Abstract
The epithelial sodium channel (ENaC) is probably a heterotrimer with three well characterized subunits (αβγ). In humans an additional δ-subunit (δ-hENaC) exists but little is known about its function. Using the Xenopus laevis oocyte expression system, we compared the functional properties of αβγ- and δβγ-hENaC and investigated whether δβγ-hENaC can be proteolytically activated. The amiloride-sensitive ENaC whole-cell current (ΔIami) was about 11-fold larger in oocytes expressing δβγ-hENaC than in oocytes expressing αβγ-hENaC. The 2-fold larger single-channel Na+ conductance of δβγ-hENaC cannot explain this difference. Using a chemiluminescence assay, we demonstrated that an increased channel surface expression is also not the cause. Thus, overall channel activity of δβγ-hENaC must be higher than that of αβγ-hENaC. Experiments exploiting the properties of the known βS520C mutant ENaC confirmed this conclusion. Moreover, chymotrypsin had a reduced stimulatory effect on δβγ-hENaC whole-cell currents compared with its effect on αβγ-hENaC whole-cell currents (2-fold versus 5-fold). This suggests that the cell surface pool of so-called near-silent channels that can be proteolytically activated is smaller for δβγ-hENaC than for αβγ-hENaC. Proteolytic activation of δβγ-hENaC was associated with the appearance of a δ-hENaC cleavage product at the cell surface. Finally, we demonstrated that a short inhibitory 13-mer peptide corresponding to a region of the extracellular loop of human α-ENaC inhibited ΔIami in oocytes expressing αβγ-hENaC but not in those expressing δβγ-hENaC. We conclude that the δ-subunit of ENaC alters proteolytic channel activation and enhances base-line channel activity.
The epithelial sodium channel (ENaC)2 is a member of the ENaC/degenerin family of nonvoltage-gated ion channels (1). It is localized in the apical membranes of sodium-absorbing epithelia like the aldosterone-sensitive distal nephron, respiratory epithelia, distal colon, and sweat and salivary ducts. In these epithelia ENaC is the rate-limiting step for sodium absorption and plays a critical role in the maintenance of body sodium balance (1–4). In addition, ENaC expression has been reported in a number of other tissues, including skin, endothelial cells, vascular smooth muscle cells, and neurons where its physiological role remains to be determined (5–12).
In epithelial tissues ENaC is believed to form a heteromeric channel composed of three homologous subunits α, β, and γ (13). Each subunit contains two transmembrane domains (M1 and M2), a large extracellular loop, and short intracellular N and C termini. With their M2 domains all subunits are thought to contribute to the channel pore (1). In the absence of a crystal structure for ENaC, its subunit stoichiometry remains a matter of debate. Nevertheless, the recently published crystal structure of the related acid-sensing ion channel ASIC1 suggests that ENaC is probably a heterotrimer (14–16).
In addition to the well characterized αβγ-subunits, a fourth ENaC subunit, δ-ENaC, has been cloned from a human kidney cDNA library with transcriptional expression in a range of tissues with highest expression levels in testis, ovary, pancreas, and brain. Small amounts of δ-ENaC-mRNA were also detected in heart, placenta, lung, liver, kidney, thymus, prostate, colon, and lymphocytes but not in small intestine and spleen (17, 18). So far little is known about the physiological role and the functional properties of this additional subunit. Genes corresponding to human δ-ENaC have been identified in chimpanzee, dog, chicken, and rabbit (19). Although it has previously been thought that δ-ENaC is absent in rat and mouse (20, 21), recent reports suggest that δ-ENaC is expressed in mouse sperm (22) and in mouse pleural tissue (23).
At the sequence level δ-ENaC is more closely related to α-ENaC (37% amino acid identity) and to the recently described ϵ-subunit of Xenopus laevis (24) than to the β- and γ-subunits of ENaC. The genes encoding δ-hENaC and α-hENaC are localized on human chromosomes 1p36.3-p36.2 (25) and 12p13 (26, 27), respectively. Thus, α-hENaC and δ-hENaC are mapped to different chromosomes, whereas β- and γ-hENaC are found within a common 400-kb fragment on chromosome 16p12 (28) and probably arise from gene duplication. Two splice variants of δ-ENaC have been described as follows: a shorter form with 638 amino acids (GI 34101282) originally cloned from a human kidney cDNA library (17) and a longer form with 704 amino acids (GI 21752051) originally cloned from human testis (29). In neuronal tissue the two isoforms have a cell-specific expression pattern (8). So far no functional differences have been observed between the two splice variants expressed in heterologous expression systems (10). In this study we used the shorter δ-ENaC isoform, which was the first one to be cloned (17).
In heterologous expression systems δ-ENaC has functional similarities with α-ENaC. Isolated expression of δ-hENaC in X. laevis oocytes results in small but significant amiloride-sensitive sodium currents (17). These currents are increased by a factor of about 50 when δ-hENaC is co-expressed together with β-hENaC and γ-hENaC. In contrast, co-expression of δβ-, δγ-, or αδ-subunits results in small amiloride-sensitive currents similar to those seen with the expression of δ-ENaC alone (17). These findings suggest that δ-ENaC preferentially assembles and functions as a δβγ-channel.
The biophysical properties of the δβγ-hENaC channel are different from those of the αβγ-channel (17). δβγ-ENaC is more than an order of magnitude less sensitive to amiloride than αβγ-ENaC for which the IC50 for amiloride inhibition is about 100 nm (17, 30–32). Additional pharmacological differences are the activating effect of capsazepine and icilin on δβγ-ENaC and its inhibition by Evans blue (21, 33, 34). Another difference is the higher single-channel Na+ conductance of δβγ-hENaC (∼12 pS) compared with αβγ-hENaC (∼5 pS) (17). Interestingly, both channels have a similar single-channel conductance for Li+ (∼7 pS). Thus, δβγ-hENaC is more permeable for Na+ than for Li+, whereas αβγ-hENaC has a higher permeability for Li+ than for Na+. Finally, δβγ-hENaC but not αβγ-hENaC has been reported to be activated by extracellular protons and may contribute to pH sensing (18, 35, 36).
There is recent evidence that proteases contribute to ENaC regulation by cleaving specific sites in the extracellular loops of the α- and γ-subunits but not the β-subunit (37–41). The channel is thought to be in its mature and active form in its cleaved state, but there is evidence for the presence of both cleaved and noncleaved channels in the plasma membrane (42). Cleavage may activate the channel by changing its conformation probably by releasing inhibitory peptides from the extracellular loops of α- and γ-ENaC (43–45). Cleavage of the γ-subunit seems to be particularly important for channel activation by extracellular proteases (39, 46). As far as we know it has not yet been shown whether the δ-subunit is also proteolytically processed and whether δβγ-ENaC can be proteolytically activated by exposing the channel to extracellular proteases.
In this study we investigated the functional properties of αβγ- and δβγ-hENaC expressed in X. laevis oocytes. Our starting point was the striking observation that the amiloride-sensitive whole-cell current (ΔIami) was significantly larger in oocytes expressing δβγ-hENaC compared with control oocytes expressing αβγ-hENaC. The aim of this study was to elucidate the underlying mechanisms by which the δ-subunit enhances ENaC activity. Furthermore, we tested the hypothesis that the δ-subunit alters proteolytic ENaC activation.
EXPERIMENTAL PROCEDURES
Plasmids
Full-length cDNAs for α-, β-, and γ-hENaC and for the short isoform of δ-hENaC (17) were kindly provided by H. Cuppens (Leuven, Belgium) and by R. Waldmann (Valbonne, France), respectively. They were subcloned into the pcDNA3.1 vector, and linearized plasmids were used as templates for cRNA synthesis (mMessage mMachine, Ambion, Austin, TX) using T7 as promotor. Site-directed mutagenesis extension overlap PCR was used to insert a FLAG tag in β-hENaC between Thr-137 and Arg-138, which corresponds to the site previously described for rat β-ENaC (47). The QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce a cysteine at the degenerin site of β-hENaC to create an MTSET-sensitive subunit βS520C (48). For the detection of α- and δ-hENaC by Western blot analysis, we generated an α- and δ-hENaC construct with an N-terminal HA tag (YPYDVPDYA) and a C-terminal V5 tag (GKPIPNPLLGLDST). Mutations were confirmed by sequence analysis (GATC Biotech, Konstanz, Germany).
Isolation of Oocytes and Injection of cRNA
Adult female X. laevis were anesthetized in 0.2% MS222 (Sigma), and oocytes were obtained by a partial ovariectomy. The oocytes were isolated from the ovarian lobes by enzymatic digestion at 19 °C on a rocking platform for 3–4 h with 600–700 units/ml type 2 collagenase from Clostridium histolyticum (CLS 2, Worthington) dissolved in calcium-free OR2 solution (in mm: NaCl 82.5, KCl 2, MgCl2 1, and HEPES 1, adjusted to pH 7.4 with Tris). Defolliculated stage V–VI oocytes were injected (Nanoject automatic injector, Drummond, Broomall, PA) with an equal amount of cRNA per ENaC subunit (injected amounts of cRNA per ENaC subunit per oocyte (ng per subunit) are given under “Results” or in the figure legends). The cRNAs were dissolved in RNase-free water, and the total volume injected into each oocyte was 46 nl. Injected oocytes were stored at 19 °C either in ND96 (high Na+) or in ND9 (low Na+). The latter solution contained (in mm) the following: NaCl 9, NMDG-Cl 87, KCl 2, CaCl2 1.8, MgCl2 1, HEPES 5 (adjusted to pH 7.4 with Tris). To prevent bacterial overgrowth, the solutions were supplemented with 100 units/ml sodium penicillin and 100 μg/ml streptomycin sulfate.
Two-electrode Voltage Clamp
Oocytes were routinely studied 2 days after injection using the two-electrode voltage clamp technique essentially as described previously (49–51). The oocytes were placed in a small experimental chamber and constantly superfused with ND96 (in mm: NaCl 96, KCl 2, CaCl2 1.8, MgCl2 1, HEPES 5, pH 7.4 with Tris) at a rate of 2–3 ml/min at room temperature. Voltage clamp experiments were performed using an OC-725C amplifier (Warner Instruments Corp., Hamden, CT) interfaced via an LIH-1600 (HEKA, Lambrecht, Germany) to a computer running PULSE software (HEKA) for data acquisition and analysis. For continuous whole-cell current recordings, oocytes were routinely clamped at a holding potential of −60 mV. Amiloride-sensitive whole-cell currents (ΔIami) were obtained by washing out amiloride (100 μm) with amiloride-free ND96 and subtracting the whole-cell currents measured in the presence of amiloride from the corresponding whole-cell currents recorded in the absence of amiloride.
Surface Labeling of Oocytes
Experiments were performed essentially as described (51, 52) using mouse monoclonal anti-FLAG M2 antibody (Sigma) as primary antibody and peroxidase-conjugated sheep anti-mouse IgG (Chemicon, Boronia Victoria, Australia) as secondary antibody. Individual oocytes were placed in a white U-bottom 96-well plate, and 50 μl of SuperSignal ELISA femto maximum sensitivity substrate (Pierce) were added to each oocyte. Chemiluminescence was quantified with a Tecan GENios microplate reader (TECAN, Crailsheim, Germany). Results are given in relative light units.
Preparation of Membrane-enriched Fractions from Oocyte Whole-cell Lysates
30 oocytes per group were homogenized with a 27-gauge needle in 1 ml of homogenization buffer (in mm: HEPES 10, NaCl 83, MgCl2 1, pH 7.9) supplemented with protease inhibitor mixture (“Complete Mini EDTA-free” protease inhibitor mixture tablets, Roche Diagnostics) and were sonicated three times for 5 s. After centrifugation at 1000 × g for 10 min, the supernatant was collected and ultracentrifuged at 100,000 × g for 1 h. The resulting pellet was resuspended in 150 μl of homogenization buffer, and equal amounts of protein (50 μg per lane in Fig. 5, A and B; 30 μg per lane in Fig. 5, C and D) were separated by SDS-PAGE.
FIGURE 5.
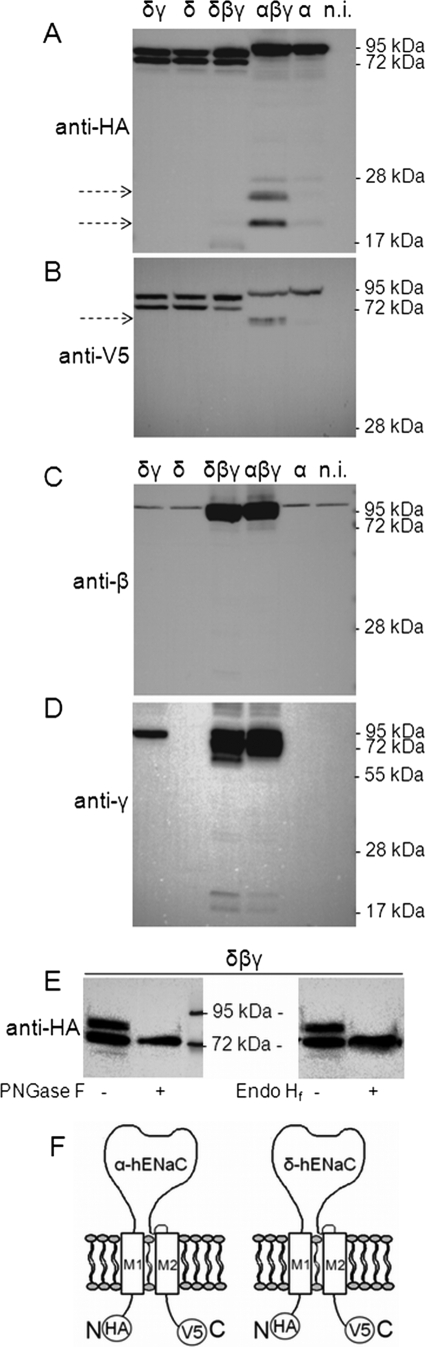
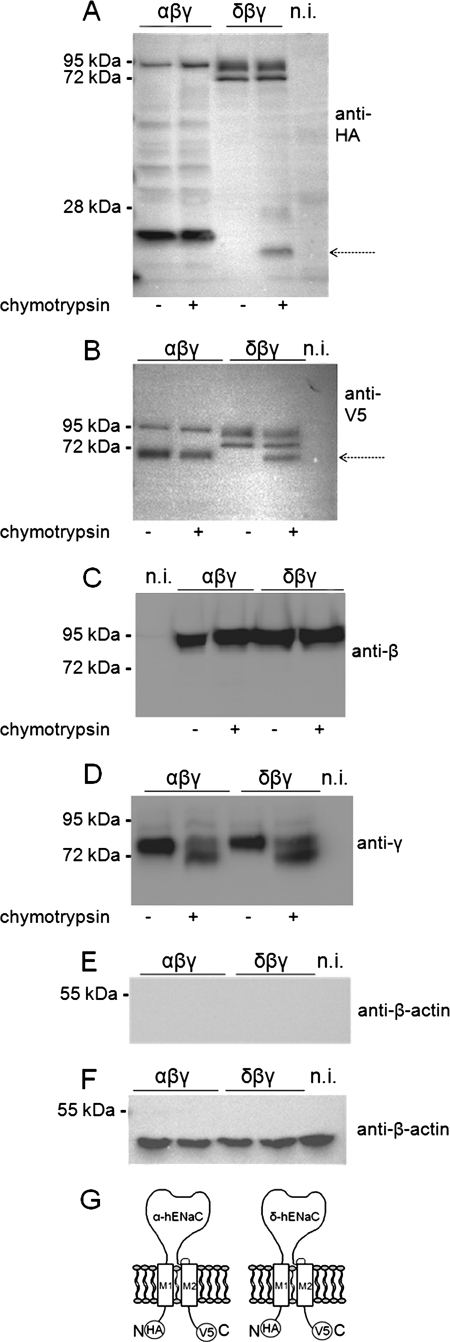
Co-expression of βγ-hENaC enhances proteolytic cleavage of the α-subunit but not of the δ-subunit. Experiments were performed in oocytes injected with cRNAs for δγ-, δ-, or α-hENaC (4 ng per subunit) or with cRNA for αβγ- or δβγ-hENaC (2 ng per subunit). N-terminally HA-tagged and C-terminally V5-tagged α- and δ-hENaC were co-expressed with wild-type β- and γ-hENaC. Western blot analysis of membrane-enriched fractions from oocyte whole-cell lysates was used to study the proteolytic processing of hENaC. Each Western blot represents one of at least five similar blots. Noninjected (n.i.) oocytes demonstrate the absence of a signal in oocytes that do not express ENaC. A and B, α- and δ-hENaC were detected by SDS-PAGE using a 10% gel and an anti-HA antibody (A) or an 8% gel and an anti-V5 antibody (B). The arrows indicate cleavage products. C and D, β- or γ-hENaC was detected by SDS-PAGE (10% gels) using specific antibodies against β- (C) or γ-hENaC (D). E, to investigate whether δ-hENaC is glycosylated, membrane-enriched fractions from oocytes expressing δβγ-hENaC were left untreated (−) or were treated (+) either with N-glycosidase F (PNGase F) or with endoglycosidase Hf (Endo Hf). δ-hENaC was detected by SDS-PAGE (10% gel) using an anti-HA antibody. F, schematics of α- and δ-hENaC showing the positions of the HA and V5 tag at the N/C terminus.
Detection of ENaC Cleavage Products at the Cell Surface
Biotinylation experiments were performed essentially as described previously (39) using 50 oocytes per group. All biotinylation steps were performed at 4 °C. In some experiments oocytes were preincubated for 5 min either in ND96 solution or in ND96 solution containing 2 μg/ml chymotrypsin. After washing the oocytes three times with ND96 solution, they were incubated in the biotinylation buffer (in mm: triethanolamine 10, NaCl 150, CaCl2 2, EZ-link sulfo-NHS-SS-Biotin (Pierce) 1 mg/ml, pH 9.5) for 15 min with gentle agitation. The biotinylation reaction was stopped by washing the oocytes twice for 5 min with quench buffer (in mm: glycine 192, Tris-Cl 25, pH 7.5). Afterward, the oocytes were lysed by passing them through a 27-gauge needle in lysis buffer (in mm: NaCl 500, EDTA 5, Tris-Cl 50, pH 7.4) supplemented with protease inhibitor mixture according to the manufacturer's instruction. The lysates were centrifuged for 10 min at 1,500 × g. Supernatants were transferred to 1.5-ml Eppendorf tubes and incubated with 0.5% Triton X-100 and 0.5% Igepal CA-630 (Sigma) for 20 min on ice. Biotinylated proteins were precipitated with 100 μl of Immunopure-immobilized Neutravidin beads (Pierce) washed with lysis buffer. After overnight incubation at 4 °C on a rotating wheel, the tubes were centrifuged for 3 min at 1,500 × g. Supernatants were removed, and beads were washed three times with lysis buffer. 100 μl of 2× SDS-PAGE sample buffer (Rotiload 1, Roth, Karlsruhe, Germany) was added to the beads.
Western Blot Analysis
After separating the proteins by SDS-PAGE, they were transferred to polyvinylidene difluoride membranes by semi-dry blotting and probed with the indicated antibodies. Chemiluminescent signals were detected using ECL Plus (GE Healthcare).
Antibodies
HA-/V5-tagged α- or δ-hENaC constructs were used to study the expression level of these subunits by Western blot analysis. Mouse monoclonal anti-V5 antibody (Invitrogen) and rabbit polyclonal anti-β-actin (Sigma) were used at a dilution of 1:5000; rat monoclonal anti-HA antibody (Roche Diagnostics) was used at a dilution of 1:1000. To detect the β- and γ-hENaC subunits in Western blot experiments, we used subunit-specific antibodies against human β- and γ-ENaC that were obtained by immunizing rabbits (Pineda Antibody Service, Berlin, Germany). The antibodies were tested for specificity as described in the supplement material. Horseradish peroxidase-labeled secondary sheep anti-mouse, goat anti-rat, and goat anti-rabbit antibodies were purchased from Sigma, Jackson ImmunoResearch (West Grove, PA), and Santa Cruz Biotechnology (Heidelberg, Germany), respectively, and used at a dilution of 1:10,000.
Solution and Chemicals
Amiloride hydrochloride that was added from an aqueous 10 mm stock solution and α-chymotrypsin TLCK-treated (1-chloro-3-tosylamido-7-amino-2-heptanone) type VII from bovine pancreas were purchased from Sigma. MTSET was obtained from Toronto Research Chemicals (Toronto, Canada). N-Glycosidase F (PNGase F) and endoglycosidase Hf were obtained from New England Biolabs (Ipswich, MA) to remove N-linked glycoproteins or the high mannose type N-glycans, respectively.
Peptide
The inhibitory 13-mer peptide (sequence LRGTLPHPLQRLR) was synthesized and purified by high performance liquid chromatography (purity >95%) by Coring System Diagnostix GmbH (Gernsheim, Germany). The peptide was modified by N-terminal acetylation and C-terminal amidation. It was dissolved in an aqueous stock solution at a concentration of 27 μm. Aliquots of this stock solution were kept at −80 °C and were added to the bath solution on the day of the experiment to give a final concentration of 2.7 μm.
Patch Clamp Experiments
Single-channel recordings in conventional outside-out patches were essentially performed as described previously using oocytes kept in low sodium ND9 solution after cRNA injection to prevent Na+ overloading (39, 53). Pipettes were filled with potassium gluconate pipette solution (in mm: potassium gluconate 90, NaCl 5, Mg-ATP 2, EGTA 2, and HEPES 10, pH 7.28 with Tris). Seals were routinely formed in a NMDG-Cl bath solution (in mm: NMDG-Cl 95, NaCl 1, KCl 4, MgCl2 1, CaCl2 1, HEPES 10, pH 7.4 with Tris). After seal formation the bath solution was changed to a NaCl bath solution in which the NMDG-Cl was replaced by 95 mm NaCl. Outside-out patches were routinely held at a holding pipette potential of −70 mV, which was close to the calculated reversal potential of Cl− (ECl = −77.2 mV) and K+ (EK = −79.4 mV) under our experimental conditions with experiments performed at room temperature. To obtain current-voltage (I-V) plots, the holding potential was varied from −100 to −10 mV in 30-mV increments. Downward current deflections correspond to cell membrane inward currents, i.e. movement of positive charge from the extracellular side to the cytoplasmic side. Single-channel current data were initially filtered at 500 Hz and sampled at 2 kHz. Single-channel current traces were re-filtered at 50 Hz to estimate the single-channel current amplitude and channel open probability (Po) using binned amplitude histograms. Liquid junction potentials occurring at the bridge/bath junction were measured using a 3 m KCl flowing boundary electrode. In NaCl bath solution the liquid junction potential averaged about 6 mV and was taken into account for data analysis.
Statistical Methods
Data are presented as mean ± S.E. and were analyzed using GraphPad Prism 4.01 for Windows (Graph Pad Software Inc., San Diego). Statistical significance was assessed by the appropriate version of Student's t test. N indicates the number of different batches of oocytes, and n indicates the number of individual oocytes studied.
RESULTS
ENaC Whole-cell Currents Are Larger in X. laevis Oocytes Expressing δβγ-hENaC Than in Those Expressing αβγ-hENaC
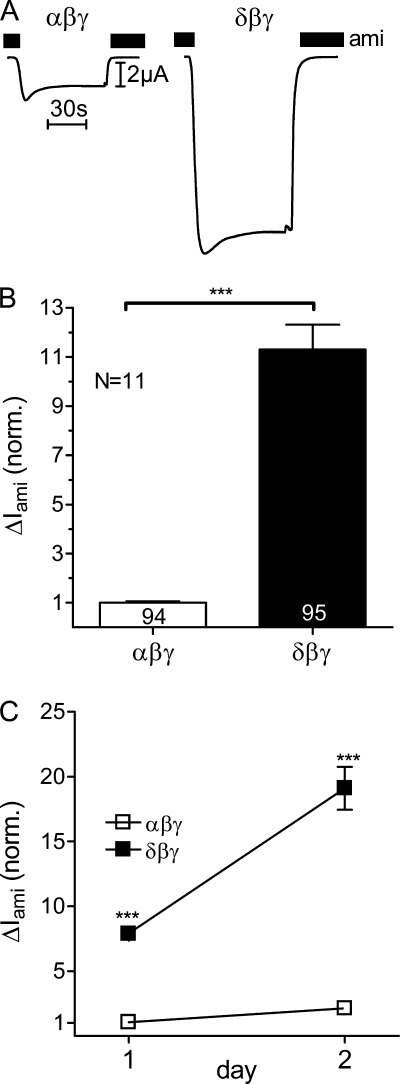
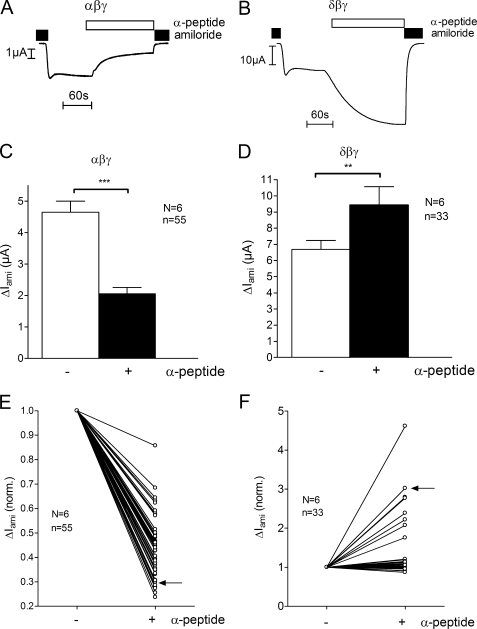
ENaC is thought to function as a heterotrimeric channel. It is likely that both the α-subunit and the δ-subunit can co-assemble with the other two subunits to form αβγ- or δβγ-ENaC. The resulting channels may have different functional properties. Therefore, we compared ENaC-mediated whole-cell currents in X. laevis oocytes heterologously expressing either αβγ- or δβγ-hENaC. Fig. 1A shows a representative whole-cell current trace from an αβγ-hENaC-expressing oocyte (left) and another trace from an oocyte expressing δβγ-hENaC (right). Recordings were started in the presence of amiloride in a concentration of 100 μm known to inhibit αβγ-ENaC and δβγ-ENaC almost completely (17, 32). At a holding potential of −60 mV washout of amiloride revealed a sizeable inward current component that corresponds to the ENaC-mediated sodium inward current. Re-addition of amiloride instantaneously returned the whole-cell current toward the initial base-line level. As illustrated by these traces and as summarized in Fig. 1B, the amiloride-sensitive whole-cell current (ΔIami) was on average about 11-fold larger in oocytes expressing δβγ-hENaC than in matched oocytes expressing αβγ-hENaC.
FIGURE 1.
ENaC whole-cell currents are larger in X. laevis oocytes expressing δβγ-hENaC than in those expressing αβγ-hENaC. Oocytes were injected with cRNAs coding for αβγ- or δβγ-hENaC (each 2 ng per subunit). Amiloride-sensitive whole-cell currents (ΔIami) were measured using the two-electrode voltage clamp technique. A, representative whole-cell current traces of an oocyte expressing αβγ-hENaC (left) or δβγ-hENaC (right). Amiloride (ami, 100 μm) was present in the bath solution during the time periods indicated by black bars. B, summary of similar experiments as shown in A performed in αβγ- and δβγ-hENaC-expressing oocytes. To pool data from 11 different batches of oocytes, individual ΔIami values were normalized to the mean ΔIami value of the corresponding αβγ-hENaC-expressing control group. Numbers inside the columns indicate the number of individual oocytes measured. N indicates number of different batches of oocytes. C, ΔIami of αβγ- and δβγ-hENaC-expressing oocytes detected 1 and 2 days after cRNA injection. Each data point represents the mean ΔIami measured in 8–10 individual oocytes of one batch. S.E. values are represented by vertical bars unless they are smaller than the symbols used. ***, p < 0.001, unpaired t test.
We routinely measured ΔIami 2 days after cRNA injection, because we know from previous studies that at this point ENaC expression is at a high level without compromising oocyte integrity. In addition, in one batch of oocytes we also measured ΔIami on day 1 after cRNA expression (Fig. 1C). As expected, in αβγ- and in δβγ-hENaC-expressing oocytes ΔIami was smaller on day 1 after cRNA injection than on day 2, which probably reflects the fact that with longer incubation periods more time is available for ENaC synthesis and channel delivery to the plasma membrane (54). Importantly, on both days ΔIami was significantly larger in oocytes expressing δβγ-hENaC than in oocytes expressing αβγ-hENaC. This confirms that the stimulatory effect of the δ-subunit on ENaC currents is a robust phenomenon. The stimulatory effect of δ-hENaC was also preserved when tagged α- and δ-hENaC constructs were used with N-terminal HA and C-terminal V5 epitopes (data not shown).
Replacing Extracellular Na+ by Li+ Increases ΔIami in αβγ-hENaC-expressing Oocytes but Decreases ΔIami in δβγ-hENaC-expressing Oocytes
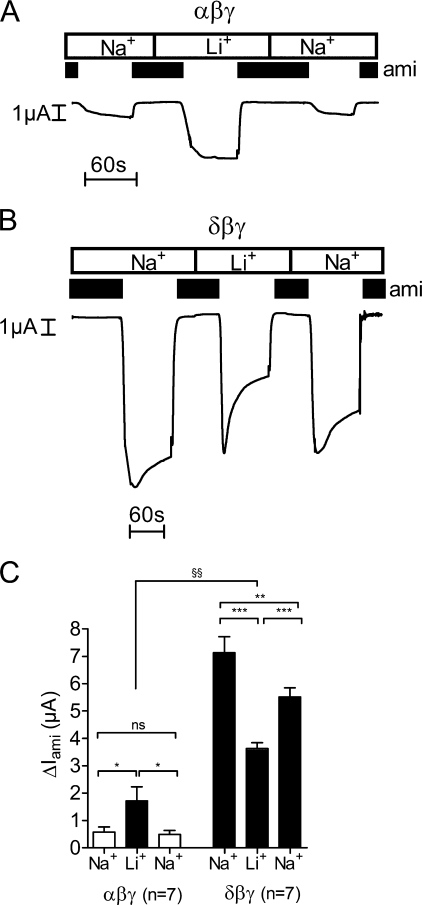
The single-channel Na+ conductance of δβγ-hENaC is known to be 2-fold larger (∼12 pS) than that of αβγ-hENaC (∼5 pS) (17). In a bath solution containing Na+ as predominant cation, this difference is likely to contribute to the phenomenon that whole-cell currents in δβγ-hENaC-expressing oocytes are larger than those in αβγ-hENaC-expressing oocytes. In contrast, for Li+ the single-channel conductance is ∼7 pS for both αβγ- and δβγ-hENaC (17). Thus, replacing extracellular Na+ by Li+ should lead to a single-channel conductance that is essentially the same in αβγ- and δβγ-hENaC-expressing oocytes. In experiments as illustrated in Fig. 2, Na+ in the bath solution was completely replaced by Li+, and ΔIami was determined in the presence of Na+ and in the presence of Li+ by washout and re-application of amiloride. As expected, changing from Na+ to Li+ in the bath solution increased ΔIami in αβγ-hENaC-expressing oocytes (Fig. 2A) and decreased it in oocytes expressing δβγ-hENaC (Fig. 2B). To test the reversibility of the effect of Li+ on ΔIami, the bath solution was subsequently switched back to a Na+-containing bath solution, and ΔIami was determined again. The slightly lower whole-cell Na+ currents in δβγ-hENaC-expressing oocytes after washout of Li+ are most likely caused by spontaneous channel “rundown,” which is a commonly observed phenomenon that is not yet well understood (55). It is usually more pronounced in oocytes expressing large currents, which may explain why it is more prominent in δβγ-hENaC-expressing oocytes than in αβγ-hENaC-expressing oocytes. Interestingly, upon changing from Na+ to Li+, the observed ∼3-fold increase of ΔIami in αβγ-hENaC-expressing oocytes was larger than predicted from an increase of the single-channel conductance from 5 to 7 pS (Fig. 2C). Thus, replacing Na+ by Li+ in the extracellular bath solution is likely to increase channel Po of αβγ-hENaC in addition to increasing its single-channel conductance. In contrast, the decrease of ΔIami in δβγ-hENaC-expressing oocytes by about 50% was close to the value predicted from the reduction of the single-channel conductance from 12 to 7 pS upon changing from Na+ to Li+ (Fig. 2C). In the presence of Li+ the peak inward current increase observed in δβγ-hENaC-expressing oocytes after washout of amiloride was usually followed by a steeper current decline than that observed in the presence of Na+ (Fig. 2B). This suggests that exposure to Li+ affects the gating of δβγ-hENaC in a complex and time-dependent manner.
FIGURE 2.
Replacing extracellular Na+ by Li+ increases ΔIami in αβγ-hENaC-expressing oocytes but decreases ΔIami in δβγ-hENaC-expressing oocytes. A and B, representative whole-cell current traces of an oocyte expressing αβγ- (A) or δβγ-hENaC (B) (each 1 ng per subunit). Na+ in the bath solution was temporarily replaced by Li+. Amiloride (ami, 100 μm) was applied as indicated by the black bars. C, mean ΔIami values from seven similar whole-cell current recordings as shown in A and B. n indicates number of individual oocytes measured from one batch. ns is not significant. *, p < 0.05; **, p < 0.01; ***, p < 0.001, paired t test, and §§, p < 0.01, unpaired t test.
Importantly, even in the presence of Li+ the average steady state ΔIami was significantly larger in δβγ-hENaC-expressing oocytes (3.64 ± 0.21 μA) than that in αβγ-hENaC-expressing oocytes (1.71 ± 0.52 μA, p < 0.01). This suggests that the channel Po or channel surface expression of δβγ-hENaC must be larger than that of αβγ-hENaC.
Channel Surface Expression Is Similar in αβγ- and in δβγ-hENaC-expressing Oocytes
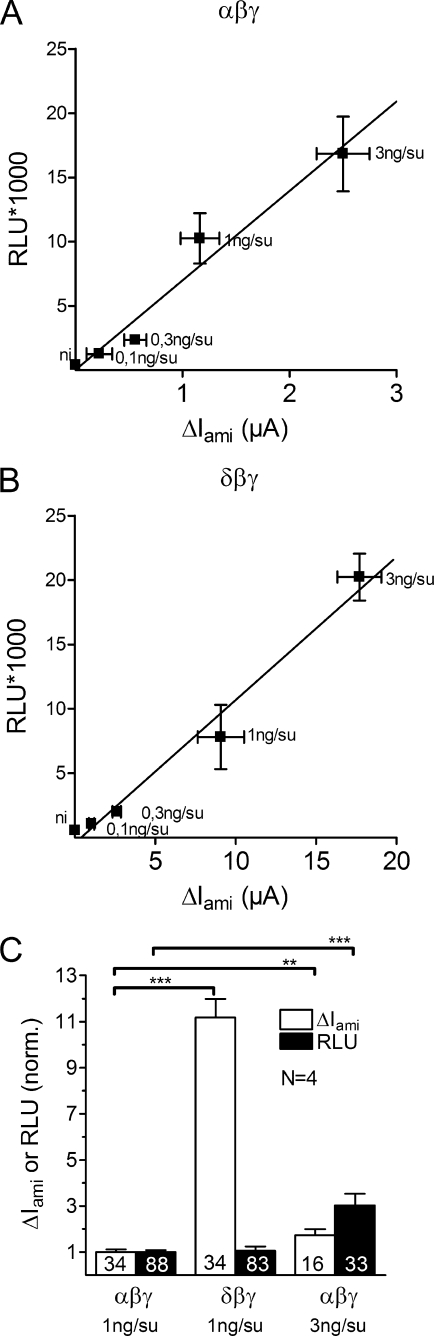
To investigate channel surface expression, we used a chemiluminescence assay (51, 52, 56). For this purpose we generated a β-hENaC construct with a FLAG reporter epitope inserted in its extracellular loop at the homologous site as reported previously for rat β-ENaC. At this site the FLAG epitope does not seem to interfere with normal channel function (47). We chose to tag the β-subunit because this subunit is not known to be proteolytically cleaved (41). To test the FLAG-tagged β-hENaC and to validate our chemiluminescence assay, we analyzed the surface expression of αβγ-hENaC-expressing oocytes using different amounts of injected cRNA (0.1, 0.3, 1, and 3 ng per subunit) (Fig. 3A). In an independent experiment we investigated the surface expression of δβγ-hENaC-expressing oocytes using the same amounts of injected cRNA (Fig. 3B). These experiments demonstrated that in αβγ- and in δβγ-hENaC-expressing oocytes a nearly linear correlation exists between ΔIami and the detected chemiluminescence signal reflecting channel surface expression.
FIGURE 3.
Channel surface expression is similar in αβγ- and in δβγ-hENaC-expressing oocytes. Channel surface expression was detected by insertion of a FLAG reporter epitope in the extracellular loop of the β-subunit and a chemiluminescence assay. A and B, relationship between channel surface expression expressed in relative light units (RLU) and ΔIami values measured in parallel in groups of αβγ- (A) and δβγ-hENaC (B)-expressing oocytes injected with different amounts of cRNA (0.1, 0.3, 1, and 3 ng per subunit). Noninjected (ni) oocytes served as negative controls and showed negligible background luminescence. Each square represents the mean of 22–24 oocytes of one batch for surface expression and 7–10 oocytes of the same batch for ΔIami. C, in four different batches of oocytes surface expression and ΔIami values were obtained in parallel in αβγ- and δβγ-hENaC-expressing oocytes. Data were normalized to the corresponding αβγ-hENaC-expressing control group. Oocytes injected with triple amount of cRNA (3 ng per subunit) served as control. Numbers inside the columns indicate the number of individual oocytes measured. N indicates number of different batches of oocytes. **, p < 0.01,***, p < 0.001, unpaired t test.
In the experiments summarized in Fig. 3C we compare surface expression and ΔIami measured in parallel in αβγ- and δβγ-hENaC-expressing oocytes. Normalized data obtained in four different batches of oocytes are shown. Although ΔIami was much larger in δβγ-hENaC-expressing oocytes compared with ΔIami in oocytes expressing αβγ-hENaC, the chemiluminescence signals were not significantly different. Thus, under the assumption that the antibody binding to the FLAG epitope of the β-subunit is similar in δβγ- and αβγ-expressing oocytes, channel surface expression is similar in the two groups of oocytes. To confirm that the chemiluminescence assay can reliably detect an increase in channel surface expression under our experimental conditions, we performed control experiments in which the amount of αβγ-cRNA injected per oocyte was increased from 1 ng per subunit to 3 ng per subunit. As expected, this significantly increased both ΔIami and the chemiluminescence signal (Fig. 3C). The finding that the relative increase in chemiluminescence was larger than the increase in ENaC whole-cell currents demonstrates that our assay is very sensitive to detect an increase in channel surface expression. Thus, our experiments clearly indicate that an increase in cell-surface expression is not the cause for the increased ΔIami in oocytes expressing δβγ-hENaC.
δβγ-hENaC Has a Higher Average Open Probability than αβγ-hENaC
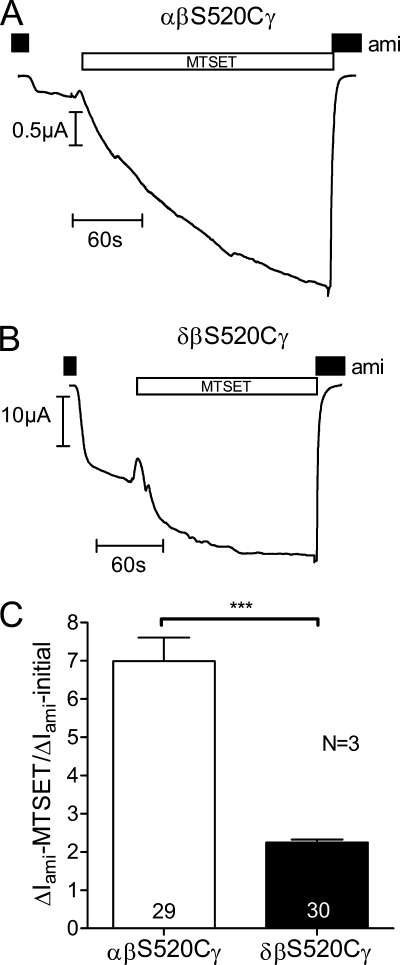
To investigate whether the average Po of δβγ-hENaC is increased, we used the S520C mutant of β-hENaC (βS520C). A channel with this mutant subunit is thought to be converted to a channel with a Po of nearly 1 by exposure to the positively charged sulfhydryl reagent MTSET (32). The chemical modification at this site destabilizes the closed state of the channel (48, 57). As illustrated by two representative current traces, application of MTSET to oocytes expressing αβS520Cγ-hENaC (Fig. 4A) or δβS520Cγ-hENaC (Fig. 4B) caused a substantial increase in the amiloride-sensitive current in both αβS520Cγ- and δβS520Cγ-hENaC-expressing oocytes. At the beginning of the experiment the bath solution contained 100 μm amiloride. Washout of amiloride revealed the presence of an amiloride-sensitive inward current component (ΔIami). Application of MTSET caused an inward current increase that approached a plateau after several minutes. Re-exposure to amiloride demonstrated that the observed current increase upon MTSET application was caused by an increase in the amiloride-sensitive current component, i.e. reflects a stimulation of ENaC. Importantly, the relative stimulatory effect of MTSET was larger in αβS520Cγ-hENaC-expressing oocytes than in those expressing δβS520Cγ (Fig. 4C). On average, in oocytes expressing δβS520Cγ-hENaC application of MTSET increased ENaC currents by a factor of about 2 consistent with an increase of the average Po from about 0.5 to 1 (Fig. 4C). In contrast, in αβS520Cγ-hENaC-expressing oocytes MTSET increased ΔIami about 7-fold. Under the assumption that MTSET increased Po essentially to 1, this indicates that the average Po prior to the application of MTSET must have been about 0.14. These data indicate that the average base-line open probability of δβS520Cγ-hENaC is 3–4-fold higher than that of αβS520Cγ-hENaC. This can explain at least in part the larger amiloride-sensitive whole-cell current observed in oocytes expressing δβγ-hENaC.
FIGURE 4.
δβγ-hENaC has a higher average Po than αβγ-hENaC. Oocytes were injected with cRNAs coding for αβS520Cγ- or δβS520Cγ-hENaC (each 1 ng per subunit). To increase the Po of these mutant channels close to 1, the sulfhydryl reagent MTSET (1 mm) was added to the bath solution. A and B, representative whole-cell current traces of an oocyte expressing αβS520Cγ-hENaC (A) or δβS520Cγ-hENaC (B). The small artifacts seen in the traces were caused by voltage step protocols. The resulting current responses were omitted from the traces for clarity. C, average ratios of ΔIami measured after MTSET to the initial ΔIami (ΔIami − MTSET/ΔIami − initial). Numbers inside the columns indicate the number of individual oocytes measured. N indicates number of different batches of oocytes. ***, p < 0.001, unpaired t test.
Co-expression of βγ-hENaC Enhances Proteolytic Cleavage of the α-Subunit but Not the δ-Subunit
It is well established that proteolytic processing along the biosynthetic pathway is important for ENaC maturation and activation (41). Thus, the increased average open probability of δβγ-hENaC may reflect a difference in proteolytic channel processing. In a recent study it has been demonstrated that proteolytic processing of rat α-ENaC by endogenous proteases requires co-expression of the β- and γ-subunits (58). Therefore, we performed experiments to compare the effects of βγ-hENaC co-expression on the proteolytic processing of α- and δ-hENaC. For this purpose we prepared membrane-enriched fractions from oocyte whole-cell lysates. To allow detection of both N- and C-terminal fragments, we used α- and δ-hENaC constructs with an N-terminal HA tag and a C-terminal V5 tag (Fig. 5F).
In α-hENaC- and in αβγ-hENaC-expressing oocytes, a band corresponding to the expected size of full-length α-hENaC was detected at about 95 kDa. As expected, co-expression with βγ-hENaC resulted in the appearance of cleaved fragments of the α-subunit that are absent when α-hENaC is expressed alone (Fig. 5, A and B). The main cleavage products detected with the HA and the V5 antibody had a size of about 25 and 68 kDa, respectively (Fig. 5, A and B). These fragments correspond well to cleavage products previously reported for rat α-ENaC co-expressed with βγ-rENaC (58). They are likely to reflect cleavage of the α-subunit at one of its putative furin cleavage sites (41). Interestingly, the antibody directed against the N-terminal HA tag of α-hENaC revealed another band of about 20 kDa, which may reflect additional cleavage of α-hENaC at a site closer to the N terminus. To our knowledge this small N-terminal fragment has not been described for rat or mouse α-ENaC (41). If both the 25- and 20-kDa N-terminal fragment resulted from cleavage of full-length α-hENaC, a second C-terminal fragment should be detectable in addition to the main 68-kDa C-terminal fragment. However, this was not the case. Therefore, the small 20-kDa N-terminal fragment probably arises from additional cleavage of the 25-kDa N-terminal fragment rather than from cleavage of full-length α-hENaC.
In oocytes expressing δ-hENaC alone or in combination with γ-hENaC or with βγ-hENaC, we detected two prominent bands at about 86 and 75 kDa (Fig. 5, A and B). Because similar size bands were detected with both the V5 and the HA antibody, they are unlikely to reflect cleavage products but probably reflect glycosylated and nonglycosylated forms of δ-hENaC. Indeed, in additional experiments using N-glycosidase F or endoglycosidase Hf, we confirmed that the larger size band represents a glycosylated form of δ-hENaC (Fig. 5E). Importantly, unlike the cleavage induced by the co-expression of α-hENaC with βγ-hENaC, we did not observe cleavage of δ-hENaC by co-expressing it with γ-hENaC or with βγ-hENaC (Fig. 5, A and B). These findings indicate that proteolytic processing of the δ-subunit differs from that of the α-subunit. Our Western blot data also demonstrate that similar overall amounts of δ-hENaC and α-hENaC protein were synthesized in the oocytes. Thus, differences in protein expression levels are unlikely to explain the differences in ENaC currents observed in αβγ- and in δβγ-hENaC-expressing oocytes.
No β-hENaC cleavage products were observed in oocytes expressing αβγ-hENaC or δβγ-hENaC (Fig. 5C). This is consistent with the concept that the β-subunit of ENaC is not proteolytically processed (41). For rat γ-ENaC it has previously been shown that in addition to the full-length 87-kDa band a 76-kDa cleavage product can be detected in membrane-enriched fractions of oocytes expressing γ-ENaC alone or in combination with α- and/or β-ENaC (58). Consistent with this we detected a broad double band probably representing full-length γ-hENaC with a predicted size of about 95-kDa and an ∼74-kDa γ-hENaC cleavage product in both αβγ-hENaC- and δβγ-hENaC-expressing oocytes. Interestingly, in δβγ-hENaC-expressing oocytes we observed an additional γ-hENaC cleavage product with a molecular mass of about 60 kDa (Fig. 5D). A γ-ENaC cleavage product of similar size has previously been reported to occur at the plasma membrane upon activation of ENaC by extracellular proteases (39, 45, 59, 60). Thus, δ-hENaC may promote cleavage of the γ-subunit in the presence of the β-subunit. This may contribute to the increased activity of δβγ-hENaC compared with that of αβγ-hENaC. No cleavage of the γ-subunit was observed in oocytes co-expressing δ- and γ-hENaC without the β-subunit. In contrast, cleaved γ-ENaC can be detected in intracellular membranes of oocytes co-expressing the α- and γ-subunits of rat ENaC (58). The two faint low molecular bands running above and below the 17-kDa marker in Fig. 5D (3rd and 4th lanes) were detected in all five experiments performed. In two out of five experiments these low molecular bands were slightly stronger in the δβγ-hENaC lane than in the αβγ-hENaC lane. These bands may represent additional proteolytic processing of the γ-subunit or nonspecific protein degradation. Essentially no γ-hENaC cleavage was observed in oocytes co-expressing δγ-hENaC without the β-subunit. Moreover, the signal for full-length γ-hENaC was somewhat reduced in δγ-hENaC-expressing oocytes compared with δβγ-expressing oocytes. This suggests that protein expression and proteolytic processing of γ-hENaC are less efficient in δγ-hENaC-expressing oocytes than in δβγ-hENaC-expressing oocytes.
Chymotrypsin Can Activate δβγ-hENaC, but the Stimulatory Effect Is Reduced Compared with That on αβγ-hENaC
Application of extracellular proteases has been shown to increase average ENaC open probability by a dual effect, i.e. by further activation of channels that are already active in the plasma membrane and by the recruitment of a population of so-called near-silent channels (39, 61, 62). It has not yet been shown whether δβγ-hENaC can be activated by extracellular proteases. Our chemiluminescence data indicate that replacing the α-subunit by the δ-subunit increases ENaC whole-cell currents without affecting channel surface expression. Therefore, the pool of near-silent channels may be smaller in δβγ-hENaC-expressing oocytes than in αβγ-hENaC-expressing oocytes. In this case the relative responsiveness of δβγ-hENaC to extracellular proteases, if present, should be reduced compared with that of αβγ-hENaC. To test this, we used chymotrypsin as a prototypical serine protease known to have a robust stimulatory effect on rat αβγ-ENaC expressed in oocytes (39, 63). Moreover, unlike trypsin, chymotrypsin does not cause a transient stimulation of a calcium-activated chloride conductance in the oocyte expression system (39, 63, 64).
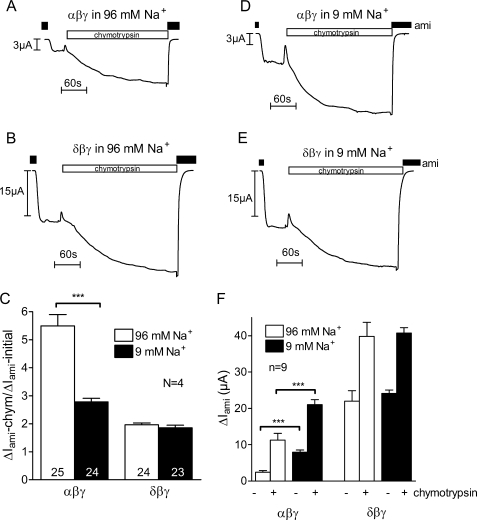
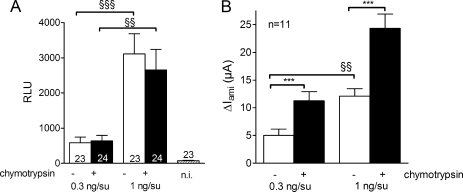
In Fig. 6, A and B, representative current traces are shown that confirm the stimulatory effect of chymotrypsin on αβγ-hENaC and demonstrate for the first time that chymotrypsin can also activate δβγ-hENaC. As expected, the relative stimulatory effect of chymotrypsin on ΔIami was significantly smaller in δβγ-hENaC-expressing oocytes than in αβγ-hENaC-expressing oocytes when they were maintained in a usual high Na+ (96 mm) bath solution after cRNA injection. Under these conditions application of chymotrypsin increased ΔIami on average more than 5-fold in αβγ-hENaC-expressing oocytes but only about 2-fold in δβγ-hENaC-expressing oocytes (Fig. 6C).
FIGURE 6.
Chymotrypsin can activate δβγ-hENaC, but the stimulatory effect is reduced compared with that on αβγ-hENaC. Oocytes were injected with cRNAs coding for αβγ- or δβγ-hENaC (each 1 ng per subunit) and incubated in 96 mm Na+ (open columns) or 9 mm Na+ solution (filled columns). A, B, D, and E, representative whole-cell current traces from αβγ- or δβγ-hENaC-expressing oocytes maintained in 96 mm Na+ solution (A and B) or in 9 mm Na+ solution (D and E) for 2 days after cRNA injection. During the recordings oocytes were superfused with 96 mm Na+ solution. The application of chymotrypsin (2 μg/ml) is indicated by the open bars. C, average ratios of ΔIami measured after chymotrypsin to the initial ΔIami (ΔIami − chymotrypsin/ΔIami − initial). Numbers inside the columns indicate the number of individual oocytes measured. N indicates number of different batches of oocytes. F, average ΔIami values measured before (−) or after (+) exposure to chymotrypsin in one representative batch of oocytes; n indicates number of individual oocytes measured. ***, p < 0.001, unpaired t test.
Reduced Na+ Feedback Inhibition May Contribute to the Increased Average Open Probability of δβγ-hENaC
When oocytes expressing ENaC are incubated in a bath solution containing a normal (i.e. high) extracellular Na+ concentration, they become severely sodium-loaded (13). In oocytes expressing αβγ-ENaC, an increase in intracellular Na+ is known to cause feedback inhibition of the channel by enhancing its retrieval from the plasma membrane (50, 54, 65). Moreover, it has been demonstrated that intracellular Na+ also inhibits αβγ-ENaC through an inhibitory effect on channel open probability (66). In the oocyte expression system it is well established that Na+ loading and hence feedback inhibition of αβγ-ENaC can be reduced by incubating oocytes in a low Na+ solution after cRNA injection (50, 54, 67). We hypothesized that in oocytes maintained in low Na+ (9 mm) the relative stimulatory effect of chymotrypsin on αβγ-hENaC should be reduced because the open probability of αβγ-hENaC should be increased under these conditions (66). As illustrated by the representative current trace in Fig. 6D and as summarized in Fig. 6C, this was indeed the case. As expected, preincubation in low Na+ significantly increased base-line ΔIami in αβγ-hENaC-expressing oocytes (Fig. 6F) and significantly reduced the relative stimulatory effect of chymotrypsin. On average chymotrypsin increased ΔIami by a factor of about 2.5 in αβγ-hENaC-expressing oocytes maintained in low Na+ compared with a factor of more than 5 in oocytes maintained in high Na+ (Fig. 6C). We also investigated the stimulatory effect of chymotrypsin on ΔIami of δβγ-hENaC-expressing oocytes incubated in low Na+ after cRNA injection (Fig. 6E). Interestingly, base-line ΔIami was not significantly increased in these oocytes (Fig. 6F). Moreover, the average 2-fold stimulatory effect of chymotrypsin on ΔIami was similar to that observed in oocytes maintained in high Na+ after cRNA injection (Fig. 6C). Collectively, these data indicate that δβγ-hENaC can be activated by extracellular proteases but that the stimulatory effect is reduced compared with that on αβγ-hENaC. A likely explanation for the reduced stimulatory effect of chymotrypsin is the increased base-line open probability of δβγ-hENaC compared with that of αβγ-hENaC. Moreover, our data suggest that one reason for the increased open probability of δβγ-hENaC is its reduced sensitivity to feedback inhibition by intracellular sodium.
δβγ-hENaC Stimulation by Chymotrypsin Is Associated with the Appearance of a Cleavage Fragment of δ-hENaC
It is not known whether cleavage of δ-hENaC contributes to channel activation by extracellular proteases. Therefore, we investigated whether δβγ-hENaC activation is associated with the appearance of δ-hENaC cleavage products at the cell surface. We used α- and δ-hENaC constructs with an N-terminal HA tag and a C-terminal V5 tag to detect biotinylated cell surface α- and δ-hENaC fragments by Western blot analysis (Fig. 7G). Western blots were re-probed for β-actin to confirm that the biotinylated proteins were not contaminated with intracellular proteins (Fig. 7, E and F). Exposure of αβγ-hENaC-expressing oocytes to chymotrypsin (2 μg/ml) for 5 min, a time sufficient for maximal ENaC current activation, did not alter the constitutive appearance of an ∼95-kDa full-length α-hENaC band together with an N-terminal cleavage fragment (∼25 kDa) and a corresponding C-terminal (∼68 kDa) cleavage fragment (Fig. 7, A and B, 1st and 2nd lanes). As shown previously for rat ENaC (58, 59) the N-terminal and C-terminal fragments correspond to typical cleavage products that arise from α-ENaC cleavage at its putative furin cleavage site when α-ENaC is co-expressed with the β- and γ-subunit. In contrast, in δβγ-hENaC-expressing oocytes, channel activation by 5 min of incubation with chymotrypsin was associated with the appearance of an N-terminal cleavage fragment with a molecular mass of about 20 kDa (Fig. 7A, 4th lane) that was not present in nontreated control oocytes expressing δβγ-hENaC (Fig. 7A, 3rd lane). A corresponding C-terminal cleavage fragment that migrates at about 65 kDa was readily detected by the anti-V5 antibody (Fig. 7B, 4th lane). Detection of biotinylated wild-type β- and γ-hENaC confirmed the previously reported finding that application of an extracellular protease causes cleavage of the γ-subunit without cleavage of the β-subunit (39, 45, 59). Moreover, our results demonstrate that replacing the α-subunit by the δ-subunit does not alter the cleavage of β- and γ-hENaC in response to chymotrypsin (Fig. 7, C and D).
FIGURE 7.
δβγ-hENaC stimulation by chymotrypsin is associated with the appearance of a cleavage fragment of δ-hENaC. α- and δ-hENaC constructs with an N-terminal HA tag and C-terminal V5 tag were used and co-expressed with wild-type β- and γ-hENaC. Oocytes were treated (+) for 5 min with chymotrypsin (2 μg/ml) or were left untreated (−). Biotinylated cell surface protein was analyzed by SDS-PAGE and Western blot using 10% gels (A and B) or 8% gels (C and D). Each blot represents one of at least seven similar blots. A–D, α-, β-, γ- and δ-hENaC were detected with anti-HA- (A), anti-β- (C), anti-γ (D), or anti-V5 antibody (B). Noninjected (n.i.) oocytes demonstrate the absence of a signal in oocytes that do not express ENaC. The arrows indicate cleavage products. E and F, to confirm that the biotinylated material was not contaminated by intracellular proteins, blots were routinely re-probed with an antibody against β-actin. One representative blot is shown for biotinylated proteins (E) and one for intracellular proteins (F); similar results were obtained for all the other blots. G, Schematics of α- and δ-hENaC showing the positions of the HA and V5 tag at the N/C terminus.
Channels with δ-hENaC Require the Presence of γ-hENaC to Be Activated by Chymotrypsin
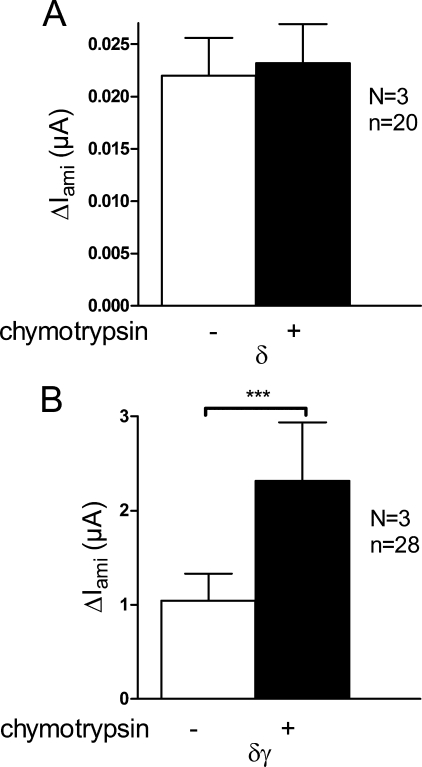
It has previously been shown that the γ-subunit is particularly important for proteolytic channel activation (39, 46, 59). Indeed, extracellular application of trypsin activates ΔIami in oocytes expressing αγ- or αβγ-ENaC but not in oocytes expressing αβ-ENaC or α-ENaC alone (39). Therefore, we tested whether γ-hENaC is required for chymotrypsin-mediated stimulation of ΔIami in δ-hENaC-expressing oocytes. As shown in Fig. 8A, we detected small but significant ENaC currents in oocytes expressing δ-hENaC alone, which is consistent with previously reported data and suggests that the δ-subunit can form functional homomeric channels (17). However, chymotrypsin failed to stimulate ΔIami in oocytes expressing δ-hENaC alone (Fig. 8A). Similarly, we did not detect a stimulatory effect of chymotrypsin on ΔIami in δβ-hENaC-expressing oocytes (data not shown). However, chymotrypsin had a significant stimulatory effect on ΔIami in oocytes co-expressing δ- and γ-hENaC (Fig. 8B). On average the stimulatory effect was about 2-fold in δγ-hENaC-expressing oocytes that is similar to that in δβγ-hENaC-expressing oocytes. These findings demonstrate that the δ-subunit alone is not sufficient to form channels that can be activated by extracellular proteases. Both the α- and the δ-subunit require co-expression of the γ-subunit to give rise to channels that can be activated by extracellular proteases.
FIGURE 8.
Channels with δ-hENaC require the presence of γ-hENaC to be activated by chymotrypsin. A and B, experiments were performed in oocytes injected with cRNAs for δ-hENaC alone or δγ-hENaC (5 ng per subunit). Average ΔIami values are shown for δ- and δγ-hENaC-expressing oocytes before (−) and after (+) exposure to chymotrypsin (2 μg/ml). N indicates number of different batches of oocytes; n indicates number of individual oocytes measured. ***, p < 0.001, paired t test.
Effect of the Synthetic Peptide α-13 on αβγ- or δβγ-hENaC
As reported previously (43), a synthetic 8-mer peptide (Leu211–Leu218) in the extracellular loop of the α-subunit of mouse ENaC reversibly inhibits αβγ-mENaC expressed in X. laevis oocytes. So far the mechanism of this inhibitory effect is unclear. In particular, it is not known whether the inhibitory effect requires the presence of the α-subunit. Therefore, we compared the effect of a homologous human peptide on αβγ- and on δβγ-hENaC currents. We used a synthetic 13-mer peptide (α-13) corresponding to amino acid residues Leu180 to Arg192 in the extracellular loop of α-hENaC. Individual whole-cell current traces from an αβγ-hENaC-expressing oocyte and from an oocyte expressing δβγ-hENaC are shown in Fig. 9, A and B, respectively. Recordings were started in the presence of amiloride, and washout of amiloride revealed the ENaC-mediated current component. Subsequent application of the synthetic peptide α-13 (2.7 μm) led to a decrease of the current in the αβγ-hENaC-expressing oocyte. This confirmed the inhibitory effect of the α-13 peptide on αβγ-hENaC. However, no inhibition but even a stimulation was observed in the δβγ-hENaC-expressing oocyte. Re-addition of amiloride returned the whole-cell current toward the initial base-line level. As summarized in Fig. 9C the peptide inhibited ΔIami on average by about 59% in αβγ-hENaC-expressing oocytes (p < 0.001). In contrast, in δβγ-hENaC-expressing oocytes (Fig. 9D) application of the 13-mer peptide led to an average stimulation of ΔIami by about 40% (p < 0.01).
FIGURE 9.
Effect of a 13-mer synthetic peptide on αβγ- or δβγ-hENaC. Oocytes were injected with cRNAs coding for αβγ- or δβγ-hENaC (each 2 ng per subunit) and were maintained in 9 mm Na+ solution after cRNA injection. A and B, individual whole-cell current trace of an oocyte expressing αβγ-hENaC (A) or δβγ-hENaC (B). Application of a 13-mer synthetic peptide (α-13) is indicated by the open bar. C and D, mean ΔIami values before (−) and after (+) application of the α-13 peptide; experiments were performed as shown in A and B. E and F, relative change of ΔIami in response to the α-13 peptide is shown for each individual oocyte tested in experiments as shown in A and B. ΔIami values were normalized to the initial ΔIami before the application of the peptide. Each line connects the ΔIami values of an individual recording from an oocyte expressing either αβγ- (E) or δβγ-hENaC (F) measured before (−) and after (+) exposure to α-13. Data points indicated by the arrows are from the individual experiments shown in A and B. N indicates number of different batches of oocytes; n indicates number of individual oocytes measured. **, p < 0.01, ***, p < 0.001, paired t test.
Interestingly, the magnitude of the effect of the peptide varied in individual oocytes (Fig. 9, E and F). In αβγ-hENaC-expressing oocytes its inhibitory effect on ΔIami was variable but present in all oocytes tested (Fig. 9E). In contrast, a sizeable (≥20%) stimulatory effect of the peptide was only observed in 8 of 33 δβγ-hENaC-expressing oocytes tested (Fig. 9F). However, we never observed a substantial inhibitory effect of the peptide on ΔIami in δβγ-hENaC-expressing oocytes. We conclude that the α-subunit is essential for mediating the inhibitory effect of the α-13 peptide. Moreover, replacing the α-subunit by the δ-subunit not only abolishes the inhibitory effect of the α-13 peptide but converts it into a stimulatory effect.
Single-channel Po of δβγ-hENaC Is High in Outside-out Patches
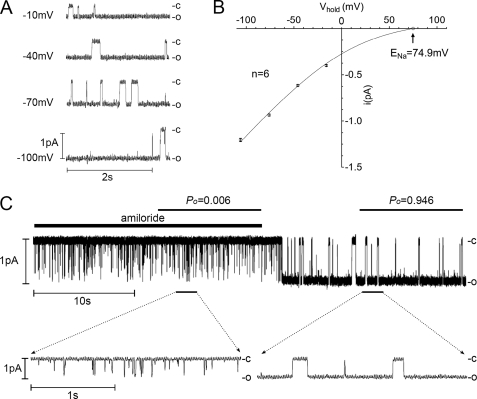
To confirm our conclusion that the Po of δβγ-hENaC is high compared with that reported for αβγ-ENaC in the literature, we performed patch clamp experiments in outside-out patches from oocytes expressing δβγ-hENaC. In these patches we were able to detect single channels with an average single-channel conductance of 11.4 ± 0.1 pS (n = 6) (Fig. 10, A and B), which is consistent with values previously reported for δβγ-hENaC (17). Ideally, single-channel Po is estimated from patches with only one active channel in the patch. Therefore, we used single-channel recordings as shown in Fig. 10 to determine the Po of δβγ-hENaC. In the presence of 10 μm amiloride, channel openings were brief and flickered as expected for an amiloride-sensitive channel. Washout of amiloride resulted in an increase of the single-channel Po. Indeed, in the absence of amiloride δβγ-hENaC spends most of its time in the open state (Fig. 10C). In five similar recordings the single-channel Po of δβγ-hENaC averaged 0.89 ± 0.04 (n = 5) in the absence of amiloride. Moreover, analysis of multichannel patches with up to five active channels also supports our conclusion that the Po of δβγ-hENaC is ∼0.9 (supplemental Table 1). Thus, the single-channel Po of δβγ-hENaC appears to be higher than the single-channel Po of about 0.5 previously reported for αβγ-ENaC in various preparations (3).
FIGURE 10.
Single-channel Po of δβγ-hENaC is high in outside-out patches. The single-channel current traces shown were obtained from an outside-out patch of an oocyte expressing δβγ-hENaC. Closed and open levels are indicated by -c and -o, respectively. A, single-channel current traces recorded at different holding potentials (Vhold). B, corresponding average single-channel I-V plot from six similar experiments as shown in A. To correct for the liquid junction potential occurring at the bridge/bath junction, the I-V plot was shifted to the left by 6 mV. S.E. values are indicated by vertical bars unless smaller than symbols. The I-V data were fitted using the Goldman-Hodgkin-Katz equation for a Na+ concentration ratio of [Na+]in/[Na+]out = 5/95 mm with a calculated reversal potential for Na+ (ENa) of 74.9 mV. C, continuous recording at Vhold = −70 mV from the same outside-out patch as shown in A. Amiloride (10 μm) was present in the bath solution as indicated above the trace by the black bar. Using amplitude histograms single-channel Po was determined in the presence and absence of amiloride during the periods indicated. The insets below show the indicated segments of the same current trace on an expanded time scale.
Chymotrypsin Does Not Increase Surface Expression of δβγ-hENaC
If single-channel Po of δβγ-hENaC is close to 1, how does chymotrypsin produce a 2-fold increase in whole-cell currents? This could be achieved by new insertion of channels into the plasma membrane or by activation of near-silent channels that are already present in the plasma membrane (39, 61, 62). To distinguish between these possibilities, we investigated the effect of chymotrypsin on channel surface expression using the chemiluminescence assay with FLAG-tagged β-hENaC. As shown in Fig. 11, application of chymotrypsin did not increase channel surface expression in oocytes expressing δβγ-hENaC (Fig. 11A). In contrast, in matched oocytes from the same batch chymotrypsin had the usual stimulatory effect on ENaC whole-cell currents (Fig. 11B). To confirm that the chemiluminescence assay can reliably detect an increase in channel surface expression, we used two different groups of oocytes injected with either 0.3 or 1 ng of cRNA per subunit (δβγ). As expected, the average base-line ΔIami and chemiluminescence values were significantly larger in oocytes injected with 1 ng of cRNA per subunit. Importantly, in both groups of oocytes chymotrypsin increased ΔIami by about 2-fold (Fig. 11B) without a concomitant increase in channel surface expression. These findings indicate that application of chymotrypsin does not increase channel surface expression but is likely to activate near-silent channels that are already present at the cell surface.
FIGURE 11.
Chymotrypsin does not increase surface expression of δβγ-hENaC. Parallel detection of surface expression (RLU) and ΔIami values in δβγ-hENaC-expressing oocytes before (−) and after (+) exposure to chymotrypsin (2 μg/ml) in a representative batch of oocytes (similar results were obtained in two other batches). A, channel surface expression was detected by insertion of a FLAG reporter epitope in the extracellular loop of the β-subunit and a chemiluminescence assay. Oocytes were injected with either 0.3 or 1 ng of cRNA per subunit (δβγ). Noninjected oocytes (n.i.) served as controls and showed negligible background luminescence. Numbers inside the columns indicate the number of individual oocytes measured. B, average ΔIami values measured in parallel to the chemiluminescence assay; n indicates number of individual oocytes measured. ***, p < 0.001, paired t test; §§, p < 0.01; §§§, p < 0.001, unpaired t test.
DISCUSSION
In this study we compared the functional properties of αβγ- and δβγ-hENaC heterologously expressed in X. laevis oocytes. The key findings of our study are the following: 1) ENaC whole-cell currents are larger in oocytes expressing δβγ-hENaC compared with those of control oocytes expressing αβγ-hENaC; 2) δβγ-hENaC can be stimulated by chymotrypsin, but the degree of stimulation and cleavage pattern differs from that of αβγ-hENaC. To our knowledge this is the first study to report evidence for proteolytic activation of δβγ-hENaC and to demonstrate that replacing the α-subunit by the δ-subunit increases ENaC activity in co-expression experiments with βγ-hENaC.
In the experiments in which we compared base-line δβγ-hENaC and αβγ-hENaC currents in matched groups of oocytes, we made every effort to ensure that the amount and quality of the injected cRNAs were comparable. Experiments were repeated in several different batches of oocytes using several different cRNA preparations. Moreover, we confirmed that the level of protein expression of the three subunits was similar in the αβγ- and δβγ-hENaC-expressing oocytes. Under these conditions ENaC currents were on average about 11-fold larger in δβγ-hENaC-expressing oocytes compared with those in αβγ-hENaC-expressing oocytes. The effect of the δ-subunit to enhance ENaC currents was robust and preserved even when we used α- and δ-subunits with tagged N and C termini. Taken together our findings indicate that the observed difference is not an artifact of our experimental design or the expression system but is caused by δ-ENaC altering the functional properties of the heteromeric channel. It is unclear why our findings differ from those of a previous study in which ENaC whole-cell currents were found to be larger in αβγ-hENaC- than in δβγ-hENaC-expressing oocytes (8). However, the latter findings are based on a small number of measurements (five oocytes per group) possibly using oocytes from different batches. It is unlikely that the discrepancy with our findings can be attributed to the longer δ-hENaC splice variant used in the latter study because no functional differences have been observed between the two splice variants (10).
Using a semi-quantitative chemiluminescence assay, we demonstrated that channel surface expression is roughly similar in αβγ- and δβγ-hENaC-expressing oocytes. Even though with our method we cannot rule out a modest effect of δ-hENaC on channel trafficking, our results indicate that an increase in channel surface expression is not the underlying cause for the 11-fold larger whole-cell currents observed in the δβγ-hENaC-expressing oocytes. The single-channel sodium conductance of δβγ-hENaC (∼12 pS) is known to be larger than that of αβγ-hENaC (∼5 pS) (17). Indeed, our ion substitution experiments, replacing Na+ by Li+ in the bath solution, indicate that this contributes to the larger whole-cell currents in δβγ-hENaC-expressing oocytes. However, the different single-channel sodium conductance cannot account for the observed 11-fold difference in the whole-cell currents. Collectively, these findings indicate that the major contributing factor is an increased average Po of δβγ-hENaC.
In outside-out patches we observed a single-channel Po of ∼0.9 for δβγ-hENaC, which is higher than the single-channel Po of ∼0.5 usually reported for αβγ-ENaC but with a great degree of variability (3). In contrast to our findings, the Po of δβγ-hENaC has been reported to be ∼0.5 in a previous study using cell-attached patch clamp recordings (17). However, this latter approach has the inherent difficulty that amiloride cannot be used to determine the current level at which all channels are closed. This can lead to an underestimation of channel Po and may explain the discrepant findings. Our observation that the Po of δβγ-hENaC observed in single-channel patch clamp recordings is probably higher than that of αβγ-ENaC supports our conclusion that the overall activity of δβγ-hENaC at the cell surface is higher than that of αβγ-ENaC. From previous studies we have to assume that different pools of ENaC with different Po values are present at the plasma membrane, including near-silent channels that can be activated by extracellular proteases (39, 61, 62). Because of their low Po of less than 0.05 (1, 47, 68), the latter channels will easily escape detection in single-channel recordings. The presence of a population of near-silent channels in addition to channels with a Po of close to 1 could explain the conundrum that we observed a 2-fold stimulatory effect of chymotrypsin on ENaC whole-cell currents in oocytes expressing δβγ-hENaC with a single-channel Po of close to 1. Indeed, if all the channels at the cell surface had a base-line Po of close to 1, this 2-fold increase could only be explained by an additional insertion of channels into the plasma membrane. However, we demonstrated that proteolytic activation of δβγ-hENaC was not associated with an increase in channel surface expression. Thus, the most likely explanation for this observed 2-fold increase in ENaC whole-cell currents is that chymotrypsin activates a pool of near-silent δβγ-hENaC that co-exists with a pool of channels with a high Po. Proteolytic activation of rat αβγ-ENaC has previously been shown to be mediated by a dual effect as follows: (i) the recruitment of near-silent channels, and (ii) an increase in Po of channels that are already active (39). This latter effect is unlikely to contribute in a major way to the proteolytic activation of δβγ-hENaC because its single-channel Po is already high. Thus, a higher base-line single-channel Po and a reduced pool of near-silent channels probably explain the reduced stimulatory effect of chymotrypsin on δβγ-hENaC.
That the base-line activity of δβγ-hENaC at the plasma membrane is higher than that of αβγ-ENaC is also supported by the reduced stimulatory effect of MTSET on δβγ-hENaC in the presence of the S520C mutant β-subunit. The S520C mutant of β-hENaC is analogous to the previously described S518C mutation in the β-subunit of rat ENaC. Single-channel recordings of the latter have shown that this mutant channel can be converted to a channel with a Po of close to 1 by MTSET (39, 48). It is assumed that the corresponding mutation in human β-hENaC behaves in a similar way (32). The mutant ENaC can be activated by MTSET only when the channel is in its open state (48). However, near-silent channels, at least occasionally, may open long enough for MTSET to act on them and to convert them to channels with a high open probability. Thus, MTSET is likely to increase the Po of both active and near-silent channels in whole-cell experiments. We found that MTSET increased δβS520Cγ-hENaC whole-cell currents about 2-fold and αβS520Cγ-hENaC whole-cell currents about 7-fold, which is in good agreement with the chymotrypsin data. Provided that MTSET increases Po of both δβS520Cγ-hENaC and αβS520Cγ-hENaC to nearly 1, these findings indicate that the average Po prior to the application of MTSET was ∼0.5 for δβγ-hENaC and ∼0.14 for αβγ-hENaC. With this assumption the average base-line Po of δβS520Cγ-hENaC can be estimated to be 3–4-fold higher than that of αβS520Cγ-hENaC. Such a difference in average base-line Po may well be a major cause for the larger whole-cell currents observed in δβγ-hENaC expression oocytes. It should be noted that the Po values estimated from the MTSET whole-cell experiments reflect an average Po of all the channels present at the cell surface, including near-silent channels. In contrast, the single-channel Po of ∼0.9 determined for δβγ-hENaC in outside-out patches represents the Po of active channels only.
Interestingly, the relative stimulatory effect of chymotrypsin was quite similar in αβγ- and δβγ-hENaC-expressing oocytes maintained in a low Na+ solution (9 mm) after cRNA injection to prevent sodium loading of the oocytes. In good agreement with previously reported data (54, 67), low Na+ preincubation significantly increased base-line ENaC currents in αβγ-hENaC-expressing oocytes. Importantly, in these oocytes the relative stimulatory effect of chymotrypsin was reduced. This may be explained by the recently reported finding that an increase in intracellular sodium not only promotes Nedd4-2-mediated channel retrieval but also reduces channel Po (66) possibly by reducing proteolytic channel activation (69). Thus, base-line Po of αβγ-hENaC is likely to be higher in oocytes maintained in a low Na+ (9 mm) bath solution than in oocytes maintained in a standard bath solution containing 96 mm Na+. This would explain why the relative stimulatory effect of chymotrypsin is reduced in the oocytes maintained in low Na+. In contrast, in δβγ-hENaC-expressing oocytes low Na+ preincubation had no significant effect on base-line ENaC currents or on the stimulatory effect of chymotrypsin. This suggests that changes in intracellular sodium have little effect on the base-line Po and surface expression of δβγ-hENaC. Interestingly, the δ-subunit lacks a so-called PY motif (17), which is highly conserved in the intracellular C termini of the α-, β-, and γ-subunits. This motif is thought to be essential for the channel interaction with its ubiquitylating protein Nedd4-2 (70), an important regulatory protein involved in many aspects of ENaC function, including its regulation by intracellular sodium and proteases (71). Na+ self-inhibition by extracellular Na+ is a prominent feature of αβγ-hENaC and is abolished after stimulating the channel with extracellular proteases (72). Interestingly, in the presence of the δ-subunit the phenomenon of Na+ self-inhibition is reduced (30). Thus, removal of Na+ self-inhibition is likely to contribute more to the chymotrypsin-induced stimulation of αβγ-hENaC than to that of δβγ-hENaC. This suggests that a combination of a reduced constitutive Na+ self-inhibition and a reduced inhibition by intracellular Na+ contributes to the increased base-line activity of δβγ-hENaC.
In this context it is of interest to consider our findings regarding the proteolytic processing of δβγ-hENaC and αβγ-hENaC in the oocyte expression system. Co-expression of α-hENaC with βγ-hENaC was associated with the appearance of cleaved fragments of the α- and the γ-subunits but not of the β-subunit, which is consistent with previous studies using mouse (73, 74) or rat ENaC (58). These fragments are thought to be the result of channel cleavage by endogenous proteases at putative furin cleavage sites in the extracellular loops of the α- and γ-subunit (41, 73). In contrast, co-expression of δ-hENaC with βγ-hENaC was not associated with cleavage of the δ-subunit, which indicates that proteolytic processing of the δ-subunit is different from that of the α-subunit. In oocytes co-expressing δβγ-hENaC, we detected cleavage of the γ-subunit that was absent in oocytes expressing δγ-hENaC. Thus, co-expressed with βγ-hENaC the δ-subunit, like the α-subunit, promotes cleavage of the γ- but not the β-subunit. However, the effect of the δ-subunit on γ-hENaC cleavage appeared to be slightly different from that of the α-subunit. In δβγ-hENaC-expressing oocytes we detected an additional ∼60-kDa cleavage product of γ-hENaC that was not observed in the whole-cell lysate of αβγ-hENaC-expressing oocytes. Interestingly, the γ-subunit seems to be particularly important for proteolytic channel activation (39, 46, 59). The underlying mechanism is thought to involve the release of an inhibitory peptide sequence from the extracellular loop of the γ-subunit by its cleavage at a prostasin (45), plasmin (40, 60), or elastase site (75). Our findings suggest that in oocytes expressing δβγ-hENaC, cleavage of the γ-subunit by endogenous proteases may be more efficient than in αβγ-hENaC-expressing oocytes. This would correspond well to our finding of an enhanced base-line activity of δβγ-hENaC.
Using biotinylation experiments, we demonstrated that channel activation by chymotrypsin was not only associated with the previously described cleavage of γ-ENaC (39, 45, 59) but also with the appearance of an ∼25/∼68-kDa (detected from N/C terminus) δ-hENaC cleavage product at the cell surface. Consistent with data in the literature (41), we did not observe α-hENaC cleavage products at the cell surface in response to channel activation by chymotrypsin. There are several putative chymotrypsin cleavage sites localized in the extracellular loop of δ-ENaC in close proximity to the first transmembrane domain. Cleavage at these sites would result in an N-terminal cleavage product of ∼18–25 kDa in good agreement with the ∼25-kDa fragment detected with the HA antibody directed against the N-terminal HA epitope of the δ-subunit. Thus, it is plausible that chymotrypsin causes cleavage of δ-hENaC at the cell surface. The functional relevance of this δ-hENaC cleavage for proteolytic channel activation remains to be determined. Importantly, we demonstrated that ENaC currents cannot be activated by chymotrypsin in oocytes expressing δ-hENaC alone in the absence of γ-hENaC. This indicates that cleavage of the δ-subunit is not sufficient for ENaC activation and confirms the important role of the γ-subunit for channel activation by extracellular proteases (39, 46).
Finally, we demonstrated that a 13-mer peptide corresponding to a putative inhibitory region within the extracellular loop of the α-subunit of human ENaC can inhibit ENaC currents in αβγ-hENaC-expressing oocytes but fails to inhibit ENaC currents in δβγ-hENaC-expressing oocytes. In fact, the peptide even increased δβγ-hENaC currents in some but not all oocytes without ever causing a substantial current inhibition. The inhibitory effect of the 13-mer peptide on αβγ-hENaC is consistent with the previously described inhibitory effect of a corresponding 8-mer peptide on mouse ENaC (43). It has been speculated that this peptide mimics the tonic inhibitory effect of the corresponding region of the extracellular loop of α-ENaC before this segment is excised by proteases. However, so far it is not known whether the peptide exerts its inhibitory effect by interacting with the α-subunit or with an adjacent region in one of the other subunits. Our findings suggest that the presence of the α-subunit is necessary for the peptide to inhibit ENaC. It will be a challenge for future studies to determine the basis of the significant albeit variable stimulatory effect of the peptide on δβγ-hENaC. Moreover, sequence alignment revealed that three amino acids are conserved in the region of the δ-subunit (180LSATVPRHEPPFH192) corresponding to the 13-mer inhibitory α-peptide. Thus, it may be of interest to test the functional effect of a peptide corresponding to this region of the δ-subunit.
In summary, we have shown that ENaC whole-cell currents are significantly larger in oocytes expressing δβγ-hENaC compared with control oocytes expressing αβγ-hENaC, and that δβγ-hENaC can be activated by chymotrypsin. Our data indicate that the larger base-line currents of δβγ-hENaC are mainly caused by an increased activity of the channels present at the plasma membrane. This may, at least in part, be a consequence of a reduced inhibitory effect of intracellular sodium on δβγ-hENaC activity. Moreover, the δ-subunit seems to promote constitutive cleavage of the γ-subunit, which may favor the presence of active channels at the cell surface and decrease the pool of near-silent channels ready to be activated by chymotrypsin. This may also explain why the relative stimulatory response to chymotrypsin is smaller in δβγ-hENaC- than in αβγ-hENaC-expressing oocytes. Activation of δβγ-hENaC is associated with the appearance of a δ-hENaC cleavage product at the cell surface. However, the δ-subunit is not sufficient to mediate proteolytic channel activation and requires the presence of the γ-subunit.
Collectively our findings indicate that replacing the α-subunit by the δ-subunit has profound effects on ENaC activity and on channel regulation. It is an emerging paradigm that the size of the stimulatory effect of extracellular proteases on ENaC depends on the degree of proteolytic pre-activation of the channel by endogenous proteases (41, 76). Thus, it is tempting to speculate that the replacement of the α-subunit by the δ-subunit in the heterotrimeric channel favors the constitutive activation of ENaC and reduces the pool of near-silent channels that can be activated by extracellular proteases. Thus, a differential expression of the α- and δ-subunit provides an additional way of regulating ENaC activity according to the needs of different tissues.
Supplementary Material
Acknowledgments
The expert technical assistance of Ralf Rinke, Jessica Ott, and Céline Harlay is gratefully acknowledged.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB423 (Kidney Injury, Pathogenesis and Regenerative Mechanisms, Project A12 (to C. K.)), by the Johannes and Frieda Marohn Stiftung (to C. K.), an Elitenetwork Bavaria fellowship (to S. H.), and the BioMedTec International Graduate School “Lead Structures of Cell Function” of the Elitenetwork Bavaria (to S. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and a figure.
- ENaC
- epithelial sodium channel
- hENaC
- human ENaC
- MTSET
- [2-(trimethylammonium)ethyl]methanethiosulfonate bromide
- HA
- hemagglutinin
- NMDG
- N-methyl-d-glucamine
- pS
- picosiemens.
REFERENCES
- 1.Kellenberger S., Schild L. (2002) Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 2.Alvarez de la Rosa D., Canessa C. M., Fyfe G. K., Zhang P. (2000) Annu. Rev. Physiol. 62, 573–594 [DOI] [PubMed] [Google Scholar]
- 3.Garty H., Palmer L. G. (1997) Physiol. Rev. 77, 359–396 [DOI] [PubMed] [Google Scholar]
- 4.Rossier B. C., Pradervand S., Schild L., Hummler E. (2002) Annu. Rev. Physiol. 64, 877–897 [DOI] [PubMed] [Google Scholar]
- 5.Drummond H. A., Furtado M. M., Myers S., Grifoni S., Parker K. A., Hoover A., Stec D. E. (2006) Am. J. Physiol. Cell Physiol. 290, C404–C410 [DOI] [PubMed] [Google Scholar]
- 6.Drummond H. A., Jernigan N. L., Grifoni S. C. (2008) Hypertension 51, 1265–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charles R. P., Guitard M., Leyvraz C., Breiden B., Haftek M., Haftek-Terreau Z., Stehle J. C., Sandhoff K., Hummler E. (2008) J. Biol. Chem. 283, 2622–2630 [DOI] [PubMed] [Google Scholar]
- 8.Giraldez T., Afonso-Oramas D., Cruz-Muros I., Garcia-Marin V., Pagel P., González-Hernández T., Alvarez de la Rosa D. (2007) J. Neurochem. 102, 1304–1315 [DOI] [PubMed] [Google Scholar]
- 9.Yamamura H., Ugawa S., Ueda T., Nagao M., Shimada S. (2008) Biochem. Biophys. Res. Commun. 373, 155–158 [DOI] [PubMed] [Google Scholar]
- 10.Yamamura H., Ugawa S., Ueda T., Nagao M., Shimada S. (2006) Biochem. Biophys. Res. Commun. 349, 317–321 [DOI] [PubMed] [Google Scholar]
- 11.Golestaneh N., Klein C., Valamanesh F., Suarez G., Agarwal M. K., Mirshahi M. (2001) Biochem. Biophys. Res. Commun. 280, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 12.Kusche-Vihrog K., Sobczak K., Bangel N., Wilhelmi M., Nechyporuk-Zloy V., Schwab A., Schillers H., Oberleithner H. (2008) Pflugers Arch. 455, 849–857 [DOI] [PubMed] [Google Scholar]
- 13.Canessa C. M., Schild L., Buell G., Thorens B., Gautschi I., Horisberger J. D., Rossier B. C. (1994) Nature 367, 463–467 [DOI] [PubMed] [Google Scholar]
- 14.Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 15.Stockand J. D., Staruschenko A., Pochynyuk O., Booth R. E., Silverthorn D. U. (2008) IUBMB Life 60, 620–628 [DOI] [PubMed] [Google Scholar]
- 16.Canessa C. M. (2007) Nature 449, 293–294 [DOI] [PubMed] [Google Scholar]
- 17.Waldmann R., Champigny G., Bassilana F., Voilley N., Lazdunski M. (1995) J. Biol. Chem. 270, 27411–27414 [DOI] [PubMed] [Google Scholar]
- 18.Yamamura H., Ugawa S., Ueda T., Nagao M., Shimada S. (2004) J. Biol. Chem. 279, 12529–12534 [DOI] [PubMed] [Google Scholar]
- 19.Brockway L. M., Zhou Z. H., Bubien J. K., Jovov B., Benos D. J., Keyser K. T. (2002) Am. J. Physiol. Cell Physiol. 283, C126–C134 [DOI] [PubMed] [Google Scholar]
- 20.Biasio W., Chang T., McIntosh C. J., McDonald F. J. (2004) J. Biol. Chem. 279, 5429–5434 [DOI] [PubMed] [Google Scholar]
- 21.Yamamura H., Ugawa S., Ueda T., Nagao M., Shimada S. (2004) J. Biol. Chem. 279, 44483–44489 [DOI] [PubMed] [Google Scholar]
- 22.Hernández-González E. O., Sosnik J., Edwards J., Acevedo J. J., Mendoza-Lujambio I., López-González I., Demarco I., Wertheimer E., Darszon A., Visconti P. E. (2006) J. Biol. Chem. 281, 5623–5633 [DOI] [PubMed] [Google Scholar]
- 23.Nie H. G., Tucker T., Su X. F., Na T., Peng J. B., Smith P. R., Idell S., Ji H. L. (2009) Am. J. Respir. Cell Mol. Biol. 40, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babini E., Geisler H. S., Siba M., Gründer S. (2003) J. Biol. Chem. 278, 28418–28426 [DOI] [PubMed] [Google Scholar]
- 25.Waldmann R., Bassilana F., Voilley N., Lazdunski M., Mattéi M. (1996) Genomics 34, 262–263 [DOI] [PubMed] [Google Scholar]
- 26.McDonald F. J., Snyder P. M., McCray P. B., Jr., Welsh M. J. (1994) Am. J. Physiol. 266, L728–L734 [DOI] [PubMed] [Google Scholar]
- 27.Voilley N., Lingueglia E., Champigny G., Mattéi M. G., Waldmann R., Lazdunski M., Barbry P. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voilley N., Bassilana F., Mignon C., Merscher S., Mattéi M. G., Carle G. F., Lazdunski M., Barbry P. (1995) Genomics 28, 560–565 [DOI] [PubMed] [Google Scholar]
- 29.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Kimura K., Makita H., Sekine M., Obayashi M., Nishi T., Shibahara T., Tanaka T., Ishii S., Yamamoto J., Saito K., Kawai Y., Isono Y., Nakamura Y., Nagahari K., Murakami K., Yasuda T., Iwayanagi T., Wagatsuma M., Shiratori A., Sudo H., Hosoiri T., Kaku Y., Kodaira H., Kondo H., Sugawara M., Takahashi M., Kanda K., Yokoi T., Furuya T., Kikkawa E., Omura Y., Abe K., Kamihara K., Katsuta N., Sato K., Tanikawa M., Yamazaki M., Ninomiya K., Ishibashi T., Yamashita H., Murakawa K., Fujimori K., Tanai H., Kimata M., Watanabe M., Hiraoka S., Chiba Y., Ishida S., Ono Y., Takiguchi S., Watanabe S., Yosida M., Hotuta T., Kusano J., Kanehori K., Takahashi-Fujii A., Hara H., Tanase T. O., Nomura Y., Togiya S., Komai F., Hara R., Takeuchi K., Arita M., Imose N., Musashino K., Yuuki H., Oshima A., Sasaki N., Aotsuka S., Yoshikawa Y., Matsunawa H., Ichihara T., Shiohata N., Sano S., Moriya S., Momiyama H., Satoh N., Takami S., Terashima Y., Suzuki O., Nakagawa S., Senoh A., Mizoguchi H., Goto Y., Shimizu F., Wakebe H., Hishigaki H., Watanabe T., Sugiyama A., Takemoto M., Kawakami B., Yamazaki M., Watanabe K., Kumagai A., Itakura S., Fukuzumi Y., Fujimori Y., Komiyama M., Tashiro H., Tanigami A., Fujiwara T., Ono T., Yamada K., Fujii Y., Ozaki K., Hirao M., Ohmori Y., Kawabata A., Hikiji T., Kobatake N., Inagaki H., Ikema Y., Okamoto S., Okitani R., Kawakami T., Noguchi S., Itoh T., Shigeta K., Senba T., Matsumura K., Nakajima Y., Mizuno T., Morinaga M., Sasaki M., Togashi T., Oyama M., Hata H., Watanabe M., Komatsu T., Mizushima-Sugano J., Satoh T., Shirai Y., Takahashi Y., Nakagawa K., Okumura K., Nagase T., Nomura N., Kikuchi H., Masuho Y., Yamashita R., Nakai K., Yada T., Nakamura Y., Ohara O., Isogai T., Sugano S. (2004) Nat. Genet. 36, 40–45 [DOI] [PubMed] [Google Scholar]
- 30.Ji H. L., Su X. F., Kedar S., Li J., Barbry P., Smith P. R., Matalon S., Benos D. J. (2006) J. Biol. Chem. 281, 8233–8241 [DOI] [PubMed] [Google Scholar]
- 31.Ji H. L., Bishop L. R., Anderson S. J., Fuller C. M., Benos D. J. (2004) J. Biol. Chem. 279, 8428–8440 [DOI] [PubMed] [Google Scholar]
- 32.Lu M., Echeverri F., Kalabat D., Laita B., Dahan D. S., Smith R. D., Xu H., Staszewski L., Yamamoto J., Ling J., Hwang N., Kimmich R., Li P., Patron E., Keung W., Patron A., Moyer B. D. (2008) J. Biol. Chem. 283, 11981–11994 [DOI] [PubMed] [Google Scholar]
- 33.Yamamura H., Ugawa S., Ueda T., Shimada S. (2005) J. Pharmacol. Exp. Ther. 315, 965–969 [DOI] [PubMed] [Google Scholar]
- 34.Yamamura H., Ugawa S., Ueda T., Nagao M., Shimada S. (2005) Mol. Pharmacol. 68, 1142–1147 [DOI] [PubMed] [Google Scholar]
- 35.Ji H. L., Benos D. J. (2004) J. Biol. Chem. 279, 26939–26947 [DOI] [PubMed] [Google Scholar]
- 36.Yamamura H., Ugawa S., Ueda T., Nagao M., Joh T., Shimada S. (2008) Eur. J. Pharmacol. 600, 32–36 [DOI] [PubMed] [Google Scholar]
- 37.Planès C., Caughey G. H. (2007) Curr. Top. Dev. Biol. 78, 23–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughey R. P., Carattino M. D., Kleyman T. R. (2007) Curr. Opin. Nephrol. Hypertens. 16, 444–450 [DOI] [PubMed] [Google Scholar]
- 39.Diakov A., Bera K., Mokrushina M., Krueger B., Korbmacher C. (2008) J. Physiol. 586, 4587–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Passero C. J., Mueller G. M., Rondon-Berrios H., Tofovic S. P., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 36586–36591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossier B. C., Stutts M. J. (2009) Annu. Rev. Physiol. 71, 361–379 [DOI] [PubMed] [Google Scholar]
- 42.Hughey R. P., Bruns J. B., Kinlough C. L., Kleyman T. R. (2004) J. Biol. Chem. 279, 48491–48494 [DOI] [PubMed] [Google Scholar]
- 43.Carattino M. D., Passero C. J., Steren C. A., Maarouf A. B., Pilewski J. M., Myerburg M. M., Hughey R. P., Kleyman T. R. (2008) Am. J. Physiol. Renal Physiol. 294, F47–F52 [DOI] [PubMed] [Google Scholar]
- 44.Carattino M. D., Sheng S., Bruns J. B., Pilewski J. M., Hughey R. P., Kleyman T. R. (2006) J. Biol. Chem. 281, 18901–18907 [DOI] [PubMed] [Google Scholar]
- 45.Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., Kleyman T. R. (2007) J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 46.Carattino M. D., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 25290–25295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Firsov D., Schild L., Gautschi I., Mérillat A. M., Schneeberger E., Rossier B. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kellenberger S., Gautschi I., Schild L. (2002) J. Physiol. 543, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konstas A. A., Mavrelos D., Korbmacher C. (2000) Pflügers Arch. 441, 341–350 [DOI] [PubMed] [Google Scholar]
- 50.Rauh R., Dinudom A., Fotia A. B., Paulides M., Kumar S., Korbmacher C., Cook D. I. (2006) Pflügers Arch. 452, 290–299 [DOI] [PubMed] [Google Scholar]
- 51.Wielpütz M. O., Lee I. H., Dinudom A., Boulkroun S., Farman N., Cook D. I., Korbmacher C., Rauh R. (2007) J. Biol. Chem. 282, 28264–28273 [DOI] [PubMed] [Google Scholar]
- 52.Konstas A. A., Bielfeld-Ackermann A., Korbmacher C. (2001) Pflügers Arch. 442, 752–761 [DOI] [PubMed] [Google Scholar]
- 53.Diakov A., Korbmacher C. (2004) J. Biol. Chem. 279, 38134–38142 [DOI] [PubMed] [Google Scholar]
- 54.Kellenberger S., Gautschi I., Rossier B. C., Schild L. (1998) J. Clin. Invest. 101, 2741–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volk T., Konstas A. A., Bassalaý P., Ehmke H., Korbmacher C. (2004) Pflügers Arch. 447, 884–894 [DOI] [PubMed] [Google Scholar]
- 56.Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. (1999) Neuron 22, 537–548 [DOI] [PubMed] [Google Scholar]
- 57.Snyder P. M., Bucher D. B., Olson D. R. (2000) J. Gen. Physiol. 116, 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris M., Garcia-Caballero A., Stutts M. J., Firsov D., Rossier B. C. (2008) J. Biol. Chem. 283, 7455–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harris M., Firsov D., Vuagniaux G., Stutts M. J., Rossier B. C. (2007) J. Biol. Chem. 282, 58–64 [DOI] [PubMed] [Google Scholar]
- 60.Svenningsen P., Bistrup C., Friis U. G., Bertog M., Haerteis S., Krueger B., Stubbe J., Jensen O. N., Thiesson H. C., Uhrenholt T. R., Jespersen B., Jensen B. L., Korbmacher C., Skøtt O. (2009) J. Am. Soc. Nephrol. 20, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caldwell R. A., Boucher R. C., Stutts M. J. (2004) Am. J. Physiol. Cell Physiol. 286, C190–C194 [DOI] [PubMed] [Google Scholar]
- 62.Caldwell R. A., Boucher R. C., Stutts M. J. (2005) Am. J. Physiol. Lung Cell. Mol. Physiol. 288, L813–L819 [DOI] [PubMed] [Google Scholar]
- 63.Chraïbi A., Vallet V., Firsov D., Hess S. K., Horisberger J. D. (1998) J. Gen. Physiol. 111, 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Durieux M. E., Salafranca M. N., Lynch K. R. (1994) FEBS Lett. 337, 235–238 [DOI] [PubMed] [Google Scholar]
- 65.Abriel H., Horisberger J. D. (1999) J. Physiol. 516, 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anantharam A., Tian Y., Palmer L. G. (2006) J. Physiol. 574, 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Konstas A. A., Shearwin-Whyatt L. M., Fotia A. B., Degger B., Riccardi D., Cook D. I., Korbmacher C., Kumar S. (2002) J. Biol. Chem. 277, 29406–29416 [DOI] [PubMed] [Google Scholar]
- 68.Rossier B. C. (2002) J. Gen. Physiol. 120, 67–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knight K. K., Wentzlaff D. M., Snyder P. M. (2008) J. Biol. Chem. 283, 27477–27482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Staub O., Abriel H., Plant P., Ishikawa T., Kanelis V., Saleki R., Horisberger J. D., Schild L., Rotin D. (2000) Kidney Int. 57, 809–815 [DOI] [PubMed] [Google Scholar]
- 71.Ruffieux-Daidié D., Poirot O., Boulkroun S., Verrey F., Kellenberger S., Staub O. (2008) J. Am. Soc. Nephrol. 19, 2170–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chraïbi A., Horisberger J. D. (2002) J. Gen. Physiol. 120, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 74.Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., Kleyman T. R. (2003) J. Biol. Chem. 278, 37073–37082 [DOI] [PubMed] [Google Scholar]
- 75.Adebamiro A., Cheng Y., Rao U. S., Danahay H., Bridges R. J. (2007) J. Gen. Physiol. 130, 611–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nesterov V., Dahlmann A., Bertog M., Korbmacher C. (2008) Am. J. Physiol. Renal Physiol. 295, F1052–F1062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.