqualate. FLAG-mGluR1a was immunoprecipitated from 500 μg of protein lysate. The blot is representative of three independent experiments.
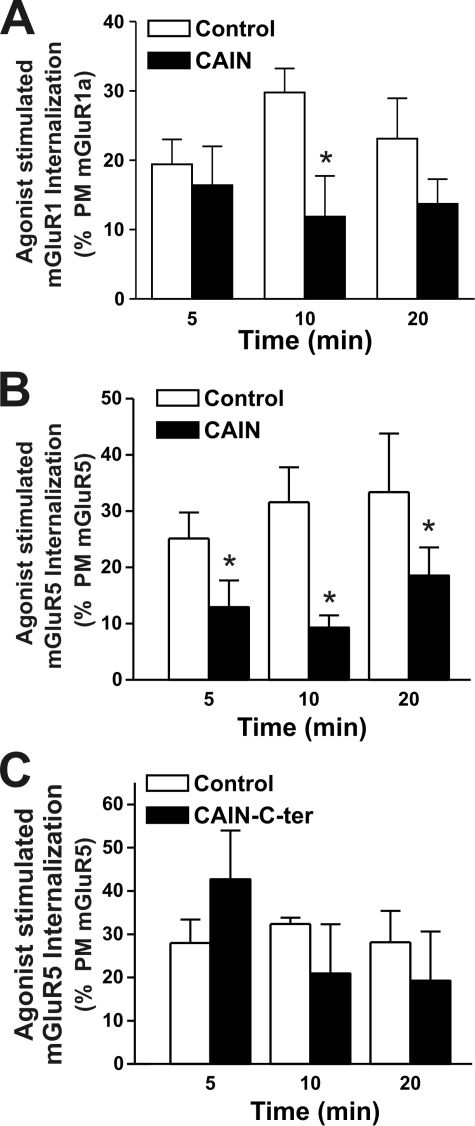
CAIN Regulates mGluR1a Internalization
CAIN was shown previously to inhibit transferrin receptor endocytosis (24), and we have demonstrated here that the amphiphysin- and calcineurin-binding domains interact with Group I mGluRs. Therefore, we tested whether CAIN and/or its C terminus might affect the agonist-stimulated internalization of Group I mGluRs. To determine the extent of mGluR1a and mGluR5a internalization, cell-surface proteins were biotinylated on ice, warmed to 37 °C, and treated with 30 μm quisqualate for 0, 5, 10, and 20 min. The internalization of mGluR1a and mGluR5a in response to agonist was determined as the proportion of total biotinylated receptor protein that was resistant to mercaptoethanesulfonic acid sodium salt stripping following agonist treatment (Fig. 5, A and B). We found that both FLAG-mGluR1a and FLAG-mGluR5a were rapidly internalized in response to agonist treatment and that the internalization of both receptors was attenuated in the presence of full-length CAIN (Fig. 5, A and B). In contrast, mGluR5a internalization was not altered by expression of the CAIN C-terminal domain (Fig. 5C). Thus, although full-length CAIN functioned to antagonize Group I mGluR endocytosis, the CAIN amphiphysin- and calcineurin-binding domains were not responsible for the CAIN-dependent blockade of Group I mGluR internalization.
FIGURE 5.
Overexpression of full-length CAIN but not the CAIN C-terminal domain decreases agonist-stimulated Group I mGluR internalization in HEK 293 cells. Shown are the internalization time courses for cell-surface biotin-labeled FLAG-mGluR1a (A) and FLAG-mGluR5 (B) in HEK 293 cells in the absence (Control) and presence of CAIN. C, shown are the internalization time courses for cell-surface biotin-labeled FLAG-mGluR5a in HEK 293 cells in the absence (Control) and presence of CAIN-CTer. The data represent the means ± S.E. of five independent experiments, taken as a percentage of the plasma membrane expression (PM) of either FLAG-mGluR1a or mGluR5 from samples kept on ice and not stripped with mercaptoethanesulfonic acid sodium salt prior to immunoprecipitation. HEK 293 cells were transfected with 3 μg of pcDNA3 plasmid cDNA encoding either FLAG-mGluR1a or FLAG-mGluR5 and 3 μg of empty pcDNA3 plasmid cDNA, pcDNA3 plasmid cDNA encoding HA-CAIN, or pcDNA3 plasmid cDNA encoding HA-CAIN-CTer. FLAG-mGluR1a and FLAG-mGluR5 were immunoprecipitated from 500–1000 μg of protein lysate. Asterisks indicate significant differences compared with the control (p < 0.05).
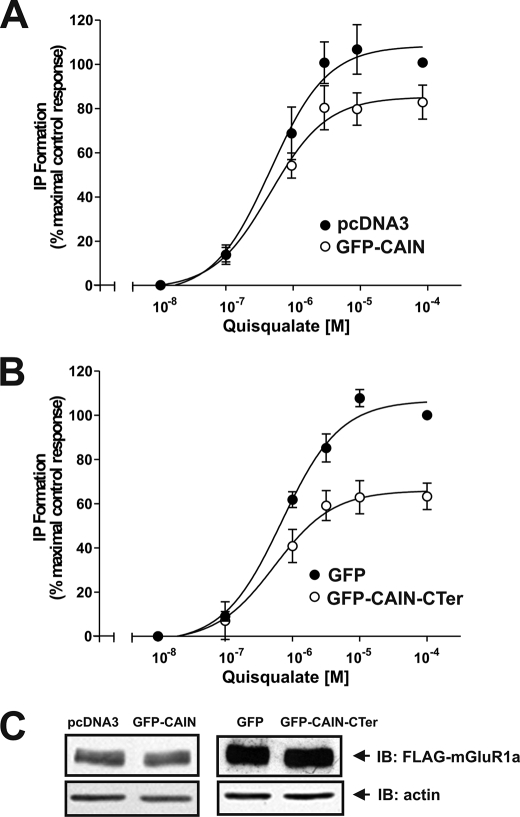
CAIN-mediated Regulation of mGluR1a Signaling
We demonstrated that IL2 represents the critical region for CAIN binding to Group I mGluRs, and this domain was shown previously to play an essential role in heterotrimeric G protein coupling of Group I mGluRs (27, 28). Therefore, we examined whether the overexpression of either full-length GFP-CAIN or the GFP-CAIN C terminus might attenuate FLAG-mGluR1a-stimulated inositol phosphate (IP) formation in HEK 293 cells. Experiments were performed only with mGluR1a, as we were unable to detect agonist-stimulated increases in IP formation in HEK 293 cells overexpressing mGluR5 due to high basal IP formation. In the absence of either full-length CAIN or the CAIN C terminus, quisqualate treatment of FLAG-mGluR1a-expressing cells resulted in a dose-dependent increase in IP formation (Fig. 6, A and B). In cells expressing full-length GFP-CAIN, the maximal response to quisqualate treatment was reduced to 79 ± 5% of the control with no apparent change in the EC50 for quisqualate (0.48 versus 0.47 μΜ, respectively) (Fig. 6A). The attenuation of FLAG-mGluR1a signaling was even more robust in the presence of the GFP-CAIN C-terminal domain construct, with the maximum response to quisqualate reduced to 59 ± 5% of the control with no apparent change in the EC50 for quisqualate (0.69 versus 0.54 μΜ, respectively) (Fig. 6B). This reduction in mGluR1a-stimulated IP formation was not the consequence of reduced receptor expression, as the overexpression of either full-length GFP-CAIN or the GFP-CAIN C-terminal domain had no effect on FLAG-mGluR1a expression compared with cells transfected with FLAG-mGluR1a and empty vector (Fig. 6C).
FIGURE 6.
CAIN attenuates mGluR1-stimulated IP formation in HEK 293 cells. A, quisqualate dose-response curves for FLAG-mGluR1a-stimulated IP formation in HEK 293 cells in the absence (control) or presence of CAIN. HEK 293 cells were transfected with 3 μg of pcDNA3 plasmid cDNA encoding FLAG-mGluR1a and 3 μg of either pEGFP-C3 plasmid cDNA or pcDNA3 plasmid cDNA encoding GFP-CAIN. The data represent the means ± S.E. of five independent experiments. B, quisqualate dose-response curves for FLAG-mGluR1a-stimulated IP formation in HEK 293 cells in the absence (control) or presence of CAIN-CTer. HEK 293 cells were transfected with 3 μg of plasmid cDNA encoding FLAG-mGluR1a and 3 μg of either pEGFP-C3 plasmid cDNA or pEGFP-C3 plasmid cDNA encoding GFP-CAIN-CTer. The data represent the means ± S.E. of five independent experiments. C, representative immunoblot (IB) showing the expression levels for FLAG-mGluR1, GFP-CAIN, and GFP-CAIN-CTer in 20 μg of protein lysate.
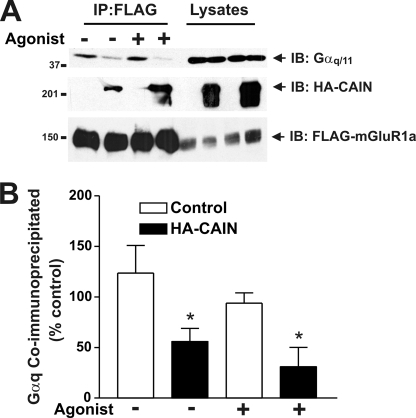
We demonstrated previously that GRK2 attenuates mGluR1a signaling by competing with Gαq/11 for binding to IL2 of mGluR1a (14). Thus, we examined whether CAIN-dependent attenuation of FLAG-mGluR1a signaling is mediated by a similar mechanism that involves the disruption of Gαq/11 interactions with FLAG-mGluR1a. In the absence of CAIN expression, endogenous Gαq/11 protein was co-immunoprecipitated with FLAG-mGluR1a, and agonist treatment resulted in a small but statistically insignificant decrease in Gαq/11 protein co-immunoprecipitated with the receptor (Fig. 7, A and B). In cells expressing CAIN, CAIN was co-immunoprecipitated with FLAG-mGluR1a, which resulted in a statistically significant decrease in Gαq/11 co-immunoprecipitated with the receptor complex in both the presence and absence of agonist (Fig. 7, A and B). Taken together, these observations indicated that CAIN contributed to the attenuation of FLAG-mGluR1a/G protein coupling by disrupting mGluR1a-Gαq/11 protein complexes.
FIGURE 7.
CAIN disrupts FLAG-mGluR1a/Gαq/11 protein complexes. A, representative immunoblot (IB) showing the co-immunoprecipitation of endogenous Gαq/11 and HA-CAIN with FLAG-mGluR1a from HEK 293 cells. HEK 293 cells were transfected with 3 μg of pcDNA3 plasmid cDNA encoding FLAG-mGluR1a and 3 μg of either empty pcDNA3 plasmid cDNA or pcDNA3 plasmid cDNA encoding HA-CAIN. FLAG-mGluR1a was immunoprecipitated from 500–1000 μg of protein lysate. Also shown is the expression of endogenous Gαq/11, HA-CAIN, and FLAG-mGluR1a from 25 μg of the corresponding HEK 293 cell lysates. B, densitometric analysis of blots showing the means ± S.E. of four independent experiments. Asterisks indicate significant differences compared with the control (p < 0.05).
DISCUSSION
In this study, we have provided evidence that the association of CAIN with Group I mGluRs represents an unexpected and new mechanism by which the agonist-dependent endocytosis and signaling of Group I mGluRs may be regulated. CAIN interacts with the second intracellular loop and distal C-terminal domains of Group I mGluRs and functions to disrupt mGluR/Gαq/11 interactions, leading to the attenuation of mGluR-stimulated IP formation. In addition, full-length CAIN inhibits the endocytosis of Group I mGluRs. However, the expression of the CAIN C-terminal domain, which encodes the CAIN amphiphysin- and calcineurin-binding domains, does not inhibit Group I mGluR endocytosis despite the fact that this CAIN domain binds to Group I mGluRs and antagonizes Group I mGluR signaling. Thus, along with GRK2 and optineurin, CAIN provides an additional regulatory protein that contributes to the control of Group I mGluR endocytosis and signaling (14, 16).
CAIN was first identified as a protein that binds to calcineurin in response to Ca2+ and PKC activation and functions to inhibit the catalytic activity of calcineurin (22, 23, 26). Calcineurin is a serine/threonine phosphatase that plays an important role in transducing intracellular Ca2+ signals into meaningful biological responses within the cell. The activation of CAIN is Ca2+/calmodulin-dependent, and both calcineurin and calmodulin have been reported to form a protein complex with mGluR5 (19, 29–31). Specifically, calcineurin is reported to co-immunoprecipitate with mGluR5 from hippocampal homogenates and to modulate mGluR5 activity by dephosphorylating intracellular PKC consensus sites that contribute to mGluR5 desensitization (19). Moreover, calmodulin is reported to regulate the intracellular trafficking of mGluR5 (31). A site for calmodulin binding to mGluR5 has been described (29, 30), but it is unknown whether calcineurin binds directly to Group I mGluRs and/or whether mGluR-calcineurin complexes are facilitated by an intermediary protein such as calmodulin or CAIN. Thus, it is possible that the interaction of CAIN with mGluRs functions to target calcineurin to these receptors and facilitates mGluR resensitization in addition to contributing to mGluR desensitization. This is mechanistically analogous to β-arrestins, which uncouple G protein-coupled receptors from heterotrimeric G proteins but target G protein-coupled receptors for endocytosis and dephosphorylation in the endosome (9, 10). The difference is that CAIN antagonizes mGluR endocytosis and that CAIN may function as phosphatase scaffold. In fact, there is also evidence that β-arrestin may also scaffold phosphatase 2A (32), which is implicated in the dephosphorylation of desensitized G protein-coupled receptors (33), but this has yet to be investigated. Nevertheless, our data suggest that CAIN may be part of a signaling complex that functions to regulate Group I mGluR endocytosis and signaling.
It is well established that the second intracellular loop domain (IL2) of Group I mGluRs is essential for heterotrimeric G protein coupling (28, 29). IL2 appears to be essential for the association of CAIN with Group I mGluRs. This domain is also essential for the interaction of a number of Group I mGluR proteins that are involved in the regulation of heterotrimeric G protein coupling and internalization, including GRK2 and optineurin (14, 16, 34). It appears that proteins that interact with this domain compete with and disrupt the formation of a Group I mGluR-Gαq/11 complex that appears to be essential for the activation of phospholipase Cβ, required for diacylglycerol and inositol 1,4,5-triphosphate formation. However, CAIN does not encode a RGS homology domain, which allows GRK2 to interact with Gαq/11, suggesting that CAIN functions primarily to disrupt Gαq/11 binding to Group I mGluRs, similar to what we have also reported to be required for GRK2-dependent regulation of Group I mGluR signaling (15).
CAIN has been identified as a functional component of the cellular endocytic machinery via its association with amphiphysin, dynamin-1, and α-adaptin (24). We have shown here that CAIN negatively regulates the endocytosis of Group I mGluRs, similar to its role in regulating the endocytosis of the transferrin receptor (24). The association of CAIN with Group I mGluRs involves the C-terminal domain of CAIN, which encodes both the CAIN amphiphysin- and calcineurin-binding domains. The association of the CAIN amphiphysin- and calcineurin-binding domains with Group I mGluRs is not sufficient to attenuate Group I mGluR endocytosis. This suggests that protein/protein interactions with the N-terminal domain of CAIN are involved in the negative regulation of Group I mGluR endocytosis by CAIN. Thus, the effect of overexpression of CAIN on Group I mGluR internalization is independent of its binding to the receptors. Despite the report that dynamin-1 and α-adaptin interact with CAIN, the localization of the CAIN binding domain(s) for these proteins has yet to be reported (24). Future structure/function analysis will be required both to localize the regions of CAIN that interact with dynamin-1 and α-adaptin and to associate these protein/protein interactions with the normal biological function of CAIN. Our data suggest that these domains are localized to the CAIN N terminus.
In summary, the association of CAIN with Group I mGluRs provides an additional mechanism by which mGluRs are uncoupled from their cognate heterotrimeric G proteins and are prevented from internalizing. Previous work has indicated that Group I mGluRs may be found in a complex with calcineurin and that calcineurin may contribute to the dephosphorylation and resensitization of PKC-phosphorylated receptors at the cell surface (19). The direct association of CAIN with Group I mGluRs may coordinate the formation of mGluR-CAIN-calcineurin protein complexes that may be essential for the spatial and temporal regulation of Group I mGluR signaling that leads to PKC oscillations (35).
Acknowledgment
We thank Dr. Solomon H. Snyder for the CAIN constructs.
This work was supported by Canadian Institutes of Health Research Grants MA-15506 and MOP-62738 (to S. S. G. F.).
- mGluR
- metabotropic glutamate receptor
- PKC
- protein kinase C
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- ORF
- open reading frame
- IL
- intracellular loop
- PBS
- phosphate-buffered saline
- GFP
- green fluorescent protein
- HBSS
- HEPES-buffered saline solution
- IP
- inositol phosphate.
REFERENCES
- 1.Choi D. W., Rothman S. M. (1990) Annu. Rev. Neurosci. 13, 171–182 [DOI] [PubMed] [Google Scholar]
- 2.Bliss T. V., Collingridge G. L. (1993) Nature 361, 31–39 [DOI] [PubMed] [Google Scholar]
- 3.Hermans E., Challiss R. A. (2001) Biochem. J. 359, 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakanishi S., Masu M. (1994) Annu. Rev. Biophys. Biomol. Struct. 23, 319–348 [DOI] [PubMed] [Google Scholar]
- 5.Conn P. J., Pin J. P. (1997) Annu. Rev. Pharmacol. Toxicol. 37, 205–237 [DOI] [PubMed] [Google Scholar]
- 6.Dhami G. K., Ferguson S. S. G. (2006) Pharmacol. Ther. 111, 260–271 [DOI] [PubMed] [Google Scholar]
- 7.Dohlman H. G., Thorner J. (1997) J. Biol. Chem. 272, 3871–3874 [DOI] [PubMed] [Google Scholar]
- 8.Siderovski D. P., Strockbine B., Behe C. I. (1999) Crit. Rev. Biochem. Mol. Biol. 34, 215–251 [DOI] [PubMed] [Google Scholar]
- 9.Krupnick J. G., Benovic J. L. (1998) Annu. Rev. Pharmacol. Toxicol. 38, 289–319 [DOI] [PubMed] [Google Scholar]
- 10.Ferguson S. S. G. (2001) Pharmacol. Rev. 53, 1–24 [PubMed] [Google Scholar]
- 11.Ferguson S. S. G. (2007) Trends Pharmacol. Sci. 28, 173–179 [DOI] [PubMed] [Google Scholar]
- 12.Lodowski D. T., Pitcher J. A., Capel W. D., Lefkowitz R. J., Tesmer J. J. G. (2003) Science 300, 1256–1262 [DOI] [PubMed] [Google Scholar]
- 13.Dhami G. K., Anborgh P. H., Dale L. B., Sterne-Marr R., Ferguson S. S. G. (2002) J. Biol. Chem. 277, 25266–25272 [DOI] [PubMed] [Google Scholar]
- 14.Dhami G. K., Dale L. B., Anborgh P. H., O'Connor-Halligan K. E., Sterne-Marr R., Ferguson S. S. G. (2004) J. Biol. Chem. 279, 16614–16620 [DOI] [PubMed] [Google Scholar]
- 15.Dhami G. K., Babwah A. V., Sterne-Marr R., Ferguson S. S. G. (2005) J. Biol. Chem. 280, 24420–24427 [DOI] [PubMed] [Google Scholar]
- 16.Anborgh P. H., Godin C., Pampillo M., Dhami G. K., Dale L. B., Cregan S. P., Truant R., Ferguson S. S. (2005) J. Biol. Chem. 280, 34840–34848 [DOI] [PubMed] [Google Scholar]
- 17.Gereau R. W., 4th, Heinemann S. F. (1998) Neuron 20, 143–151 [DOI] [PubMed] [Google Scholar]
- 18.Sato M., Tabata T., Hashimoto K., Nakamura K., Nakao K., Katsuki M., Kitano J., Moriyoshi K., Kano M., Nakanishi S. (2004) Eur. J. Neurosci. 20, 947–955 [DOI] [PubMed] [Google Scholar]
- 19.Alagarsamy S., Saugstad J., Warren L., Mansuy I. M., Gereau R. W., 4th, Conn P. J. (2005) Neuropharmacology 49, 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai M. M., Hong J. J., Ruggiero A. M., Burnett P. E., Slepnev V. I., De Camilli P., Snyder S. H. (1999) J. Biol. Chem. 274, 25963–25966 [DOI] [PubMed] [Google Scholar]
- 21.Lai M. M., Burnett P. E., Wolosker H., Blackshaw S., Snyder S. H. (1998) J. Biol. Chem. 273, 18325–18331 [DOI] [PubMed] [Google Scholar]
- 22.Sun L., Youn H. D., Loh C., Stolow M., He W., Liu J. O. (1998) Immunity 8, 703–711 [DOI] [PubMed] [Google Scholar]
- 23.Jang H., Cho E. J., Youn H. D. (2007) Biochem. Biophys. Res. Commun. 359, 129–135 [DOI] [PubMed] [Google Scholar]
- 24.Lai M. M., Luo H. R., Burnett P. E., Hong J. J., Snyder S. H. (2000) J. Biol. Chem. 275, 34017–34020 [DOI] [PubMed] [Google Scholar]
- 25.Ferguson S. S., Ménard L., Barak L. S., Koch W. J., Colapietro A. M., Caron M. G. (1995) J. Biol. Chem. 270, 24782–24789 [DOI] [PubMed] [Google Scholar]
- 26.Youn H. D., Sun L., Prywes R., Liu J. O. (1999) Science 286, 790–793 [DOI] [PubMed] [Google Scholar]
- 27.Francesconi A., Duvoisin R. M. (1998) J. Biol. Chem. 273, 5615–5624 [DOI] [PubMed] [Google Scholar]
- 28.Gomeza J., Joly C., Kuhn R., Knöpfel T., Bockaert J., Pin J. P. (1996) J. Biol. Chem. 271, 2199–2205 [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa K., Nash S. R., Nishimune A., Neki A., Kaneko S., Nakanishi S. (1999) Genes Cells 4, 381–390 [DOI] [PubMed] [Google Scholar]
- 30.Minakami R., Jinnai N., Sugiyama H. (1997) J. Biol. Chem. 272, 20291–20298 [DOI] [PubMed] [Google Scholar]
- 31.Lee J. H., Lee J., Choi K. Y., Hepp R., Lee J. Y., Lim M. K., Chatani-Hinze M., Roche P. A., Kim D. G., Ahn Y. S., Kim C. H., Roche K. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12575–12580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaulieu J. M., Sotnikova T. D., Marion S., Lefkowitz R. J., Gainetdinov R. R., Caron M. G. (2005) Cell 122, 2612–2673 [DOI] [PubMed] [Google Scholar]
- 33.Krueger K. M., Daaka Y., Pitcher J. A., Lefkowitz R. J. (1997) J. Biol. Chem. 272, 5–8 [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro F. M., Ferreira L. T., Paquet M., Cregan T., Ding Q., Gros R., Ferguson S. S. G. (2009) J. Biol. Chem. 284, 23444–23453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dale L. B., Babwah A. V., Bhattacharya M., Kelvin D. J., Ferguson S. S. G. (2001) J. Biol. Chem. 276, 35900–35908 [DOI] [PubMed] [Google Scholar]