Abstract
ADAMs (a disintegrin and metalloproteinases) are a family of multidomain transmembrane glycoproteins with diverse roles in physiology and diseases, with several members being drug targets for cancer and inflammation therapies. The spatial organization of the ADAM extracellular segment and its influence on the function of ADAMs have been unclear. Although most members of the ADAM family are active zinc metalloproteinases, 8 of 21 ADAMs lack functional metalloproteinase domains and are implicated in protein-protein interactions instead of membrane protein ectodomain shedding. One of such non-proteinase ADAMs, ADAM22, acts as a receptor on the surface of the postsynaptic neuron to regulate synaptic signal transmission. The crystal structure of the full ectodomain of mature human ADAM22 shows that it is a compact four-leaf clover with the metalloproteinase-like domain held in the concave face of a rigid module formed by the disintegrin, cysteine-rich, and epidermal growth factor-like domains. The loss of metalloproteinase activity is ensured by the absence of critical catalytic residues, the filling of the substrate groove, and the steric hindrance by the cysteine-rich domain. The structure, combined with calorimetric experiments, suggests distinct roles of three putative calcium ions bound to ADAM22, with one in the metalloproteinase-like domain being regulatory and two in the disintegrin domain being structural. The metalloproteinase-like domain contacts the rest of ADAM22 with discontinuous, hydrophilic, and poorly complemented interactions, suggesting the possibility of modular movement of ADAM22 and other ADAMs. The ADAM22 structure provides a framework for understanding how different ADAMs exert their adhesive function and shedding activities.
The ADAM2 family includes over 20 multidomain type I transmembrane glycoproteins that have diverse functions in cell adhesion/signaling and ectodomain shedding of cell-surface receptors or ligands (1, 2). They are broadly implicated in various physiological processes including sperm-egg interactions, development and function of the nervous system (e.g. cell-fate determination, axon guidance, and myelination), immune responses, and embryogenesis (2, 3). Dysregulation of the ADAM family is linked to a wide variety of pathological states including cancer, cardiovascular disease, asthma, Alzheimer disease, and inflammation (3–5). Several ADAMs have been pursued as therapeutic targets (6, 7).
ADAMs, together with their phylogenic relatives, the P- III class snake venom metalloproteinases (SVMPs) and ADAMTSs (ADAM with thrombospondin type-1 motif), constitute a subgroup of the metzincin clan of zinc proteinases (8, 9). The extracellular segments of ADAMs contain a prodomain that gets cleaved off during secretion, a metalloproteinase-like domain, a disintegrin domain, and a cysteine-rich domain, which are shared by SVMPs and ADAMTSs, and a unique epidermal growth factor-like domain preceding the transmembrane segment. All ADAMs contain metalloproteinase-like domains, but in humans, only 13 of the 21 members in the family possess the complete zinc binding environment (the HEXGHXXGXXHD sequence motif and the Met turn) in the domain (10). Although these proteolytically active ADAMs can shed cell-surface proteins from the plasma membrane, the other ADAMs are suggested to be non-enzymatic cell adhesion molecules (11, 12). Several ADAMs have been reported to interact with integrins, and the disintegrin-like domains of ADAMs have been suggested for this interaction (13). Despite these suggestions, structural proof that the ADAMs without canonical zinc-binding motif lack enzymatic activities has been absent, and it remains unclear how these molecules are structurally configured to support protein-protein interaction instead of ectodomain shedding.
ADAM22 (also named MDC2), one of such postulated non-catalytic ADAMs, was recently identified to serve as the postsynaptic receptor for the secreted neurotransmission modulator LGI-1 at neural synapses (14). The study supports that some ADAMs can function as adhesion molecules rather than metalloproteinases. ADAM22 is predominantly expressed in the nervous systems (15, 16). The Adam22−/− mice suffered from hypomyelination of peripheral nerves, leading to ataxia, and died before weaning (17). At the synapse, LGI-1 and ADAM22 form a tertiary complex with postsynaptic density-95 (PSD-95), a major scaffolding protein localized to the postsynaptic density of brain synapses, which is associated with α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor and other signaling proteins (14). In this complex, the extracellular domain of ADAM22 interacts with LGI-1, whereas its cytoplasmic PDZ-binding motif recruits PSD-95. The link of ADAM22 and LGI-1 to α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor established their roles in glutamate neurotransmission, consistent with genetic data that all these molecules are associated with epilepsy (17–19). Recently, it was further demonstrated that LGI-1 and LGI-4 bind to ADAM22, ADAM23, and ADAM11 (20).
Although ADAMs are functionally important as sheddases or adhesion receptors, the structural information about the ADAM family is limited to only isolated domains, such as the metalloproteinase domains of ADAM17 and ADAM33 and the incomplete disintegrin cysteine-rich domains of ADAM10 (21–23). Their relatives, SVMPs from the snake venom, including VAP-1, VAP-2, and RVV-X (24–26), have revealed a “C”-shaped molecular architecture. These SVMP structures and partial ADAM structures, along with those of the ADAMTS family proteins (27–29), shed light on the general mechanisms of substrate recognition and cleavage by the proteinase-type ADAMs. However, there is little structural information on those non-catalytic ADAMs, which serve as adhesion receptors. In addition, despite a low resolution electron microscopic (EM) analysis of the soluble form of pro-ADAM12, which suggested that the prodomain represents one of the leaves of the four-leaf clover-shaped ADAM12 (30), the structure of a complete ADAM ectodomain, being catalytic or non-catalytic, has been lacking. Here we report the crystal structure of the entire ectodomain of mature ADAM22.
EXPERIMENTAL PROCEDURES
Cell Culture, Cloning, and Baculovirus Generation
Sf9 cells were maintained in suspension in HyQ SFX media containing 10% (v/v) heat-inactivated fetal bovine serum; High Five cells (Invitrogen) were maintained in HyQ SFX without serum. A cDNA fragment encoding the ectodomain of human ADAM22 (residue 26–736), attached to a C-terminal 7-His tag, was subcloned into the baculovirus transfer vector pAcGP67A using restriction sites BamHI and NotI. The construct and the BacVector-3000 baculovirus DNA (EMD Chemicals) were used to co-transfect sf9 cells in 6-well plates in the presence of Insect GeneJuice transfection reagent (EMD Chemicals). After incubation of the transfected cells at 27 °C for 5 days, the resulted low titer virus stock was harvested. High titer viruses were generated by infecting 200 ml of Sf9 cells at 2 × 106 cells/ml at 0.1 multiplicity of infection. The amplified viruses were harvested when all cells showed cytopathic effects.
Protein Expression and Purification
The amplified viruses were used to infect 6 liters of High Five cells at a density of 1.8 × 106 cells/ml and at a multiplicity of infection of 10. 68 h after transfection, the conditioned media were harvested, concentrated, and buffer-exchanged into HBS (10 mm HEPES, pH 7.5, 150 mm NaCl, 0.05% w/v NaN3). The proteins were captured by nickel-nitrilotriacetic acid-Sepharose resin (Qiagen), washed extensively with HBS, and eluted with 300 mm imidazole. For crystallization, the proteins were deglycosylated by endo-β-N-acetylglucosaminidase F3, and then treated with bovine carboxypeptidase A overnight at room temperature for His tag removal. The digested products were further purified by gel filtration chromatography with a Superdex 200 column (Amersham Biosciences) pre-equilibrated and eluted with HBS. The fractions containing mature ADAM22 were pooled and concentrated to 10 mg/ml.
Crystallization
Crystallization was performed using the sitting-drop vapor diffusion method. Crystals were obtained from drops composed of 0.5 μl of reservoir solution and 0.5 μl of protein solution equilibrated against 1 ml of 12% (w/v) polyethylene glycol 8000, 0.1 m Tris, pH 7.5, and 5% (v/v) ethylene glycol. Crystals grew to their maximum dimensions (∼0.15 × 0.10 × 0.05 mm3) in 10 days.
X-ray Diffraction and Data Processing
Crystals were cryoprotected in the presence of 25% ethylene glycol in the mother liquor and immediately flash-cooled in liquid nitrogen. To prepare a heavy ion derivative, crystals were quick-soaked with a cryosolution containing 0.3 m sodium iodide before being flash-cooled. Data sets were collected at 100 K at the LS-CAT beamline 21-ID-D, the Advanced Photon Source (APS), Argonne, IL. The data were processed with HKL2000 (31). The statistics are summarized in Table 1.
TABLE 1.
Crystallographic statistics
Values in parentheses are for the highest resolution shell.
| Native | NaI derivative | |
|---|---|---|
| Data collection | ||
| Space group | R3 | R3 |
| Cell dimensions | ||
| a, b, c (Å) | 122.09, 122.09, 208.69 | 122.45, 122.45, 207.88 |
| α, β, γ (deg.) | 90, 90, 120 | 90, 90, 120 |
| Wavelength (Å) | 0.97856 | 1.5498 |
| Resolution range (Å) | 20.0–2.3 (2.4–2.3) | 20.0–3.2 (3.3–3.2) |
| Unique reflections | 49760 | 37630 |
| Completeness (%) | 96.6 (91.5) | 99.3 (96.7) |
| I/σ(I) | 11.2 (2.4) | 9.9 (2.3) |
| Redundancy | 3.3 | 4.5 |
| Rr.i.ma (%) | 8.0 (41.5) | 14.8 (52.6) |
| SIRAS phasing | ||
| Resolution range (Å) | 20.0–3.2 | |
| Numbers of heavy ions | 3 | |
| Ranomalous (%) | 0.09 | |
| Risomorphous (%) | 0.157 | |
| Figure of merit | 0.39 | |
| Refinement | ||
| Resolution range (Å) | 50–2.36 (2.51–2.36) | |
| Rworkb (%) | 24.5 (42.0) | |
| Rfreec (%) | 27.3 (43.4) | |
| Average B-values | ||
| 7456 protein atoms (Å2) | 65.4 | |
| 369 water molecules (Å2) | 58.9 | |
| 6 Ca2+ ions (Å2) | 58.2 | |
| r.m.s. deviation bonds (Å) | 0.009 | |
| r.m.s. deviation angles (degrees) | 1.2 | |
| Ramachandran (%) (favored, allowed, generously allowed, disallowed)d | 83.8, 15.4, 0.7, 0 | |
aRr.i.m refers to redundancy-independent merging R factor (43).
b Rwork = Σ|Fobs − Fcalc|/ΣFobs, where Fobs and Fcalc are the observed and the calculated structure factors, respectively.
c Rfree is calculated using 5% of reflections sequestered before refinement.
Structure Determination and Refinement
The phases of ADAM22 crystal were determined using the single isomorphous replacement with anomalous scattering (SIRAS) technique with the help of molecular replacement. The program PHASER (32) was used to locate the ADAM22 metalloproteinase-like domains in the asymmetric unit using the ADAM33 metalloproteinase domain (Protein Data Bank (PDB) ID: 1R54) as the searching model. To complete the model, partial molecular phases were calculated with the program CNS (33) for locating three iodide ions in the asymmetric unit of the NaI derivative using the anomalous difference Fourier method. SIRAS phases with an overall figure of merit of 0.39 for the reflections in the 20–3.3 Å resolution range were calculated with CNS. Following density modification with 2-fold non-crystallographic symmetry averaging, the electron density map allowed the tracing of the remaining domains. The models were rebuilt using the software COOT (34) and subjected to simulated annealing, minimization, and group B factor refinements with CNS. The calcium ions and the carbohydrate moieties were modeled as guided by the SIGMAA-weighted Fo − Fc omit map. Water molecules were automatically introduced using CNS and manually edited. A summary of the refinement statistics is given in Table 1.
Structural Analysis
Backbones of protein structures were superimposed with the program TOP3D in CCP4 (35). Buried surface areas were calculated in CNS. Surface complementarity coefficients were calculated with the program Sc in CCP4 (35).
Isothermal Titration Calorimetry
Calorimetric titrations were carried out on a VP-ITC calorimeter (MicroCal, Northhampton, MA) at 30 °C. Fully glycosylated ADAM22 with no His tag was incubated with 5 mm EDTA for 30 min for calcium stripping and then loaded to a Superdex 200 size exclusion column to separate protein aggregates and excessive EDTA. The protein concentration was determined by the bicinchoninic acid assay with bovine serum albumin as the standard (Pierce). 0.012 mm ADAM22 was placed in the reaction chamber, and deionized water was placed in the reference chamber. 0.5 mm CaCl2, the titrant, was prepared with the identical lot of HBS used for ADAM22 purification. Both the titrant and the protein sample were thoroughly degassed before titration. The titrant was then injected into the reaction chamber in 3-μl increments at 5-min intervals with stirring at 286 rpm. The titration data, measured over 35 consecutive injections, were processed with the MicroCal Origin software, version 5.0.
RESULTS
ADAM22 Expression and Propeptide Processing
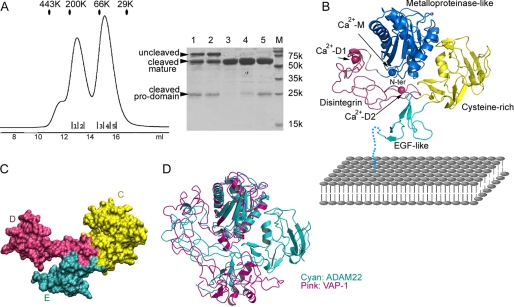
The entire ectodomain of ADAM22, including both the N-terminal prosequence and the C-terminal mature sequence, was cloned into the baculovirus transfer vector for expression in insect cells. The recombinant proteins were captured from conditioned media by nickel-nitrilotriacetic acid binding to the C-terminal His tag of ADAM22. As shown in Fig. 1A, ADAM22 was secreted as a mixture of three peptides: the uncleaved full-length peptide, the mature ADAM22, and the propeptide. The cleaved propeptide has no affinity tag but was still pulled down together with the mature peptide, indicating a direct, non-covalent binding between the propeptide and mature ADAM22. The cleaved propeptide associated with mature ADAM22 had the same elution profile as the uncleaved full-length ADAM22, suggesting that the binding geometry between the propeptides and the mature peptides does not change much upon cleavage. A standalone population of mature ADAM22 existed, indicating that the propeptide can be lost over the time course of expression. Mature ADAM22 elutes in gel filtration at an apparent size of ∼60 kDa, in line with the apparent size in SDS-PAGE, supporting that it is monomeric in solution. The cleaved propeptide was stable in gel filtration, consistent with the previous report that the prodomain of ADAM22 expressed in Escherichia coli could be folded and had its own structure (36). The propeptide likely exists as a dimer as its apparent molecular weight in gel filtration is only slightly less than that of mature, monomeric ADAM22. In support of this, the propeptide-containing ADAM22, cleaved or uncleaved, also appears as a dimer as it elutes at an apparent size (∼180 kDa) much larger than the monomeric size as shown in SDS-APGE (∼90 kDa). The dimerization of propeptide-containing ADAM22 is likely mediated by the propeptide but not the mature ADAM22.
FIGURE 1.
Structure of the ADAM22 ectodomain. A, gel filtration profile in a calibrated Superdex 200 column showing that secreted ADAM22 ectodomain exists as a mixture of three forms: the uncleaved propeptide-containing ADAM22, the non-covalent complex between propeptide and mature ADAM22, and the mature ADAM22 without propeptide. The eluted peaks were analyzed by SDS-PAGE. Lane M, molecular size. B, ribbons diagram showing the topology of the ADAM22 ectodomain. The M, D, C, and E domains are shown in blue, magenta, yellow, and cyan, respectively. The three putative calcium ions are colored the same as their host domains. EGF, epidermal growth factor. C, surface diagram of the D, C, and E domains showing that these three domains have seamless interfaces and are an integral module. D, superimposition of ADAM22 and VAP-1 based on overlapping their M domains showing a significant difference in spatial relation between domain M and the rest of the molecule.
To verify the role of the propeptide in ADAM22, an ADAM22 fragment without the propeptide (residues 226–736), immediately following the putative furin cleavage site RSKR (residues 221–224), was constructed for expression in insect cells, but no expression was observed. This suggests that the propeptide of ADAM22 is essential for ADAM22 secretion and may play an intramolecular chaperone function, similar to the cases of ADAM17 and ADAM12 (37, 38). In physiological conditions, the propeptide is likely cleaved by furin-like proprotein convertases and may fall off mature ADAM22 more easily than in the baculovirus expression system as the physiological expression level is much lower. When overexpressed, however, the propeptide may get processed incompletely due to the limitation of processing capacity intrinsic to the cells. Therefore, the propeptide-containing ADAM22 in the extracellular space that we observed may not represent a functional form of ADAM22. The non-covalent association of the propeptide with the mature protein has also been observed for ADAM12 when it is highly expressed in pregnancy serum or in transfected mammalian cells, but the propeptide was suggested to be an integral part of mature ADAM12 (30), in contrast to the previous observation that mature ADAM12 lacking the propeptide was the predominant form present at the cell surface (39). Hence, for ADAM12 and other ADAM family members, it remains to be further clarified whether their propeptides are essential to the functional form of proteins.
The Overall Structure of Mature ADAM22
The structure of mature ADAM22 without the propeptide, determined with the SIRAS method, contains two ADAM22 molecules in the asymmetric unit related by a non-crystallographic two-fold axis. The interface between the two molecules is relatively small and hydrophilic, suggesting that the mature ADAM22 ectodomain is monomeric but not dimeric, in agreement with the gel filtration results.
Viewed from the front, the ADAM22 ectodomain has four domains assembled together like a four-leaf clover, and each leaf represents one of the four domains, including the metalloproteinase-like domain (domain M, residues 233–435), the disintegrin domain (domain D, residues 445–529), the cysteine-rich domain (domain C, residues 530–676), and the epidermal growth factor-like domain (domain E, residues 677–718) (Fig. 1B). Viewed from the top, the molecule is roughly flat. The overall dimensions of ADAM22 are 80 × 70 × 40 Å. The largest domain in the four-leaf clover, domain M, is most likely distal to the cell membrane. Following domain M, domain D and domain C zigzagged to domain E in a compact, but not extended, fashion. A 15-amino acid linker, not seen in the structure, leads the C terminus of domain E to the membrane. Each ADAM22 contains three putative calcium ions, one in domain M and two in domain D, and three N-linked glycans at Asn-519, Asn-634, and Asn-675. There are numerous interdomain interactions in ADAM22 reinforcing the compact overall structure. Notably, the D, C, and E domains have a continuous hydrophobic core and appear to be an integral module (hereafter termed the DCE module) mediated by extensive and seamless interfaces between domains (Fig. 1C). Interdomain segmental flexibility is highly unlikely within this module. Domain M is held in a concave face of the rigid DCE module, supported by a long stretch of interactions between domain M and domain D, and a small, round patch between domain M and domain C. An ∼10-amino acid-long linker peptide connects domain M and the DCE module. There is no direct contact between domain M and domain E.
The locations of the M, D, and C domains in ADAM22 roughly resemble the locations of these domains in the SVMPs including VAP-1, VAP-2, and RVV-X (24–26). However, overlaying the SVMP structures to ADAM22 revealed that there is a large position shift of domain D and domain C in ADAM22 relative to the corresponding domains in the SVMPs (Fig. 1D and supplemental Fig. S1). VAP-1 is an open, C-shaped molecule with no interaction between domain M and domain C; its interactions between domain M and domain D are limited to the domain junction, where the two domains are much closer to each other than in ADAM22. The C-shaped conformation of VAP-1 is therefore enabled entirely by the extensive interactions at the domain boundary. VAP-2 is similar to VAP-1. RVV-X has a closed C-shaped structure in which nonspecific contacts are found between domain M and domain C, and its D–C arm is positioned far differently from the counterpart in ADAM22 and is docked on a different side of domain M from that in ADAM22 (supplemental Fig. S1).
The clover-shaped mature ADAM22 may, to some extent, be similar to the clover-shaped, propeptide-bound ADAM12 as revealed by EM (30). The EM study placed the ADAM12 prodomain at roughly the position of ADAM22 domain C. Therefore, if the prodomain was taken out from the reconstructed EM envelope, the overall shape of mature ADAM12 would be dramatically different from mature ADAM22. The sequence identity between ADAM12 and ADAM22 is moderate (∼35%). It remains to be clarified whether the overall structures of ADAMs are highly diverse or not.
The Catalytically Inactive Metalloproteinase-like Domain
The backbone of domain M of ADAM22 is similar to that of the metalloproteinase domains in other ADAMs (e.g. ADAM33 and ADAM17) and SVMPs (e.g. VAP-1, as shown in Fig. 1D) (23–26). These domains can be superimposed to the ADAM22 domain M with an r.m.s. deviation of 1.1–1.7 Å for Cα atoms. The main structural differences occur in loop regions connecting helices and strands, especially those located in the N-terminal part of the small subdomain.
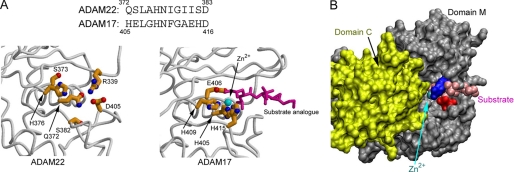
Although ADAM22 contains a metalloproteinase-like domain, it lacks the consensus zinc-binding motif (HEXGHXXGXXHD), which directly participates in catalysis (Fig. 2A). Indeed, our examination of the entire asymmetric unit of ADAM22, including the equivalent position to the catalytic site in other metalloproteinases, found no zinc ion. However, the other highly conserved structural feature for catalytic function, the Met turn, a typical marker of the metzincin family of metalloproteinases (10), appears intact in ADAM22. Comparing the catalytically inactive ADAM22 domain M with the catalytically active ADAM17 domain M revealed several features that ensure the loss of zinc metalloproteinase activity in ADAM22. Firstly, most of the zinc-chelating histidines are replaced in ADAM22. In ADAM17, the zinc is held by three imidazole groups from three histidines, whereas in ADAM22, only His-376, corresponding to the middle histidine of the three consensus histidines, remains. Secondly, the glutamate after the first histidine, essential for catalysis as a general base (22), is replaced in ADAM22 (ADAM17 Glu-406 versus ADAM22 Ser-373). Notably, histidine and serine are often observed at the active site of serine proteases, but the positions of ADAM22 His-376 and Ser-373 are inconsistent with the serine protease catalytic triad geometry. Thirdly, the zinc-coordinated hydrolysis requires a groove for holding the substrate, which is not available in ADAM22. In active zinc metalloproteinases such as ADAM17, a 20 Å long groove runs across the domain, allowing an extended peptide to access the catalytic site. In ADAM22, one end of this groove is filled by a locking salt bridge between Arg-339 and Asp-405, as well as the backbone of the Arg-339-harboring loop (Fig. 2, A and B). Fourthly, in ADAM22, a steep wall, formed by one side of domain C, sits right by the pseudo catalytic site (Fig. 2B), serving as a steric hindrance for activities near the site. Other zinc metalloproteinases either do not have domain C or likely configure this domain to other spaces when the enzymatic function is to be implemented.
FIGURE 2.
Structural features ensuring domain M to be catalytically inactive. A, a comparison between the ADAM17 catalytic site (PDB ID 1BKC) and the equivalent position in ADAM22 showing that zinc binding and substrate cleavage are disabled in ADAM22 due to the replacement of critical residues. The zinc ion is depicted as a cyan ball, and the substrate analogue is depicted as magenta sticks in ADAM17. B, modeling the zinc ion and the substrate analogue of the ADAM17 structure into ADAM22 based on domain M superimposition showing that substrate binding is disabled in ADAM22 by the salt bridge between Arg-339 (blue) and Asp-405 (red), the main chain of the Arg-339-harboring loop, and the cysteine-rich domain nearby (yellow).
The Hypervariable Region in the Cysteine-rich Domain
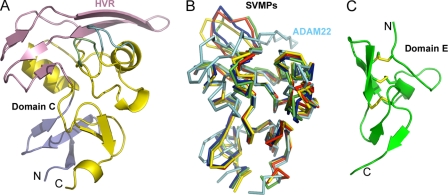
Domain C has been proposed as a major mediator of the adhesion function for ADAMs (26). The ADAM22 domain C has a backbone similar to SVMPs for the most part except for the distal end adjacent to domain M, which has been designated as the hypervariable region, or the HVR (Fig. 3, A and B). This region includes a long loop containing little secondary structure and a two-stranded β-sheet. The entire HVR appears like a large hat, flatly packed above the core domain and tightly stabilized by hydrophobic interactions. In comparison, the HVRs of SVMPs are more protruding and less ordered. Most ADAMs have an HVR region of similar length to that of ADAM22, but the HVRs of SVMPs are ∼15 amino acids shorter than those of ADAM22. In addition, the HVR sequences of ADAMs are similar to each other but are very different from SVMPs. Therefore, most ADAMs should have an HVR structure similar to ADAM22. The HVR adopts a well accessible location and presents a large surface in the overall ADAM structure, serving well for its putative role in adhesion. In addition, its variability may be desirable for coding specificity. Nevertheless, assigning functions to this structural feature of ADAMs may still require more specific studies.
FIGURE 3.
The C and E domains of ADAM22. A, ribbons diagram of domain C showing that the HVR (pink) serves as a hat for domain C. The HVR is partially supported by two flap-like loops (in cyan and green). The wrist subdomain, referred to as Cw (26), is colored in slate. N, N terminus; C, C terminus. B, comparison of the ADAM22 domain C (cyan) with SVMPs (other colors) showing that the HVR harbors large variability. C, ribbons diagram of domain E of ADAM22.
The Epidermal Growth Factor-like Domain
The membrane-proximal domain E of ADAM22 can be divided into an N-subdomain (residues 677–704) and a C-subdomain (705–718). The N-subdomain contains a loop region (residues 677–687), a 310 helix (residues 688–690), and an antiparallel β-sheet (residues 692–694 and 700–702). The C-subdomain contains a short antiparallel β-sheet (residues 705–707 and 713–715) and a C-terminal flexible segment. Three disulfide bonds, Cys-679–Cys-694, Cys-688–Cys-700, and Cys-702–Cys-711, cross-link the above secondary structural elements (Fig. 3C). Sequence alignments of the ADAM family show that the loop region (residues 677–687) is the most divergent, being variable in length as well as the position of Cys-679 (numbered in ADAM22) (supplemental Fig. S2). Nevertheless, all ADAMs should have a generally similar domain E.
Distinct Roles of the Three Putative Calcium Ions in ADAM22
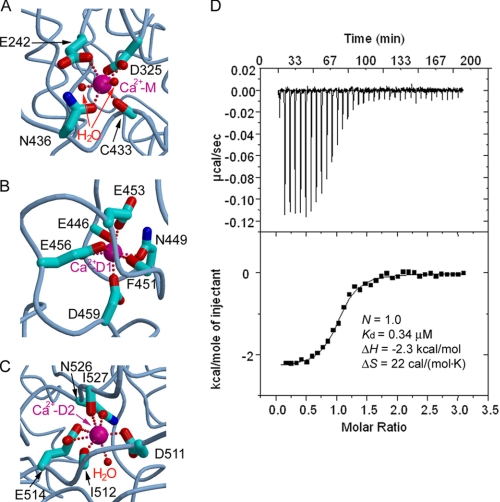
Three apparent metal cation-binding sites in each ADAM22 molecule were detected in the Fo − Fc electron density map. The chemical environment and the coordination geometry suggest that they are most likely calcium-binding sites. The putative calcium ions in ADAM22, one in domain M (designated Ca2+-M) and two in domain D (designated Ca2+-D1 and Ca2+-D2, respectively), all display pentagonal bipyramidal coordination, similar to the case of VAP-1 (26). Ca2+-M is located at the tip of domain M that interacts with the DCE module. It is coordinated by two water molecules and five oxygen atoms from domain M. One carboxyl oxygen atom of Glu-242 (Oϵ1) and the main-chain carbonyl oxygen of Cys-433 occupy the axial positions, whereas two carboxyl oxygen atoms of Asp-325, the side-chain oxygen of Asn-436, and two water molecules form the equatorial plane (Fig. 4A). Ca2+-D1 is located at the entrance of domain D and is coordinated by side-chain oxygen atoms of residues Asn-449 (Oδ1), Glu-453 (Oϵ1), Glu-456 (Oϵ1 and Oϵ2), Asp-459 (Oδ2), and main-chain carbonyl oxygen atoms of residues Phe-451 and Glu-461. No water molecule is used for Ca2+-D1 coordination (Fig. 4B). Ca2+-D2 is located near the end of domain D. It is coordinated by two carboxyl oxygen atoms of Glu-514 (Oϵ1 and Oϵ2), the side-chain oxygen atom of Asn-526 (Oδ1), one carboxyl oxygen atom of Asp-511 (Oδ2), the backbone carbonyl oxygen atoms of Ile-512 and Ile-527, and one water molecule (Fig. 4C).
FIGURE 4.
Distinct binding at the three putative calcium-binding sites. A–C, the coordination of the calcium ions (Ca2+-M, Ca2+-D1, and Ca2+-D2) by ADAM22 residues and water molecules. D, thermodynamic profile of the binding between calcium and the EDTA-treated ADAM22 protein showing a 1:1 stoichiometry.
The three putative calcium ions may have distinct roles in ADAM22. Both Ca2+-D1 and Ca2+-D2 are deeply embedded within domain D, and their absence would cause repulsion between charged residues buried at the domain core, likely resulting in the unfolding of domain D. Ca2+-M, in comparison, is bound to the surface. It is located in a pocket formed between the main body of domain M and a long stretch of peptide linking domain M and domain D. The absence of both Ca2+-M and the linker peptide (residues 435–445) would not severely compromise the integrity of domain M.
To test the roles of the putative calcium ions, we stripped calcium off from ADAM22 using a strong chelating reagent, EDTA, and then measured the binding of calcium to the emptied cation-binding sites using isothermal titration calorimetry (Fig. 4D). The EDTA treatment did not cause ADAM22 to unfold. The titration curve shows a 1:1 stoichiometry between ADAM22 and calcium, indicating that only one calcium ion was stripped off from and re-bound to ADAM22. Given the deeply buried nature and the tight coordination of Ca2+-D1 and Ca2+-D2, we reason that it is Ca2+-M, which is bound to the surface and is ligated to the most water molecules (two for Ca2+-M versus 0 for Ca2+-D1 and one for Ca2+-D2), that was striped off by EDTA. The binding strength of Ca2+-M to domain M is moderate, with an affinity of 0.34 μm. The affinity of the hexadentate chelator EDTA to calcium (∼20 pm) is sufficiently high for stripping Ca2+-M off its binding site. However, EDTA did not strip off Ca2+-D1 and Ca2+-D2, suggesting that Ca2+-D1 and Ca2+-D2 bind to their respective sites at even higher affinities than to EDTA. Such high affinities are consistent with structural roles as the calcium ions likely stay unchanged once domain D is folded. In comparison with the essential roles of Ca2+-D1 and Ca2+-D2 in folding and structural integrity, Ca2+-M is more likely to play a regulatory role, probably in line with the large external fluctuations of calcium concentration in the synaptic cleft (40), where ADAM22 is predominantly expressed.
The regulatory role of Ca2+-M versus the structural roles of Ca2+-D1 and Ca2+-D2 are also consistent with the fact that the residues coordinating Ca2+-D1 and Ca2+-D2 are absolutely conserved through the ADAM family, but the residues coordinating Ca2+-M are less conserved (26). It is likely that not all ADAMs are calcium-regulated.
DISCUSSION
In this report, we have determined the first structure of an ADAM ectodomain containing all the extracellular domains in the mature protein. As the ADAMs are a large family of over 20 proteins with significant homology (1, 2), the ADAM22 structure can be used to model many, if not all, ADAM family members. The ADAM22 is likely a better structural template than SVMPs for postulating other ADAMs, given the similar domain structure and the higher sequence similarity among ADAMs. This is supported by the previous EM study showing that the propeptide-containing ADAM12 is a four-leaf clover similar to ADAM22 despite that the position of the propeptide in the EM envelope may need to be further clarified (30).
The four-leaf-clover structure of ADAM22 has its domain M and the DCE module compactly associated with each other, unlike SVMPs. The difference between SVMPs and ADAM22 in the relative positions between domain M and the rest of the structure raises a question whether some ADAMs may have dynamic structures, allowing opening and closing between domain M and the DCE module. Indeed, the early EM study has revealed that although the majority of ADAM12 molecules appear as compact clovers, a few adopt extended conformation (30). ADAM12 is a catalytically active proteinase that can shed substrates such as Delta-like ligand-1, fibronectin, and insulin-like growth factor-binding proteins (3). If ADAM12 adopts a similar conformation to ADAM22, its catalytic site in domain M may be sterically hindered by the proximal domain C, as shown in Fig. 2C for ADAM22. Hence, a conformational change that allows the DCE module to move away from domain M may be needed for catalytically active ADAMs. Similarly, the HVR at the distal end of domain C, proposed as a protein recognition module (26), may also be sterically hindered by domain M, instead of being unrestrictedly open, for its adhesion function because of the proximity between domain M and the HVR. Therefore, moving the DCE module away from domain M may also be needed for catalytically inactive ADAMs.
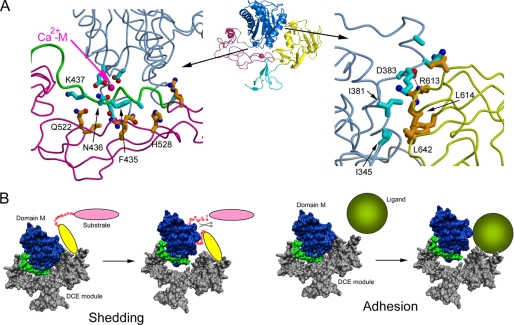
Examining the ADAM22 structure suggests that the movement of the DCE module from domain M is possible. The interaction between domain M and the DCE module is mostly hydrophilic and discontinuous and has low complementarity. The two patches of non-junction contacts between domain M and the DCE module bury surface areas of 1050 and 950 Å2, respectively, both with a calculated surface complementarity coefficient (Sc) of 0.68, which is in the lower range for protein interfaces (41). The first patch (Fig. 5A, left) is between the Ca2+-M-bearing tip of domain M and the concave face of domain D. This long and thin patch has no obvious hydrophobic interactions. Domain M uses its C-terminal peptide fragment (residues 428–437), mostly the domain M-domain D linker, to contact domain D with van der Waals interactions and four hydrogen bonds. The second patch (Fig. 5A, right) is between one end of the domain M pseudo substrate-binding cleft and one side of the domain C HVR. The patch contains a small hydrophobic cluster, including Ile-381, Ile-345, Leu-641, and Leu-642, and a salt bridge between Asp-383 and Arg-613. Most of the residues at this patch are located in surface loops, and due to enthalpy/entropy compensation, it is unlikely that their interactions contribute substantially to free energy change. Because both patches are not highly specific interactions, they probably collaborate with each other and with the domain M-domain D junction to enable a closed conformation between domain M and the DCE module. It is unlikely that breaking the two patches apart will require high energy. It needs to be noted that due to the leverage effect, the hinge between domain M and the DCE module may only need small, subtle changes to allow the opening at the tips (the pseudo site of domain M and the HVR of domain C) by over 10 Å.
FIGURE 5.
Putative dynamic relationship between domain M and the DCE module. A, the two patches of non-junctional interactions between domain M and the DCE module showing that the interactions are mostly hydrophilic and low in specificity and complementarity. B, a model of modular movement for ADAM function. For catalytic ADAMs, substrate access to the hydrolysis center in domain M may be autoinhibited by domain C; the substrate may bind to domain C first, allowing domain C, as part of the DCE module, to move away from domain M so that the peptide to be cleaved can enter the catalytic site. For adhesion ADAMs, the ligand may compete with domain M in binding domain C, forcing domain M to move away from domain C and the DCE module.
If ADAM22 and other ADAMs do employ such modular movement, calcium may have a role in its regulation, most likely as an allosteric modulator. As shown in Fig. 5A, Ca2+-M is sandwiched between domain M and the domain M-domain D linker. It tethers the linker, preparing it for closely contacting the concave face of domain D. It is conceivable that depending on the presence versus the absence of Ca2+-M tethering, the linker may have twisting differences that can result in favorable or unfavorable interdomain interactions. This putative role of calcium may be supported by a recent report showing that calcium has an influence in ADAM12 catalysis (42). As Ca2+-D1 and Ca2+-D2 are extremely tightly bound and unable to be modulated, it is likely that only Ca2+-M was affected in the study. Given that Ca2+-M is located far from the catalytic site of ADAM12, it may act through changing modular movement equilibrium but not through direct involvement in catalysis. The reported effect of calcium on ADAM12 activity is only mild (42), and further studies are needed to confirm the roles of calcium in other enzymatic ADAMs. In addition, with the current structural information, it is not completely clear whether the increasing of calcium concentration moves the equilibrium from the close conformation to the open conformation, or the opposite.
Based on the above observations, we hypothesize a model for ADAMs to implement their catalysis/adhesion functions as regulated by modular movement (Fig. 5B). For catalytic ADAMs, when domain C is sterically incompatible with substrate access to the catalytic site, a protein recognition module of the substrate protein may bind ADAMs first, resulting in a conformational change of ADAMs so that domain C, as part of the DCE module, moves away from the catalytic site, allowing the peptide to be cleaved to enter the catalytic groove (Fig. 5B). For ADAMs as adhesion receptors, when their domain M positions are sterically incompatible with their adhesive sites, the ligands may be able to compete domain M away to occupy the adhesion site (Fig. 5B). This closed-to-open model is only a simplified model, and there are two cautions to be taken. First, for both catalytic and adhesive ADAMs, it is possible that the substrate/ligand may not be the driving force for conformational change but may only selectively bind one conformation or another, changing the equilibrium between two conformations. Second, the conformational change of ADAMs may not be from closed to open, but may be from open to closed or from one mode of docking to another mode of docking, as long as the steric hindrance is released upon the conformational change, allowing hydrolysis or adhesion to happen.
Our modular movement model may be generally applicable for ADAMs, especially when assuming the HVR of domain C as an important recognition site. However, for an individual ADAM with specific adhesion substrates, specific patches from the vast surface area of ADAMs can be utilized. For instance, the well known function of ADAM22 is to act as a receptor on the surface of the postsynaptic neuron and to bind LGI-1 for increasing synaptic signal transmission (14); this ligand spectrum for ADAM22 may only be shared by its close relatives ADAM11 and ADAM23 but not by other ADAMs (20). LGI-1 is reported to bind ADAM22 through its EPTP domains, and an ADAM22 mutant (D509N) failed to bind to LGI-1 (14). Asp-509 is located in the “disintegrin loop,” a 14-amino acid stretch implicated in interactions between ADAMs and integrins (12) (supplemental Fig. S3). The side chain of Asp-509 forms a hydrogen bond with the backbone amide nitrogen of Asn-536 from C-domain. Mutation of Asp-509 to asparagine may not disrupt the interdomain hydrogen bond; it is also unlikely to cause a global conformational change of ADAM22. However, the electrochemical surface potential around the position 509 may be changed upon this mutation. The LGI-1 EPTP domain (residues 224–557) responsible for ADAM22 recognition is positively charged, with a calculated isoelectrical point of 8.8. It is possible that electrostatic attraction is important in ADAM22-LGI-1 interaction and is disrupted by the D509N mutation. A clear understanding of the association between ADAM22 and LGI1 awaits the structure determination of their complex.
CONCLUSION
We have reported the first structure of a full mature ectodomain of ADAMs, presenting a framework for understanding how the different domains of ADAMs interplay to exert their adhesive function and shedding activities. The structure indicates that the non-catalytic ADAM22 not only has lost the zinc-binding structural features but also has a filled substrate groove and steric hindrance to the pseudo site. The large surface of ADAMs provides wide possibilities for adhesion, but the HVR in domain C, which shows large differences between ADAM22 and SVMPs, may be poised to serve as a hotspot in ligand/substrate binding to ADAMs. Cross-talk between domain M and domain C, in particular the HVR, may be a general mechanism for both catalytic and non-catalytic ADAMs, which likely involves modular movement and/or calcium for its regulation. Although the ADAM22 structure is likely a faithful template for many ADAMs, especially for their close relatives ADAM11 and ADAM23, a large variability may exist between diverse ADAMs. More structural information of the ADAM family proteins as well as their complexes with substrates or adhesive partners is necessary to adequately address the wide spectrum of functions for this important protein family.
Acknowledgments
We thank Xiaoyan Chen for assistance in cell culture and Zdzislaw Wawrzak and Pamela Focia for support in x-ray data collection and reading the manuscript. The Structural Biology Facility is supported by the R. H. Lurie Comprehensive Cancer Center of Northwestern University. Data were collected at the Life Sciences Collaborative Access Team (LS-CAT) beamline 21-ID-D at APS, Argonne National Laboratory. Use of the APS is supported by the U.S. Department of Energy under Contract W-31-109-Eng-38.
The atomic coordinates and structure factors (code 3G5C) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ADAM
- a disintegrin and metalloproteinase
- ADAMTS
- ADAM with thrombospondin type-1 motif
- SVMP
- snake venom metalloproteinases
- EM
- electron microscopic
- SIRAS
- single isomorphous replacement with anomalous scattering
- HVR
- hypervariable region
- r.m.s.
- root mean square deviation.
REFERENCES
- 1.Primakoff P., Myles D. G. (2000) Trends Genet 16, 83–87 [DOI] [PubMed] [Google Scholar]
- 2.Seals D. F., Courtneidge S. A. (2003) Genes Dev. 17, 7–30 [DOI] [PubMed] [Google Scholar]
- 3.Edwards D. R., Handsley M. M., Pennington C. J. (2008) Mol. Aspects Med. 29, 258–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiss K., Ludwig A., Saftig P. (2006) Pharmacol. Ther. 111, 985–1006 [DOI] [PubMed] [Google Scholar]
- 5.Rocks N., Paulissen G., El Hour M., Quesada F., Crahay C., Gueders M., Foidart J. M., Noel A., Cataldo D. (2008) Biochimie 90, 369–379 [DOI] [PubMed] [Google Scholar]
- 6.Duffy M. J., Lynn D. J., Lloyd A. T., O'Shea C. M. (2003) Thromb. Haemost. 89, 622–631 [PubMed] [Google Scholar]
- 7.Moss M. L., Bartsch J. W. (2004) Biochemistry 43, 7227–7235 [DOI] [PubMed] [Google Scholar]
- 8.Gomis-Rüth F. X. (2003) Mol. Biotechnol. 24, 157–202 [DOI] [PubMed] [Google Scholar]
- 9.Gomis-Rüth F. X. (2009) J. Biol. Chem. 284, 15353–15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bode W., Gomis-Rüth F. X., Stöckler W. (1993) FEBS Lett. 331, 134–140 [DOI] [PubMed] [Google Scholar]
- 11.Pruessmeyer J., Ludwig A. (2009) Semin. Cell Dev. Biol. 20, 164–174 [DOI] [PubMed] [Google Scholar]
- 12.White J. M. (2003) Curr. Opin. Cell Biol. 15, 598–606 [DOI] [PubMed] [Google Scholar]
- 13.Lu X., Lu D., Scully M. F., Kakkar V. V. (2007) Cardiovasc. Hematol. Agents Med. Chem. 5, 29–42 [DOI] [PubMed] [Google Scholar]
- 14.Fukata Y., Adesnik H., Iwanaga T., Bredt D. S., Nicoll R. A., Fukata M. (2006) Science 313, 1792–1795 [DOI] [PubMed] [Google Scholar]
- 15.Novak U. (2004) J. Clin. Neurosci. 11, 227–235 [DOI] [PubMed] [Google Scholar]
- 16.Sagane K., Yamazaki K., Mizui Y., Tanaka I. (1999) Gene 236, 79–86 [DOI] [PubMed] [Google Scholar]
- 17.Sagane K., Hayakawa K., Kai J., Hirohashi T., Takahashi E., Miyamoto N., Ino M., Oki T., Yamazaki K., Nagasu T. (2005) BMC Neurosci. 6, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicoll R. A., Tomita S., Bredt D. S. (2006) Science 311, 1253–1256 [DOI] [PubMed] [Google Scholar]
- 19.Steinlein O. K. (2004) Nat. Rev. Neurosci. 5, 400–408 [DOI] [PubMed] [Google Scholar]
- 20.Sagane K., Ishihama Y., Sugimoto H. (2008) Int. J. Biol. Sci. 4, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janes P. W., Saha N., Barton W. A., Kolev M. V., Wimmer-Kleikamp S. H., Nievergall E., Blobel C. P., Himanen J. P., Lackmann M., Nikolov D. B. (2005) Cell 123, 291–304 [DOI] [PubMed] [Google Scholar]
- 22.Maskos K., Fernandez-Catalan C., Huber R., Bourenkov G. P., Bartunik H., Ellestad G. A., Reddy P., Wolfson M. F., Rauch C. T., Castner B. J., Davis R., Clarke H. R., Petersen M., Fitzner J. N., Cerretti D. P., March C. J., Paxton R. J., Black R. A., Bode W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3408–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orth P., Reichert P., Wang W., Prosise W. W., Yarosh-Tomaine T., Hammond G., Ingram R. N., Xiao L., Mirza U. A., Zou J., Strickland C., Taremi S. S., Le H. V., Madison V. (2004) J. Mol. Biol. 335, 129–137 [DOI] [PubMed] [Google Scholar]
- 24.Igarashi T., Araki S., Mori H., Takeda S. (2007) FEBS Lett. 581, 2416–2422 [DOI] [PubMed] [Google Scholar]
- 25.Takeda S., Igarashi T., Mori H. (2007) FEBS Lett. 581, 5859–5864 [DOI] [PubMed] [Google Scholar]
- 26.Takeda S., Igarashi T., Mori H., Araki S. (2006) EMBO J. 25, 2388–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerhardt S., Hassall G., Hawtin P., McCall E., Flavell L., Minshull C., Hargreaves D., Ting A., Pauptit R. A., Parker A. E., Abbott W. M. (2007) J. Mol. Biol. 373, 891–902 [DOI] [PubMed] [Google Scholar]
- 28.Mosyak L., Georgiadis K., Shane T., Svenson K., Hebert T., McDonagh T., Mackie S., Olland S., Lin L., Zhong X., Kriz R., Reifenberg E. L., Collins-Racie L. A., Corcoran C., Freeman B., Zollner R., Marvell T., Vera M., Sum P. E., Lavallie E. R., Stahl M., Somers W. (2008) Protein Sci. 17, 16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shieh H. S., Mathis K. J., Williams J. M., Hills R. L., Wiese J. F., Benson T. E., Kiefer J. R., Marino M. H., Carroll J. N., Leone J. W., Malfait A. M., Arner E. C., Tortorella M. D., Tomasselli A. (2008) J. Biol. Chem. 283, 1501–1507 [DOI] [PubMed] [Google Scholar]
- 30.Wewer U. M., Mörgelin M., Holck P., Jacobsen J., Lydolph M. C., Johnsen A. H., Kveiborg M., Albrechtsen R. (2006) J. Biol. Chem. 281, 9418–9422 [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 32.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 34.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 36.Sørensen H. P., Jacobsen J., Nielbo S., Poulsen F. M., Wewer U. M. (2008) Protein Expr. Purif. 61, 175–183 [DOI] [PubMed] [Google Scholar]
- 37.Leonard J. D., Lin F., Milla M. E. (2005) Biochem. J. 387, 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loechel F., Overgaard M. T., Oxvig C., Albrechtsen R., Wewer U. M. (1999) J. Biol. Chem. 274, 13427–13433 [DOI] [PubMed] [Google Scholar]
- 39.Cao Y., Kang Q., Zhao Z., Zolkiewska A. (2002) J. Biol. Chem. 277, 26403–26411 [DOI] [PubMed] [Google Scholar]
- 40.Egelman D. M., Montague P. R. (1999) Biophys. J. 76, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence M. C., Colman P. M. (1993) J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 42.Jacobsen J., Visse R., Sørensen H. P., Enghild J. J., Brew K., Wewer U. M., Nagase H. (2008) Biochemistry 47, 537–547 [DOI] [PubMed] [Google Scholar]
- 43.Weiss M. S. (2001) J. Appl. Crystallogr. 34, 130–135 [Google Scholar]
- 44.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]