Abstract
Benzophenone metabolism provides a number of plant natural products with fascinating chemical structures and intriguing pharmacological activities. Formation of the carbon skeleton of benzophenone derivatives from benzoyl-CoA and three molecules of malonyl-CoA is catalyzed by benzophenone synthase (BPS), a member of the superfamily of type III polyketide synthases. A point mutation in the active site cavity (T135L) transformed BPS into a functional phenylpyrone synthase (PPS). The dramatic change in both substrate and product specificities of BPS was rationalized by homology modeling. The mutation may open a new pocket that accommodates the phenyl moiety of the triketide intermediate but limits polyketide elongation to two reactions, resulting in phenylpyrone formation. 3-Hydroxybenzoyl-CoA is the second best starter molecule for BPS but a poor substrate for PPS. The aryl moiety of the triketide intermediate may be trapped in the new pocket by hydrogen bond formation with the backbone, thereby acting as an inhibitor. PPS is a promising biotechnological tool for manipulating benzoate-primed biosynthetic pathways to produce novel compounds.
Introduction
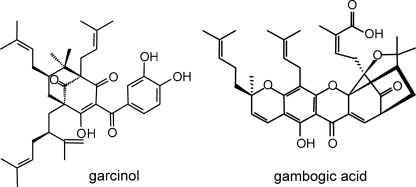
Benzophenone derivatives constitute a class of plant secondary metabolites, which includes a number of chemically complex and pharmacologically active constituents. Bridged polycyclic compounds result from the stepwise prenylation of the benzophenone nucleus and concomitant intramolecular cyclization reactions of the attached C5 and C10 isoprenoid units (1). To date, no chemical synthesis has been reported for any of these caged molecules. A well studied compound is garcinol (Fig. 1), which exhibits anti-tumoral and anti-inflammatory activities (2, 3). Polyprenylated benzoyl and acylphloroglucinol derivatives are widely distributed in the plant family Clusiaceae (= Guttiferae) where they serve as floral UV pigments and defense compounds against herbivores (4). The class of benzophenone derivatives also includes xanthones, which are the products of regioselective oxidative phenol coupling reactions catalyzed by cytochrome P450 enzymes (5). An example of a bridged polycyclic xanthone is gambogic acid (Fig. 1), which induces apoptosis independently of the cell cycle by a novel mechanism of caspase activation (6). Active benzoylphloroglucinol derivatives may be potential lead compounds for designing new drugs.
FIGURE 1.
Examples of polyprenylated polycyclic benzophenone derivatives with caged skeletons.
The carbon skeleton of benzophenone derivatives is formed by benzophenone synthase (BPS).4 The enzyme from Hypericum androsaemum (Clusiaceae) catalyzes the iterative condensation of benzoyl-CoA with three molecules of malonyl-CoA to give a linear tetraketide intermediate, which is subsequently cyclized into 2,4,6-trihydroxybenzophenone via intramolecular Claisen condensation (7). In contrast, BPS from Centaurium erythraea (Gentianaceae) prefers 3-hydroxybenzoyl-CoA as a starter substrate and catalyzes the formation of 2,3′,4,6-tetrahydroxybenzophenone (8). These two BPS products are the precursors of all prenylated benzoylphloroglucinols and xanthones.
BPS is a type III polyketide synthase (PKS) (7). This superfamily of enzymes generates an array of secondary metabolites in various plants and microorganisms (9). The most common type III PKS in higher plants is chalcone synthase (CHS), which initiates flavonoid biosynthesis (10). A number of functionally divergent plant PKSs arose via gene duplication and diversification (11). They differ from CHS in their preference for starter substrates (aliphatic or aromatic units), the number of acetyl additions catalyzed (one to seven), and the mechanism of ring formation used to cyclize linear polyketide intermediates (Claisen condensation, aldol condensation, or heterocyclic lactone formation) (9). Conversely, type III PKSs share a homodimeric architecture, a similar subunit molecular mass, and a conserved polyketide extension mechanism. Furthermore, each type III PKS monomer utilizes a Cys-His-Asn catalytic triad within a buried active site cavity, as first revealed in the crystal structure of alfalfa CHS (12). A narrow CoA-binding tunnel provides access to this large bi-lobed internal cavity termed the initiation/elongation cavity. One lobe of this catalytic center forms the starter unit binding pocket and the other accommodates the growing polyketide chain (13).
The aim of this work was to diversify BPS activity by introducing mutations into the residues lining the active site cavity of the enzyme from H. androsaemum. A point mutation was found to alter both substrate and product specificities of BPS without decreasing the catalytic efficiency. This dramatic functional variation was rationalized by homology modeling. The BPS mutant, which was termed phenylpyrone synthase (PPS), has so far not been found as a naturally occurring type III PKS variation. Interestingly, derivatives of the PPS product are highly potent anti-human immunodeficiency virus agents (14).
EXPERIMENTAL PROCEDURES
Chemicals
2,4,6-Trihydroxybenzophenone and 6-phenyl-4-hydroxy-2H-pyran-2-one were purchased from ICN (Meckenheim, Germany) and Aurora Fine Chemicals (Graz, Austria). 2,3′,4,6-Tetrahydroxybenzophenone was synthesized as described previously (5). Benzoyl-CoA, malonyl-CoA, and acetyl-CoA were obtained from Sigma. 2-Hydroxybenzoyl-CoA, 3-hydroxybenzoyl-CoA, 4-hydroxybenzoyl-CoA, cinnamoyl-CoA, 2-coumaroyl-CoA, 3-coumaroyl-CoA, and 4-coumaroyl-CoA were synthesized as described previously (8, 15).
Protein Expression and Affinity Purification
Wild-type BPS and enzyme mutants were heterologously expressed as N-terminally His6-tagged proteins in Escherichia coli and purified on a nickel-nitrilotriacetic acid affinity matrix, as described previously (16).
Enzyme Assays, Determination of Kinetic Constants, and Inhibition Experiments
Enzyme assays (250 μl) contained 15 μm starter CoA ester, 65 μm malonyl-CoA, 0.1 m potassium phosphate buffer, pH 7.0, and 2–5 μg of protein. After incubation at 35 °C for 20 min, the reaction mixture was acidified by addition of acetic acid (5%) and extracted twice with 250 μl of ethyl acetate. The combined organic phase was dried under vacuum, and the residue was dissolved in 50 μl of methanol (50%). Kinetic data were determined, as described previously (7). Inhibition experiments included a 5-min preincubation with varying concentrations of 3-hydroxybenzoyl-CoA and acetyl-CoA (2.7–21.6 μm) in the presence of malonyl-CoA (21.7 μm). Control assays contained only protein or protein and malonyl-CoA. After preincubation, benzoyl-CoA (7.8 μm) was added, and the formation of phenylpyrone was determined over a 20-min incubation period.
Mutagenesis
Site-directed mutants were generated using the QuikChange (Stratagene) protocol and verified by DNA sequencing.
Analytical Methods
HPLC-DAD analysis of the products from enzyme assays was carried out as described previously (16), except for the following modifications: gradient from 50 to 90% (v/v) methanol in water over 20 min; column Hypersil Gold (5 μm, 4.6 × 150 mm; Thermo). GC-MS was performed using two configurations. Configuration I has been described previously (16). For configuration II, a Hewlett-Packard 6890N gas chromatograph was equipped with a 30-m × 0.32-mm (film thickness 0.25 μm) analytical column (ZB-5, Agilent, Waldbronn, Germany). The capillary column was directly coupled to a GCMate II mass spectrometer (Jeol, Japan). The conditions applied were as follows: injector and transfer line were set at 260 and 280 °C, respectively; the GC oven program used was 70 °C for 3 min, increased by 10 °C/min to a final temperature of 310 °C for 3 min; the injection volume was 1 μl; the split ratio was varied from 1:2 to 1:20, depending on the concentration of the analytes; the constant carrier gas flow was 1.5 ml min−1 helium; the mass spectra were recorded at 70 eV.
The identity of individual compounds was confirmed by comparison of the MS data with spectra in the NIST mass spectral library version 2.0a and with reference compounds. The retention index (RI) on ZB-1 and ZB-5 columns was calculated with the aid of a set of hydrocarbons (even numbered C8 to C28) by linear interpolation for 2,4,6-tri-(trimethylsiloxy)-benzophenone as follows: RI (ZB-1) 2204, RI (ZB-5) 2250; GC-EI-MS (70 eV), m/z (%), 446 (4) [M+], 433 (16), 432 (35), 431 (100), 369 (6), 359 (4), 269 (4), 208 (5), 147 (7), 135 (4), 105 (29), 77 (9), 74 (4), 73 (55), and 45 (9); 2,3′,4,6-tetra-(trimethylsiloxy)-benzophenone as follows: RI (ZB-5) 2436; GC-EI-MS (70 eV)/ m/z (%)/ 534 (9) [M+], 523 (3), 522 (8), 521 (26), 520 (48), 519 (100), 369 (7), 357 (3), 193 (19), 165 (3), 147 (4), 74 (3), 73 (43), and 45 (4); 6-phenyl-4-(trimethylsiloxy)-2H-pyran-2-one as follows: RI (ZB-1) 2065, RI (ZB-5) 2135; GC-EI-MS (70 eV), m/z (%), 260 (91) [M+], 245 (24), 233 (18), 232 (100), 217 (4), 203 (7), 189 (12), 158 (10), 145 (4), 115 (6), 105 (33), 99 (5), 77 (21), 75 (8), 73 (30), and 45 (6); and 6-(3′-trimethylsiloxyphenyl)-4-(trimethylsiloxy)-2H-pyran-2-one as follows: RI (ZB-5) 2453; GC-EI-MS (70 eV), m/z (%), 348 (77) [M+], 334 (7), 333 (28), 322 (11), 321 (29), 320 (100), 305 (8), 246 (5), 193 (18), 159 (10), 99 (4), 75 (9), 74 (7), 73 (79), and 45 (8).
Molecular Modeling
A homology model of H. androsaemum BPS was constructed based on the crystal structure of alfalfa (Medicago sativa) CHS2 (12) complexed with resveratrol (Protein Data Bank code 1CGZ) using the Swiss-Model server (17, 18). The conserved CoA-binding sites and catalytic residues were correctly aligned, which indicates that the generated model is reliable. Modeling of protein-substrate complexes was performed using the program MOLOC and the implemented united atom force field MAB (19). The CoA molecule was placed according to the reference structure of alfalfa CHS2 (12) complexed with malonyl-CoA (Protein Data Bank code 1CML). Models for the full elongation cycle, including the hemithioketal covalently bound to Cys-167, were constructed based on the known catalytic mechanism of type III PKSs. All generated models were initially minimized and subsequently relaxed by molecular dynamics simulations for 1 ns at 300 K. We used the MAB force field (20) with implicit solvation and a hydrogen bond weight of 1.786. Modeling of point mutants was performed on the basis of the previously constructed model of wild-type BPS to investigate the perturbation introduced by the mutations in the active site cavity.
RESULTS
Generation of BPS Mutants
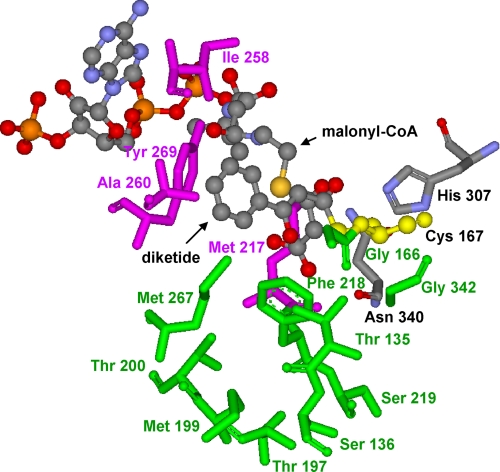
Based on the crystal structure of M. sativa CHS2 complexed with resveratrol (12), a homology model of BPS was generated using the Swiss-Model server. CHS and BPS share 59.1% amino acid sequence identity (7). The sequential polyketide intermediates of the phlorbenzophenone biosynthesis reaction from monoketide through tetraketide were modeled into the BPS active site cavity as Cys-167-tethered polyketides using the program Moloc (Fig. 2). Cys-167 serves as the attachment site of the growing polyketide chain, as verified by mutation of the cysteine residue into an alanine, resulting in enzyme inactivation (7). Benzoyl-CoA and malonyl-CoA as the starter and extender substrates, respectively, and phlorbenzophenone as the BPS product were also modeled into the catalytic center.
FIGURE 2.
CHS-based homology model of the BPS active site cavity complexed with both the benzoyl-primed diketide that is covalently bound to the catalytic cysteine 167 and malonyl-CoA that is used for the next chain elongation reaction. Amino acids of the initiation and elongation pockets are highlighted in pink and green, respectively. The catalytic triad is displayed in yellow (Cys-167) and gray (His-307 and Asn-340).
Residues of the BPS initiation/elongation cavity were altered by site-directed mutagenesis (supplemental Table S1). The resultant enzyme mutants were expressed in E. coli as N-terminally His6-tagged proteins and purified by affinity chromatography on a nickel-nitrilotriacetic acid matrix. The isolated proteins migrated on an SDS-polyacrylamide gel with a monomeric molecular mass of 43 kDa. The enzymes were assayed for changes in substrate and product specificities by HPLC-DAD analysis of their products using an array of aromatic CoA-linked starter substrates and acetyl-CoA in combination with malonyl-CoA as an extender.
A first series of mutations aimed at the replacement of BPS active site residues with the corresponding CHS active site residues. Seven amino acids are different (Table 1). None of the seven single mutants generated accepted CoA thioesters of cinnamic acids, such as 4-coumaroyl-CoA and cinnamoyl-CoA. The majority of the subsequently created double to 5-fold mutants were either inactive or inhibited the growth of their host E. coli cells. The remaining active mutants functionally resembled the wild-type enzyme. Thus, the functional conversion of BPS into CHS failed. Previously, H. androsaemum CHS was transformed by triple mutation into an enzyme with a preference for benzoyl-CoA over 4-coumaroyl-CoA. However, the catalytic efficiency of the triple mutant was 10-fold lower than that of wild-type BPS (7).
TABLE 1.
Corresponding active site cavity residues of crystallized alfalfa CHS2 (12) and H. androsaemumBPS
| CHS M. sativa | BPS H. androsaemum |
|---|---|
| Initiation pocket | |
| Leucine 214 | Methionine 217 |
| Isoleucine 254 | Isoleucine 258 |
| Glycine 256 | Alanine 260 |
| Phenylalanine 265 | Tyrosine 269 |
| Elongation pocket | |
| Threonine 132 | Threonine 135 |
| Serine 133 | Serine 136 |
| Threonine 194 | Threonine 197 |
| Valine 196 | Methionine 199 |
| Threonine 197 | Threonine 200 |
| Glycine 216 | Serine 219 |
| Leucine 263 | Methionine 267 |
| Serine 338 | Glycine 342 |
| Catalytic triad | |
| Cysteine 164 | Cysteine 167 |
| Histidine 303 | Histidine 307 |
| Asparagine 336 | Asparagine 340 |
In further rounds of mutations, BPS active site residues were replaced with amino acids present at the corresponding positions in type III PKSs other than CHS, such as acridone synthase and 2-pyrone synthase (2PS), which prefer bulkier (N-methylanthraniloyl) and smaller (acetyl) starter units, respectively (21, 22). In addition, a number of mutations were created on the basis of the CHS-based homology model of the BPS active site cavity complexed with the sequential reaction intermediates. However, the resulting enzyme mutants were either inactive, did not produce any soluble protein, or functionally resembled the wild-type enzyme, albeit with reduced catalytic activity. The only dramatic change in the functional behavior of BPS was observed with the substitution of the Thr-135 residue for a leucine. A threonine residue is also present in CHS and 2PS at the corresponding position, whereas acridone synthase has a serine.
Functional Characterization of the T135L Mutant
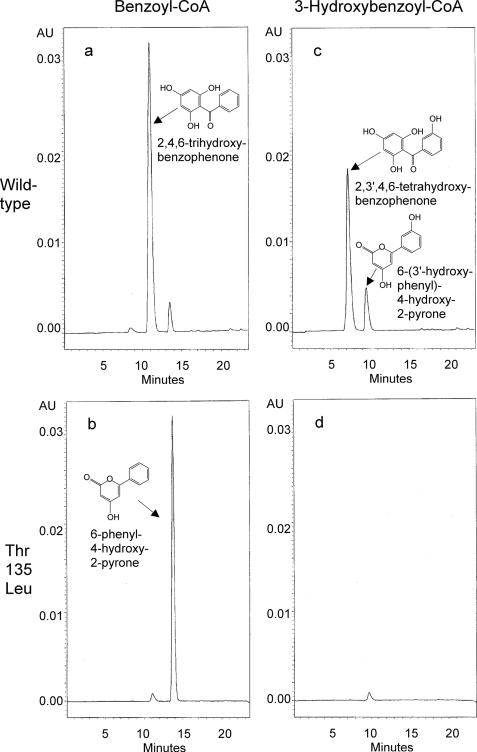
Wild-type BPS formed 2,4,6-trihydroxybenzophenone as a major product when incubated with benzoyl-CoA and malonyl-CoA, along with small amounts of 6-phenyl-4-hydroxy-2-pyrone (phenylpyrone) and traces of benzoyltriacetic acid lactone as derailment products (Fig. 3a). In contrast, the T135L mutant formed phenylpyrone as a major product and only traces of 2,4,6-trihydroxybenzophenone (Fig. 3b). The products were identified by GC-MS after silylation in comparison with authentic reference compounds.
FIGURE 3.
HPLC analysis of the products formed by wild-type BPS (a and c) and the T135L mutant (b and d) with benzoyl-CoA (a and b) and 3-hydroxybenzoyl-CoA (c and d) as starter substrates. AU, absorption units.
Kinetic characterization of the wild-type enzyme and the T135L mutant demonstrated that the catalytic efficiencies (kcat/Km) for benzoyl-CoA were similar (Table 2). The 8-fold lower turnover number (kcat) of the T135L mutant was compensated for by an 8-fold improvement in Km. Both enzymes exhibited pH and temperature optima of 7.5 and 35–40 °C, respectively. Previously, wild-type BPS was expressed as a glutathione S-transferase fusion protein that had a 4.5-fold higher catalytic efficiency for benzoyl-CoA (kcat/Km = 28,345 m−1 s−1, kcat = 9.66 min−1, Km = 5.7 μm) compared with the His6-tagged protein used in this study (7). This remarkable discrepancy, mainly in turnover rate, was confirmed by careful reinvestigation of the kinetic properties of glutathione S-transferase-BPS and might be attributed to the effect of the different affinity tags on either protein stability or protein determination.
TABLE 2.
Steady-state kinetic parameters for wild-type BPS and the T135L mutant
| Benzoyl-CoA |
Malonyl-CoA |
||||
|---|---|---|---|---|---|
| kcat | Km | kcat/Km | Km | ||
| min−1 | μm | m−1s−1 | μm | ||
| Wild-type BPS | 3.31 | 8.6 | 6414 | 31.3 | |
| T135L mutant | 0.41 | 1.0 | 6833 | 8.7 | |
Wild-type BPS also accepted 3-hydroxybenzoyl-CoA as a starter molecule and formed 2,3′,4,6-tetrahydroxybenzophenone as a major product and 6-(3′-hydroxyphenyl)-4-hydroxy-2-pyrone (hydroxyphenylpyrone) as a side product in the presence of malonyl-CoA (Fig. 3c). In contrast, the T135L mutant was almost inactive with 3-hydroxybenzoyl-CoA (Fig. 3d). It formed only trace amounts of hydroxyphenylpyrone, and 2,3′,4,6-tetrahydroxybenzophenone was not observed. The wild-type enzyme and the T135L mutant were both inactive with 2- and 4-hydroxybenzoyl-CoA, acetyl-CoA, and the CoA thioesters of cinnamic acids (Table 3).
TABLE 3.
Substrate specificities of wild-type BPS and the T135L mutant
| Substrate | Enzyme activity |
|
|---|---|---|
| Wild-type BPS | T135L mutant | |
| % of maximum each | ||
| Benzoyl-CoA | 100 | 100 |
| 3-Hydroxybenzoyl-CoA | 57 | 4 |
| 2-Hydroxybenzoyl-CoA | 0 | 0 |
| 4-Hydroxybenzoyl-CoA | 0 | 0 |
| Cinnamoyl-CoA | 0 | 0 |
| 2-Coumaroyl-CoA | 0 | 0 |
| 3-Coumaroyl-CoA | 0 | 0 |
| 4-Coumaroyl-CoA | 0 | 0 |
| Acetyl-CoA | 0 | 0 |
Interestingly, the replacement of the Thr-135 residue with a glycine, alanine, valine, isoleucine, asparagine, and tyrosine, instead of a leucine, gave catalytically inactive proteins. SDS-PAGE demonstrated that all of these mutant proteins were successfully expressed in E. coli and were highly purified by affinity chromatography. The mutants T135S and T135F functionally resembled the wild-type enzyme, albeit with reduced catalytic activities.
Rationalization of the Functional Behavior of the T135L Mutant by Homology Modeling
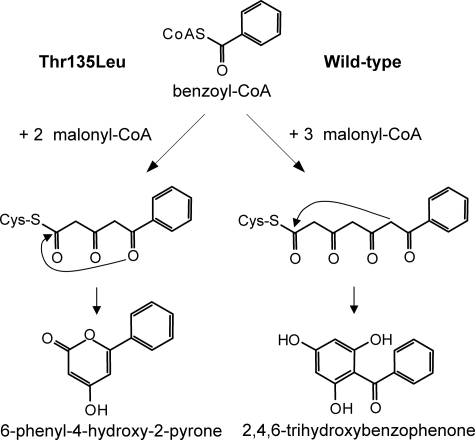
Wild-type BPS catalyzes the iterative condensation of benzoyl-CoA with three malonyl-CoAs to give a linear tetraketide that undergoes intramolecular C-6 → C-1 Claisen condensation to yield phlorbenzophenone (Fig. 4). In contrast, the T135L mutant catalyzes the addition of only two acetyl groups to the benzoyl starter unit. The triketide is the final linear intermediate and cyclizes into phenylpyrone via C-5 keto-enol oxygen → C-1 lactonization. These alternative reaction mechanisms were rationalized by homology modeling. Up to the diketide stage (Fig. 2), no differences in the accommodation of the reaction intermediates in the initiation/elongation cavity were observed between the wild-type enzyme and the T135L mutant. At the triketide stage, however, there were dramatic changes in the guidance and the conformation of the polyketide chain.
FIGURE 4.
Postulated reaction mechanisms of wild-type BPS and the T135L mutant. The mutant enzyme catalyzes the condensation of benzoyl-CoA with two malonyl-CoAs to give an intermediate triketide, which cyclizes into phenylpyrone via intramolecular C-5 keto-enol oxygen → C-1 lactonization. The wild-type enzyme adds three acetyl units from malonyl-CoA to form an intermediate tetraketide, which cyclizes into phlorbenzophenone via intramolecular C-6 → C-1 Claisen condensation.
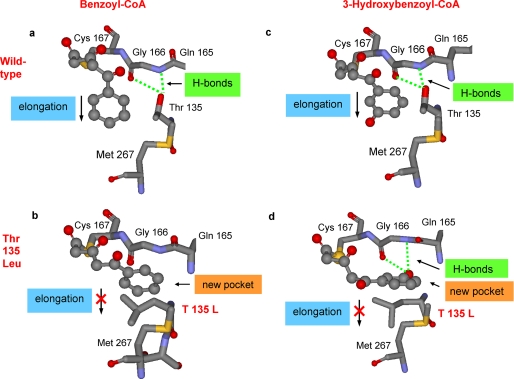
In wild-type BPS, the hydroxyl group of the Thr-135 residue forms hydrogen bonds with the polar backbone groups (carbonyl oxygen and amide hydrogen) of Gly-166 (Fig. 5a). The side chain of the threonine residue does not hinder the extension of the polyketide chain into the large elongation cavity that can readily accommodate the ultimate tetraketide. In the T135L mutant, however, the bulky and lipophilic side chain of the introduced leucine residue protrudes into the elongation cavity and may block the path of the growing polyketide chain (Fig. 5b). Instead, the T135L substitution opens a new pocket, the entrance of which is blocked in the wild-type enzyme by hydrogen bond formation between the threonine side chain and the backbone. Because of the interaction of the lipophilic side chain of the introduced leucine residue with the phenyl group of the growing polyketide chain, the triketide in the active site cavity of the T135L mutant may be redirected into the new pocket. The model suggests that the volume of the new cavity is 168 Å3, which is sufficient to accommodate a phenyl group (144 Å3). However, it does not allow for a third acetyl addition to the growing polyketide chain and the accommodation of the resultant tetraketide. Instead, the triketide undergoes heterocyclic lactone formation to give phenylpyrone. The total solvent-accessible volume of the catalytic cavity of wild-type BPS is 1264 Å3, which is increased to 1337 Å3 in the T135L mutant by opening of the new pocket. This counterintuitive volume increase is a consequence of the structural reorientation of the slightly longer Leu-135 side chain, observed in the molecular dynamics studies. Leucine cannot form a hydrogen bond toward the backbone carbonyl and amide groups of Gly-166, and it has no substituent in the β position. Thus, it can open access to a small extra cavity while hindering access to the large elongation pocket. In our modeling study, isoleucine and valine did not show this behavior, as corroborated by the results of the mutation experiments.
FIGURE 5.
CHS-based homology models of the active site cavities of wild-type BPS (a and c) and the T135L mutant (b and d). The benzoyl-primed (a and b) and 3-hydroxybenzoyl-primed triketides (c and d) are covalently attached to the catalytic cysteine 167. The wild-type enzyme catalyzes another acetyl addition to the intermediate triketides. In the mutant enzyme, however, the triketides may be redirected into a new pocket where no further chain elongation takes place.
When 3-hydroxybenzoyl-CoA serves as a starter substrate, the wild-type enzyme catalyzes the stepwise addition of three acetyl groups to the starter unit inside the large initiation/elongation cavity to give a tetraketide, which cyclizes into 2,3′,4,6-tetrahydroxybenzophenone (Fig. 5c). In the active site cavity of the T135L mutant, however, no tetraketide is formed. The aryl moiety of the triketide intermediate may enter the new pocket whose small size does not allow for a third chain extension reaction (Fig. 5d). Interestingly, the 3-hydroxyl group of the aryl moiety of the triketide intermediate resides in the position that is occupied in the wild-type enzyme by the hydroxyl group of the Thr-135 side chain. As a consequence, the 3-hydroxyl group of the triketide now forms hydrogen bonds with the backbone carbonyl and amide groups of Gly-166. Thus, the 3-hydroxybenzoyl-primed triketide may be trapped in the new pocket of the active site cavity of the T135L mutant. It acts as an inhibitor, explaining why the mutant enzyme is almost inactive with 3-hydroxybenzoyl-CoA as a starter substrate.
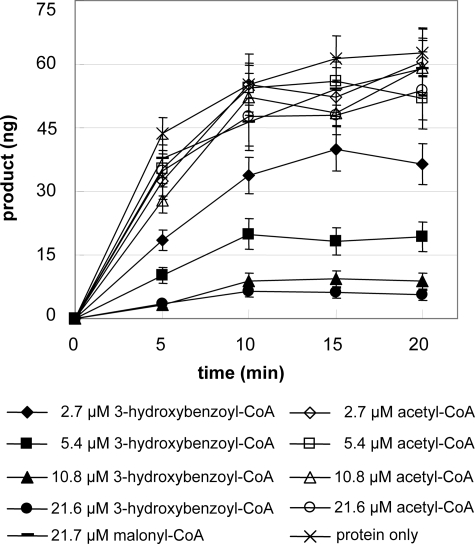
This conclusion was supported by inhibition experiments. The 3-hydroxybenzoyl-primed triketide as an inhibitor was formed by preincubation of the T135L mutant with malonyl-CoA and varying concentrations of 3-hydroxybenzoyl-CoA. After addition of benzoyl-CoA following preincubation, decreasing amounts of phenylpyrone correlated with increasing concentrations of 3-hydroxybenzoyl-CoA in preincubation (Fig. 6). In contrast, similar increases in phenylpyrone formation were observed after preincubation of the mutant enzyme with malonyl-CoA and increasing concentrations of acetyl-CoA, which is not accepted as a starter substrate, and malonyl-CoA only.
FIGURE 6.
Phenylpyrone formation by the T135L mutant after preincubation with malonyl-CoA and increasing concentrations of 3-hydroxybenzoyl-CoA or acetyl-CoA. Control assays contained only protein or protein and malonyl-CoA in preincubation.
DISCUSSION
The functional behavior of BPS was dramatically altered by a single amino acid substitution in the active site cavity, which transformed BPS into PPS. The point mutation modulated between the formation of a tetraketide product derived from Claisen condensation and a triketide product derived from lactone formation. Although a number of functionally diverse type III PKSs have been detected and characterized (9), an enzyme accomplishing a complete derailment of the BPS reaction has not yet been identified. A small proportion of the BPS reaction intermediates on the path to benzophenone become permanently derailed, leading to the formation of phenylpyrone as a side product. Increasing amounts of this truncated reaction product were found with increasing dithiothreitol concentrations in enzyme assays (7). Similar lactones as by-products of in vitro reactions are also formed by other type III PKSs (9).
The condensation of benzoyl-CoA, which is a rare starter substrate for PKSs (23), with two acetyl units from malonyl-CoA is also catalyzed by 2PS from Gerbera hybrida (22). However, the physiological starter molecule for this enzyme is acetyl-CoA, resulting in the formation of 6-methyl-4-hydroxy-2-pyrone (methylpyrone), which is converted by downstream enzymes to gerberin and parasorboside. Unlike 2PS, PPS and BPS are inactive with acetyl-CoA, nor do they catalyze malonyl-CoA decarboxylation to generate acetyl-CoA, which can serve as starter substrate for methylpyrone formation (22). Acetyl-CoA is a central intermediate of primary metabolism, whereas benzoyl-CoA is a component of secondary metabolism. Benzoic acid biosynthesis branches from cinnamic acid from the general phenylpropanoid pathway (24).
Replacement of the polar residue Thr-135 of BPS with the apolar amino acid leucine in PPS may open the gate to a buried pocket, into which the triketide intermediate is redirected during the elongation process of the diketide. Compared with the initiation/elongation cavity of BPS, the size of the newly accessible pocket in PPS is smaller and does not allow for a third extension step with malonyl-CoA, thereby preventing the formation of a tetraketide. Kinetic characterization of BPS and PPS demonstrated that the T135L mutation increased the affinity for the starter substrate, benzoyl-CoA, but decreased the turnover number to the same extent, resulting in similar catalytic efficiencies (kcat/Km). In stilbene synthase (STS), the Thr-135 residue (which corresponds to Thr-132 in M. sativa CHS) is involved in the transition from the Claisen cyclization mechanism to the aldol cyclization mechanism (25). Displacement of the Thr-135 side chain hydroxyl group is critical for the STS intramolecular aldol cyclization specificity mediated by a thioesterase-like hydrogen bonding network termed the aldol switch.
Single residues in the catalytic center of type III PKSs were previously found to determine polyketide chain length and product specificity. In Aloe arborescens pentaketide chromone synthase (PCS), a point mutation, M207G (Met-207 corresponds to Thr-197 in M. sativa CHS), opens the gate to two hidden pockets behind the catalytic center and expands the volume of this cavity, thereby converting PCS into octaketide synthase (26). The M207G substitution occurs naturally in an octaketide synthase from the same plant species, and a G207M mutation converted this enzyme into pentaketide synthase (27). Both enzymes use malonyl-CoA as a sole substrate. The steric bulk of residue 207 also controlled the polyketide chain length in heptaketide-producing aloesone synthase (28). When the corresponding residue of BPS (Thr-200) was replaced with a glycine, the resulting mutant was inactive. In contrast, the mutant containing an alanine (T200A) functionally resembled the wild-type enzyme, albeit with reduced catalytic activity. In M. sativa CHS, mutation of the threonine residue at this position into a leucine (T197L) caused a derailment of the regiospecific Claisen condensation reaction of the tetraketide intermediate, resulting in the exclusive formation of 4-coumaroyltriacetic acid lactone (13). The corresponding mutant of BPS (T200L) was functionally similar to the wild-type enzyme, and the mutants T200C and T200F were inactive. Recently, the polyketide elongation tunnel of the M207G mutant of PCS was further widened by the simultaneous replacement of the aromatic residues Phe-80 and Tyr-82 at the bottom of the new pocket with an alanine residue each (29). The resulting triple mutant formed low yields of a novel nonaketide naphthopyrone that is the longest polyketide chain formed by a type III PKS.
Further results that functionally link the size of the active site cavity to chain length determination were obtained with M. sativa CHS. The side chain volume of position 256 influenced both polyketide chain-length determination and intramolecular cyclization of the polyketide intermediates (30). Although the G256A and G256V mutations altered the regiospecific cyclization reaction, leading to increased coumaroyltriacetic acid lactone formation, the G256L and G256F mutations resulted in a derailment of the reaction at the triketide step, with bis-noryangonin and methylpyrone being released as products. When the corresponding residue of BPS (Ala-260) was replaced with valine, the phlorbenzophenone:phenylpyrone ratio was shifted from 1:0.08 (wild-type) to 1:0.39 (A260V). The A260L mutant was inactive with the starter substrates used.
The T135L mutation not only changed the product specificity but also the substrate specificity of BPS. 3-Hydroxybenzoyl-CoA is the second best starter substrate for wild-type BPS but a poor starter molecule for PPS. In this enzyme mutant, the 3-hydroxybenzoyl-primed triketide may be trapped in the new pocket by hydrogen bonds of the 3-hydroxyl group with the backbone, thereby acting as an inhibitor. Preincubation of the T135L mutant with malonyl-CoA and increasing 3-hydroxybenzoyl-CoA concentrations resulted in decreasing rates of phenylpyrone formation from benzoyl-CoA and malonyl-CoA. So far, changes in starter molecule selectivity of type III PKSs were attributed to alterations in the volume of the initiation/elongation cavity caused by specific amino acid substitutions. A combination of three mutations (T197L/G256L/S338I) converted M. sativa CHS into 2PS by reducing the volume of the active site cavity from 923 to 274 Å3 (13). The constricted catalytic centers of 2PS and the functionally identical CHS triple mutant sterically excluded the bulky 4-coumaroyl starter unit but allowed access of the smaller acetyl unit. When the same amino acid substitutions were introduced at the corresponding positions of BPS (T200L, G342I, and A260L), the resulting triple mutant was inactive with both benzoyl-CoA and acetyl-CoA, nor did the three single mutants use acetyl-CoA to form methylpyrone. Conversely, in M. sativa CHS the mutation of Phe-215, which is a gatekeeper to the active site, into a serine widened the active site entrance, allowing for productive binding of N-methylanthraniloyl-CoA, which is too bulky to be accepted as a starter substrate by wild-type CHS (31). Contrary to acridone synthase that naturally uses N-methylanthraniloyl-CoA as starter, the CHS F215S mutant did not catalyze the regiospecific Claisen cyclization reaction, resulting in release of N-methylanthraniloyltriacetic acid lactone as a novel alkaloid.
Derivatives of the PPS product are known as natural products. A 4′-hydroxylated phenylpyrone named psilotinin was found in the pteridophyte Psilotum nudum, which is the most primitive vascular plant (32). cDNAs encoding CHS, STS, and valerophenone synthase were cloned from this species; however, a PPS cDNA was not identified (33). Feeding experiments suggested that psilotinin is formed by condensation of 4-coumaroyl-CoA with one molecule of malonyl-CoA followed by lactonization (34). The same one-step condensation is catalyzed by benzalacetone synthase that, however, decarboxylates the diketide intermediate (35). Recently, phenylpyrones have also been isolated from some Aloe species (36, 37). It will be interesting to isolate a PPS cDNA and to compare the structural and kinetic properties of the naturally occurring enzyme optimized by evolution with the T135L mutant created in vitro. PPS is a promising biotechnological tool for manipulating benzoate-primed biosynthetic pathways, such as benzophenone and biphenyl biosyntheses in Clusiaceae and the Maloideae, respectively. The latter rosaceous subfamily includes economically important fruit trees, such as apple and pear. Metabolically engineered plants expressing PPS might produce new natural products as either constitutive metabolites or pathogen-induced phytoalexins.
Supplementary Material
Acknowledgment
We thank Dr. E. J. Stauber for linguistic help.
This work was supported by the Priority Programme “Evolution of Metabolic Diversity” (SPP 1152) of the Deutsche Forschungsgemeinschaft.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- BPS
- benzophenone synthase
- CHS
- chalcone synthase
- GC-EI-MS
- gas chromatography-electron ionization-mass spectrometry
- HPLC-DAD
- high performance liquid chromatography-diode array detection
- PCS
- pentaketide chromone synthase
- PKS
- polyketide synthase
- PPS
- phenylpyrone synthase
- 2PS
- 2-pyrone synthase
- STS
- stilbene synthase
- RI
- retention index.
REFERENCES
- 1.Hu L. H., Sim K. Y. (2000) Tetrahedron 56, 1379–1386 [Google Scholar]
- 2.Yoshida K., Tanaka T., Hirose Y., Yamaguchi F., Kohno H., Toida M., Hara A., Sugie S., Shibata T., Mori H. (2005) Cancer Lett. 221, 29–39 [DOI] [PubMed] [Google Scholar]
- 3.Hong J., Sang S., Park H. J., Kwon S. J., Suh N., Huang M. T., Ho C. T., Yang C. S. (2006) Carcinogenesis 27, 278–286 [DOI] [PubMed] [Google Scholar]
- 4.Gronquist M., Bezzerides A., Attygalle A., Meinwald J., Eisner M., Eisner T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13745–13750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters S., Schmidt W., Beerhues L. (1998) Planta 204, 64–69 [Google Scholar]
- 6.Zhang H. Z., Kasibhatla S., Wang Y., Herich J., Guastella J., Tseng B., Drewe J., Cai S. X. (2004) Bioorg. Med. Chem. 12, 309–317 [DOI] [PubMed] [Google Scholar]
- 7.Liu B., Falkenstein-Paul H., Schmidt W., Beerhues L. (2003) Plant J. 34, 847–855 [DOI] [PubMed] [Google Scholar]
- 8.Beerhues L. (1996) FEBS Lett. 383, 264–266 [DOI] [PubMed] [Google Scholar]
- 9.Austin M. B., Noel J. P. (2003) Nat. Prod. Rep. 20, 79–110 [DOI] [PubMed] [Google Scholar]
- 10.Schröder J. (1999) in Comprehensive Natural Products Chemistry (Sankawa U. ed) Vol. 1, pp. 749–771, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 11.Helariutta Y., Kotilainen M., Elomaa P., Kalkkinen N., Bremer K., Teeri T. H., Albert V. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9033–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrer J. L., Jez J. M., Bowman M. E., Dixon R. A., Noel J. P. (1999) Nat. Struct. Biol. 6, 775–784 [DOI] [PubMed] [Google Scholar]
- 13.Jez J. M., Austin M. B., Ferrer J., Bowman M. E., Schröder J., Noel J. P. (2000) Chem. Biol. 7, 919–930 [DOI] [PubMed] [Google Scholar]
- 14.Vara Prasad J. V. N., Lunney E. A., Ferguson D., Tummino P. J., Rubin J. R., Reyner E. L., Stewart B. H., Guttendorf R. J., Domagala J. M., Suvorov L. I., Gulnik S. V., Topol I. A., Bhat T. N., Erickson J. W. (1995) J. Am. Chem. Soc. 117, 11070–11074 [Google Scholar]
- 15.Abd El-Mawla A. M., Schmidt W., Beerhues L. (2001) Planta 212, 288–293 [DOI] [PubMed] [Google Scholar]
- 16.Liu B., Raeth T., Beuerle T., Beerhues L. (2007) Planta 225, 1495–1503 [DOI] [PubMed] [Google Scholar]
- 17.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 18.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 19.Gerber P. R., Müller K. (1995) J. Comput. Aided Mol. Des. 9, 251–268 [DOI] [PubMed] [Google Scholar]
- 20.Gerber P. R. (1998) J. Comput. Aided Mol. Des. 12, 37–51 [DOI] [PubMed] [Google Scholar]
- 21.Junghanns K. T., Kneusel R. E., Baumert A., Maier W., Gröger D., Matern U. (1995) Plant Mol. Biol. 27, 681–692 [DOI] [PubMed] [Google Scholar]
- 22.Eckermann S., Schröder G., Schmidt J., Strack D., Edrada R. A., Helariutta Y., Elomaa P., Kotilainen M., Kilpelainen I., Proksch P., Teeri T. H., Schröder J. (1998) Nature 396, 387–390 [Google Scholar]
- 23.Moore B. S., Hertweck C. (2002) Nat. Prod. Rep. 19, 70–99 [DOI] [PubMed] [Google Scholar]
- 24.Abd El-Mawla A. M., Beerhues L. (2002) Planta 214, 727–733 [DOI] [PubMed] [Google Scholar]
- 25.Austin M. B., Bowman M. E., Ferrer J. L., Schröder J., Noel J. P. (2004) Chem. Biol. 11, 1179–1194 [DOI] [PubMed] [Google Scholar]
- 26.Morita H., Kondo S., Oguro S., Noguchi H., Sugio S., Abe I., Kohno T. (2007) Chem. Biol. 14, 359–369 [DOI] [PubMed] [Google Scholar]
- 27.Abe I., Oguro S., Utsumi Y., Sano Y., Noguchi H. (2005) J. Am. Chem. Soc. 127, 12709–12716 [DOI] [PubMed] [Google Scholar]
- 28.Abe I., Watanabe T., Lou W., Noguchi H. (2006) FEBS J. 273, 208–218 [DOI] [PubMed] [Google Scholar]
- 29.Abe I., Morita H., Oguro S., Noma H., Wanibuchi K., Kawahara N., Goda Y., Noguchi H., Kohno T. (2007) J. Am. Chem. Soc. 129, 5976–5980 [DOI] [PubMed] [Google Scholar]
- 30.Jez J. M., Bowman M. E., Noel J. P. (2001) Biochemistry 40, 14829–14838 [DOI] [PubMed] [Google Scholar]
- 31.Jez J. M., Bowman M. E., Noel J. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McInnes A. G., Yoshida S., Towers G. H. N. (1965) Tetrahedron 21, 2939–2946 [Google Scholar]
- 33.Yamazaki Y., Suh D. Y., Sitthithaworn W., Ishiguro K., Kobayashi Y., Shibuya M., Ebizuka Y., Sankawa U. (2001) Planta 214, 75–84 [DOI] [PubMed] [Google Scholar]
- 34.Leete E., Muir A., Towers G. H. N. (1982) Tetrahedron Lett. 23, 2635–2638 [Google Scholar]
- 35.Abe I., Takahashi Y., Morita H., Noguchi H. (2001) Eur. J. Biochem. 268, 3354–3359 [DOI] [PubMed] [Google Scholar]
- 36.Rebecca W., Kayser O., Hagels H., Zessin K. H., Madundo M., Gamba N. (2003) Phytochem. Anal. 14, 83–86 [DOI] [PubMed] [Google Scholar]
- 37.Durì L., Morelli C. F., Crippa S., Speranza G. (2004) Fitoterapia 75, 520–522 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.