Abstract
Ether-type inositol phospholipids are ubiquitously distributed in Archaea membranes. The present paper describes a novel biosynthetic pathway of the archaeal inositol phospholipid. To study the biosynthesis of archaetidylinositol in vitro, we prepared two possible substrates: CDP-archaeol, which was chemically synthesized, and myo-[14C]inositol 1-phosphate, which was enzymatically prepared from [14C]glucose 6-phosphate with the inositol 1-phosphate (IP) synthase of this organism. The complete structure of the IP synthase reaction product was determined to be 1l-myo-inositol 1-phosphate, based on gas liquid chromatography with a chiral column. When the two substrates were incubated with the Methanothermobacter thermautotrophicus membrane fraction, archaetidylinositol phosphate (AIP) was formed along with a small amount of archaetidylinositol (AI). The two products were identified by fast atom bombardment-mass spectrometry and chemical analyses. AI was formed from AIP by incubation with the membrane fraction, but AIP was not formed from AI. This finding indicates that archaeal AI was synthesized from CDP-archaeol and d-glucose 6-phosphate via myo-inositol 1-phosphate and AIP. Although the relevant enzymes were not isolated, three enzymes are implied: IP synthase, AIP synthase, and AIP phosphatase. AIP synthase was homologous to yeast phosphatidylinositol synthase, and we confirmed AIP synthase activity by cloning the encoding gene (MTH1691) and expressing it in Escherichia coli. AIP synthase is a newly found member of the enzyme superfamily CDP-alcohol phosphatidyltransferase, which includes a wide range of enzymes that attach polar head groups to ester- and ether-type phospholipids of bacterial and archaeal origin. This is the first report of the biosynthesis of ether-type inositol phospholipids in Archaea.
Introduction
The structures and composition of membrane polar lipids of Methanothermobacter thermautotrophicus cells have been reported (1, 2). The major polar lipids of the archaeon are l-serine, ethanolamine, or myo-inositol-containing di- and tetraether-type phospholipids and phosphoglycolipids and gentiobiose-containing di- and tetraether-type glycolipids. The biosynthetic pathways of diether-type serine-phospholipid (archaetidylserine) and diether-type glycolipid (gentiobiosyl archaeol) were previously elucidated in vitro (3, 4). The pathway begins with the formation of the enantiomeric glycerophosphate (sn-glycerol 1-phosphate) from dihydroxyacetone phosphate (5). After two ether bonds are formed on sn-glycerol 1-phosphate with two molecules of geranylgeranyl pyrophosphate (6, 7), the product, 2,3-di-O-geranylgeranyl-sn-glycerol 1-phosphate (or unsaturated archaetidic acid), is activated by CTP to form CDP-archaeol (8). Archaetidylserine is synthesized by replacing the CMP group of CDP-archaeol with l-serine. The enzyme catalyzing this reaction (archaetidylserine synthase) is homologous to bacterial type-II phosphatidylserine synthase. An extensive BLAST search by Daiyasu et al. (9) revealed a great number of homologous gene-encoding enzymes that attach polar head groups of several kinds of phospholipids in Archaea and Bacteria. A possible archaetidylinositol synthase was also included in the homologous group. Koga and Morii (10) hypothesized that the polar head group-attaching enzymes, which belong to an enzyme superfamily, CDP-alcohol phosphatidyltransferase, were derived from a common ancestor enzyme and existed in a common ancestor of Archaea and Bacteria. The only enzyme in Archaea that belongs to the CDP-alcohol phosphatidyltransferase family to have been studied in vitro is archaetidylserine synthase. The hypothesis has not been supported by in vitro experiments of archaeal ether-type inositol lipids, however, because nothing is known about biosynthesis of the lipids.
Many Archaea contain inositol lipids (11, 12). Membrane phospholipids in many Archaea are composed solely of inositol phospholipids. To date, the only known Archaea that do not possess inositol lipids are organisms of Methanococcales, Methanomicrobiales, and Halobacteriales (12). In M. thermautotrophicus, inositol lipids are the most predominant polar lipids (1, 2, 13). Moreover, archaetidylinositol (AI)2 is essential for glycolipid biosynthesis in Archaea (3). Therefore, the in vitro mechanism of AI biosynthesis is one of the most important topics in lipid biosynthesis in Archaea.
CDP-archaeol was expected to be the common precursor of biosynthesis of the phospholipids, analogous to CDP-diacylglycerol in bacteria. Therefore, we investigated the AI synthetic pathway from CDP-archaeol as the starting material.
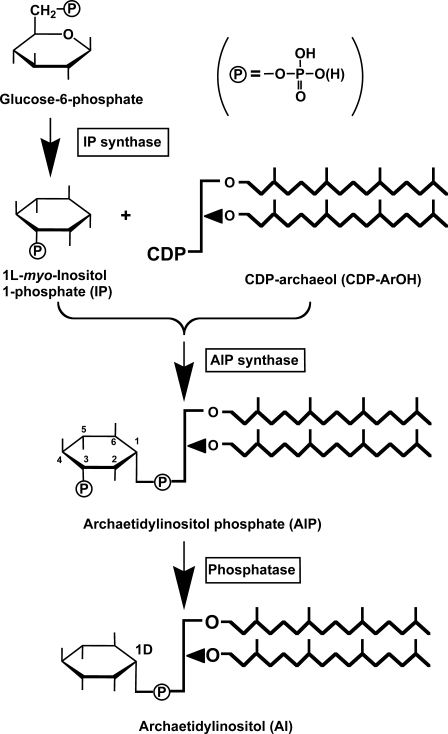
The structure of archaetidyl-myo-inositol (myo is often omitted in this paper) is reported to be an archaeol linked with 1d-myo-inositol 1-phosphate by a phosphodiester linkage (see Fig. 1) (2). This configuration of myo-inositol 1-phosphate is identical to that of yeast or soybean phosphatidyl-myo-inositol (14). Eukaryotic phosphatidylinositol is synthesized from CDP-diacylglycerol and myo-inositol (15), which is generated from d-glucose 6-phosphate (16, 17).
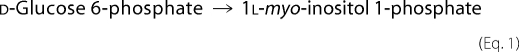 |
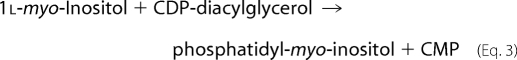 |
Reactions 1 and 3 are catalyzed by 1l-myo-inositol 1-phosphate (IP) synthase and phosphatidylinositol synthase, respectively. A candidate gene (MTH1691) of AI synthase in the M. thermautotrophicus genome was detected by a data base search and is homologous to the yeast phosphatidylinositol synthase gene (9). Therefore, we tried to induce an in vitro AI synthase reaction with CDP-archaeol, myo-inositol, and M. thermautotrophicus cell homogenates under conditions similar to those of the yeast phosphatidylinositol synthase reaction (18). Several trials using modified reaction conditions, however, were unsuccessful. The reaction sequence in the archaeon eventually proceeded after Reaction 1 as follows (Fig. 1),
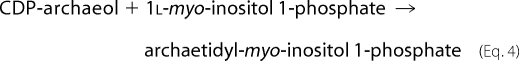 |
 |
FIGURE 1.
Proposed biosynthetic pathway of AI in M. thermautotrophicus.
The natural product of the IP synthase of M. thermautotrophicus was confirmed to be 1l-myo-inositol 1-phosphate. Here, we identified the reaction sequence and the chemical structure of the reaction products. The nomenclature for archaeal ether lipids proposed by Nishihara et al. (19) is used throughout the present paper.
EXPERIMENTAL PROCEDURES
Materials
β-d-Glucose 6-phosphate was purchased from Sigma. [U-14C]Glucose 6-phosphate (3.7 MBq/ml) was obtained from Moravek Biochemicals, Inc. (Brea, CA). 1d-myo-Inositol 1-phosphate and myo-inositol 2-phosphate were purchased from Sigma. A mixture of 1d- and 1l-myo-inositol 1-phosphate was prepared by heating myo-inositol 2-phosphate at 80 °C for 4 h in 1 m HCl (20), a condition at which the phosphoryl group at the 2 position of inositol migrated to the 1 (1l) and 3 (1d) positions. Bicine was obtained from Dojin Laboratories (Kumamoto, Japan). 2,3-Di-O-phytanyl-sn-glycerol (archaeol) was prepared from the total lipid of Halobacterium salinarum as described previously (8); briefly, the polar head groups of H. salinarum total lipids were removed by hydrolysis and purified by preparative thin layer chromatography. CDP-2,3-di-O-phytanyl-sn-glycerol (CDP-archaeol) was chemically synthesized from archaeol as described previously (8). AI was isolated from the total lipid of M. thermautotrophicus by TLC using solvent B (see below).
Growth of Microorganisms
Cells of M. thermautotrophicus (DSM 1053) and H. salinarum (JCM 8981) were grown and harvested at the log phase as described previously (8). Cells of M. thermautotrophicus were washed with buffer A (50 mm phosphate buffer (pH 7) containing 1 mm Na2EDTA and 10 mm 2-mercaptoethanol). The pelleted cell paste was stored at −20 °C until use.
Preparation of Cell-free Homogenates
Frozen cells (∼9 g wet weight) of M. thermautotrophicus were thawed in 10 ml of buffer A (see above) containing 1 mg DNase I (Sigma) and passed through a French pressure cell operated at 1400 kg/cm2. This process was repeated three times. Cell debris and unbroken cells were removed by centrifugation (10,000 × g) for 10 min. The resultant homogenates were centrifuged at 100,000 × g for 2 h to separate the supernatant and membrane fractions. The membrane fraction was washed once and resuspended with the same buffer. The washed membranes and supernatant fraction were saved at −20 °C until use.
Measurement of IP Synthase Activity
myo-Inositol 1-phosphate synthase activity was assayed essentially based on the measurement of inorganic phosphate (Pi) released by periodate oxidation of myo-inositol 1-phosphate, as described by Barnett et al. (21). The complete reaction mixture (final volume, 0.5 ml) contained the supernatant fraction of M. thermautotrophicus homogenates (7–9 mg of protein), 50 mm Bicine buffer (pH 8.5), 0.5 mm d-glucose 6-phosphate, 5 mm NAD, 1 mm dithiothreitol, 1 mm ammonium acetate. After incubation at 60 °C for 2 h, the reaction was stopped by the addition of 0.2 ml of 20% (w/v) trichloroacetic acid. The precipitated protein was removed by centrifugation, and the supernatant was incubated at 37 °C for 1 h with 0.2 m NaIO4. Then 1 m Na2SO3 was added to destroy excess NaIO4, and liberated Pi was measured using the method of Barnett (21). The net Pi value was obtained by subtraction of the blank value of the reaction mixture that received Na2SO3 before the addition of NaIO4. For structure identification of the reaction product, an IP synthase preparation partially purified by ammonium sulfate fractionation (50–70%) was used because low molecular weight compounds in the cell homogenates may interfere with gas liquid chromatography (GLC) analyses of the products.
Identification of the IP Synthase Reaction Product
To obtain the IP synthase reaction product in an amount large enough for structural analysis, the volume of the reaction mixture (containing the ammonium sulfate-fractionated enzyme preparation as described above) was increased 8 times (concentration of the reactants was maintained at the same level), and the incubation time was prolonged to 4 h. After incubation, the reaction mixture was filtered using an Ultrafilter (USY-1 Mr = 10,000, Advantec, Tokyo Roshi Kaisha, Japan) and then dried under a nitrogen stream. The resulting residue was trimethylsilylated with a mixture of pyridine, N,O-bis(trimethylsilyl)trifluoroacetamide, and trimethylchlorosilane (10:2:1 (v/v)) at room temperature for 24 h. This procedure resulted in the formation of completely trimethylsilylated IP, myo-inositol-2,3,4,5,6-penta-O-trimethylsilyl-1-O-di(trimethylsilyl) phosphate (IP(TMS)7). A packed glass column of 3% OV-17 was used to identify the trimethylsilyl derivatives of the reaction products by comparison to the standard IP. The enantiomeric configuration of the IP synthase reaction product was determined on a 25-m capillary column with a chiral liquid phase cp-Chirasil-l-Val (Varian, Palo Alto, CA) after conversion of IP(TMS)7 to myo-inositol-2,3,4,5,6-penta-O-trimethylsilyl-1-O-di(methyl) phosphate (IP(TMS)5Me2) as described by Sherman et al. (22) using trimethylsilyldiazomethane (Tokyo Kasei Kogyo Co., Ltd.) in place of diazomethane. Eluates from the columns were recorded with an electron-impact-mass detector or a flame photometric detector.
Production of [14C]IP
1-l-myo- [14C]Inositol 1-phosphate, which was not commercially available, was enzymatically prepared with the IP synthase of the methanogen from d-[14C]glucose 6-phosphate (148 Bq/nmol) in the same reagent composition as described above, except that a radiolabeled substrate was used. Because IP synthase from the methanogen was rather unstable, ammonium sulfate fractionation was omitted, and the supernatant fraction was used. After incubation for 2 h at 60 °C, the reaction mixture was filtered with an Ultrafilter (USY-1 Mr = 10,000, Advantec) to remove macromolecular compounds. The filtrate contained [14C]inositol 1-phosphate, unreacted [14C]glucose 6-phosphate, and other low molecular weight compounds in the reaction mixture. This filtrate (hereafter referred to as “[14C]inositol 1-phosphate”) was used without further purification as a substrate for the archaetidylinositol phosphate (AIP) synthase reaction.
Measurement of AIP Synthase Activity
The complete assay mixture (final volume, 0.2 ml) contained 50 μl of the “[14C]inositol 1-phosphate” (containing 19 nmol, 2800 Bq), 40 nmol of CDP-archaeol, 0.1 m Bicine buffer (pH 8.0), 4 mm MnCl2, 20 mm MgCl2, 5 mm 2-mercaptoethanol, 0.5% (w/v) Triton X-100, and the membrane fraction (180 μg of protein) of M. thermautotrophicus cell homogenate. In some experiments the homogenate of Escherichia coli pET21a-MTH1691, in which the gene encoding AIP synthase had been expressed, was used as the AIP synthase preparation. CDP-archaeol was dispersed with the aqueous components of the reaction mixture except for Triton X-100, “[14C]inositol 1-phosphate,” and the enzyme preparation in a 1.5-ml microtube by continuous sonication at room temperature for 15 min using a Bransonic 1210 (Branson, Danbury, CT) bath. After the addition of Triton X-100, “[14C]inositol 1-phosphate,” and the enzyme preparation, the reaction mixture was incubated at 60 °C for 30 min. The assay using the membrane fraction of Pyrococcus furiosus only was carried out at 70 °C. The reaction was stopped by adding 1 ml of 0.1 m HCl in methanol, and the mixture was transferred to a 10-ml Teflon-lined, screw-capped glass tube with 1.5 ml of 0.1 m HCl in methanol and 2.5 ml CHCl3. Finally, 2.15 ml of 1 m MgCl2 (pH 2) was added to the mixture to partition the reactants and the products into aqueous and organic layers. After washing twice with 0.1 m HCl, methanol, 1 m MgCl2 (pH 2) (1:0.8, v/v), the organic layer was evaporated to dryness, and radioactivity in the fraction was counted.
Identification of the Products (AIP and AI) of the AIP Synthase Reaction
To obtain the AIP synthase reaction products in an amount large enough for structural analysis, the reaction conditions were modified as follows; the volume of the reaction mixture was increased 40 times (concentration of the reactants was not changed), and the incubation time was prolonged to 13.5 h. The concentration of Triton X-100 was decreased to 0.1%. Each product (AIP and AI) was purified by TLC with solvent B and analyzed by fast atom bombardment (FAB)-mass spectrometry (MS). The purified product was hydrolyzed with 5% HCl, methanol at 100 °C for 2 h. Equal volumes of chloroform and water were added to the reaction mixture, and the two layers were separated. Chloroform-soluble products were analyzed by TLC with solvent A. A portion of the aqueous methanol-soluble product was dried under a stream of nitrogen. The resulting residue was subjected to strong acid hydrolysis with 2 m HCl at 125 °C for 48 h to obtain inositol and then dried under a nitrogen stream. The residue was acetylated in pyridine/acetic anhydride (1:1) at 80 °C for 1 h. The acetylated inositol was identified and quantified by GLC on a 2-m glass column 20% OV-11 using n-hexacosane as an internal standard for quantification.
TLC
TLC of lipids was performed on a Silica Gel 60 plate (Merck) with the following solvents: solvent A, light petroleum (bp 30–70 °C), diethyl ether, and acetic acid (50:50:1); solvent B, chloroform, methanol, acetic acid, and water (80:30:20:10). Phospholipid spots were visualized by spraying acid molybdate reagent. Non-polar lipid spots were detected by charring after spraying 30% (v/v) aqueous H2SO4. Radioactive spots on the TLC plate were recorded by a Fujifilm FLA-5000 fluor-image analyzer with an imaging plate (Fujifilm type BAS-MS, Japan).
Analytical Methods and Physical Measurements
Phosphate (23) and protein (24) were determined as described previously. FAB-MS was recorded using a mass spectrometer (JEOL JMS DX-303) with a matrix of glycerol plus 15-crown-5 in a negative mode. Radioactivity was counted using a liquid scintillation spectrometer (Aloka LSC-3500E) with Aquasol-2 (Packard Instrument Co.) as the scintillator.
Construction of the Expression Plasmid for the MTH1691 Gene
The MTH1691 gene was amplified by polymerase chain reaction directly from M. thermautotrophicus genomic DNA using the oligonucleotides 5′-dGCGCCATATGCCAGATATAAATGAGTCCAT-3′ (the NdeI recognition sequence is underlined) and 5′-dGCGCGGATCCTTATTTCAGCCTTTGCCACA-3′ (the BamHI recognition sequence is underlined) as the forward and reverse primers, respectively. The amplified gene was cloned into the pGEM-T Easy vector (Promega, Boston, MA), and its nucleotide sequence was confirmed. The cloned gene was digested by NdeI-BamHI and inserted into the corresponding sites of the expression vector, pET21a (Novagen). The resultant plasmid was designated pET21a-MTH1691.
Expression of AIP Synthase
To analyze the production of recombinant MTH1691 protein, E. coli BL21-CodonPlus (DE3)-RIL cells (Stratagene, La Jolla, CA) carrying pET21a-MTH1691 were grown at 37 °C in 5 ml of Luria Bertani medium containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol. When culture absorbance at 600 nm was ∼0.5, expression of MTH1691 gene was induced by adding isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mm and continuing the culture for 3 h. After cultivation, the cells were harvested and disrupted by sonication in 500 μl of buffer containing 50 mm Tris-HCl (pH 8.0), 0.5 mm dithiothreitol, 0.1 mm EDTA, and 10% glycerol. The soluble and insoluble materials (membrane fraction) were separated by centrifugation. Each fraction was analyzed by SDS-PAGE. When AIP synthase activity in the recombinant E. coli cells was analyzed, the strain was cultivated in 5 liters of the same medium, and the harvested cells were processed the same as described above except that buffer A was used as the suspending buffer. In some experiments the suspending buffer was replaced with 0.1 m Bicine buffer (pH 8.0) containing 10 mm 2-mercaptoethanol using a HiTrap desalting column (5 ml; GE Healthcare).
RESULTS
Because eukaryotic phosphatidylinositol is synthesized from CDP-diacylglycerol and myo-inositol, an analogous reaction was first attempted; that is, CDP-archaeol (a diether analog of CDP-diacylglycerol) was incubated with myo-inositol in the presence of M. thermautotrophicus cell homogenates. Several attempts under various reaction conditions, however, were all unsuccessful. Therefore, we attempted to use other substrates for the reaction. First, we used myo-inositol 1-phosphate instead of myo-inositol.
Activity of IP Synthase
Because radioactively labeled IP is not commercially available, we synthesized it by IP synthase. The activity of the enzyme was detected in the supernatant fraction of M. thermautotrophicus homogenates, although this enzyme had not been studied in this organism. Because the enzyme was rather unstable, the supernatant fraction of M. thermautotrophicus cell homogenates was used for the synthesis of inositol 1-phosphate without purification, except for when determining the structure of the product. The apparent specific activity of this crude enzyme was 18 nmol h−1 mg of protein−1. Thus, it was sufficiently active to synthesize myo-inositol 1-phosphate for the study of AIP synthesis.
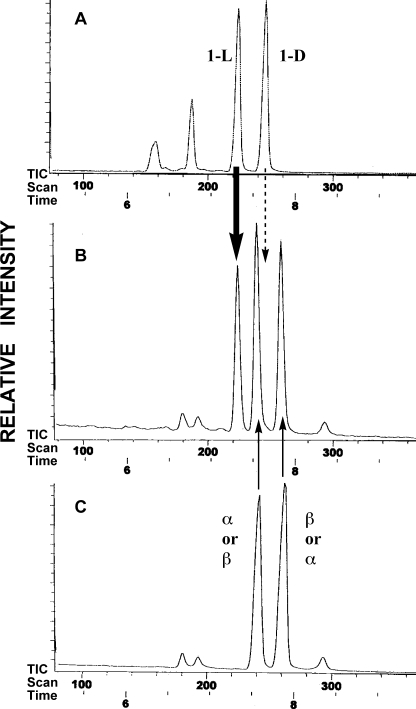
The structure of myo-inositol 1-phosphate prepared with this enzyme was confirmed to be IP(TMS)7 by GLC after trimethylsilylation (data not shown). Furthermore, the absolute stereochemical configuration of myo-inositol 1-phosphate was determined by GLC of IP(TMS)5Me2 using a chiral capillary column. The main peak in the GLC profile of the reaction mixture coincided with authentic 1l-myo-inositol 1-phosphate (Fig. 2B). Two other major peaks in the reaction mixture represented unreacted α- and β-glucose 6-phosphate (we did not determine which was α or β; Fig. 2). In addition, electron impact-mass spectrometry of the first main peak gave a signal of m/z 633 (M-CH3)+, which is consistent with the molecular weight of IP(TMS)5Me2. The second and third main peaks were confirmed to be glucose 6-phosphate(TMS)4Me2 by electron impact-mass spectrometry (m/z 561 (M-CH3)+). Thus, the product of the IP synthase reaction was decisively determined to be 1l-myo-inositol 1-phosphate. The unfractionated “[14C]inositol 1-phosphate” also included α- and β-[14C]glucose 6-phosphate. The configuration of 1l-myo-inositol 1-phosphate is identical to that of yeast (25), an animal (26), and a higher plant (27). This configuration of myo-inositol 1-phosphate may form archaetidylinositol with the natural configuration by reacting with CDP-archaeol because the hydroxyl group at the 3 (1d) position remains free to react with the phosphate group of CDP-archaeol (Fig. 1). Therefore, “[14C]inositol 1-phosphate” was used as the substrate for the AIP synthase reaction.
FIGURE 2.
Gas liquid chromatograms of trimethylsilyl-methyl derivatives of IP synthase reaction products on a cp-Chirasil-l-Val capillary column. Shown are standard mixtures of 1d- and 1l-myo-inositol 1-phosphate (A), IP synthase reaction products (B), and a IP synthase reaction mixture without M. thermautotrophicus homogenates (C).
Activity of AIP Synthase
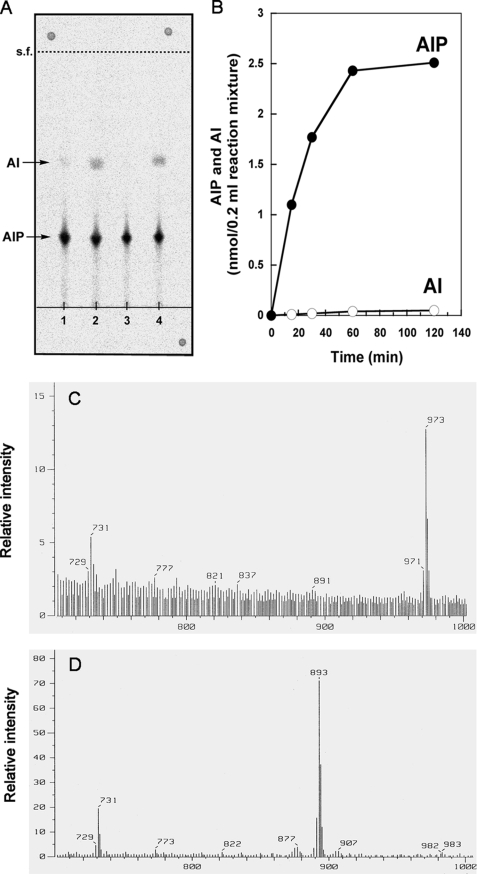
When CDP-archaeol was incubated with “[14C]inositol 1-phosphate” in the presence of the membrane fraction of M. thermautotrophicus, a significant amount of radioactivity was incorporated into chloroform-soluble materials. The apparent specific activity of this crude enzyme was 20 nmol h−1 mg of protein−1. The activity was completely dependent on the presence of CDP-archaeol and “myo-inositol 1-phosphate” and required the addition of either Mg2+ or Mn2+ ions (Table 1). When one of the two kinds of ions was added, partial AIP synthase activity was recovered. Full activity required both ions. One major spot and one minor spot were detected by TLC of the chloroform-soluble products with solvent B (Fig. 3). The major spot (Rf = 0.27) migrated slightly slower than standard diglucosyl caldarchaetidylinositol. The other minor radioactive spot comigrated with standard AI (Rf = 0.56). The two products were identified as AIP and AI by FAB-MS and chemical analysis (see below). The time course of AIP and AI synthesis from CDP-archaeol is shown in Fig. 3. Six percent of CDP-archaeol and 13% of [14C]inositol-1-phosphate were converted to the products (AIP + AI) during a 1-h reaction. AIP synthase activity was greatly stimulated by 0.5% Triton X-100. At this concentration most products were AIP, whereas more AI was formed when the concentration of the detergent was decreased to 0.1% (Fig. 3).
TABLE 1.
Incorporation of “[14C]inositol 1-phosphate” into lipid (AIP and AI) under various AIP synthase reaction conditions
The reaction mixture of the AIP synthase reaction is described under “Experimental Procedures.” After incubation at 60 °C for 30 min, radioactivity in the chloroform-soluble materials was counted with the exception that the assay using the P. furiosusmembrane was carried out at 70 °C.
| “[14C]Inositol 1-phosphate” | Source of enzyme | Other constituents in the reaction mixturea | Relative incorporation |
|---|---|---|---|
| % | |||
| +b | M. thermautotrophicus membranec | Complete | 100 |
| + | −CDP-ArOH | 4 | |
| + | −Mg2+, Mn2+ | 2 | |
| + | −Mg2+ | 91 | |
| + | −Mn2+ | 52 | |
| + | −Triton X-100 | 6 | |
| + | P. furiosusmembraned | Complete | 145 |
| + | None | Complete | 1 |
| + | E. coli pET21a-MTH1691e | Complete | 281 |
| + | E. coli pET21af | Complete | 1 |
| −g | M. thermautotrophicus. membranec | Complete | 1 |
a Complete mixture other than the enzyme preparation and “[14C]inositol 1-phosphate” contained 40 nmol of CDP-archaeol, 0.1 m Bicine buffer (pH 8.0), 4 mm MnCl2, 20 mm MgCl2, 5 mm 2-mercaptoethanol, and 0.5% (w/v) Triton X-100.
b [14C]Inositol 1-phosphate was synthesized from [14C]glucose 6-phosphate in a complete IP synthase reaction mixture with the supernatant fraction of the M. thermautotrophicus homogenate.
c The membrane fraction of M. thermautotrophicus homogenates (180 μg of protein/assay).
d The membrane fraction of P. furiosus homogenates (180 μg of protein/assay).
e Homogenate of E. coli pET21a-MTH1691 (180 μg of protein/assay).
f Homogenate of E. coli pET21a (180 μg of protein/assay).
g The IP synthase reaction mixture was incubated without the addition of the M. thermautotrophicussupernatant fraction. The solution included only [14C]glucose 6-phosphate as radiolabeled material.
FIGURE 3.
Analysis of the reaction products of AIP synthase. A, shown is a thin-layer chromatogram of 14C-labeled products of the AIP synthase reaction. The source of AIP synthase preparation was the membrane fraction of M. thermautotrophicus cell homogenates (lanes 1 and 2) or homogenates of E. coli pET21a-MTH1691 (lanes 3 and 4) in the presence of 0.5% Triton X-100 (lanes 1 and 3) or 0.1% Triton X-100 (lanes 2 and 4). The products were extracted and developed on TLC. Radioactive spots were detected by autoradiography. s.f., solvent front. B, shown is time-course of AIP synthase reaction. AIP and AI were formed from “[14C]inositol 1-phosphate” and CDP-archaeol catalyzed by the membrane fraction of M. thermautotrophicus cell homogenates. The reaction was stopped at the indicated time points by the addition of HCl, chloroform-soluble products were separated by TLC, and radioactivity of each spot was counted. The ratio of AIP and AI was determined by autoradiography after TLC development of the chloroform-soluble products. C and D, shown are negative ion FAB-mass spectra of AIP (C) and AI (D) enzymatically synthesized from CDP-archaeol and “myo-inositol 1-phosphate” (non-radiolabeled).
AIP synthase activity was detected not only in M. thermautotrophicus but also in the membrane fraction of another hyperthermophilic archaeon P. furiosus. The apparent specific activity of the latter organism was higher when measured at 70 °C than that of the former (Table 1).
Identification of the AIP Synthase Reaction Products
The lipid products of the AIP synthase reaction from CDP-archaeol and non-radiolabeled “inositol 1-phosphate” were purified by TLC with solvent B. The FAB-MS of the major lipid product gave signals of m/z 973 (M-H)− and 731 (archaetidic acid) (Fig. 3), which is consistent with the molecular weight and structure of AI + HPO3 (AIP; Fig. 1). The minor lipid product showed signals of m/z 893 (M-H)− and 731 on FAB-MS, identical to AI (28). The two products were completely hydrolyzed by 5% HCl, methanol at 100 °C for 2 h without forming any chloroform-soluble, phosphate-containing material. Chloroform-soluble materials of the both hydrolysates coincided with authentic archaeol on TLC with solvent A (data not shown). Strong acid hydrolysis of water-soluble methanolysate resulted in free inositol. It was proven to be myo-inositol on GLC of TMS derivatives, and the amounts were quantitatively measured. The ratio of phosphate to inositol in AIP and AI were 2.3: 1 (n = 3) and 1.3: 1 (n = 2), respectively. These results indicate that the major slower moving lipid has the structure of diphytanyl-glycerophospho-myo-inositol phosphate (AIP), and the minor faster migrating lipid is diphytanyl-glycerophospho-myo-inositol (AI) (Fig. 1).
Cloning and Expression of an AIP Synthase Gene in E. coli
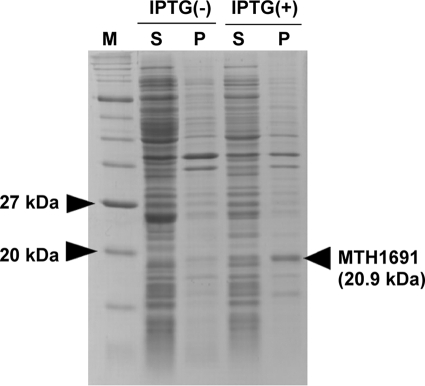
Daiyasu et al. (9) predicted the MTH1691 gene as a candidate for archaetidylinositol synthase of M. thermautotrophicus by BLAST search. The amino acid sequence of the MTH1691 gene product is homologous to that of the phosphatidylinositol synthase in Saccharomyces cerevisiae (supplemental Fig. S1). We, therefore, cloned and expressed the MTH1691 gene in E. coli cells. SDS-PAGE of the homogenates of the expressed E. coli cells clearly showed a new dense band corresponding to a molecular mass 20.9 kDa (Fig. 4), which coincided with the molecular weight (20,764) of the MTH1691 gene product calculated from the deduced amino acid sequence (29). However, our trial to detect AI synthase activity in the E. coli cell homogenates, in which the MTH1691 gene was expressed, failed. The E. coli-containing pET21a-MTH1691 cell homogenates did display considerably higher AIP synthase activity than the M. thermautotrophicus membrane. E. coli pET21a cell homogenates carrying an empty vector possessed little activity for the AIP synthesis (Table 1). Therefore, we concluded that MTH1691 is a gene that encodes AIP synthase but not AI synthase, and AIP synthesis from CDP-archaeol and myo-inositol 1-phosphate is catalyzed by a single enzyme. AIP synthase encoded by the MTH1691 gene is composed of 195 amino acid residues and has a molecular weight of 20,764. Homologous genes from several archaeal species P. furiosus, Sulfolobus solfataricus, and Aeropyrum pernix, a bacterium, Mycobacterium tuberculosis, and a eukaryote, S. cerevisiae, were aligned (supplemental Fig. S1). A typical sequence of a CDP-alcohol phosphatidyltransferase motif was found by comparing to the motif proposed by Williams and McMaster (DGXXARXXXXXXXXGXXXDXXXD) (30). A phylogenic tree of the enzymes of the CDP-alcohol phosphatidyltransferase family has been published (9). These genes belong to the CDP-alcohol phosphatidyltransferase family along with archaetidylserine synthase and phosphatidylserine synthase.
FIGURE 4.
Production of the MTH1691 gene product in E. coli. Soluble (S) and insoluble (P) fractions of E. coli cells in which the MTH1691gene was expressed were subjected to 15% SDS-PAGE, and the proteins were stained by Coomassie Brilliant Blue. The lanes shown as IPTG(−) include cell extracts from the culture without the addition of IPTG as a negative control of the gene expression. Lane M contains size marker proteins (Protein Marker, Broad range, New England Biolabs).
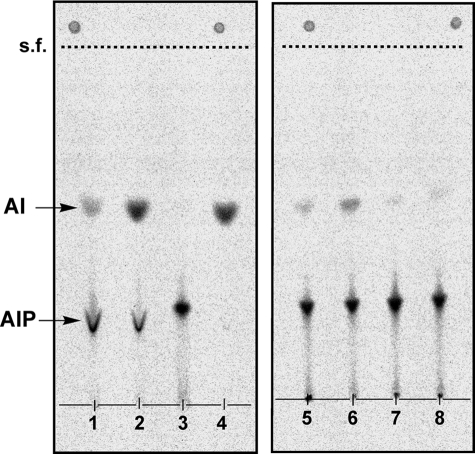
Formation of AI from AIP (AIP Phosphatase Activity)
To determine the reaction sequence of the synthesis of AIP and AI, [14C]AIP and [14C]AI were synthesized from CDP-archaeol and “[14C]inositol 1-phosphate” for 4 h under conditions similar to the AIP synthase reaction, except that the concentration of Triton X-100 was decreased to 0.1%, a concentration of Triton X-100 at which more AI was produced compared with that at 0.5% Triton X-100. The products were isolated by TLC. The purified [14C]AIP was converted to [14C]AI by the actions of the membrane fraction of M. thermautotrophicus cell homogenate but not by the supernatant fraction (Fig. 5). On the other hand, [14C]AIP was not formed from [14C]AI (Fig. 5, lane 4). The AIP phosphatase activity was localized in the membrane fraction of M. thermautotrophicus cells and inhibited by 8–20 mm phosphate buffer (Fig. 5). Although it was expected that the use of E. coli homogenate containing the recombinant AIP synthase instead of M. thermautotrophicus membranes in the same experiments would prevent the conversion of AIP to AI because the cells contained only AIP synthase as a methanogen protein, a small amount of AI was nevertheless also formed in this case.
FIGURE 5.
Conversion of AIP to AI by the membrane fraction of M. thermautotrophicus cell homogenates or homogenates of E. coli pET21a-MTH1691. [14C]AIP and [14C]AI were prepared from CDP-archaeol and “[14C]inositol 1-phosphate” under the conditions described under “Experimental Procedures.” The purified [14C]AIP (lanes 1, 2, 3, 5, 6, 7, and 8) or [14C]AI (lane 4) was incubated at 60 °C for 2 h with either enzyme preparation (500 μg protein/30–80 μl), the membranes of M. thermautotrophicus homogenate (lanes 1, 2, and 4), the supernatant fraction of M. thermautotrophicus homogenates (lane 3), the homogenates of E. coli pET21a-MTH1691 (lanes 5 and 6), or the homogenates of E. coli pET21a carrying an empty vector plasmid (lanes 7 and 8) in the presence of 0.1% Triton X-100. Other constituents in the reaction mixture were the same as in the AIP synthase reaction. The enzyme preparation was dissolved in 50 mm phosphate buffer (lanes 1, 4, 5, and 7), in aqueous dithiothreitol solution (lanes 2 and 3), and in Bicine buffer (lanes 6 and 8). After the reaction, lipids were extracted, separated by TLC, and recorded by autoradiography. s.f., solvent front.
DISCUSSION
An inositol-containing ether-type phospholipid in Archaea was synthesized from CDP-archaeol and 1l-myo-inositol 1-phosphate via AIP, which was dephosphorylated to the final product, AI (Fig. 1). Three enzyme activities, IP synthase, AIP synthase, and a possible phospholipid-phosphate phosphatase, were detected at the crude homogenate level. This pathway was also confirmed by identification of the chemical structures of the intermediates. This is a new biosynthetic pathway of inositol-phospholipid, either the ester type or ether-type phospholipid. This reaction sequence of attachment of the archaetidyl group and dephosphorylation is the reverse compared with eukaryotic inositol phospholipid synthesis.
Because this pathway, characterized by the presence of AIP as the essential intermediate, was also synthesized by the membrane fraction of the other extremely thermophilic archaeon P. furiosus, and homologous genes encoding AIP synthase were found in two other hyperthermophilic Crenarchaeota, S. solfataricus and A. pernix, the pathway is not unique to M. thermautotrophicus but rather seems to be generally functional in Archaea.
This reaction sequence of AI synthesis and the stereo configuration of the IP synthase product are compatible with construction of AI with the natural configuration of 1d-myo-inositol 1-phosphate (Fig. 1). Incidentally, the compatible solute of Archaeoglobus fulgidus (di-myo-inositol phosphate) is synthesized from 1l-myo-inositol 1-phosphate (31).
AIP is an important intermediate in this new pathway (see above). The structure was identified as archaetidylinositol phosphate by FAB-MS and chemical analyses. Although the exact position of the inositol where the distal phosphate was bound has not been experimentally determined, the phosphoryl group is the most likely to be located at the position shown in Fig. 1. In animal tissues phosphatidylinositol 4,5-bisphosphate generates a powerful second messenger, inositol 1,4,5-trisphosphate. The distal phosphate group of AIP is linked at a position different from that phosphatidylinositol 4,5-bisphosphate.
Bachhawat and Mande (32) showed that Archaea (M. thermautotrophicus etc.) contains an IP synthase gene (MTH1105) that has significant homology to the eukaryotic INO1 gene. MTH1105 is most likely an IP synthase structural gene. Because IP synthase activity was detected at the enzyme level in this work, the MTH1105 gene is more likely to be the gene encoding IP synthase, although the gene has not been isolated and the sequence of the enzyme has not been determined. There is a significant difference, however, in the length of the enzymes; the monomer polypeptide of the archaeal IP synthase deduced from the gene consists of 350–420 residues, whereas the eukaryotic enzyme comprises more than 550 residues. Nevertheless, the reaction catalyzed by the two groups of enzymes is the same. Although IP synthase has been studied in A. fulgidis as a member of a new osmolyte metabolism (33), this is the first report on the enzymatic analysis of IP synthase and the structural determination of IP as the enzyme reaction product in the methanoarchaeon.
AIP synthase is a new member of the CDP-alcohol phosphatidyltransferase superfamily. This superfamily includes enzymes that catalyze the transfer of phosphatidyl (archaetidyl) groups to an alcohol to form phosphatidyl (archaetidyl)-alcohol, in which the CDP-bound substrates do not depend on ester/ether linkages, the stereochemical glycerophosphate backbone structure, or straight chain fatty acid/highly methyl-branched isoprenoid chains. A broad group of enzymes with activity toward such a variety of substrates suggests that the enzyme existed in a common ancestor before the differentiation of Archaea and bacteria and that diverse enzymes of phospholipid synthesis evolved from the ancestral enzyme.
At the last step of AI synthase, AI is produced from AIP by a phosphatase. Because the homogenate of E. coli pET21a-MTH1691, in which only AIP synthase was expressed, showed AIP dephosphorylation activity, the possibility that AIP synthase also has AIP phosphatase activity cannot be excluded. Because the homogenate of E. coli pET21a carrying an empty vector also exhibited weak dephosphorylation activity, some phosphatases in E. coli cells might act to dephosphorylate AIP; for example, phosphatidyl glycerophosphate phosphatase. In fact, an M. thermautotrophicus gene, MTH798, is homologous to the E. coli pgpB gene that encodes a protein belonging to a phospholipid-phosphate phosphatase group. AIP dephosphorylation remains to be clarified. A modified pathway of lipid biosynthesis in Archaea, such as inversion of the transfer reaction and the dephosphorylation in AI synthesis, can be observed in another example, a modified mevalonate pathway for isopentenyl pyrophosphate synthesis found in Methanocaldococcus jannaschii (34).
The new pathway is similar to that of phosphatidylglycerol synthesis of E. coli. Phosphatidylglycerol of E. coli is synthesized from CDP-diacylglycerol by two steps (35). Phosphatidyl glycerophosphate is first synthesized by the transfer of a phosphatidyl group from CDP-diacylglycerol to sn-glycerol-3-phosphate. Phosphatidyl glycerophosphate is then rapidly dephosphorylated to produce phosphatidylglycerol. Transfer of a phosphatidyl group to a phosphorylated polyol followed by dephosphorylation markedly resembled archaeal inositol phospholipid biosynthesis as described in this paper. This feature is not only observed in E. coli but is also seen in archaetidylglycerol synthesis in Halobacterium (36). Three types of phospholipids (ester or ether type) with ethanolamine, glycerol, and myo-inositol as polar head groups are all synthesized from serine, glycerophosphate, and inositol 1-phosphate phospholipids by the removal of small acidic groups (COOH or phosphate groups). These might be vestiges of ancient membrane phospholipid metabolism in a surface metabolism where a strong acidic group of phospholipids firmly bound to a pyrite surface, according to the surface metabolism hypothesis (37). Once the acidic group is removed, the phospholipids are likely to be released from the surface and might form phospholipid bilayer membranes.
Supplementary Material
Acknowledgments
We are grateful to Professor K. Matsumoto (Saitama University) and to Dr. H. Daiyasu (Osaka University) for kindness in supplying valuable information of genes of bacterial phospholipid biosynthesis.
This work was supported in part by Grant-in-aid for Scientific Research (KAKENHI) C16580067 from the Japan Society for the Promotion of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- AI
- archaetidylinositol
- AIP
- AI phosphate
- GLC
- gas liquid chromatography
- IP
- 1l-myo-inositol 1-phosphate
- IP(TMS)7
- myo-inositol-2,3,4,5,6-penta-O-trimethylsilyl-1-O-di(trimethylsilyl) phosphate
- FAB
- fast atom bombardment
- MS
- mass spectrometry
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1.Nishihara M., Koga Y. (1987) J. Biochem. 101, 997–1005 [DOI] [PubMed] [Google Scholar]
- 2.Nishihara M., Morii H., Koga Y. (1989) Biochemistry 28, 95–102 [Google Scholar]
- 3.Morii H., Eguchi T., Koga Y. (2007) J. Bacteriol. 189, 4053–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morii H., Koga Y. (2003) J. Bacteriol. 185, 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishihara M., Koga Y. (1995) J. Biochem. 117, 933–935 [DOI] [PubMed] [Google Scholar]
- 6.Zhang D. L., Daniels L., Poulter C. D. (1990) J. Am. Chem. Soc. 112, 1264–1265 [Google Scholar]
- 7.Zhang D. L., Poulter C. D. (1993) J. Am. Chem. Soc. 115, 1270–1277 [Google Scholar]
- 8.Morii H., Nishihara M., Koga Y. (2000) J. Biol. Chem. 275, 36568–36574 [DOI] [PubMed] [Google Scholar]
- 9.Daiyasu H., Kuma K., Yokoi T., Morii H., Koga Y., Toh H. (2005) Archaea 1, 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koga Y., Morii H. (2007) Microbiol. Mol. Biol. Rev. 71, 97–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koga Y., Akagawa-Matsushita M., Ohga M., Nishihara M. (1993) Syst. Appl. Microbiol. 16, 342–351 [Google Scholar]
- 12.Koga Y., Nakano M. (2008) Syst. Appl. Microbiol. 31, 169–182 [DOI] [PubMed] [Google Scholar]
- 13.Morii H., Koga Y. (1993) FEMS Microbiol. Lett. 109, 283–288 [Google Scholar]
- 14.Ballou C. E., Pizer L. I. (1960) J. Am. Chem. Soc. 82, 3333–3335 [Google Scholar]
- 15.Steiner M. R., Lester R. L. (1972) Biochim. Biophys. Acta 260, 222–243 [DOI] [PubMed] [Google Scholar]
- 16.Chen I. W., Charalampous F. C. (1965) Biochem. Biophys. Res. Commun. 19, 144–149 [DOI] [PubMed] [Google Scholar]
- 17.Eisenberg F., Jr. (1967) J. Biol. Chem. 242, 1375–1382 [PubMed] [Google Scholar]
- 18.Carman G. M., Fischl A. S. (1992) Methods Enzymol. 209, 305–312 [DOI] [PubMed] [Google Scholar]
- 19.Nishihara M., Morii H., Koga Y. (1987) J. Biochem. 101, 1007–1015 [DOI] [PubMed] [Google Scholar]
- 20.Pizer F. L., Ballou C. E. (1959) J. Am. Chem. Soc. 81, 915–921 [Google Scholar]
- 21.Barnett J. E., Brice R. E., Corina D. L. (1970) Biochem. J. 119, 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman W. R., Leavitt A. L., Honchar M. P., Hallcher L. M., Phillips B. E. (1981) J. Neurochem. 36, 1947–1951 [DOI] [PubMed] [Google Scholar]
- 23.Bartlett G. R. (1959) J. Biol. Chem. 234, 466–468 [PubMed] [Google Scholar]
- 24.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 25.Chen I. W., Charalampous F. C. (1966) Arch. Biochem. Biophys. 117, 154–157 [Google Scholar]
- 26.Sherman W. R., Stewart M. A., Zinbo M. (1969) J. Biol. Chem. 244, 5703–5708 [PubMed] [Google Scholar]
- 27.Loewus M. W., Loewus F. A., Brillinger G. U., Otsuka H., Floss H. G. (1980) J. Biol. Chem. 255, 11710–11712 [PubMed] [Google Scholar]
- 28.Morii H., Nishihara M., Koga Y. (1988) Agric. Biol. Chem. 52, 3149–3156 [Google Scholar]
- 29.Smith D. R., Doucette-Stamm L. A., Deloughery C., Lee H., Dubois J., Aldredge T., Bashirzadeh R., Blakely D., Cook R., Gilbert K., Harrison D., Hoang L., Keagle P., Lumm W., Pothier B., Qiu D., Spadafora R., Vicaire R., Wang Y., Wierzbowski J., Gibson R., Jiwani N., Caruso A., Bush D., Safer H., Patwell D., Prabhakar S., McDougall S., Shimer G., Goyal A., Pietrokovski S., Church G. M., Daniels C. J., Mao J. I., Rice P., Nolling J., Reeve J. N. (1997) J. Bacteriol. 179, 7135–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams J. G., McMaster C. R. (1998) J. Biol. Chem. 273, 13482–13487 [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues M. V., Borges N., Henriques M., Lamosa P., Ventura R., Fernandes C., Empadinhas N., Maycock C., da Costa M. S., Santos H. (2007) J. Bacteriol. 189, 5405–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachhawat N., Mande S. C. (2000) Trends Genet. 16, 111–113 [DOI] [PubMed] [Google Scholar]
- 33.Neelon K., Wang Y., Stec B., Roberts M. F. (2005) J. Biol. Chem. 280, 11475–11482 [DOI] [PubMed] [Google Scholar]
- 34.Grochowski L. L., Xu H., White R. H. (2006) J. Bacteriol. 188, 2836–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanfer J., Kennedy E. P. (1964) J. Biol. Chem. 239, 1720–1726 [PubMed] [Google Scholar]
- 36.Moldoveanu N., Kates M. (1988) Biochim. Biophys. Acta 960, 164–182 [Google Scholar]
- 37.Wächtershäuser G. (1988) Microbiol. Rev. 52, 452–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.