Abstract
Notch signaling is controlled by ligand binding, which unfolds a negative control region to induce proteolytic cleavage of the receptor. First, a membrane-proximal cleavage is executed by a metalloprotease, removing the extracellular domain. This allows γ-secretase to execute a second cleavage within the Notch transmembrane domain, which releases the intracellular domain to enter the nucleus. Here we show that the ADAM10 metalloprotease Kuzbanian, but not ADAM17/tumor necrosis factor α-converting enzyme, plays an essential role in executing ligand-induced extracellular cleavage at site 2 (S2) in cells and localizes this step to the plasma membrane. Importantly, genetic or pharmacological inhibition of metalloproteases still allowed extracellular cleavage of Notch, indicating the presence of unknown proteases with the ability to cleave at S2. Gain of function mutations identified in human cancers and in model organisms that map to the negative control region alleviate the requirement for ligand binding for extracellular cleavage to occur. Because cancer-causing Notch1 mutations also depend on (rate-limiting) S2 proteolysis, the identity of these alternative proteases has important implications for understanding Notch activation in normal and cancer cells.
Introduction
The Notch signaling pathway plays multiple essential functions during metazoan development and in adult tissues where it controls homeostatic self-renewal, differentiation, proliferation, and apoptosis (1). Notch receptors are type I transmembrane glycoproteins that undergo furin cleavage at site 1 (S1)2 during transit to the cell surface. S1-cleaved Notch proteins accumulate at the plasma membrane as heterodimeric polypeptides composed of the Notch extracellular domain (NECD) and a transmembrane and intracellular domain held together by the heterodimerization domain (HD). In the absence of ligand, a negative regulatory region (NRR) composed of the globular HD domain and the overlaying Lin12/Notch repeats (LNR) prevents access of proteases and thus prevents activation of Notch (2–4). Ligand binding to Notch receptors unfolds the NRR permitting cleavage by a metalloprotease at a site close to the membrane (S2). This removes NECD (5) producing a short lived NH2-terminal fragment that becomes a substrate for the aspartyl protease presenilin, a component of the γ-secretase complex (6, 7). γ-Secretase executes an intramembrane cleavage at site 3 (S3), which releases the Notch intracellular domain (NICD). NICD translocates to the nucleus and mediates target gene transcription after it associates with the CSL protein (8) (Fig. 1A). S3 cleavage is essential for canonical Notch signaling in vivo (9–11).
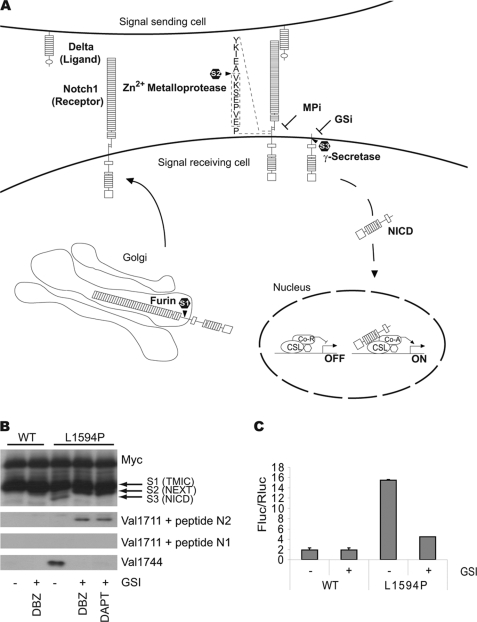
FIGURE 1.
A, diagram depicting S1, S2, and S3 cleavage steps leading to NICD production and activity; see text for details. Boxed area indicates immunization peptide sequence. B, immunoblot showing expression of wild type (WT) and T-ALL mutant (L1594P) LNR 6Myc proteins transfected in HEK293 cells. Upper panel, anti-Myc immunoblot showing equal expression levels of transfected constructs. S1, S2, and S3 cleavage products are indicated. Lower panel, Val1744 immunoblot for S3-cleaved Notch1 showing NICD formation in L1594P but not in the inactive wild type. NICD production and Val1744 staining is blocked by inhibition of γ-secretase by GSIs DAPT and dibenzazepine. Middle panels, accumulation of S2-cleaved Notch1 detected by Val1711 only seen upon GSI treatment and concomitant loss of Val1744/NICD in panel below. Val1711 only observed in active L1594P mutant and not in wild type. Note accumulation of S2-cleaved Notch1 fragments is also observed in Myc immunoblot for L1594P upon GSI treatment. Incubation with the immunization peptide (N1) prevents detection of S2-cleaved Notch1 by Val1711, and control peptide against hNotch2 (N2) does not block immunoreactivity. C, Notch-CSL transcription reporter assay in U2OS cells showing wild type LNR 6Myc compared with LNR L1594P 6Myc, which is 5-fold more active. Notch1 activity is attenuated using GSI. TMIC, transmembrane and intracellular domain; DBZ, dibenzazepine.
The rate-limiting step in Notch activation appears to be NRR unfolding (2, 3), which facilitates S2 cleavage directly or after HD dissociation at S1 (12). Several lines of evidence support this sequence of events. First, Notch proteins lacking NECD are constitutively cleaved by γ-secretase (5), causing cancer in mice and humans (13, 14). Second, receptors lacking only the epidermal growth factor repeats but containing the NRR are functionally inert. Third, NRR mutations relax autoinhibition acting as γ-secretase-dependent gain of function mutations (2, 10, 15–17). Fourth, missense mutations in the HD domain destabilize the NRR and occur at high frequency in human T-cell acute lymphocytic leukemia (T-ALL) (18). It has been well established that HD domain mutations cause ligand-independent S2 cleavage (5, 17); however, the identity of the enzymes involved is poorly understood.
Notch1 S2 cleavage is sensitive to the metalloprotease inhibitor 1,10-phenanthroline (5) and attenuated in cells lacking the ADAM17 protease tumor necrosis factor α-converting enzyme (TACE) (19). ADAMs or disintegrin and metalloproteases are membrane-bound zinc-dependent enzymes of the metzincin clan that have diverse roles in adhesion and proteolytic cleavage of numerous cell surface signaling molecules in normal homeostasis and disease (20). In vitro, TACE cleaves Notch1 just outside the transmembrane region between residues Ala1710 and Val1711 (19); the same site is used for cleavage of Notch1 in transfected cells (5). However, Tace-deficient mice or flies do not phenocopy the Notch1 phenotype suggesting that TACE is either redundant with another protease or is a tissue-specific Notch1 S2-protease in vivo (21, 22). In contrast, mice lacking Adam10 die at day 9.5 of embryogenesis with reduced neuronal Hes5 expression resembling Notch1-null embryos (24), and T-cell-specific deletion of Adam10 in vivo phenocopied the Notch1 null phenotype during thymocyte development (25). However, mouse embryonic fibroblasts lacking Adam10 have no apparent defect in ligand-independent Notch1 processing (5, 24). In contrast to this ambiguity in vertebrates, in flies the ADAM10 homolog Kuzbanian (Kuz) binds dNotch directly and is the major enzyme involved in Notch cleavage and signaling (22, 23).
Understanding the precise role of ADAM10 in Notch signaling has been further complicated by the fact that Kuz has also been reported to cleave Notch ligands in flies (26) and mammalian cells (27–29). Whereas in flies this task is shared with an ADAM10/Kuz homolog (Kuz-like or Kul) that is dedicated to cleavage of the Notch ligand Delta (30), no Kul homolog has been identified in mammals thus far. Therefore, the phenotypes attributed to ADAM10 loss in mammals could reflect compound phenotypes due to defects in the cleavage of Notch, Delta, or both.
Because the identity of enzyme(s) cleaving Notch1 at S2 remains controversial, we characterized Notch1 cleavage in ligand-dependent and -independent signaling and mapped the amino acids required for cleavage. We find that ADAM10, but not ADAM17/TACE, is essential for catalyzing ligand-induced S2 cleavage. This step occurs at the plasma membrane, suggestive of a similar localization for the subsequent cleavage by γ-secretase. Importantly, genetic or pharmacological inhibition of metalloproteases still permits S2 cleavage, indicating that multiple proteases have the ability to cleave Notch1. Our findings provide further insight into the mechanism of Notch1 activation in normal and cancer cells. Elucidating the proteolytic machinery leading to Notch1 activation is important because inhibition of this rate-limiting step using targeted drugs may offer novel treatment options in Notch1-related diseases.
EXPERIMENTAL PROCEDURES
Plasmids and Vectors
All mNotch1 plasmids were initially cloned into pCS2+6Myc as described (16). Notch1 LNR and Notch1 ΔE (supplemental Fig. 1A) were constructed as described previously. Notch1 Δice (supplemental Fig. 1B) is made by deleting part of the NICD and inserting a 6Myc tag after Gly1755 (31). Notch1 GV16 fusion proteins are made by inserting a Gal4VP16 lacking an internal ATG via a unique SgrAI site after the transmembrane domain. Notch1 mCherry fusions were made by PCR-directed cloning and removal of the 6Myc for replacement with the mCherry gene. Site-directed mutagenesis was performed using the QuikChange kit according to the manufacturer's instructions (Stratagene). All mutant constructs were sequence-verified. A construct containing 4xCSL synthetic binding sites in tandem (PJA23) was used for Notch transcription assays (kindly provided by D. Hayward). For Notch1 GV16 cleavage assays, an FR-Luc construct containing 5xGal4 DNA-binding sites (Stratagene) was used. For normalization of the luciferase assays, a cytomegalovirus-driven Renilla luciferase (Promega) was used. The LZRS Jagged1 construct was a gift from B. Blom. A plasmid expressing stable HIF1α was described earlier (32).
Antibodies
The Notch1 Val1711 antibody was generated by immunizing chickens with the peptide VKSEPVE. Total IgY was isolated from yolks using the IgY purification kit (Pierce), and the antibody was affinity-purified using the SulfoLink immobilization kit for peptides (Thermo Scientific). Eluted antibody was dialyzed in PBS. Antibodies used in Western analysis are as follows: Notch1 C-20 1:500 (Santa Cruz Biotechnology); Val1711 1:1000, Val1744 1:1000 (Cell Signaling); Myc 9E10 1:5000, Jag1 H-114 1:1000 (Santa Cruz Biotechnology); Dll1 C-20 1:1000 (Santa Cruz Biotechnology); and TACE 1:2000 (GeneTex).
Cell Lines
OP9-wt, OP9-Jag1, and OP9-Dll1 (33) (a gift from B. Blom AMC, Amsterdam, The Netherlands) were cultured in α-minimum Eagle's medium (Invitrogen) supplemented with 20% fetal calf serum and β-mercaptoethanol (100 μm). Adam10, Adam17, Adam9/12/15 wild type and knock-out cells (5, 21, 34), U2OS, HeLa, Phoenix, and HeLaN1 cells (35) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum and 100 μm β-mercaptoethanol. HeLaN1 cells were maintained in the same medium with G418 (800 μg/ml). HEK293 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 5% fetal calf serum. NIH-3T3 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% normal calf serum. Co-cultures were in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Transfections of HEK293, HeLa, and U2OS were performed using polyethyleneimine (Polysciences Inc.). NIH-3T3 were transfected by calcium phosphate using BBS buffer.
Western Blotting
Cells were lysed in RIPA buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris pH 8.0) for 20 min at 4 °C. Lysates were spun down for 15 min at 14,000 rpm at 4 °C. Supernatant was added to 2× Laemmli buffer. Samples were boiled for 5 min and loaded onto SDS-PAGE.
Immunoprecipitation
Cells were lysed in CoIP buffer (200 mm KCl, 25 mm Hepes pH 7.4, 20 mm NaF, 1% Nonidet P-40, and 0.2 mm EGTA, protease inhibitor mixture (Roche Applied Science)) for 20 min at 4 °C. DNA was sheared using a syringe and centrifuged for 15 min at 4 °C. Input samples were taken, and 2 μg of Notch1 C-20 antibody was added for 6 h at 4 °C to precipitate endogenous Notch1. Protein A-Sepharose 4B beads (Zymed Laboratories Inc.) were added overnight to retrieve antibody-bound protein. Samples were washed three times in lysis buffer, 3× PBS, and were taken in 2× Laemmli buffer with 50 mm dithiothreitol. Samples were run on SDS-PAGE, and proteins were detected by immunoblotting and ECL (Amersham Biosciences).
Chemicals
Drugs were added 24 h after transfection for 16 h. APMA (250 μm), PMA (50 ng/ml), E-64 (10 μm), α1-antitrypsin (3 μg/ml), chymostatin (50 μm), antipain (50 μg/ml), and Ser-Cys protease inhibitor mixture (P1860) were from Sigma. GM6001 (50 μm), TAPI-2 (100 μm), DAPT (1 μm), and β-secretase inhibitor (1 μg/ml) were from Calbiochem. GW280264 and GI254023 (10 μm) were gifts from A. Ludwig (Aachen, Germany). BB94 (10 μm) and dibenzazepine (0.2 μm) were from Syncom (Groningen, The Netherlands).
EDTA Stimulation
Cells were washed twice in 1× Hanks' balanced salt solution buffer (Invitrogen) and incubated in 1× Hanks' balanced salt solution buffer containing 5 mm EDTA for 15 min at 37 °C in the presence or absence of GSI. Buffer was replaced with medium and incubated for 60 min at 37 °C.
Immunohistochemistry
Cells were fixed with 4% paraformaldehyde for 10 min and incubated in 20 mm ammonium chloride for 10 min. Cells were blocked in NET-gel buffer (50 mm Tris pH 7.4, 150 mm NaCl, 5 mm EDTA, 0.05% Nonidet P-40, 0.25% gelatin), and Val1711 and Val1744 antibodies were incubated at 1:100 dilution in NET-gel buffer. Secondary donkey anti-chicken fluorescein isothiocyanate (Jackson ImmunoResearch) and swine anti-rabbit fluorescein isothiocyanate (Dako) were used at 1:100 in NET-gel buffer. Nuclei were stained with TO-PRO3. Slides were analyzed on a Leica SP2 CLSM.
Luciferase Assays
Cells were transfected with firefly-luciferase reporters containing 4xCSL-binding sites and cytomegalovirus promoter driven Renilla luciferase as a transfection control. S3 cleavage reporter assays were performed using Notch1 GV16 fusion constructs and a firefly-luciferase reporter pFR-luc containing 5xGal4 DNA-binding sites (Stratagene). Cytomegalovirus-Renilla luciferase was used as a transfection control, and signals are given as fold firefly/Renilla corrected for background.
Viral Transduction
Phoenix ecotropic receptor-expressing cells were transfected with LZRS-Jag1 using PEI. After 24 h medium was replaced, and 24 h later supernatant with Polybrene (1:1000; Sigma) was used to infect target cells, already pretreated with Polybrene (1:1000). The following day cells were washed, trypsinized, and used for co-culture experiments.
Surface Biotinylation
Transfected cells were washed with ice-cold PBS supplemented with 1 mm MgCl2 and 0.1 mm CaCl2 (PBS++) on ice and incubated with 0.5 mg/ml NHS-biotin (Pierce) for 20 min. Reaction was stopped by addition of 1 volume of 100 mm glycine/PBS++ and washed twice with 50 mm glycine/PBS++. Cells were lysed in RIPA buffer; DNA was sheared using a syringe, and the lysate was spun down at 14,000 rpm at 4 °C. After input, samples were taken, and streptavidin-coupled agarose beads (Pierce) were added to the supernatant and incubated for 90 min. Samples were washed three times in RIPA and three times in PBS, boiled in Laemmli buffer, and analyzed by PAGE.
RESULTS
Notch1 Cleavage in Vivo Occurs at Val1711
Using constitutively active Notch1 proteins, the S2 cleavage site was identified previously in cultured cells (5) and in vitro (19) to lie between residues Ala1710 and Val1711. To determine whether during Notch1 signaling NECD cleavage occurred at this position, we generated a peptide-specific antibody (αVal1711) against the seven NH2-terminal residues (NH2-VKSEPVE) exposed after cleavage of mouse Notch1 in cell-free assays.
The Val1711 antibody was first validated in HEK293 cells transiently transfected with a ligand-independent mouse Notch1 construct lacking the epidermal growth factor repeats and containing the HD gain of function mutation L1594P found in T-ALL (LNR; supplemental Fig. S1A) (18). Immunoblotting of transfected cell lysates using the S3 cleavage-specific antibody Val1744 indicated that L1594P mutation-containing proteins were cleaved by γ-secretase, whereas wild type LNR proteins were not. RBP-Jk/CSL-luciferase reporter assays confirmed that inserting the L1594P mutation resulted in ligand-independent activation (Fig. 1, B and C). Val1744 cleavage can be blocked by the γ-secretase inhibitors (GSI) dibenzazepine and DAPT, leading to loss of luciferase activity in cells expressing L1594P. SDS-PAGE analysis of lysates from GSI-treated cells followed by anti-Myc immunoblotting revealed accumulation of a fragment with lower mobility migrating just below the transmembrane and intracellular domain fragment, consistent with NEXT (the S2 cleavage product; Fig. 1B) (5). Probing these blots for Val1711 confirmed that cells expressing mutant (active) but not wild type (inactive) Notch1 LNR proteins contained the epitope detectable by αVal1711. Inclusion of the immunization peptide completely blocked Val1711 detection, whereas a control Notch2-derived peptide had no effect, establishing specificity. Importantly, this fragment was identified only when cells were cultured in the presence of GSIs, confirming the following: 1) that proteolysis at Val1711 precedes γ-secretase cleavage at Val1744, and 2) demonstrates a very short half-life for the S2-cleaved NEXT fragment suggesting S2 and S3 may occur in the same cellular compartment (5).
It is possible that mutant Notch1 molecules are cleaved at a different scissile bond than ligand-activated receptors. We therefore investigated whether ligand-dependent Notch1 signaling produced the Val1711 epitope by co-culturing HeLaN1 cells, stably expressing mNotch1 (35), with the OP9 bone marrow stromal cell line stably expressing Jagged1 or Delta1 (33). Ligand stimulation of HeLaN1 leads to S3 cleavage of Notch1, readily detected with αVal1744 (Fig. 2A). GSI treatment blocked ligand-induced transcriptional reporter activation and resulted in the accumulation of αVal1711-detectable S2 fragments in both Jagged1 and Dll1 stimulated cells with similar efficiency. Thus, ligand-induced Notch1 signaling leads to NECD cleavage at Val1711.
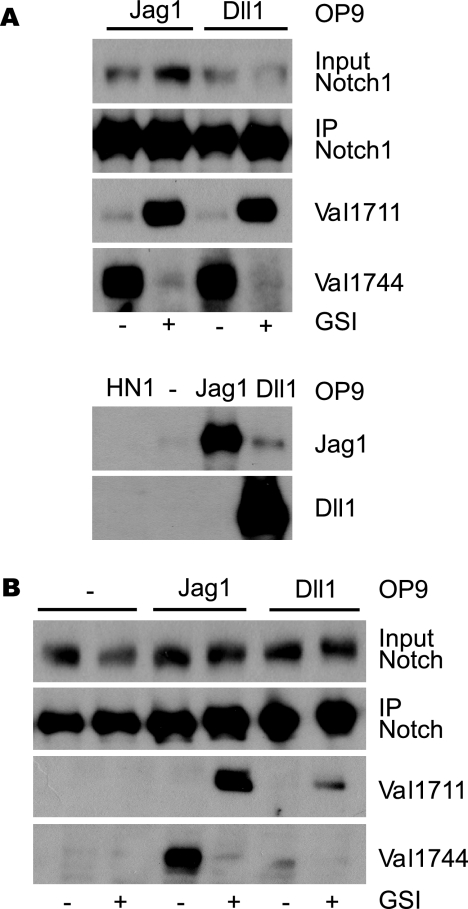
FIGURE 2.
Ligand-induced Notch1 signaling leads to Val1711 S2 cleavage. A, co-culture of wild type-, Jagged1- (Jag1), and Delta1 (Dll1)-expressing murine OP9 cells with mouse Notch1-expressing HeLaN1 (HN1). Immunoprecipitation (IP) for Notch1 shows Notch1 receptor expression. Immunoblot for Val1744 and Val1711 shows that both Jag1 and Dll1 induce NICD, which is blocked by GSIs. Both Dll1 and Jag1 induce S2 cleavage at Val1711 seen upon GSI treatment. Lower panel, expression of Jagged1 and Delta1 in OP9-transduced cells. No ligand expression is observed in HN1 cells. B, activation of endogenous Notch1 signaling by ligand stimulation in OP9 cells proceeds through Val1711 proteolysis. Immunoprecipitation shows OP9 ligand-expressing cells also express endogenous Notch1 receptor. OP9-Jag1 and OP9-Dll1 cultures undergo ligand-dependent Notch1 signaling to produce S2-cleaved Notch1 at Val1711. OP9 cells not expressing ligand do not activate Notch1 cleavage at Val1711 or Val1744.
During the course of these experiments, we found that OP9 cells express low levels of Notch1 that could be efficiently immunoprecipitated with Notch1 antibodies (Fig. 2B). We reasoned that OP9 ligand-expressing cells may therefore undergo endogenous ligand-dependent Notch1 signaling (36). Extracts from confluent OP9-Jagged1 cells contain Notch1 receptors cleaved at Val1744 (Fig. 2B). OP9-control cells expressed Notch1 that was not cleaved in the absence of ligand. Furthermore, upon GSI treatment, in both OP9-Jag1 and OP9-Dll1, but not in control OP9 cells, endogenous Notch1 receptors were cleaved at Val1711 (Fig. 2B). Thus, ligand stimulation of endogenous Notch1 also proceeds through consecutive cleavage at Val1711 and Val1744 in mammalian cells.
Next, we investigated if cleavage at Val1711 occurred in constitutively active Notch1 proteins harboring other HD mutations found in human T-ALL patients (17, 18) or in gain of function alleles identified in genetic screens in flies (10) and worms (15). These mutations were introduced into the inactive mouse N1LNR-6Myc expression construct and transfected in HEK293 cells (Fig. 3 and supplemental Fig. 2). All LNR mutant proteins were expressed at comparable levels, and all were furin-cleaved (albeit to a different extent) as shown by anti-Myc immunoblots from cell lysates (17). In the absence of HD mutations, LNR molecules were only weakly active (37). In contrast, LNR molecules containing HD mutations were more active in reporter assays and produced Val1744-cleaved NICD in a γ-secretase-dependent manner (Fig. 3, A and B, and supplemental Fig. 2, A and B). Upon GSI treatment, all mutants accumulate at least some αVal1711-reactive fragments. We found no clear correlation between transcriptional output and the extent of Val1711 cleavage in Notch1 LNR S1597N, suggesting cleavage may occur also at a different site not detected with our antibody.
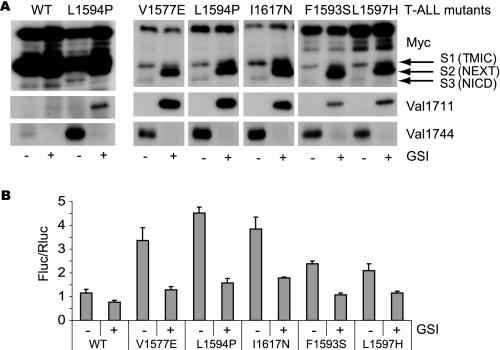
FIGURE 3.
T-ALL ligand-independent Notch1 LNR molecules are cleaved at Val1711. A, upper panel, T-ALL LNR 6Myc HD mutants transfected in HEK293 cells. S1, S2, and S3 cleavage products are indicated. Lower panel, Val1744 immunoblot shows all T-ALL mutants but not wild type (WT) LNR produce NICD, which is inhibited by GSI. Middle panel shows Val1711 cleavage in T-ALL mutants but not wild type upon GSI treatment. B, transcriptional reporter activity of LNR T-ALL mutants on CSL-fLuc reporter in HeLa cells. Shown is fold Firefly luciferase (Fluc) activity corrected for Renilla luciferase (Rluc) transfection of LNR mutants over background. Note that the less efficient S2 and S3 cleavage of F1593S and L1597H in A is also reflected in the reduced activity in the CSL-fluc reporter assay. Luciferase assays are representative of at least two independent experiments in triplicate; all mutants are significantly (p < 0.05) more active compared with wild type. TMIC, transmembrane and intracellular domain.
Collectively, these results demonstrate that endogenous Notch1 activation by either Jagged1 or Delta1 ligands triggers cleavage at Val1711, as do mutations in the NRR that lead to ligand-independent activity. S2-cleaved Notch1 proteins form excellent γ-secretase substrates and are thus rapidly converted to S3-cleaved molecules in mammalian cells. These data confirm that the αVal1711 antibody detects ligand-induced Notch1 cleavage in mammalian cells.
No Strict Requirement for Specific Amino Acids at the S2 Site
To further investigate if the amino acid composition as S2 was important for proteolysis, we inserted mutations at the P1-P1′ cleavage site S2. Notch1 proteins with a mutant S2 site (A1710V/V1711H) are not cleaved in vitro by TACE (19). To investigate if these mutations also impaired Notch1 signaling, we introduced them into full-length Notch1 molecules and transfected the mutant cDNA into HEK293 or U2OS cells (only HEK293 shown). We artificially induced heterodimer dissociation with EDTA (38) to induce Notch activity. After EDTA treatment, wild type Notch1 was cleaved at S3, and this step could be blocked by GSI (Fig. 4A). As seen with ligand stimulation (Fig. 2) (12), EDTA treatment induced S2 cleavage at Val1711 in GSI-treated cells (Fig. 4A). Unexpectedly, Notch1 carrying the AV → VH mutation was also S3-cleaved prior to EDTA stimulation, consistent with ligand-independent activation. Cleavage of the various Notch molecules was also tested in a sensitive assay based on replacement of NICD with a fusion protein between the Gal4-DNA binding domain and the VP16 transactivation domain (39). Upon γ-secretase cleavage, the GV16 is released from the membrane to activate luciferase transcription from a promoter containing artificial Gal4-binding sites. The Notch1-GV16 cleavage assay was used to evaluate protease activity (Fig. 4, B and C). Notch1 full-length molecules carrying the L1594P mutation were already active and cleaved at Val1711 and Val1744 in the absence of ligand or EDTA (Fig. 4A, lanes 9 and 10). Only a modest increase could be seen after EDTA addition (Fig. 4A, lanes 11 and 12). Notch1-GV16 containing AV → VH was active in the absence of a stimulus (Fig. 4, B and C), confirming ligand-independent cleavage. Interestingly, although neither cleavage nor activity of AV → VH could be further induced by EDTA (Fig. 4, A and B), ligand stimulation could further activate AV→ VH but not L1594P-containing molecules (Fig. 4C).
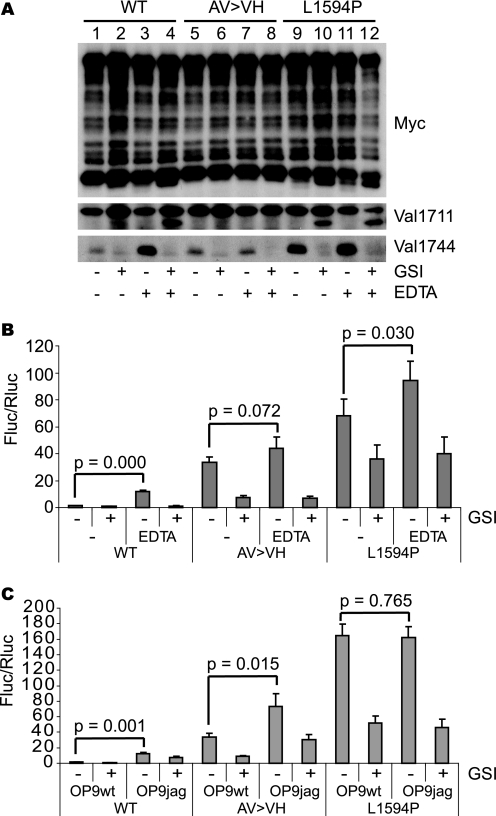
FIGURE 4.
Val1711 cleavage site mutation leads to ligand-independent Notch1 cleavage and activity. A, Myc immunoblot of HEK293 cell lysates transfected with wild type (WT), S2 cleavage mutant (AV → VH), and L1594P full-length Notch1 6Myc constructs. Receptor dissociation and cleavage stimulated by EDTA induce Val1744 and Val1711 cleavage in wild type and L1594P but not in AV → VH S2 cleavage mutant. L1594P and AV → VH mutants are already Val1744-cleaved in the absence of EDTA. Val1711 cleavage in AV → VH cannot be monitored because of mutation of epitope. B, Notch1-GV16 cleavage reporter assay in HeLa cells showing EDTA-induced cleavage of WT, AV → VH, and L1594P mutants. AV → VH and L1594P are already highly active in the absence of EDTA compared with wild type. C, Notch1-GV16 cleavage assay in HeLa cells co-cultured with OP9 wild type or Jagged1 cells showing that the basal activity of AV → VH mutant and L1594P T-ALL mutant in absence of ligand is severalfold higher than wild type. Whereas the activity of the AV → VH mutant can be stimulated with Jagged1, L1594P T-ALL is not further stimulated. Shown is fold Firefly luciferase (Fluc) activity corrected for Renilla luciferase (Rluc) transfection of Notch1 6Myc constructs over background. Figure is representative of at least two independent experiments in triplicate; p values are shown and calculated using a Student's t test.
Next, we introduced the AV → VH mutation into the truncated ligand-independent Notch1 molecules, LNR and ΔE (the latter lacking the NRR domain). In cells transfected with LNR AV → VH, basal RBP-Jk/CSL reporter activity and S3 cleavage were increased compared with wild type LNR (supplemental Fig. 2). In Notch1 ΔE, S2 cleavage still occurred at Val1711 (supplemental Fig. 3A). However, the AV → VH mutation had no effect on NICD production or transcriptional activity when placed in the context of Notch1 ΔE (supplemental Fig. 3, A and C). Notch1 Δice, a minimal Notch1 mutant in which the extracellular domain is missing and the entire intracellular domain is replaced by 6Myc epitope tags (31), was also cleaved at Val1711 (supplemental Fig. 3B) indicating that the minimal sequence requirements for S2 cleavage are contained within the 15 juxtamembrane amino acids encompassing Val1711 and possibly the Notch1 TMD. These results are consistent with a model in which S2 cleavage is not sequence-dependent; rather, the substitution V1711H may have destabilized the NRR leading to promiscuous cleavage. As seen with γ-secretase (40), the exact composition of the scissile bond at S2 is not an important determinant of cleavage.
Above, we speculated that the lack of correlation between transcriptional output and the extent of S2 cleavage in LNR S1597N could reflect cleavage away from S2. To examine this possibility, we generated the LNR mutant A1710E in which Val1711 remained intact allowing detection with the αVal1711 antibody to monitor cleavage between Glu1710 and Val1711. Like the AV → VH molecules, the AV→ EV mutant protein was furin-cleaved and constitutively S3-cleaved at Val1744 (supplemental Fig. 4). Whereas L1594P-transfected cells treated with GSI induced accumulation of S2-cleaved Notch1 detectable by αVal1711 and αMyc antibodies (see also Fig. 3A), AV → EV-transfected cells treated with GSI showed no accumulation of a NEXT fragment (detected with αMyc) or of the Val1711 epitope (supplemental Fig. 4). NEXT accumulation was not seen in AV → VH as well (immunoreactivity with Val1711 was not expected because the epitope is lacking).
Collectively, these data show that HD mutations enhance ligand-independent cleavage at a preferred site (Val1711), but when this site is mutated, proteases cleave other scissile bonds. Alternatively, γ-secretase cleavage occurs in the absence of S2 cleavage, perhaps because of dissociation of the heterodimerization domain (12).
S2 Proteolysis Requires ADAM10/Kuz
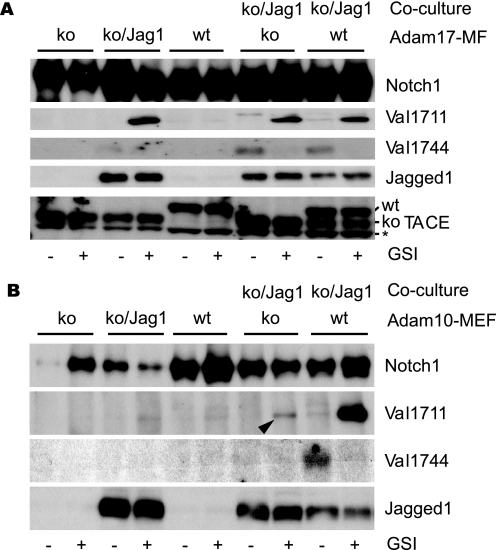
The ADAM metalloprotease ADAM17 (TACE) and ADAM10 (Kuz) have been implicated in ectodomain shedding of Notch1 in mammals and flies, respectively. To examine which protease is responsible for cleaving Notch1 at Val1711 upon ligand stimulation, we analyzed Notch1 receptor cleavage in cells lacking ADAM proteases. Adam17 knock-out mouse fibroblasts were co-cultured with Adam17 knock-out cells transduced with a Jagged1-expressing retrovirus. In the absence of Adam17, endogenous Notch1 activation by ligand was similar to wild type cells, and S2 cleavage occurred at Val1711 and was followed by S3 cleavage at Val1744 (Fig. 5A). Furthermore, ligand-induced Notch1 proteolysis proceeded normally in cells lacking Adam9, -12, and -15 (supplemental Fig. 5). In contrast, in cells lacking Adam10, S2 cleavage at Val1711 was dramatically reduced when compared with wild type controls, resulting in reduced S3 cleavage (Fig. 5B). As expected, Adam10-deficient, ligand-expressing cells were competent to induce Notch1 activation when co-cultured with wild type, Notch1-expressing cells. These results implicate ADAM10 as the main protease responsible for Notch1 cleavage following DSL ligand stimulation under physiological conditions. However, because residual S2 cleavage was still observed in the absence of ADAM10 (Fig. 5B, arrowhead), other protease(s) have the ability to cleave Notch1 at Val1711, albeit inefficiently, in Adam10-deficient cells.
FIGURE 5.
ADAM10 is essential for endogenous Notch1 Val1711 cleavage. A, Adam17-deficient cells are transduced with Jagged1 ligand and used in co-culture experiments to stimulate wild type (WT) and Adam17-deficient cells that express endogenous Notch1 receptor. Immunoblotting for Notch1, Val1711, and Val1744 shows no defect is observed in Notch1 cleavage in the absence of ADAM17. ko, knock-out. B, in contrast in cells lacking ADAM10-expressing endogenous Notch1 receptor, S2 and S3 cleavages are severely impaired compared with wild type cells. Jagged1 and TACE/ADAM17 expression is shown. Asterisk indicates nonspecific reaction of TACE antibody.
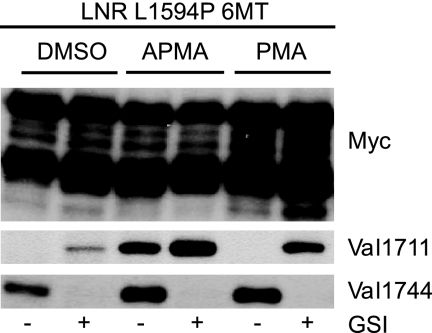
The constitutive activity of ADAM proteases in unstimulated cells can be further induced through activation of protein kinase C using phorbol ester PMA or the mercuric compound APMA (41, 42). We hypothesized that constitutive shedding of ligand-independent Notch1 can also be further induced by both compounds in cell-based assays. Both compounds enhanced shedding of LNR L1594P mutant molecules, which increased S3 cleavage (Fig. 6). Importantly, APMA dramatically enhanced detection of the Val1711 epitope even in the absence of GSI, demonstrating that NEXT accumulation is not an indirect effect of GSI and that local γ-secretase activity could be saturated in vivo by excess substrate (43).
FIGURE 6.
Constitutive and regulated cleavage of Notch1 at S2. HEK293 cells transfected with LNR L1594P 6Myc. Upper panel, Myc immunoblot. Lower panel shows that the phorbol ester PMA and mercuric compound APMA stimulate constitutive Val1711 and Val1744 cleavage compared with vehicle-treated cells. Note Val1711 cleavage is observed even in the absence of GSI with APMA, indicating Val1711 cleavage is not indirectly caused by GSI.
S2 Cleavage Occurs at the Cell Surface and Not in an Endocytic Organelle
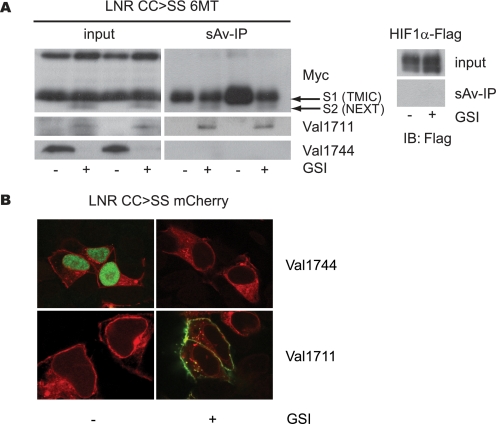
ADAM proteases are membrane-bound proteases thought to cleave substrates at the cell surface. To investigate where Val1711 cleavage occurred, we performed biotin labeling and streptavidin pulldown from LNR CC → SS (C1675S/C1682S)-transfected cells. This was efficiently enriched for transmembrane and intracellular domain and S2-Val1711 Notch1 fragments, the latter labeled on Lys1712 (Fig. 7A). Full-length Notch1 (present only in endoplasmic reticulum and early Golgi) and S3-cleaved Notch1 (intracellular) were not enriched by biotinylation. As an additional control, we show that the cytoplasmic protein HIF1α could not be precipitated after biotinylation, demonstrating that biotin did not pass the plasma membrane and label cellular proteins. Furthermore, S2-Val1711 enrichment was absent in non-biotin-treated cells. On Myc immunoblots of input lysates, uncleaved full-length Notch1 and furin-processed Notch1 were detected as well as Val1711 and Val1744 cleavage products. These results are consistent with S2 cleavage at the plasma membrane.
FIGURE 7.
Notch1 S2 cleavage occurs at the cell surface. A, upper panel, Myc immunoblot of surface-biotinylated and streptavidin-precipitated U2OS cells transfected with the active LNR CC → SS 6Myc. Left upper panel shows input, and right panel streptavidin pulldown (sAv-IP) of biotinylated cells. Lower panels show corresponding Val1711 and Val1744 immunoblots. Streptavidin pulldown demonstrates S2-cleaved fragments enriched at the cell surface compared with Val1744-cleaved Notch present in input but not detected by surface biotinylation. Streptavidin pulldown on U2OS cells transfected with FLAG-tagged HIF1α protein shows biotin did not label cytoplasmic proteins. B, U2OS cells transfected with LNR CC → SS mCherry are fixed and permeabilized and used for immunofluorescent staining for Val1711- and Val1744-cleaved Notch1 (green). Val1744 staining is only present in absence of GSI, whereas Val1711-cleaved molecules are only present in GSI-treated cells predominantly located at the cell membrane. Red fluorescence shows total Notch1 expression. Val1711-positive vesicular structures are observed near the cell surface. TMIC, transmembrane and intracellular domain.
Next, we used immunofluorescence to visualize the subcellular compartment where Notch1 cleavage occurred in cells transfected with constitutively active LNR CC → SS-tagged with monomeric Cherry (mCherry) fluorescent protein (supplemental Fig. 1B). In the absence of GSI, Val1744 cleavage led to nuclear accumulation of the Val1744 epitope in transfected cells (Fig. 7B), and no Val1711 staining was detected. Upon GSI inhibition, Val1744 nuclear staining and nuclear mCherry were lost, but Val1711 staining was gained decorating the plasma membrane in permeabilized cells. Although we observed co-localization of LNR CC → SS-mCherry and Val1711 staining in a few cytoplasmic vesicles in the presence of GSI, the abundance of cell surface staining suggests that internalization of S2-cleaved Notch1 was secondary to S2 cleavage and not its primary location. Similar results were obtained using L1594P, and no staining was observed using purified preimmune IgY antibodies (data not shown). Combined, these experiments show for the first time that S2 cleavage of Notch1 receptors occurs at the cell surface and strongly suggest that S3 cleavage follows in the same compartment.
In Molecules Activated by T-ALL Mutations, ADAM10 Is No Longer Required for S2
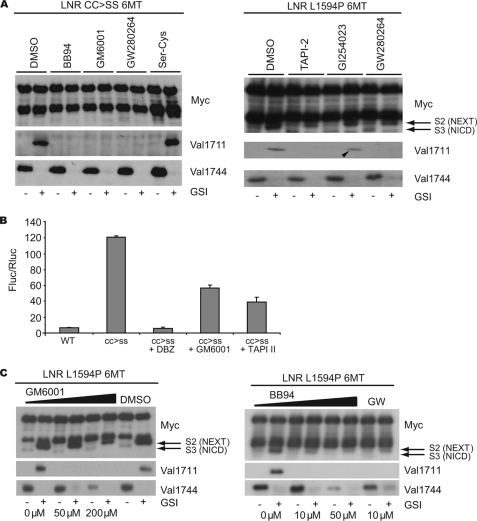
Above, we detected hints that multiple enzymes can cleave Notch1 molecules once the NRR is destabilized by T-ALL mutations. Next, we investigated whether hydroxamate-based metalloprotease inhibitors (MPi) could block the cleavage of cancer-causing Notch1 mutants. Constitutive shedding of LNR L1594P at Val1711 could be completely inhibited by the broad spectrum inhibitor GM6001 (Galardin) as well as the more ADAM-specific inhibitors BB94 (Batimastat) and TAPI-2 (IC-3), and the ADAM10/17-specific inhibitor GW280264X (44) (Fig. 8A). Unexpectedly, the reported ADAM10-selective inhibitor GI254023X (44) could not inhibit Notch1 processing (Fig. 8A, arrowhead). Similar results were obtained using LNR molecules carrying anti-neurogenic mutations found in flies (i.e. CC → SS, S1597N, A1695T; data not shown). Remarkably, although Val1711 cleavage was completely blocked by all hydroxamate inhibitors, this had little effect on NEXT production and thus subsequent Val1744 cleavage detected by αMyc immunoblot (Fig. 8A). Accordingly, transcriptional reporter assays displayed a modest 50% reduction in transcriptional output in the presence of GM6001 and TAPI-2 at concentrations sufficient for complete inhibition of Val1711 cleavage (Fig. 8B). Increasing the concentrations to achieve significant Notch inhibition by GM6001 (50–200 μm), BB94 (10–50 μm), and GW280264X (10–30 μm; data not shown) led to a clear inhibition of S3 cleavage (Fig. 8C), consistent with extracellular shedding at sites other than Val1711 by enzymes only weakly affected by these inhibitors. To identify which type of protease(s) display this activity, we tested a wide range of broad spectrum cysteine (E-64) and serine protease inhibitors (antipain, chymostatin, and α1-antitrypsin); none could inhibit Val1711 or Val1744 proteolysis (supplemental Fig. 6). The β-secretase inhibitor also did not affect Notch1 cleavage (supplemental Fig. 6).
FIGURE 8.
Metalloprotease inhibitors block Val1711 cleavage but not S2 cleavage. A, broad spectrum metalloprotease inhibitors and ADAM-specific inhibitors show complete inhibition of Val1711 cleavage but not of Val1744-NICD production in LNR L1594P 6Myc-transfected HEK293 cells. The ADAM10-specific inhibitor GI254023 shows minimal inhibition of Val1711 cleavage (arrowhead). Serine-cysteine protease inhibitor mixture (Ser-Cys) does not affect S2 or S3 cleavage. B, Notch1-GV16 reporter assay in transfected NIH-3T3 cells showing only partial inhibition of Notch1 S3 cleavage measured by GV16 release with MPi GM6001 and TAPI-2 on constitutively active CC → SS constructs. Shown is fold Firefly luciferase (Fluc) activity corrected for Renilla luciferase (Rluc) transfection of Notch1 wild type and CC → SS-GV16 constructs over background. C, increased concentrations of MPi lead to a reduction of Val1744-NICD production. Note in Myc immunoblots S2-like products still accumulate in GSI- and MPi-treated cells, indicating that S2 cleavage occurs elsewhere when Val1711 cleavage is blocked. TMIC, transmembrane and intracellular domain; DBZ, dibenzazepine.
DISCUSSION
Previously, we established that mammalian Notch1 undergoes two successive proteolytic cleavages upon ligand binding leading to release of NICD. First the extracellular domain of Notch1 is shed by a Zn+-dependent metalloprotease (5) followed by a presenilin-dependent γ-secretase cleavage at an intramembranous site (S3 at Val1744 (7)). Genetic analysis in flies has demonstrated an important role for Kuz upstream of γ-secretase activity at the level of S2 cleavage where Kuz has been shown to directly bind and cleave Notch (22, 23). Both flies and knock-out mice lacking Kuzbanian/ADAM10 resemble Notch1 phenotypes in a cell autonomous manner (22, 24, 25, 46, 47). Here we use a novel epitope-specific antibody recognizing the NH2 terminus of Val1711 cleaved mNotch1 to show that endogenous Notch1 receptors are invariably cleaved at Val1711 upon ligand binding. We demonstrate that like in flies this cleavage occurs at the cell surface by ADAM10 but not by ADAM17/TACE. Notably, the substrate requirements for S2- Val1711 cleavage are contained within the first membrane-proximal amino acids. Our study confirms the identity of the protease involved in ligand-dependent S2 cleavage in mammalian cells to be ADAM10. However, because a complete block of Val1711 proteolysis with MPi did not block the formation of NEXT or NICD from Notch1 receptors harboring gain of function T-ALL mutations, we conclude that unknown proteases, insensitive to hydroxamic acid inhibitors, can cleave Notch1 with NRR mutations. We noted that the constitutive cleavage of ligand-independent Notch1 at Val1711 could be enhanced by known inducers of growth factor receptor/cytokine shedding by ADAMs and matrix metalloproteinases (APMA and PMA) (48). Regulated shedding is a common modulator of growth factor receptors; our observations argue that cellular signaling cascades may be activated/inhibited to modulate endogenous Notch1 S2 cleavage.
Similar to the AV site present in the canonical TACE substrate tumor necrosis factor α (49, 50), in vitro studies confirm a requirement for Ala1710 and Val1711 for TACE/ADAM17-mediated cleavage of Notch1 (19). These studies used recombinant Notch1 protein in cell-free extracts and found cleavage to be dependent on TACE but not on ADAM10. Moreover, cleavage of Notch1 proteins with mutated S2 sites (AV → VH and AV → ED) was abrogated in these in vitro assays. In vivo, Notch1 receptors with a mutated S2 cleavage site act as ligand-independent gain of function mutants and were cleaved at an unknown residue instead. This could be explained because substitution of Val1711 into His or Asp introduces charged bulky amino acids that likely interfere with formation of the compact pocket protecting S2 and allowing partial unfolding of the NRR, exposing different scissile bonds (3, 4, 17) that can be cleaved by other proteases. This hypothesis could be experimentally tested using urea unfolding experiments (17). The differential requirement for TACE in cleavage of purified polypeptides and of ADAM10 for cleavage of endogenous Notch1 may reflect the important relationship between the structural conformation of substrates and their proteases or the spatial distribution of a Notch “cleavosome” containing ADAM10 and γ-secretase. The saturation of γ-secretase by APMA treatment supports the idea that S2 cleavage occurs at a site with limiting amounts of γ-secretase but with predominance of ADAM10.
Whereas endogenous ligand-dependent Notch1 signaling in cells requires ADAM10, perhaps due to a privileged cellular location where ADAM10 concentration is optimal, our studies indicate that ligand-independent Notch1 proteins no longer depend on a single enzyme. The identity of proteases that access mutant Notch1 molecules are of interest; in this respect it is worth noting that ligand-independent cleavage of wild type Notch1 can be induced by overexpression of TACE and ADAM10 in Drosophila (37). In addition, we show that different metalloprotease inhibitors can completely block Val1711 cleavage of Notch1 at low micromolar concentrations; however, under these conditions S3 cleavage of Notch1 is not fully inhibited. These results also suggest that Notch1 activation via S3 cleavage and NICD release proceeds with assistance of BB94-insensitive proteases or, alternatively, that γ-secretase can cleave such molecules after they unfold at S1. We favor the former possibility as we still observe NEXT fragments, even at the highest MPi concentrations. Apparently, mutant Notch1 molecules are still cleaved at another site (i.e. S2*). We tested several cysteine, serine, and aspartyl protease inhibitors without success, indicating that multiple hydroxamic acid-insensitive proteases can cleave mutant Notch1 proteins.
Interestingly, ADAM10-deficient cells display residual Val1711 cleavage reminiscent of that observed in MPi-treated cells. We cannot rule out that the closely related ADAM17/TACE can cleave Notch1 in the absence of ADAM10. This hypothesis is supported by the observation that Val1711 cleavage of T-ALL mutant Notch1 proteins is strongly inhibited by the ADAM10/17-specific inhibitor GW280264 but not by the ADAM10-specific inhibitor GI254023 (44). Furthermore, in Caenorhabditis elegans ADAM17/TACE (ADM-4) and ADAM10 (SUP17) orthologs function redundantly in Lin12/Notch signaling (51). Of note, the neurogenic phenotype of ADAM10 in flies is less severe than the dNotch null phenotype, also pointing to additional S2 proteases (23). Altogether these data argue that multiple proteases exist that can participate in Notch1 S2 cleavage, but ADAM10 is the most efficient/best localized, and thus, in its absence, a severe loss of function phenotype is observed.
Surface biotinylation and immunostaining with αVal1711 antibodies indicate that S2 cleavage occurred at the cell surface in GSI-treated cells. We noted internalization of some S2-cleaved fragments into cytoplasmic vesicles. At present, the identity of these vesicles is unknown, but they could represent γ-secretase-containing vesicles that produce distinct NICD species of different stability and signaling activity (40). It is interesting to speculate that the alternative S2 cleavage we observed may influence the γ-secretase cleavage position and thus NICD stability and activity (40, 52).
Taken together, the data presented here illustrate a common mechanism for extracellular domain cleavage of wild type and oncogenic forms of Notch1 by the membrane-bound metalloprotease ADAM10/Kuzbanian. The application of γ-secretase inhibitors to treat Notch-dependent malignancies is hampered by their toxic side effects (53, 54) and by acquired resistance (55, 56). Novel approaches partly overcome intestinal toxicity (57), but Notch1 inhibitors more specific to Notch1 NRR mutants are desirable. We propose that prevention of the rate-limiting S2 cleavage is another interesting strategy that could be exploited (45). Cell-based parallel screens with NRR mutants and ligand-dependent Notch proteins should assist in identifying new drugs targeting aberrant Notch S2 proteolysis to enhance current strategies for Notch inhibition.
Supplementary Material
Acknowledgments
We thank D. Holtzman and J. Cirrito (Washington University, St. Louis) for advice and antibody production (National Institutes of Health AG13956 to D. H.); C. Blobel (Hospital for Special Surgery, New York) for providing Adam9/12/15-deficient cell lines; D. J. Pan (University of Texas Southwestern Medical Center, Dallas) for Adam10−/− cell lines; R. Black (Amgen) for Adam17ΔZn/ΔZn cell lines; B. Blom (AMC, Amsterdam, The Netherlands) for providing OP9 ligand-expressing cells; and A. Ludwig (Rheinisch-Westfälische Technische Hochschule Aachen University) for providing hydroxamate inhibitors.
This work was supported by National Institutes of Health Grant GM55479 (to R. K.) and by Dutch Cancer Society Grant KWF UU2006-3623 (to M. V. and G. v. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- S1
- site 1
- S2
- site 2
- S3
- site 3
- NECD
- Notch extracellular domain
- NEXT
- Notch extracellular truncation
- NICD
- Notch intracellular domain
- HD
- heterodimerization domain
- NRR
- negative regulatory region
- LNR
- Lin12/Notch repeat
- ADAM
- A disintegrin and metalloprotease
- PBS
- phosphate-buffered saline
- APMA
- 4-amino-phenylmercuric acetate
- T-ALL
- T-cell acute lymphocytic leukemia
- TACE
- tumor necrosis factor α-converting enzyme
- MPi
- metalloprotease inhibitor
- GSI
- γ-secretase inhibitor
- DAPT
- N-(N-(3,5-difluorophenacetyl-l-alanyl))-S-phenylglycine-t-butyl ester
- PMA
- phorbol 12-myristate 13-acetate.
REFERENCES
- 1.Wilson A., Radtke F. (2006) FEBS Lett. 580, 2860–2868 [DOI] [PubMed] [Google Scholar]
- 2.Sanchez-Irizarry C., Carpenter A. C., Weng A. P., Pear W. S., Aster J. C., Blacklow S. C. (2004) Mol. Cell. Biol. 24, 9265–9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon W. R., Vardar-Ulu D., Histen G., Sanchez-Irizarry C., Aster J. C., Blacklow S. C. (2007) Nat. Struct. Mol. Biol. 14, 295–300 [DOI] [PubMed] [Google Scholar]
- 4.Gordon W. R., Roy M., Vardar-Ulu D., Garfinkel M., Mansour M. R., Aster J. C., Blacklow S. C. (2009) Blood 113, 4381–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) Mol. Cell 5, 197–206 [DOI] [PubMed] [Google Scholar]
- 6.De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J. S., Schroeter E. H., Schrijvers V., Wolfe M. S., Ray W. J., Goate A., Kopan R. (1999) Nature 398, 518–522 [DOI] [PubMed] [Google Scholar]
- 7.Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 8.Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995) Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- 9.Huppert S. S., Le A., Schroeter E. H., Mumm J. S., Saxena M. T., Milner L. A., Kopan R. (2000) Nature 405, 966–970 [DOI] [PubMed] [Google Scholar]
- 10.Lieber T., Kidd S., Alcamo E., Corbin V., Young M. W. (1993) Genes Dev. 7, 1949–1965 [DOI] [PubMed] [Google Scholar]
- 11.Struhl G., Adachi A. (1998) Cell 93, 649–660 [DOI] [PubMed] [Google Scholar]
- 12.Nichols J. T., Miyamoto A., Olsen S. L., D'Souza B., Yao C., Weinmaster G. (2007) J. Cell Biol. 176, 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girard L., Hanna Z., Beaulieu N., Hoemann C. D., Simard C., Kozak C. A., Jolicoeur P. (1996) Genes Dev. 10, 1930–1944 [DOI] [PubMed] [Google Scholar]
- 14.Ellisen L. W., Bird J., West D. C., Soreng A. L., Reynolds T. C., Smith S. D., Sklar J. (1991) Cell 66, 649–661 [DOI] [PubMed] [Google Scholar]
- 15.Greenwald I., Seydoux G. (1990) Nature 346, 197–199 [DOI] [PubMed] [Google Scholar]
- 16.Kopan R., Schroeter E. H., Weintraub H., Nye J. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malecki M. J., Sanchez-Irizarry C., Mitchell J. L., Histen G., Xu M. L., Aster J. C., Blacklow S. C. (2006) Mol. Cell. Biol. 26, 4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng A. P., Ferrando A. A., Lee W., Morris J. P., 4th, Silverman L. B., Sanchez-Irizarry C., Blacklow S. C., Look A. T., Aster J. C. (2004) Science 306, 269–271 [DOI] [PubMed] [Google Scholar]
- 19.Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) Mol. Cell 5, 207–216 [DOI] [PubMed] [Google Scholar]
- 20.Reiss K., Saftig P. (2009) Semin. Cell Dev. Biol. 20, 126–137 [DOI] [PubMed] [Google Scholar]
- 21.Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., Boyce R. W., Nelson N., Kozlosky C. J., Wolfson M. F., Rauch C. T., Cerretti D. P., Paxton R. J., March C. J., Black R. A. (1998) Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 22.Lieber T., Kidd S., Young M. W. (2002) Genes Dev. 16, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sotillos S., Roch F., Campuzano S. (1997) Development 124, 4769–4779 [DOI] [PubMed] [Google Scholar]
- 24.Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., Saftig P. (2002) Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 25.Tian L., Wu X., Chi C., Han M., Xu T., Zhuang Y. (2008) Int. Immunol. 20, 1181–1187 [DOI] [PubMed] [Google Scholar]
- 26.Qi H., Rand M. D., Wu X., Sestan N., Wang W., Rakic P., Xu T., Artavanis-Tsakonas S. (1999) Science 283, 91–94 [DOI] [PubMed] [Google Scholar]
- 27.LaVoie M. J., Selkoe D. J. (2003) J. Biol. Chem. 278, 34427–34437 [DOI] [PubMed] [Google Scholar]
- 28.Six E., Ndiaye D., Laabi Y., Brou C., Gupta-Rossi N., Israel A., Logeat F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7638–7643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra-Gorur K., Rand M. D., Perez-Villamil B., Artavanis-Tsakonas S. (2002) J. Cell Biol. 159, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sapir A., Assa-Kunik E., Tsruya R., Schejter E., Shilo B. Z. (2005) Development 132, 123–132 [DOI] [PubMed] [Google Scholar]
- 31.Vooijs M., Schroeter E. H., Pan Y., Blandford M., Kopan R. (2004) J. Biol. Chem. 279, 50864–50873 [DOI] [PubMed] [Google Scholar]
- 32.Groot A. J., Gort E. H., van der Wall E., van Diest P. J., Vooijs M. (2008) Cell Oncol. 30, 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dontje W., Schotte R., Cupedo T., Nagasawa M., Scheeren F., Gimeno R., Spits H., Blom B. (2006) Blood 107, 2446–2452 [DOI] [PubMed] [Google Scholar]
- 34.Sahin U., Weskamp G., Kelly K., Zhou H. M., Higashiyama S., Peschon J., Hartmann D., Saftig P., Blobel C. P. (2004) J. Cell Biol. 164, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarriault S., Le Bail O., Hirsinger E., Pourquié O., Logeat F., Strong C. F., Brou C., Seidah N. G., Israel A. (1998) Mol. Cell. Biol. 18, 7423–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luty W. H., Rodeberg D., Parness J., Vyas Y. M. (2007) J. Immunol. 179, 819–829 [DOI] [PubMed] [Google Scholar]
- 37.Delwig A., Rand M. D. (2008) Cell. Mol. Life Sci. 65, 2232–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rand M. D., Grimm L. M., Artavanis-Tsakonas S., Patriub V., Blacklow S. C., Sklar J., Aster J. C. (2000) Mol. Cell. Biol. 20, 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlström H., Bergman A., Lendahl U., Näslund J., Lundkvist J. (2002) J. Biol. Chem. 277, 6763–6766 [DOI] [PubMed] [Google Scholar]
- 40.Tagami S., Okochi M., Yanagida K., Ikuta A., Fukumori A., Matsumoto N., Ishizuka-Katsura Y., Nakayama T., Itoh N., Jiang J., Nishitomi K., Kamino K., Morihara T., Hashimoto R., Tanaka T., Kudo T., Chiba S., Takeda M. (2008) Mol. Cell. Biol. 28, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merlos-Suárez A., Ruiz-Paz S., Baselga J., Arribas J. (2001) J. Biol. Chem. 276, 48510–48517 [DOI] [PubMed] [Google Scholar]
- 42.Arribas J., Coodly L., Vollmer P., Kishimoto T. K., Rose-John S., Massagué J. (1996) J. Biol. Chem. 271, 11376–11382 [DOI] [PubMed] [Google Scholar]
- 43.Schroeter E. H., Ilagan M. X., Brunkan A. L., Hecimovic S., Li Y. M., Xu M., Lewis H. D., Saxena M. T., De Strooper B., Coonrod A., Tomita T., Iwatsubo T., Moore C. L., Goate A., Wolfe M. S., Shearman M., Kopan R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13075–13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., Kallen K. J., Rose-John S., Ludwig A. (2003) Blood 102, 1186–1195 [DOI] [PubMed] [Google Scholar]
- 45.Li K., Li Y., Wu W., Gordon W. R., Chang D. W., Lu M., Scoggin S., Fu T., Vien L., Histen G., Zheng J., Martin-Hollister R., Duensing T., Singh S., Blacklow S. C., Yao Z., Aster J. C., Zhou B. B. (2008) J. Biol. Chem. 283, 8046–8054 [DOI] [PubMed] [Google Scholar]
- 46.Pan D., Rubin G. M. (1997) Cell 90, 271–280 [DOI] [PubMed] [Google Scholar]
- 47.Manilay J. O., Anderson A. C., Kang C., Robey E. A. (2005) J. Immunol. 174, 6732–6741 [DOI] [PubMed] [Google Scholar]
- 48.Merlos-Suárez A., Fernández-Larrea J., Reddy P., Baselga J., Arribas J. (1998) J. Biol. Chem. 273, 24955–24962 [DOI] [PubMed] [Google Scholar]
- 49.Moss M. L., Jin S. L., Milla M. E., Bickett D. M., Burkhart W., Carter H. L., Chen W. J., Clay W. C., Didsbury J. R., Hassler D., Hoffman C. R., Kost T. A., Lambert M. H., Leesnitzer M. A., McCauley P., McGeehan G., Mitchell J., Moyer M., Pahel G., Rocque W., Overton L. K., Schoenen F., Seaton T., Su J. L., Becherer J. D. (1997) Nature 385, 733–736 [DOI] [PubMed] [Google Scholar]
- 50.Black R. A., Rauch C. T., Kozlosky C. J., Peschon J. J., Slack J. L., Wolfson M. F., Castner B. J., Stocking K. L., Reddy P., Srinivasan S., Nelson N., Boiani N., Schooley K. A., Gerhart M., Davis R., Fitzner J. N., Johnson R. S., Paxton R. J., March C. J., Cerretti D. P. (1997) Nature 385, 729–733 [DOI] [PubMed] [Google Scholar]
- 51.Jarriault S., Greenwald I. (2005) Dev. Biol. 287, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okochi M., Steiner H., Fukumori A., Tanii H., Tomita T., Tanaka T., Iwatsubo T., Kudo T., Takeda M., Haass C. (2002) EMBO J. 21, 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Es J. H., van Gijn M. E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D. J., Radtke F., Clevers H. (2005) Nature 435, 959–963 [DOI] [PubMed] [Google Scholar]
- 54.Milano J., McKay J., Dagenais C., Foster-Brown L., Pognan F., Gadient R., Jacobs R. T., Zacco A., Greenberg B., Ciaccio P. J. (2004) Toxicol. Sci. 82, 341–358 [DOI] [PubMed] [Google Scholar]
- 55.Palomero T., Sulis M. L., Cortina M., Real P. J., Barnes K., Ciofani M., Caparros E., Buteau J., Brown K., Perkins S. L., Bhagat G., Agarwal A. M., Basso G., Castillo M., Nagase S., Cordon-Cardo C., Parsons R., Zúñiga-Pflücker J. C., Dominguez M., Ferrando A. A. (2007) Nat. Med. 13, 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Neil J., Grim J., Strack P., Rao S., Tibbitts D., Winter C., Hardwick J., Welcker M., Meijerink J. P., Pieters R., Draetta G., Sears R., Clurman B. E., Look A. T. (2007) J. Exp. Med. 204, 1813–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Real P. J., Tosello V., Palomero T., Castillo M., Hernando E., de Stanchina E., Sulis M. L., Barnes K., Sawai C., Homminga I., Meijerink J., Aifantis I., Basso G., Cordon-Cardo C., Ai W., Ferrando A. (2009) Nat. Med. 15, 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.