Abstract
DNA strand passage through an enzyme-mediated gate is a key step in the catalytic cycle of topoisomerases to produce topological transformations in DNA. In most of the reactions catalyzed by topoisomerases, strand passage is not directional; thus, the enzyme simply provides a transient DNA gate through which DNA transport is allowed and thereby resolves the topological entanglement. When studied in isolation, the type IA topoisomerase family appears to conform to this rule. Interestingly, type IA enzymes can carry out directional strand transport as well. We examined here the biochemical mechanism for directional strand passage of two type IA topoisomerases: reverse gyrase and a protein complex of topoisomerase IIIα and Bloom helicase. These enzymes are able to generate vectorial strand transport independent of the supercoiling energy stored in the DNA molecule. Reverse gyrase is able to anneal single strands, thereby increasing linkage number of a DNA molecule. However, topoisomerase IIIα and Bloom helicase can dissolve DNA conjoined with a double Holliday junction, thus reducing DNA linkage. We propose here that the helicase or helicase-like component plays a determinant role in the directionality of strand transport. There is thus a common biochemical ground for the directional strand passage for the type IA topoisomerases.
Introduction
DNA topoisomerases are important and indispensable biochemical tools to solve the problems of DNA entanglement that arise during processes of DNA transaction, including replication, transcription, recombination, and repair (reviewed in Refs. 1–3). These enzymes accomplish this impressive feat with simple yet elegant chemistry of reversible transesterification reactions. A tyrosine residue at the enzyme's active site serves as a nucleophile to initiate the transesterification reaction, resulting in a DNA chain scission and the formation of an enzyme-DNA adduct. The reversal of this reaction then restores the integrity of the DNA backbone and active-site tyrosine. The topological transformation is accomplished by the passage of the DNA strand through this transient strand break. There are two types of topoisomerases, distinguishable on both mechanistic and structural grounds. Type I enzymes make a single-strand break at a time, whereas type II enzymes make a concerted double-strand break. The DNA transported through the reversible single-strand break in type I enzymes is usually single-stranded, the strand complementary to the scissile one, but it is possible to have a double strand transported as well. The DNA passing through the transient double-strand breaks in type II enzymes is another duplex segment. The intramolecular strand passage events will lead to reactions that change DNA supercoiling or tying and untying topological knots, whereas the intermolecular strand passage results in catenation and decatenation reactions.
The type IA topoisomerases are ubiquitous enzymes found in every living organism with the exception of viruses. This family includes bacterial topo3 I and topo III as well as eukaryotic topo III. In higher eukaryotes, there are two isoforms of topo III, designated α and β, with evidence mounting that topo IIIα is imported into the mitochondria for roles yet to be defined in addition to its roles in the nucleus (4). In lower organisms, deletion of the type IA enzymes generally results in viable cells, albeit with genomic instability phenotypes and, in the case of Saccharomyces cerevisiae, complete inhibition of sporulation (5). In higher eukaryotes, deletion of topo IIIα is early embryonic lethal, whereas topo IIIβ deletions are viable but with genomic instability apparent in the germ line (6, 7).
Structural and biochemical analysis of the type IA topoisomerases indicates that these enzymes function by binding and creating a transient break in one strand of DNA, forming a gate through which another strand (or strands) of DNA can pass into an interior channel of the enzyme (8). The DNA break is then resealed, and the enzyme disassociates from the DNA, releasing both the bound and trapped strands. This mechanism has two implications for these enzymes: first, the enzyme requires single-stranded DNA upon which to function, and second, the reaction possesses no energy source except for the energy stored in the topology (supercoiling) of the DNA substrate. Consequently, when these enzymes are studied in isolation on supercoiled circular substrates containing permanently denatured regions, they show no preference for the direction of strand passage and function equally well in adding or removing linkages to the substrate until all of the supercoiling energy has been exhausted, leaving the substrate in the topologically relaxed state (9, 10). On DNA that does not contain a permanently denatured region, these enzymes are most active on highly negatively supercoiled DNA, as these topological conditions favor the formation of the single-stranded regions required for protein binding. The affinity of a particular type IA enzyme for single-stranded DNA then determines the extent of relaxation of the substrate, as the DNA duplex becomes more stable when negative supercoiling energy is removed from the molecule. Because of this, no relaxation of positively supercoiled circular DNA (without a permanently denatured region) is seen with these enzymes because, under these conditions, the DNA duplex is too stable to become single-stranded for any of the known type IA topoisomerases to bind.
However, several type IA topoisomerases that appear to violate these “rules” have recently emerged. It appears that these enzymes have been cast in roles that require them to bind and actively add or remove topological linkages from DNA independent of the supercoiling energy stored in the molecules. Here, we explore the potential mechanisms of two such type IA topoisomerase family members.
Reverse Gyrase: A DNA Renaturase
Reverse gyrase was first discovered to be an enzyme in a hyperthermophilic archaebacterium capable of introducing positive DNA supercoiling dependent upon ATP hydrolysis and was named based on this activity (which is exactly opposite of DNA gyrase) (11). However, in contrast to DNA gyrase, reverse gyrase is a type IA topoisomerase with two closely linked domains: a helicase-like domain and a topoisomerase domain (12). Reverse gyrase may play an essential role in the growth of hyperthermophiles in harsh environments because the reverse gyrase gene is unique to the genomes of these organisms (13). This hypothesis is supported by the following two observations as well. A hyperthermophilic bacterium with the reverse gyrase gene deleted shows defects in growth at extreme temperatures (14). A moderately thermophilic bacterium growing near hydrothermal vents needs to adapt to extreme fluctuations in growth temperature and contains a reverse gyrase gene (15). In addition, the expression of reverse gyrase is significantly induced when the growth temperature shifts higher. The idea that reverse gyrase has an important function in stabilizing the genome at extreme temperatures is reinforced by its biochemical activities as a DNA renaturase (16) and as a DNA chaperone (17).
The biochemical mechanism of how reverse gyrase carries out the directional strand passage remains an area under active investigation. There are several mechanisms proposed. One is based on differential relaxation of DNA supercoils (12). If the helicase domain is able to translocate along DNA, it would generate positive supercoiling in front of the moving helicase and negative supercoiling in its wake (18). The preferential relaxation of negative supercoils by the type IA topoisomerase domain will lead to a net accumulation of positive supercoils in a circular DNA molecule. However, there has not been any detectable helicase activity in either full-length reverse gyrase or the isolated helicase domain (19).
Another proposed mechanism is that, upon unwinding of DNA by reverse gyrase, it can segregate DNA into two separate topological domains: one with the unwound DNA and the other with positive DNA supercoils induced by unwinding (19). Preferential rewinding of the unwound region can lead to retention of positive supercoils. The mechanistic basis for topological segregation and the switching between the unwinding and rewinding actions by reverse gyrase is not addressed in this model.
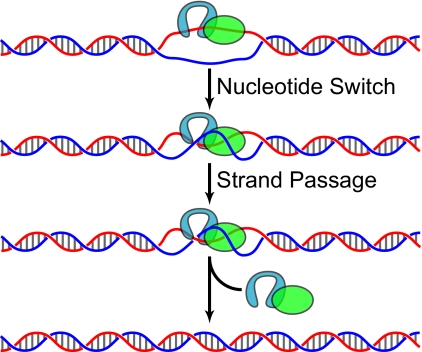
A third proposed mechanism is based on the hypothesis that, although the helicase domain cannot translocate and unwind DNA, it has a preferential binding activity for either single- or double-stranded DNA depending on the state of the bound nucleotide (20, 21). We envision the mechanism of action as a sequence of the following reaction steps (Fig. 1). If the temperature of the environment in which the organism is living rises high enough, regions of the genome will begin to denature spontaneously. The enzyme bound with a particular form of nucleotide (e.g. ATP) has a high affinity for the single-stranded region and associates with the denatured bubble in the duplex DNA. There is a switch in the bound nucleotide (e.g. from ATP to ADP) and consequentially a change in affinity with a preference for double-stranded DNA, thus promoting the rewinding of the denatured bubble. The topoisomerase domain engages in strand passage and increases the linkage (linking number) between the DNA strands. The protein recycles and can promote further renaturation if single-stranded DNA persists. The strand passage in these reaction cycles increases the DNA linking number and thus favors generation of positive supercoiling.
FIGURE 1.
Mechanistic steps of rewinding action catalyzed by reverse gyrase. Reverse gyrase is depicted as separate domains of topo IA (cyan) and helicase (green), bound to a single-stranded bubble. Through a nucleotide switch in the helicase domain, reverse gyrase promotes strand annealing, followed by strand passage to increase the DNA linking number. Cycles of these reactions result in the renaturation of the single-stranded bubble and positive supercoiling.
A number of experimental evidences support such a mechanism. Positive supercoiling of plasmid DNA by reverse gyrase is relatively inefficient and depends on the enzyme/DNA stoichiometry (21). The generation of a single-stranded region depends on the higher temperature at which reactions occur and the amount of enzyme that can preferentially bind it. Because the positive supercoiling is limited by the amount of single-stranded region formed under such conditions, it is dependent on the enzyme/DNA stoichiometry. This also suggests that, for a DNA with a permanent denatured bubble, the reaction would be more efficient because it is not limited by the available single-stranded region derived from unwinding in the base-paired region. Indeed, for a plasmid DNA with a bubble size of >25 nucleotides, positive supercoiling is highly efficient (16). Also as expected from this proposed mechanism, DNA supercoiling depends on the bound nucleotides: ATP promotes positive supercoiling, and a non-hydrolyzable analog of ATP (AMPPNP) induces negative supercoiling (21). This is presumably because the enzyme with bound AMPPNP can preferentially associate with a single-stranded region and induce the formation of a denatured bubble. With the relaxation of positive supercoils induced by the generation of single-stranded bubbles, the DNA will become negatively supercoiled once the enzyme is removed and the temperature is reduced, renaturing the single-stranded bubbles. There is also direct evidence suggesting a switch in DNA binding preference dependent upon the bound nucleotide for the recombinant helicase domain (20). However, the holoenzyme appears to have lost the nucleotide-dependent specificity in DNA binding, possibly because of the attenuation from the topoisomerase domain.
Topoisomerase IIIα/Bloom Helicase: Dissolution of Double Holliday Junctions
Whereas reverse gyrase provides a DNA machine for directional strand passage to increase DNA strand linkage, topo IIIα and the Bloom syndrome helicase Blm are able to reduce it (22, 23). A dHJ has long been proposed as an intermediate of homologous recombination that gives rise to the crossover and non-crossover products of this pathway (24), and this structure presents some very unique topological challenges to the cell (2). Once formed, this structure joins two DNA duplexes via the intertwining of the component single strands into hybrid duplexes, which we will refer to as heteroduplexes, between the two HJs. This generates a topological linking number between the two DNA duplexes that is a function of the length of the heteroduplex region, which should be approximately two linkages/10.5 bp of DNA separating the two HJs (one linkage/10.5 bp in each of the two arms of the heteroduplex). Each heteroduplex is a quasi-isolated topological domain; whereas the topology of the heteroduplexes will be influenced by the topology of the DNA outside of the dHJ, no action of a topoisomerase outside of the dHJ can change the number of linkages holding the two DNA duplexes together. In the classical resolution of a dHJ by resolvases, this problem is overcome by simply breaking the covalent bonds in a pair of DNA single strands at each HJ. However, there is a growing body of genetic evidence that indicates that a second pathway for HJ resolution exists in which the dHJ is dissolved by a type IA topoisomerase working in conjunction with a RecQ family helicase, resulting exclusively in non-crossover recombination outcomes (25). This implies that a type IA topoisomerase, which shows no inherent directionality of strand passage in isolation, is catalyzing directional strand passage in this context to reduce the number of linkages within the heteroduplex regions to zero, separating the two DNA duplexes. This pathway has gained further support with the discovery that model dHJs can be dissolved in vitro with human (23) or Drosophila (22) topo IIIα/Blm.
Mechanistically, topo IIIα seems ill suited for a role in dHJ linkage removal. Blm was shown to be highly active on oligonucleotide-based single HJ substrates and in HJ formation via fork regression on substrates mimicking stalled replication forks (reviewed in Ref. 26). However, if Blm were to attempt to migrate a dHJ convergently, there would be positive supercoiling stress within the heteroduplex region, making strand separation more difficult. As discussed above, type IA topoisomerases require a single-stranded region for strand passage activity. This raises the question of how topo IIIα efficiently removes the linkages within a dHJ when that region is likely to be under positive supercoiling stress, thus reducing the likelihood of single-stranded DNA formation.
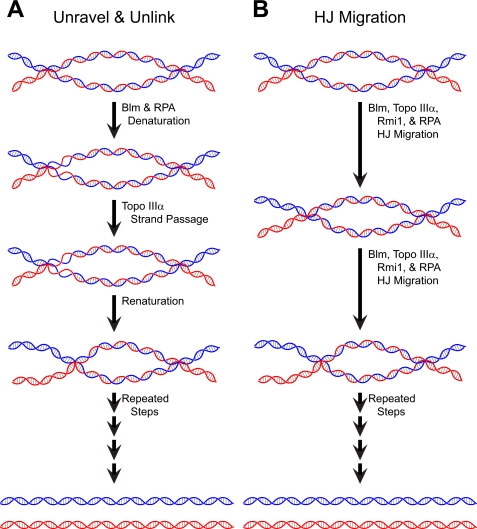
There are two possible mechanistic models for the dissolution of a dHJ by topo IIIα and Blm, with the first being an “unravel and unlink” model. In this model, with the aid of single-stranded binding proteins, Blm binds to a HJ and denatures a region of the heteroduplex, providing the preferred substrate (single-stranded DNA) for topo IIIα binding and strand passage. Most, if not all, of the single-stranded DNA-binding proteins would then dissociate, and the denatured bubbles would rewind with the exchanged strand (Fig. 2A). For this model to work, there would have to be some coordination between two reactions, each occurring on opposing heteroduplexes and on the same side of the HJ, for the HJ to move forward efficiently. The size of the region denatured by Blm would define a step size for the reaction, which, in practical terms, could range from one turn of DNA to the entire heteroduplex region. If the step size for this reaction were smaller than the entire heteroduplex, however, a protein “bookmark” would have to be left behind at the conclusion of each round of reactions to ensure that the next round would occur in the same direction as the first. Failure to do so would mean that the dissolution of a dHJ would occur via a random walk model, which would make resolution of a large dHJ (relative to the step size) potentially inefficient. If the entire heteroduplex were unwound by Blm, the problem of backtracking by the helicase would not exist, and the uncoupled action of topo IIIα will lead to the eventual dissipation of the topological linkages in the heteroduplex region.
FIGURE 2.
Diagrammatic representation of two proposed models for dissolution of dHJs. For simplification, these diagrams show only the migration of the left HJ, whereas the right remains stationary. In addition, representations of the proteins have been omitted. A, unravel and unlink model; B, HJ migration model. RPA, replication protein A.
A similar mode of action has been proposed for RecQ- and topo III-catalyzed segregation of a late replication intermediate, two nearly replicated daughter circles linked through the unreplicated region (27). The late replication intermediate was biochemically prepared by blocking the convergence of the bidirectional replication forks. The RecQ helicase, topo III, and single-stranded DNA-binding protein of Escherichia coli can resolve these interlinked circular molecules and yield two gapped circles. Biochemical analysis of the unlinking reaction suggests that RecQ helicase first unwinds the duplex DNA between the converging replication forks and that topo III then carries out strand passage in the entangled single-stranded region to segregate the conjoined circles. In this process, topo III, in collaboration with RecQ helicase, can reduce DNA linkages by first generating an extensively unwound region before actual strand passage events occur. In other words, the unwinding and unlinking do not act in concert. The mechanistic basis of this reaction is similar to that of the catenation reaction by RecQ/topo III (28). Both reactions, segregation and catenation, are initiated by extensive unwinding of a segment of duplex DNA by RecQ helicase. However, another plausible model exists in which DNA unlinking is not preluded with extensive DNA unwinding.
The second model, which we have simply dubbed “HJ migration,” envisions a much more coordinated and processive mechanism by which a dHJ can be dissolved. In this model, Blm “pushes” a HJ with a bound topo IIIα positioned to perform coordinated strand passage (unlinking) on each heteroduplex (Fig. 2B). Blm might accomplish this HJ migration by coupling its intrinsic helicase and single-stranded DNA annealing activities to split the heteroduplex DNA ahead of the HJ while annealing the duplex behind the HJ (29). The concerted unwinding and rewinding activities would be able to push a HJ along the DNA with a minimum of single-stranded DNA generation. Replication protein A may be incorporated into the complex and help stabilize a limited single-stranded DNA region necessary for topo IIIα catalysis, explaining the specificity of the stimulation of the reaction for replication protein A over the other single-stranded DNA-binding proteins tested (22). Because this model proposes that one protein complex removes the linkages from both heteroduplexes, it would remove the linkages from each heteroduplex at the same pace, which is required for concomitant strand exchange when a HJ migrates.
The actual differences between these two mechanisms would be manifested by distinct characteristics of the in vitro reactions. In contrast to the unravel and unlink model, the enzymatic activities of the helicase and topoisomerase in the HJ migration model are tightly coupled, requiring physical interaction between the two proteins for efficient dHJ dissolution. In addition, the two models presented here differ in the amount of single-stranded DNA generated and are therefore expected to differ in their tolerance for sequence heterologies between the heteroduplexes. In the actual dissolution of a dHJ, the action of these two proposed mechanistic models may not be exclusive of each other. It is possible that topo IIIα/Blm may alternate the use of these two modes of action or use them at different stages of the dissolution reaction.
With these criteria, studies using a substrate consisting of two double-stranded DNA circles conjoined by a dHJ with 165-bp heteroduplexes (linking number of ∼32) indicate that the overall dissolution reaction occurs via the HJ migration model (22). The enzymatically almost identical Drosophila topo IIIβ could not substitute for Drosophila topo IIIα, suggesting that a physical interaction with Drosophila melanogaster Blm is required for the efficient dissolution of this substrate. In addition, this substrate was dissolved only by the unlinking of the homologous regions of the substrate; no reaction products consistent with the unlinking of the heterologous regions were detected, indicating that the reaction is sensitive to sequence heterologies between the two HJs. However, dissolution reactions using an oligonucleotide-based substrate (23) demonstrate that E. coli topo I and topo III can substitute for human topo IIIα, suggesting that species-specific protein-protein interactions may not be a strict requirement for this reaction (30).
It is unclear at this point what biochemical role Rmi1/BLAP75 plays in this reaction. Rmi1/BLAP75 may act to coordinate the helicase and topoisomerase activities, as this protein is known to have a function in organizing the topo IIIα-Blm complex. Both genetic and biochemical evidence showed that Rmi1 in S. cerevisiae stabilizes the topo III-Sgs1 complex (31), yeast homologs of topo IIIα-Blm. BLAP75, the human homolog of Rmi1, was originally identified by its binding affinity for Blm (32), and it can greatly promote the activity of the topo IIIα-Blm complex in dissolution of the oligonucleotide-based dHJ (30, 33, 34). More mechanistic studies will be required to probe the molecular basis of the functional roles for the individual components in this molecular complex.
Conclusions
Although it is sufficient for some topoisomerases to simply eliminate the topological stress that accumulates as a result of DNA metabolism and to bring the DNA to an energetic minimum, there is also an emerging role for the type IA topoisomerases to act upon particular DNA structures and to add or remove topological linkages in a vectorial manner independent of the supercoiling energy stored in the DNA molecules. The type IA topoisomerases capable of such reactions share two common features: their binding to DNA is regulated by helicases or helicase-like domains, and their function is coupled to ATP hydrolysis. At this point, it seems likely that the structural coordination of the topoisomerase (in relation to the DNA) by the helicase domain is what provides these reactions the directionality of strand passage, whereas the coupling of ATP hydrolysis provides an energy source for reactions that are not necessarily driven by the topological energy of the DNA. Despite these common themes, the elucidation of the complex mechanisms utilized by these molecular machines, as well as those yet to be discovered, will be an area of intense interest over the coming years.
Supplementary Material
This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
- topo
- topoisomerase
- AMPPNP
- 5′-adenylyl imidodiphosphate
- dHJ
- double Holliday junction.
REFERENCES
- 1.Schoeffler A. J., Berger J. M. (2008) Q. Rev. Biophys. 41, 41–101 [DOI] [PubMed] [Google Scholar]
- 2.Wang J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 3.Champoux J. J. (2001) Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Lyu Y. L., Wang J. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gangloff S., de Massy B., Arthur L., Rothstein R., Fabre F. (1999) EMBO J. 18, 1701–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwan K. Y., Moens P. B., Wang J. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2526–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Wang J. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1010–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lima C. D., Wang J. C., Mondragón A. (1994) Nature 367, 138–146 [DOI] [PubMed] [Google Scholar]
- 9.Kirkegaard K., Wang J. C. (1985) J. Mol. Biol. 185, 625–637 [DOI] [PubMed] [Google Scholar]
- 10.Plank J. L., Chu S. H., Pohlhaus J. R., Wilson-Sali T., Hsieh T. S. (2005) J. Biol. Chem. 280, 3564–3573 [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi A., Asai K. (1984) Nature 309, 677–681 [DOI] [PubMed] [Google Scholar]
- 12.Confalonieri F., Elie C., Nadal M., de La Tour C., Forterre P., Duguet M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4753–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forterre P. (2002) Trends Genet. 18, 236–237 [DOI] [PubMed] [Google Scholar]
- 14.Atomi H., Matsumi R., Imanaka T. (2004) J. Bacteriol. 186, 4829–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell B. J., Smith J. L., Hanson T. E., Klotz M. G., Stein L. Y., Lee C. K., Wu D., Robinson J. M., Khouri H. M., Eisen J. A., Cary S. C. (2009) PLoS Genet. 5, e1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh T. S., Plank J. L. (2006) J. Biol. Chem. 281, 5640–5647 [DOI] [PubMed] [Google Scholar]
- 17.Kampmann M., Stock D. (2004) Nucleic Acids Res. 32, 3537–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L. F., Wang J. C. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Déclais A. C., Marsault J., Confalonieri F., de La Tour C. B., Duguet M. (2000) J. Biol. Chem. 275, 19498–19504 [DOI] [PubMed] [Google Scholar]
- 20.del Toro Duany Y., Jungblut S. P., Schmidt A. S., Klostermeier D. (2008) Nucleic Acids Res. 36, 5882–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh T. S., Capp C. (2005) J. Biol. Chem. 280, 20467–20475 [DOI] [PubMed] [Google Scholar]
- 22.Plank J. L., Wu J., Hsieh T. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L., Hickson I. D. (2003) Nature 426, 870–874 [DOI] [PubMed] [Google Scholar]
- 24.Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. (1983) Cell 33, 25–35 [DOI] [PubMed] [Google Scholar]
- 25.Ira G., Malkova A., Liberi G., Foiani M., Haber J. E. (2003) Cell 115, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheok C. F., Bachrati C. Z., Chan K. L., Ralf C., Wu L., Hickson I. D. (2005) Biochem. Soc. Trans. 33, 1456–1459 [DOI] [PubMed] [Google Scholar]
- 27.Suski C., Marians K. J. (2008) Mol. Cell 30, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harmon F. G., DiGate R. J., Kowalczykowski S. C. (1999) Mol. Cell 3, 611–620 [DOI] [PubMed] [Google Scholar]
- 29.Cheok C. F., Wu L., Garcia P. L., Janscak P., Hickson I. D. (2005) Nucleic Acids Res. 33, 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L., Bachrati C. Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G. W., Hickson I. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullen J. R., Nallaseth F. S., Lan Y. Q., Slagle C. E., Brill S. J. (2005) Mol. Cell. Biol. 25, 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin J., Sobeck A., Xu C., Meetei A. R., Hoatlin M., Li L., Wang W. (2005) EMBO J. 24, 1465–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bussen W., Raynard S., Busygina V., Singh A. K., Sung P. (2007) J. Biol. Chem. 282, 31484–31492 [DOI] [PubMed] [Google Scholar]
- 34.Raynard S., Zhao W., Bussen W., Lu L., Ding Y. Y., Busygina V., Meetei A. R., Sung P. (2008) J. Biol. Chem. 283, 15701–15708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.