Abstract
Allosteric kinase inhibitors hold promise for revealing unique features of kinases that may not be apparent using conventional ATP-competitive inhibitors. Here we explore the activity of a previously reported allosteric inhibitor of BCR-Abl kinase, GNF-2, against two cellular isoforms of Abl tyrosine kinase: one that carries a myristate in the N terminus and the other that is deficient in N-myristoylation. Our results show that GNF-2 inhibits the kinase activity of non-myristoylated c-Abl more potently than that of myristoylated c-Abl by binding to the myristate-binding pocket in the C-lobe of the kinase domain. Unexpectedly, indirect immunofluorescence reveals a translocation of myristoylated c-Abl to the endoplasmic reticulum in GNF-2-treated cells, whereas GNF-2 has no detectable effect on the localization of non-myristoylated c-Abl. These results indicate that GNF-2 competes with the NH2-terminal myristate for binding to the c-Abl kinase myristate-binding pocket and that the exposed myristoyl group accounts for the localization to the endoplasmic reticulum. We also demonstrate that GNF-2 can inhibit enzymatic and cellular kinase activity of Arg, a kinase highly homologous to c-Abl, which is also likely to be regulated through intramolecular binding of an NH2-terminal myristate lipid. These results suggest that non-ATP-competitive inhibitors, such as GNF-2, can serve as chemical tools that can discriminate between c-Abl isoform-specific behaviors.
The catalytic activity of a protein kinase can be modulated by binding of a ligand to a site distant from the active site, also referred to as the allosteric site (1). The ligand is referred to as an allosteric kinase inhibitor and induces a protein conformation that is not compatible with kinase activity. Allosteric inhibitors can potentially be exploited to elucidate kinase functions not discovered using ATP-competitive inhibitors, because they can exploit binding sites and regulatory mechanisms that are unique to a particular kinase.
The c-Abl and Arg (Abl-related gene) proteins comprise the Abl family of non-receptor tyrosine kinases. Each family member has two isoforms: one that is myristoylated in the N terminus (1b or IV) and the other that is deficient in N-myristoylation due to an alternative splicing of the first exon (1a or I) (Fig. 1A). N-Myristoylation often serves as a mechanism for targeting proteins to cellular membranes. However, Abl family members localize to multiple subcellular compartments; whereas Arg is mostly found in the cytoplasm, c-Abl shuttles between the nucleus and the cytoplasm, where it localizes to the cytosol, endoplasmic reticulum, and mitochondria (2).
FIGURE 1.
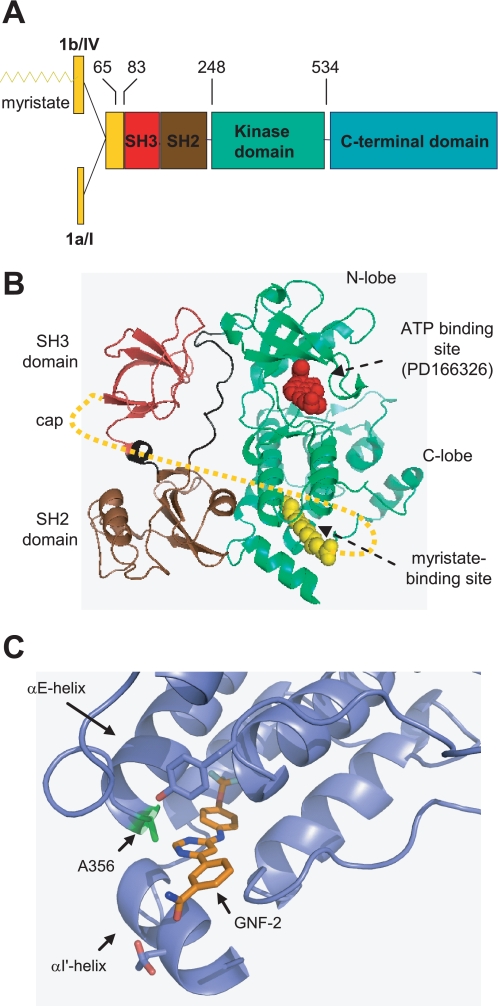
A, domain structure of Abl family members (5). The numbers indicate amino acid residues in c-Abl 1b, and the recombinant protein constructs used in this study encompass amino acids 65–534, 83–534, and 248–531. B, ribbon representation of the c-Abl kinase NH2-terminal half residues, including the SH3, SH2, and kinase domains (Protein Data Bank code 1OPK) (7). The NH2-terminal cap (amino acids 2–79) is indicated by dotted lines (8). The myristate-binding site and ATP binding pocket are indicated by arrows. C, ribbon representation of an enlarged view of GNF-2 (colored gold) bound to the c-Abl myristate binding site. The location of Ala356 is indicated.
The Abl family members share a high degree of sequence identity (∼90%) in the NH2-terminal half residues, including the SH3,2 SH2, and kinase domains (3). The kinase domain is followed by proline-rich motifs that serve as binding sites for SH3 domains. A range of proteins are reported to bind directly or indirectly to the SH3, SH2, and proline-rich domains of c-Abl and are implicated in the proper regulation of the kinase activities of Abl family members in the cytoplasm (4–6). In addition, as revealed by recent crystallographic analyses of inactive and assembled form of recombinant Abl, the kinase activity of c-Abl is modulated by the intrinsic binding of the N-myristoyl residue to a hydrophobic pocket in the C-lobe of the kinase domain, which induces conformational changes in the kinase domain and subsequently allows the SH3 and SH2 domains to pack against the kinase domain (7, 8). Altogether, these observations suggest that the kinase activities of Abl family members in normal cells are tightly regulated by both intra- and intermolecular interactions (2, 9). Disruption of these strong regulatory mechanisms results in deregulated kinase activity, as illustrated by the BCR-Abl and v-Abl oncoproteins.
Recent years have seen great advances in pharmacological inhibition of deregulated c-Abl kinase activity. Among the small molecule inhibitors targeting BCR-Abl kinase are imatinib (STI-571; Gleevec), nilotinib (AMN 107), and dasatinib (BMS-354825) (10). These small molecules have been used not only for clinical intervention in patients with leukemia but also as chemical tools to further dissect BCR-Abl kinase-linked signaling pathways in tissue culture cells (11). However, efforts to analyze the effects of monospecific inhibition of BCR-Abl kinase have been complicated by cross-reactivity of ATP-competitive Abl inhibitors with other kinases. For example, in addition to inhibiting c-Abl and BCR-Abl, STI-571 and nilotinib also potently inhibit c-Kit, platelet-derived growth factor receptor, and DDR1, whereas dasatinib potently inhibits all of these kinases as well as the Src family, Tec family, and KDR kinases (12). The multitargeted nature of these ATP-competitive inhibitors makes it difficult to assign a particular biological effect to inhibition of a specific kinase target.
We previously reported the discovery of the first non-ATP site-monoselective BCR-Abl inhibitor (GNF-2), which targets not only wild type BCR-Abl but also many clinically relevant STI-571-resistant mutants either alone or in combination with other BCR-Abl inhibitors (13). Molecular modeling, site-directed mutagenesis, competition assays, NMR spectroscopy, and protein crystallography were used to determine that GNF-2 binds to a myristate-binding site in the C-lobe of the c-Abl kinase domain (Fig. 1, B and C) (3). The discovery of GNF-2 was the first demonstration that c-Abl kinase activity could be pharmacologically modulated by an inhibitor that binds outside the ATP or substrate binding sites. Although it remained unclear how GNF-2 is capable of inhibiting c-Abl upon binding to the myristate-binding site, we speculated that GNF-2 probably mimics the function of the N-myristoyl residue in c-Abl. Here, we investigated the effects of GNF-2 on Abl family members with the goals of providing further insights into the mechanism of GNF-2 function and laying the foundation to utilize GNF-2 as a tool to investigate c-Abl- and Arg-linked cellular processes.
EXPERIMENTAL PROCEDURES
In Vitro Tyrosine Kinase Assay
In vitro kinase assays were performed according to three different formats as follows.
ELISA-based Assay
Recombinant proteins (100 nm for each construct) or immunoprecipitated proteins were diluted in kinase buffer (20 mm HEPES (pH 7.4), 50 mm KCl, 0.1% CHAPS, 30 mm MgCl2, 2 mm MnCl2, 1 mm DTT, and 1% glycerol). Aliquots of the diluted proteins were preincubated with either DMSO or compounds for 30 min at room temperature and then added to K-LISA PTK EAY reaction plates (EMD Chemicals). The kinase reaction was initiated by adding 0.1 mm ATP and was allowed to proceed for 30 min at room temperature. The levels of tyrosine phosphorylation were monitored by following the manufacturer's protocol.
Continuous Spectrophotometric Assays
Activity of the protein kinases toward c-Abl substrate peptide (14) (sequence EAIYAAPFAKKK) was determined as described previously (15, 16).
Labeling with 32P-Labeled Inorganic Phosphate
Recombinant Abl and Arg proteins (100 nm for each protein) were diluted in kinase buffer (20 mm Tris-Cl (pH 7.4), 50 mm KCl, and 1 mm DTT) and were preincubated with either DMSO or compounds for 30 min at room temperature. Aliquots of the diluted proteins were mixed with kinase buffer containing 50 μm ATP, 15 mm MgCl2, 1 μCi of [γ-32P]ATP, and 1 μg of GST-Abltide (Upstate Biotechnology). The reactions were allowed to proceed for 30 min at room temperature and stopped by adding SDS-sample buffer. The phosphorylation of GST-Abltide was monitored by SDS-PAGE and phosphorimaging analysis or autoradiography.
Establishing Polyclonal Populations of Cells Stably Expressing c-Abl Proteins
Phoenix− Ampho cells (4 × 106 cells/10-cm dish) were transfected with 10 μg of DNA by using FuGENE6 (Roche Applied Science). Twenty-four h after transfection, the culture medium was replaced with DMEM containing 10% fetal bovine serum, and then the cells were further incubated for 48 h. The conditioned medium was filtered through a 0.45-μm filter, mixed with 4 μg/ml Polybrene, and then added to abl−/−arg−/− 3T3 fibroblast culture for 24 h. The abl−/−arg−/− cells were then divided at a 1:5 dilution and further incubated in medium containing 200 μg/ml hygromycin.
Recombinant Protein Expression and Purification
The human c-Abl kinase domain (amino acids 248–531, c-Abl 1b numbering), c-Abl Cap-SH3-SH2-kinase domain (amino acids 65–534), and c-Abl SH3-SH2-kinase domain (amino acids 83–534) were co-expressed with YopH phosphatase in bacteria and purified as described (16). The murine Arg SH3-SH2-kinase domain (amino acids 110–558, Arg 1b numbering) was cloned into pET28a vector (Novagen) and expressed in BL21 CodonPlus (DE3) RIPL (Stratagene). Cells were grown at 37 °C to an A600 nm of 1.0, cooled for 1 h at 18 °C, and then induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside for 12 h at 18 °C. The harvested cells were resuspended in lysis buffer (20 mm Tris-Cl (pH 8.0), 500 mm KCl, 5% glycerol, 20 mm imidazole, and 1 mm DTT) and subject to lysis by French press. After centrifugation at 40,000 × g for 30 min, the supernatant was loaded onto an Ni2+-nitrilotriacetic acid Superflow column (Qiagen). The column was extensively washed with the lysis buffer as above except with 50 mm imidazole, and the His6-tagged Arg protein was eluted with 20 mm Tris-Cl (pH 8.0), 500 mm KCl, 5% glycerol, 200 mm imidazole, and 1 mm DTT. The eluted protein was dialyzed against buffer containing 20 mm Tris-Cl (pH 7.4), 100 mm KCl, 5% glycerol, and 1 mm DTT.
Affinity Pull-down Experiment
3T3 fibroblasts were collected in PBS and lysed in buffer containing 25 mm HEPES (pH 7.4), 0.3% CHAPS, 150 mm NaCl, 1 mm EDTA, 1 mm sodium vanadate, 10 mm β-glycerophosphate, and protease inhibitor mixture (Roche Applied Science). One mg of cleared lysates, in a total volume of 1 ml, was incubated with Sepharose-immobilized compounds (50 μl of 50% suspension) at 4 °C for 1 h. After extensive washing with the lysis buffer, the bound proteins were eluted in 25 μl of SDS-sample buffer, and half of the eluates were analyzed by Western blot. About 10% of input was loaded onto a gel to compare binding efficiency.
Indirect Immunofluorescence
3T3 fibroblasts were plated on coverslips and grown in DMEM containing 10% fetal bovine serum. The cells were treated with either DMSO or compound for 1 h, washed with ice-cold PBS containing 3.7% formaldehyde, and fixed in PBS containing 3.7% formaldehyde for 10 min. After washing with PBS, the cells were covered with methanol and incubated at −20 °C for 10 min and then permeabilized with 0.2% Triton X-100 in PBS for 5 min. Primary and secondary antibodies were diluted in PBS containing 5% normal goat serum and used for staining. The antibodies are as follows: 10 μg/ml of mouse anti-c-Abl (8E9; BD Bioscience); 1:500 dilution of rabbit anti-protein-disulfide isomerase (Sigma); 2 μg/ml Alexa Fluor 488 goat anti-mouse IgG (Invitrogen); 2 μg/ml tetramethylrhodamine goat anti-rabbit IgG (Invitrogen). Wide field fluorescence images were acquired using a Nikon ECLIPSE TE2000-E equipped with a Hamamatsu ORCA camera and captured with NIS Element AR 2.30 software (Nikon). Confocal fluorescence images were acquired using a Nikon TE2000U microscope with Spinning Disk Confocal Head, Hamamatsu ORCA-ER camera and Metamorph software.
RESULTS
GNF-2 Inhibits c-Abl Kinase Activity in Vitro
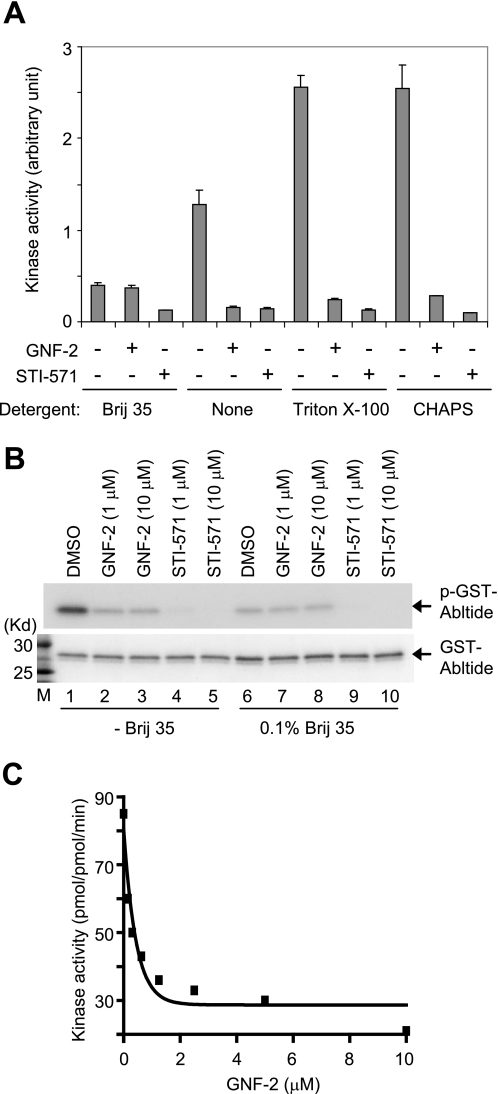
In a previous study, we failed to observe inhibition of BCR-Abl kinase activity by GNF-2 in an in vitro kinase reaction with multiple different protein constructs (13). Here we determined the activity of recombinant Abl (amino acids 65–534) using an ELISA-based assay that measures phosphotransfer to an immobilized peptide substrate. To our surprise, GNF-2 was capable of inhibiting recombinant Abl in this format with an IC50 of <1 μm. We examined differences between the ELISA-based kinase assay and the previously used radioenzymatic assay that is frequently used to measure the kinase activity of both recombinant Abl and immunoprecipitated BCR-Abl with imatinib (STI-571; Gleevec) or other Abl inhibitors (17). One difference was the presence of the non-ionic detergent “polyoxyethylene lauryl ether” (Brij 35) in the radioenzymatic kinase assay buffer but not in the ELISA-based assay. To determine whether the addition of Brij 35 could prevent inhibition of recombinant Abl kinase activity by GNF-2 in the ELISA-based format, we repeated the assay with the non-ionic detergent. The result showed that 10 μm GNF-2 clearly inhibited recombinant Abl (amino acids 65–534) kinase activity in the presence of other non-ionic detergents, such as Triton X-100 and CHAPS, but was unable to inhibit recombinant Abl kinase activity in the presence of Brij 35 (Fig. 2A). To extend the analysis, we tested whether removal of Brij 35, in the radioenzymatic kinase assay, restores inhibition of recombinant Abl kinase activity by GNF-2. As expected, GNF-2 was capable of inhibiting the kinase activity of recombinant Abl (amino acids 65–534) in the absence of Brij 35 (Fig. 2B). The addition of Brij 35 to the kinase assay buffer likewise resulted in a decrease in recombinant Abl kinase activity in both the ELISA-based assay and the radioenzymatic kinase assay (compare lanes 1 and 6 in Fig. 2B).
FIGURE 2.
GNF-2 inhibits recombinant Abl kinase activity in vitro. A, recombinant Abl (amino acids 65–534) was diluted in kinase buffer containing 0.1% of the indicated detergents, incubated with either DMSO or 10 μm compounds for 30 min, and then its tyrosine kinase activity was measured by an ELISA-based assay. It is of note that Brij 35 interferes with recombinant Abl kinase activity as well as the ability of GNF-2 to inhibit recombinant Abl kinase activity. B, recombinant Abl (amino acids 65–534) was diluted in kinase assay buffer in the absence or presence of 0.1% Brij 35, incubated with either DMSO or compounds for 30 min, and then its tyrosine kinase activity was assessed by radioenzymatic assay. The phosphorylation of GST-Abltide was monitored by SDS-PAGE and autoradiography (top). The total substrate used (GST-Abltide) was visualized by Coomassie staining (bottom). M, molecular weight marker. C, a continuous spectrophotometric assay confirms the GNF-2 inhibition of recombinant Abl kinase activity. The recombinant Abl (amino acids 65–534) was diluted to a final concentration of 50 nm in kinase buffer (50 mm Tris-Cl (pH 7.4), 50 mm KCl, 10 mm MgCl2) in the absence or presence of GNF-2. Kinase activity was measured with 300 μm substrate and 500 μm ATP for 20 min and expressed as pmol/pmol·min.
In order to examine how Brij 35 inhibits recombinant Abl kinase activity, we employed a “continuous spectrophotometric assay” (15). This assay measures ATP depletion through a coupled enzyme cascade involving the enzymes pyruvate kinase and lactate dehydrogenase, which ultimately convert NADH to NAD, and has allowed investigation of enzymatic properties, including kcat, Km for ATP, and Km for substrate. Monitoring the recombinant Abl kinase activity with increasing concentrations of ATP revealed that Brij 35 reduces the recombinant Abl catalytic rate without affecting ATP affinity of recombinant Abl kinase (Table 1 and supplemental Fig. S1A). The substrate titration experiment demonstrated that Brij 35 increases Km of recombinant Abl for a peptide substrate (EAIYAAPFAKKK) (Table 1 and supplemental Fig. S1B).
TABLE 1.
Effect of Brij 35 on enzymatic activity of recombinant Abl (residues 65–534)
|
kcat |
Km |
|||
|---|---|---|---|---|
| None | Brij 35 (0.1%) | None | Brij 35 (0.1%) | |
| min−1 | μm | |||
| ATP | 120 ± 12 | 23 ± 9.1 | 39 ± 0.2 | 37 ± 19.2 |
| Substrate | 228 ± 15 | 126 ± 13.4 | 323 ± 42.3 | 827 ± 47.3 |
We next examined the potency of GNF-2 on the kinase activity of recombinant Abl (amino acids 65–534) using the continuous spectrophotometric assay. A GNF-2 titration experiment demonstrated that GNF-2 inhibits recombinant Abl kinase activity with an IC50 of 0.24 μm in this assay (Fig. 2C), which is in approximate agreement with the data from the BCR-Abl kinase activity-dependent cellular assay (EC50 = 138 nm) (13). In order to support the previous observation that GNF-2 inhibits BCR-Abl kinase activity in a non-ATP-competitive manner, we monitored the catalytic rate of recombinant Abl in the presence of increasing concentrations of ATP. Incubation of recombinant Abl kinase with GNF-2 resulted in a decrease in Vmax while not affecting Km for ATP (Table 2 and supplemental Fig. 2A), indicating that GNF-2 is indeed a non-ATP-competitive inhibitor. However, as was observed with Brij 35, GNF-2 treatment resulted in an increase in Km of recombinant Abl for the peptide substrate (Table 2 and supplemental Fig. S2B), suggesting that GNF-2 binding to the myristate site leads to a decrease in the affinity of the peptide substrate and, as a result, reduces the catalytic rate of recombinant Abl kinase.
TABLE 2.
Effect of GNF-2 on enzymatic activity of recombinant Abl (residues 65–534)
|
kcat |
Km |
|||
|---|---|---|---|---|
| DMSO | GNF-2 (1 μm) | DMSO | GNF-2 (1 μm) | |
| min−1 | μm | |||
| ATP | 108 ± 3.5 | 15 ± 2 | 42 ± 3.6 | 40 ± 14 |
| Substrate | 252 ± 49.5 | 95 ± 2.9 | 295 ± 3.5 | 933 ± 112.4 |
GNF-2 Requires Abl SH3 and/or SH2 Domains to Inhibit Kinase Activity
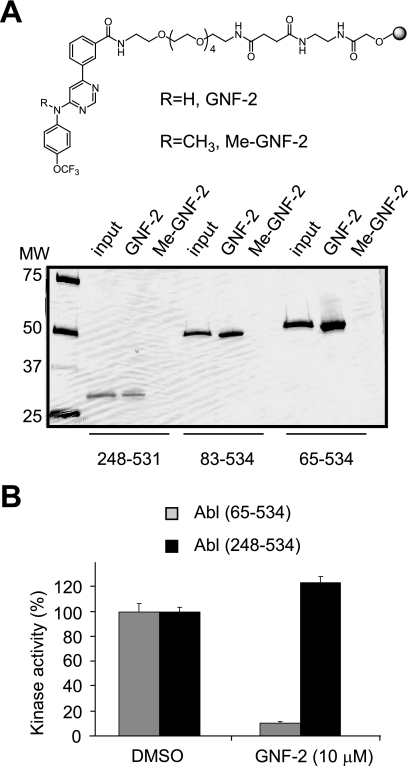
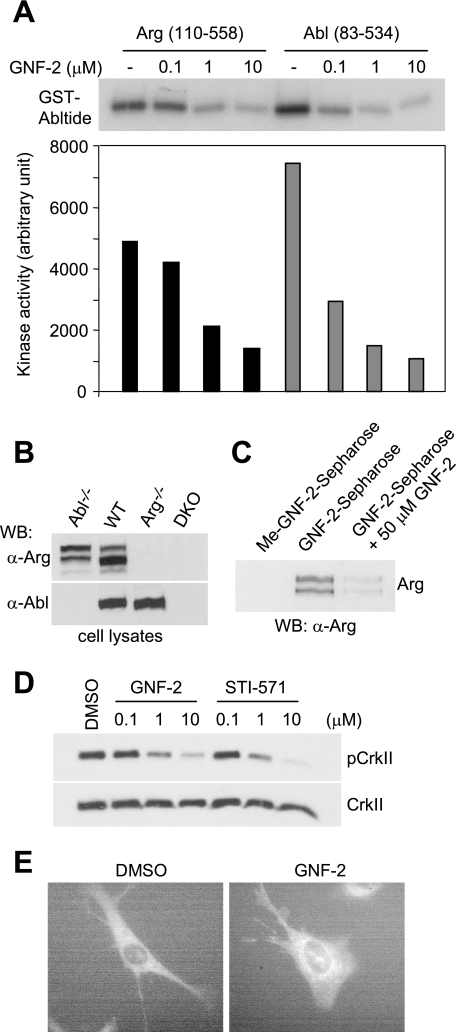
GNF-2 failed to inhibit the proliferation of Ba/F3 cells that express NPM-Abl or Tel-Abl fusion proteins (13). These fusion proteins lack either the SH3/SH2 domains or the SH3 domain of the c-Abl protein. The results suggested that GNF-2 might require BCR and/or the c-Abl SH3 and/or SH2 domains to inhibit BCR-Abl-dependent cell proliferation. The in vitro ELISA-based kinase assays that were established in this study revealed that the BCR domain is not required for inhibition of c-Abl kinase activity by GNF-2 (Fig. 2). Next we tested the ability of GNF-2 to bind and inhibit the tyrosine kinase activity of recombinant Abl (amino acids 248–531) lacking the SH3 and SH2 domains. Affinity pull-down experiments with Sepharose-immobilized GNF-2 demonstrated binding of GNF-2 to recombinant Abl (amino acids 248–531) (Fig. 3A). In addition, methylation of the aniline nitrogen at the C4 position of the pyrimidine (Me-GNF-2) abolished the interaction, indicating binding specificity of GNF-2 for recombinant Abl proteins. However, in vitro kinase assays showed that GNF-2 fails to inhibit kinase activity of recombinant Abl lacking the SH3 and SH2 domains (Fig. 3B), supporting the previous observation that GNF-2 requires the presence of these SH3 and/or SH2 domains to exert its inhibitory effect.
FIGURE 3.
A, indicated recombinant Abl proteins were incubated with Sepharose-immobilized GNF-2 or Me-GNF-2 (top), and the bound proteins were visualized by silver staining (bottom). B, the SH3 and/or SH2 domains in Abl are required for GNF-2 to exert its activity. Indicated recombinant Abl proteins were incubated with either DMSO or 10 μm of GNF-2, and then its tyrosine kinase activity was monitored by an ELISA-based assay. Data are expressed as a percentage of DMSO controls (mean ± S.D.; n = 3).
GNF-2 Inhibits c-Abl Tyrosine Kinase Activity in Tissue Culture Cells
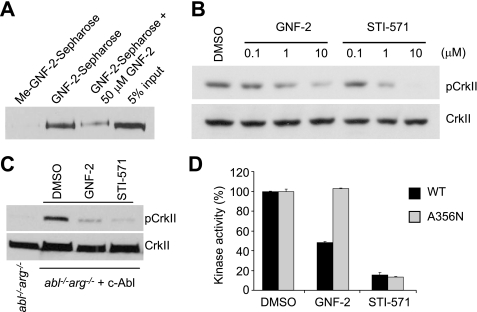
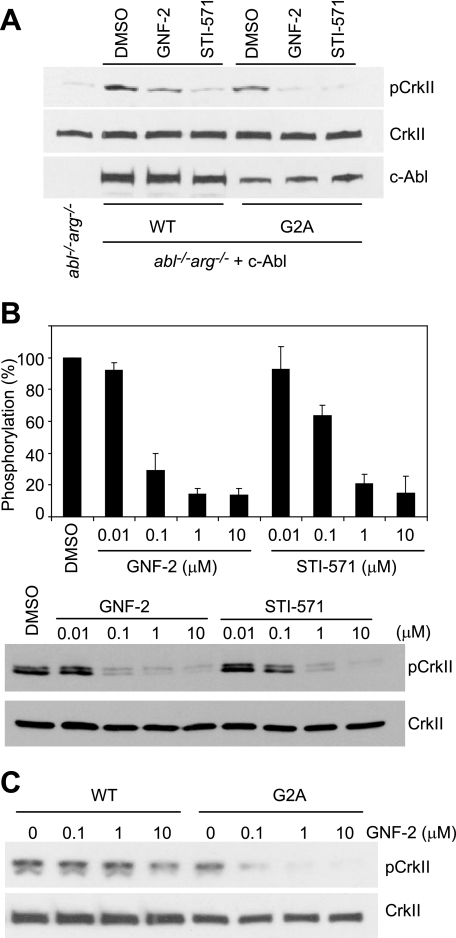
With Sepharose-immobilized GNF-2, we previously demonstrated that GNF-2 directly interacts with cellular BCR-Abl and recombinant Abl proteins in vitro (13). Structural evidence based upon NMR spectroscopy and protein crystallography indicate that GNF-2 binds the myristate-binding pocket in the C-lobe of recombinant Abl kinase domain.3 These observations led us to investigate whether GNF-2 interacts with c-Abl in tissue culture cells. Using affinity chromatography, we determined that GNF-2, but not a methylated GNF-2 analog, binds c-Abl in cellular extracts derived from 3T3 fibroblasts (Fig. 4A). To test whether GNF-2 inhibits the tyrosine kinase activity of c-Abl in tissue culture cells, we incubated 3T3 cells with increasing concentrations of GNF-2 and analyzed tyrosine phosphorylation levels of a well characterized c-Abl substrate, CrkII, by Western blot (6). GNF-2, in a dose-dependent manner, clearly inhibited tyrosine phosphorylation of CrkII (Fig. 4B). In order to test if GNF-2 directly targets c-Abl and whether the inhibition of CrkII phosphorylation by GNF-2 is mediated through c-Abl, we generated a polyclonal population of cells that stably express c-Abl (mouse type IV) using retroviral mediated gene transfer to 3T3 fibroblasts, which were established from abl−/−arg−/− embryonic day 9.5 mouse embryos (18). The abl−/−arg−/− 3T3 cells showed barely detectable levels of phosphorylated CrkII; however, a significant increase in CrkII phosphorylation was observed upon expression of c-Abl (Fig. 4C). The phosphorylation of CrkII in c-Abl-reconstituted 3T3 cells was inhibited by both GNF-2 and STI-571, indicating that GNF-2 indeed inhibits c-Abl kinase activity in tissue culture cells. Next, we investigated whether GNF-2 inhibition of c-Abl kinase activity is mediated through its binding to the myristate-binding pocket. To test this hypothesis, we introduced a point mutation, A356N, which is located in the αE-helix and was previously demonstrated to induce resistance of BCR-Abl to GNF-2 by sterically interfering with inhibitor binding (13). The A356N mutation was introduced into c-Abl and transiently transfected into HEK293T cells, which were then used for immunoprecipitation kinase assays. The kinase assay with the immunoprecipitated c-Abl revealed that the A356N mutation in the myristate-binding site rendered c-Abl resistant to inhibition by GNF-2, whereas the mutation had no effect on STI-571 inhibition of the kinase activity (Fig. 4D). In vitro pull down of c-Abl from the lysates of abl−/−arg−/− 3T3 cells reconstituted with either WT or A356N c-Abl confirmed that the mutation in the myristate-binding pocket in c-Abl compromises the ability of GNF-2 to bind c-Abl (supplemental Fig. S3).
FIGURE 4.
GNF-2 targets c-Abl in tissue culture cells. A, GNF-2 binds to c-Abl in 3T3 fibroblasts. The lysates of 3T3 WT fibroblasts were incubated with either Sepharose-immobilized Me-GNF-2 or Sepharose-immobilized GNF-2 in the absence or presence of GNF-2, and the bound proteins were analyzed by Western blot with an anti-Abl antibody (8E9). B, GNF-2 inhibits the phosphorylation of CrkII. 3T3 WT fibroblasts were treated with either DMSO or various concentrations of compounds for 1 h, and then total lysates were analyzed by Western blot. C, GNF-2 inhibits c-Abl-induced phosphorylation of CrkII. The abl−/−arg−/− 3T3 cells and abl−/−arg−/− cells reconstituted with c-Abl were treated with either DMSO or 10 μm compounds for 1 h, and then total lysates were analyzed by Western blot. D, the point mutation in the myristate-binding site renders c-Abl resistant to inhibition by GNF-2. The indicated c-Abl proteins were immunoprecipitated following transfection into HEK293T cells, treated with either DMSO or 10 μm compounds, and then their kinase activities were assessed by ELISA-based assay. Data are expressed as a percentage of DMSO controls (mean ± S.D.; n = 3 for a representative experiment).
It is of interest to note that GNF-2 was not as potent as STI-571 with respect to inhibition of kinase activity of endogenous c-Abl (Fig. 4B), despite the observation that GNF-2 inhibited the kinase activity of recombinant Abl (amino acids 65–534) and proliferation of Ba/F3 cells expressing BCR-Abl with potencies equal to that of STI-571. Since recombinant Abl and the BCR-Abl fusion proteins both lack the N-myristoyl modification, we speculated that the N-myristoyl group might interfere with the ability of GNF-2 to inhibit c-Abl kinase activity. To address this question, we established a polyclonal population of abl−/−arg−/− 3T3 cells that stably express c-Abl (mouse type IV) carrying alanine at position 2 instead of glycine (G2A mutation). This G2A mutation results in the expression of a non-myristoylated kinase. Reconstitution of c-AblG2A in abl−/−arg−/− cells resulted in an increase in the phosphorylation level of CrkII, as was seen with c-AblWT, and the phosphorylation of CrkII was inhibited by both GNF-2 and STI-571 (Fig. 5A and supplemental Fig. S4A). It is of note that the phosphorylation level of CrkII in c-AblG2A-expressing cells is comparable with that of CrkII in c-AblWT-expressing cells. GNF-2 titration experiments showed that GNF-2 (IC50 = 0.051 μm) is more potent than STI-571 (IC50 = 0.160 μm) at inhibiting the phosphorylation of CrkII in c-AblG2A-expressing cells (Fig. 5B). Furthermore, the phosphorylation of CrkII in c-AblG2A-expressing cells was inhibited by concentrations of GNF-2 as low as 0.1 μm, whereas GNF-2 at concentrations of 0.1 and 1 μm had little effect on the phosphorylation of CrkII in c-AblWT-expressing cells (Fig. 5C and supplemental Fig. S4B). These results strongly indicate that c-Abl protein lacking a myristate group is more sensitive to GNF-2 and that GNF-2 competes with the myristate in the c-Abl N terminus for binding to the myristate-binding pocket in tissue culture cells. Our previous results indeed showed a competition of GNF-2 with a myristoylated peptide corresponding to N-terminal amino acids 2–16 of c-Abl 1b in vitro (13).
FIGURE 5.
N-Myristoyl group in c-Abl affects the ability of GNF-2 to inhibit c-Abl kinase activity. A, reconstitution of c-AblG2A in abl−/−arg−/− 3T3 cells increases the phosphorylation of CrkII, and GNF-2 inhibits c-AblG2A-induced phosphorylation of CrkII. The abl−/−arg−/− 3T3 cells and abl−/−arg−/− cells reconstituted with either c-AblWT or c-AblG2A were treated with either DMSO or 5 μm compounds for 1 h, and then total lysates were analyzed by Western blot with an anti-phospho-CrkII antibody (top) or anti-c-Abl antibody (8E9) (bottom). The blot was reprobed with an anti-CrkII antibody to show equal loading of proteins (middle). B, GNF-2 is more potent than STI-571 with respect to inhibition of c-AblG2A. The abl−/−arg−/− cells reconstituted with c-AblG2A were treated with either DMSO or various concentrations of compounds for 1 h, and then total lysates were analyzed by Western blot with an anti-phospho CrkII antibody. The data from three independent experiments were averaged for each condition and presented in the form of bar graphs (top). Data are expressed as a percentage of DMSO controls (mean ± S.D.; n = 3). One representative Western blot is shown (bottom). C, substitution of the myristoylation site (Gly2) in c-Abl with alanine renders c-Abl highly susceptible to inhibition by GNF-2. The abl−/−arg−/− cells reconstituted with either c-AblWT or c-AblG2A were treated with either DMSO or various concentrations of compounds for 1 h, and then total lysates were analyzed by Western blot.
N-Myristoylated c-Abl Localizes to the Endoplasmic Reticulum upon Interaction with GNF-2
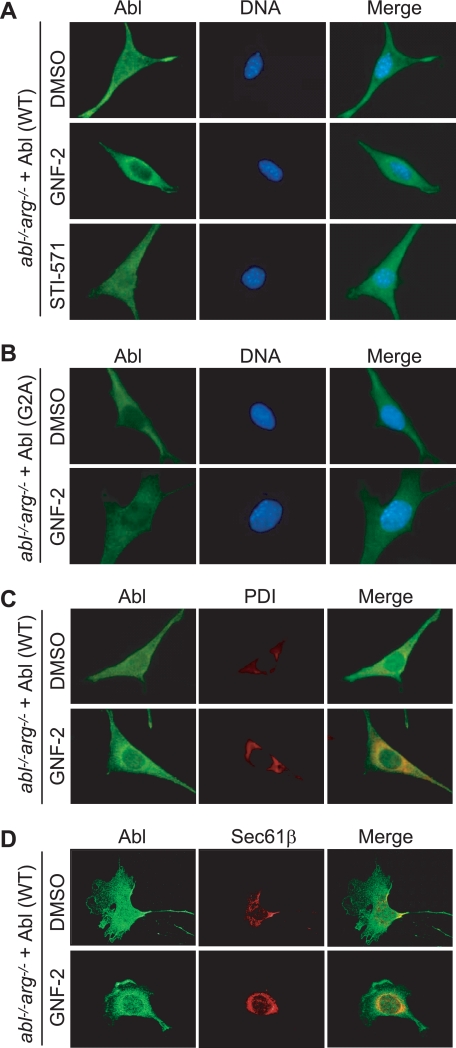
Since the myristoyl group is expected to be displaced from the c-Abl myristate-binding pocket in the presence of GNF-2, we hypothesized that GNF-2 may influence the cellular localization of c-Abl potentially by targeting c-Abl to cellular membranes. We employed indirect immunofluorescence to determine the intracellular localization of c-Abl in abl−/−arg−/− cells reconstituted with the c-Abl. This system has the advantage that a specific form of c-Abl can be analyzed in isolation. The immunostaining results demonstrated that c-AblWT is localized both in the cytoplasm and the nucleus in the absence of GNF-2 treatment (Fig. 6A). Unexpectedly, although STI-571 had no detectable effect on the localization of c-AblWT, GNF-2 treatment induced a fraction of the c-AblWT in 60–70% of the cells to localize to the perinuclear region in a manner that is reminiscent of endoplasmic reticulum (ER)-resident proteins (Fig. 6A). The observed pattern of c-Abl localization was also detected in wild type 3T3 fibroblasts treated with GNF-2 (data not shown). To test whether the N-terminal myristoylation is required for the observed pattern of c-AblWT localization, we investigated localization of c-AblG2A in abl−/−arg−/− cells reconstituted with the c-AblG2A. The G2A mutant was found mostly in the cytoplasm, and GNF-2 had little effect on the localization of c-AblG2A (Fig. 6B). c-Abl-deficient 3T3 fibroblast showed no detectable staining with an anti-c-Abl antibody (8E9), confirming specificity of the antibody used in this study (supplemental Fig. 5A).
FIGURE 6.
GNF-2 induces translocation of the myristoylated c-Abl to the ER. A and B, the indicated cells were plated on coverslips and grown in DMEM containing 10% fetal bovine serum. The cells were treated with either DMSO or 10 μm compound for 1 h, and c-Abl proteins were visualized by indirect immunofluorescence using an anti-c-Abl antibody (8E9) and Alexa Fluor 488 goat anti-mouse IgG. 4′,6-Diamidino-2-phenylindole was used to stain the nucleus. C, wide field fluorescence images. The cells were double-stained for c-Abl (green) and protein-disulfide isomerase (red) following treatment with either DMSO or 10 μm GNF-2 for 1 h. D, confocal fluorescence images. The cells were double-stained for c-Abl (green) and Sec61β (red) following treatment with either DMSO or 10 μm GNF-2 for 1 h. It is evident that the intense green fluorescence around the nucleus in GNF-2-treated cells colocalizes with the red fluorescence.
To investigate whether N-myristoylated c-Abl localizes to the ER upon interaction with GNF-2, we first detected the localization of ER-resident proteins, protein-disulfide isomerase and Sec61β (19, 20), by indirect immunofluorescence and then compared the localization of c-AblWT with that of protein-disulfide isomerase and Sec61β in GNF-2-treated cells. The results showed that a pool of c-AblWT in GNF-2-treated cells colocalizes with protein-disulfide isomerase (Fig. 6C) and Sec61β (Fig. 6D), indicating a translocation of c-AblWT to the ER upon binding of GNF-2. In order to confirm the association of c-AblWT with the ER in GNF-2-treated cells by an independent method, we isolated microsome fractions that are enriched for the ER from abl−/−arg−/− cells reconstituted with the c-AblWT. The level of c-Abl in the microsome fraction from GNF-2-treated cells was indeed 3-fold higher than that of DMSO control (supplemental Fig. S5B).
GNF-2 Interacts with Arg and Inhibits Kinase Activity
The amino acids residues that constitute the myristoyl binding pocket in c-Abl are conserved in Arg (7), suggesting that GNF-2 may interact with Arg and inhibit its kinase activity as well. In an effort to demonstrate that GNF-2 inhibits the kinase activity of Arg in vitro, we expressed a recombinant Arg protein containing the SH3, SH2, and kinase domains (amino acids 110–558, murine Arg 1b numbering). This construct shares 90% sequence identity with its c-Abl counterpart (amino acids 83–534) and is expected to represent the minimal region for Arg regulation, as is the case for c-Abl regulation. In vitro kinase assays with recombinant proteins showed that GNF-2 indeed inhibited Arg tyrosine kinase activity (IC50 = 0.67 μm), although Arg was slightly less sensitive to inhibition by GNF-2 than c-Abl (IC50 = 0.07 μm) (Fig. 7A).
FIGURE 7.
GNF-2 inhibits Arg kinase activity in vitro and in tissue culture cells. A, GNF-2 inhibits tyrosine kinase activity of recombinant Arg. Recombinant Arg (amino acids 110–581) and Abl (amino acids 83–534) were incubated with various concentrations of compounds, and then their tyrosine kinase activity was monitored by radioenzymatic kinase assay with [γ-32P]ATP and GST-Abltide as a substrate. The phosphorylation of GST-Abltide was visualized (top) and quantified (bottom) by phosphorimaging analysis. B, Western blot analyses of total lysates of the indicated 3T3 cells. It is evident that the anti-Arg antibody does not cross-react with c-Abl. C, GNF-2 interacts with Arg in abl−/− 3T3 cells. The lysates of abl−/− 3T3 fibroblasts were incubated with either Sepharose-immobilized Me-GNF-2 or Sepharose-immobilized GNF-2 in the absence or presence of GNF-2, and the bound proteins were analyzed by Western blot with an anti-Arg antibody. D, GNF-2 inhibits the phosphorylation of CrkII in abl−/− 3T3 cells. The abl−/− 3T3 cells were treated with either DMSO or various concentrations of compounds for 1 h, and then total lysates were analyzed by Western blot. E, The arg−/− 3T3 cells reconstituted with Arg-yellow fluorescent protein (mouse 1b) were treated with either DMSO or GNF-2 (10 μm). The cells were imaged live using a Nikon TE2000U microscope, Hamamatsu ORCA camera, and Metamorph software (version 7.6.0) in the Nikon Imaging Center at Harvard Medical School.
To investigate whether GNF-2 is capable of inhibiting cellular Arg activity, we incubated lysates of abl−/− 3T3 cells with Sepharose-immobilized GNF-2 either in the presence or absence of free GNF-2 (50 μm) as a competitor. Western blot analysis of the bound proteins with an anti-Arg antibody (18), which does not cross-react with c-Abl (Fig. 7B), demonstrated that GNF-2, but not a methylated GNF-2 analog, binds Arg and that GNF-2 competes with Sepharose-immobilized GNF-2 for Arg interaction (Fig. 7C). To test whether GNF-2 inhibits the tyrosine kinase activity of Arg in tissue culture cells, we incubated abl−/− 3T3 cells with GNF-2 and analyzed tyrosine phosphorylation levels of CrkII by Western blot. GNF-2 clearly inhibited tyrosine phosphorylation of CrkII in a dose-dependent manner with an EC50 of ∼1 μm (Fig. 7D).
In order to test whether cellular Arg also translocates to the ER upon binding to GNF-2, we acquired images of yellow fluorescence from arg−/− fibroblasts reconstituted with a previously described Arg-yellow fluorescent protein fusion protein (21). As was observed with c-Abl, Arg-yellow fluorescent protein in GNF-2-treated cells localized to the perinuclear region in a manner that is reminiscent of ER-resident proteins (Fig. 7E).
Effect of GNF-2 on the Kinase Activity of Src Family Members
The NH2-terminal core region of c-Abl (including the SH3, SH2, and kinase domains) shares a high degree of structural similarity with N-myristoylated Src family kinases, and recent structural studies of c-Src suggested the formation of a potential myristate-binding pocket in the C-lobe of the Src kinase domain (22). Therefore, we tested the possibility that GNF-2 also inhibits the kinase activity of Src family kinases in vitro. We observed that GNF-2 had no detectable inhibitory effect on the Src family kinases Hck, Lyn, Lck, and c-Src while clearly inhibiting the activity of recombinant Abl using the Z′-LYTE kinase assay (supplemental Fig. S6). These results indicate that GNF-2 can distinguish between the myristate-binding sites in Abl family members versus Src family kinases and provide new support for the monospecificity of these inhibitors for c-Abl and Arg.
DISCUSSION
Our results demonstrate that GNF-2 inhibits the Abl family of tyrosine kinases in tissue culture cells and in vitro by binding to a hydrophobic pocket located in the C-lobe of the kinase domain, which is the binding site for an N-terminal myristate group. Absence of the N-myristoyl residue in the c-Abl type IV isoform renders it ∼100-fold more sensitive to inhibition by GNF-2 in tissue culture cells, indicating that GNF-2 competes with the N-myristoyl residue for binding to the myristate-binding pocket. Upon binding to this pocket, GNF-2 requires the SH3 and/or SH2 domains in c-Abl to inhibit kinase activity but does not require additional cellular cofactors, as determined by three different types of in vitro kinase assays.
We also discovered that the ability of GNF-2 to inhibit recombinant Abl (amino acids 65–534) in an in vitro kinase assay is compromised by Brij 35 (polyoxyethyleneglycol dodecyl ether), a non-ionic detergent that is commonly used in in vitro tyrosine kinase assays (Fig. 2A). It consists of a polyoxyethylene “headgroup” and a 12-carbon unsaturated hydrocarbon “tail” that might be capable of mimicking the N-myristoyl residue in c-Abl and binding to the myristate-binding pocket. Although the addition of Brij 35 does diminish recombinant Abl kinase activity, it does so with significantly reduced efficacy and potency relative to GNF-2. We speculate that Brij 35 can act as a competitive antagonist of GNF-2 by occupying the myristate-binding site and preventing access. Detergents have been previously demonstrated to bind to lipid binding sites in protein kinases, as exemplified by binding of n-octyl-β-glucopyranoside to p38α MAPK (23), although it is not known whether n-octyl-β-glucopyranoside modulates the kinase activity of p38α MAPK.
The kinase activities of non-myristoylated isoforms of Abl family members are also tightly controlled in tissue culture cells (24). In this study, we also noticed that the kinase activity of the c-AblG2A (murine type IV) is comparable with basal kinase activity of c-AblWT (murine type IV), as determined by the level of CrkII phosphorylation in cell types expressing WT and G2A c-Abl (Fig. 5). This observation differs with a previous report in which transiently transfected G2A mutant exhibited higher kinase activity than WT in HEK293 cells and in vitro (8). This difference may be a result of different experimental conditions. Our study employed a polyclonal population of cells stably expressing G2A mutant, whereas previous studies used transient overexpression. Because many cellular cofactors, in addition to myristoylation, that represent the “autoinhibition” mechanism of c-Abl are also implicated in “co-inhibition” of c-Abl kinase activity in tissue culture cells (9), one could hypothesize that the c-AblG2A-expressing cells evolve to express a sufficient amount of cellular cofactors that bind the SH3 and proline-rich domains in c-Abl such that the kinase activity of the G2A mutant is well regulated despite the lack of myristoylation. In addition, this study employed 3T3 fibroblasts, whereas the previous study used HEK293 cells. Further study would be required to determine how these experimental differences may impact the kinase activity of G2A mutant c-Abl.
The ability of GNF-2 to inhibit c-Abl and Arg kinase activity appears to be highly dependent on the conditions and protein construct that are used (25). The following preliminary observations support this hypothesis: 1) although c-AblG2A was sensitive to inhibition by GNF-2 in tissue culture cells and immunoprecipitated c-AblG2A showed sensitivity toward STI-571, the kinase activity of immunoprecipitated c-AblG2A was not inhibited by 10 μm GNF-2 (data not shown); 2) the lysis of bacteria by French press in the presence of non-ionic detergents resulted in a recombinant Arg (amino acids 110–581) being insensitive to inhibition by GNF-2 (10 μm), whereas this form of Arg was sensitive to inhibition by STI-571. These observations suggest that cell lysis in the presence of detergents could lead to a disruption of kinase conformations that are maintained by the coordinated action of the SH3, SH2, and kinase domains.
A pool of c-Abl was previously shown to localize to the ER, and the ER-associated c-Abl is involved in the cellular response to ER stress (26). However, the mechanism by which c-Abl associates with the ER has not been investigated. Our results suggest that N-myristoylated c-Abl (type IV) is targeted to the ER via exposure of its myristate residue to the cytoplasm upon displacement by GNF-2 from the myristate-binding pocket. This is based on two main observations. First, c-Abl protein carrying a myristate group is much less sensitive to inhibition by GNF-2 than the one lacking a myristate, suggesting that GNF-2 competes with the N-terminal myristate of c-Abl for binding to the myristate-binding pocket in tissue culture cells. Second, indirect immunofluorescence and immunoblot analysis of microsome fractions showed an enrichment of N-myristoylated c-Abl in the ER in GNF-2-treated cells. However, it remains to be elucidated why the N-myristoylated c-Abl in GNF-2-treated cells preferentially localizes to the ER rather than to the plasma membrane.
The observation that recombinant Arg is slightly less sensitive than recombinant Abl to inhibition by GNF-2 raises a question of how GNF-2 discriminates between the myristate-binding sites in Arg and c-Abl. A close comparison of c-Abl and Arg amino acid residues that make up the myristate-binding site pinpoints some differences in the residues forming αE (Asn355 and Gln357), αH (Arg479 and Glu485), and αI′-helices (Glu526 and Lys527) (supplemental Fig. S7). These amino acid differences between c-Abl and Arg could alter the conformation of their respective myristoyl binding sites, suggesting that it may be possible to develop myristate-targeted agents that can discriminate between these two kinases.
Activation of c-Abl has been reported in some types of human breast cancer cell lines and human cell lines of non-small cell lung cancer (27–30). The biological relevance of activated c-Abl kinase in cancer cell proliferation was validated by the use of the small molecule inhibitor, STI-571. Although STI-571 is a good chemical tool in this respect, it shows equal potency in inhibiting both non-myristoylated and myristoylated c-Abl kinases and inhibits other tyrosine kinases, such as platelet-derived growth factor receptor, c-Kit, and DDR1. GNF-2, however, appears to be selective for c-Abl/Arg kinase and exhibits no detectable inhibitory activity toward the closely related Src family of tyrosine kinases. In addition, GNF-2 is more effective against non-myristoylated c-Abl kinase than against myristoylated c-Abl, providing an excellent tool compound to discriminate the role of each isoform in the analyses of c-Abl- and Arg-linked pathways. However, when testing GNF-2 at an organism level, we cannot exclude the possibility of toxic effects due to the consequences of localization of c-Abl and Arg to the ER, which may be different from those that result from simply targeting the catalytic activity through binding at the ATP site.
Supplementary Material
Acknowledgments
We thank Prof. Jean Wang for providing c-Abl cDNA (31) and helpful comments throughout this study. We thank Prof. John Kuriyan for providing recombinant Abl protein. We thank Pamela Woodring, Priscilla Yang, and Sonal Jhaveri-Schneider for helpful discussions. Live cell imaging and confocal microscopy were performed in the Nikon Imaging Center at Harvard Medical School with help from Jennifer Waters and Cassandra Rogers.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA130876-01A1; NS39475 and MH77306 (to A. J. K.); and R01 CA101828 (to T. E. S.). This work was also supported by the Novartis Institute of Biomedical Research (to N. S. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
J. Zhang, F. J. Adrián, W. Jahnke, S. W. Cowan-Jacob, A. G. Li, R. E. Iacob, T. Sim, J. Powers, C. Dierks, F. Sun, G. R. Guo, Q. Ding, B. Okram, Y. Choi, A. Wojciechowski, X. Deng, G. Liu, G. Fendrich, A. Strauss, N. Vajpai, S. Grzesiek, T. Tuntland, Y. Liu, B. Bursulaya, M. Azam, P. W. Manley, J. R. Engen, G. Q. Daley, M. Warmuth, N. S. Gray, submitted for publication.
- SH3
- Src homology 3
- SH2
- Src homology 2
- ELISA
- enzyme-linked immunosorbent assay
- BCR
- break point cluster region
- ER
- endoplasmic reticulum
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- DTT
- dithiothreitol
- PBS
- phosphate-buffered saline
- WT
- wild type.
REFERENCES
- 1.Goodey N. M., Benkovic S. J. (2008) Nat. Chem. Biol. 4, 474–482 [DOI] [PubMed] [Google Scholar]
- 2.Pendergast A. M. (2002) Adv. Cancer Res. 85, 51–100 [DOI] [PubMed] [Google Scholar]
- 3.Kruh G. D., Perego R., Miki T., Aaronson S. A. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5802–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen S. T., Van Etten R. A. (1997) Genes Dev. 11, 2456–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hantschel O., Superti-Furga G. (2004) Nat. Rev. Mol. Cell Biol. 5, 33–44 [DOI] [PubMed] [Google Scholar]
- 6.Woodring P. J., Hunter T., Wang J. Y. (2003) J. Cell Sci. 116, 2613–2626 [DOI] [PubMed] [Google Scholar]
- 7.Nagar B., Hantschel O., Young M. A., Scheffzek K., Veach D., Bornmann W., Clarkson B., Superti-Furga G., Kuriyan J. (2003) Cell 112, 859–871 [DOI] [PubMed] [Google Scholar]
- 8.Hantschel O., Nagar B., Guettler S., Kretzschmar J., Dorey K., Kuriyan J., Superti-Furga G. (2003) Cell 112, 845–857 [DOI] [PubMed] [Google Scholar]
- 9.Wang J. Y. (2004) Nat. Cell Biol. 6, 3–7 [DOI] [PubMed] [Google Scholar]
- 10.Weisberg E., Manley P. W., Cowan-Jacob S. W., Hochhaus A., Griffin J. D. (2007) Nat. Rev. Cancer 7, 345–356 [DOI] [PubMed] [Google Scholar]
- 11.Wong S., McLaughlin J., Cheng D., Zhang C., Shokat K. M., Witte O. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17456–17461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaman M. W., Herrgard S., Treiber D. K., Gallant P., Atteridge C. E., Campbell B. T., Chan K. W., Ciceri P., Davis M. I., Edeen P. T., Faraoni R., Floyd M., Hunt J. P., Lockhart D. J., Milanov Z. V., Morrison M. J., Pallares G., Patel H. K., Pritchard S., Wodicka L. M., Zarrinkar P. P. (2008) Nat. Biotechnol. 26, 127–132 [DOI] [PubMed] [Google Scholar]
- 13.Adrián F. J., Ding Q., Sim T., Velentza A., Sloan C., Liu Y., Zhang G., Hur W., Ding S., Manley P., Mestan J., Fabbro D., Gray N. S. (2006) Nat. Chem. Biol. 2, 95–102 [DOI] [PubMed] [Google Scholar]
- 14.Songyang Z., Carraway K. L., 3rd, Eck M. J., Harrison S. C., Feldman R. A., Mohammadi M., Schlessinger J., Hubbard S. R., Smith D. P., Eng C. (1995) Nature 373, 536–539 [DOI] [PubMed] [Google Scholar]
- 15.Barker S. C., Kassel D. B., Weigl D., Huang X., Luther M. A., Knight W. B. (1995) Biochemistry 34, 14843–14851 [DOI] [PubMed] [Google Scholar]
- 16.Seeliger M. A., Young M., Henderson M. N., Pellicena P., King D. S., Falick A. M., Kuriyan J. (2005) Protein Sci. 14, 3135–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foulkes J. G., Chow M., Gorka C., Frackelton A. R., Jr., Baltimore D. (1985) J. Biol. Chem. 260, 8070–8077 [PubMed] [Google Scholar]
- 18.Koleske A. J., Gifford A. M., Scott M. L., Nee M., Bronson R. T., Miczek K. A., Baltimore D. (1998) Neuron 21, 1259–1272 [DOI] [PubMed] [Google Scholar]
- 19.Kalies K. U., Rapoport T. A., Hartmann E. (1998) J. Cell Biol. 141, 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Görlich D., Rapoport T. A. (1993) Cell 75, 615–630 [DOI] [PubMed] [Google Scholar]
- 21.Miller A. L., Wang Y., Mooseker M. S., Koleske A. J. (2004) J. Cell Biol. 165, 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowan-Jacob S. W., Fendrich G., Manley P. W., Jahnke W., Fabbro D., Liebetanz J., Meyer T. (2005) Structure 13, 861–871 [DOI] [PubMed] [Google Scholar]
- 23.Diskin R., Engelberg D., Livnah O. (2008) J. Mol. Biol. 375, 70–79 [DOI] [PubMed] [Google Scholar]
- 24.Nagar B., Hantschel O., Seeliger M., Davies J. M., Weis W. I., Superti-Furga G., Kuriyan J. (2006) Mol. Cell 21, 787–798 [DOI] [PubMed] [Google Scholar]
- 25.Zhou V., Gao X., Han S., Brinker A., Caldwell J. S., Gu X. J. (2008) Anal. Biochem. 385, 300–308 [DOI] [PubMed] [Google Scholar]
- 26.Ito Y., Pandey P., Mishra N., Kumar S., Narula N., Kharbanda S., Saxena S., Kufe D. (2001) Mol. Cell Biol. 21, 6233–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J., Arlinghaus R. (2008) Oncogene 27, 4385–4391 [DOI] [PubMed] [Google Scholar]
- 28.Srinivasan D., Sims J. T., Plattner R. (2008) Oncogene 27, 1095–1105 [DOI] [PubMed] [Google Scholar]
- 29.Sirvent A., Boureux A., Simon V., Leroy C., Roche S. (2007) Oncogene 26, 7313–7323 [DOI] [PubMed] [Google Scholar]
- 30.Lin J., Sun T., Ji L., Deng W., Roth J., Minna J., Arlinghaus R. (2007) Oncogene 26, 6989–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodring P. J., Hunter T., Wang J. Y. (2001) J. Biol. Chem. 276, 27104–27110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.