Abstract
Loss of CDKN2A/p16INK4A in hematopoietic stem cells is associated with enhanced self-renewal capacity and might facilitate progression of damaged stem cells into pre-cancerous cells that give rise to leukemia. This is also reflected by the frequent loss of the INK4A locus in acute lymphoblastic T-cell leukemia. T-cell acute lymphoblastic leukemia cells designed to conditionally express p16INK4A arrest in the G0/G1 phase of the cell cycle and show increased sensitivity to glucocorticoid- and tumor necrosis factor receptor superfamily 6-induced apoptosis. To investigate the underlying molecular mechanism for increased death sensitivity, we interfered with specific steps of apoptosis signaling by expression of anti-apoptotic proteins. We found that alterations in cell death susceptibility resulted from changes in the composition of pro- and anti-apoptotic BCL2 proteins, i.e. repression of MCL1, BCL2, and PMAIP1/Noxa and the induction of pro-apoptotic BBC3/Puma. Interference with Puma induction by short hairpin RNA technology or retroviral expression of MCL1 or BCL2 significantly reduced both glucocorticoid- and FAS-induced cell death in p16INK4A-reconstituted leukemia cells. These results suggest that Puma, in concert with MCL1 and BCL2 repression, critically mediates p16INK4A-induced death sensitization and that in human T-cell leukemia the deletion of p16INK4A confers apoptosis resistance by shifting the balance of pro- and anti-apoptotic BCL2 proteins toward apoptosis protection.
Introduction
The INK4A gene locus on chromosome 9p21 codes for the two functionally unrelated tumor suppressor genes p16INK4A and p14ARF (1, 2). p16INK4A acts as a G0/G1 cell cycle inhibitor, whereas p14ARF interacts with MDM2 and thereby prevents TP53/p53 degradation. Inactivation of the INK4A gene locus frequently occurs in primary tumor cells of T-cell acute lymphoblastic leukemia (T-ALL)2 and predicts relapse in children with ALL, suggesting a critical role of this locus in disease development (3–5). More recently, evidence has been provided that down-regulation of p16INK4A is associated with enhanced self-renewal and proliferative capacity of hematopoietic stem cells and that the inactivation of this tumor suppressor in immature pre-cancerous cells might allow them to overcome replicative senescence or apoptosis (6).
p16INK4A binds to and inhibits the activity of the CCND1/cyclin D-dependent kinases CDK4 and CDK6, which are critical for G1 progression and G1/S transition. The activity of these serine/threonine protein kinases is further regulated by mitogenic hormones and by additional cyclin-dependent kinase inhibitors (7, 8). Active CDK4/6 complexes phosphorylate and inactivate retinoblastoma protein and its family members RBL1/p107 and RBL2/p130, thus promoting the activity of E2F transcription factors and the expression of genes essential for the onset of S phase and mitosis (9).
Apoptosis is initiated by a number of signals that either activate membrane death receptors (extrinsic pathway) and/or intracellular pathways controlled by members of the BCL2 family via the mitochondria (intrinsic pathway) (10, 11). In the extrinsic apoptosis pathway death receptor ligands such as FASLG/FAS ligand bind to their cognate receptors, thereby inducing the formation of the death-inducing signaling complex that contains the adaptor molecule Fas-associated death domain (FADD) and procaspase-8. Autocatalytic cleavage of procaspase-8 leads to activation of a downstream caspase cascade. In some cells, caspase-8 also connects to the intrinsic pathway through cleavage of pro-apoptotic BID and cleavage of the anti-apoptotic BCL2 protein MCL1 (12), thereby providing a cross-talk between extrinsic and mitochondrial death pathways.
Mitochondria are central executioners of programmed cell death that integrate apoptotic signals such as DNA damage, growth factor withdrawal, GC treatment, and anoikis. These stimuli induce apoptosis either by directly regulating genes controlling cell survival or via (de)regulating gene networks leading to cellular distress that in turn triggers apoptosis. In both scenarios, members of the large family of pro- and anti-apoptotic BCL2 proteins, referred to as the “BCL2 rheostat,” might be involved either as direct targets or as sensors for cellular stress. In addition, the status of the BCL2 rheostat, regardless of whether directly affected by a specific treatment, might define sensitivity to, and kinetics of, cell death induction.
BCL2 proteins can be divided into multidomain and BH3-only proteins. The multidomain proteins, such as the pro-apoptotic proteins BAX and BAK1/Bak, contain three BCL2 homology domains, and the anti-apoptotic proteins BCL2, BCL2L2/Bcl-w, BCL2L1/Bcl-xL, BCL2A1/A1, and MCL1 contain four BH domains (11). Two models have been proposed for apoptosis induction by BH3-only proteins as follows. In the “direct activator/de-repressor model” (13) strong BH3-only proteins such as BCL2L11/Bim, BBC3/Puma, and truncated BID act as direct activators of BAX, and in the “displacement model” (14, 15) these three proteins are potent neutralizers of all five BCL2-like pro-survival proteins. Weak BH3-only proteins such as PMAIP1/Noxa act as sensitizers by inactivating specific pro-survival BCL2 proteins. Oligomerization of BAX or Bak in the mitochondrial outer membrane causes cytochrome c release from mitochondria, which then binds to APAF1 and activates procaspase-9.
Recently, it was proposed that the deletion of the INK4A locus and the loss of the tumor suppressors p16INK4A and p19ARF enhance lymphoid progenitor proliferation and thereby critically contribute to the development of lymphoblastic leukemia. We therefore studied the effect of conditional reconstitution of p16INK4A in human T-ALL cells, and we observed that protracted p16INK4A expression in addition to its anti-proliferative function also exert a profound effect on apoptosis regulation by affecting the expression of pro- and anti-apoptotic BCL2 family members.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture Conditions, and Reagents
Subclones of the CCRF-CEM T-ALL 2C8 cell line (16) that express p16INK4A under the control of the tetracycline-regulated transactivators CEM-6E2/p16 and CEM-1D2/p16 (17) and PhoenixTM packaging cells for helper-free production of amphotropic retroviruses (kindly provided by G. P. Nolan) (18) were maintained in RPMI 1640 medium containing 10% fetal calf serum (Invitrogen), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine (Invitrogen) at 5% CO2 and at 37 °C in saturated humidity. All cell lines were tested for and found to be free of mycoplasma infection. The authenticity of all cell lines was verified by DNA fingerprinting, as detailed previously (19). Dexamethasone (Dex) was stored as a 10 mm stock solution in 100% ethanol, and doxycycline (Dox) was maintained as a 10 mm solution dissolved in phosphate-buffered saline. All reagents were from Sigma, unless indicated otherwise. The anti-FAS/CD95 antibody (clone CH11) was purchased from Werfen (Vienna, Austria). For each experiment, mid-log phase cultures were seeded in fresh medium.
Construction of Retroviral Vectors
Retroviral vectors coding for dnFADD, BCL2 (20), and vectors for the expression of Puma-specific shRNA (21) have been described. For construction of pLIB-dnBid-iresPuro, the coding region of dnBID was amplified from pLZRS-dnBid-iresGFP (22) and inserted into the EcoRI and BamHI sites of pLIB-MCS2-iresPuro (23). For tetracycline-regulated expression of p16INK4A in Molt4 cells, a conditional retroviral expression system was developed (supplemental Fig. 2S). cDNA sequences were verified by sequencing.
Production of Retroviruses and Retroviral Infection
8 × 105 Phoenix packaging cells were transfected with 3 μg of pLIB-Bcl2-iresPuro, pLIB-dnFADD-iresPuro, pLIB-dnBid-iresPuro, or pMSCV-shRNA-PUMA2-SV40-Puro using Lipofectamine2000 (Invitrogen). Control cell lines were generated by infecting CEM/p16 (CEM-6E2/p16 and CEM-1D2/p16) or parental CEM/Ctr cells (CEM-2C8/Ctr) with the appropriate empty control vector (pLIB-MCS2-iresPuro). After transfection, cells were cultured for 48 h in RPMI 1640 medium. The retrovirus-containing supernatants were filtered through 0.22 μm syringe filters (Sartorius, Goettingen, Germany) and used to infect CEM/p16 cells. For statistical presentation, CEM-6E2/p16 and CEM-1D2/p16 cell lines were pooled and termed CEM/p16. In the same way, bulk-selected sublines of CEM-6E2/p16 and CEM-1D2/p16 infected with retroviral vectors coding for dnFADD, dnBID, MCL1, or BCL2 were pooled and termed CEM/p16-dnFADD, CEM/p16-dnBid, CEM/p16-Mcl1, and CEM/p16-Bcl2, respectively (supplemental Fig. 1S).
Quantitative RT-PCR
To quantify FasL, FAS, Puma, and BCL2 mRNA levels, “real time” RT-PCR assays using glyceraldehyde-3-phosphate dehydrogenase as reference gene were performed. CCRF-CEM cells were cultured in the presence or absence of 250 ng/ml Dox for the times indicated. Total RNA was isolated from 5 × 106 cells using TRIzolTM reagent (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized from 1 μg of total RNA using the RevertAidTM first strand cDNA synthesis kit (MBI Fermentas, St. Leon-Rot, Germany). The oligonucleotides for FasL, Puma, and glyceraldehyde-3-phosphate dehydrogenase have been described previously (20). Primers for FAS (GCTCTTTCACTTCGGAGGATTGC and GCCTTCCAAGTTCTGAGTCTCAAC) and BCL2 (AGATGTCCAGGCAGCTGCAC and CACAGGGCGATGTTGTCCAC) were obtained from Sigma. Amplification efficiency was determined by serial log2 dilutions. All reactions were conducted in triplicate. Real time RT-PCR was run on the iCycler instrument (Bio-Rad) using a thermal profile of an initial 3-min melting step at 95 °C, followed by 40 cycles at 95 °C for 20 s and 55 °C for 45 s. To verify the presence of only one amplicon, a melting curve was processed after each run. After normalization on glyceraldehyde-3-phosphate dehydrogenase expression, regulation was calculated between treated and untreated cells.
Determination of Apoptosis
Apoptosis was determined by quantification of propidium iodide (PI)-stained nuclei (24) and forward/sideward scatter analysis using a Cytomics FC-500 (Werfen, Austria). Briefly, 2 × 105 cells were centrifuged at 700 × g for 5 min and resuspended in hypotonic PI solution containing 0.1% Triton X-100. Cellular debris and small particles were excluded from FACS analysis, and stained nuclei in the sub-G1 marker window were considered to represent apoptotic cells. Statistical analysis was performed using GraphPad Prism 4.0 software.
Immunoblotting
Identical numbers of cells were lysed on ice in CelLyticTM-M mammalian cell lysis/extraction reagent containing a protease inhibitor mixture (Sigma) and centrifuged at 17,000 × g. The supernatant was mixed with 2× SSB containing 10% β-mercaptoethanol and boiled. Samples were separated by SDS-PAGE on 15% polyacrylamide gels, transferred to nitrocellulose membranes by a Hoeffer semi-dry transfer apparatus, and stained with Ponceau red. The membranes were blocked with Tris-buffered saline containing 1% Tween 20 and 5% nonfat dry milk; incubated with primary antibodies specific for human BCL2, Bak, BID, Bcl-xL, BAX, Bcl-w CoxIV (Cell Signaling Technology), MCL1, Bim, FADD, caspase-8 (Pharmingen), Noxa (Alexis Corp., Lausen, Switzerland), Puma (Sigma), and α-tubulin (Oncogene Research, Cambridge, MA); washed; and incubated with anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences). The blots were developed by enhanced chemiluminescence (Amersham Biosciences) according to the manufacturer's instructions and analyzed in an AutoChemi chemiluminescence detection system (UVP, Cambridge, United Kingdom).
Subcellular Fractionation Assays
Cytoplasm and mitochondrial extracts were prepared with ApoAlert® cell fractionation kit (Clontech) according to the manufacturer's instructions. In brief, 5 × 107 cells were washed with 1 ml of ice-cold Wash Buffer. The cell pellet was lysed in 0.8 ml of ice-cold Cell Fractionation Buffer for 1 h at 4 °C. The cell lysates were then centrifuged for 30 min at 20,000 × g. Proteins from the supernatant (cytosolic fraction) and pellet (membrane fraction) were mixed with 4× SSB containing 20% β-mercaptoethanol and separated by SDS-PAGE on 15% polyacrylamide gels.
RESULTS
p16INK4A Expression Sensitizes T-ALL Cells to FAS-induced Apoptosis, Induces FASLG/FasL Expression, and Increases Cleavage of Caspase-8 and BID
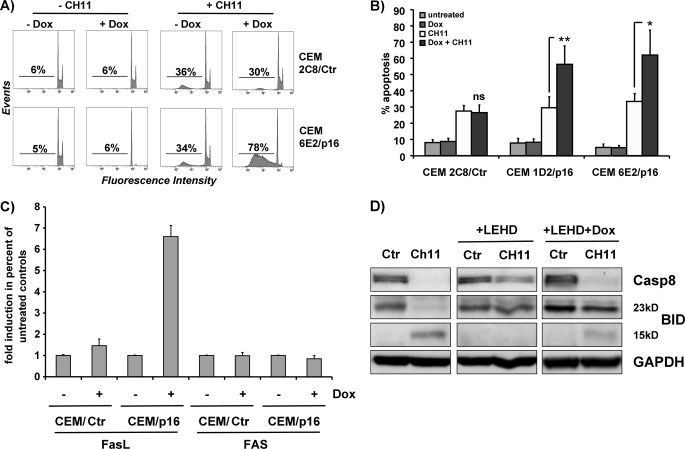
We have shown previously that Dox-induced p16INK4A expression stably arrested CCRF-CEM T-ALL cells in the G1 phase of the cell cycle (25) and sensitized these cells to physiologic concentrations of cortisol, which was associated with induction of the GC receptor (17). GC-induced apoptosis is generally thought to depend on the intrinsic pathway (26); however, the extrinsic apoptosis pathway has also been implicated in GC-induced cell death (27). To determine whether p16INK4A might affect the extrinsic pathway as well, CEM/p16 cells (Dox-inducible human p16INK4A) were treated with 250 ng/ml Dox for 24 h and for another 4 h with 0.1 μg/ml FAS-activating CH11 antibody. Similar to GC-induced cell death, FAS-triggered apoptosis was significantly increased in p16INK4A-expressing leukemia cells (Fig. 1, A and B). Interestingly, p16INK4A reconstitution increased FasL mRNA expression up to 6-fold, whereas FAS steady state levels remained unaltered (Fig. 1C).
FIGURE 1.
p16INK4A sensitizes CEM leukemia cells to FAS-induced apoptosis, induces FasL expression and increases caspase-8 and BID cleavage. A and B, CEM/p16 cells were cultured in the absence or presence of 250 ng/ml Dox for 24 h and then treated with 0.1 μg/ml FAS-activating CH11 antibody for 4 h. The percentage of apoptotic cells was determined by FACS analysis of PI-stained nuclei. Shown is the mean ± S.E. of three independent experiments. *, p = 0.045; **, p = 0.0086 CH11-treated cells compared with CH11 + Dox cells (unpaired t test). C, for quantification of FAS and FasL transcripts, T-ALL cells were treated for 24 h with or without 250 ng/ml Dox and then subjected to “real time” RT-PCR analysis. Data of CEM-6E2/p16 and CEM-1D2/p16 cells were pooled for graphical presentation and labeled CEM/p16. D, CEM-6E2/p16 cells were cultured in the presence or absence of 250 ng/ml Dox for 24 h, treated with CH11 antibody for 4 h, and/or the caspase-9-inhibitor LEHD-fluoromethyl ketone, as indicated. Processing of pro-caspase-8 and of the BH3-only protein BID was assessed by immunoblot, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as loading control (Ctr).
Activation of caspase-8 by FAS either directly leads to cell killing via cleavage of caspase-3 or requires caspase cascade amplification via caspase-8-mediated activation of the pro-apoptotic BH3-only protein BID (28). In cells that require BID cleavage, cytochrome c release and caspase-9 activation are essential for death receptor-induced apoptosis. We therefore assessed whether pro-caspase-8 processing and BID cleavage in response to FAS activation were affected by the inhibition of caspase-9 and whether this pathway was modified by reconstitution of the cell cycle inhibitor p16INK4A. CEM-6E2/p16 cells were cultured for 24 h in the presence or absence of Dox and treated with CH11 antibody with or without the caspase-9 inhibitor LEHD-fluoromethyl ketone for another 4 h. As shown by immunoblot in Fig. 1D, processing of pro-caspase-8 and BID was partly reduced by LEHD treatment suggesting involvement of caspase-9. In cells re-expressing p16INK4A, however, the inhibitory effect of LEHD was attenuated.
To address whether increased death ligand expression constitutes an additional apoptosis signal that may sensitize leukemia cells to death receptor- and GC-induced apoptosis, we next interfered with death-inducing signaling complex formation and cross-talk to mitochondria by retroviral expression of dominant-negative FADD (dnFADD) and dominant-negative BID (dnBID).
Cross-talk between Extrinsic and Intrinsic Death Pathways via BID Is Critical for Increased Apoptosis Sensitivity of p16INK4A-expressing Leukemia Cells
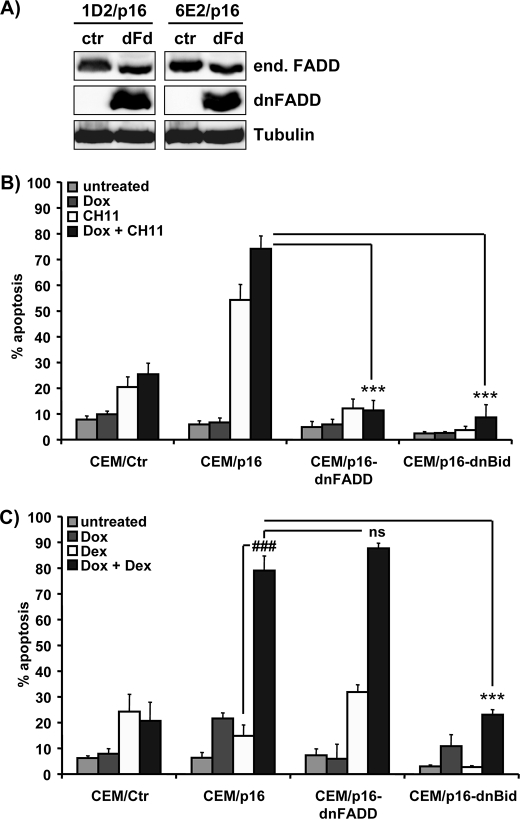
CEM-6E2/p16 and CEM-1D2/p16 ALL cells were infected with retroviruses coding for dnFADD (CEM/p16-dnFADD) or caspase cleavage-resistant dnBID (CEM/p16-dnBid). dnFADD lacks the death effector domain that is necessary for the interaction with caspase-8/-10, whereas dnBID carries point mutations (D59E and D75E) in the caspase-8/-10 cleavage sites. dnFADD, which was detectable as a faster migrating band in the Western blot (Fig. 2A), efficiently prevented FAS-induced cell death (Fig. 2B), whereas dnFADD did not affect GC-induced apoptosis (Fig. 2C). This result rules out increased FasL expression as a cause for increased death sensitivity. In contrast to dnFADD, dnBID efficiently blocked both FAS- and GC-induced cell death (Fig. 2, B and C) suggesting that death receptor-induced apoptosis is regulated by BCL2 proteins in this system. The fact that caspase-resistant dnBID diminished p16INK4A-induced sensitization to GCs (p < 0.0001) further implied that BID and upstream caspases (e.g. caspase-8) are part of a GC sensitivity regulating the amplification loop that requires mitochondria. To study how this amplification loop is controlled by proteins of BCL2 rheostat and how p16INK4A might influence it, we next investigated the expression and subcellular distribution of pro- and anti-apoptotic BCL2 proteins in CEM/p16 cells.
FIGURE 2.
Effect of dnFADD and dnBID on FAS- and GC-induced apoptosis in CEM/p16 leukemia cells. A, CEM-1D2/p16 and CEM-6E2/p16 cells were infected with the plasmid pLIB-dnFADD-iresPuro (dFd) or empty control vector (ctr). Puromycin-resistant cells were subjected to immunoblot analysis using a monoclonal anti-FADD antibody that detects both the endogenous (end.) FADD and truncated dnFADD protein. α-Tubulin served as a loading control. B and C, CEM/Ctr, CEM/p16-ctr, and CEM/p16-dnFADD cells as well as dnBID-expressing CEM/p16-dnBid cells were cultured with/without 250 ng/ml Dox for 24 h and then treated with anti-FAS antibody (0.1 μg/ml CH11 clone) for 4 h or with 10 nm Dex for 40 h, respectively. Apoptosis induction was measured by PI-FACS analysis. Shown is the mean ± S.E. of four independent experiments. Statistical analysis was performed using unpaired t test. B, ***, p < 0.0001 CEM/p16 + Dox + CH11-treated cells compared with CEM/p16-dnFADD or CEM/p16-dnBid cells + Dox + CH11. C, ###, p < 0.0001 compared with CEM/p16 + Dex + Dox. ns, not significant. ***, p < 0.0001 CEM/p16 + Dox + Dex-treated cells compared with CEM/p16-dnFADD or dnBid cells + Dox + Dex.
p16INK4A Causes Repression of BCL2, MCL1, and Noxa and Induces Puma
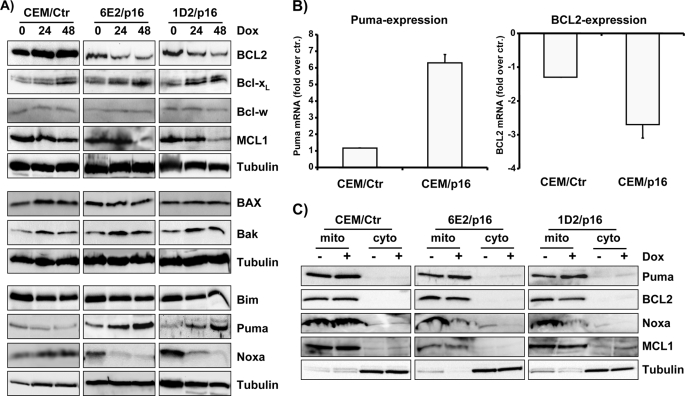
p16INK4A reconstitution did not significantly affect the protein levels of Bcl-w (Fig. 3A, top panels) and BAX and Bak (Fig. 3A, middle panels) within 48 h. In contrast, BCL2 and MCL1 were markedly reduced after 48 h of p16INK4A expression, whereas Bcl-xL slightly increased (Fig. 3A, top panels). Because MCL1 and BCL2 sequester different subsets of pro-apoptotic BCL2 proteins, the concerted repression of MCL1 and BCL2 might lower the apoptosis-protecting capacity of BCL2 rheostat in p16INK4A-expressing cells. Moreover, loss of MCL1 and BCL2 may not be fully compensated by the changes observed for Bcl-xL. In parallel to protein steady state expression, BCL2 mRNA levels also declined, suggesting that p16INK4A reconstitution at least in part exerts its effects via changes in mRNA expression (Fig. 3B, right panel).
FIGURE 3.
p16INK4A-induced G0/G1 arrest is associated with repression of BCL2, MCL1, and Noxa and induction of Puma. A, CEM/Ctr, CEM-1D2/p16, and CEM-6E2/p16 cells were treated with 250 ng/ml Dox for 24 and 48 h. The lysates were subjected to immunoblot analyses using specific antibodies directed against BCL2, Bcl-xL, Bcl-w, MCL1, BAX, Bak, Bim, Puma, Noxa, and α-tubulin. B, for RT-PCR analysis, RNA was prepared from CEM/Ctr, CEM-6E2/p16, and CEM-1D2/p16 cells after treatment with 250 ng/ml Dox for 24 h. mRNA steady state levels of Puma and BCL2 were determined by quantitative real time RT-PCR. The bars represent fold-induction over control (untreated cells) of three independent experiments each performed in triplicate. C, mitochondrial (mito) and cytoplasmic (cyto) extracts were prepared from CEM/Ctr, CEM-6E2/p16, and CEM-1D2/p16 cells cultured in the absence or presence of 250 ng/ml Dox for 24 h. The expression of Puma, BCL2, Noxa, MCL1, and α-tubulin in subcellular fractions was detected by immunoblot.
Next, we analyzed the expression of the BH3-only proteins Bim, Puma, and Noxa (Fig. 3A, bottom panels). Bim and Puma are strong apoptosis inducers that bind and inactivate all anti-apoptotic BCL2 proteins, whereas Noxa preferentially binds to A1 and MCL1 (29). p16INK4A reconstitution was found to significantly affect the cellular BH3-only protein pool, and although Bim steady state levels did not change, Noxa was down-regulated already at 24 h post-addition of Dox. In contrast, the Puma mRNA (Fig. 3B) and protein levels (Fig. 3A, bottom panels) were strongly induced within 48 h of p16INK4A expression.
Although endogenous Puma protein strongly accumulated within 48 h, the leukemia cells did not undergo apoptotic cell death. To study whether the lack of spontaneous apoptosis was due to impaired recruitment of Puma to the mitochondria, we analyzed the subcellular distribution of Puma in p16INK4A-expressing and -nonexpressing cells. Puma, like BCL2 and MCL1, was found exclusively in the membrane fraction suggesting that the majority of Puma protein was localized at the mitochondrial membrane presumably in complex with pro-survival BCL2 proteins. As expected from changes in total protein expression, mitochondrial Noxa diminished after 24 h of Dox treatment (Fig. 3C). These results demonstrate that re-expression of p16INK4A induces the accumulation of Puma at the mitochondria, which in turn might lower the pro-survival capacity of anti-apoptotic BCL2 proteins.
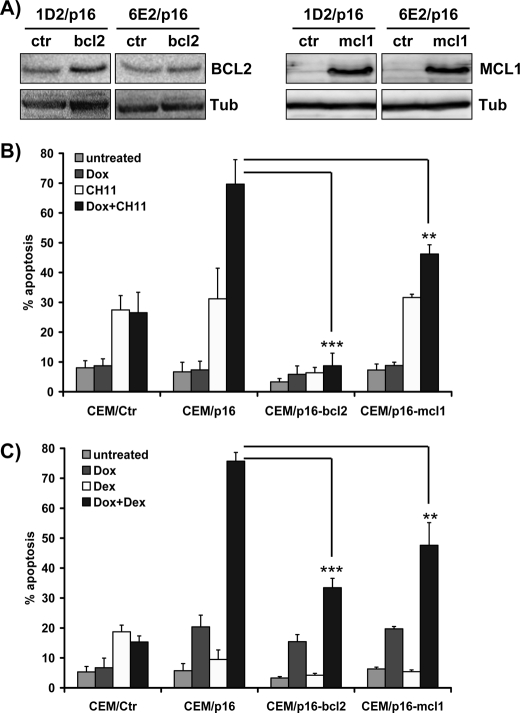
Transgenic BCL2 and MCL1 Both Inhibit p16INK4A Effects on FAS- and GC-induced Cell Death
CEM/p16 cells were infected with retroviruses coding for human BCL2 (CEM/p16-bcl2) and MCL1 (CEM/p16-mcl1) to generate leukemia cells with conditional p16INK4A and stable BCL2 or MCL1 expression. Bulk-selected leukemia cells were analyzed by immunoblotting to verify increased expression of the transgenes (Fig. 4A). Expression of ectopic BCL2 almost completely prevented FAS-induced apoptosis (Fig. 4B, CEM/p16 + Dox versus CEM/p16-bcl2 + Dox, p < 0.0001) and also significantly reduced GC-induced cell death to about 50% of controls (p < 0.0001). However, the sensitizing effect of p16INK4A was still visible with about 2-fold higher apoptosis rates in p16INK4A-expressing CEM/p16-bcl2 cells compared with controls (Fig. 4C). Transgenic MCL1 also exerted a significant inhibitory effect on FAS-triggered apoptosis in p16INK4A-expressing cells (p < 0.0001) and lowered p16INK4A-induced death sensitization to GC (p < 0.01). The combined data suggest that repression of both MCL1 and BCL2 contributes directly or indirectly to p16INK4A-induced death sensitization.
FIGURE 4.
Transgenic BCL2 and MCL1 inhibit FAS- and GC-induced apoptosis in G0/G1-arrested CEM/p16 cells. A, ectopic expression of BCL2 and MCL1 in CEM-1D2/p16-bcl2, CEM-6E2/p16-bcl2, CEM-1D2/p16-mcl1, and CEM-6E2/p16-mcl1 cells was determined by immunoblot analyses. Equal protein loading was confirmed by α-tubulin (Tub) staining. B and C, CEM/Ctr, CEM/p16-ctr, CEM/p16-bcl, and CEM/p16-mcl1 cells were incubated with 250 ng/ml Dox for 24 h and then treated with 0.1 μg/ml anti-FAS antibody for 4 h or with 10 nm Dex for 40 h. Apoptosis induction was measured by PI-FACS analysis. Shown is the mean ± S.E. of four independent experiments. Statistical analysis was performed using unpaired t test. **, p < 0.01; ***, p < 0.0001 for Bcl2- or Mcl1-transgenic CEM/p16-cells compared with CEM/p16 controls (ctr).
Puma Is Critical for Increased Apoptosis Sensitivity of p16INK4A-expressing Leukemia Cells
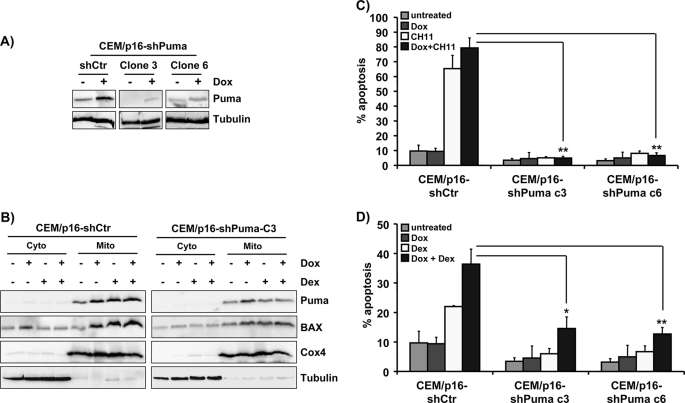
To directly assess whether elevated Puma expression accounts for increased death sensitivity of p16INK4A-expressing human leukemia cells, we infected CEM/p16 cells with retroviruses coding for Puma-specific shRNA (21) and selected cell clones that failed to induce endogenous Puma (Fig. 5A) upon p16INK4A expression. Cells carrying a control short hairpin vector (CEM/p16-shCtr) or shPuma clones (CEM/p16-shPuma clones 3 and 6) were cultured in the presence or absence of 250 ng/ml Dox and then treated with/without 10 nm Dex. Mitochondrial and cytoplasmic extracts were analyzed for the content of Puma, BAX, Cox4, and tubulin. In CEM/p16-shPuma cells, the accumulation of Puma was attenuated in the mitochondrial fraction upon p16INK4A induction (Fig. 5B, right panels). In both CEM/p16-shCtr and CEM/p16-shPuma cells, the activation of p16INK4A and the concomitant Puma accumulation were associated with an increase of BAX in the membrane fraction. However, the amount of BAX in Puma knockdown cells was reduced compared with controls suggesting that Puma steady state levels might directly affect accumulation of BAX at the mitochondria (Fig. 5B). Like in CEM/p16 cells expressing dnBID (Fig. 2B), BCL2, or MCL1 (Fig. 4B), the expression of Puma-shRNAs significantly reduced FAS-induced cell death after 4 h in presence of CH11 antibody (p < 0.01) as shown in Fig. 5C. This indicates that the induction of Puma in response to p16INK4A expression is critical for increased death receptor-induced apoptosis in this leukemia model. Similarly, the GC-sensitizing effect of p16INK4A was reduced by Puma gene knockdown but not to the same extent as FAS-induced apoptosis. This was in concordance with the observation that transgenic dnBID (Fig. 2C) and BCL2 and MCL1 (Fig. 4C) exerted a similar effect when treated with GC. The combined data demonstrate that the increased sensitivity to FAS- and GC-induced apoptosis observed after re-expression of p16INK4A in leukemia cells is controlled by a concerted change in the expression of BCL2, MCL1, and Puma.
FIGURE 5.
Knockdown of Puma by shRNA prevents sensitization to FAS- and Dex-induced apoptosis. A, CEM-6E2/p16 cells were infected with pMSCV-shPuma or the empty control vector (shCtr). Single cell clones were isolated by limiting dilution from bulk-selected CEM/p16-shPuma cells. To determine the knockdown of endogenous Puma, CEM/p16-shPuma cells were cultured in the absence or presence of 250 ng/ml Dox for 24 h, and lysates were analyzed by immunoblot. Two clones that induced p16INK4A but not endogenous Puma were selected. Equal protein loading was confirmed by α-tubulin staining. B, mitochondrial (mito) and cytoplasmic (cyto) extracts were prepared from CEM/p16-shCtr and CEM/p16-shPuma cells (clone 3) that were cultured in absence or presence of 250 ng/ml Dox for 24 h and treated with 10 nm Dex for another 40 h. The distribution of Puma, BAX, Cox4, and α-tubulin in subcellular fractions was detected by immunoblot. C and D, CEM/p16-shCtr and the shPuma-expressing clones CEM/p16-shPuma clone 3 and 6 were cultured with 250 ng/ml Dox for 24 h and then treated with 0.1 μg/ml anti-FAS antibody for 4 h or with 10 nm Dex for 24 h, respectively. Apoptosis induction was measured by PI-FACS analysis. Shown is the mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01 for CEM/p16-shPuma clones treated either with +CH11 + Dox or +Dex + Dox compared with identically treated CEM/p16-shCtr control cells.
DISCUSSION
In this study we show that p16INK4A-sensitized leukemia cells to both FAS- and GC-induced cell death, suggesting that survival/death regulators that are critical for both extrinsic and intrinsic apoptosis pathways might be affected by p16INK4A expression. GC-induced apoptosis has been shown to be regulated by the BCL2 rheostat (30) but can also be modulated by components of the extrinsic pathway (27), and FAS-triggered apoptosis can be integrated and amplified via the mitochondria. Because death receptor and BH3-only protein-triggered death pathways converge at the level of mitochondria in CEM ALL cells (31), we sought to exclude death receptor signaling as an apoptosis-sensitizing cause in p16INK4A-expressing leukemia cells. Therefore, inducing the formation of the death-inducing signaling complex and the cross-talk between the extrinsic and the intrinsic apoptosis pathways were prevented by retroviral expression of dnFADD and dnBID. Expression of dnFADD inhibited FAS- but not GC-induced apoptosis, which demonstrates that FADD serves as an essential adaptor for the recruitment of caspase-8/-10. However, dnFADD did not affect GC-induced cell death excluding the hypothesis that elevated death receptor signaling contributes to apoptosis sensitization in p16INK4A-expressing leukemia cells. dnBID, in contrast, reduced GC apoptosis suggesting that BID cleavage, presumably via an amplification loop that involves mitochondria and caspase-8/-10, modulates GC sensitivity of CEM/p16 leukemia cells. This protective effect of dnBID showed that both FAS- and GC-induced apoptosis pathways converge at mitochondria and implied that components of the BCL2 rheostat as critical death regulators are regulated by p16INK4A expression.
Immunoblot analyses revealed that BCL2, MCL1, and Noxa were repressed, whereas Puma accumulated in p16INK4A transgenic cells (Fig. 3A and supplemental Fig. 3S). The molecular basis for these regulations is currently unclear. E2F1 has been demonstrated to induce the transcription of Noxa, Bim, and Puma in NIH3T3 cells (32). We have previously shown that p16INK4A reconstitution results in retinoblastoma protein and p130 activation and the repression of E2F targets in CEM leukemia cells (25). However, only Noxa might fit into the scheme of an E2F1 target as it was repressed within 24 h of p16INK4A expression. In contrast, Bim steady state levels were not affected, whereas Puma mRNA and protein was strongly induced by p16INK4A suggesting an E2F-independent regulation. Puma was originally identified as a direct transcriptional target of p53 (33–35) mediating at least part of the pro-apoptotic effects of p53 in hematopoietic cells (36). Similar to p53, the p53-related proteins TP63/p63 and TP73/p73 also bind to the Puma promoter. Although p53 is mutated and nonfunctional in CEM leukemia cells (37), p63 and/or p73 may well contribute to the induction of Puma in response to p16INK4A expression. However, an activation of p63 or p73 by p16INK4A or forced G0/G1 arrest has not been reported to date. The transcription factor FOXO3/FKHRL1 was recently identified as a transcriptional upstream regulator of Puma (38). We tested this hypothesis by expressing a tamoxifen-inducible FKHRL1 allele in CEM cells and found strong induction of Bim but no effect on Puma (data not shown). This excludes FKHRL1 as a p16INK4A-activated upstream regulator of Puma induction. The exact mechanism how Puma is induced by p16INK4A in leukemic T-cells therefore remains unclear.
Despite being a potent pro-apoptotic BH3-only protein, Puma accumulated at the mitochondria without triggering apoptosis per se suggesting that pro-survival BCL2 members neutralize excess of Puma. The simultaneous decrease in Noxa expression might free some pro-survival capacity and thereby prevent spontaneous cell death. Nevertheless, the strong induction of Puma seems to suffice to reduce the capacity of the BCL2 rheostat to cope with additional death signals. In support, when BCL2 or MCL1 are overexpressed or Puma induction is prevented by shRNA expression, the pro-survival capacity is restored as shown in Fig. 4, B and C, and Fig. 5, C and D. Therefore, the induction of Puma in p16INK4A-expressing cells appears to be critical for sensitizing leukemia cells to FAS- and GC-induced apoptosis.
Further studies will have to address whether the induction of Puma and the changes in steady state expression of BCL2 and MCL1 are directly caused by forced cell cycle arrest in G0/G1 or are the result of a senescent program initiated by p16INK4A in leukemia cells. The observation that p16INK4A similar to its alternative reading frame product p14ARF causes induction of pro-apoptotic Puma in p53-deficient leukemia cells suggests that loss of the INK4A gene locus during leukemia development confers resistance to certain apoptotic stimuli.
Supplementary Material
Acknowledgments
We thank S. W. Lowe and J. Borst for donating vectors and C. Kitzbichler and M. Wille for excellent technical assistance.
This work was supported by grants from “Kinderkrebshilfe Tirol and Vorarlberg,” “Provita Kinderleukämie Stiftung,” “Kinderkrebshilfe Südtirol-Regenbogen,” “SVP-Frauen,” “Medizinischer Forschungsfond,” “Südtiroler Krebshilfe,” Austrian Science Fund SFB-F021, and the COMET Center ONCOTYROL. “The Tyrolean Cancer Research Institute is supported by the “Tiroler Landeskrankenanstalten Ges.m.b.H. Tiroler Landeskrankenanstalten Ges.m.b.H.,” Tyrolean Cancer Society, and the Department of Health-Care, Autonomy of South Tyrol.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1S–3S.
- T-ALL
- T-cell acute lymphoblastic leukemia
- ALL
- acute lymphoblastic leukemia
- GC
- glucocorticoid
- Dex
- dexamethasone
- Dox
- doxycycline
- FACS
- fluorescence-activated cell sorter
- dn
- dominant-negative
- RT
- reverse transcription
- PI
- propidium iodide
- shRNA
- short hairpin RNA
- BH
- BCL2-homology
- FAS
- tumor necrosis factor receptor superfamily 6
- FADD
- FAS-associated death domain.
REFERENCES
- 1.Serrano M., Hannon G. J., Beach D. (1993) Nature 366, 704–707 [DOI] [PubMed] [Google Scholar]
- 2.Quelle D. E., Zindy F., Ashmun R. A., Sherr C. J. (1995) Cell 83, 993–1000 [DOI] [PubMed] [Google Scholar]
- 3.Kees U. R., Burton P. R., Lü C., Baker D. L. (1997) Blood 89, 4161–4166 [PubMed] [Google Scholar]
- 4.Fizzotti M., Cimino G., Pisegna S., Alimena G., Quartarone C., Mandelli F., Pelicci P. G., Lo Coco F. (1995) Blood 85, 2685–2690 [PubMed] [Google Scholar]
- 5.Okuda T., Shurtleff S. A., Valentine M. B., Raimondi S. C., Head D. R., Behm F., Curcio-Brint A. M., Liu Q., Pui C. H., Sherr C. J., et al. (1995) Blood 85, 2321–2330 [PubMed] [Google Scholar]
- 6.Mimeault M., Batra S. K. (2009) Ageing Res. Rev. 8, 94–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherr C. J., Roberts J. M. (1999) Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 8.Ekholm S. V., Reed S. I. (2000) Curr. Opin. Cell Biol. 12, 676–684 [DOI] [PubMed] [Google Scholar]
- 9.Classon M., Harlow E. (2002) Nat. Rev. Cancer 2, 910–917 [DOI] [PubMed] [Google Scholar]
- 10.Borner C. (2003) Mol. Immunol. 39, 615–647 [DOI] [PubMed] [Google Scholar]
- 11.Strasser A. (2005) Nat. Rev. Immunol. 5, 189–200 [DOI] [PubMed] [Google Scholar]
- 12.Han J., Goldstein L. A., Gastman B. R., Rabinowich H. (2006) J. Biol. Chem. 281, 10153–10163 [DOI] [PubMed] [Google Scholar]
- 13.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 14.Labi V., Erlacher M., Kiessling S., Villunger A. (2006) Cell Death Differ. 13, 1325–1338 [DOI] [PubMed] [Google Scholar]
- 15.Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 16.Löffler M., Ausserlechner M. J., Tonko M., Hartmann B. L., Bernhard D., Geley S., Helmberg A., Kofler R. (1999) Oncogene 18, 4626–4631 [DOI] [PubMed] [Google Scholar]
- 17.Ausserlechner M. J., Obexer P., Wiegers G. J., Hartmann B. L., Geley S., Kofler R. (2001) J. Biol. Chem. 276, 10984–10989 [DOI] [PubMed] [Google Scholar]
- 18.Pear W. S., Nolan G. P., Scott M. L., Baltimore D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parson W., Kirchebner R., Mühlmann R., Renner K., Kofler A., Schmidt S., Kofler R. (2005) FASEB J. 19, 434–436 [DOI] [PubMed] [Google Scholar]
- 20.Obexer P., Geiger K., Ambros P. F., Meister B., Ausserlechner M. J. (2007) Cell Death Differ. 14, 534–547 [DOI] [PubMed] [Google Scholar]
- 21.Hemann M. T., Zilfou J. T., Zhao Z., Burgess D. J., Hannon G. J., Lowe S. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9333–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner A. B., de Vries E., Tait S. W., Bontjer I., Borst J. (2002) J. Biol. Chem. 277, 40760–40767 [DOI] [PubMed] [Google Scholar]
- 23.Ausserlechner M. J., Obexer P., Deutschmann A., Geiger K., Kofler R. (2006) Mol. Cancer Ther. 5, 1927–1934 [DOI] [PubMed] [Google Scholar]
- 24.Nicoletti I., Migliorati G., Pagliacci M. C., Grignani F., Riccardi C. (1991) J. Immunol. Methods 139, 271–279 [DOI] [PubMed] [Google Scholar]
- 25.Ausserlechner M. J., Obexer P., Geley S., Kofler R. (2005) Leukemia 19, 1051–1057 [DOI] [PubMed] [Google Scholar]
- 26.Distelhorst C. W. (2002) Cell Death Differ. 9, 6–19 [DOI] [PubMed] [Google Scholar]
- 27.Marchetti M. C., Di Marco B., Cifone G., Migliorati G., Riccardi C. (2003) Blood 101, 585–593 [DOI] [PubMed] [Google Scholar]
- 28.Yin X. M., Wang K., Gross A., Zhao Y., Zinkel S., Klocke B., Roth K. A., Korsmeyer S. J. (1999) Nature 400, 886–891 [DOI] [PubMed] [Google Scholar]
- 29.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 30.Hartmann B. L., Geley S., Löffler M., Hattmannstorfer R., Strasser-Wozak E. M., Auer B., Kofler R. (1999) Oncogene 18, 713–719 [DOI] [PubMed] [Google Scholar]
- 31.Scaffidi C., Fulda S., Srinivasan A., Friesen C., Li F., Tomaselli K. J., Debatin K. M., Krammer P. H., Peter M. E. (1998) EMBO J. 17, 1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hershko T., Ginsberg D. (2004) J. Biol. Chem. 279, 8627–8634 [DOI] [PubMed] [Google Scholar]
- 33.Han J., Flemington C., Houghton A. B., Gu Z., Zambetti G. P., Lutz R. J., Zhu L., Chittenden T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11318–11323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano K., Vousden K. H. (2001) Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 35.Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001) Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]
- 36.Villunger A., Michalak E. M., Coultas L., Müllauer F., Böck G., Ausserlechner M. J., Adams J. M., Strasser A. (2003) Science 302, 1036–1038 [DOI] [PubMed] [Google Scholar]
- 37.Cheng J., Haas M. (1990) Mol. Cell. Biol. 10, 5502–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You H., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T. W. (2006) J. Exp. Med. 203, 1657–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.