Abstract
Spiders spin high performance threads that have diverse mechanical properties for specific biological applications. To better understand the molecular mechanism by which spiders anchor their threads to a solid support, we solubilized the attachment discs from black widow spiders and performed in-solution tryptic digests followed by MS/MS analysis to identify novel peptides derived from glue silks. Combining matrix-assisted laser desorption ionization tandem time-of-flight mass spectrometry and cDNA library screening, we isolated a novel member of the silk gene family called pysp1 and demonstrate that its protein product is assembled into the attachment disc silks. Alignment of the PySp1 amino acid sequence to other fibroins revealed conservation in the non-repetitive C-terminal region of the silk family. MS/MS analysis also confirmed the presence of MaSp1 and MaSp2, two important components of dragline silks, anchored within the attachment disc materials. Characterization of the ultrastructure of attachment discs using scanning electron microscopy studies support the localization of PySp1 to small diameter fibers embedded in a glue-like cement, which network with large diameter dragline silk threads, producing a strong, adhesive material. Consistent with elevated PySp1 mRNA levels detected in the pyriform gland, MS analysis of the luminal contents extracted from the pyriform gland after tryptic digestion support the assertion that PySp1 represents one of the major constituents manufactured in the pyriform gland. Taken together, our data demonstrate that PySp1 is spun into attachment disc silks to help affix dragline fibers to substrates, a critical function during spider web construction for prey capture and locomotion.
All spiders (order Araneae, ∼40,000 described species) spin silk, but members of the highly diverse suborder Araneomorphae (∼37,000 species) spin multiple high performance fibers that enable them to perform a wide range of functions, including prey capture, locomotion, and protection of developing offspring (1, 2). Araneomorph spiders use specialized abdominal glands to express up to seven different fibroins, protein-based silks/glues, that have diverse mechanical properties.
Over the past 20 years, seven distinct members of the silk gene family have been identified and characterized at the molecular level, which include the silk proteins MaSp1 and MaSp2 (dragline silk)(3–5), AcSp1 or AcSp1-like (wrapping silk and egg case silk)(6,7), TuSp1 (egg case silk)(8–11), MiSp1 and MiSp2 or MiSp1-like (temporary spiral capture silk or web reinforcement silk)(12,13), and Flag silk (capture spiral silk in orb weavers)(14). These spider fibroins have revealed that they share a number of distinctive properties, including four fundamental amino acid repeat motifs that characterize the majority of the family: (i) alternating glycine alanine couplets (GAn), (ii) polyalanine blocks (An), (iii) GGX (X = subset of residues, which include Leu, Ile, and Ala), and (iv) GPGXn. Biophysical studies support that repeated GA couplets or polyalanine blocks form β sheet structures that correspond to the crystalline regions in the spun fibers, which provide much of the high tensile strengths for these silk types (15–17), whereas the GGX motifs give rise to a non-structured, amorphous region. GPGXn repeats, which are found in MaSp2 and Flag silk, have been hypothesized to form β turn structures, which provide extensibility to the threads (14). To date, the only reported full-length gene sequences from the spider silk family include masp1, masp2, and cysp1 (11, 18). These gene sequences are known to encode large molecular mass proteins, with their corresponding predicted molecular masses representing 250, 311, and 277 kDa, respectively.
Although much emphasis has been placed on studying the major and minor ampullate glands, as well as the tubuliform glands of araneomorph spiders, little, if any information regarding the chemical properties of the pyriform gland has been reported. Based upon observational and histological data, the pyriform gland has been implicated in the production of attachment disc silks, which function to affix dragline silk to substrates (19–21). SEM data support the spigots of the pyriform gland as being relatively small and numerous, located near the major ampullate spigots on the same spinneret (22). Attachment discs have an important biological function for the spider, as they serve to cement dragline silks to a solid support, anchoring the web to wood, concrete, or other surfaces during web construction for prey capture. Additionally, dragline silk, which is often referred to as a “safety line” for the spider has been observed to fuse with attachment disc silk, providing dragline silk with a secure anchor point for locomotor functions to assist prey capture and predator evasion. Although the current hypothesis supports the pyriform gland in the production of fibroins that are spun into short, sticky threads, no glue silk fibroin family members have been identified and characterized at the molecular level that match this description. Moreover, given the nature of attachment disc silks, which are spun in a gluey liquid material that dries to facilitate affixing dragline silk, it is uncertain whether the biochemical properties of the attachment disc silk fibroins would be highly divergent from traditional silk family members.
To investigate the hypothesis that the pyriform gland manufactures silk fibroins to help anchor dragline silk to solid surfaces, we solubilized the attachment discs with chaotropic agents, followed by in-solution tryptic digests and MS/MS analysis to search for peptides potentially derived from the sticky threads. MS3 MS analysis of 55 distinct peptides, coupled with cDNA library screening, led to the identification of a cDNA that encodes a new member of the spider silk gene family. Molecular and biochemical studies indicated that the pyriform gland was chiefly responsible for the expression of this gene, and it was subsequently named pyriform spidroin 1 (pysp1). Translation of the PySp1 cDNA revealed internal iterated blocks with a high degree of polar amino acid residues (Gln, Glu, Arg, and Lys) adjacent to triplet alanine repeats, with substantial divergence relative to repeat modules found within traditional fibroin family members. Translation of the PySp1 cDNA revealed the highest levels of alanine relative to other reported fibroin family members; however, surprisingly, little, if any, glycine was found in the PySp1 protein. Phylogenetic analysis of the C terminus revealed that PySp1 was a member of the spider silk family, but also showed that it was a highly divergent fibroin with distinctive internal molecular features. Collectively, these data suggest the unique chemical properties of PySp1 likely facilitate its ability to be spun into a sticky liquid environment that is well suited to cement dragline silk to solid surfaces. Given the specialized chemical properties of PySp1, these findings have substantial impact for structural biologists and material scientists seeking to engineer synthetic silks that have diverse properties for applications that include body armor, ropes and cords, tissue engineering, and drug delivery.
EXPERIMENTAL PROCEDURES
Scanning Electron Microscopy
Attachment disc silks were collected by anchoring the backside of SEM stubs to the walls of glass jars using tape. After several days, the spiders would spin attachment disc silks on the surface of the stubs. After detaching the top portion of the anchored scaffolding fibers embedded in the attachment disc silks from the SEM stubs using scissors, the attachment disc structures were analyzed using previously described procedures (23).
Collection of Attachment Disc Materials from Spiders
Spiders were placed into glass jars (∼8 inches tall and 3 inches in diameter) along with a small wooden stick for 2 weeks. Each spider was fed one cricket every 2 weeks. Prior to solubilization of the attachment discs, the scaffolding fibers were carefully severed with scissors. Using a sterile surgical blade, >200 attachment discs were scraped off the walls of glass jars. Materials on the blade were collectively dissolved in less than 50 μl of 9 m LiBr and pooled in a single microcentrifuge tube. After solubilization of the attachment discs, the solution was brought up to a 100-μl final reaction volume. To facilitate solubilization, the sample was heated for 20 min at 95 °C and then vortexed for an additional 20 min. Following vortexing, the sample was diluted from 9 to 1 m LiBr using 50 mm ammonium bicarbonate. LiBr was selected because it has been shown to be an effective solvent in dissolving fibroins (24). Samples were digested and the peptides extracted and desalted as previously described (6).
Mass Spectrometric Analysis
Conventional mass spectra were obtained with a MALDI-TOF-TOF mass spectrometer (4800 Proteomics Analyzer, Applied Biosystems, Foster City, CA) operated in reflectron mode. MS/MS spectra were obtained by operating this same instrument in MS/MS mode. De novo peptide sequences were obtained by manual interpretation of the high energy collision-activated dissociation spectra. Samples from the in-solution tryptic digest of attachment discs were separated by high pressure liquid chromatography and analyzed by mass spectrometry as previously described (6).
For the mass spectrometric analysis involving the luminal contents isolated from the pyriform gland, the proteins were extracted as described in the amino acid composition section. Following extraction of the proteins, the sample was re-suspended in buffer, digested with trypsin, and analyzed by MALDI-TOF as outlined above.
Cloning of the PySp1 cDNA
A composite cDNA library was prepared from the seven different silk-producing glands of the black widow spider. Hundreds of distinct clones were randomly isolated and subject to DNA sequencing as previously described (25). After DNA sequencing, a single clone was identified that carried a ∼2.4-kb cDNA insert that contained a long open reading frame (ORF) with a predicted region that showed similarity to the conserved, non-repetitive C-terminal region common to spider silk gene family members. This cDNA fragment was subsequently used to re-screen the cDNA library using conventional nucleic acid-nucleic acid hybridization to obtain a larger ∼3.6-kb cDNA fragment.
Phylogenetic Analysis
The C terminus DNA sequence for pysp1 was aligned with sequences obtained from GenBankTM representing all known members of the spidroin family except misp2, which was not available. Preliminary alignments were created in MUSCLE (26) and were heavily modified in MacClade version 4.08 (27) by eye with reference to the translated amino acid sequence. The data matrix consisted of 55 taxa that each contained 342 base pairs. The data were analyzed using the maximum likelihood criterion and a stochastic genetic algorithm with 25 replications as implemented in GARLI version 0.96 (28). Data were analyzed using (a) the GTR+G model for nucleotide data, (b) the Goldman and Yang (29) model for codons with F3 × 4 codon frequencies, and (c) the WAG+G+I+F model for amino acids (30). Models of evolution were indicated by the AIC criterion in ModelTest for nucleotide data (31) and ProtTest for amino acid data (32). One hundred nonparametric bootstrap replicates were also performed in GARLI for each evolutionary model.
Amino Acid Composition of Pyriform and Flagelliform Gland Contents
Luminal fluids were collected by separately treating each of the flagelliform, aggregate, tubuliform, and pyriform glands using an established protocol (6) and the contents were subjected to amino acid analysis at the Protein Chemistry Laboratory of Texas A & M University as previously described (33).
Real Time PCR Analysis
Total RNA was isolated from each of the seven different silk-producing glands, as well as fat tissue, using TRIzolTM according to the manufacturer's instructions. Reverse transcription reactions and real-time PCR analyses were performed as previously described (34). Oligonucleotides used for the analysis of PySp1 were the forward and reverse primers 5′-AGC TGC TGC CAA AGC TAA AA-3′ and 5′-GGA GGA AGC ACT TTC ACT GC-3′, respectively. Primers used for normalization were designed using the housekeeping gene, β-actin (6).
RESULTS
Attachment Disc Silk Is Embedded in a Glue-like Matrix That Anchors Dragline Silk
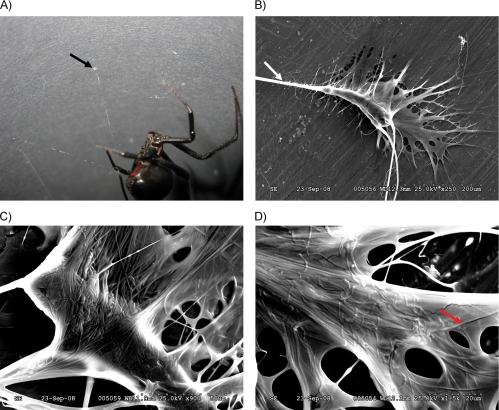
To investigate the physical structure of attachment discs produced by the black widow spider, female spiders were allowed to spin attachment discs on the surface of either cardboard paper (Fig. 1A) or SEM stubs. Analyses of the attachment discs spun directly on stubs for SEM revealed the presence of large diameter fibers that penetrated into a glue-like matrix, with the sticky material making several distinct contacts to the surface with finger-like projections (Fig. 1B). These large diameter threads represent dragline silk fibers anchored to the surface, which are embedded in a glue-like substance containing smaller diameter fibers (Fig. 1C). Complex networks of the smaller diameter fibers were observed that formed a mesh-like structure (Fig. 1D). Larger diameter fibers measuring 4 ± 0.2 μm, a size consistent with major ampullate silks from the black widow spider, were observed leading into the attachment disc structure (Fig. 1B). Smaller diameter fibers were 1 order of magnitude smaller relative to the large diameter fibers, measuring 0.4 ± 0.02 μm.
FIGURE 1.
Physical structure of an attachment disc collected from a black widow spider. A, black widow spiders use attachment disc material (black arrow) to anchor dragline silk to substrates to facilitate locomotion and web-building functions. This image was captured with a Canon EOS 40D digital camera. B, SEM of an attachment disc at ×250 magnification; white arrow denotes dragline silk fibers leading into attachment disc. C, SEM of an attachment disc at ×900. D, SEM of an attachment disc at ×1500; red arrow represents an attachment disc fiber.
MS/MS Analysis of Tryptic Digest Products from Solubilized Attachment Discs Reveals Novel Peptide Sequences
To identify the major structural proteins within the attachment discs, we solubilized the attachment discs using LiBr and performed biochemical analyses on the dissolved proteins. SEM was used to monitor the effectiveness of the solubilization process (data not shown). MALDI-TOF analysis of the in-solution tryptic digests from the solubilized attachment discs revealed 55 distinct peptide masses (data not shown). All 55 peptides were sequenced using high energy collision-activated dissociation. Analysis of the peptide sequences against the nrNCBI protein data base using BLASTp revealed that 23 peptides were similar to MaSp1 or MaSp1 variants (identities ranged from 78 to 100% with peptide lengths spanning from 11 to 29 residues), 1 peptide matched MaSp2 (100% identity for 19 amino acid residues) and 29 peptides showed no significant matches to other proteins in the data base. Six of the peptides derived from the MaSp molecules, those with m/z values of 1319.6, 1611.8, 1622.8, 2372.1, 3115.5, and 3297.5, are shown in Table 1. Of the 29 unknown peptides, peptide ions 864.4, 986.5, 1014.5, 1553.7, 1595.8, and 2267.1 are reported in Table 1. The product ion spectra for the peptides MH+ 986.5 and 1563.8 are shown in Fig. 2, A and B, respectively. The N termini of ions 1553.7, 2372.1, 3115.5, and 3297.5 could not be successfully retrieved, but a significant amount of amino acid sequence was obtained (Table 1). Two remaining peptide ions, those with m/z values of 1206.6 and 1222.7, contained sequences that were 100 and 90% identical, respectively, to regions found within SCP-1, a glue-coating peptide previously reported on gumfooted lines, scaffolding threads, and egg sacs in the black widow spider (35). Collectively, these data support the presence of the dragline silk fibroins MaSp1 and MaSp2, along with SCP1 and other uncharacterized proteins, embedded within attachment disc material.
TABLE 1.
MS/MS analyses of tryptic fragments generated from the in-solution digest of solubilized attachment discs from L. hesperus supports the presence of the dragline silk fibroins MaSp1 and MaSp2, silk coating peptide SCP-1, as well as other proteins
The N termini of the peptides generating ions at m/z 1553.7, 2372.1, 3115.5, and 3297.5 could not be successfully retrieved by the MS/MS analysis. Parentheses for ions 1319.6 and 1611.8 indicate residues that could not be conclusively determined by the MS/MS analysis. Bold text, along with the asterisks, indicates peptides that were later found within the new attachment silk fibroin after translation of the PySp1 cDNA sequence (Fig. 3A).
|
De novo sequences obtained by mass spectrometry | ||
|---|---|---|
| Peptide mass (M+H) | Sequence | Identity |
| 864.4 | AYAEALAR | PySp1* |
| 986.5 | AQAQAEAAAR | PySp1* |
| 1014.5 | AQAQVEAAAR | PySp1* |
| 1206.6 | AVHHYEVPVR | SCP1 |
| 1222.7 | AVHHYEVLVR | SCP1 |
| 1319.6 | AAAGGAG(AG/Q)GGLGGYGR | MaSp1 |
| 1553.7 | …LAPYYGNYYR | Unknown |
| 1563.8 | LASLSQAMSSVLSGGR | PySp1* |
| 1595.8 | TTVPLYLQFTEQR | Unknown |
| 1611.8 | (N/GG)AGQGGAAAAAAAAAGAGQR | MaSp1 |
| 1622.8 | GPGGSGAAAAAAAAAGGAGGPR | MaSp2 |
| 2267.1 | YLYLDDNVLNGAQNWLLSR | Unknown |
| 2372.1 | …GGQGAGQGGAGAAAAAAAAGGAGQGGQR | MaSp1 |
| 3115.5 | …DNGVTTNALADALTSAFYQTTGVVNSR | MaSp1 |
| 3297.5 | …GGYGQGGYGQGGAGQGGAGAAAAAAAAAGGAGQGGYGR | MaSp1 |
FIGURE 2.
MALDI-TOF tandem analyses of precursor peptide ions generated from an in-solution tryptic digestion of solubilized attachment disc materials. A, high energy collision-activated dissociation spectrum of precursor ion with m/z 986.5 (MH+, monoisotopic). The sequence of this peptide was found to be AQAQAEAAR. B, high energy collision-activated dissociation spectrum of precursor ion with m/z 1563.8. This peptide ion had the derived sequence LASLSQAMSSVLSGGR. The N- or C-terminal leucine residues could be any combination of leucine and/or isoleucine. The middle leucine residue was confirmed with the existence of the w13 ion.
A New Member of the Spider Silk Gene Family, PySp1, Contains Distinct Molecular Block Repeats
During a random cDNA library screen for silk genes, we fortuitously isolated a ∼2.4-kb cDNA sequence that, after translation, indicated the fragment contained a long ORF. Analysis of the ORF indicated that the fragment encoded the C-terminal region of a putative silk fibroin. Using the 2.4-kb cDNA fragment as a probe to screen our cDNA library by conventional nucleic acid-nucleic acid hybridization, we isolated a longer cDNA sequence that was ∼3.6 kb (ORF; GenBank accession FJ973621; Fig. 3A). Translation of this cDNA sequence revealed no in-frame translational start codon, suggesting this cDNA represents a partial sequence. The longest ORF encodes a 1153-amino acid protein, with a predicted molecular mass of 116.5 kDa. Based upon the sequences of full-length silk family members, which predict masses ranging from ∼250 to 310 kDa, this implies our cDNA sequence likely corresponds to ∼50% of the PySp1 gene sequence. Protein-protein BLAST searches against the nrNCBI data base using the last 122 amino acids, which corresponds to the highly conserved, non-repetitive C-terminal region in the spider silk gene family, revealed similarity to other members of the spider gene family. Alignment of the C-terminal 122 amino acid residues of this clone to fibroins MaSp1, MaSp2, AcSp1-like, MiSp1-like, and TuSp1 from L. hesperus revealed conservation at specific residues, but divergence in others (Fig. 3B). However, when the 3.6-kb translated cDNA sequence was analyzed using BLASTp against the protein data base, there were no strong matches to any of the spider gene family members. The top matches corresponded to a hypothetical protein from Streptococcus pneumoniae (GenBank accession ZP02717830; E-value 8 × 10−83 with 370/1059 identities), a kinetoplast-associated protein-like protein from Leishmania braziliensis (GenBank accession XP_001565786; E-value 1 × 10−74 with 448/909 identities), and a neurofilament protein from Trichomonas vaginalis (GenBank accession EAY15892.1; E-value 6 × 10−72 with 333/892 identities). Manual inspection of the translated cDNA revealed novel short block repeats. These repeats consisted of the sequences AAARAQAQAEARAKAE and AAARAQAQAE, which were iterated many times within our translated PySp1 cDNA. Interestingly, scattered throughout the amino acid sequence there were short blocks of alanine that consisted of 3 residues (Fig. 3A). Three larger 78-amino acid iterations were found to interrupt runs of the AAARAQAQAEARAKAE and AAARAQAQAE motifs; these 78-amino acid iterations were exceptionally hydrophilic, containing over 14% histidine, 22% serine, 14% threonine, 10% glutamine, and 7% glutamic acid (Fig. 3A). Theoretical digestion of the translated cDNA sequence with trypsin revealed two peptides that contained identical sequences to those found for ions 986.5 and 1563.8; these ions were found in the in-solution tryptic digest of the attachment discs (compare Table 1 to Fig. 3A). Peptide ion 986.5, which was found 10 times within the translated amino acid sequence, represents part of a novel molecular module that is absent in other spidroin family members (Fig. 3A). Additionally, peptide ion 1563.8, which localized to the non-repetitive conserved region, was unique to our translated cDNA and was not found in the C-terminal regions of other family members (Fig. 3B). Phylogenetic analysis of a 342-base pair alignment of the C terminus of spider fibroin genes strongly support that PySp1 is a member of the fibroin gene family. Both the codon and amino acid models indicate PySp1 is closely related to the TuSp group (Fig. 3C), whereas the nucleotide model placed the pysp1 sister to the flag genes, with this grouping closest to the tusp gene (data not shown). Bootstrap values for these basal nodes were low.
FIGURE 3.
The amino acid sequence of L. hesperus PySp1 shows similarity to spider fibroin family members. A, translation of the nucleotide sequence from the PySp1 cDNA contains an ORF. Peptide sequences determined by MS/MS analysis (in-solution tryptic digest products from LiBr solubilized attachment disc silks) that are found within the ORF are underlined. Red and blue denote the block repeats with either the consensus AAARAQAQAEARAKAE or AAARAQAQAE. The last 122 amino acids represent the non-repetitive C terminus. B, alignment of the PySp1 C-terminal sequence reveals similarity to other fibroin family members. Alignments were performed with the last C-terminal 122 amino acids of the fibroins using the computer algorithm CLUSTALW. Amino acids are represented by one-letter abbreviations, gaps are indicated by dashes. Shaded gray areas represent residues that are found in the highest frequency at that position for the fibroin family members. The blue underlined region represents a segment that is 100% identical to the peptide ion at m/z 1563.8; this region is unique to PySp1. Purple denotes a 78-amino acid region that is rich in histidine and other polar residues. GenBank accession codes for the L. hesperus fibroin family members are listed: MaSp1 (DQ409057), MaSp2 (DQ409058), MiSp1-like (EU394445.1), PySp1 (FJ973621), AcSp1-like (EU025854.1), and TuSp1 (DQ109035). C, maximum likelihood tree of the nucleotide sequence encoding the conserved, non-repetitive C-terminal region of spider fibroin genes from all known members of the gene family except MiSp2 using the codon model of Goldman and Yang (29) with F3 × 4 codon frequencies. The model of evolution used was GTR+G. PySp1 apparently arose through duplication of the ancestral gene that also gave rise to tusp genes. Although this placement is not robust to bootstrapping, the same phylogenetic arrangement was found when the data were analyzed by an amino acid model (WAG+I+G+F). D, predicted amino acid composition profile from the translated cDNAs L. hesperus MaSp1 (EF595246.1), MaSp2 (EF595245.1), TuSp1 (AY953070.1), MiSp1-like (EU394445.1), AcSp1-like (EU025854.1), and PySp1 (FJ973621). The translated gene sequences used for MaSp1 and MaSp2 represent full-length protein sequences, whereas TuSp1, MiSp1-like, AcSp1-like, and PySp1 represent partial protein sequences.
To compare the theoretical amino acid composition of PySp1 to other fibroin family members, we translated their sequences and analyzed their amino acid percentages. Translation of the PySp1 cDNA revealed that this fibroin contained the highest amount of alanine but the lowest amount of glycine relative to other fibroin family members (Fig. 3D). PySp1 also contained the highest amount of glutamic acid, as well as significant amounts of the polar amino acid residues serine, threonine, arginine, lysine, and glutamine. Relative to the other spidroins, PySp1 displayed the largest percentages of amino acid residues with R groups with either positive (Lys and Arg, Fig. 3D) or negative charges (Glu and Asp, Fig. 3D). Although the protein sequences for TuSp1, MiSp1-like, AcSp1-like, and PySp1 represent partial sequences, their amino acid composition profiles are assumed to be representative of the full-length sequences because silk protein family members are largely comprised of block repeat modules. Overall, these data support the assertion that the PySp1 protein is unique and has distinctive molecular properties relative to other fibroin family members, representing a novel fibroin member.
Amino Acid Composition Analyses of the Luminal Contents of the Spider Silk Producing Glands
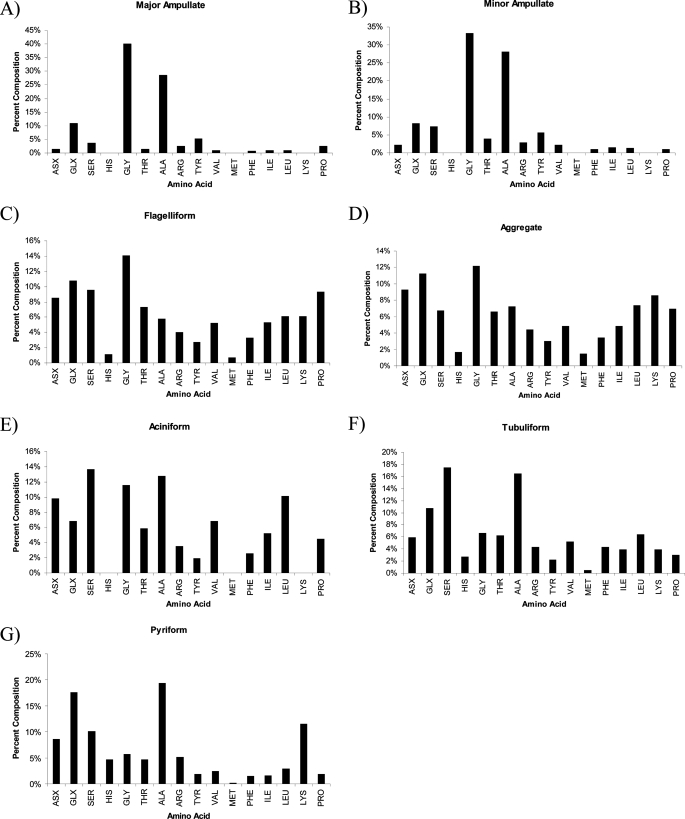
Recent studies have shown that spider silk glands are specialized structures designed to store large quantities of silk proteins (7). In many cases, it has been shown that a single silk family member is stored within one silk-producing gland. To determine whether PySp1 was the major constituent stored within the pyriform gland, we extracted the luminal contents and hydrolyzed the proteins with acid for an amino acid composition analysis. Comparative analysis of the predicted amino acid composition of PySp1 to the hydrolyzed luminal contents revealed remarkably similar amino acid percentages for serine and glutamine (Table 2). Despite the similarities for several amino acid residues, alanine levels were substantially higher in the translated PySp1 cDNA relative to the stored contents within the pyriform gland (Table 2). To compare the amino acid composition profiles for the luminal proteins stored within all seven silk producing glands from L. hesperus, we extracted the proteins and performed acid hydrolysis, followed by quantification of the residues (Fig. 4). Amino acid composition profiles for the luminal contents extracted from the major and minor ampullate glands revealed large amounts of glycine and alanine (Fig. 4, A and B), whereas the flagelliform and aggregate glands displayed a more complex mixture of different residues (Fig. 4, C and D). Proteins stored in the aciniform glands were rich in serine, glycine, and alanine (Fig. 4E). Both the tubuliform and pyriform glands contained similar amounts of alanine and glutamic acid and/or glutamine (denoted as Glx), but the pyriform gland contained higher amounts of lysine (Fig. 4, F and G).
TABLE 2.
Predicted amino acid composition of PySp1, as well as the experimentally determined composition of proteins extracted from the pyriform glands from L. hesperus
The results are expressed as an average of multiple samples (n = 2). The values are expressed as mol % per 100 residues.
| Gly | Ala | Glx | Tyr | Pro | Arg | Asx | Leu | Ser | Ile | Thr | Val | Phe | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicted PySp1 of L. hesperusa | 0.3 | 42.5 | 23.1 | 0.9 | 0.3 | 8.5 | 0.8 | 8.2 | 1.6 | 1.3 | 4.3 | 2.2 | 0.3 |
| Luminal contents from the pyriform gland | 5.7 | 19.4 | 17.6 | 1.9 | 1.9 | 5.2 | 8.6 | 10.2 | 2.9 | 1.6 | 4.7 | 2.5 | 1.5 |
a Data are derived from the predicted amino acid sequence of L. hesperus PySp1 using the computer algorithm, ProtParam. Lys (1.9%), His (3.6%), Met (0.1%), and Trp (0%) are not shown in the table.
FIGURE 4.
Luminal contents of the pyriform gland contain proteins with large amounts of alanine and glutamine. Proteins stored in the lumen of the seven different silk-producing glands from L. hesperus were extracted and subjected to acid hydrolysis. A, major ampullate; B, minor ampullate; C, flagelliform; D, aggregate; E, aciniform; F, tubuliform; and G, pyriform. Data from the major and minor ampullate glands, as well as the aciniform gland were previously determined (6, 12).
Expression of PySp1 Is Restricted to the Pyriform Gland
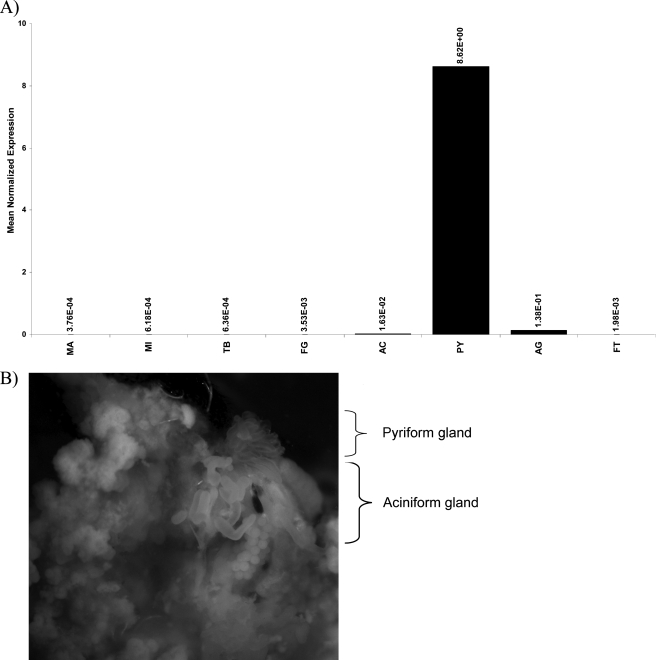
Because the silk community has hypothesized that the pyriform gland produces attachment disc silks, we investigated the expression pattern of our novel fibroin using quantitative real time PCR analysis. All seven silk-producing glands were dissected from the abdominal region of the black widow spider and total RNA was isolated for real time quantitative reverse transcription-PCR analysis. Our novel fibroin mRNA levels were the highest in the pyriform gland, with virtually no mRNA being detected in the major and minor ampullate glands, tubuliform, flagelliform, and aciniform glands (Fig. 5A). Expression of the novel fibroin mRNA was found to be >500-fold higher in the pyriform gland relative to these other tissues. Fat tissue, which was used as a negative control, also showed no significant levels of expression (Fig. 5A). Aggregate tissue, which showed the second highest level of mRNA production, displayed 60-fold lower amounts of mRNA compared with the pyriform gland (Fig. 5A). Similar to the reported morphological features of pyriform glands from orb weaver spiders, the pyriform glands from L. hesperus were also small, slightly elongated, and irregularly shaped (Fig. 5B). Based upon the mRNA expression pattern, we have named this new fibroin PySp1.
FIGURE 5.
Pyriform glands are fingerlike structures that express PySp1 mRNA at high levels. A, real time quantitative PCR analysis was used to determine the mRNA expression pattern of the novel fibroin in a variety of different tissues. Total RNA was isolated from the major ampullate gland (MA), minor ampullate gland (MI), tubuliform (TB), flagelliform (FL), aciniform (AC), pyriform (PY), aggregate (AG), and fat tissues (FT). Equivalent amounts of total RNA were reversed transcribed using Superscript III and aliquots used for quantitative reverse transcriptase-PCR. Reactions were performed in triplicate and normalized internally using the black widow actin mRNA. Data are representative of experimental results obtained from two independent trials. B, pyriform glands were dissected from black widow spiders (upper right, pyriform gland removed from a single spider and photographed on a Leica MZ16 dissecting microscope at ×10 magnification; lower left, aciniform tissue is also shown in the field of view; fat tissue is surrounding both glands).
To confirm that the PySp1 mRNA was translated and the protein was stored within the luminal contents of the pyriform gland, we dissected the pyriform gland from the spider and extracted the stored protein contents from the tissue. Following extraction of the luminal proteins from the tissue, we subjected the extracted protein lysate to an in-solution tryptic digestion and analyzed the peptide masses using MS analysis. MS analysis of the digest revealed 11 different peptide ion masses that were identical to PySp1 products after theoretical digestion of the translated PySp1 sequence with trypsin, which included ion masses 864.4, 986.5, 995.5, 1014.5, 1040.5, 1222.6, 1548.7, 1563.8, 1836.8, 2582.2, and 2899.3 (Fig. 6 and Table 3). Based upon our translated PySp1 cDNA sequence, this corresponds to ∼33% sequence coverage (Figs. 3A and 6 and Table 3). Several other peptide ions that were relatively abundant were not found to match products after theoretical digestion of the translated PySp1 sequence, suggesting these ions were derived from the missing N-terminal region of PySp1 or other proteins found within the luminal contents. These ions corresponded to masses 1179.5, 1277.6, 1308.6, 1427.7, 1475.6, 1636.7, 1729.6, 1792.7, and 2384.5. Collectively, these data support the pyriform gland as the principal tissue responsible for the production of the PySp1 protein.
FIGURE 6.
MALDI-TOF mass spectrum of an in-solution trypsin digestion of luminal contents extracted from the pyriform gland of L. hesperus. Peptide ions from the protein PySp1 are labeled with #, with the exception of peptide ion 995.5. Trypsin autolysis peptide signals are labeled with an asterisk. The spectrum was recorded using a CHCA matrix and the instrument was operated in reflectron mode.
TABLE 3.
Observed m/z values of [M+H]+ PySp1 peptides from tryptic digestion of the luminal proteins extracted from the pyriform gland from the black widow spider, L. hesperus, which give a coverage of ∼33% over the translated PySp1 sequence
| PySp1 peptide | [M + H]theor+ | [M + H]obs+ | |
|---|---|---|---|
| m/z | |||
| 1 | AYAEALAR | 864.4 | 864.4 |
| 2 | AQAQAEAAAR | 986.5 | 986.4 |
| 3 | AHAQAEAAAR | 995.5 | 995.5 |
| 4 | AQAQVEAAAR | 1014.5 | 1014.5 |
| 5 | AQAEAHSQAK | 1040.5 | 1040.5 |
| 6 | AQAHAQAEAVAR | 1222.6 | 1222.6 |
| 7 | AQAEAAALAHAQAVAR | 1548.8 | 1548.7 |
| 8 | IASLSQAMSSVISGGR | 1563.8 | 1563.8 |
| 9 | SPGSEENFVESVLQAFP | 1836.8 | 1836.8 |
| 10 | VNYAALSSSLSGLASEIQNESNLSK | 2582.3 | 2582.2 |
| 11 | AHTEADTYSQAQSAAHTIAAAQAQAETR | 2899.4 | 2899.3 |
DISCUSSION
PySp1 is the first spider glue silk cDNA to be isolated and characterized at the molecular level and its product could be best described as “Spider Man's glue silk.” Analysis of the amino acid sequence of PySp1 reveals it lacks conventional subrepeat modules that are common characteristics of other spider silk fibroin family members. No glycine or proline residues were identified within the internal repeat block units of PySp1, which is strikingly different from major and minor ampullate, tubuliform, and aciniform silks (7, 8, 13, 18). New block motifs were also identified within the translated amino acid sequence of PySp1, which include the motifs AAARAQAQAEARAKAE and AAARAQAQAE. Additionally, although the translated cDNA sequence predicted substantial amounts of alanine, there were no long polyalanine stretches similar to block repeats found in MaSp1 or MaSp2 (18). Instead, short runs of 3 consecutive alanine residues appeared in a regular pattern within the PySp1 sequence. Consistent with belonging to the spider silk family, PySp1 was found to contain the non-repetitive, conserved C terminus that is characteristic of fibroin family members (36–38). Although our PySp1 cDNA represents a partial sequence, it is likely representative of the full-length sequence because silk proteins are known to be modular in nature and consist of block repeats. Obtaining full-length spider silk cDNA sequences has been very challenging for the silk community, given their repetitive nature, long lengths, and rich GC contents. Overall, our clone represents one of the longer cDNAs published by the silk community.
Previous histological studies reveal the pyriform silks are extruded from small diameter spigots located on the same spinneret adjacent to large diameter spigots that produce major ampullate silks (22). This observation is consistent with the ultrastructure of attachment discs obtained by our SEM studies, which revealed networks of similarly sized small diameter fibers embedded within a cement-like substance. Based upon the diameter sizes of the pyriform spigots, the small diameter fibers embedded in the cement are consistent with extrusion from the pyriform spigots. Therefore, our data support the assertion that PySp1 represents the fibrillar protein component of the mesh-like material that forms the small diameter fibers observed in attachment disc silks.
Our MS/MS analysis of solubilized attachment discs after tryptic digestion revealed two peptides that were 100% identical to regions found within the translated PySp1 cDNA sequence. Both of the peptides are unique to PySpl and are not found in the translated sequences of the other fibroin family members. These peptides corresponded to ions 986.5 and 1563.8. A third tryptic peptide, AQAQVEAAAR (mass 1014.5), was virtually identical to peptide AQAQAEAAAR (mass 986.5), except it contained a valine instead of an alanine at the fifth residue. More specifically, peptide ion 1563.8, which was 16 amino acids in length, mapped to within the non-repetitive, conserved C-terminal region of the family. However, despite the conservation of the family within this region, alignments of the C-terminal regions of all the family members and BLASTp searches against the protein data base clearly demonstrated this peptide was derived from PySp1 and is unique. Additionally, MS and MS/MS analyses of egg cases, dragline silk, gumfooted lines, and scaffolding threads following fiber solubilization and in-solution tryptic digest revealed no indication of the presence of PySp1 peptide fragments in these fiber types (data not shown). Collectively, this supports a specialized role for PySp1 in attachment disc structures.
Although our translated PySp1 cDNA sequence predicted around 43% alanine, the amino acid composition of the luminal contents stored within the pyriform gland revealed 19% alanine. This discrepancy would seem to support the assertion that other protein(s) are also present within the pyriform gland that either lack or have lower alanine levels. Although 4 of the 29 unknown peptides from the MS/MS analysis could be located within the translated PySp1, the remaining 25 peptide sequences searched against the nrNCBI protein data base acid revealed no significant similarity to any proteins, including MaSp1 or MaSp2. Based upon this observation, our data suggest that these peptides could be derived from other proteins secreted into the attachment discs or they could correspond to the missing N-terminal region of PySp1. This is also consistent with detection of peptide ion masses in the in-solution tryptic digest of the pyriform luminal protein extract that could not be accounted for within the translated PySp1 cDNA sequence, which implies that additional protein(s) are found in the luminal contents of the pyriform gland and likely extruded during the spinning process (Fig. 6). Further biochemical and molecular work will need to be performed to resolve the molecular identity of these constituents.
Twenty-four of the 55 peptides obtained by tryptic digestion of the solubilized attachment discs after sequencing by MS/MS analysis were clearly derived from MaSp1 or MaSp2, which is consistent with the supposition that dragline silks are embedded in attachment discs. Our studies also detected the presence of two peptides from SCP-1 in attachment disc materials. Previously, we have demonstrated that SCP-1 can be removed from scaffolding joints, gumfooted lines, and egg sacs after water treatment, supporting the assertion of the polar nature of this molecule (35). More extensive expression studies have also demonstrated that the flagelliform gland is capable of producing large amounts of SCP-1, which raises an interesting question regarding whether the flagelliform gland in black widow spiders is extruding materials that are constituents of attachment discs. Because water treatment can solubilize SCP-1, it seems unlikely this molecule is serving to anchor the fibers to the substrate. Biochemical analyses have shown that SCP-1 has the intrinsic ability to bind metal ions, but the precise biological significance of this property remains unclear (35). Furthermore, whether SCP-1 serves some antimicrobial function, or perhaps, facilitates the folding or assembly of extruded proteins remains to be elucidated.
Relative to the other spider silk family members, PySp1 may have evolved to have specialized molecular properties that are optimized for being spun into a liquid gluey substance that undergoes rapid solidification. The high degree of polar and charged amino acid residues embedded in its sequence would seem to support this assertion. Currently, the molecular mechanism that governs adhesion of the attachment disc silk fibers to the solid surface is unclear. Although PySp1 is spun into the attachment disc, we have also detected the presence of several small organic molecules (m/z ∼100–250 Da) in this structure using direct analysis in real time mass spectrometry, but additional chemical analyses will need to be conducted to determine their identities and biological functions.
One of the current obstacles for using dragline or Flag silks (only reported from orb-weavers) for commercial or military applications is that immersion of these fibers in water results in a process known as supercontraction (39). Under these conditions, the fibers shrink and swell, acting more like a rubber, which is not a desirable feature for materials exposed to wet environments (40). Because PySp1 contains block modules that have a high degree of polarity and charge, it will be of substantial value for materials scientists to determine whether attachment disc fibers display a lower degree of supercontraction when exposed to water or high humidity. Additionally, given the higher degree of hydrophilic residues embedded within the PySp1 amino acid sequence relative to other spider silk family members, PySp1 could prove more suitable for developing expression systems that achieve high amounts of soluble protein needed for the spinning of artificial spider silks for a wide range of different commercial and military purposes. Future experimental studies will help elucidate the unique intrinsic properties of PySp1 as well as its potential use for engineering applications.
Acknowledgments
We appreciate the critical reading of the manuscript by Keshav Vasanthavada (Roche, Palo Alto, CA). We are also thankful to Pat Jones for assistance with the direct analysis in real time mass spectrometer (University of the Pacific, Department of Chemistry, Stockton, CA).
This work was supported by National Science Foundation RUI Grant MCB-0544087.
- MS
- mass spectrometry
- MALDI-TOF
- matrix-assisted laser desorption ionization-time of flight
- SEM
- scanning electronmicrograph
- MaSp1 and MaSp2
- major ampullate spidroin 1 and 2
- MiSp1-like
- minor ampullate spidroin 1-like
- AcSp1-like
- aciniform spidroin 1-like
- PySp1
- pyriform spidroin 1
- ORF
- open reading frame.
REFERENCES
- 1.Foelix R. (1996) Biology of Spiders, Oxford University Press, New York [Google Scholar]
- 2.Schultz J. W. (1987) Biol. Rev. 62, 89–113 [Google Scholar]
- 3.Xu M., Lewis R. V. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 7120–7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerette P. A., Ginzinger D. G., Weber B. H., Gosline J. M. (1996) Science 272, 112–115 [DOI] [PubMed] [Google Scholar]
- 5.Hinman M. B., Lewis R. V. (1992) J. Biol. Chem. 267, 19320–19324 [PubMed] [Google Scholar]
- 6.Vasanthavada K., Hu X., Falick A. M., La Mattina C., Moore A. M., Jones P. R., Yee R., Reza R., Tuton T., Vierra C. (2007) J. Biol. Chem. 282, 35088–35097 [DOI] [PubMed] [Google Scholar]
- 7.Hayashi C. Y., Blackledge T. A., Lewis R. V. (2004) Mol. Biol. Evol. 21, 1950–1959 [DOI] [PubMed] [Google Scholar]
- 8.Hu X., Lawrence B., Kohler K., Falick A. M., Moore A. M., McMullen E., Jones P. R., Vierra C. (2005) Biochemistry 44, 10020–10027 [DOI] [PubMed] [Google Scholar]
- 9.Tian M., Lewis R. V. (2005) Biochemistry 44, 8006–8012 [DOI] [PubMed] [Google Scholar]
- 10.Garb J. E., Hayashi C. Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11379–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao A. C., Zhao T. F., Nakagaki K., Zhang Y. S., Sima Y. H., Miao Y. G., Shiomi K., Kajiura Z., Nagata Y., Takadera M., Nakagaki M. (2006) Biochemistry 45, 3348–3356 [DOI] [PubMed] [Google Scholar]
- 12.La Mattina C., Reza R., Hu X., Falick A. M., Vasanthavada K., McNary S., Yee R., Vierra C. A. (2008) Biochemistry 47, 4692–4700 [DOI] [PubMed] [Google Scholar]
- 13.Colgin M. A., Lewis R. V. (1998) Protein Sci. 7, 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi C. Y., Lewis R. V. (1998) J. Mol. Biol. 275, 773–784 [DOI] [PubMed] [Google Scholar]
- 15.Simmons A., Ray E., Jelinski L. W. (1994) Macromolecules 27, 5235–5237 [Google Scholar]
- 16.van Beek J. D., Hess S., Vollrath F., Meier B. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhe A. D., Seeley S. K., Gardner K., Thompson L., Lewis R. V. (1997) J. Mol. Recognit. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 18.Ayoub N. A., Garb J. E., Tinghitella R. M., Collin M. A., Hayashi C. Y. (2007) PLoS ONE 2, e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovoor J., Zylberberg L. (1980) Tissue Cell 12, 547–556 [DOI] [PubMed] [Google Scholar]
- 20.Kovoor J., Zylberberg L. (1982) Tissue Cell 14, 519–530 [DOI] [PubMed] [Google Scholar]
- 21.Vollrath F., Knight D. P. (2001) Nature 410, 541–548 [DOI] [PubMed] [Google Scholar]
- 22.Moon M. J., An J. S. (2006) Entomological Res. 36, 133–138 [Google Scholar]
- 23.Hu X., Kohler K., Falick A. M., Moore A. M., Jones P. R., Sparkman O. D., Vierra C. (2005) J. Biol. Chem. 280, 21220–21230 [DOI] [PubMed] [Google Scholar]
- 24.Sponner A., Vater W., Monajembashi S., Unger E., Grosse F., Weisshart K. (2007) PLoS ONE 2, e998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell B., Mugiya M., Youngblom J., Funes-Duran M., Miller R., Ezpeleta J., Rigby N., Vierra C. (2000) Biochim. Biophys. Acta 1492, 320–329 [DOI] [PubMed] [Google Scholar]
- 26.Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddison D. R., Maddison W. P. (2005) MacClade Version 4.08, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 28.Zwickl D. J. (2006) Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Likelihood Criterion, Version 0.95, Doctoral dissertation, The University of Texas, Austin, TX [Google Scholar]
- 29.Goldman N., Yang Z. (1994) Mol. Biol. Evol. 11, 725–736 [DOI] [PubMed] [Google Scholar]
- 30.Whelan S., Goldman N. (2001) Mol. Biol. Evol. 18, 691–699 [DOI] [PubMed] [Google Scholar]
- 31.Posada D., Crandall K. A. (1998) Bioinformatics 14, 817–818 [DOI] [PubMed] [Google Scholar]
- 32.Abascal F., Zardoya R., Posada D. (2005) Bioinformatics 21, 2104–2105 [DOI] [PubMed] [Google Scholar]
- 33.Casem M. L., Turner D., Houchin K. (1999) Int. J. Biol. Macromol. 24, 103–108 [DOI] [PubMed] [Google Scholar]
- 34.Nguyen L., Round J., O'Connell R., Geurts P., Funes-Duran M., Wong J., Jongeward G., Vierra C. A. (2001) Nucleic Acids Res. 29, 4423–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X., Yuan J., Wang X., Vasanthavada K., Falick A. M., Jones P. R., La Mattina C., Vierra C. A. (2007) Biochemistry 46, 3294–3303 [DOI] [PubMed] [Google Scholar]
- 36.Challis R. J., Goodacre S. L., Hewitt G. M. (2006) Insect Mol. Biol. 15, 45–56 [DOI] [PubMed] [Google Scholar]
- 37.Sponner A., Unger E., Grosse F., Weisshart K. (2004) Biomacromolecules 5, 840–845 [DOI] [PubMed] [Google Scholar]
- 38.Hu X., Vasanthavada K., Kohler K., McNary S., Moore A. M., Vierra C. A. (2006) Cell Mol. Life Sci. 63, 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Work R. W. (1977) Textile Res. J. 47, 650–662 [Google Scholar]
- 40.Gosline J. M., Denny M. W., DeMont M. E. (1984) Nature 309, 551–552 [Google Scholar]