Abstract
Despite the small size and conserved tertiary structure of defensins, little is known at a molecular level about the basis of their functional versatility. For insight into the mechanism(s) of defensin function, we prepared enantiomeric pairs of four human defensins, HNP1, HNP4, HD5, and HBD2, and studied their killing of bacteria, inhibition of anthrax lethal factor, and binding to HIV-1 gp120. Unstructured HNP1, HD5, and HBD3 and several other human α- and β-defensins were also examined. Crystallographic analysis showed a plane of symmetry that related LHNP1 and DHNP1 to each other. Either d-enantiomerization or linearization significantly impaired the ability of HNP1 and HD5 to kill Staphylococcus aureus but not Escherichia coli. In contrast, LHNP4 and DHNP4 were equally bactericidal against both bacteria. d-Enantiomers were generally weaker inhibitors or binders of lethal factor and gp120 than their respective native, all-l forms, although activity differences were modest, particularly for HNP4. A strong correlation existed among these different functions. Our data indicate: (a) that HNP1 and HD5 kill E. coli by a process that is mechanistically distinct from their actions that kill S. aureus and (b) that chiral molecular recognition is not a stringent prerequisite for other functions of these defensins, including their ability to inhibit lethal factor and bind gp120 of HIV-1.
Defensins are 2–5 kDa, disulfide-stabilized cationic peptides found in the leukocytes and epithelial tissues of mammals (1–5). On the basis of disulfide topology, defensins are classified into three structural families: α, β, and θ. To date, six human α-defensin peptides, also known as human neutrophil peptides (HNPs)4 1–4 and enteric defensins 5–6 (HD5 and HD6) have been identified. Many more human β-defensins (HBDs) exist and are believed to be expressed predominantly in epithelia. However, only a few of these have been characterized thus far at the protein level (6). Macrocyclic θ-defensins are expressed in the leukocytes and bone marrow of certain nonhuman primates, but not of humans (7). Despite differences in amino acid composition, Cys connectivity and tissue distribution, mammalian α- and β-defensins are structurally conserved, adopting a three-stranded β-sheet core structure stabilized by three intramolecular disulfide bonds. At the functional level, however, defensins exert extremely diverse effects.
Originally identified as “natural peptide antibiotics” (8, 9), defensins act early in innate immune defenses against potentially infectious bacteria, fungi, and viruses. It is generally accepted that bacterial killing by defensins is initiated by a “fatal attraction” between the cationic defensins and the anionic microbial membrane that culminates in target cell death elicited by microbial membrane disruption and leakage of cellular contents (10, 11). An array of molecular mechanisms has been suggested to account for the ability of defensins to inhibit both enveloped and non-enveloped viruses. For HIV-1 alone, at least six distinct modes of action have been proposed, including direct inactivation of virions (12–15), interference with protein kinase C signaling and viral replication (12), up-regulation of CC-chemokines (16), inhibition of gp41-membrane fusion (17), inhibition of CD4-gp120 interactions (18), and down-regulation of HIV co-receptors (14). Adding to this complexity is a recent finding that HD5 and HD6 enhance HIV-1 infection during viral entry, acting via some unknown mechanism (19).
Defensins also appear to be important immunomodulators, capable of acting on a variety of cellular receptors and host proteins (3, 20, 21). For example, defensins chemoattract and activate different types of immune cells (22–25), regulate cytokine production (26, 27), interact with components of the complement system (28, 29), and participate in wound healing by promoting epithelial cell migration and proliferation (30, 31). More recently, β-defensins have been shown to bind with high affinity to melanocortin receptors to signal pigment-type switching in dogs (32).
Defensins also neutralize many secreted bacterial toxins (33), including anthrax lethal toxin (LeTx), a binary complex of two bacterial proteins secreted by Bacillus anthracis, protective antigen and lethal factor (LF) (34, 35). LF is a Zn2+-dependent metalloprotease, which, upon entering macrophages, cleaves important cellular proteins, induces cell death, and is the primary virulence factor in the pathogenesis of anthrax. Kaufmann and colleagues first reported that HNP1–3 non-competitively inhibited LF and protected cells as well as experimental animals against killing induced by B. anthracis LeTx (36). B. anthracis spores engulfed by human neutrophils germinated intracellularly, only to then be killed effectively by HNPs (37). Inhibition of LF and killing of vegetative cells of B. anthracis by retrocyclins, putative hominid homologues of rhesus monkey θ-defensins encoded by human pseudogenes, have also been reported (38).
Aside from their ability to interact with bacterial membranes and a variety of proteins, many defensins also bind carbohydrates, nucleic acids, and lipids. Retrocyclins, for example, inhibit influenza virus infection by cross-linking glycoproteins on the (host) membrane surface, thus preventing hemagglutinin-mediated viral fusion and entry (39). Some antiviral activities of defensins appear to be associated with their lectin properties (40–42). How such small peptides have acquired functional versatility or promiscuity at the molecular level remains obscure. To better understand defensin functionality in innate and adaptive immunity, we compared HNP1, HNP4, HD5, and HBD2 with their enantiomeric counterparts, made up entirely of d-amino acids, with respect to bacterial killing, LF inhibition, and HIV-1 gp120 binding. High resolution crystal structures of the enantiomeric pair of HNP1 were determined. Hoping to gain additional mechanistic insights, we examined five other human defensins (HNP2, HNP3, HD6, HBD1, and HBD3), as well as linearized analogs of HNP1, HD5, and HBD3 whose six Cys residues were all replaced by either Ala or α-aminobutyric acid.
EXPERIMENTAL PROCEDURES
Materials
Synthesis of HNP1–4, HD5–6, and HBD1–3 was done as described previously (43–45). The d-enantiomeric defensins DHNP1, DHNP4, DHD5, and DHBD2 were prepared similarly to their natural counterparts using d-amino acids and custom-made 4-hydroxymethylphenylacetamidomethyl resins. The three unstructured/linearized defensin analogs, Ala-HNP1, α-aminobutyric acid-HD5, and α-aminobutyric acid-HBD3, were synthesized on an ABI 433A peptide synthesizer using the published 2-(1H-benzotriazolyl)-1,1,3,3-tetramethyluroniumhexafluorophosphate activation/N,N-diisopropylethylamine in situ neutralization protocol for t-butoxycarbonyl chemistry (46). All peptides were purified to homogeneity by C18 reversed-phase high-performance liquid chromatography, and their molecular masses were verified by electrospray ionization mass spectrometry. Quantification of defensins was done by UV measurements at 280 nm using molar extinction coefficients calculated from a published algorithm (47).
Recombinant anthrax lethal factor and protective antigen were purchased from List Biological Laboratories, Inc. A sequence-optimized chromogenic substrate of lethal factor, Ac-NleKKKKVLP-p-nitroaniline, was synthesized essentially as described (48). In brief, a peptide acid precursor, Ac-NleKKKKVL-OH, whose lysine side chains were orthogonally protected by t-butoxycarbonyl, was first synthesized on 2-chlorotrityl chloride resin using a standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry and cleaved in a mixture of acetic acid/trifluoroethanol/dichloromethane (1:2:7). Coupling of Pro-p-nitroaniline to the C terminus of the precursor peptide was achieved using 2-(1H-benzotriazolyl)-1,1,3,3-tetramethyluroniumhexafluorophosphate and N,N-diisopropylethylamine in dimethylformamide for 2 h. The product was precipitated and washed by ice water, deprotected by trifluoroacetic acid, and purified to homogeneity by reversed-phase high-performance liquid chromatography. HIVBaL gp120, expressed in T-RExTM-293 cells and affinity-purified, was a generous gift from Profectus Biosciences, Inc.
LF Inhibition Kinetics
The inhibition of LF by various defensins was quantified using an enzymatic kinetic assay (36, 48). Briefly, freshly prepared LF at a final concentration of 1 μg/ml (∼10 nm) was incubated at 37 °C for 30 min with a 2-fold dilution series of defensin in 20 mm HEPES buffer containing 1 mm CaCl2 and 0.5% Nonidet P-40, pH 7.2. 20 μl of LF substrate (1 mm in the buffer) was added to each well to a final concentration of 100 μm in a total volume of 200 μl. The enzyme activity, characterized as a time-dependent absorbance increase at 405 nm due to the release of p-nitroaniline, was monitored at 37 °C over a period of 5 min on a 96-well Vmax microplate reader (Molecular Dynamics, Inc.). Data are presented in a plot showing percent inhibition versus defensin concentration, from which IC50 values (the concentration of defensin that reduced the enzymatic activity of LF by 50%) were derived by a non-linear regression analysis.
Surface Plasmon Resonance-based LF and gp120 Binding
Experiments were performed on a BIAcore T100 System (BIAcore, Inc., Piscataway, NJ), unless stated otherwise, at 25 °C in 10 mm HEPES, 150 mm NaCl, 0.05% surfactant P20, pH 7.4 (±3 mm EDTA). LF was immobilized on a CM5 sensor chip at a level of 2500 response units (RU) by the amine-coupling protocol. HIV gp120 chips were prepared at 2830 and 3200 RU. Analytes were introduced into the flow-cells at 30 μl/min in the running buffer. Association and dissociation were assessed for 5 and 10 min, respectively. Resonance signals were corrected for nonspecific binding by subtracting the background of the control flow-cell. After each analysis, the sensor chip surfaces were regenerated with 10 mm glycine solution (pH 2.0) and 50 mm NaOH for LF or 10 mm NaOH for gp120 and equilibrated with the buffer before next injection. Binding isotherms were analyzed with BIAevaluation software and/or GraphPad Prism.
Virtual Colony Count
Antimicrobial assays against Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213 (Microbiologics) were conducted using a previously detailed 96-well turbidimetric method dubbed “virtual colony counting” (49). A 2-fold dilution series of defensin, ranging from 256 to 1 μg/ml in 10 mm sodium phosphate, pH 7.4, was incubated at 37 °C for 2 h with E. coli or S. aureus (1 × 106 CFU/ml), followed by addition of twice-concentrated Mueller-Hinton broth (2×) and kinetic measurements of bacterial growth at 650 nm over 12 h. To increase the sensitivity of bacterial killing by some defensins, 1% tryptic soy broth (TSB) was added to the phosphate buffer during the 2-h incubation period. The virtual LD50 (vLD50), vLD90, vLD99, and vLD99.9 were reported as the defensin concentration that resulted in survival rates of 0.5, 0.1, 0.01, and 0.001, respectively.
Crystallization and Data Collection
Crystallization screenings were conducted at room temperature using the hanging-drop, vapor diffusion method and the commercially available crystallization Sparse Matrix Screens (Hampton Research). The drops were generated by mixing 0.5 μl of defensin solution (prepared at 20 mg/ml in water) with 0.5 μl of reservoir solution, and placed over 0.8 ml of reservoir solution. HNP1 crystals were grown from mother liquor containing 0.1 m imidazole and 1.0 m sodium acetate trihydrate, pH 6.5, whereas DHNP1 crystals from 0.1 m sodium citrate tribasic dehydrate, 0.1 m HEPES sodium, and 20% (v/v) isopropanol, pH 7.5. In both cases the crystals appeared after 1 day and grew to the final sizes within a week.
Prior to freezing in a 100 K stream of nitrogen, crystals were briefly soaked in crystallization solutions with 20% glycerol (w/v) added. X-ray diffraction data were collected from flash-frozen crystals mounted on a rotating anode x-ray generator Rigaku-MSC Micromax 7 equipped with a Raxis-4++ image plate detector (at the x-ray Crystallography Core Facility, University of Maryland, Baltimore). Crystal diffraction images were indexed, integrated, scaled and merged using the HKL2000 package (50). Both defensins crystallized in the orthorhombic form and belong to the space group P21212. The data collection statistics are shown in supplemental Table S1.
Structure Determination and Refinement
The structure of HNP1 was solved by molecular replacement using the program Phaser from the CCP4 suite (51) with the HNP3 molecule (1DFN (52)) as a search model. The refined structure of HNP1 was subsequently used as an initial model for DHNP1 structure determination with Phaser. The structures were refined to 1.56 Å with the program Refmac (53), and the models were corrected by manually re-fitting into the electron density and rebuilt using the program COOT (54). The results of refinement are summarized in supplemental Table S1. The coordinates and structure factors have been deposited in the Protein Data Bank with accession codes of 3gny and 3go0 for HNP1 and DHNP1, respectively. Molecular graphics were generated using PyMOL (DeLano Scientific LLC, San Carlos, CA).
RESULTS
Structural Studies
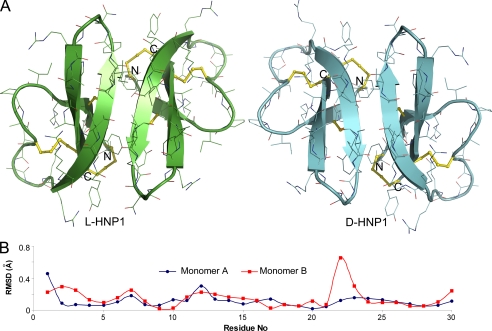
Ideally, an unnatural protein that is composed entirely of d-amino acids is the mirror image of its native form comprising only l-amino acids in the same sequence. Two mirror image proteins (enantiomers), in identical amounts, rotate the plane of polarized light equally, but in opposite directions. When the paired defensin enantiomers, LHNP1/DHNP1, LHNP4/DHNP4, LHD5/DHD5, and LHBD2/DHBD2, were examined by CD spectroscopy, the spectra of each enantiomeric defensin pair appeared symmetrical about the x-axis (supplemental Fig. S1), characteristic of two optically active chiral molecules related to one another by a plane of symmetry. LHNP1 and DHNP1 were chosen for further characterization by x-ray crystallography. Both defensins crystallized rapidly in the orthorhombic system and diffracted to 1.56 Å on our home x-ray source with the final values of R (Rfree) of 0.171 (0.185) and 0.191 (0.199), respectively (supplemental Table S1). The crystal structures of LHNP1 and DHNP1, with two defensin molecules present in one asymmetric unit, are shown in Fig. 1A. As expected, LHNP1 adopts the canonical three-stranded β-sheet fold arranged in a dimeric form, which is conserved in the α-defensin family (55), whereas DHNP1 shows a nearly perfect mirror image of its native l-counterpart. The vast majority of the side-chain conformations, with a few exceptions, are also preserved symmetrically between LHNP1 and DHNP1. When the dimers (60 residues) of LHNP1 and an inverted DHNP1 were superimposed, the root mean square deviations were found to be 0.17 Å for Cα atoms and 0.5 Å for all atoms (Fig. 1B). In addition to the flexible termini, the loop region connecting the second and third β strands of monomer B (residues Gln-22 and Gly-23) constitutes a notable local structural difference between LHNP1 and DHNP1, characterized by a larger than normal root mean square deviation. Because the β2–β3 loop of DHNP1 is involved in substantial crystal contacts, the observed difference likely resulted from the effect of crystal packing rather than intrinsic properties of the backbone.
FIGURE 1.
Crystal structures of LHNP1 and DHNP1 related to one another by a plane of symmetry. A, ribbon diagrams of HNP1 dimers in the asymmetric unit of LHNP1 and DHNP1 crystals. The three conserved disulfide bridges are shown as yellow sticks. B, root mean square deviations of Cα atoms between two enantiomeric monomers of HNP1. Monomers A and B of LHNP1 were compared with monomers A and B of inverted DHNP1, respectively.
Synthetic HNP1 Neutralizes LeTx
For validation purposes, we characterized the ability of our synthetic HNP1 to neutralize cytotoxicity of anthrax LeTx in murine macrophages (supplemental Fig. S2). We measured in vitro protective activity of HNP1 against LeTx (400 ng/ml LF and 1600 ng/ml protective antigen) using the same assay protocols as described by Kim et al. (36). Shown in supplemental Fig. S2 is HNP1 dose-dependent protection of RAW 264.7 cells against cytolysis by LeTx assayed in RPMI medium 1640 supplemented with 5% fetal calf serum. Three independent experiments were performed, giving rise to a highly reproducible EC50 value of 15 ± 1 μm (effective concentration of HNP1 at which 50% cell viability is observed), in quantitative agreement with the value reported by Kim et al. (36). Consistent with these findings, a Trypan blue cell staining experiment showed that 20 μm synthetic LHNP1 fully protected murine macrophages from LeTx-induced cytolysis with no apparent cytotoxicity.
d-Defensins Are Weaker Inhibitors of LF Than l-Defensins
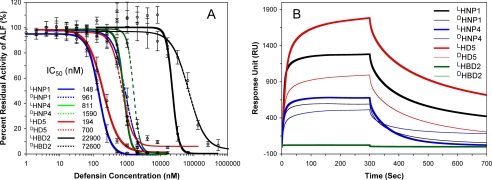
LF inhibition by a 2-fold, 11-point dilution series of LHNP1 (0, 1, 2, 4, 8, 16, 32, 64, 128, 256, 512, and 1024 nm) was quantified at 37 °C using an enzyme kinetic assay protocol tailored from the published procedures developed by Kaufmann and colleagues (36). Shown in Fig. 2A is a percent residual LF activity plot versus defensin concentration on a log scale, obtained from 24 independent measurements. Complete inhibition of 10 nm LF was achieved by HNP1 at ≥1024 nm. A non-linear regression analysis yielded an IC50 value of 148 ± 11 nm, similar to the IC50 value of 190 ± 33 nm reported by Kim et al. (36). We also quantified LF inhibition by DHNP1, LHNP4/DHNP4, LHD5/DHD5, and LHBD2/DHBD2 (Fig. 2A). On the basis of their IC50 values, the four l-defensins ranked in the following order of activity: LHNP1 (148 nm) ≈ LHD5 (194 nm) > LHNP4 (811 nm) ≫ LHBD2 (22.9 μm). LHBD2 was substantially weaker than the three Lα-defensins. The inhibition curve of HNP4 was relatively steep, with percent residual LF activity dropping from ∼90% to almost zero as the defensin concentration increased from 516 to 2048 nm. By contrast, LHD5, despite a significantly lower IC50 value, did not cause complete LF inhibition even at 2048 nm. Notably, the order of activity of d-defensins remained roughly the same, i.e. DHNP1 (961 nm) ≈ DHD5 (700 nm) > DHNP4 (1.59 μm) ≫ DHBD2 (72.6 μm). Enantiomerization impacted defensin activity differentially, as evidenced by an almost 7-fold increase in IC50 of DHNP1, a more modest 3- to 4-fold increase in IC50 of DHD5 or DHBD2, and a merely 2-fold increase in IC50 of DHNP4.
FIGURE 2.
Inhibition and binding of LF by enantiomeric defensins. A, inhibition of LF activity by different concentrations of l- (solid line) or d-defensin (dotted line). The data are averages of three independent enzyme kinetic measurements, except for LHNP1, for which 24 independent measurements were performed. A Student t-test was used to calculate the p values for statistical significance: p = 0.0084 for LHNP1/DHNP1; p = 0.066 for LHNP4/DHNP4; p = 0.014 for LHD5/DHD5; and p = 0.17 for LHBD2/DHBD2. B, representative sensorgrams of l (thick lines)- and d (thin lines)-defensins, each at 1 μm, on 2500 RU of immobilized LF.
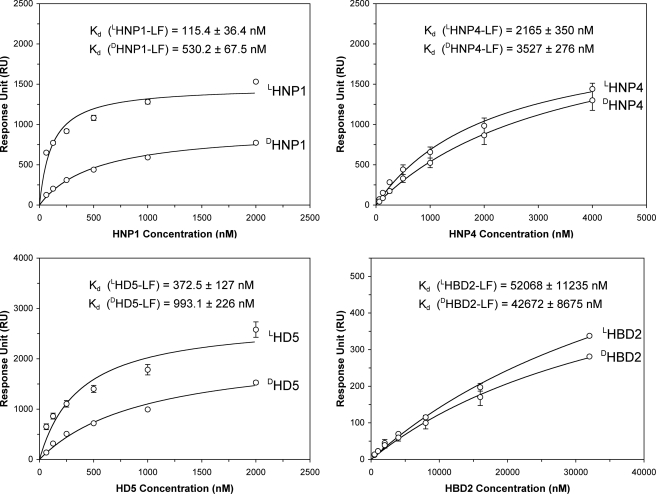
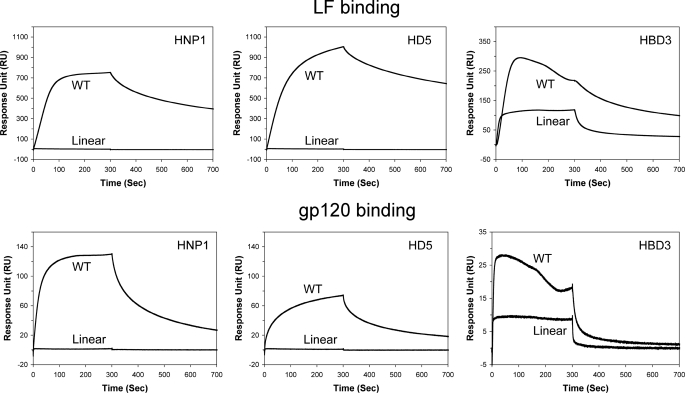
Using SPR, we compared LF binding kinetics of l- and d-defensins at various peptide concentrations (62.5, 125, 250, and 500 nm and 1 and 2 μm for HNP1 and HD5; 62.5, 125, 250, and 500 nm and 1, 2, and 4 μm for HNP4; and 0.5, 1, 2, 4, 8, 16, and 32 μm for HBD2). Representative sensorgrams of the eight defensins at 1 μm each on 2500 RU of LF are shown in Fig. 2B. Unlike the enantiomeric β-defensins LHBD2 and DHBD2, the six α-defensins, and LHNP1 and LHD5 in particular, bound well to LF. The three native Lα-defensins showed stronger LF binding than their corresponding d-enantiomers, although the differences measured by the RU values at 300 s of association were by and large within a factor of 2. Fitting of steady-state kinetic data is presented in Fig. 3, yielding the Kd values of the eight defensins for LF generally in line with the IC50 values determined by enzyme inhibition assays. For both l- and d-defensins, the order of LF-binding activity was: HNP1 ≈ HD5 > HNP4 ≫ HBD2. Importantly, the binding of d-defensins to LF decreased uniformly as compared with their corresponding l-forms. The -fold increase in Kd was ∼5, 2, and 3 for DHNP1, DHNP4, and DHD5, respectively, consistent with the aforementioned changes in IC50. LHBD2 and DHBD2 were excluded from the comparison, because their LF binding failed to approach saturation at the highest concentration used. Taken together, the LF inhibition and binding data suggest that d-defensins are weaker inhibitors of LF than their native forms, with the largest disparity seen for HNP1 and the smallest for HNP4.
FIGURE 3.
Enantiomeric defensins bound to 2500 RUs of immobilized LF as a function of concentration. The RU values collected at t = 300 s from three independent SPR measurements were fitted to a one-site binding model (Y = Bmax·X/[Kd + X]) using Graphpad Prism version 4.0. The p values were calculated using a Student's t-Test: p < 0.001 for LHNP1/DHNP1; p = 0.0019 for LHNP4/DHNP4; p < 0.001 for LHD5/DHD5; p = 0.09 for LHBD2/DHBD2.
d-Defensins Are Weaker Lectins Than l-Defensins
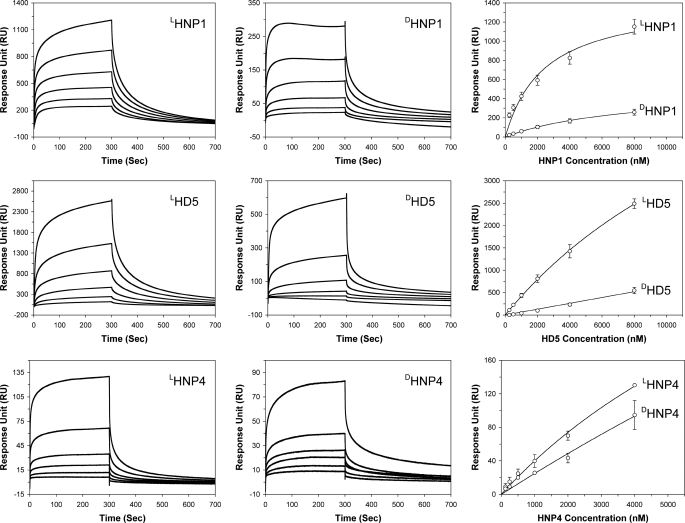
HNP1–3 and HD5, but not HNP4, HD6, and β-defensins, are known lectins that are capable of binding at high nanomolar affinities to HIV-1 gp120, a heavily glycosylated protein. To better understand lectin properties of enantiomeric defensins, we immobilized HIVBaL gp120 (2830 RU for HNP1 and HD5, 3200 RU for HNP4) on a CM5 sensor chip and analyzed association and dissociation kinetics of various defensins at different concentrations. HNP1 and HD5 exhibited, from 250 nm to 8 μm, dose-dependent binding to gp120 (Fig. 4). No appreciable binding was observed with HBD2 at the highest concentration of 32 μm used (data not shown). DHNP1 and DHD5 were weaker than their corresponding l-forms. For direct comparison, the RU values at 300 s of all sensorgrams for HNP1 and HD5 were plotted versus defensin concentration. As shown in Fig. 4, the disparity between LHNP1 and DHNP1 in their ability to bind HIV gp120 varied from 10- to 5-fold as the defensin concentration increased from 250 nm to 8 μm. A similar disparity varying from 16- to 5-fold was found between LHD5 and DHD5 within the same concentration range. We did not try to fit the kinetic data to any mathematical model to obtain Kd values due to known mechanistic complexities associated with multivalent carbohydrate-binding and binding-induced self-oligomerization of HNP1 and HD5 on the surface of glycoproteins (56).
FIGURE 4.
Binding of enantiomeric defensins to HIV gp120. Left and middle columns: representative sensorgrams of enantiomeric defensins at different concentrations (from 250 nm to 8 μm for HNP1 and HD5, and from 125 nm to 4 μm for HNP4). A sensor chip with 2830 RUs of gp120 was used for HNP1 and HD5 binding, and 3200 RUs of gp120 were immobilized for HNP4 measurements. Right column: RU values at 300 s of association from three independent SPR measurements were fitted to a one-site binding model (Y = Bmax·X/[Kd + X]) using GraphPad Prism version 4.0. The p values for statistical significance are: p = 0.003 for LHNP1/DHNP1, p = 0.022 for LHD5/DHD5, and p = 0.024 for LHNP4/DHNP4.
HNP4 showed greatly reduced ability to bind gp120 compared with HNP1 and HD5, consistent with our previous observations (57). Further, unlike HNP1 and HD5, DHNP4 was only slightly weaker than LHNP4 in gp120 binding within the concentration range tested (125 nm to 4 μm). Despite a much-reduced capacity to bind gp120 compared with their native molecules, DHNP1 and DHD5 were still better lectins than LHNP4. Interestingly, when HNP4 concentration was increased to 8 μm and above, a sudden surge in reference cell binding ensued, resulting in irregularly shaped sensorgrams drifting into the negative RU region (data not shown). A similar artifact occurred with DHNP4 at 8 μm and above, making it impossible to compare LHNP4 and DHNP4 binding to gp120 at higher concentrations. The anomalous behavior of HNP4 at high concentrations may reflect distinct oligomerization properties of this defensin.
Bactericidal Activities of d-Defensins Are Both Strain-dependent and Peptide-dependent
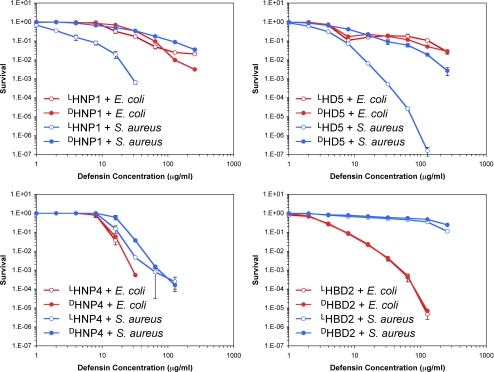
Membrane permeabilization as a possible mechanism for bacterial killing by cationic antimicrobial peptides received early support from the studies that demonstrated a non-receptor mediated pathway, initially evidenced by the finding that some antimicrobial peptides consisting exclusively of all d-amino acids were equally active as their natural l-form counterparts (58). To investigate whether enantiomeric defensins kill bacteria equivalently, we quantified bactericidal activities of LHNP1/DHNP1, LHNP4/DHNP4, LHD5/DHD5, and LHBD2/DHBD2 against E. coli and S. aureus using a previously established assay protocol termed virtual colony count (49). Defensin dose-dependent survival of both strains are plotted in Fig. 5. As expected, the α-defensins, except for HNP4, killed the Gram-positive strain much more efficiently, whereas the β-defensin HBD2 exhibited strong bactericidal activity against the Gram-negative strain.
FIGURE 5.
Survival curves of E. coli ATCC 25922 (red) and S. aureus ATCC 29213 (blue) exposed to l- (empty circles) and d-defensin (filled circles). Strains were exposed to the peptides at concentrations varying 2-fold from 1 to 256 μg/ml. Each curve is the mean of two (HNP1, HD5, and HBD2) or three (HNP4) separate experiments, where the error bars represent the ±S.D. of the measurements. Points scored as zero survival could not be plotted.
As anticipated, each of the four enantiomeric defensin pairs showed nearly identical killing activity against E. coli, consistent with the premise that induction of bacterial cell lysis by cationic antimicrobial peptides does not involve proteinaceous receptors on the cell membrane. Surprisingly and importantly, the validity of a unitary, membrane-only model was seriously undermined when we examined the killing of S. aureus by HNP1 and HD5. Here, DHNP1 and DHD5 displayed significantly reduced bactericidal activity compared with LHNP1 and LHD5, respectively. Upon l → d inversion of HNP1, the vLD50 value increased from 1.5 μg/ml to 16 μg/ml, while the vLD90 value increased by a factor of 19 (6 μg/ml to 115 μg/ml). At 64 μg/ml, LHNP1 quantitatively killed S aureus; bacterial growth could not be measured after the 12-h incubation time. In contrast, DHNP1 at the highest concentration of 256 μg/ml reduced S. aureus survival by <2 logs. Similar, but smaller differences were obtained with HD5. For HD5, the vLD50 value increased from 2.5 μg/ml for the l-form to 6.4 μg/ml for the d-form, whereas the vLD90 value increased only by a factor of 4.6 (from 7.3 μg/ml to 33 μg/ml).
Enantiomerization of HNP4 resulted in little change in its activity and selectivity toward both strains tested. This finding coincides with the earlier observation that inversion of HNP4 to its d-enantiomer exerted the least deleterious effect on HNP4 binding or inhibition of LF and gp120. HBD2 was extremely weak against S. aureus, and it was not possible to determine if its bactericidal activity against the Gram-positive strain was sensitive to enantiomerization.
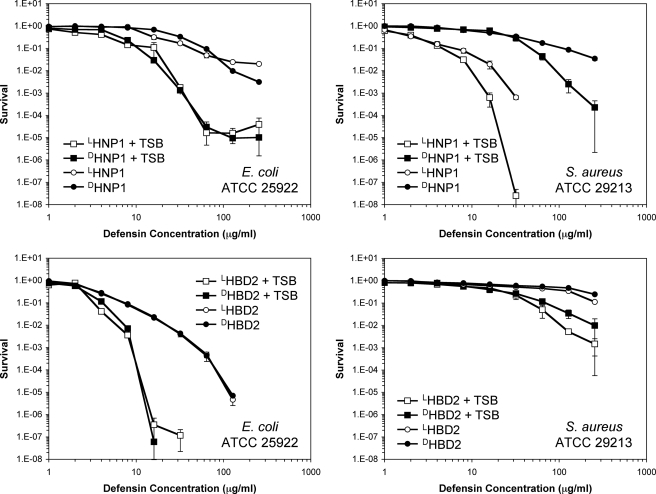
It was recognized that target cell growth and metabolism greatly sensitized microbes to the killing of less cationic defensins such as HNP-1 and -2 and rabbit defensin NP-5 (as opposed to highly cationic defensins such as rabbit NP-1 and NP-2) (11, 59, 60). Consistent with these older observations, the addition of 1% TSB to the phosphate buffer during the initial 2-h incubation period in our vCC assay produced lower survival at a given concentration of HNP1 compared with that in the absence of TSB (Figs. 5 and 6). LHNP1 and DHNP1 showed much enhanced and nearly identical bactericidal activity against E. coli in the presence of 1% TSB. Providing nutrients that allowed bacterial growth enhanced the killing of S. aureus by both LHNP1 and DHNP1, without eliminating the much greater susceptibility of the Gram-positive strain to the l-enantiomer, as was also observed in the absence of TSB. The presence of 1% TSB made E. coli considerably more susceptible than S. aureus to HBD2. In their totality, the data related to chirality in Figs. 5 and 6 indicate that the mechanism whereby α-defensins HNP1 and HD5 kill E. coli is distinct from the mechanism that they use to kill S. aureus, because only the latter is chirality-dependent. The staphylococcal partner in this chiral interaction remains to be identified.
FIGURE 6.
Survival curves of E. coli ATCC 25922 and S. aureus ATCC 29213 exposed to l- (empty symbols) and d-defensin (filled symbols) in the presence (squares) and absence (circles) of 1% TSB. Strains were exposed to the peptides at concentrations varying 2-fold from 1 to 256 μg/ml. Each curve is the mean of two separate experiments, where the error bars represent the ±S.D. of the measurements. Points scored as zero survival could not be plotted.
Other Human Defensins
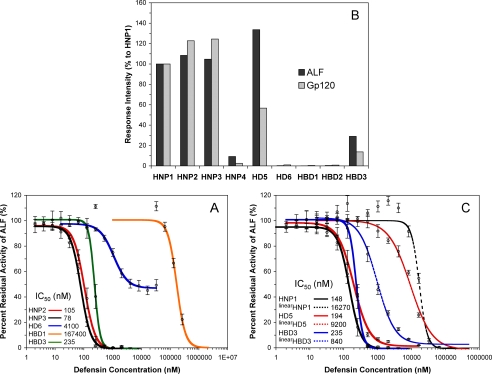
We also tested HNP2, HNP3, HD6, HBD1, and HBD3 with respect to their inhibition of LF activity (Fig. 7A). HNP2 and HNP3 differ from HNP1 by one amino acid residue at the N terminus, and, as expected, had similar IC50 values to that of HNP1. HD6 was the weakest among the six α-defensins, with its maximal inhibitory activity leveling off at ∼50% inhibition of LF. The reason that HD6 failed to progress beyond this plateau remains to be determined, but may be related to its ability to oligomerize differently from other human α-defensins (55). HBD1 had the highest IC50 value of 167 μm in the panel of nine native defensins tested. Notably, HBD3 is as active as HD5 against LF, and two orders of magnitude more potent than HBD2. These inhibitory activity data are in general agreement with the results obtained from SPR-based binding studies on immobilized LF and gp120 (Fig. 7B). The nine native defensins can be classified into three categories on the basis of their LF inhibition and gp120 binding activity: strong (HNP1, HNP2, HNP3, HD5 and HBD3), medium (HNP4), and weak (HBD1, HBD2 and HD6).
FIGURE 7.
Inhibition and binding of LF by other defensins. A, inhibition of LF activity by different concentrations of HNP2 (red), HNP3 (black), HD6 (blue), HBD1 (orange), and HBD3 (green). B, percent RU, relative to HNP1, at 300 s of association of 100 nm defensin on 2500 RUs of immobilized LF or 2830 RUs of immobilized gp120. C, inhibition of LF activity by linearized defensins (dotted lines): linearHNP1 (black), linearHD5 (red), and linearHBD3 (blue). Each inhibition curve is the mean of three independent enzyme kinetic measurements. The p values for statistical significance are: p = 0.0012 for HNP1/linearHNP1, p = 0.0046 for HD5/linearHD5, and p = 0.0076 for HBD3/linearHBD3.
Defensin Tertiary Structure in Relation to LF Inhibition, HIV gp120 Binding, and Bacterial Killing
Kaufmann and colleagues reported that dithiothreitol-reduced HNP1 failed to inhibit LF (36). Reduction of the three disulfides followed by S-alkylation in retrocyclin-1 also dramatically reduced the ability of the θ-defensin to inhibit LF (38). To better understand the effect of disulfide bonding on LF inhibition, we characterized the unstructured forms of HNP1, HD5, and HBD3, in which all six Cys residues were simultaneously replaced by either Ala (in HNP1) or α-aminobutyric acid (in HD5 and HBD3). Dose-dependent LF inhibition by the three linearized defensins, designated as linearHNP1, linearHD5, and linearHBD3, is shown in Fig. 7C. For comparison, the inhibition curves of native HNP1, HD5, and HBD3 are also plotted in the same figure. For the two α-defensins, loss of their tertiary structure was clearly detrimental to LF inhibition. A reduction in LF inhibition by 110-fold, characterized by an increase in IC50 from 148 nm to 16.3 μm, was seen with HNP1, whereas the inhibitory activity of HD5 was weakened by 47-fold (an increase in IC50 from 194 nm to 9.2 μm). By contrast, loss of disulfide bonding in HBD3 had a much smaller effect on LF inhibition, as evidenced by a <4-fold increase in IC50 from 235 to 840 nm. The three unstructured defensins were also analyzed along with wild-type HNP1, HD5, and HBD3 with respect to their binding to LF, and, as shown in Fig. 8 (top panel), the SPR data fully agreed with the above findings. Further, disulfide bonding was also much less important for HBD3 than for HNP1 and HD5 in HIV gp120 binding (Fig. 8, bottom panel).
FIGURE 8.
Comparison of native HNP1, HD5, and HBD3 with their corresponding unstructured forms, each at 100 nm, in LF (top panels) and gp120 (bottom panels) binding. Sensor chips with 2500 RUs of LF and 2830 RUs of gp120 were used for the SPR measurements.
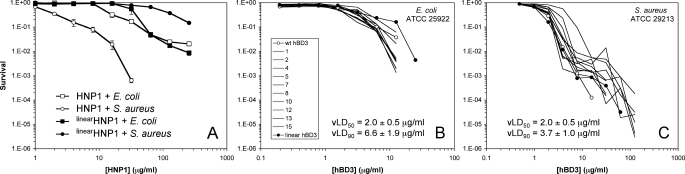
Using virtual colony count, we have previously shown that linearHD5 was significantly less active than HD5 in killing S. aureus, but largely indistinguishable from HD5 in the killing of E. coli (61). We have extended the same observation to linearHNP1 and HNP1 under identical assay conditions. Shown in Fig. 9A are plots of E. coli and S. aureus survival versus defensin concentration. Both HNP1 and linearHNP1 had similarly weak killing activity against E. coli. By contrast, although HNP1 quantitatively killed S. aureus at 64 μg/ml, linearHNP1 was barely active at the same concentration. The survival of S. aureus was reduced by less than one log by linearHNP1 at its highest concentration of 256 μg/ml tested. Clearly, native α-defensin structure, while dispensable in the killing of E. coli, is required for efficient killing of S. aureus, reminiscent of DHNP1 and DHD5.
FIGURE 9.
Survival curves of E. coli ATCC 25922 and S. aureus ATCC 29213 exposed to HNP1 and linearHNP1 (A), and, to HBD3, linearHBD3 and 10 disulfide analogs of HBD3 (B and C). Strains were exposed to the peptides at concentrations varying 2-fold from 1 to 256 μg/ml (HNP1 and linearHNP1), 0.195 to 50 μg/ml (HBD3 and E. coli), or 0.488 to 125 μg/ml (HBD3 and S. aureus). Each curve is the mean of two (HNP1 and linearHNP1) or three (HBD3 and analogs) separate experiments. Points scored as zero survival could not be plotted.
Due to an unusually high number of cationic charges, the bactericidal activity of HBD3 is generally insensitive to loss of or topological change in disulfide bonding (44, 62, 63). To extend our previous observation (44), we quantified dose-dependent killing of E. coli and S. aureus by HBD3, linearHBD3, and 10 disulfide analogs (each with a unique three-disulfide connectivity different from the native S–S pairing (Cys1–Cys5, Cys2–Cys4, and Cys3–Cys6). As shown in Fig. 9 (B and C), HBD3 and all its disulfide analogs, regardless of whether or not and how their disulfide bridges are paired, efficiently killed both strains of bacteria, with vLD90 values clustering around 6.6 ± 1.9 μg/ml for E. coli and 3.7 ± 1.0 μg/ml for S. aureus, and an identical average vLD50 value of 2.0 ± 0.5 μg/ml for both strains. At 25 μg/ml, complete killing of E. coli was achieved by all disulfide-bridged defensins, representing a reduction of the number of colonies by at least six orders of magnitude. By contrast, an average reduction of the number of colonies of S. aureus was approximately three orders of magnitude at the same defensin concentration.
DISCUSSION
Defensins are inherently effective multitaskers. How such small peptides have acquired functional versatility in innate and adaptive immunity is not well understood at the molecular level. The mechanism whereby HNP 1–3 killed E. coli was described in 1989 (11). The authors noted that, under conditions that supported bactericidal activity, HNP-1 sequentially permeabilized the outer membrane and inner membrane of E. coli and that, coincident with these events, bacterial synthesis of DNA, RNA, and protein ceased and the colony count fell.
Merrifield and colleagues first used enantiomers to probe mechanisms of antimicrobial peptides (58) and reported in 1990 that the l- and d-enantiomers of three α-helical peptides (cecropin, magainin, or melittin) were equally active in polarizing planar lipid bilayers, killing Gram-positive and -negative strains of bacteria, and lysing erythrocytes. They suggested that chiral target cell molecules are not involved in the action of these antimicrobial peptides. We also noted functional equivalence of l- and d-enantiomers of protegrin-1, in studies performed with bacteria and Candida albicans (64–66). In contrast, when Tempst and colleagues studied enantiomers of apidaecins, proline- and arginine-rich antimicrobial peptides of insect origin that act preferentially on Gram-negative bacteria, the d-enantiomer proved to be much less potent (67, 68). They concluded that the peptides acted via a mechanism that included stereoselective elements but was completely devoid of any pore-forming activity. These earlier reports induced us to compare the functional properties of several paired d- and l-defensins. As discussed below, we gained some unexpected insights into the molecular basis for defensin function.
First, DHNP1 and DHD5 were significantly less active than their native l-forms in the killing of S. aureus, but the d- and l-enantiomers of HNP1 or HD5 were equally bactericidal against E. coli. This strain-dependent activity profile of d-defensins has unveiled a yet-to-be-recognized mechanistic complexity of bacterial killing by defensins, likely arising from differences in the chemical composition and structure of the bacterial cell wall between E. coli and S. aureus.
For S. aureus, the cell wall consists primarily of a single but thick layer of peptidoglycan covered with (lipo)teichoic acid, whereas in E. coli, it is composed of a thin layer of peptidoglycan surrounded by a thin outer membrane whose outer leaflet is largely composed of lipopolysaccharide (69). Cationic antimicrobial peptides can associate with the negatively charged lipopolysaccharide or teichoic acid, an event thought to be important not only for antimicrobial selectivity but also for peptide uptake across the bacterial cell wall (70). It is plausible that an unidentified cellular component of S. aureus, possibly of chiral nature, preferably interacts with native defensins, thus contributing a great deal to bacterial killing. Consistent with this hypothesis, loss of the structure of HNP1 and HD5 dramatically reduced their bactericidal activity against S. aureus but not against E. coli. Our bactericidal data on HNP1 and HD5 as well as their linearized and d-enantiomeric analogs suggest that the membrane of S. aureus is not the sole lethal target for certain defensins and that an alternative mode of action exists in microbial killing.
Membrane-independent mechanisms have been proposed for bacterial killing by other classes of cationic antimicrobial peptides based on a poor correlation between their abilities to permeate model membranes and to kill bacteria (71). A poor correlation has also been established for the six human α-defensins between their membrane activity and bactericidal activity.5 Hancock and colleagues argue that an alternative mode of action exists that likely involves internalization of cationic peptides and targeting of intracellular molecules (72, 73). The internal targeting hypothesis is supported by the observation that certain antimicrobial peptides interact with nuclear acids and interfere with protein synthesis but do not cause permanent membrane depolarization. Although it remains unclear how efficiently HNP1 and HD5 traverse the cytoplasmic membrane, the possibility of endocytic internalization cannot be ruled out. Alternatively, various effector molecules anchored on the cell wall surface may interact in the uptake process of certain defensins, thus facilitating or attenuating subsequent membrane permeabilization.
Nisin, an amphiphilic antibiotic peptide produced by various strains of Lactococcus lactis, is relatively weak in its ability to induce liposomal leakage but potently active in killing Gram-positive bacteria (74). Because the ability of nisin to induce liposomal leakage markedly increased by several orders of magnitude in the presence of Lipid II, a membrane-bound peptidoglycan precursor, it was suggested that nisin specifically binds to the pyrophosphate moiety of Lipid II for enhanced bacterial membrane permeabilization (75). More recently, nisin has been reported to displace Lipid II from the cell division site to block cell wall synthesis (76), a bacterial killing mechanism reminiscent of vancomycin (77). These findings raise an intriguing possibility, that is, HNP1 and HD5 may use some components of the bacterial cell wall or of the cytoplasmic membrane as docking molecules for enhanced membrane interaction and/or for inhibiting cell wall synthesis. Because HNP1 and HD5 are the strongest among the six human α-defensins in binding glycosylated proteins and peptidoglycans, the possibility merits further investigation. In this regard, lectin-like properties of HNP1 and HD5 may be functionally relevant to the killing of S. aureus.
A second surprising finding was the effective inhibition of LF by DHNP1 and DHD5 at high nanomolar concentrations. In nature, the l-amino acids are by far the predominant enantiomers (78). Whereas d-amino acids are commonly synthesized and incorporated into antibiotic peptides by prokaryotes, the animal kingdom restricts their appearance to post-translationally modified diastereomeric peptides in lower species, such as amphibians (79, 80). In humans, however, there exist no known d-amino acids. It is well established in the literature that the d-enantiomer of a native protein does not recognize the protein partners of the l-enantiomer or vice versa due to steric incompatibility. For example, a chemically synthesized d-HIV-1 protease does not hydrolyze natural substrates of the native viral protease but cleaves the d-enantiomer of a natural substrate as efficiently as l-HIV-1 protease cleaves the l-substrate (81). This chiral mode of molecular recognition is stringently maintained in interacting protein systems such as enzyme-substrate and enzyme-inhibitor. For these reasons, d-defensins were not expected to “specifically” interact with LF with high affinity. The fact that d-defensins were effective inhibitors of LF, as was the case with d-retrocyclins (38), suggests that defensin association with LF and many other proteins may be driven by forces that act in a nonspecific and non-directional fashion.
Defensins are small enough that both cationic and hydrophobic residues are solvent-exposed, a circumstance that makes the peptides unusually “sticky.” It is plausible that these surface electrostatic and hydrophobic forces, in combination with a disulfide-stabilized molecular scaffold, enable multiple defensin molecules to bind a single target protein molecule. In fact, both HNP1 and HD5 have been shown to bind bacterial toxins and HIV gp120 at a high molar ratio (56, 82). Further, defensins can dimerize and potentially form higher order soluble aggregates in solution (6, 55). It has been demonstrated that binding of HNP1 or HD5 to gp120 or bacterial toxins promotes defensin self-aggregation (56, 82). Defensin oligomerization affords additional molecular complexity at the quaternary structural level, thereby contributing to enhanced structural diversity and functional versatility.
Finally, we observed that LF inhibition, gp120 binding, and S. aureus killing were strongly correlated among the various defensins studied. In general, the strongest inhibitors of LF were also the most effective bactericidal agents of S. aureus and often the best lectins. This is surprising given the different nature of the three defensin targets: a bacterial enzyme, a viral glycoprotein, and a bacterium. Does this correlation hint at a common molecular mechanism that allows defensins to intervene in so many diverse biological processes? Presently, we are exploring the possibility that events at the quaternary structural level, such as in situ oligomerization, may provide a common driving force that is ultimately responsible for modulating the interaction of defensins with diverse targets and endowing them with so broad an array of functional properties.
Several conserved structural elements in α-defensins have been investigated, including a salt bridge, an invariant Gly residue, and disulfide bonding (83–87). However, these studies do not address the structural determinants that make some α-defensins considerably more potent than others in accomplishing a given task.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI072732 and AI061482 (to W. L.).
The atomic coordinates and structure factors (codes 3gny and 3go0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
P. Zeng, C. Xie, B. Ericksen, Z. Wu, X. Li, W.-Y. Lu, and W. Lu, unpublished results.
- HNP
- human neutrophil peptide
- HBD
- human β-defensin
- HIV-1
- human immunodeficiency virus, type 1
- RU
- response units
- TSB
- tryptic soy broth
- LeTx
- lethal toxin
- LF
- lethal factor
- SPR
- surface plasmon resonance.
REFERENCES
- 1.Zasloff M. (2002) Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 2.Selsted M. E., Ouellette A. J. (2005) Nat. Immunol. 6, 551–557 [DOI] [PubMed] [Google Scholar]
- 3.Lehrer R. I. (2004) Nat. Rev. Microbiol. 2, 727–738 [DOI] [PubMed] [Google Scholar]
- 4.Ganz T. (2003) Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 5.Bevins C. L. (2006) Biochem. Soc. Trans. 34, 263–266 [DOI] [PubMed] [Google Scholar]
- 6.Pazgier M., Hoover D. M., Yang D., Lu W., Lubkowski J. (2006) Cell Mol. Life Sci. 63, 1294–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y. Q., Yuan J., Osapay G., Osapay K., Tran D., Miller C. J., Ouellette A. J., Selsted M. E. (1999) Science 286, 498–502 [DOI] [PubMed] [Google Scholar]
- 8.Ganz T., Selsted M. E., Szklarek D., Harwig S. S., Daher K., Bainton D. F., Lehrer R. I. (1985) J. Clin. Invest. 76, 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selsted M. E., Harwig S. S., Ganz T., Schilling J. W., Lehrer R. I. (1985) J. Clin. Invest. 76, 1436–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagan B. L., Selsted M. E., Ganz T., Lehrer R. I. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehrer R. I., Barton A., Daher K. A., Harwig S. S., Ganz T., Selsted M. E. (1989) J. Clin. Invest. 84, 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang T. L., Vargas J., Jr., DelPortillo A., Klotman M. E. (2005) J. Clin. Invest. 115, 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L., Finnegan C. M., Kish-Catalone T., Blumenthal R., Garzino-Demo P., La Terra Maggiore G. M., Berrone S., Kleinman C., Wu Z., Abdelwahab S., Lu W., Garzino-Demo A. (2005) J. Virol. 79, 14318–14329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quiñones-Mateu M. E., Lederman M. M., Feng Z., Chakraborty B., Weber J., Rangel H. R., Marotta M. L., Mirza M., Jiang B., Kiser P., Medvik K., Sieg S. F., Weinberg A. (2003) AIDS 17, F39–F48 [DOI] [PubMed] [Google Scholar]
- 15.Mackewicz C. E., Yuan J., Tran P., Diaz L., Mack E., Selsted M. E., Levy J. A. (2003) AIDS 17, F23–F32 [DOI] [PubMed] [Google Scholar]
- 16.Guo C. J., Tan N., Song L., Douglas S. D., Ho W. Z. (2004) AIDS 18, 1217–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallo S. A., Wang W., Rawat S. S., Jung G., Waring A. J., Cole A. M., Lu H., Yan X., Daly N. L., Craik D. J., Jiang S., Lehrer R. I., Blumenthal R. (2006) J. Biol. Chem. 281, 18787–18792 [DOI] [PubMed] [Google Scholar]
- 18.Furci L., Sironi F., Tolazzi M., Vassena L., Lusso P. (2007) Blood 109, 2928–2935 [DOI] [PubMed] [Google Scholar]
- 19.Klotman M. E., Rapista A., Teleshova N., Micsenyi A., Jarvis G. A., Lu W., Porter E., Chang T. L. (2008) J. Immunol. 180, 6176–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D., Biragyn A., Hoover D. M., Lubkowski J., Oppenheim J. J. (2004) Annu Rev. Immunol. 22, 181–215 [DOI] [PubMed] [Google Scholar]
- 21.Rehaume L. M., Hancock R. E. (2008) Crit. Rev. Immunol. 28, 185–200 [DOI] [PubMed] [Google Scholar]
- 22.Funderburg N., Lederman M. M., Feng Z., Drage M. G., Jadlowsky J., Harding C. V., Weinberg A., Sieg S. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18631–18635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Territo M. C., Ganz T., Selsted M. E., Lehrer R. (1989) J. Clin. Invest. 84, 2017–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D., Chen Q., Chertov O., Oppenheim J. J. (2000) J. Leukoc. Biol. 68, 9–14 [PubMed] [Google Scholar]
- 25.Yang D., Chertov O., Bykovskaia S. N., Chen Q., Buffo M. J., Shogan J., Anderson M., Schröder J. M., Wang J. M., Howard O. M., Oppenheim J. J. (1999) Science 286, 525–528 [DOI] [PubMed] [Google Scholar]
- 26.van Wetering S., Mannesse-Lazeroms S. P., van Sterkenburg M. A., Hiemstra P. S. (2002) Inflamm. Res. 51, 8–15 [DOI] [PubMed] [Google Scholar]
- 27.Shi J., Aono S., Lu W., Ouellette A. J., Hu X., Ji Y., Wang L., Lenz S., van Ginkel F. W., Liles M., Dykstra C., Morrison E. E., Elson C. O. (2007) J. Immunol. 179, 1245–1253 [DOI] [PubMed] [Google Scholar]
- 28.van den Berg R. H., Faber-Krol M. C., van Wetering S., Hiemstra P. S., Daha M. R. (1998) Blood 92, 3898–3903 [PubMed] [Google Scholar]
- 29.Panyutich A. V., Szold O., Poon P. H., Tseng Y., Ganz T. (1994) FEBS Lett. 356, 169–173 [DOI] [PubMed] [Google Scholar]
- 30.Sørensen O. E., Cowland J. B., Theilgaard-Mönch K., Liu L., Ganz T., Borregaard N. (2003) J. Immunol. 170, 5583–5589 [DOI] [PubMed] [Google Scholar]
- 31.Aarbiou J., Ertmann M., van Wetering S., van Noort P., Rook D., Rabe K. F., Litvinov S. V., van Krieken J. H., de Boer W. I., Hiemstra P. S. (2002) J. Leukoc. Biol. 72, 167–174 [PubMed] [Google Scholar]
- 32.Candille S. I., Kaelin C. B., Cattanach B. M., Yu B., Thompson D. A., Nix M. A., Kerns J. A., Schmutz S. M., Millhauser G. L., Barsh G. S. (2007) Science 318, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehrer R. I. (2007) Curr. Opin Hematol. 14, 16–21 [DOI] [PubMed] [Google Scholar]
- 34.Moayeri M., Leppla S. H. (2004) Curr. Opin. Microbiol. 7, 19–24 [DOI] [PubMed] [Google Scholar]
- 35.Collier R. J., Young J. A. (2003) Annu. Rev. Cell Dev. Biol. 19, 45–70 [DOI] [PubMed] [Google Scholar]
- 36.Kim C., Gajendran N., Mittrücker H. W., Weiwad M., Song Y. H., Hurwitz R., Wilmanns M., Fischer G., Kaufmann S. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4830–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer-Scholl A., Hurwitz R., Brinkmann V., Schmid M., Jungblut P., Weinrauch Y., Zychlinsky A. (2005) PLoS Pathog. 1, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W., Mulakala C., Ward S. C., Jung G., Luong H., Pham D., Waring A. J., Kaznessis Y., Lu W., Bradley K. A., Lehrer R. I. (2006) J. Biol. Chem. 281, 32755–32764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leikina E., Delanoe-Ayari H., Melikov K., Cho M. S., Chen A., Waring A. J., Wang W., Xie Y., Loo J. A., Lehrer R. I., Chernomordik L. V. (2005) Nat. Immunol. 6, 995–1001 [DOI] [PubMed] [Google Scholar]
- 40.Münk C., Wei G., Yang O. O., Waring A. J., Wang W., Hong T., Lehrer R. I., Landau N. R., Cole A. M. (2003) AIDS Res. Hum. Retroviruses 19, 875–881 [DOI] [PubMed] [Google Scholar]
- 41.Wang W., Owen S. M., Rudolph D. L., Cole A. M., Hong T., Waring A. J., Lal R. B., Lehrer R. I. (2004) J. Immunol. 173, 515–520 [DOI] [PubMed] [Google Scholar]
- 42.Wang W., Cole A. M., Hong T., Waring A. J., Lehrer R. I. (2003) J. Immunol. 170, 4708–4716 [DOI] [PubMed] [Google Scholar]
- 43.Wu Z., Ericksen B., Tucker K., Lubkowski J., Lu W. (2004) J. Pept. Res. 64, 118–125 [DOI] [PubMed] [Google Scholar]
- 44.Wu Z., Hoover D. M., Yang D., Boulègue C., Santamaria F., Oppenheim J. J., Lubkowski J., Lu W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8880–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Z., Powell R., Lu W. (2003) J. Am. Chem. Soc. 125, 2402–2403 [DOI] [PubMed] [Google Scholar]
- 46.Schnölzer M., Alewood P., Jones A., Alewood D., Kent S. B. (1992) Int. J. Pept. Protein Res. 40, 180–193 [DOI] [PubMed] [Google Scholar]
- 47.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min D. H., Tang W. J., Mrksich M. (2004) Nat. Biotechnol. 22, 717–723 [DOI] [PubMed] [Google Scholar]
- 49.Ericksen B., Wu Z., Lu W., Lehrer R. I. (2005) Antimicrob. Agents Chemother. 49, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 51.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 52.Hill C. P., Yee J., Selsted M. E., Eisenberg D. (1991) Science 251, 1481–1485 [DOI] [PubMed] [Google Scholar]
- 53.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 54.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 55.Szyk A., Wu Z., Tucker K., Yang D., Lu W., Lubkowski J. (2006) Protein Sci. 15, 2749–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lehrer R. I., Jung G., Ruchala P., Andre S., Gabius H. J., Lu W. (2009) J. Immunol. 183, 480–490 [DOI] [PubMed] [Google Scholar]
- 57.Wu Z., Cocchi F., Gentles D., Ericksen B., Lubkowski J., Devico A., Lehrer R. I., Lu W. (2005) FEBS Lett. 579, 162–166 [DOI] [PubMed] [Google Scholar]
- 58.Wade D., Boman A., Wåhlin B., Drain C. M., Andreu D., Boman H. G., Merrifield R. B. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4761–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lehrer R. I., Ganz T., Szklarek D., Selsted M. E. (1988) J. Clin. Invest. 81, 1829–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ganz T., Selsted M. E., Lehrer R. I. (1990) Eur. J. Haematol. 44, 1–8 [DOI] [PubMed] [Google Scholar]
- 61.de Leeuw E., Burks S. R., Li X., Kao J. P., Lu W. (2007) FEBS Lett. 581, 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor K., Clarke D. J., McCullough B., Chin W., Seo E., Yang D., Oppenheim J., Uhrin D., Govan J. R., Campopiano D. J., MacMillan D., Barran P., Dorin J. R. (2008) J. Biol. Chem. 283, 6631–6639 [DOI] [PubMed] [Google Scholar]
- 63.Klüver E., Schulz-Maronde S., Scheid S., Meyer B., Forssmann W. G., Adermann K. (2005) Biochemistry 44, 9804–9816 [DOI] [PubMed] [Google Scholar]
- 64.Kokryakov V. N., Harwig S. S., Panyutich E. A., Shevchenko A. A., Aleshina G. M., Shamova O. V., Korneva H. A., Lehrer R. I. (1993) FEBS Lett. 327, 231–236 [DOI] [PubMed] [Google Scholar]
- 65.Cho Y., Turner J. S., Dinh N. N., Lehrer R. I. (1998) Infect. Immun. 66, 2486–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyasaki K. T., Iofel R., Oren A., Huynh T., Lehrer R. I. (1998) J. Periodontal Res. 33, 91–98 [DOI] [PubMed] [Google Scholar]
- 67.Casteels P., Tempst P. (1994) Biochem. Biophys. Res. Commun. 199, 339–345 [DOI] [PubMed] [Google Scholar]
- 68.Castle M., Nazarian A., Yi S. S., Tempst P. (1999) J. Biol. Chem. 274, 32555–32564 [DOI] [PubMed] [Google Scholar]
- 69.Brock T. D., Madigan M. T. (1991) Biology of Microorganisms, Sixth Ed., pp. 54–62, Prentice-Hall, Englewood Cliffs, NJ [Google Scholar]
- 70.Hancock R. E. (1997) Lancet 349, 418–422 [DOI] [PubMed] [Google Scholar]
- 71.Brogden K. A. (2005) Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]
- 72.Wu M., Maier E., Benz R., Hancock R. E. (1999) Biochemistry 38, 7235–7242 [DOI] [PubMed] [Google Scholar]
- 73.Hancock R. E., Rozek A. (2002) FEMS Microbiol. Lett. 206, 143–149 [DOI] [PubMed] [Google Scholar]
- 74.Breukink E., Wiedemann I., van Kraaij C., Kuipers O. P., Sahl H., de Kruijff B. (1999) Science 286, 2361–2364 [DOI] [PubMed] [Google Scholar]
- 75.Hsu S. T., Breukink E., Tischenko E., Lutters M. A., de Kruijff B., Kaptein R., Bonvin A. M., van Nuland N. A. (2004) Nat. Struct. Mol. Biol. 11, 963–967 [DOI] [PubMed] [Google Scholar]
- 76.Hasper H. E., Kramer N. E., Smith J. L., Hillman J. D., Zachariah C., Kuipers O. P., de Kruijff B., Breukink E. (2006) Science 313, 1636–1637 [DOI] [PubMed] [Google Scholar]
- 77.Breukink E., de Kruijff B. (2006) Nat. Rev. Drug Discov. 5, 321–332 [DOI] [PubMed] [Google Scholar]
- 78.Kreil G. (1997) Annu. Rev. Biochem. 66, 337–345 [DOI] [PubMed] [Google Scholar]
- 79.Heck S. D., Siok C. J., Krapcho K. J., Kelbaugh P. R., Thadeio P. F., Welch M. J., Williams R. D., Ganong A. H., Kelly M. E., Lanzetti A. J., et al. (1994) Science 266, 1065–1068 [DOI] [PubMed] [Google Scholar]
- 80.Kreil G. (1994) Science 266, 996–997 [DOI] [PubMed] [Google Scholar]
- 81.Milton R. C., Milton S. C., Kent S. B. (1992) Science 256, 1445–1448 [DOI] [PubMed] [Google Scholar]
- 82.Lehrer R. I., Jung G., Ruchala P., Wang W., Micewicz E. D., Waring A. J., Gillespie E. J., Bradley K. A., Ratner A. J., Rest R. F., Lu W. (2009) Infect. Immun. 77, 4028–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu Z., Li X., de Leeuw E., Ericksen B., Lu W. (2005) J. Biol. Chem. 280, 43039–43047 [DOI] [PubMed] [Google Scholar]
- 84.Xie C., Prahl A., Ericksen B., Wu Z., Zeng P., Li X., Lu W. Y., Lubkowski J., Lu W. (2005) J. Biol. Chem. 280, 32921–32929 [DOI] [PubMed] [Google Scholar]
- 85.Rajabi M., de Leeuw E., Pazgier M., Li J., Lubkowski J., Lu W. (2008) J. Biol. Chem. 283, 21509–21518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maemoto A., Qu X., Rosengren K. J., Tanabe H., Henschen-Edman A., Craik D. J., Ouellette A. J. (2004) J. Biol. Chem. 279, 44188–44196 [DOI] [PubMed] [Google Scholar]
- 87.Rosengren K. J., Daly N. L., Fornander L. M., Jönsson L. M., Shirafuji Y., Qu X., Vogel H. J., Ouellette A. J., Craik D. J. (2006) J. Biol. Chem. 281, 28068–28078 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.