Abstract
Hypoxia-inducible factor (HIF) plays an important role in cell survival by regulating iron, antioxidant defense, and mitochondrial function. Pharmacological inhibitors of the iron-dependent enzyme class prolyl hydroxylases (PHD), which target α subunits of HIF proteins for degradation, have recently been demonstrated to alleviate neurodegeneration associated with stroke and hypoxic-ischemic injuries. Here we report that inhibition of PHD by 3,4-dihydroxybenzoate (DHB) protects against 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced nigral dopaminergic cell loss and up-regulates HIF-1α within these neurons. Elevations in mRNA and protein levels of HIF-dependent genes heme oxygenase-1 (Ho-1) and manganese superoxide dismutase (Mnsod) following DHB pretreatment alone are also maintained in the presence of MPTP. MPTP-induced reductions in ferroportin and elevations in nigral and striatal iron levels were reverted to levels comparable with that of untreated controls with DHB pretreatment. Reductions in pyruvate dehydrogenase mRNA and activity resulting from MPTP were also found to be attenuated by DHB. In vitro, the HIF pathway was activated in N27 cells grown at 3% oxygen treated with either PHD inhibitors or an iron chelator. Concordant with our in vivo data, the MPP+-elicited increase in total iron as well as decreases in cell viability were attenuated in the presence of DHB. Taken together, these data suggest that protection against MPTP neurotoxicity may be mediated by alterations in iron homeostasis and defense against oxidative stress and mitochondrial dysfunction brought about by cellular HIF-1α induction. This study provides novel data extending the possible therapeutic utility of HIF induction to a Parkinson disease model of neurodegeneration, which may prove beneficial not only in this disorder itself but also in other diseases associated with metal-induced oxidative stress.
Parkinson disease (PD)2 is a neurodegenerative disorder primarily associated with loss of dopaminergic (DAergic) neurons of the pars compacta region of the substantia nigra (SNpc). Dopaminergic neurons are particularly prone to oxidative damage due to high levels of inherent reactive oxygen species that are produced during dopamine synthesis or its breakdown by monoamine oxidases or autoxidation to quinones (1–3). Importantly, iron bound to neuromelanin within DAergic neurons can subsequently react with metabolically liberated hydrogen peroxide through the Fenton reaction to produce extremely toxic hydroxyl radicals. If not properly buffered, hydroxyl radicals can stimulate protein oxidation and lipid peroxidation, which is thought to contribute to macromolecular injury and neuronal death. Iron is the most abundant metal in the brain and some degree of accessible reactive iron is necessary for brain viability as it serves as a cofactor in DNA, RNA, and protein synthesis and for heme and non-heme enzymes involved in both mitochondrial respiration and neurotransmitter synthesis (4). Although iron deficiencies early in life are known to result in impairments in brain development (5), high concentrations of iron may result in cellular toxicity (6) in part due to its ability to catalyze the production of toxic oxygen radicals.
An important family of enzymes that require iron as an essential cofactor are the prolyl 4-hydroxylases (PHDs), which serve to hydroxylate proline residues situated within hypoxia-inducible factor proteins (HIFs) (7). Under hypoxic or iron-lacking conditions, PHDs are prevented from hydroxylating proline residues within the alpha (α) subunits of the HIF protein, preventing the ubiquitination and proteasomal degradation of the protein. Stabilization of HIFα results in its accumulation within the cytosol and translocation to the nucleus where it binds HIFβ and then to hypoxia response elements found on a variety of genes including heme oxygenase-1 (Ho-1) and manganese superoxide dismutase (Mnsod).
Previous studies have demonstrated that deferoxamine, an iron chelator, can activate HIF-1α and prevent neuronal death in both in vitro and in vivo models of ischemia likely via inhibition of PHDs (8, 9). PHD inhibitors have been demonstrated to prevent oxidative cell death and ischemic injury via HIF pathway activation (10). More recently, it has been shown that inactivation of HIF-1α in specific cortical and striatal neurons exacerbated tissue damage in a mouse model of ischemia (11). With increasing evidence of the protective effects of induction of HIF-dependent gene products involved in iron regulation, cell survival, and energy metabolism, PHD inhibitors have been implicated as targets for neuroprotection in the central nervous system. We demonstrate here that PHD inhibition increases induction of HIF and HIF-related genes, functionally impacts on parameters of iron homeostasis and metabolic function, and, most importantly, significantly reduces the extent of DAergic nigrostriatal injury observed in the well established murine MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) PD model.
MATERIALS AND METHODS
Mouse Studies
Male 10-week-old C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME) used in this study were housed according to standard animal care protocols, kept on a 12-h light/dark cycle and maintained in a pathogen-free environment in the Buck Institute vivarium. All experiments were approved by local IACUC review and conducted according to current NIH policies on the use of animals in research. For 3,4-dihydroxybenzoate (DHB) studies (n = 20 per group), 3,4-dihydroxybenzoate was diluted to a final dose of 100 mg/kg (in 5% ethanol) and administered intraperitoneally to 10-week-old male C57BL/6 mice 6 h prior to 2 consecutive intraperitoneal injections of either saline vehicle or 20 mg/kg of MPTP given 12 h apart. For clioquinol (CQ) studies (n = 12 per group), another subset of mice were dosed daily by oral gavage with either saline or CQ, 30 mg/kg, for 2 weeks prior to the subacute MPTP treatment paradigm (2 × 20 mg/kg MPTP, 12 h apart). For VEGF studies (n = 4 per group), transgenic mice expressing human VEGF driven by the rat neuron-specific enolase promoter were maintained on a C57BL/6 background (kindly provided by Dr. David Greenberg (12)). Age-matched non-transgenic controls (10 weeks of age) were also treated with either 2 × 20 mg/kg MPTP or saline, 12 h apart. Seven days following the final MPTP or saline injection in the 3 different paradigms, mice were sacrificed for either tissue harvest for biochemical assays or brain fixation via intra-cardiac perfusion for immunohistochemistry.
Stereological Assessments
A subset of mice (n = 4 per condition) were subject to cardiac perfusion with phosphate-buffered saline followed by 4% paraformaldehyde. Brains were then removed, dehydrated in 30% sucrose, and sectioned at 20 μm. Immunohistochemistry was performed using antibody against tyrosine hydroxylase (1:500 TH; Chemicon, Temecula, CA) followed by biotin-labeled secondary antibody and development using 3,3′ diaminobenzidine (Vector Labs, Burlingame, CA) to immunostain DAergic neurons throughout the SNpc. TH-positive cells were counted stereologically using the optical fractionator method (13). Striatal sections stained with TH primary antibodies were subject to densitometric analysis using Scion Image.
HPLC Assay for Dopamine and Metabolites
Dissected striata were sonicated and centrifuged in chilled 0.1 m perchloric acid (about 100 μl/mg tissue). The supernatants were taken for measurements of dopamine and its metabolites 3,4-dihydroxyphenylacetic and homovanillic acid by HPLC as described in our previously described method (14, 15). Briefly, 15 μl of supernatant was isocratically eluated through an 80 × 4.6-mm C18 column (ESA, Inc., Chelmsford, MA) with a mobile phase containing 0.1 m LiH2PO4, 0.85 mm 1-octanesulfonic acid, and 10% (v/v) methanol and detected by a 2-channel Coulochem II electrochemical detector (ESA, Inc.). Concentrations of dopamine, 3,4-dihydroxyphenylacetic, and homovanillic acid are expressed as nanograms per milligram of protein. The protein concentrations of tissue homogenates were measured according to the Bio-Rad protein analyze protocol (Bio-Rad Laboratories) and PerkinElmer Life Sciences BioAssay Reader (Norwalk, CT).
Dynamic Array RT-PCR
Using the standard animal tissue protocol from Qiagen, each experimental sample was prepped for two-step RT-PCR (Ambion, Foster City, CA) and pre-amplification of each cDNA (Applied Biosystem, Foster City, CA). Using the Roche Universal ProbeLibrary Assay Design Center, forward and reverse primers for each gene of interest were designed (Operon). Two experimental samples were chosen as normalizing standards to be used on every reaction plate for the experiment. Potential housekeeping genes were profiled against 15 samples spanning the experimental set, using the 48.48 Dynamic Array “BioMark” Real Time PCR Plate Set-up and Protocol (Fluidigm, South San Francisco, CA). From this, 2 genes that showed little variance across the samples were chosen as the “housekeepers” for the experiment (ribosomal protein S20 and signal recognition particle 14). For the experiment, each gene (“assay”) of interest was represented by a UPL probe (Universal Probe Library, Roche), plus the Roche designed forward and reverse primers from the ×100 primer stock. Thermal cycle conditions for the PCR reaction were: 1 cycle at 50 °C for 2 min; 1 cycle at 95 °C for 10 min; and 40 cycles at 95 °C for 15 s, 70 °C for 5 s, and 60 °C for 1 min. Normalization was then carried out using both the inter-run calibration (standards), as well as the two housekeeping genes to derive the relative normalized quantity for each gene.
Immunoblot and Immunohistochemical Analyses
Midbrain sections containing the substantia nigra (SN) were dissected out and homogenized in phosphate-buffered saline containing 0.3% Triton, a mixture of protease inhibitors (Sigma), and 5 mm EDTA and processed with the NE/PER nuclear and cytosolic protein extraction kit (Pierce). Protein concentrations were determined using the Bradford assay (Bio-Rad). 100-μg protein samples were separated on 4–12% bisacrylamide gels before transfer to polyvinylidene difluoride membranes. Membranes were blocked with 5% powdered milk solution in 0.3% Triton/phosphate-buffered saline solution before incubation with 1:1000 HIF-1α (Upstate, Lake Placid, NY), 1:2000 HO-1 (Stressgen, Ann Arbor, MI), 1:2000 MnSOD (Novus, Littleton, CO), and 1:1000 vascular endothelial growth factor (VEGF, Santa Cruz Biotechnology, Santa Cruz, CA). Actin (1:2000, Sigma) was used as a loading control. Protein bands were detected via chemiluminescence substrate for horseradish peroxidase (Amersham Biosciences) and resulting bands were scanned and densitometry measured by Scion Image. To determine co-localization of various proteins, 7-μm sections of the SN from paraffin-embedded brains were cut and processed for staining. Sections were mounted onto slides and processed in a 10 mm citrate buffer for enhancement of antigen retrieval. After blocking with 10% donkey serum for 1 h, specific primary antibodies (1:200 HIF-1α, Novus; 1:1000 HIF-2α, Novus; 1:1000 TH, Chemicon; 1:200 ferroportin (FPT), Alpha Diagnostic, San Antonio, TX; 1:500 MnSOD, Novus; 1:500 VEGF, Santa Cruz Biotechnology) were applied to the sections for overnight incubation at 4 °C. Alexa-conjugated secondary antibodies were used for fluorescence detection of proteins. For staining of dopaminergic nerve terminals, striatal sections were stained with 1:1000 TH followed by biotinylated secondary antibody and 3,3′ diaminobenzidine processing. Images were captured on a Zeiss LSM 510 confocal microscope.
Perl Iron Staining
7-μm sections of SN from paraffin-embedded brains were used for Perls iron staining. SN sections were incubated for 2 h in 7% potassium ferrocyanide, in aqueous hydrochloric acid (3%) at 37 °C and subsequently incubated in 0.75 mg/ml of 3,3′-diaminobenzidine and 0.015% H2O2 for 5 min.
Inductively Coupled Plasma-Mass Spectrometry (ICP-MS)
All sample processing and analyses were conducted using trace metal-clean techniques under HEPA-filtered air conditions to minimize contamination of the samples. Prior to analyses, brain samples (n = 3 per condition) were thawed, transferred to Teflon® vials, dried, and digested for 4–8 h in 2 ml of hot 16 n QHNO3, evaporated to dryness, and redissolved in 1 n QHNO3 for analyses (16). Total iron was measured using a Finnigan MAT Element magnetic sector ICP-MS.
Pryruvate Dehydrogenase Activity
Striatal tissue was resuspended in ice-cold isolation buffer (320 mm sucrose, 5 mm TES, and 1 mm EGTA, pH 7.4) followed by homogenization in a Dounce homogenizer. The homogenates were centrifuged at 1,000 × g for 5 min at 4 °C. Supernatant containing mitochondria was saved and the pellet resuspended in isolation buffer and re-homogenized. Centrifugation of the resuspended homogenate was repeated and the supernatants were pooled. The pooled supernatant was centrifuged at 10,000 × g for 10 min at 4 °C. The pellet containing the mitochondrion was resuspended in 50 μl of isolation buffer for pyruvate dehydrogenase (PDH) enzyme activity assay (MitoSciences, Eugene, OR) whereby the PDH enzyme was immunocaptured within the wells of the 96-well plate and the reduction of NAD+ to NADH was followed at 340 nm.
In Vitro Studies
Rat N27 SN DA-derived cells were cultured at 3% O2 to mimic physiological oxygen tension and treated with various PHD inhibitors (DHB and dimethyloxaloylglycine (DMOG), 200 μm) or the iron chelator (salicylaldehyde isonicotinoyl hydrazone (SIH), 100 μm) in the presence and absence of a subtoxic concentration of MPP+ (400 μm). Cells were fixed with 4% paraformaldehyde after 24 h co-treatment and processed for immunocytochemistry to determine nuclear translocation of HIF-1α. To determine intracellular iron accumulation, another subset of cells were labeled with 57Fe for 24 h, treated with PHD inhibitors ± MPP+, and then processed for ICP-MS analysis. Cell viability was determined by treating cells with a toxic concentration of MPP+ (800 μm) in the presence or absence of DHB and was measured via the microplate 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay whereby the formation of water-insoluble formazan salt is indicative of metabolic activity (Roche).
Statistical Analyses
Analyses were made by comparing means between treatment groups using Student's t-tests for paired data, with p < 0.05 considered significant.
RESULTS
PHD Inhibition Protects against MPTP-induced DAergic Cell Loss and Striatal Denervation
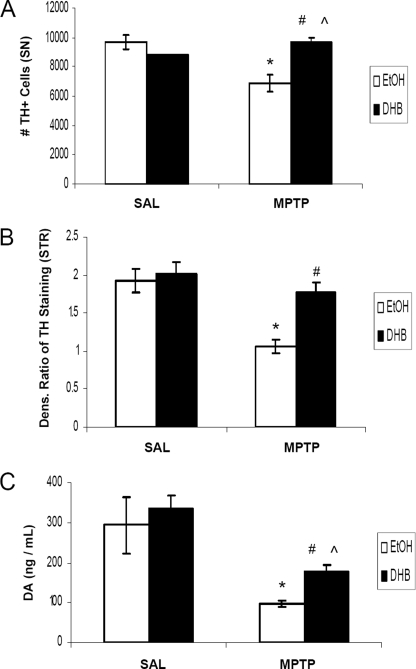
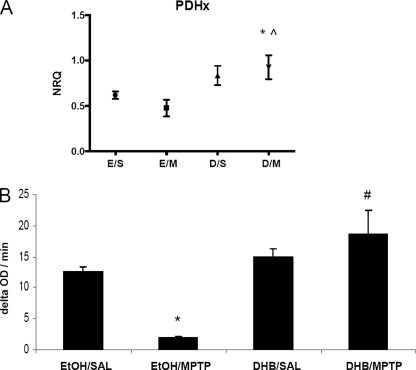
Based on the hypothesis that an inhibition of iron-dependent PHD activity could result in the accumulation of HIF-1α and a HIF-mediated activation of pro-survival gene products, it was postulated that DHB may reduce the extent of DAergic injury induced by systemic MPTP treatment in young adult mice. DHB is a low molecular weight inhibitor of PHD that displaces 2-oxoglutarate or ascorbate, co-factors required for PHD activity (17), and has been shown to reduce infarct volume induced by middle cerebral artery occlusion (10). Tyrosine hydroxylase (TH) is the rate-limiting enzyme involved in DA synthesis and positive staining of the cell for TH distinguishes DAergic neurons from other cellular populations. In our study, subacute MPTP administration (2 × 20 mg/kg body weight, 12 h apart) was found to result in a 30% loss in SNpc DAergic neurons in mice receiving only ethanol vehicle pre-treatment (Fig. 1A). Pretreatment with DHB for 6 h prior to initial MPTP administration, however, was found to result in complete protection against SN TH+ cell loss (9691.3 ± 483, EtOH/SAL; 6894.2 ± 566.8, EtOH/MPTP; 8765 ± 41, DHB/SAL; 9686.9 ± 252, DHB/MPTP; n = 4, *, p < 0.005 between EtOH/SAL and EtOH/MPTP; ⋀, p < 0.001 between DHB/SAL and DHB/MPTP; #, p < 0.001 between EtOH/MPTP and DHB/MPTP). Pre-treatment with DHB was also demonstrated to protect against loss of TH+ striatal terminals induced by MPTP (densitometric ratio depicted in Fig. 1B; 1.98 ± 0.15, EtOH/SAL; 1.06 ± 0.1, EtOH/MPTP; 2.01 ± 0.17, DHB/SAL; 1.77 ± 0.14, DHB/MPTP; *, p < 0.05 between EtOH/SAL and EtOH/MPTP; #, p < 0.05 between EtOH/MPTP and DHB/MPTP). Although DHB pre-treatment prior to MPTP did not completely revert ST DA to control levels, there was a significant increase in ST DA levels, measured by HPLC, compared with MPTP treatment alone (Fig. 1C; 294.10 ± 70.4 ng/ml, EtOH/SAL; 96.78 ± 7.35 ng/ml, EtOH/MPTP; 336.67 ± 32.71 ng/ml, DHB/SAL; 179.62 ± 13.74 ng/ml, DHB/MPTP; n = 7, *, p < 0.0001 between EtOH/SAL and EtOH/MPTP; ⋀, p < 0.0001 between DHB/SAL and DHB/MPTP; #, p < 0.0001 between EtOH/MPTP and DHB/MPTP).
FIGURE 1.
PHD inhibition protects against MPTP-induced DAergic cell loss and striatal denervation. Male C57BL/6 mice were treated with 5% ethanol (EtOH) as a vehicle control or 100 mg/kg of DHB 6 h prior to saline (SAL) or 2 × 20 mg/kg of MPTP (intraperitoneally, 12 h apart). Mice were sacrificed 7 days post-MPTP and processed for various assays measuring nigrostriatal DAergic cell integrity. A, stereological quantification of TH-positive DAergic cell counts within the SN. *, p < 0.05 between EtOH/SAL and EtOH/MPTP; ⋀, p < 0.05 between DHB/SAL and DHB/MPTP; #, p < 0.05 between EtOH/MPTP and DHB/MPTP (mean ± S.E.; n = 4). B, quantification of TH immunoreactivity in striatum sections following treatments. *, p < 0.05 between EtOH/SAL and EtOH/MPTP; #, p < 0.05 between EtOH/MPTP and DHB/MPTP (mean ± S.E.; n = 3). C, neurochemical analysis of DA within ST terminals. *, p < 0.0001 between EtOH/SAL and EtOH/MPTP; ⋀, p < 0.0001 between DHB/SAL and DHB/MPTP; #, p < 0.0001 between EtOH/MPTP and DHB/MPTP (mean ± S.E.; n = 7).
HIF-1α Levels Are Increased in DAergic SN Neurons by DHB Pretreatment and Maintained following MPTP
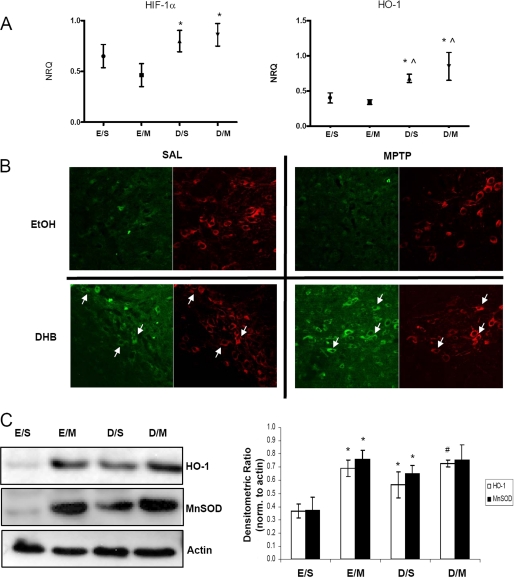
RT-PCR analyses of multiple genes of interest were performed on midbrain SN tissues obtained from mice at various time points following the various treatments on a microfluidics system (Fluidigm, San Francisco, CA). We observed the most dramatic changes in gene expression at 6–12 h post-treatment. At this time point, DHB pre-treatment alone or in combination with MPTP was found to result in increased expression of several genes related to energy metabolism, mitochondrial function, iron regulation, or transcriptional control compared with controls (SAL or MPTP alone). Gene expression of Hif-1α and its downstream target Ho-1 was found to be elevated under conditions of DHB pre-treatment and maintained following MPTP administration (Fig. 2A). Immunohistochemical analyses of brains from mice harvested 24 h post-treatment was performed to determine whether HIF-1α protein levels increased within DAergic SN neurons following DHB +/− MPTP (Fig. 2B). In the absence of DHB and/or MPTP, very little basal HIF-1α staining was observed. However, following pre-treatment with DHB alone or in conjunction with MPTP administration, HIF-1α was found to increase within DAergic (TH+) SN neurons including in the nucleus (data not shown). To further assess activation of the HIF pathway by DHB, midbrain SN protein homogenates from the various experimental conditions (8 days post-treatment) were assessed via Western blot analysis. Protein levels of HO-1 were observed to be similarly elevated following MPTP, in the presence and absence of DHB pretreatment, even at 8 days following the last MPTP injection (Fig. 2B; for HO-1, *, p < 0.05 significantly different from EtOH/SAL; #, p < 0.05 significantly different from DHB/SAL).
FIGURE 2.
HIF-1α and its downstream target HO-1 are up-regulated by DHB in the SN; up-regulation is maintained in the presence of MPTP. A, RT-PCR analysis of Hif-1α and its downstream target Ho-1 were performed on midbrain SN tissue obtained 6 h post-treatment; means ± S.D. are shown. EtOH/SAL (E/S), EtOH/MPTP (E/M), DHB/SAL (D/S), and DHB/MPTP (D/M). *, p < 0.05 significantly different from EtOH/MPTP; ⋀, p < 0.05 significantly different from EtOH/SAL (n = 4). B, 7-μm paraffin-embedded sections of SN, obtained from mice harvested at 24 h post-treatment, were stained for HIF-1α (green) and TH (red); white arrows point to HIF-1α localized within representative DAergic SN neurons. C, 100 μg of total protein extracts prepared from midbrain SN homogenates were loaded onto 4–12% BisTris gels and electrophoresed prior to transfer onto polyvinylidene difluoride membranes and subsequently immunoblotted for HO-1 and MnSOD; actin was used as a loading control, a representative blot is shown in the left panel and quantification of at least 3 separate immunoblots (means ± S.E.) is shown in right panel. *, p < 0.05 significantly different from EtOH/SAL; #, p < 0.05 significantly different from DHB/SAL.
Up-regulation of HIF-2α and MnSOD Levels
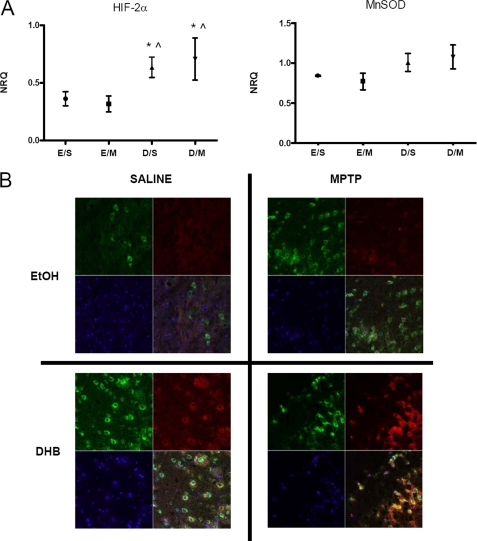
Hif-2α (encoded by the Epas1 gene) is a structurally related isoform of Hif-1α (18). Although both isoforms are stabilized by hypoxia, they have different sets of target genes and as such confer different biological roles (19–21). Similar to Hif-1α, we found that gene expression of Hif-2α was elevated under conditions of DHB pre-treatment and maintained following MPTP administration (Fig. 3A). Although mRNA levels of midbrain SN Ho-1 were found to be significantly up-regulated with DHB pre-treatment in the presence and absence of MPTP (Fig. 2A), alterations in the mRNA levels of Mnsod were found to only be approaching significance (p < 0.08) (Fig. 3A). Mice that received DHB pre-treatment showed significant elevations of HIF-2α and MnSOD protein in the SN that was maintained or augmented in the presence of MPTP administration (see Figs. 2C and 3B).
FIGURE 3.
HIF-2α and its downstream target MnSOD, are up-regulated by DHB in the SN; up-regulation is maintained in the presence of MPTP. A, RT-PCR analyses of Hif-2α and mitochondrial Mnsod were performed on midbrain SN tissue obtained 6 h post-treatment; means ± S.D. are shown. EtOH/SAL (E/S), EtOH/MPTP (E/M), DHB/SAL (D/S), and DHB/MPTP (D/M). *, p < 0.05 significantly different from EtOH/MPTP; ⋀, p < 0.05 significantly different from EtOH/SAL (n = 4). B, 7-μm paraffin-embedded sections of SN, obtained from mice harvested at 24 h post-treatment, were stained for MnSOD (green), HIF-2α (red), and nuclear DAPI (blue); yellow staining indicates co-localization of MnSOD and HIF-2α within the same cell.
PHD Inhibition by DHB Prevents MPTP-induced Losses in SN Levels of Ferroportin and Accumulation of Nigral and Striatal Iron
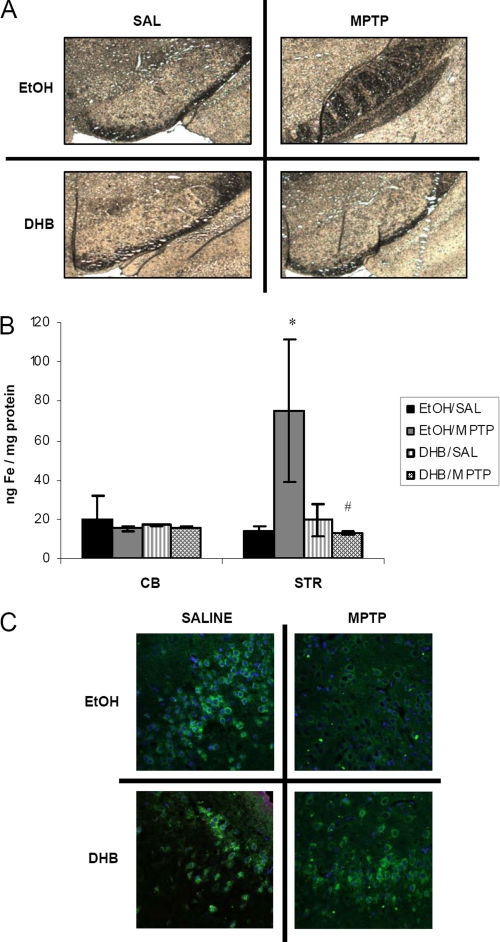
Dopaminergic cell loss in the SNpc is reported to be accompanied by overaccumulation of unsequestered iron (22, 23). To assess whether alterations in iron regulatory proteins as observed following DHB in our MPTP model coincide with any functional consequences, iron levels in the SN, striatum, and cerebellum were assessed in the various experimental conditions. Nigral ferric iron levels were assessed by Perl staining (Fig. 4A). Consistent with previous findings (13), we observed an elevation of iron in MPTP-treated mice that was attenuated in the presence of DHB. Because the striatum also includes DA nerve terminals that project from the SN, iron levels of the ST were also measured by ICP-MS (Fig. 4B). Although, as previously demonstrated (24, 25), MPTP treatment alone was found to result in a significant increase in striatal iron levels, this was found to be abolished in the presence of DHB pretreatment. An absence of alterations in iron levels in the cerebellum confirms the regional specificity of the effects of MPTP neurotoxicity on the nigrostriatal system in vivo.
FIGURE 4.
PHD inhibition by DHB attenuates MPTP-induced accumulation of nigrostriatal iron and decreases in ferroportin. A, nigral sections were used for Perls iron staining using 7% potassium ferrocyanide. B, striatal (STR) and cerebellar (CB) tissue were analyzed for total iron levels by ICP-MS (mean ± S.D.; n = 4). *, p < 0.05 between EtOH/SAL and EtOH/MPTP; #, p < 0.05 between EtOH/MPTP and DHB/MPTP. C, 7-μm paraffin-embedded sections of SN obtained from treated mice harvested at 24 h post-treatment were immunostained for ferroportin (green) and 4′,6-diamidino-2-phenylindole (blue).
Another important protein known to be involved in regulation of cellular iron levels is the iron export protein FPT, which is indirectly regulated by HIF. In the presence of cytosolic HIFα, hepcidin is down-regulated and prevented from binding to and internalizing FPT (26, 27), thereby increasing the levels of the iron exporter. MPTP treatment alone was observed to result in a decrease of ferroportin levels within the SN (Fig. 4C). However, in the presence of DHB pre-treatment, protein expression of FPT was found to return to control levels.
DHB Prevents MPTP-mediated Reduction in the Activity of the Mitochondrial Tricarbocylic Acid Cycle Enzyme PDH
PDH is responsible for the conversion of pyruvate to acetyl-CoA as substrate for the tricarbocylic acid cycle necessary for continued mitochondrial respiration (28, 29). We observed in conjunction with our gene expression analyses that MPTP reduces the message levels of Pdh 6–12 h post-treatment and that this is restored by DHB pre-treatment (Fig. 5A, *, p < 0.05 between EtOH/SAL and DHB/MPTP; ⋀, p < 0.05 between EtOH/MPTP and DHB/MPTP). To assess the possible long-term functional consequences, we measured PDH activity at 8 days post-MPTP. MPTP alone was found to result in a significant reduction in PDH activity levels, whereas pre-treatment with DHB completely attenuated this decrease (Fig. 5B, *, p < 0.0001 between EtOH/SAL and EtOH/MPTP; #, p < 0.05 between EtOH/MPTP and DHB/MPTP).
FIGURE 5.
DHB prevents MPTP-mediated reduction in pyruvate dehydrogenase mRNA and activity levels. A, RT-PCR analysis of Pdh was performed on midbrain SN tissue obtained 12 h post-treatment; means ± S.D. are shown. EtOH/SAL (E/S), EtOH/MPTP (E/M), DHB/SAL (D/S), and DHB/MPTP (D/M). *, p < 0.05 significantly different from EtOH/SAL; ⋀, p < 0.05 significantly different from EtOH/MPTP (n = 4). B, PDH activity was measured in ST homogenates obtained 8 days post-treatment (means ± S.E.). *, p < 0.0001 between EtOH/SAL and EtOH/MPTP; #, p < 0.05 between EtOH/MPTP and DHB/MPTP (n = 3).
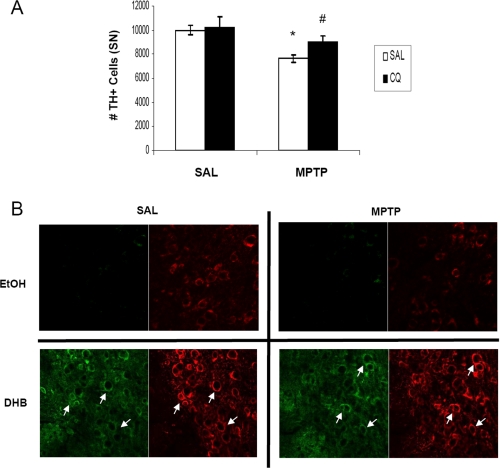
Short-term Pre-treatment with the Bioavailable Iron Chelator CQ Results in Increased HIF-1α within DAergic Neurons and Protects against MPTP-induced Nigral Cell Death
Iron is an important co-factor required for proper functioning of PHD to regulate HIF-α levels (30). Chelation of iron has been shown to inhibit PHD activity and elevate cytosolic accumulation of HIF-α (31). Previous studies in our laboratory have demonstrated that pre-treatment with the iron chelating compound CQ prevents MPTP-induced neurotoxicity by sequestering redox-active iron (13). To determine whether pharmacological iron chelation by CQ could be acting in part via activation of the HIF pathway, mice were pretreated for a shorter regime (2 weeks) of CQ administration than previously published (2 months) prior to MPTP treatment. Compared with SAL-fed mice, CQ-fed mice lost 50% less SN TH+ neurons in response to MPTP (Fig. 6A, 9993.5 ± 401, SAL/SAL; 7643.5 ± 325, SAL/MPTP; 10243.5 ± 830.7, CQ/SAL; 9006.7 ± 531, CQ/MPTP; n = 4, *, p < 0.01, #, p < 0.05). Similar to DHB treatment in vivo (Fig. 2B), CQ administration alone and in the presence of MPTP elicited stabilization of cytosolic HIF-1α (Fig. 6B) suggesting that activation of the HIF pathway by CQ could in part underlie the protection against DAergic cell loss observed in various studies utilizing iron chelation therapy in MPTP PD models (13, 32).
FIGURE 6.
The bioavailable iron chelator clioquinol protects against MPTP-induced nigral cell death and stabilizes HIF-1α accumulation within DAergic neurons. A, quantification of TH-positive cell counts. *, p < 0.05 between SAL/SAL and SAL/MPTP; #, p < 0.05 between CQ/SAL and CQ/MPTP (mean ± S.E.; n = 4). B, 20-μm sections of SN obtained from control and CQ-fed mice harvested at 8 days post-SAL/MPTP were stained for HIF-1α (green) and TH (red); white arrows point to HIF-1α co-localized to SN DAergic neurons.
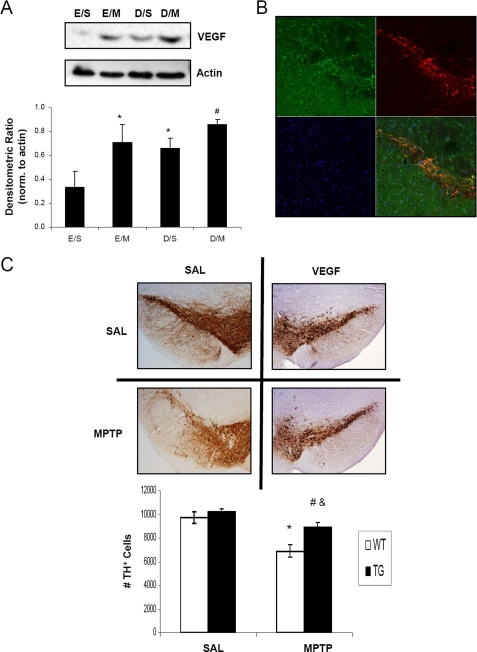
VEGF Overexpression Protects against MPTP-induced Nigral Cell Death
VEGF is an important downstream target of HIF-1α that has been implicated not only for angiogenic processes in stroke models (33, 34) but also in the protection of motor neurons in an amyotrophic lateral sclerosis mouse model (12). Direct intra-striatal infusion and implantation of capsules containing VEGF (35) and gene transfer of VEGF using adenoassociated viral vectors (36) have also been demonstrated to rescue DAergic neurons from 6-hydroxydopamine toxicities in rodent models of PD. In our studies, we utilized a mouse model of pan-neuronal overexpression of VEGF to determine whether VEGF would also provide protection against MPTP toxicity in our model. VEGF protein levels were observed, by immunoblot analysis of midbrain SN homogenates, to be elevated under conditions of MPTP treatment and PHD inhibition by DHB (Fig. 7A), consistent with the effects of HIF-1α on VEGF (37, 38). VEGF protein was observed to co-localize to nigral DAergic neurons in the transgenic mice (Fig. 7B) and the overexpression of VEGF was also able to protect against MPTP neurotoxicity (Fig. 7C, 9691.3 ± 483, SAL/SAL; 6894.2 ± 566, SAL/MPTP; 10239.9 ± 240, VEGF/SAL; 8931.2 ± 351, VEGF/MPTP; n = 4; *, p < 0.01 between SAL/SAL and SAL/MPTP; #, p < 0.05 between VEGF/SAL and VEGF/MPTP; &, p < 0.05 SAL/MPTP and VEGF/MPTP).
FIGURE 7.
VEGF overexpression protects against MPTP-induced nigral cell death. A, 100 μg of total protein extracts prepared from midbrain SN homogenates were loaded onto 4–12% BisTris gels and electrophoresed prior to transfer onto polyvinylidene difluoride membranes and subsequent immunoblotting for VEGF; actin was used as a loading control, a representative blot is shown in upper panel and quantification of at least 3 separate immunoblots (means ± S.E.) is shown in the lower panel. *, p < 0.05 significantly different from EtOH/SAL; #, p < 0.05 significantly different from DHB/SAL. B, 20-μm sections of SN obtained from VEGF transgenic mice were stained for VEGF (green) and TH (red); overlay of VEGF and TH staining demonstrates co-localization of VEGF in DAergic neurons (yellow-orange). C, brain scans show TH-stained DAergic neurons of saline (SAL)-treated mice from the wild-type and VEGF transgenic groups (upper scans). Bottom scans show the mice from the corresponding groups treated with MPTP and then allowed to recover for 7 days before sacrifice. Quantification of TH-positive cell counts is depicted in the graph. *, p < 0.05 between SAL/SAL and SAL/MPTP; &, p < 0.05 between VEGF WT/MPTP and VEGF TG/MPTP; #, p < 0.05 between VEGF TG/SAL and VEGF TG/MPTP (mean ± S.E.; n = 4).
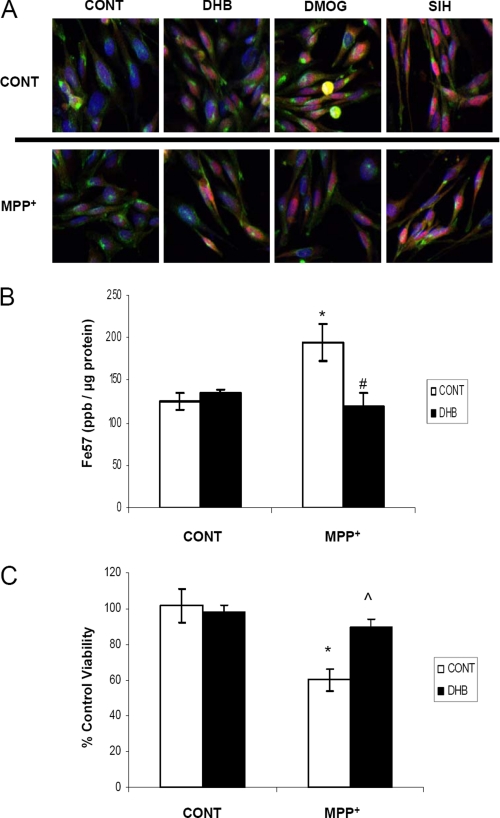
In Vivo Results Are Recapitulated in an SN DA-derived Cell Model
Nigrostriatal homogenates used for RT-PCR, Western blot, and biochemical analyses contain a mixed population of cell types and nerve terminals. To verify that the effects we observed in vivo following DHB administration in our MPTP PD model coincide with alterations within DAergic cell types, we turned to an in vitro midbrain-derived dopaminergic cell line, N27. Additionally, although many in vitro experiments utilize cells grown in 21% O2 in humidified incubators, cultivation of cells at 3% O2 represents a more physiologically relevant paradigm because the oxygen tension in most mammalian tissues range from 1 to 6% O2 and so we chose to grow our N27 cells under these conditions (39). Dopaminergic N27 cells were treated with a subtoxic concentration of 400 μm MPP+ (the bioactive metabolite of MPTP) either in the presence or absence of PHD inhibitors (DHB and DMOG) or an iron chelator (SIH). DHB, DMOG, and SIH all were found to elicit translocation of HIF-1α into the nucleus and this was sustained in the presence of MPP+ (Fig. 8A). Concurrent with these data, we observed that N27 cells transfected with hypoxia responsive element-luciferase reporter plasmid also displayed HIF activation following treatment with PHD inhibitors in both the presence and absence of MPP+ (data not shown). Measurement of total iron levels by ICP-MS in N27 cells grown in normoxic conditions demonstrated, as observed in striatal homogenates in vivo, that whereas MPP+ elicited an increase in total intracellular iron, this was attenuated in the presence of DHB (Fig. 8B, *, p < 0.05 between control/control and DHB/control; #, p < 0.05 between control/MPP+ and DHB/MPP+). Although iron chelators have been shown to be protective against MPP+ neurotoxicity (24, 40), it was determined that PHD inhibition by DHB was also able to attenuate cell death associated with a toxic concentration of MPP+ (Fig. 8C, *, p < 0.005 between control/control and control/MPP+; ⋀, p < 0.001 between control/MPP+ and DHB/MPP+), results that parallel those observed in vivo (Fig. 1A).
FIGURE 8.
In vitro studies in SN DA-derived N27 cells. A, N27 cells grown in 3% O2 were treated with 200 μm DHB, 200 μm DMOG, or 100 μm SIH in the presence of 400 μm MPP+ for 24 h. Cells were then fixed with 4% paraformaldehye and stained with transferrin receptor (green), HIF-1α (red), and 4′,6-diamidino-2-phenylindole (nuclear marker, blue); pink staining represents nuclear localization of HIF-1α. B, cells were labeled with 57Fe for 24 h, treated with 200 μm DHB ± 400 μm MPP+, and then digested with nitric acid for ICP-MS analysis of total iron measurements (mean ± S.D.; n = 4). Asterisk denotes p < 0.05 between control/control and DHB/control; # denotes p < 0.05 between control/MPP+ and DHB/MPP+. C, cells were treated with 800 μm MPP+ for 24 h in the presence and absence of 200 μm DHB. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was used as a measure of cell viability. *, p < 0.005 between control/control and control/MPP+; ⋀, p < 0.001 between control/MPP+ and DHB/MPP+ (mean ± S.D.; n = 4).
DISCUSSION
MPTP is a pro-toxicant whose systemic administration results in selective destruction of DAergic SNpc neurons in both primates and rodents resulting in an acute Parkinsonism phenotype; it is currently a widely used model for the disease (41). MPTP has been proposed to exert its neurotoxic effects via selective inhibition of mitochondrial complex I activity resulting in both a reduction in ATP synthesis and accumulation of reactive oxygen species (42). It is also accompanied by the specific accumulation of SN iron levels similar to that observed in the human disease state (43). In earlier studies from our laboratory (13), we demonstrated that iron chelation was effective in significantly attenuating MPTP-induced DAergic nigral neurodegeneration via overexpression of the iron-storage protein ferritin in DAergic neurons and pharmacological chelation of iron by the blood-brain barrier-miscible metal chelator clioquinol. Specific potential mechanism(s) involved in the protective effects of iron, however, were not explored. Prolyl hydroxylases are iron-dependent enzymes that reduce levels of HIFs via their ability to hydroxylate these proteins, marking them for subsequent degradation by the ubiquitin-proteasome system. Reduction in HIF signaling pathways by PHDs results in decreased transcription of several genes whose protein products have been shown to be protective against either oxygen depletion or oxidative stress within the brain (10, 44, 45). Because PHDs require the presence of iron, it is possible that iron chelation therapy is in part protective against MPTP neurotoxicity by resulting in decreased PHD activity, which can contribute to the induction of protective HIF-dependent genes including those involved in cellular iron regulation and protection against oxidative stress and mitochondrial dysfunction. To test this hypothesis, we assessed the impact of the general PHD inhibitor DHB on MPTP neurotoxicity. We report here that pre-treatment with this agent results in not only maintained up-regulation of the Hif-1α message and protein and HIF-induced gene products in the presence of MPTP administration but, importantly, prevents MPTP-induced DAergic nigral and striatal neurodegeneration.
Transgenic mice overexpressing MnSOD have previously been shown to be more resistant to MPTP toxicity (49). MnSOD, well characterized to be involved in elimination of mitochondrial superoxide and demonstrated to be HIF-induced (50), was found in this study to be up-regulated by PHD inhibition, whereby the co-localization of MnSOD with HIF-2α was observed in the SN even in the presence of MPTP (Fig. 3B). Although HO-1 activation has been associated with both protective and detrimental consequences, the effects of HO-1 are highly dependent upon the duration, intensity of protein induction, and the specific cellular microenvironment. HO-1-deficient mice and fibroblasts are less protected against oxidative stress from paraquat and hydrogen peroxide (51) and overexpression of HO-1 has been shown to protect against hyperoxia-mediated cellular injury (52) and cisplatin-induced nephrotoxicity (53). HO-1 has been demonstrated to be involved in cellular iron efflux (54). Indeed in our study, elevations in HO-1 levels via DHB treatment (Fig. 2C) corresponds with an attenuation in nigral and striatal iron accumulation as a result of MPTP administration (Fig. 4, A and B). Iron efflux by ferroportin also contributes to the homeostatic regulation of intracellular iron levels (55–57). Similar to recent findings (58), MPTP was found to reduce expression of ferroportin within the SN (Fig. 4C). The reduction in ferroportin and its ability to export iron probably contributes to the MPTP-induced accumulation of iron observed in previous MPTP studies. Given that ferroportin is intricately regulated by hepcidin (27), the stabilization of HIFα by DHB, even in the presence of MPTP, could promote iron export and prevent neurodegeneration associated with MPTP. Similar to previous studies, divalent metal transporter-1 was observed to be up-regulated by MPTP (59) and this increase was sustained even in the presence of DHB (data not shown). Recent studies suggest that overexpression of divalent metal transporter-1 alone is associated with intracellular iron accumulation (60) and MPTP-induced apoptosis (61), however, these detrimental effects appear to be overridden by other HIF-related events in our system.
The activity of the mitochondrial PDH complex (PDC) is regulated by pyruvate dehydrogenase kinase, a direct target of HIF-1α (62). Stabilization of HIF-1α can activate pyruvate dehydrogenase kinase to phosphorylate and inactivate the PDC complex. In this way, a metabolic switch occurs whereby oxidative phophorylation is shunted to a glycolytic pathway to maintain energy demands while preventing reactive oxygen species production (28). Interestingly, in our model, PDH activity is inhibited by MPTP treatment and this inhibition is restored by DHB treatment (Fig. 5), which would presumably act to contribute to restoration of mitochondrial function. As part of our RT-PCR analysis we have preliminary evidence that expression of other genes involved in mitochondrial function including uncoupling protein 2 (Ucp2) and peroxisome proliferator-activated receptor-γ coactivator 1α are also down-regulated in the presence of MPTP but restored to above basal levels when pre-treated with DHB. Of particular interest is the involvement of proliferator-activated receptor-γ coactivator 1α in the activation of mitochondrial biogenesis and respiration (63), which could perhaps provide a potential mechanism underlying the restoration of PDH activity by DHB treatment.
Pharmacological chelation of iron can effectively sequester co-factor that is necessary for PHD activity and subsequent induction of HIF as demonstrated via our in vivo studies using the iron chelator CQ. Activation of the HIF pathway by iron chelation may also be one of the mechanisms underlying the neuroprotection against MPTP observed in other studies including following ferritin overexpression in DAergic SN neurons (13). Because of its role in angiogenesis, VEGF has primarily been considered to elicit a protective effect in such conditions as stroke, tumor growth, and tissue grafting via its involvement in vascular remodeling (64, 65). However, previous studies have implicated VEGF in the rescue of motor neuron function in an amyotrophic lateral sclerosis mouse model (12) as well as protecting DAergic neurons from 6-hydroxydopamine toxicity in rat models of PD (36). Although it is not surprising that neuronal overexpression of VEGF is able to protect mice against MPTP neurotoxicity (Fig. 7C), it is interesting to note that although VEGF expression is principally regulated by HIF, VEGF can also regulate HIF-1α levels as shown recently in cultured endothelial cells via superoxide-mediated signaling (66, 67). Thus, the utility of CQ-fed and VEGF transgenic mice in this study provided additional evidence that the HIF pathway may be activated in the absence of direct pharmacological inhibitors of PHD.
Depletion of HIF proteins in vivo has recently been demonstrated to also impact on catecholamine synthesis and mitochondrial metabolism suggesting that PHD inhibition may have pleiotropic protective effects as a consequence of HIF induction (46, 47). Recently, a set of elegant in vitro experiments set forth the premise that there are indeed cell-type specific roles for HIF-1α (48). Using co-cultures of neurons and astrocytes obtained from transgenic mice, it was demonstrated that selective depletion of neuronal HIF-1α resulted in reduced viability induced by hypoxia, whereas a selective depletion of astrocytic HIF-1α during hypoxia protected neurons.
Currently, some PHD inhibitors that activate the HIF pathway are being pursued for use in the treatment of anemia and tissue injury resulting from ischemia (33). To our knowledge, this study is the first to demonstrate, in both in vitro and in vivo systems, the beneficial aspects of PHD inhibition in a neurodegenerative model of PD, representing a novel therapeutic pathway in treatment of this disorder. Findings of these studies are illustrated in Fig. 9, The stabilization of HIFα transcription factors resulting from PHD inhibition can lead to the up-regulation of several proteins involved in promoting iron efflux (FPT, HO-1), DA synthesis (TH), and mitochondrial integrity and bioenergetics (MnSOD, PDH, proliferator-activated receptor-γ coactivator 1α), combating the neurotoxic effects associated with MPTP. Additional mechanistic studies are necessary to fully elucidate the potential players involved in the protective effect of PHD inhibition as well the relative contributions of HIF-1α and HIF-2α in conferring protection in the MPTP administration model.
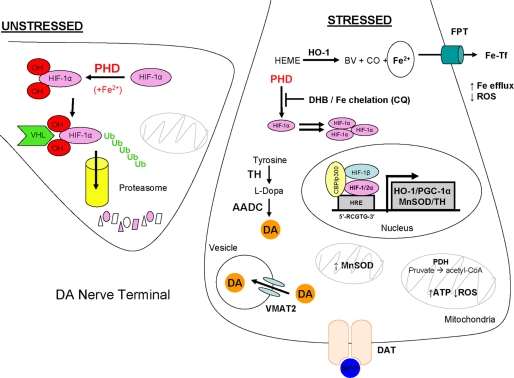
FIGURE 9.
Proposed model. Manipulation of the HIF pathway is proposed to result in induction of gene products that are neuroprotective against MPTP toxicity. Under conditions of non-stress and substrate availability, PHD hydroxylates proline residues on HIFα transcription factors to be targeted for proteasomal degradation. Stabilized HIFα arise from the inhibition of PHD brought about by either pharmacological sequestration of PHD substrates by DHB or chelation of iron as co-factor by CQ. Consequently, induction of genes containing HIF-responsive elements result in elevated expression of proteins responsible for maintaining catecholamine biosynthesis (TH), mitochondrial and/or respiratory function (peroxisome proliferator-activated receptor-γ coactivator, PGC-1α; MnSOD, PDH), intracellular iron homeostasis and oxidative stress (HO-1 and ferroportin (FPT)), cellular functions that are compromised in Parkinson disease and recapitulated in MPTP models of DA neurotoxicity.
Acknowledgments
We thank Krysta Felkey and Dr. Simon Melov (Buck Institute Genomics Core) for RT-PCR analyses on the Biomark.
This work was supported, in whole or in part, by National Institutes of Health Grant NS041264 (to J. K. A.). This work was also supported by the American Parkinson's Disease Association and Larry L. Hillblom Foundation fellowships (to D. W. L.).
- PD
- Parkinson disease
- HIF
- hypoxia inducible factor
- DA
- dopamine
- MPTP
- 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- DMOG
- dimethyloxaloylglycine
- DHB
- dihydroxybenzoate
- SIH
- salicylaldehyde isonicotinoyl hydrazone
- PHD
- prolyl 4-hydroxylase
- HO-1
- heme oxygenase-1
- SN
- substantia nigra
- ST
- striatum
- VEGF
- vascular endothelial growth factor
- MnSOD
- manganese superoxide dismutase
- TH
- tyrosine hydroxylase
- MPTP
- 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- CQ
- clioquinol
- HPLC
- high pressure liquid chromatography
- RT
- reverse transcriptase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- FPT
- ferroportin
- ICP-MS
- inductively coupled plasma-mass spectrometry
- PDH
- pyruvate dehydrogenase
- SNpc
- substantia nigra pars compacta.
REFERENCES
- 1.Haavik J., Almås B., Flatmark T. (1997) J. Neurochem. 68, 328–332 [DOI] [PubMed] [Google Scholar]
- 2.Cohen G., Farooqui R., Kesler N. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4890–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham D. G., Tiffany S. M., Bell W. R., Jr., Gutknecht W. F. (1978) Mol. Pharmacol. 14, 644–653 [PubMed] [Google Scholar]
- 4.Ward R. J., Dexter D., Florence A., Aouad F., Hider R., Jenner P., Crichton R. R. (1995) Biochem. Pharmacol. 49, 1821–1826 [DOI] [PubMed] [Google Scholar]
- 5.Connor J. R., Menzies S. L. (1995) J. Neurol. Sci. 134, (suppl.) 33–44 [DOI] [PubMed] [Google Scholar]
- 6.Tanaka M., Sotomatsu A., Kanai H., Hirai S. (1991) J. Neurol. Sci. 101, 198–203 [DOI] [PubMed] [Google Scholar]
- 7.Hirsilä M., Koivunen P., Günzler V., Kivirikko K. I., Myllyharju J. (2003) J. Biol. Chem. 278, 30772–30780 [DOI] [PubMed] [Google Scholar]
- 8.Hurn P. D., Koehler R. C., Blizzard K. K., Traystman R. J. (1995) Stroke 26, 688–694; discussion 694–685 [DOI] [PubMed] [Google Scholar]
- 9.Hamrick S. E., McQuillen P. S., Jiang X., Mu D., Madan A., Ferriero D. M. (2005) Neurosci. Lett. 379, 96–100 [DOI] [PubMed] [Google Scholar]
- 10.Siddiq A., Ayoub I. A., Chavez J. C., Aminova L., Shah S., LaManna J. C., Patton S. M., Connor J. R., Cherny R. A., Volitakis I., Bush A. I., Langsetmo I., Seeley T., Gunzler V., Ratan R. R. (2005) J. Biol. Chem. 280, 41732–41743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baranova O., Miranda L. F., Pichiule P., Dragatsis I., Johnson R. S., Chavez J. C. (2007) J. Neurosci. 27, 6320–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Mao X. O., Xie L., Banwait S., Marti H. H., Greenberg D. A., Jin K. (2007) J. Neurosci. 27, 304–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaur D., Yantiri F., Rajagopalan S., Kumar J., Mo J. Q., Boonplueang R., Viswanath V., Jacobs R., Yang L., Beal M. F., DiMonte D., Volitaskis I., Ellerby L., Cherny R. A., Bush A. I., Andersen J. K. (2003) Neuron 37, 899–909 [DOI] [PubMed] [Google Scholar]
- 14.Beal M. F., Matson W. R., Swartz K. J., Gamache P. H., Bird E. D. (1990) J. Neurochem. 55, 1327–1339 [DOI] [PubMed] [Google Scholar]
- 15.Beal M. F., Matson W. R., Storey E., Milbury P., Ryan E. A., Ogawa T., Bird E. D. (1992) J. Neurol. Sci. 108, 80–87 [DOI] [PubMed] [Google Scholar]
- 16.Smith D. R., Flegal A. R. (1992) Toxicol. Appl. Pharmacol. 116, 85–91 [DOI] [PubMed] [Google Scholar]
- 17.Majamaa K., Günzler V., Hanauske-Abel H. M., Myllylä R., Kivirikko K. I. (1986) J. Biol. Chem. 261, 7819–7823 [PubMed] [Google Scholar]
- 18.Luo G., Gu Y. Z., Jain S., Chan W. K., Carr K. M., Hogenesch J. B., Bradfield C. A. (1997) Gene Expr. 6, 287–299 [PMC free article] [PubMed] [Google Scholar]
- 19.Wiesener M. S., Turley H., Allen W. E., Willam C., Eckardt K. U., Talks K. L., Wood S. M., Gatter K. C., Harris A. L., Pugh C. W., Ratcliffe P. J., Maxwell P. H. (1998) Blood 92, 2260–2268 [PubMed] [Google Scholar]
- 20.Talks K. L., Turley H., Gatter K. C., Maxwell P. H., Pugh C. W., Ratcliffe P. J., Harris A. L. (2000) Am. J. Pathol. 157, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warnecke C., Zaborowska Z., Kurreck J., Erdmann V. A., Frei U., Wiesener M., Eckardt K. U. (2004) FASEB J. 18, 1462–1464 [DOI] [PubMed] [Google Scholar]
- 22.Dexter D. T., Wells F. R., Lees A. J., Agid F., Agid Y., Jenner P., Marsden C. D. (1989) J. Neurochem. 52, 1830–1836 [DOI] [PubMed] [Google Scholar]
- 23.Martin W. R., Wieler M., Gee M. (2008) Neurology 70, 1411–1417 [DOI] [PubMed] [Google Scholar]
- 24.Lan J., Jiang D. H. (1997) J. Neural. Transm. 104, 469–481 [DOI] [PubMed] [Google Scholar]
- 25.He Y., Thong P. S., Lee T., Leong S. K., Mao B. Y., Dong F., Watt F. (2003) Free Radic. Biol. Med. 35, 540–547 [DOI] [PubMed] [Google Scholar]
- 26.Lesbordes-Brion J. C., Viatte L., Bennoun M., Lou D. Q., Ramey G., Houbron C., Hamard G., Kahn A., Vaulont S. (2006) Blood 108, 1402–1405 [DOI] [PubMed] [Google Scholar]
- 27.Viatte L., Lesbordes-Brion J. C., Lou D. Q., Bennoun M., Nicolas G., Kahn A., Canonne-Hergaux F., Vaulont S. (2005) Blood 105, 4861–4864 [DOI] [PubMed] [Google Scholar]
- 28.Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 29.Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C. (2006) Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 30.Martín-Puig S., Temes E., Olmos G., Jones D. R., Aragonés J., Landázuri M. O. (2004) J. Biol. Chem. 279, 9504–9511 [DOI] [PubMed] [Google Scholar]
- 31.Callapina M., Zhou J., Schnitzer S., Metzen E., Lohr C., Deitmer J. W., Brüne B. (2005) Exp. Cell Res. 306, 274–284 [DOI] [PubMed] [Google Scholar]
- 32.Youdim M. B., Stephenson G., Ben Shachar D. (2004) Ann. N.Y. Acad. Sci. 1012, 306–325 [DOI] [PubMed] [Google Scholar]
- 33.Ratan R. R., Siddiq A., Aminova L., Lange P. S., Langley B., Ayoub I., Gensert J., Chavez J. (2004) Stroke 35, Suppl. 1, 2687–2689 [DOI] [PubMed] [Google Scholar]
- 34.Mu D., Jiang X., Sheldon R. A., Fox C. K., Hamrick S. E., Vexler Z. S., Ferriero D. M. (2003) Neurobiol. Dis. 14, 524–534 [DOI] [PubMed] [Google Scholar]
- 35.Yasuhara T., Shingo T., Kobayashi K., Takeuchi A., Yano A., Muraoka K., Matsui T., Miyoshi Y., Hamada H., Date I. (2004) Eur. J. Neurosci. 19, 1494–1504 [DOI] [PubMed] [Google Scholar]
- 36.Tian Y., Sun S., Tang C., Wang J., Chen X., Qiao X. (2006) J. Huazhong Univ. Sci. Technolog. Med. Sci. 26, 670–673 [DOI] [PubMed] [Google Scholar]
- 37.Pichiule P., Agani F., Chavez J. C., Xu K., LaManna J. C. (2003) Adv. Exp. Med. Biol. 530, 611–617 [DOI] [PubMed] [Google Scholar]
- 38.Rivard A., Berthou-Soulie L., Principe N., Kearney M., Curry C., Branellec D., Semenza G. L., Isner J. M. (2000) J. Biol. Chem. 275, 29643–29647 [DOI] [PubMed] [Google Scholar]
- 39.Wright W. E., Shay J. W. (2006) Nat. Protoc. 1, 2088–2090 [DOI] [PubMed] [Google Scholar]
- 40.Molina-Holgado F., Gaeta A., Francis P. T., Williams R. J., Hider R. C. (2008) J. Neurochem. 105, 2466–2476 [DOI] [PubMed] [Google Scholar]
- 41.Tetrud J. W., Langston J. W., Garbe P. L., Ruttenber A. J. (1989) Neurology 39, 1483–1487 [DOI] [PubMed] [Google Scholar]
- 42.Cosi C., Marien M. (1998) Brain Res. 809, 58–67 [DOI] [PubMed] [Google Scholar]
- 43.Dexter D. T., Carayon A., Javoy-Agid F., Agid Y., Wells F. R., Daniel S. E., Lees A. J., Jenner P., Marsden C. D. (1991) Brain 114, 1953–1975 [DOI] [PubMed] [Google Scholar]
- 44.Hochachka P. W., Buck L. T., Doll C. J., Land S. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9493–9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang G. L., Semenza G. L. (1993) Blood 82, 3610–3615 [PubMed] [Google Scholar]
- 46.Tian H., Hammer R. E., Matsumoto A. M., Russell D. W., McKnight S. L. (1998) Genes Dev. 12, 3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruber M., Hu C. J., Johnson R. S., Brown E. J., Keith B., Simon M. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vangeison G., Carr D., Federoff H. J., Rempe D. A. (2008) J. Neurosci. 28, 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klivenyi P., St. Clair D., Wermer M., Yen H. C., Oberley T., Yang L., Flint Beal M. (1998) Neurobiol. Dis. 5, 253–258 [DOI] [PubMed] [Google Scholar]
- 50.Scortegagna M., Ding K., Oktay Y., Gaur A., Thurmond F., Yan L. J., Marck B. T., Matsumoto A. M., Shelton J. M., Richardson J. A., Bennett M. J., Garcia J. A. (2003) Nat. Genet. 35, 331–340 [DOI] [PubMed] [Google Scholar]
- 51.Poss K. D., Tonegawa S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10925–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee P. J., Alam J., Wiegand G. W., Choi A. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10393–10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agarwal A., Balla J., Alam J., Croatt A. J., Nath K. A. (1995) Kidney Int. 48, 1298–1307 [DOI] [PubMed] [Google Scholar]
- 54.Ferris C. D., Jaffrey S. R., Sawa A., Takahashi M., Brady S. D., Barrow R. K., Tysoe S. A., Wolosker H., Barañano D. E., Doré S., Poss K. D., Snyder S. H. (1999) Nat. Cell. Biol. 1, 152–157 [DOI] [PubMed] [Google Scholar]
- 55.Wu L. J., Leenders A. G., Cooperman S., Meyron-Holtz E., Smith S., Land W., Tsai R. Y., Berger U. V., Sheng Z. H., Rouault T. A. (2004) Brain Res. 1001, 108–117 [DOI] [PubMed] [Google Scholar]
- 56.Rouault T. A., Cooperman S. (2006) Semin. Pediatr. Neurol. 13, 142–148 [DOI] [PubMed] [Google Scholar]
- 57.Huang E., Ong W. Y., Connor J. R. (2004) Neuroscience 128, 487–496 [DOI] [PubMed] [Google Scholar]
- 58.Wang J., Xu H. M., Yang H. D., Du X. X., Jiang H., Xie J. X. (2009) Neurochem. Int. 54, 43–48 [DOI] [PubMed] [Google Scholar]
- 59.Salazar J., Mena N., Hunot S., Prigent A., Alvarez-Fischer D., Arredondo M., Duyckaerts C., Sazdovitch V., Zhao L., Garrick L. M., Nuñez M. T., Garrick M. D., Raisman-Vozari R., Hirsch E. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18578–18583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H. M., Jiang H., Wang J., Luo B., Xie J. X. (2008) Neurochem. Int. 52, 1044–1051 [DOI] [PubMed] [Google Scholar]
- 61.Zhang S., Wang J., Song N., Xie J., Jiang H. (2009) Neurobiol. Aging 30, 1466–1476 [DOI] [PubMed] [Google Scholar]
- 62.Holness M. J., Sugden M. C. (2003) Biochem. Soc. Trans. 31, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 63.St-Pierre J., Drori S., Uldry M., Silvaggi J. M., Rhee J., Jäger S., Handschin C., Zheng K., Lin J., Yang W., Simon D. K., Bachoo R., Spiegelman B. M. (2006) Cell 127, 397–408 [DOI] [PubMed] [Google Scholar]
- 64.Greenberg D. A., Jin K. (2004) Trends Mol. Med. 10, 1–3 [DOI] [PubMed] [Google Scholar]
- 65.Greenberg D. A., Jin K. (2005) Nature 438, 954–959 [DOI] [PubMed] [Google Scholar]
- 66.Yasuhara T., Shingo T., Muraoka K., Kameda M., Agari T., Wen Ji Y., Hayase H., Hamada H., Borlongan C. V., Date I. (2005) Brain Res. 1053, 10–18 [DOI] [PubMed] [Google Scholar]
- 67.Deudero J. J., Caramelo C., Castellanos M. C., Neria F., Fernández-Sánchez R., Calabia O., Peñate S., González-Pacheco F. R. (2008) J. Biol. Chem. 283, 11435–11444 [DOI] [PubMed] [Google Scholar]