Abstract
Members of the RecQ family of proteins are highly conserved DNA helicases that have important functions in the maintenance of genomic stability. Deficiencies in RecQ4 have been linked to human diseases including Rothmund-Thomson, RAPADILINO, and Baller-Gerold syndromes, all of which are characterized by developmental defects, tumor propensity, and genetic instability. However, there are conflicting results shown in the literature regarding the DNA helicase activity of RecQ4. We report here the expression of Drosophila melanogaster RecQ4 with a baculoviral vector and its purification to near homogeneity. The purified protein has a DNA-dependent ATPase activity and is a 3′-5′ DNA helicase dependent on hydrolysis of ATP. The presence of 5′-adenylyl-β,γ-imidodiphosphate (AMPPNP), a nonhydrolyzable ATP analog, promotes stable complex formation between RecQ4 and single-stranded DNA. Drosophila RecQ4 can also anneal complementary single strands; this activity was reduced in the presence of AMPPNP, possibly because of the stable protein-DNA complex formed under such conditions. A point mutation of the highly conserved lysine residue in the helicase domain, although retaining the wild type level of annealing activity, inactivated ATPase and helicase activities and eliminated stable complex formation. These results suggest that the helicase domain alone is responsible for the DNA unwinding action of the Drosophila enzyme. We generated a null recq4 mutant that is homozygous lethal, which we used to test the genetic function of the helicase-dead mutant in flies. Complementation tests showed that the helicase-dead mutant recq4 transgenes are incapable of rescuing the null mutation, demonstrating that the helicase activity has an essential biological function.
Introduction
DNA helicases are critical to almost all aspects of DNA metabolism. These enzymes unwind double-stranded DNA using the energy derived from the hydrolysis of ATP or other nucleotide triphosphates. The RecQ family of helicases is of particular interest, as its members not only unwind DNA but also anneal complementary DNA strands (reviewed in Refs. 1–3). The combination of these two activities can be employed to form and resolve DNA structures that are involved in replication, repair, and recombination. Mutations in this highly conserved family are characterized by increased genetic instability. Three of the five RecQ helicases (Blm, Wrn, and RecQ4) in humans are associated with clinical syndromes. Bloom syndrome, caused by deficiencies in Blm, is defined by abnormally high levels of sister chromatid exchange leading to an increased occurrence of cancer. Mutations in Wrn lead to Werner syndrome, a severe form of progeria. Defects in RecQ4 are responsible for three different syndromes with a diverse set of symptoms. The remaining human RecQ helicases, RecQ1 and RecQ5, are not associated with any diseases but are also implicated in suppressing inappropriate sister chromatid exchange, perhaps serving as backups to Blm (4).
The least characterized of the five human RecQ helicases is RecQ4. Discovered in 1998 (5), RecQ4 is now known to be associated with Rothmund-Thomson syndrome, RAPADILINO, and Baller-Gerold syndrome (reviewed in Refs. 6–8). There is significant overlap between the symptoms of these three syndromes, with patients exhibiting some combination of the following: poikiloderma, cataracts, sparse hair, radial ray hypoplasia, gastrointestinal defects, growth retardation, craniosynostosis, palatal abnormalities, joint dislocation, and a higher risk of osteosarcoma and lymphoma. It has been difficult to establish correlations between individual mutations and specific disease symptoms. Nonetheless, certain trends can be observed. Many of the mutations found in Rothmund-Thomson syndrome, RAPADILINO, and Baller-Gerold syndrome patients with defects in the RecQ4 gene occur in the helicase domain, often truncating it or excluding it entirely (6). Mouse mutants with partial or complete deletions of the helicase domain recapitulate to varying degrees the symptoms of Rothmund-Thomson syndrome and RAPADILINO patients, including small stature, skin abnormalities, radial ray defects, and cancer predisposition (9, 10). Thus it is possible that defects within the helicase domain may be responsible for many of the phenotypes found in RecQ4-related syndromes.
The cellular functions of RecQ4 remain to be elucidated. In Drosophila, the enzyme plays an essential role in replication, where homozygous null mutants do not survive past the first instar because of severe defects in replication, and a hypomorphic mutant is defective in chorion gene amplification at the stage of replication initiation (11). The N-terminal 200 residues are homologous to Sld2 in Saccharomyces cerevisiae; this domain is necessary for in vitro replication initiation in Xenopus egg extracts (12, 13). RecQ4 has been reported in the nucleus (14, 15), in both the nucleus and cytoplasm (16–18), and primarily in the cytoplasm (19). This apparent discrepancy may be because of the cell-specific or cell cycle-dependent differential localization of RecQ4. In mitotic cells during early Drosophila embryogenesis, RecQ4 is localized to the nucleus in interphase, dispersed to the cytoplasm during prophase after nuclear envelope breakdown, and then reimported to the nucleus in telophase (11). Recent results showed that post-translational modification such as acetylation can promote the cytoplasmic localization of RecQ4 (20). The enzyme has also been observed to relocate from the cytoplasm to nuclear foci in response to hydrogen peroxide treatment, and Rothmund-Thomson syndrome-derived fibroblasts fail to recover after induction of cell growth arrest (19). RecQ4 is also known to participate in the repair of double strand breaks and in recovery after treatment with UV light or hydroxyurea (15, 17, 21). Taken together, these results suggest that RecQ4 is critically involved in many aspects of DNA repair and replication.
There have been conflicting reports concerning the activity of the RecQ4 helicase domain. Both Yin et al. (16) and Macris et al. (22) have demonstrated DNA-dependent hydrolysis of ATP by human RecQ4. Neither group detected helicase activity, although Macris et al. (22) did observe single strand annealing. Recently, Xu and Liu (23) observed that RecQ4 simultaneously unwinds and anneals DNA but found this activity to be derived mostly from the Sld2 domain rather than the helicase domain. Furthermore, these activities may be better described as DNA strand exchange. Robust helicase activity in the RecQ4 holoenzyme has yet to be established.
In this work, we demonstrate that Drosophila RecQ4 is a 3′-5′ helicase. We also demonstrate single-stranded annealing and DNA-stimulated ATPase activities in RecQ4. We have generated and purified a helicase-dead mutant protein, which establishes that the DNA unwinding activity is a result of the RecQ4 helicase domain. In vivo, this mutation is unable to rescue viability for null mutant flies, proving that the helicase activity is important to RecQ4 biological activity. This work constitutes the first clear demonstration of helicase activity for RecQ4 from any species.
EXPERIMENTAL PROCEDURES
Proteins and DNA
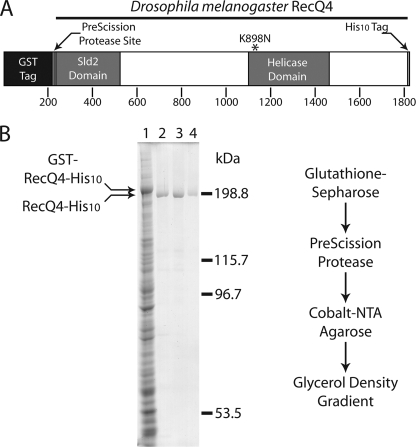
RecQ4 was expressed using the Bac-to-Bac baculovirus expression system (Invitrogen). Drosophila RecQ4 cDNA was cloned into the pFastbac 1 vector with an N-terminal glutathione S-transferase (GST)2 tag (separated from the enzyme by a PreScission Protease recognition site) and a C-terminal tag consisting of 10 histidines (His10). A diagrammatic representation of the recombinant RecQ4 construct is shown in Fig. 1A. The RecQ4-K898N mutant was generated by site-directed mutagenesis of the pFastbac 1 recombinant vector using the Stratagene QuikChange Lightning system.
FIGURE 1.
A, schematic diagram of Drosophila melanogaster RecQ4 construct. An N-terminal GST tag is attached to the enzyme by a PreScission Protease site and linker region. Also indicated are the C-terminal His10 tag and lysine 898, which is mutated to asparagine in the ATPase-dead mutant (K898N). B, Coomassie-stained SDS-PAGE of samples from purification stages with a flow chart diagramming the purification strategy. Lane 1, soluble fraction; lane 2, RecQ4 after PreScission Protease treatment; lane 3, RecQ4 after elution from Co2+-nitrilotriacetic acid (NTA) resin; lane 4, RecQ4 peak from glycerol density gradient.
From the pFastbac 1-RecQ4 vector, the Bac-to-Bac system was used to create baculovirus encoding the RecQ4 construct under control of the p10 promoter. Sf9 cells were grown to a density of 1 × 106 cells/ml and infected with a multiplicity of infection of 1. Virus-infected cells were grown for an additional 48 h, pelleted, snap-frozen, and stored at −80 °C. Pelleted cells were lysed by resuspension with a Dounce homogenizer in lysis buffer (10 mm Tris HCl, pH 7.8, 20 mm KCl, 25% glycerol, 2 mm EDTA). NaCl was added to a final concentration of 350 mm, and the lysate was incubated on ice for 20 min with stirring. All subsequent steps were performed at 4 °C. The soluble fraction was isolated by centrifugation at 20,000 × g for 20 min. Soluble material was incubated with glutathione-Sepharose 4B resin (GE Healthcare) for 1 h, allowing the GST-tagged enzyme to bind. Glutathione resin was washed three times with buffers of 50 mm Tris HCl, pH 7.8, 10% glycerol, and 0.02% Triton X-100 containing, successively: 350 mm NaCl, 2 mm EDTA, 1 m NaCl, and 150 mm NaCl. To remove the GST tag, the enzyme was incubated for 2 h with 40 units of PreScission Protease (GE Healthcare) per 1 liter of culture. Cleaved enzyme was then passed over a Co2+ IMAC (Clontech) column and washed with buffer containing 50 mm Tris HCl, pH 7.8, 150 mm NaCl, 10% glycerol, and 0.02% Triton X-100. The same buffer, with 400 mm imidazole added, was used for step elution. Peak fractions from the IMAC column, as determined by the Bradford assay, were combined and applied to a glycerol density gradient from 30 to 60%. Gradients were centrifuged for 20 h at 60,000 rpm in a rotor TV-865B and collected in 0.5-ml fractions. The gradient peak was determined by ATPase assay for wtRecQ4 (see below) or SDS-PAGE analysis for RecQ4-K898N. The purified enzyme was flash-frozen and stored at −80 °C.
High pressure liquid chromatography-purified DNA oligonucleotides were obtained from Integrated DNA Technologies (sequences shown in supplemental Table 1A). Oligonucleotides were resuspended in TE (10 mm Tris HCl, pH 7.9, 0.1 mm EDTA) buffer and 5′-radiolabeled with 32P using γ-32P-labeled ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (New England Biolabs). Radiolabeled oligonucleotides were annealed to nonlabeled conjugate strands at a ratio of at least 1:3 in 10 mm Tris HCl, pH 7.9, 100 mm KCl, and 5 mm MgCl2 by a controlled temperature decrease in a thermocycler from 95 to 25 °C. Annealed substrates were separated on a polyacrylamide gel (8 or 15%) in TBE (89 mm Tris borate, pH 8.3, 2 mm EDTA). Substrates were extracted from excised gel fragments by electroelution with the S&S Elutrap system (Whatman). Purified substrates were stored at −20 °C in TBE with 5 mm MgCl2.
Helicase and Annealing Assays
For helicase assays, enzyme was incubated with 0.2 nm radiolabeled substrate for 30 min at 27 °C in 2.5 mm Tris acetate, pH 7.9, 2.5 mm potassium acetate, 8 mm magnesium acetate, 50 μg/ml bovine serum albumin, 1 mm dithiothreitol, and 1 mm ATP (or the indicated nucleotide cofactor) in a total reaction volume of 20 μl. Reactions were stopped by the addition of 4 μl of stop mix to a final concentration of 0.2% SDS, 5% sucrose, 10 mm EDTA, 0.1 mg/ml bromphenol blue, and 0.1 mg/ml xylene cyanol. Reaction products were separated on a 15% TBE-polyacrylamide gel and subjected to phosphorimaging analysis.
Annealing assays were conducted as detailed for helicase assays, except that enzyme was incubated with 0.2 nm single-stranded radiolabeled oligonucleotide and 0.2 nm nonlabeled complementary oligonucleotide. Radiolabeled oligonucleotide was purified as described previously. Complementary nonlabeled oligonucleotide was resuspended in TE buffer upon receipt from Integrated DNA Technologies. Radiolabeled and complementary nonlabeled oligonucleotides were heated separately for 5 min at 80 °C before being added to the reaction.
ATPase Assays
Enzyme was incubated with 1 mm ATP (3.8 nm γ-32P-labeled ATP) for 60 min at 27 °C in 2.5 mm Tris acetate, pH 7.9, 2.5 mm potassium acetate, 8 mm magnesium acetate, 50 μg/ml bovine serum albumin, 1 mm dithiothreitol, and 50 μg/ml pBSK+ single-stranded circular DNA in a total volume of 20 μl. At the indicated time points of 0, 5, 10, 15, 20, 30, 40, 50 and 60 min, 1 μl of the reaction was spotted on Polygram CEL 300 PEI thin layer chromatography plates (Macherey-Nagel) that had been prerun in distilled water. Plates were soaked in methanol for 5 min, dried, and developed in 0.5 m LiCl, 1 m formic acid to separate inorganic phosphate from ATP. Dried plates were subjected to phosphorimaging analysis.
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assays were conducted as described for helicase assays with the following modifications. After 30 min, 4 μl of nondenaturing loading mix was added to each reaction to a final concentration of 5% sucrose, 0.1 mg/ml bromphenol blue, and 0.1 mg/ml xylene cyanol, and reactions were placed on ice. Reaction products were separated on an 8% Tris borate-polyacrylamide gel and subjected to phosphorimaging analysis.
Double Filter Binding Assays
Double filter binding assays were done with DE81 paper and nitrocellulose according to Wong and Lohman (24) with binding buffer (25 mm Tris acetate, pH 7.9, 25 mm potassium acetate, 8 mm magnesium acetate, 1 mm dithiothreitol). Filters were prewashed with 500 μl of binding buffer drawn through at 30 kilopascals of vacuum. Reactions (50 μl) were carried out in binding buffer with 3.4 nm RecQ4, 0.25 nm radiolabeled DNA substrate (as indicated), 1 mm nucleotide cofactor (as indicated) and 50 μg/ml bovine serum albumin at 27 °C for 30 min. Each reaction was drawn through at 30 kilopascals of vacuum and then washed with 500 μl of binding buffer with 500 mm potassium acetate. Nitrocellulose and DE81 sheets were dried and subjected to phosphorimaging analysis.
Generation of Transgenes
Genomic DNA transgene P[RecQ4-V5] was generated by replacing green fluorescent protein with the V5 epitope in P[RecQ4-GFP] (11), which encodes a protein with an in-frame V5 fusion at the C terminus of RecQ4. The K898N point mutation transgene was generated by employing a method similar to that used in making the point mutant in the baculoviral expression vector. The DNA sequences of the constructs were confirmed by sequencing before microinjection, which was carried out with the published procedure (25). Genetic rescue of the null recq419 mutant with the transgenic RecQ4 lines was carried out in a manner similar to that described previously (11).
RESULTS
Purification of Drosophila RecQ4
Drosophila RecQ4, with an N-terminal GST tag and a C-terminal His10 tag (GST-RecQ4-His10; see Fig. 1A), was expressed in Sf9 cells using the Bac-to-Bac baculovirus expression system. The two tags allowed for a double affinity purification strategy (see flowchart in Fig. 1B). Soluble material was first incubated with glutathione-Sepharose beads, which were bound by the GST tag of GST-RecQ4-His10. The GST tag was removed from RecQ4 by PreScission Protease, resulting in RecQ4-His10. The enzyme was further purified and concentrated by Co2+ affinity chromatography. The final purification step was accomplished by ultracentrifugation across a 30–60% glycerol density gradient. Peak fractions (Fig. 1B) as determined by ATPase activity and SDS-PAGE analysis were more than 96% pure based on densitometric tracing. RecQ4 and RecQ4-K898N peak fractions from glycerol density gradients consistently co-sediment with lactate dehydrogenase (a tetramer of polypeptides, each 36,000 Mr, for a total of 144,000 Mr). This suggests that RecQ4 exists predominantly as a monomer in solution. Because sedimentation velocity experiments alone do not provide a rigorous measurement of molecular weight, future experimentation will be necessary to further address the oligomerization state of RecQ4 in solution.
Drosophila RecQ4 Has Proficient Helicase Activity
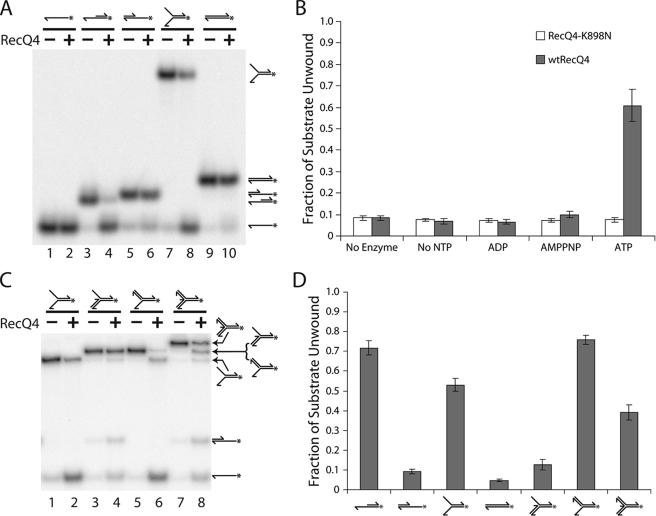
Previous studies have been unsuccessful in detecting any significant helicase activity from the human RecQ4 protein either purified by immunoprecipitation (16) or expressed as a recombinant protein in Escherichia coli (22). Recently, using the same heterologous E. coli expression system, a limited DNA unwinding activity has been detected (23). However, the reaction that we observed is more akin to DNA strand exchange than the helicase activity commonly found in other RecQ homologs. A double-stranded molecule with blunt ends was unwound only in the presence of an excessive amount of complementary single strand. These results were unexpected given that all other RecQ family members exhibit a robust helicase activity with a preference for 3′ single-stranded ends. Although the sequence of RecQ4 diverges significantly from the rest of the family, the helicase domain maintains all of the key helicase domain motifs. To investigate the helicase activity of the purified Drosophila RecQ4 protein, we incubated RecQ4 with a 3′ extension DNA substrate (see Fig. 2A, lanes 3 and 4, and B and supplemental Table 1B). Helicase activity was detected only in the presence of ATP, with no activity observed in the absence of nucleotide cofactor or in the presence of ADP or a nonhydrolyzable homolog, AMPPNP.
FIGURE 2.
RecQ4 shows 3′ to 5′ helicase activity. A, RecQ4 unwinds substrates with a 3′ single-stranded region. Radiolabeled oligonucleotide substrates were incubated in the presence (even-numbered lanes) or absence (odd-numbered lanes) of RecQ4 (13.4 nm), and separated by PAGE. RecQ4 has no effect on single-stranded substrate (compare lanes 1 and 2) or double-stranded substrate (lanes 9 and 10). RecQ4 can unwind substrate with a single-stranded region 3′ (lanes 3 and 4, 7, and 8) but not 5′ (lanes 5 and 6), of a double-stranded region. B, RecQ4 (shaded bars) unwinds the 3′ extension substrate (see A, lanes 3 and 4) in the presence of ATP. This activity is dependent on ATP hydrolysis, as it is not seen the absence of ATP or in the presence of AMPPNP. This activity is also due to RecQ4 and not a contaminant, because RecQ4-K898N (open bars) did not unwind the 3′ extension substrate under any of the conditions tested. Helicase assays were conducted as seen in A, except using 7.4 nm enzyme, and were quantified by phosphorimaging analysis. C, RecQ4 unwinds the 3′ flap substrate (lanes 5 and 6) but not the 5′ flap substrate (lanes 3 and 4). The assay was conducted as in A. The duplex fork substrate (lanes 7 and 8) was also unwound by RecQ4. D, quantification of RecQ4 activity on substrates. Helicase assays as seen in A and C were quantified by phosphorimaging analysis. The fraction of fully annealed substrate lost after incubation without enzyme was subtracted from the fraction of fully annealed substrate lost after incubation with RecQ4. Error bars in these experiments indicate S.D. (n = 3).
To ensure that the observed DNA unwinding was because of RecQ4 rather than a contaminating helicase, lysine 898 in the Walker A motif (GSGKS) was mutated to asparagine (K898N). This mutation is known to reduce ATPase activity greatly, thus preventing helicase activity (26). The expression and purification of the mutant protein are identical to those of the wild type including ultracentrifugation through a glycerol density gradient, suggesting that the mutant is neither unfolded nor aggregated. As expected, RecQ4-K898N did not exhibit helicase activity under any conditions (Fig. 2B). This establishes that the helicase activity observed is intrinsic to purified wild type RecQ4 and not attributable to a co-purifying contaminant.
To further characterize this helicase activity, we incubated RecQ4 with several substrates constructed from oligonucleotides (relative substrate preferences are summarized in Fig. 2D; substrate sequences and makeup are given in supplemental Table 1). RecQ4 does not unwind a completely duplex substrate (Fig. 2A, lanes 9 and 10) but will unwind a fork substrate (lanes 7 and 8). This indicates that a single-stranded region is necessary for RecQ4 helicase activity. Helicases in general exhibit distinct directionality preferences and may unwind DNA moving either 3′ to 5′ or 5′ to 3′. RecQ family members have been observed primarily acting in a 3′ to 5′ direction. To determine the directionality of RecQ4 helicase activity, a comparison was made between the 3′ and 5′ extension substrates (Fig. 2A, lanes 3–6). Significant helicase activity was detected on the substrate with a 3′ extension but not on the one with a 5′ extension. This indicates that a single-stranded region at the 3′-end of the double-stranded region is necessary for RecQ4 helicase activity and suggests the enzyme moves in a 3′ to 5′ direction.
We also examined the activity of RecQ4 on several variations of the fork, which resemble key intermediates in replication and recombination. We first tested 3′ and 5′ flap substrates. These are similar to the fork substrate, except that the indicated arm is single-stranded and the other is double-stranded. RecQ4 does not demonstrate appreciable helicase activity on the 5′ flap substrate (Fig. 2C, lanes 3 and 4). The contrasting 3′ flap substrate is unwound almost to completion by RecQ4 (Fig. 2C, lanes 5 and 6). Both single-stranded DNA and simple fork substrates were detected as products, indicating that RecQ4 unwinds the substrate in both directions available to it (down the center of the substrate and along the 5′ double-stranded branch). Interestingly, RecQ4 unwinds a duplex fork substrate (a fork with both the 3′ and 5′ branches double-stranded) (Fig. 2C, lanes 7 and 8). The presence of partial reaction products, including 3′ or 5′ flap substrates, a fork substrate, and 3′ or 5′ extension substrates, suggests that RecQ4 may enter at the three-way junction and unwind along any of the three duplex arms. Indeed, unwinding a three-junction substrate has been observed for other RecQ helicases such as RecQ5 (27), Wrn (28), and RecQ1 (29). Therefore, these data suggest that Drosophila RecQ4 has an efficient helicase activity with substrate specificity similar to that of other members in the RecQ family of proteins.
Drosophila RecQ4 Exhibits DNA-dependent ATPase Activity
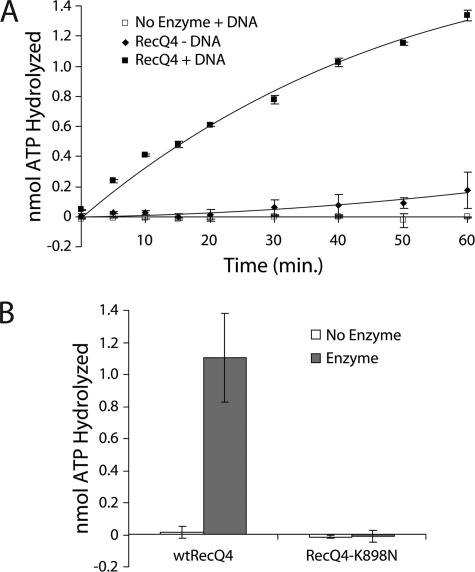
A hallmark activity for any DNA helicase is DNA-dependent ATP hydrolysis (30). To characterize the ATPase activity of RecQ4, we incubated RecQ4 with γ-32P ATP in the presence or absence of single-stranded circular DNA and monitored the liberation of phosphate using thin layer chromatography. The presence of DNA greatly stimulated the hydrolysis of ATP by RecQ4 (Fig. 3A). Plasmid DNA also stimulated RecQ4 ATPase activity (data not shown). In contrast, the K898N mutant showed no detectable amount of ATP hydrolysis above background (Fig. 3B). This confirms that the K898N mutation abolishes ATP binding/hydrolysis in RecQ4.
FIGURE 3.
RecQ4 exhibits DNA-stimulated ATP hydrolysis. A, ATP hydrolysis by RecQ4 is stimulated in the presence of single-stranded DNA. In the absence of RecQ4, ATP hydrolysis is not observed, independently of the presence of DNA. The reaction time course was performed by taking 1-μl samples from a total reaction volume of 20 μl at the indicated times. For each time point of each trial, the background was removed by subtracting the amount of ATP hydrolyzed in the absence of both enzyme and DNA. Enzyme concentration is 13.4 nm. B, RecQ4-K898N does not hydrolyze ATP. 1-μl samples were taken from 20-μl reactions after 60 min and analyzed by TLC and phosphorimaging analysis. For each trial, a 0 min time point was also taken. Background was removed by subtracting the amount ATP hydrolyzed at 0 min from that at 60 min. Enzyme concentration was 7.4 nm. Error bars in these experiments indicate S.D. (n = 3).
Stable Binding of RecQ4 with Single-stranded DNA in the Presence of AMPPNP
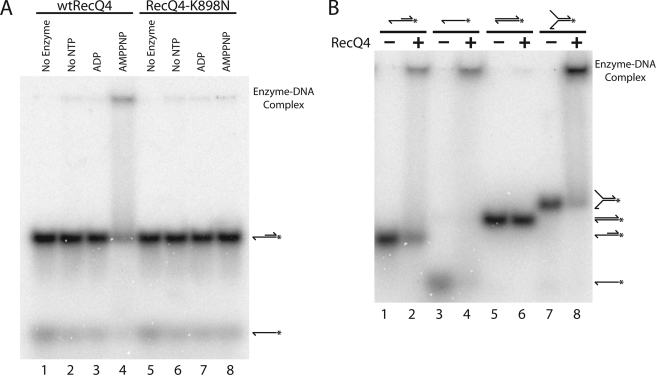
The mechanism of action of DNA helicases may require recognition of different DNA secondary structures and differential enzyme-substrate complex formation depending on the state of the bound nucleotide cofactors. Thus we examined RecQ4 DNA binding as a function of the nucleotide cofactor. Wild type RecQ4 was incubated with the 3′ extension substrate in the absence of nucleotide cofactor or in the presence of ADP or AMPPNP, and reaction products were analyzed by electrophoresis on a native polyacrylamide gel (Fig. 4A). Complex formation evidenced by electrophoretic mobility shift was observed in the presence of AMPPNP (Fig. 4A, lane 4) but not in the presence of ADP (lane 3) or in the absence of nucleotide cofactor (lane 2). The importance of ATP binding in promoting the stable complex formation was corroborated by experiments using RecQ4-K898N. The mutant, which is defective in both ATPase and helicase activities, did not cause a shift under any of these conditions (Fig. 4A, lanes 6–8). This demonstrates that a stable enzyme-substrate complex is formed only with bound nucleotide triphosphate.
FIGURE 4.
RecQ4 forms a stable complex with single-stranded DNA in the presence of AMPPNP. A, wtRecQ4 binds DNA in the presence of AMPPNP. wtRecQ4 (lanes 2–4) and RecQ4-K898N (lanes 6–8) at 3.7 nm were incubated with the 3′ extension substrate in the absence of nucleotide (lanes 2 and 6) or in the presence of ADP (lanes 3 and 7) or AMPPNP (lanes 4 and 8). Significant binding was seen only with wtRecQ4 and AMPPNP. B, RecQ4 forms a stable complex with single-stranded DNA but not double-stranded DNA. Substrates were incubated with AMPPNP in the presence (even-numbered lanes) or absence (odd-numbered lanes) of RecQ4 (3.3 nm). Stable complexes were formed by RecQ4 with the 3′ extension substrate (compare lanes 1 and 2), single-stranded substrate (lanes 3 and 4), and the fork substrate (lanes 7 and 8), all of which contain single-stranded DNA regions. No stable complex was detected with RecQ4 and the duplex substrate (lanes 5 and 6).
The 3′ extension substrate has both single-stranded and double-stranded regions. To determine whether the critical determinant for stable RecQ4 binding is single-stranded DNA, double-stranded DNA, or the junction between the two, the enzyme was incubated with AMPPNP and single-stranded, duplex, and fork substrates (Fig. 4B, lanes 3–8). RecQ4 was not observed to bind the duplex substrate, although it bound both the single-stranded and fork substrates. Thus RecQ4 forms a stable complex with single-stranded DNA, but not with double-stranded DNA, in the presence of AMPPNP.
Because the binding specificity of RecQ4 for single-stranded DNA in the presence of ATP is of direct relevance to its biochemical mechanism, we corroborated these results by using a double filter binding assay. This assay demonstrated that RecQ4 did not form a stable complex with duplex DNA under any of the conditions examined (supplemental Fig. 1). Under the conditions of this assay, single-stranded DNA is retained by RecQ4 in the absence of nucleotide cofactor and in the presence of ADP and AMPPNP. However, the presence of AMPPNP renders this complex significantly more resistant to washing with 500 mm potassium acetate than the absence of nucleotide cofactor or the presence of ADP. This indicates greater stability of the enzyme-DNA complex in the presence of AMPPNP.
Binding assays were also conducted with 3′ and 5′ flap substrates as well as the three-way junction substrate (data not shown). RecQ4 bound the 3′ and 5′ flap substrates equally but did not significantly bind the three-way junction substrate. This confirms that RecQ4 requires single-stranded DNA for binding. It also suggests that the low level helicase activity observed with the duplex fork substrate (Fig. 2C, lanes 7 and 8) is a consequence of the transient binding of the enzyme to the junction region, from which the enzyme is capable of unwinding the duplex arms provided ATP is present.
DNA Annealing Activity of Drosophila RecQ4
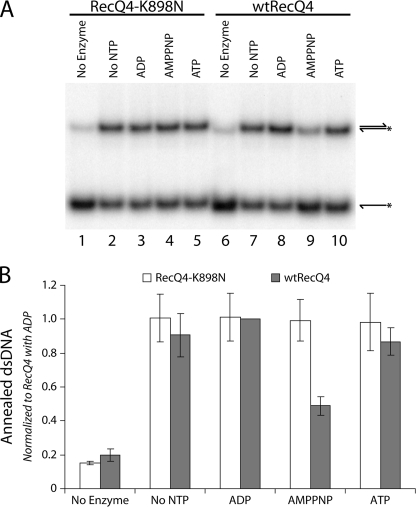
DNA strand annealing activity has been exhibited by all human RecQ helicases (22, 31–33). This strand rewinding activity may coordinate with the helicase activity to play an important role in the formation and resolution of recombination intermediates. Human RecQ4 was demonstrated to have strand annealing activity (22); it has been suggested that this interferes with detection of the helicase activity in this enzyme (23). As Drosophila RecQ4 has robust helicase activity, it would be interesting to examine whether it also possesses annealing activity like other RecQ family members. We incubated enzyme with complementary single-stranded DNA oligonucleotides, which when renatured form a 60-bp duplex. The absence of a single-stranded region in the annealed reaction product prevents it from being a substrate for RecQ4 helicase activity. Annealing reactions were performed in the absence of nucleotide and in the presence of ADP, ATP, or AMPPNP (Fig. 5A, lanes 7–10). RecQ4 demonstrated annealing activity under all conditions tested, although in the presence of AMPPNP annealing was reduced by ∼50% (Fig. 5B). This indicates that annealing is partially inhibited under the same conditions necessary to form a stable enzyme-DNA complex (compare with Fig. 4A). It is possible that stable association of RecQ4 with single-stranded DNA interferes with its ability to rewind the complementary strands. This notion of annealing activity regulation as a function of bound nucleotide cofactor can be tested using the ATPase-deficient mutant. Because RecQ4-K898N does not form a stable complex in the presence of AMPPNP (Fig. 4A, lane 8), its annealing activity should not be sensitive to AMPPNP. Indeed, the K898N mutant protein was able to anneal complementary single strands equally well under all conditions tested, including in the presence of AMPPNP (Fig. 5A, lanes 2–5; quantified in Fig. 5B). This finding supports the notion that stable complex formation between RecQ4 and single-stranded DNA may inhibit annealing with the complementary strand. This also demonstrates that the K898N mutant protein is not grossly misfolded or aggregated, because it shows strand annealing activity comparable with the wild type enzyme.
FIGURE 5.
RecQ4 exhibits annealing activity. A, AMPPNP reduces the annealing activity of RecQ4. wtRecQ4 (lanes 7–10) and RecQ4-K898N (lanes 2–5) at 7.4 nm were each incubated with two complementary single strands of DNA in the absence of nucleotide (lanes 2 and 7) or in the presence of ADP (lanes 3 and 8), AMPPNP (lanes 4 and 9), or ATP (lanes 5 and 10). Annealing was observed in all of these conditions, with wtRecQ4 exhibiting decreased annealing in the presence of AMPPNP. B, quantification of the annealing activity of RecQ4. Phosphorimaging analyses of the annealing assays were quantified to determine the fraction of duplex product formed by wtRecQ4 (shaded bars) and RecQ4-K898N (open bars). For each trial, results were normalized to those from wtRecQ4 and ADP. Error bars indicate S.D. (n = 3).
RecQ4 Helicase Activity Is Required for Its Biological Functions
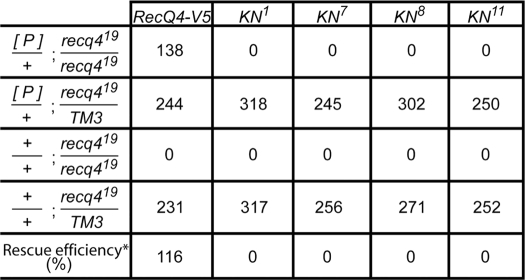
We showed earlier that a null mutation in Drosophila RecQ4 results in lethality, suggesting that RecQ4 is essential for the viability of Drosophila (11). This provides a system for testing the essentiality of helicase activity to RecQ4 function. To investigate the biological consequences of abolishing the DNA helicase activity of RecQ4, we generated a transposition vector of a RecQ4 transgene with the K898N point mutation and characterized four independent transgenic lines. Like the wild type control, P[RecQ4-V5], all RecQ4-K898N transgenic lines expressed similar levels of a protein with the expected size, which was detectable by Western blotting with V5 antibody (supplemental Fig. 2). However, complementation experiments showed that none of these K898N transgenic lines could rescue the viability of RecQ4 null mutants, recq419, whereas the wild type control could fully rescue the null mutant (Table 1). These data indicate that the DNA helicase activity of RecQ4 is essential for its biological functions.
TABLE 1.
Table indicating the results of genetic complementation experiments
Rescue experiments were carried out by crossing [P]/+;recq419/TM3 males (where [P] is the specific transgene indicated at the top of each column) with recq419/TM3 virgins. *, rescue efficiency was calculated for each transgene by dividing the numbers of the first row by the expected numbers based on the Mendelian ratio (one-fourth of the sum of the second and fourth rows).
DISCUSSION
We expressed and purified Drosophila RecQ4. The enzyme exhibits helicase activity in a 3′ to 5′ direction and requires single-stranded DNA to act. The ATPase-dead RecQ4-K898N mutant was unable to unwind DNA, demonstrating that the helicase domain is solely responsible for the observed helicase activity. The helicase activity is essential in Drosophila, because RecQ4-K898N cannot rescue the viability of homozygous null mutant flies. RecQ4 in the presence of AMPPNP formed a stable complex with single-stranded, but not double-stranded, DNA. Like its human counterpart, Drosophila RecQ4 engages in strand annealing activity. Reduced annealing was observed in the presence of AMPPNP, indicating that the stable enzyme-DNA complex may be inhibitory to annealing. RecQ4-K898N acted as an annealase at levels comparable with the wild type enzyme, but it was not inhibited by AMPPNP. Similarly, it did not form stable enzyme-DNA complexes even in the presence of AMPPNP. These results suggest that the K898N mutation does not disrupt the structure of RecQ4 but instead renders the enzyme incapable of binding nucleotide triphosphate cofactors.
Until recently, previous studies had not been able to detect helicase activity in human RecQ4 (16, 22). When helicase activity was finally reported, a 25-fold excess cold strand was required as a reaction product trap, presumably because strong annealing activity renders helicase activity undetectable (23), making this better described as strand exchange activity. The helicase activity we observed here is evident in the absence of any DNA trap, similar to assays used for other helicases. This contrast may be because of differences between species. The Drosophila enzyme is 371 residues longer than human RecQ4, and the enzymes as a whole share 42% similarity, although the helicase domains are 70% similar. Another source of difference may be the expression system employed. Macris et al. (22) and Xu and Liu (23) expressed human RecQ4 in E. coli, whereas we have expressed Drosophila RecQ4 in insect cells derived from Spodoptera frugiperda, presumably closer to a native expression environment.
RecQ4 is necessary for viability, but there are conflicting data concerning the necessity of the helicase domain. Removal of most of the helicase domain in mice leads to severe defects in development and life span but does not affect viability at birth (10). However, here we report that the K898N helicase-dead mutant was unable to rescue viability in Drosophila lacking RecQ4. This may reflect a species-specific difference in the biological role or the presence of a redundancy for RecQ4 helicase activity in mammals. Further analysis of RecQ4 mutants from Drosophila and mice may provide new insight into their cellular functions.
Helicase and annealase activities have now been observed for every RecQ helicase in humans. These appear to be central to the varied roles that the enzymes play in maintaining genomic stability. The motifs responsible for helicase activity in RecQ4 are common to all superfamily II helicases and thus are easily identified. Determination of the annealing motifs is more challenging, although evidence suggests they may reside in the N terminus (23). Annealing and helicase activities are inherently competitive, and our results indicate that RecQ4 cannot carry out both activities simultaneously. The following mechanistic sequence presents itself: 1) RecQ4 binds ATP and single-stranded DNA, forming a stable complex which inhibits annealing; 2) ATP is hydrolyzed, causing RecQ4 to unwind double-stranded DNA 5′ of the binding site; 3) ADP is released, and the process is restarted with newly bound ATP preventing reannealing. The fact that Drosophila RecQ4 has both helicase and annealing activities, which can readily be assayed by biochemical methods, will allow us to probe the distinct biochemical functions for these separate activities, specifically in terms of forming and resolving intermediates involved in DNA transactions.
Supplementary Material
Acknowledgments
We thank Jody Plank for discussions and the initial construction of the expression vector and Michael Webb for advice on helicase assays. We also thank Lored Asllani and Ravi Iyer for assistance with the double filter binding assays.
This work was supported, in whole or in part, by National Institutes of Health Grant GM29006.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- GST
- glutathione S-transferase
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate.
REFERENCES
- 1.Bachrati C. Z., Hickson I. D. (2008) Chromosoma 117, 219–233 [DOI] [PubMed] [Google Scholar]
- 2.Seki M., Otsuki M., Ishii Y., Tada S., Enomoto T. (2008) Cell Cycle 7, 2472–2478 [DOI] [PubMed] [Google Scholar]
- 3.Bohr V. A. (2008) Trends Biochem. Sci. 33, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W., Seki M., Narita Y., Nakagawa T., Yoshimura A., Otsuki M., Kawabe Y., Tada S., Yagi H., Ishii Y., Enomoto T. (2003) Mol. Cell. Biol. 23, 3527–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitao S., Ohsugi I., Ichikawa K., Goto M., Furuichi Y., Shimamoto A. (1998) Genomics 54, 443–452 [DOI] [PubMed] [Google Scholar]
- 6.Siitonen H. A., Sotkasiira J., Biervliet M., Benmansour A., Capri Y., Cormier-Daire V., Crandall B., Hannula-Jouppi K., Hennekam R., Herzog D., Keymolen K., Lipsanen-Nyman M., Miny P., Plon S. E., Riedl S., Sarkar A., Vargas F. R., Verloes A., Wang L. L., Kääriäinen H., Kestilä M. (2009) Eur. J. Hum. Genet. 17, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larizza L., Magnani I., Roversi G. (2006) Cancer Lett. 232, 107–120 [DOI] [PubMed] [Google Scholar]
- 8.Van Maldergem L., Siitonen H. A., Jalkh N., Chouery E., De Roy M., Delague V., Muenke M., Jabs E. W., Cai J., Wang L. L., Plon S. E., Fourneau C., Kestilä M., Gillerot Y., Mégarbané A., Verloes A. (2006) J. Med. Genet. 43, 148–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoki Y., Araki R., Fujimori A., Ohhata T., Koseki H., Fukumura R., Nakamura M., Takahashi H., Noda Y., Kito S., Abe M. (2003) Hum. Mol. Genet. 12, 2293–2299 [DOI] [PubMed] [Google Scholar]
- 10.Mann M. B., Hodges C. A., Barnes E., Vogel H., Hassold T. J., Luo G. (2005) Hum. Mol. Genet. 14, 813–825 [DOI] [PubMed] [Google Scholar]
- 11.Wu J., Capp C., Feng L., Hsieh T. S. (2008) Dev. Biol. 323, 130–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sangrithi M. N., Bernal J. A., Madine M., Philpott A., Lee J., Dunphy W. G., Venkitaraman A. R. (2005) Cell 121, 887–898 [DOI] [PubMed] [Google Scholar]
- 13.Matsuno K., Kumano M., Kubota Y., Hashimoto Y., Takisawa H. (2006) Mol. Cell. Biol. 26, 4843–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitao S., Shimamoto A., Goto M., Miller R. W., Smithson W. A., Lindor N. M., Furuichi Y. (1999) Nat. Genet. 22, 82–84 [DOI] [PubMed] [Google Scholar]
- 15.Fan W., Luo J. (2008) J. Biol. Chem. 283, 29037–29044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin J., Kwon Y. T., Varshavsky A., Wang W. (2004) Hum. Mol. Genet. 13, 2421–2430 [DOI] [PubMed] [Google Scholar]
- 17.Petkovic M., Dietschy T., Freire R., Jiao R., Stagljar I. (2005) J. Cell Sci. 118, 4261–4269 [DOI] [PubMed] [Google Scholar]
- 18.Burks L. M., Yin J., Plon S. E. (2007) Gene 391, 26–38 [DOI] [PubMed] [Google Scholar]
- 19.Werner S. R., Prahalad A. K., Yang J., Hock J. M. (2006) Biochem. Biophys. Res. Commun. 345, 403–409 [DOI] [PubMed] [Google Scholar]
- 20.Dietschy T., Shevelev I., Pena-Diaz J., Huhn D., Kuenzle S., Mak R., Miah M. F., Hess D., Fey M., Hottiger M. O., Janscak P., Stagljar I. (2009) J. Cell Sci. 122, 1258–1267 [DOI] [PubMed] [Google Scholar]
- 21.Park S. J., Lee Y. J., Beck B. D., Lee S. H. (2006) DNA Cell Biol. 25, 696–703 [DOI] [PubMed] [Google Scholar]
- 22.Macris M. A., Krejci L., Bussen W., Shimamoto A., Sung P. (2006) DNA Repair (Amst.) 5, 172–180 [DOI] [PubMed] [Google Scholar]
- 23.Xu X., Liu Y. (2009) EMBO J. 28, 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong I., Lohman T. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5428–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin G. M., Spradling A. C. (1982) Science 218, 348–353 [DOI] [PubMed] [Google Scholar]
- 26.Hall M. C., Matson S. W. (1999) Mol. Microbiol. 34, 867–877 [DOI] [PubMed] [Google Scholar]
- 27.Ozsoy A. Z., Ragonese H. M., Matson S. W. (2003) Nucleic Acids Res. 31, 1554–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brosh R. M., Jr., Waheed J., Sommers J. A. (2002) J. Biol. Chem. 277, 23236–23245 [DOI] [PubMed] [Google Scholar]
- 29.Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R. M., Jr. (2005) J. Biol. Chem. 280, 28072–28084 [DOI] [PubMed] [Google Scholar]
- 30.Lohman T. M., Bjornson K. P. (1996) Annu. Rev. Biochem. 65, 169–214 [DOI] [PubMed] [Google Scholar]
- 31.Machwe A., Xiao L., Groden J., Matson S. W., Orren D. K. (2005) J. Biol. Chem. 280, 23397–23407 [DOI] [PubMed] [Google Scholar]
- 32.Cheok C. F., Wu L., Garcia P. L., Janscak P., Hickson I. D. (2005) Nucleic Acids Res. 33, 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muftuoglu M., Kulikowicz T., Beck G., Lee J. W., Piotrowski J., Bohr V. A. (2008) Biochemistry 47, 10247–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.