Abstract
The simultaneous activation of many distinct G protein-coupled receptors (GPCRs) and heterotrimeric G proteins play a major role in various pathological conditions. Pan-inhibition of GPCR signaling by small molecules thus represents a novel strategy to treat various diseases. To better understand such therapeutic approach, we have characterized the biomolecular target of BIM-46187, a small molecule pan-inhibitor of GPCR signaling. Combining bioluminescence and fluorescence resonance energy transfer techniques in living cells as well as in reconstituted receptor-G protein complexes, we observed that, by direct binding to the Gα subunit, BIM-46187 prevents the conformational changes of the receptor-G protein complex associated with GPCR activation. Such a binding prevents the proper interaction of receptors with the G protein heterotrimer and inhibits the agonist-promoted GDP/GTP exchange. These observations bring further evidence that inhibiting G protein activation through direct binding to the Gα subunit is feasible and should constitute a new strategy for therapeutic intervention.
G protein-coupled receptors (GPCRs)3 represent the largest superfamily of signaling proteins with a very high impact on drug discovery (1). Approximately 30% of the current drug targets are indeed GPCRs and these latter are involved in all major disease areas (2). The classical drug discovery process selects and optimizes compounds that interact selectively with a specific receptor (1), but recent reports show that certain critical conditions such as cancer (3) or pain (4) are driven by the concomitant activation of many different GPCRs (5). Novel therapeutic strategies could therefore emerge from the simultaneous blockade of the various GPCRs involved in such pathologies. The GPCR signaling downstream cascade triggers several protein/protein interactions that may be blocked or modulated by small molecules (6). Such protein/protein interactions involve the GPCR transmembrane domain and the heterotrimeric G protein complex, composed of an α subunit (Gα) and a βγ dimer (Gβγ), which interact sequentially with several partners (e.g. guanine nucleotides, effectors, and regulatory proteins) (7). This offers multiple possibilities to develop small molecules controlling heterotrimeric G protein signaling (6, 8, 9). For example, Higashijima et al. (10, 11) showed that Mastoparan, a peptide toxin from wasp venom, directly acts on G proteins to mimic the role played by the activated receptors. The anti-helminthic drug Suramin and some analogs represent a second class of compounds that directly interact with G proteins and interfere with nucleotide exchange (12–14). Small molecules modulating regulator of G protein signaling proteins have also been proposed for drug development (15). More recently, Bonacci et al. (16) have described fluorescein analogs that display central pain relief activity via binding to the Gβγ subunits. From our own group, we have reported in vivo inhibition of the GPCR signaling pathway by two closely related imidazopirazine containing small molecules, displaying potent antiproliferative activity (BIM-46174) (17) and potent pain relief activity (BIM-46187) (18).
Here, we examined the molecular mechanisms underlying the biological activity of BIM-46187 with the various constituents of the GPCR signaling pathways. We report that this small molecule prevents GPCR-G protein signaling through a selective binding to the Gα protein subunit. Our results support the concept of targeting and inhibiting the heterotrimeric G protein complex as an approach to treat certain pathologies involving simultaneous activation of several GPCRs and/or heterotrimeric G proteins.
EXPERIMENTAL PROCEDURES
Chemicals, Reagents, and Plasmids
The cDNA of PAR1, PAR2, LPAR1, and β2-adrenergic receptors in pcDNA3.1 were purchased from cDNA Resources Center (Rolla, MO). Plasmids encoding for 5-HT4a, 5-HT2c, and V2 receptors were generously provided by Dr. S. Claeysen, Dr. P. Marin, and Dr. T. Durroux, respectively (Institut de Génomique Fonctionnelle, Montpellier, France). Serotonin, GABA, LPA, and forskolin were purchased from Sigma. Isoproterenol is from Tocris Cookson Inc. (Ellisville, MO). AVP is from Bachem (Bubendorf, Switzerland). Thrombin was from Calbiochem Merck. FUB132 was prepared as described by Breitweg-Lehmann et al. (19).
Plasmid Construction
For the BRET assay between receptors and G proteins, we used protease-activated receptors (PAR1 and PAR2) and V2 vasopressin receptors fused to YFP at their C terminus. These constructs were previously reported and shown to have similar pharmacological and functional properties as the wild-type receptors (20) (21). For the Gα-Rluc fusion proteins, the Renilla reniformis luciferase (Rluc) was fused to the G protein α subunits Gαi1, Gαo, Gα12, and Gαs in the loop between helices αA and αB in the h-α domain, after residues Ile93, Val93, Ile117, and Ala188, respectively, as previously described for Gαi1 (20). Such tag insertion at that position in Gαi1 (22), Gαo (23), and Gαs (22) was previously shown not to affect their activation by GPCRs nor their action on effectors. The same was also observed with Gα12.4 For BRET between G protein subunits, we used the Venus-tagged Gγ2 subunit generously provided by Dr. C. Galès (INSERM U858, Toulouse, France) (24) and YFP-tagged Gβ1 generated by creating the EcoRI restriction site 5′ of the pcDNA.1-Gβ1 plasmid (form cDNA Resources Center), and then the PCR product of YFP cDNA bearing EcoRI sites on 5′ and 3′ was inserted at the EcoRI site. Changes in intrinsic tryptophan fluorescence of Gαi upon the addition of AlF4− were carried out as described by Medkova et al. (25).
BRET Measurements
COS-7 cells were transiently transfected by electroporation with the indicated constructs, and 24 h after transfection they were washed with PBS and preincubated or not 2 h at 37 °C with BIM-46187 at the indicated concentrations in serum-free DMEM. The cells were then washed and resuspended in PBS, and BRET measurements were performed after the addition of drugs at the indicated concentrations and the luciferase substrate, Coelenterazine h (5 μm), as previously reported (20). BRET signals were expressed in milliBRET units of BRET ratio as previously described (26). The data represent the BRET ratio for each concentration of BIM-46187 under basal and agonist-stimulated conditions. The curves were fitted with a nonlinear regression and sigmoid dose-response equation using Prism GraphPad software (San Diego, CA).
In Vitro Assays Using Purified G Protein/Receptor
The BLT1 receptor was produced and reconstituted in asolectin vesicles as recently described (27). The Gαi2 and βγ proteins were also purified as previously described (28). The nucleotide exchange assay was carried out as described by Oldham et al. (29). The basal rate of GTPγS binding was determined by monitoring the relative increase in the intrinsic fluorescence (λex = 300 nm, λem = 345 nm) of Gαi in the presence of BLT1 and βγ in buffer containing 10 mm MOPS (pH 7.2), 130 mm NaCl, and 2 mm MgCl2 for 40 min at 15 °C after the addition of 10 mm GTPγS. Similarly, the receptor-catalyzed rate was measured under the same conditions in the presence of 50 μm LTB4. The data were normalized to the fluorescence maximum (100%). For the FRET experiments, BLT1 was labeled with Alexa-488 at a unique reactive cysteine located at position 51 in the first intracellular loop, whereas the Gαi2 protein used with a unique reactive cysteine at position 3 and recombinant β-arrestin-1 with a single reactive cysteine at position 172 (30) were labeled with Alexa-568 (the production and characterization of these mutants will be described elsewhere).5 We used the procedure described by Hickerson and Cunningham (31) for derivatization of these cysteine residues. Fluorescence spectra were recorded at 25 °C between 500 and 750 nm on a Cary Eclipse spectrofluorimeter with an excitation at 480 or 570 nm. Buffer contributions were systematically subtracted. All of the experiments were carried out in the presence of saturating concentrations in LTB4 (1 μm). Nonspecific FRET was taken into account by subtracting from the observed signal that measured under the same conditions when using Gαs labeled at its N terminus with Alexa-568 instead of Gαi2.
cAMP Production Measurement
The determination of the cAMP accumulation in COS-7 cells was performed in black 96-well microplates using the cAMP Dynamic kit according to the manufacturer's instructions (CisBio International, Bagnols sur Cèze, France). Briefly, the cells were pretreated with the indicated concentrations of BIM-46187 for 2 h at 37 °C and stimulated 30 min at 37 °C with the indicated agonists in diluent buffer (50 mm phosphate buffer, pH 7.0, 0.2% bovine serum albumin, 0.02% NaN3, and preservatives). The reaction was stopped by 0.5% Triton X-100 containing HTRF® assay reagents: the Europium Cryptate-labeled anti-cAMP antibody and the d2-labeled cAMP. The assay was incubated for 1 h at 4 °C, and Europium Cryptate fluorescence and the time resolved FRET signal at 620 and 665 nm were measured 50 μs after excitation at 337 nm using a RubyStar instrument (BMG Labtechnologies, Champigny-sur-Marne, France). The data represent cAMP levels in pmol/well calculated from the calibration curve generated with the increasing concentrations of cAMP added in the assay according to the manufacturer's instructions. Dose-response curves were fitted using Prism GraphPad software.
IP1 Production Measurement
The determination of the IP1 accumulation in COS-7 cells was performed in black 96-well microplates. The cells were incubated 1 h at 37 °C in the stimulation buffer (10 mm Hepes, pH 7.4, 1 mm CaCl2, 0.5 mm MgCl2, 4 mm KCl, 146 mm NaCl, 5.5 mm glucose, and 50 mm LiCl) containing the indicated agonists. The cells were then lysed by adding the HTRF® assay reagents, the Europium Cryptate-labeled anti-IP1 antibody, and the d2-labeled IP1 analog, previously diluted in a the lysis buffer containing 1% Triton X-100. The assay was incubated for 1 h at room temperature, and Europium Cryptate fluorescence and the time resolved FRET signal were measured 50 μs after excitation at 337, 620, and 665 nm, respectively, using a RubyStar instrument (BMG Labtechnologies, Champigny, France). The data represent IP1 levels in pmol/well calculated from the calibration curve generated with the increasing concentrations of IP1 added in the assay according to the manufacturer's instructions. Dose-response curves were fitted using Prism GraphPad software.
Intracellular Calcium Release Assay
The FLIPR calcium assay kit (Molecular Devices), a fluorescent assay, has been used for detecting changes in intracellular calcium in intact cells measured on FLIPR (fluorometric imaging plate reader) according to the manufacturer's instructions (Molecular Devices). Briefly, human melanoma cancer cells A2058 were plated in 96-well plates at a density of 300,000 cells/well in DMEM containing 10% fetal calf serum, 1% glutamine, and antibiotics at 37 °C in 5% CO2 humidified atmosphere. When reaching confluent monolayer, the cells were preincubated for 30 min with BIM-46187 as previously described (17).
Small G Protein Activity and Tyrosine Phosphorylation of Focal Adhesion Kinase (FAK)
The activation status of the small G protein Rac1 was quantified by pull-down assays in human colon cancer cells HCT8/S11 using PAK-1 immunoprecipitates as described (32, 33). GTP-bound and total levels of Rac1 were then detected by immunoblotting using the corresponding antibodies. The relative intensity of the autoradiographic bands was determined with the National Institutes of Health Image software system. Tyrosine phosphorylation of FAK at Tyr925 by the epidermal growth factor receptor signaling pathways in HCT8/S11 cells treated with epidermal growth factor was monitored by Western blots using the phospho-FAK Tyr925 polyclonal antibody (Alsace Biovalley, Illkirch, France), as described (33).
Assessment of Agonist-stimulated SRE-Luc Activity
24 h post-transfection of the serum-responsive element-luciferase gene reporter (SRE-Luc) without or with PAR1, COS-7 cells were washed with PBS and switched to serum-free DMEM overnight. The cells were then pretreated or not with: pertussis toxin (200 ng/ml, overnight), U73122 (10 μm for 2 h), Y27632 (10 μm for 2 h), or BIM-46187 (100 μm for 2 h), as indicated. For the last 6 h, the cells were stimulated with Me2SO, thrombin, or serum (10%) in the serum-free DMEM at 37 °C. The luciferase activity in cell extracts was then measured using the luciferase assay system (Promega Corp.) following the manufacturer's protocol using the Mithras LB 940 instrument.
Circular Dichroism Measurement
The CD spectra are the averages of five scans using a bandwidth of 2 nm, a step width of 0.2 nm, and a 0.5-s averaging time/point. The cell path lengths were 1.00 mm and 2.00 cm (far- and near-UV measurements, respectively). [θ] are mean residue (Mr = 115) molar ellipticities. Absorbance values in the 1 range were used to avoid any saturation of the photomultiplicator.
RESULTS
BIM-46187 Inhibits Heterotrimeric G Protein Downstream Signaling in Whole Cells
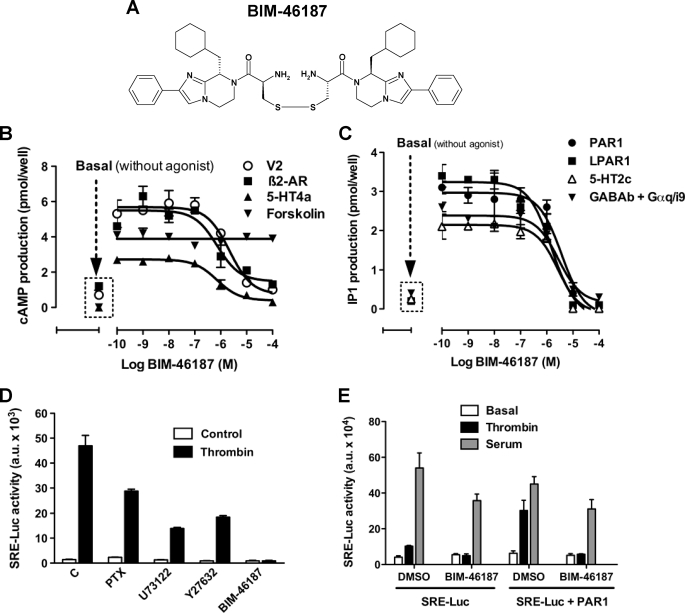
The effects of BIM-46187 (Fig. 1A) on the downstream signaling of various GPCR-G protein complexes were observed in whole cells, monitored by cyclic AMP production and IP1 production. Preincubation of cells with BIM-46187 dramatically inhibited both agonist-promoted cAMP production (Fig. 1B) and IP1 (Fig. 1C) mediated by all tested GPCR agonists. BIM-46187 activity was found to be concentration-dependent (Table 1), irrespective of any specific G protein-mediated events or any given activated GPCR. On the other hand, BIM-46187 was ineffective on forskolin-induced cAMP production (Fig. 1B). This diterpene is known to act directly on the adenylyl cyclase, suggesting that BIM-46187 acts directly on the receptor-G protein complex rather than on any downstream signaling proteins.
FIGURE 1.
BIM-46187 inhibits several GPCR downstream signaling pathways. A, chemical structure of BIM-46187. B and C, BIM-46187 inhibits cAMP accumulation (B) or IP1 (C) induced by activation of the indicated GPCR transiently expressed in COS-7 cells. Agonist concentrations used were: AVP (1 μm for V2 vasopressin), isoproterenol (10 μm for β2-adrenergic), serotonin (10 μm for 5HT4a), forskolin (10 μm on mock cells), thrombin (5 units/ml for PAR1), LPA (10 μm for LPA1), and GABA (100 μm for GABAb coexpressed with Gαq/i9). The data are the means ± S.E. from triplicate determinations from a representative experiment of three independent experiments. D, SRE-Luc gene reporter assay on cells transiently expressing PAR1 and stimulated by thrombin (5 units/ml) for 6 h after pretreatment overnight with Pertussis toxin (200 ng/ml) or 2 h with U73122 (10 μm), Y27632 (10 μm), or BIM-46187 (10 μm). E, SRE-Luc gene reporter assay on cells co-expressing SRE-Luc gene reporter without or with PAR1 and stimulated with thrombin (5 units/ml) or serum (10%) for 6 h after their pretreatment for 2 h with BIM-46187 (10 μm). The data are the means ± S.E. from triplicate determinations from a representative experiment of three independent experiments.
TABLE 1.
Effect of BIM-46187 on GPCR-G protein signaling
IC50 values of BIM-46187 on GPCR agonist-induced cAMP or IP1 production determined from experiments shown in Fig. 1.
| BIM-46187-inhibited IP1 production |
BIM-46187-inhibited cAMP production |
||
|---|---|---|---|
| Receptors | IC50 (n = 3) | Receptors | IC50 (n = 3) |
| μm | μm | ||
| PAR1 | 3.0 ± 0.7 | V2 | 2.7 ± 0.7 |
| LPAR1 | 1.6 ± 0.2 | β2-AR | 3.0 ± 0.8 |
| 5-HT2c | 2.0 ± 0.7 | 5-HT4a | 1.0 ± 0.2 |
| GABAb | 2.1 ± 1.0 | ||
To further demonstrate the pan-inhibitory effect of BIM-46187 on G protein-mediated signaling, we used the SRE-Luc gene reporter assay combined to the PAR1, which is known to activate many different G proteins including Gαi/o, Gαq, and Gα12/13 (34, 35). By using selective inhibitors of G proteins or their downstream effectors (pertussis toxin for Gαi/o, U73122 for Gαq/PLC, and Y27632 for Gα12/ROK), we confirmed that PAR1 can activate the SRE-Luc reporter through different G protein-mediated signaling pathways because these selective inhibitors have only a partial effect on thrombin-stimulated SRE-Luc activity (Fig. 1D). In contrast, BIM-46187 completely blocked the effect of thrombin, indicating that BIM-46187 is able on its own to block simultaneously all of these G protein pathways (Fig. 1D). Such an effect of BIM-46187 was specific on PAR1-mediated G protein activation pathway because it has not been observed when the SRE-Luc was induced by serum (Fig. 1E).
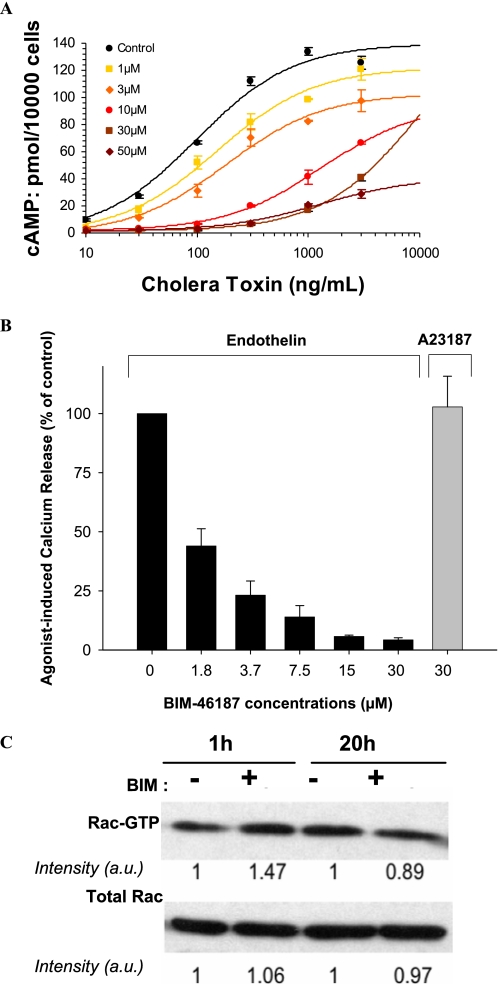
To ensure the specificity of BIM-46187 on the heterotrimeric G protein-mediated signaling pathway, Fig. 2 shows that BIM-46187 fully prevents the cAMP production directly activated by the G protein subunit α agonist cholera toxin in the human tumoral cells MCF-7 (Fig. 2A). In addition, BIM-46187 inhibited the endothelin-stimulated calcium release in human melanoma cells, whereas no effect was observed when the calcium release was activated by a G protein-independent stimulus, such as the ionophore A23187 (Fig. 2B). Third, we investigated the possible interference of the GPCR signaling inhibitors on the activation status of the Rac1 small G protein in HCT8/S11 cells. Specifically, BIM-46174 was ineffective at reducing the activated form of Rac1-GTP in HCT8/S11 cells (Fig. 2C). In addition, BIM-46187 showed no interference on the epidermal growth factor receptor-induced tyrosine phosphorylation of FAK at the Tyr925 motif involved in cancer cell motility, invasion, and angiogenesis (supplemental Fig. S1). Consistently, BIM-46174 does not affect the formation of capillary-like structures by human umbilical vein endothelial cells stimulated with the tyrosine kinase receptor ligands vascular endothelial growth factor and fibroblast growth factor (data not shown). Taken together, these data indicate that BIM-46187 is a specific pan-inhibitor of GPCR/heterotrimeric G protein signaling in living cells.
FIGURE 2.
BIM-46187 specifically inhibits heterotrimeric G protein downstream pathways. A, BIM-46187 inhibits the cyclic AMP accumulation in the MCF-7 cells treated with increasing concentrations of the G protein α subunit activator cholera toxin. B, BIM-46187 specifically inhibits the endothelin-induced calcium release in the A2058 cells without effects on the ionophore induced calcium release. C, BIM-46174 was ineffective to down-regulate the activation status of Rac1-GTP in human colon cancer cells HCT8/S11 cultured for 1 or 20 h in the presence of the G protein inhibitor. Similar data were obtained with BIM-46187 (1 mm, 10 min of incubation) in HCT8/S11 cells cultured under standard conditions. The data are representative of three separate experiments.
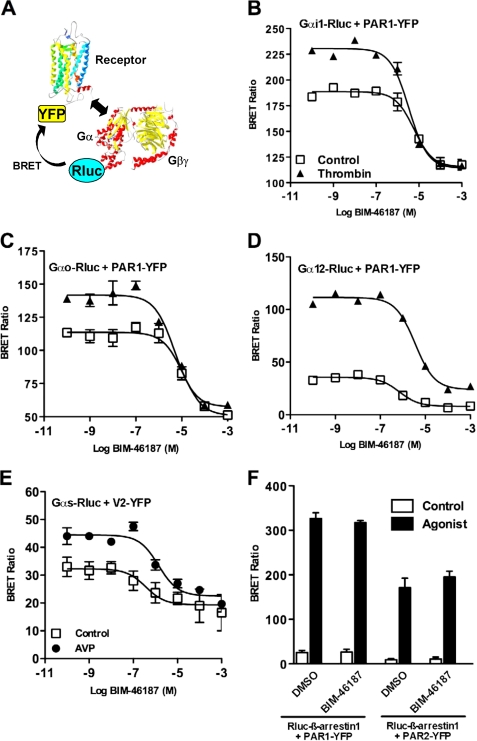
BIM-46187 Prevents GPCR-mediated G Protein Activation in Living Cells as Monitored by BRET
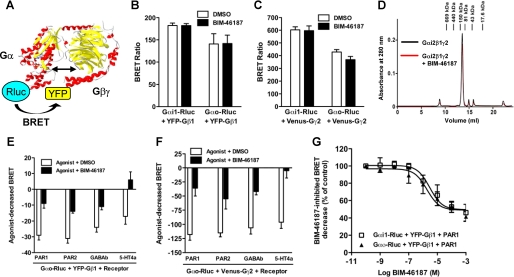
The pan-inhibitory nature of BIM-46187 implies that this compound would act directly on the G protein heterotrimer. To confirm this mode of action, we used a recently described BRET approach to directly monitor G protein activation in living cells (20, 24). This assay is based on the conformational change of the receptor-G protein complex upon activation. We examined the effects of BIM-46187 on the GPCR-mediated activation of four types of G proteins, tested in two distinct models: 1) PAR1 and its coupling to Gαi1 or Gαo, or Gα12 and 2) V2 vasopressin receptor coupled to Gαs. BRET measurements were performed on cells co-expressing PAR1-YFP or V2-YFP and Gα-Rluc fusion proteins, before and after receptor activation with thrombin or AVP, respectively (Fig. 3A). In all cases (Fig. 3, B–E), pretreatment of the cells with BIM-46187 totally inhibited the agonist-promoted BRET changes in a concentration-dependent manner (Table 2). Interestingly, even at high concentration, BIM-46187 only partially inhibited the basal BRET signal measured between the receptor and the Gα subunit. This indicates that BIM-46187 does not prevent the receptor/G protein preassembly responsible for the basal BRET signal (24). Such an effect of BIM-46187 seems highly specific to the BRET signal measured between a receptor and the G protein because no effect was observed on PAR1- or PAR2-mediated β-arrestin 1 recruitment (30) (Fig. 3F). To ensure that there is no interference between the BRET methodology and BIM-46187, we have shown that BIM-46187 has no impact on the BRET signal measured between the membrane GABAb subunits, GB1 and GB2, obligated to form an heterodimer (supplemental Fig. S2). These data are further consistent with a direct effect of BIM-46187 on the receptor-G protein complex in living cells to prevent G protein activation.
FIGURE 3.
BIM-46187 inhibits agonists mediated effect on receptor-G protein complexes in living cells. A, for BRET experiments, G proteins and receptors were fused to energy donor (Rluc) and energy acceptor (YFP), respectively. B–F, BRET experiments were performed in COS-7 cells transiently co-expressing PAR1-YFP and Gαi1-Rluc (B), Gαo-Rluc (C), Gα12-Rluc (D), and Rluc-β-arrestin 1 (F) or V2-YFP and Gαs-Rluc (E). The cells were first pretreated with BIM-46187 and then stimulated with PBS (□), Thrombin 5 units/ml (▲), or AVP 10 μm (●). The data are the means ± S.E. from triplicate determinations from a representative experiment of at least three independent experiments. DMSO, dimethyl sulfoxide.
TABLE 2.
Effects of BIM-46187 on BRET between PAR1 and Gα proteins
The table summarizes data represented in Fig. 3 including the effect of BIM-46187 (100 μm) on the basal BRET and thrombin-promoted BRET increase between Gα -Rluc and PAR1-YFP.
| BRET couple | Basal BRET (n = 3) |
Thrombin-induced BRET increase (n = 3)a |
||||
|---|---|---|---|---|---|---|
| Control | BIM-46187 (100 μm) | IC50 | Control | BIM-46187 (100 μm) | IC50 | |
| μm | μm | |||||
| Gαi1-Rluc/PAR1-YFP | 156 ± 16 | 99 ± 17 | 18 ± 7.3 | 51 ± 3 | 6 ± 3 | 7 ± 2.4 |
| Gαo-Rluc/PAR1-YFP | 136 ± 18 | 79 ± 16 | 14 ± 3.5 | 27 ± 1.5 | 6 ± 1 | 5 ± 0.3 |
| Gα12-Rluc/PAR1-YFP | 41 ± 5 | 20 ± 6 | 0.6 ± 0.3 | 48 ± 11 | 10 ± 1 | 2.4 ± 0.8 |
a Thrombin-induced BRET increase = BRET in the presence of thrombin − basal BRET.
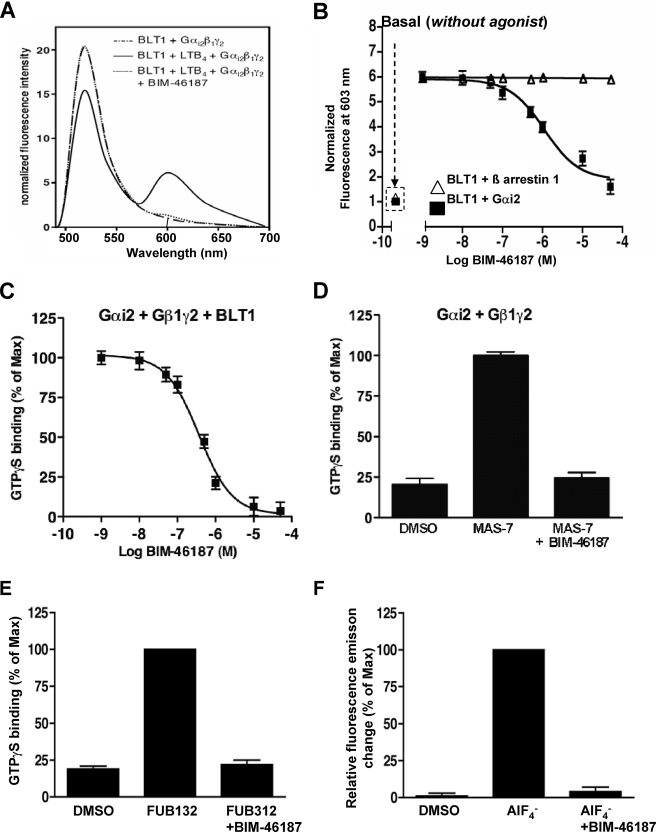
BIM-46187 Inhibits the Physical Interaction between a Ligand-activated GPCR and the Heterotrimeric G Protein in Vitro
We then used a reconstituted receptor-G protein complex to directly monitor the action of BIM-46187 (28). These experiments were carried out with a BLT1 receptor carrying a fluorescence donor (Alexa-488) in its first intracellular loop and a Gαi2β1γ2 complex where the α subunit was labeled with the acceptor (Alexa-568) at the level of Cys3 in the N-terminal region (Fig. 4A). Under these conditions, a significant FRET signal was measured between the agonist-activated receptor and the G protein trimer (Fig. 4B). BIM-46187 completely inhibited this FRET signal (absence of the peak at 603 nm) (Fig. 4B) in a concentration-dependent manner (IC50 = 3.9 × 10−7 m) (Fig. 4C), suggesting that BIM-46187 directly affects the topology of the complex between the G protein and the agonist-activated BLT1 receptor. Again, the specificity of such an effect on receptor-G protein interaction was nicely illustrated by the complete absence of BIM-46187 effect on the direct interaction between the purified BLT1 receptor and β-arrestin using a similar FRET-based assay (Fig. 4C). Similar data were obtained using a light scattering-based assay (data not shown). These results are concordant with the BRET data obtained in living cells and confirm that BIM-46187 selectively prevents proper assembly of the activated receptor with its associated G protein and blocks of the signal transduction.
FIGURE 4.
BIM-46187 inhibits the physical interaction between GPCR and the heterotrimeric G protein and the GDP/GTP exchange. A, fluorescence emission spectra of the purified Alexa-488-labeled BLT1 receptor mixed with the G protein trimer Gαi2β1γ2 where the Gαi2 subunit is labeled with Alexa-568 in the presence of LTB4 (1 μm) and in the absence or presence of BIM-46187. The absence of nonspecific FRET was assessed by using Gαsβ1γ2 instead of Gαi2β1γ2. B, changes in the intensity of the 603-nm band as a function of BIM-46187 concentration (■). Open triangles (△) correspond to the changes when using Alexa-568-labeled β-arrestin 1 instead of Gαi2β1γ2. C, inhibition of BLT1-catalyzed GTPγS binding on the purified Gαi2β1γ by increasing concentrations in BIM-46187. D, effects of BIM-46187 on GTP binding induced by the GPCR peptidomimetic Mastoparan-7. The data are the means ± S.E. of three independent experiments. E, effects of BIM-46187 on FUB132-induced GTP binding to isolated Gαi2. The data are normalized to the maximal effect observed in the absence of BIM-46187. F, effects of BIM-46187 on AlF4−-induced fluorescence changes of Gαi2. The data are normalized to the maximal effect observed in the absence of BIM-46187. The data are the means ± S.E. of three independent experiments. DMSO, dimethyl sulfoxide.
BIM-46187 Inhibits GDP/GTP Exchange on the Isolated Subunit α
To further decipher the action of BIM-46187 on G protein signaling, we again used a cell-free assay where the purified BLT1 receptor, reconstituted with purified trimeric Gαi2β1γ2 proteins, triggers G protein activation in an LTB4-dependent manner. G protein activation is monitored by measuring GTPγS incorporation in Gαi2 (28). As shown in Fig. 4C, BIM-46187 suppressed BLT1-dependent GTPγS binding in a concentration-dependent manner (IC50 = 3.6 × 10−7 m), indicating that this compound directly acts on the receptor-catalyzed G protein activation. The IC50 value observed in this cell-free assay is similar to that deduced from the FRET data shown in Fig. 4C, indicating that both processes are likely to be closely related. The effect of BIM-46187 on G protein complex activation results from a direct action on the G protein, because it also fully inhibits the activating effect of mastoparan (37), a direct activator of heterotrimeric G proteins (Fig. 4D). This inhibition is concentration-dependent with an IC50 of 4.6 × 10−7 m, similar to that obtained while inhibiting the receptor-catalyzed G protein activation. To finally assess whether this effect is due to a direct action on Gα, we monitored the effects of BIM-46187 on FUB132-induced GTPγS binding using the isolated Gαi subunit. FUB132 is a selective receptor mimetic for Gi/o that stimulates GTP binding independently of the Gβγ dimer or lipids (19). As clearly shown in Fig. 4E, BIM-46187 (50 μm) completely inhibited FUB132-induced GTPγS binding, indicating a direct effect on Gα. This was confirmed by the inhibition of AlF4−-induced conformational changes of the purified Gαi intrinsic fluorescence observed in the presence of BIM-46187 (Fig. 4F). Our data are consistent with our biological studies showing that ALF4−-dependent collagen type-1 invasion of tumoral cells is blocked by the monomer BIM-46174 (17).
BIM-46187 Prevents Molecular Rearrangements within Gα-Gβγ Trimer Promoted by GPCRs
To investigate more in depth the molecular mechanism of BIM-46187, we used the BRET approach to examine the association between Gα, Gβ, and Gγ subunits and their molecular rearrangements following GPCR activation (Fig. 5A). First, we found that BIM-46187 did not affect the high basal BRET signal measured between Gαi1 or Gαo and either Gβ1 (Fig. 5B) or Gγ2 (Fig. 5C) subunits, indicating that BIM-46187 does not prevent the constitutive formation of the trimer Gαβγ. The absence of dissociating effect of BIM-46187 on Gαβγ trimer was also observed in vitro. Indeed, a similar size exclusion chromatography elution profile was observed with a major peak corresponding to the heterotrimeric G protein complex whether BIM-46187 was present or not (Fig. 5D). Next, we examined the effect of BIM-46187 on the BRET changes between G protein subunits induced by various activated GPCRs. Indeed, as reported by Galès et al. (24) activation of the receptor induced a change in the BRET signal measured between Gα and Gβ or Gγ subunits (either an increase or a decrease, depending on the respective position of the BRET partners in the G protein subunits), reflecting a conformational rearrangement between the G protein subunits. We found that BIM-46187 largely inhibited the agonist-decreased BRET between Gαο-Rluc and YFP-Gβ1 (Fig. 5E) or between Gαο-Rluc and Venus-Gγ2 (Fig. 5F) with all of the receptors tested (PAR1, PAR2, GABAb, and 5-HT4a). The effect of BIM-46187 on the agonist-induced change in BRET between Gα-Rluc and YFP-Gβ1 was dose-dependent with IC50 values of 4.7 ± 1.9 and 4.3 ± 2.4 μm for Gαi1 and Gαο, respectively (Fig. 5G), similar to those obtained for BRET between Gα-Rluc and Venus-Gγ2 (data not shown). These data are compatible with those obtained with the effect of BIM-46187 on BRET signal between Gα-Rluc and the receptor PAR1-YFP (Table 2). Taken together, these data indicate that BIM-46187 prevents the molecular rearrangement between Gα and Gβγ subunits associated with their activation by a GPCR.
FIGURE 5.
BIM-46187 does not prevent Gα-Gβγ subunit association, but it blocks their molecular rearrangements upon agonist activation. A, the interaction between Gα, Gβ1, and Gγ2 protein subunits was studied in living cells by BRET approach. For this, COS-7 cells were transiently cotransfected with Gαi1-Rluc or Gαo-Rluc and either YFP-Gβ1 (B) or Venus-Gγ2 (C) subunits, as indicated. The cells were then pretreated (■) or not (□) with BIM-46187 (100 μm) for 2 h at 37 °C, and BRET signal was measured. D, difference near-UV circular dichroism spectra obtained by subtracting from the spectra of a Gαi2-BIM-46187 or Gβ1γ2-BIM-46187 mixture the contribution of the individual components (Gαi2 and BIM-46187 or Gβ1γ2 and BIM-46187, respectively). E and F, for the activation of G proteins, the cells were transiently cotransfected with Gαo-Rluc and either YFP-Gβ1 (E) or Venus-Gγ2 (F) subunits in the presence of various GPCRs as indicated. The cells were pretreated (■) or not (□) with BIM-46187 for 2 h and then stimulated with 5 units/ml thrombin (for PAR1), 10 μm PAR2-Amide peptide (for PAR2), 100 μm GABA (for GABAb), and 10 μm serotonin (for 5-HT4a), and BRET was immediately measured. G, dose response of BIM-46187 on PAR1-promoted BRET changes between either Gαi1-Rluc (□) or Gαo-Rluc (▲) and YFP-Gβ1 after thrombin stimulation. The data are the means ± S.E. of three independent experiments. DMSO, dimethyl sulfoxide.
BIM-46187 Specifically Binds to the α Subunit Changing Its Structural Features
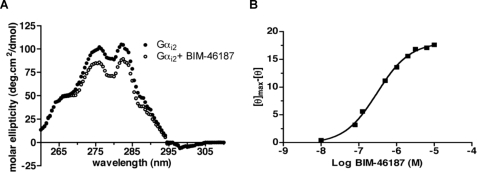
We monitored the possible binding of BIM-46187 to the G protein subunits and any associated conformational changes using CD. Adding BIM-46187 did not affect the far-UV CD profile of any of the G protein subunits, indicating that this compound does not affect their secondary structures (data not shown). In contrast, BIM-46187 induced a marked effect on the near-UV dichroic properties of Gαi2 (Fig. 6A). This represents direct evidence for a rearrangement of the Gα three-dimensional structural features that certainly results from a direct binding of BIM-46187 to this subunit. In agreement with this assumption, titration of the changes in the ellipticity as a function of BIM-46187 concentration gave a well defined saturation plot (Fig. 6B). The affinity value inferred from this titration plot, i.e. 3.3 10−7 m, is close to that measured for the inhibition of GTPγS binding and the modulation of the BLT1/G protein FRET signal (Fig. 4). In contrast, no effect on the CD properties of the Gβγ dimer was observed, indicating that BIM-46187 does not affect its structure. The near-UV CD changes obtained with the whole G protein trimer in the presence of BIM-46187 were strictly identical to those reported above for the isolated Gαi2 (data not shown), indicating that the binding and the subsequent conformational changes were identical whether the isolated Gα protein was alone or engaged in the αβγ trimer. Finally, as expected on the basis of the FRET and BRET experiments reported above, BIM-46187 induced no change in the CD properties of purified arrestin (data not shown). Altogether, these data indicate that BIM-46187 directly binds to the Gα subunit in the Gαβγ trimer and leads to conformational changes of this subunit.
FIGURE 6.
Conformational changes of Gαi2 following BIM-46187 binding. A, near-UV circular-dichroism spectra of Gαi2 in the absence (closed circles) or the presence (open circles) of BIM-46187. B, variations in the molar ellipticity at 280 nm as a function of BIM-46187 concentration. [θ] is the ellipticity at a given BIM-46187 concentration, [θ]max the ellipticity in the absence of BIM-46187.
DISCUSSION
Heterotrimeric G proteins have for a long time been considered as potential drug targets (6, 13). Recently, antitumor activities and pain relief activity have been shown in animal models using compounds interacting with Gβγ subunits (16) or the inhibitors of heterotrimeric G protein signaling (17, 18). Here, we characterize the molecular target of BIM-46187, previously shown to efficiently reduce tumor progression (17) and pain (18). By combining biochemical, biophysical, and pharmacological approaches, we demonstrate that BIM-46187 blocks GPCR signaling mediated by all heterotrimeric G proteins (Gs, Gi/o, Gq, and G12). We showed using BRET and FRET approaches that BIM-46187 affects the way GPCRs and heterotrimeric G proteins interact both in living cells and in a purified and reconstituted GPCR-G protein complex. Finally, we bring evidence that BIM-46187 directly interacts with the Gα subunit blocking the GDP/GTP exchange on the Gα subunit within the entire complex or on the isolated subunit. This binding prevents the dissociation and/or molecular rearrangements of Gαβγ trimer without affecting the constitutive preassembly between G protein subunits.
BRET data in living cells clearly indicate that BIM-46187 prevents the receptor-mediated G protein activation and affects the topology of preassembled receptor-G protein complexes (20, 24, 38). This conclusion is also supported by the decrease in the BRET signal induced by BIM-46187 and measured between various types GPCRs and Gβγ subunits (data not shown). These results were confirmed with an in vitro reconstituted system composed of the purified LTB4 receptor BLT1 and its cognate G protein partners (Gαi2β1γ2) where BIM-46187 fully inhibited agonist-dependent receptor-catalyzed Gα activation. Consistent with the data obtained by BRET techniques in living cells, in vitro FRET experiments with reconstituted system indicate that this inhibitory effect could be due to a change in the geometrical features of the activated GPCR-G protein complex. It is interesting to note that the concentrations for BIM-46187 activity are comparable in the various cellular assays reported here. Lower concentrations were required in in vitro reconstituted systems, possibly because the compound does not have to cross the cell membrane.
Although inducing a change in the conformation of the Gα subunit and thus preventing its activation, BIM-46187 did not prevent the formation of the heterotrimeric Gαi2-Gβ1γ2 complex in vitro, as shown by size exclusion chromatography experiments. This observation was confirmed by BRET measurements in living cells, where BIM-46187 had no effect on the basal signal between the three G protein subunits but blocked the agonist-mediated changes in the BRET signal within active GPCR-G protein complexes. Taken together, our data clearly indicate that BIM-46187 directly affects the primary effects of GPCRs on their cognate G protein heterotrimers.
Our measurements indicate that it involves a direct binding to the Gα subunit leading to a BIM-46187-induced conformational change, as shown by circular dichroism. However, as expected for a direct interaction of BIM-46187 with the G protein subunit, the compound had no effect on β-arrestin recruitment by GPCR both in vivo and in vitro. The conformational changes induced by BIM-46187 on Gαi2 protein could directly affect the molecular mechanism of Gαi2 activation or, alternatively, any required relative movement of the Gβγ subunits relative to Gα associated with G protein activation. Our observation that BIM-46187 had no effect on GDP binding to Gα indicates that BIM-46187 does not bind to the GDP-binding site but may rather prevent required conformational changes in Gα, necessary for GTP binding (data not shown). Further studies are required to precisely map BIM-46187 binding site, but because of the nondiscriminating action of BIM-46187 in all heterotrimeric G proteins tested, one may assume that this site is well conserved in most G proteins.
Various heterotrimeric G protein inhibitors or antagonists have been reported in the literature. Suramin has been reported to be partially selective for Gs (13), and more selective compounds for Gs have been identified by Hohenegger et al. (39). Pertussis toxin is a selective inhibitor of the Gi/o protein family as well as small molecules reported by Breitweg-Lehmann et al. (19). YM-254890 has recently been found to selectively block the GTP/GDP exchange on Gq/11 proteins (36, 40). By contrast to these selective inhibitors, our data indicate that BIM-46187 can be used as a pan-inhibitor of most heterotrimeric G proteins. This pan-effect is broadly observed on all tested G protein families including Gαs and Gα12 with no specific inhibitor known so far. In addition, BIM-46187 could be very useful to further investigate G protein-mediated events of GPCRs. Indeed we showed here that BIM-46187 did not affect β-arrestin recruitment and could therefore be used to specifically distinguish between G protein-dependent and -independent signaling pathways.
Regarding the chemical structure of BIM-46187, this compound contains an intramolecular disulfide bridge, which begs the question, is the reversible disulfide or free sulfhydryl required for activity? Because no irreversible disulfide can be generated preserving the intact molecule, only indirect arguments can be provided. The fact that the monomer BIM-46174 by itself displayed a G protein signaling inhibition (17) supports an active role for the free sulfhydryl form. However, the presence of the intramolecular disulfide bridge in BIM-46187 certainly modifies the kinetic of action with a prodrug behavior compared with the monomer.
BIM-46187 has shown promising activities in animal models that should support further therapeutic applications (17, 18). As mentioned above, Bonacci et al. (16) have identified compounds that bound to Gβγ subunits and showed that G protein subunits have potential as a target for therapeutic treatment of a number of diseases. Here, we bring additional evidence that the direct modulation of G protein signaling may represent a new therapeutic strategy for a number of diseases by using small molecules such as BIM-46187.
Supplementary Material
Acknowledgments
We thank Professor Joel Bockaert (University of Montpellier, France), Dr. John Wright, Dr. Susan Holbeck (Cancer Therapy Evaluation Program, NCI, National Institutes of Health), Dr. Marie-Odile Lonchampt, Dr. Michel Auguet, Dr. Christophe Thurieau (IPSEN), Dr. Philip Kasprzyk (IPSEN), Dr. Barry Morgan (GlaxoSmithKline, Boston, MA), and Dr. Thomas Gordon (Boston, MA) for very helpful discussions and strong continuous support for this new field of research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
M. A. Ayoub, E. Trinquet, and J.-P. Pin, manuscript in preparation.
M. Damian, A. Martin, J.-P. Pin, and J.-L. Banères, manuscript in preparation.
- GPCR
- G protein-coupled receptor
- BRET
- bioluminescence resonance energy transfer
- FRET
- fluorescence resonance energy transfer
- GABA
- γ-aminobutyric acid
- Rluc
- R. reniformis luciferase
- YFP
- yellow fluorescent protein
- PBS
- phosphate-buffered saline
- DMEM
- Dulbecco's modified Eagle's medium
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- MOPS
- 4-morpholinepropanesulfonic acid
- IP1
- inositol phosphate
- FAK
- focal adhesion kinase
- PAR
- protease-activated receptor
- AVP
- arginine vasopressin.
REFERENCES
- 1.Lundstrom K. (2006) Curr. Protein Pept. Sci. 7, 465–470 [DOI] [PubMed] [Google Scholar]
- 2.Overington J. P., Al-Lazikani B., Hopkins A. L. (2006) Nat. Rev. Drug Discov. 5, 993–996 [DOI] [PubMed] [Google Scholar]
- 3.Dorsam R. T., Gutkind J. S. (2007) Nat. Rev. Cancer 7, 79–94 [DOI] [PubMed] [Google Scholar]
- 4.Ahmad S., Dray A. (2004) Curr. Opin. Investig. Drugs 5, 67–70 [PubMed] [Google Scholar]
- 5.Cotecchia S., Fanelli F., Costa T. (2003) Assay. Drug Dev. Technol. 1, 311–316 [DOI] [PubMed] [Google Scholar]
- 6.Freissmuth M., Waldhoer M., Bofill-Cardona E., Nanoff C. (1999) Trends Pharmacol. Sci. 20, 237–245 [DOI] [PubMed] [Google Scholar]
- 7.Gilman A. G. (1995) Biosci. Rep. 15, 65–97 [DOI] [PubMed] [Google Scholar]
- 8.Höller C., Freissmuth M., Nanoff C. (1999) Cell Mol. Life Sci. 55, 257–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tesmer J. J. (2006) Science 312, 377–378 [DOI] [PubMed] [Google Scholar]
- 10.Higashijima T., Uzu S., Nakajima T., Ross E. M. (1988) J. Biol. Chem. 263, 6491–6494 [PubMed] [Google Scholar]
- 11.Higashijima T., Burnier J., Ross E. M. (1990) J. Biol. Chem. 265, 14176–14186 [PubMed] [Google Scholar]
- 12.Beindl W., Mitterauer T., Hohenegger M., Ijzerman A. P., Nanoff C., Freissmuth M. (1996) Mol. Pharmacol. 50, 415–423 [PubMed] [Google Scholar]
- 13.Freissmuth M., Boehm S., Beindl W., Nickel P., Ijzerman A. P., Hohenegger M., Nanoff C. (1996) Mol. Pharmacol. 49, 602–611 [PubMed] [Google Scholar]
- 14.Nanoff C., Kudlacek O., Freissmuth M. (2002) Methods Enzymol. 344, 469–480 [DOI] [PubMed] [Google Scholar]
- 15.Neubig R. R., Siderovski D. P. (2002) Nat. Rev. Drug Discov. 1, 187–197 [DOI] [PubMed] [Google Scholar]
- 16.Bonacci T. M., Mathews J. L., Yuan C., Lehmann D. M., Malik S., Wu D., Font J. L., Bidlack J. M., Smrcka A. V. (2006) Science 312, 443–446 [DOI] [PubMed] [Google Scholar]
- 17.Prévost G. P., Lonchampt M. O., Holbeck S., Attoub S., Zaharevitz D., Alley M., Wright J., Brezak M. C., Coulomb H., Savola A., Huchet M., Chaumeron S., Nguyen Q. D., Forgez P., Bruyneel E., Bracke M., Ferrandis E., Roubert P., Demarquay D., Gespach C., Kasprzyk P. G. (2006) Cancer Res. 66, 9227–9234 [DOI] [PubMed] [Google Scholar]
- 18.Favre-Guilmard C., Zeroual-Hider H., Soulard C., Touvay C., Chabrier P. E., Prevost G., Auguet M. (2008) Eur. J. Pharmacol. 594, 70–76 [DOI] [PubMed] [Google Scholar]
- 19.Breitweg-Lehmann E., Czupalla C., Storm R., Kudlacek O., Schunack W., Freissmuth M., Nürnberg B. (2002) Mol. Pharmacol. 61, 628–636 [DOI] [PubMed] [Google Scholar]
- 20.Ayoub M. A., Maurel D., Binet V., Fink M., Prézeau L., Ansanay H., Pin J. P. (2007) Mol. Pharmacol. 71, 1329–1340 [DOI] [PubMed] [Google Scholar]
- 21.Terrillon S., Durroux T., Mouillac B., Breit A., Ayoub M. A., Taulan M., Jockers R., Barberis C., Bouvier M. (2003) Mol. Endocrinol. 17, 677–691 [DOI] [PubMed] [Google Scholar]
- 22.Galés C., Rebois R. V., Hogue M., Trieu P., Breit A., Hébert T. E., Bouvier M. (2005) Nat. Methods 2, 177–184 [DOI] [PubMed] [Google Scholar]
- 23.Azpiazu I., Gautam N. (2004) J. Biol. Chem. 279, 27709–27718 [DOI] [PubMed] [Google Scholar]
- 24.Galés C., Van Durm J. J., Schaak S., Pontier S., Percherancier Y., Audet M., Paris H., Bouvier M. (2006) Nat. Struct. Mol. Biol. 13, 778–786 [DOI] [PubMed] [Google Scholar]
- 25.Medkova M., Preininger A. M., Yu N. J., Hubbell W. L., Hamm H. E. (2002) Biochemistry 41, 9962–9972 [DOI] [PubMed] [Google Scholar]
- 26.Ayoub M. A., Couturier C., Lucas-Meunier E., Angers S., Fossier P., Bouvier M., Jockers R. (2002) J. Biol. Chem. 277, 21522–21528 [DOI] [PubMed] [Google Scholar]
- 27.Damian M., Martin A., Mesnier D., Pin J. P., Banères J. L. (2006) EMBO J. 25, 5693–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banères J. L., Parello J. (2003) J. Mol. Biol. 329, 815–829 [DOI] [PubMed] [Google Scholar]
- 29.Oldham W. M., Van Eps N., Preininger A. M., Hubbell W. L., Hamm H. E. (2006) Nat. Struct. Mol. Biol. 13, 772–777 [DOI] [PubMed] [Google Scholar]
- 30.Sommer M. E., Smith W. C., Farrens D. L. (2005) J. Biol. Chem. 280, 6861–6871 [DOI] [PubMed] [Google Scholar]
- 31.Hickerson M. J., Cunningham C. W. (2000) Mol. Biol. Evol. 17, 639–644 [DOI] [PubMed] [Google Scholar]
- 32.Rodrigues S., Van Aken E., Van Bocxlaer S., Attoub S., Nguyen Q. D., Bruyneel E., Westley B. R., May F. E., Thim L., Mareel M., Gespach C., Emami S. (2003) FASEB J. 17, 7–16 [DOI] [PubMed] [Google Scholar]
- 33.Grijelmo C., Rodrigue C., Svrcek M., Bruyneel E., Hendrix A., de Wever O., Gespach C. (2007) Cell Signal. 19, 1722–1732 [DOI] [PubMed] [Google Scholar]
- 34.Arora P., Ricks T. K., Trejo J. (2007) J. Cell Sci. 120, 921–928 [DOI] [PubMed] [Google Scholar]
- 35.Nguyen Q. D., Faivre S., Bruyneel E., Rivat C., Seto M., Endo T., Mareel M., Emami S., Gespach C. (2002) FASEB J. 16, 565–576 [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi M., Nagai K., Arao N., Kawasaki T., Saito T., Moritani Y., Takasaki J., Hayashi K., Fujita S., Suzuki K., Tsukamoto S. (2003) J. Antibiot. 56, 358–363 [DOI] [PubMed] [Google Scholar]
- 37.Danilenko M., Worland P., Carlson B., Sausville E. A., Sharoni Y. (1993) Biochem. Biophys. Res. Commun. 196, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 38.Nobles M., Benians A., Tinker A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18706–18711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hohenegger M., Waldhoer M., Beindl W., Böing B., Kreimeyer A., Nickel P., Nanoff C., Freissmuth M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 346–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takasaki J., Saito T., Taniguchi M., Kawasaki T., Moritani Y., Hayashi K., Kobori M. (2004) J. Biol. Chem. 279, 47438–47445 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.