Abstract
Thymus-derived lymphocytes protect mammalian hosts against virus- or cancer-related cellular alterations through immune surveillance, eliminating diseased cells. In this process, T cell receptors (TCRs) mediate both recognition and T cell activation via their dimeric αβ, CD3ϵγ, CD3ϵδ, and CD3ζζ subunits using an unknown structural mechanism. Here, site-specific binding topology of anti-CD3 monoclonal antibodies (mAbs) and dynamic TCR quaternary change provide key clues. Agonist mAbs footprint to the membrane distal CD3ϵ lobe that they approach diagonally, adjacent to the lever-like Cβ FG loop that facilitates antigen (pMHC)-triggered activation. In contrast, a non-agonist mAb binds to the cleft between CD3ϵ and CD3γ in a perpendicular mode and is stimulatory only subsequent to an external tangential but not a normal force (∼50 piconewtons) applied via optical tweezers. Specific pMHC but not irrelevant pMHC activates a T cell upon application of a similar force. These findings suggest that the TCR is an anisotropic mechanosensor, converting mechanical energy into a biochemical signal upon specific pMHC ligation during immune surveillance. Activating anti-CD3 mAbs mimic this force via their intrinsic binding mode. A common TCR quaternary change rather than conformational alterations can better facilitate structural signal initiation, given the vast array of TCRs and their specific pMHC ligands.
Introduction
The T cell receptor (TCR)3 is a multimeric transmembrane complex composed of a disulfide-linked antigen binding clonotypic heterodimer (αβ or γδ) in non-covalent association with the signal-transducing CD3 subunits (CD3ϵγ, CD3ϵδ, and CD3ζζ) (reviewed in Ref. 1). TCR signaling via CD3 dimers evokes T cell lineage commitment and repertoire selection during development, maintains the peripheral T cell pool, and further differentiates naïve T cells into effector or memory cell populations upon immune stimulation (2–5). The interaction between an Fab-like αβ TCR heterodimer and an antigenic peptide bound to a major histocompatibility complex molecule (pMHC) initiates a cascade of downstream signaling events via the immunoreceptor tyrosine-based activation motif elements in the cytoplasmic tails of the associated CD3 subunits (6–9). The length of these CD3 cytoplasmic tails is substantial, relative to those of the TCR α and β chains (6, 7).
How recognition of pMHC by a weakly interacting (∼1–100 μm Kd) clonotypic heterodimer on the T cell surface evokes intracellular signaling via the adjacent CD3 components remains undefined (1). Solution structures of CD3ϵγ and CD3ϵδ heterodimers reveal a unique side-to-side hydrophobic interface with conjoined β-sheets involving the G-strands of the two Ig-like ectodomains of the pair (10, 11). The squat and rigid CD3 connecting segments contrast sharply with the long and flexible TCR α and β connecting peptides linking their respective constant domains to the transmembrane segments.
To investigate the basis of signal transduction involving the ectodomain components within the TCR membrane complex, we have focused on three anti-CD3ϵ mAbs (2C11, 500A2, and 17A2) and mapped those interactions using NMR techniques. We present detailed information on antibody recognition sites on surfaces of these T cell elements, revealing surfaces not engaged in interactions with the αβ or γδ heterodimer. We characterize the functional consequences of their binding on αβ and γδ T cells. TCR triggering is not linked to mAb affinity or CD3 binding stoichiometry per TCR. Rather, our results emphasize the critical role of the TCR β-CD3ϵγ module in αβ T cells, reveal a rationale for site-specific binding topology essential for anti-CD3 mAb-induced signaling, and demonstrate that a dynamic αβ TCR complex quaternary structure is essential for T cell activation by pMHC ligands. In this regard, an external torque on the TCR complex following pMHC ligation during T cell scanning of antigen presenting cells (APCs) can serve as the source of energy for directional force, defining the TCR as a mechanosensor.
EXPERIMENTAL PROCEDURES
Construction and Expression of an scCD3ϵγ Heterodimer
The scCD3ϵγ construct encoding a murine CD3γ fragment (residue codes 23 to 103 of Swiss-Prot P11942), a 26-residue flexible linker, and a murine CD3ϵ fragment (residue codes 22 to 100 of Swiss-Prot P22646) were cloned into a pET11a expression vector (New England BioLabs Inc.). Recombinant CD3ϵγ was produced as inclusion bodies in Escherichia coli BL21(DE3) and dissolved in 6 m guanidine HCl as described (11). Refolding efficiency in eight different conditions was monitored by 17A2 binding via surface plasmon resonance and confirmed by gel filtration chromatography. The optimal refolding buffer contained 55 mm MES, pH 6.5, 264 mm NaCl, 11 mm KCl, 2.2 mm MgCl2, 2.2 mm CaCl2, 440 mm sucrose, 0.1 mm reduced glutathione, 1 mm oxidized glutathione, and 0.5 × complete protease inhibitor mixture (Roche Applied Sciences). Soluble and monomeric CD3 proteins were purified by gel filtration on a Superdex-75.
Antibodies and Flow Cytometric Analysis
The following fluorochrome-labeled mAbs were used for surface receptor analysis by flow cytometry: FITC anti-CD3 (2C11) and Alexa 647 anti-CD3 (17A2), Pacific Blue-CD4 (H129.19), pacific orange anti-CD8α (53-6.7), FITC-conjugated anti-TCR Cβ (H57), phycoerythrin Cy7-conjugated anti-CD69 (H1.2F3), and allophycocyanin conjugated anti-CD5 (53-7.3) (BD Pharmingen). For flow cytometry, single-cell suspensions of thymocytes or lymph node (LN) cells were prepared at 5 × 106 cells/ml in phosphate-buffered saline containing 2% fetal calf serum and 0.05% NaN3. Those cells were triple or five-color stained with the Abs at saturating concentrations according to standard procedures. The phosphorylation state of ERK (Alexa 647 anti-pERK) or MAPK (phycoerythrin-anti-pMAPK) was determined by intracellular staining using BD Phosflow Lyse/Fix buffer BD and Phosflow Perm Buffer III after incubation with the immobilized anti-CD3 mAb. A FACScan or FACSAria (BD Biosciences) was used for flow cytometric measurements. Data analysis was performed using FlowJo software (Tree Star) after excluding dead cells by forward and side scatter gating. Measured F/P ratios for FITC-labeled 17A2 and 2C11 mAbs were 2.89 and 2.96, respectively.
Antibody Purification and Fab Preparation
The hybridoma cell lines expressing 17A2 (from R. MacDonald, Switzerland), 2C11, and 500A2 were used for antibody production (see supplemental Fig. S5 for the sequences of the variable regions). Each mAb was purified from culture supernatant using Gammabind plus Sepharose Fast Flow chromatography (Amersham Biosciences). After concentration using centrifugal filter devices with a 10-kDa molecular mass cut-off (Millipore), gel filtration using Superdex-75 (Amersham Biosciences) was performed for further purification and buffer change. Antibody fragments, Fabs, were generated using an Immunopure® Fab Preparation Kit following the manufacturer's instructions (Pierce). An additional Superdex-75 gel filtration was performed followed by concentration.
Mice
C57BL/6 and TCRβ−/− mice were purchased from Taconic (Germanton, NY) and Jackson Laboratory, respectively. CD3γ−/− mice have been described in detail elsewhere (12). N15 TCR transgenic (tg) mice on the RAG2−/− background bear T cells with a TCR specific for a vesicular stomatitis virus nuclear protein octapeptide (VSV8, RGYVYQGL) bound to the H-2Kb molecule (13). Mice were maintained and bred under specific pathogen-free conditions in the animal facility of the Dana-Farber Cancer Institute under a protocol reviewed and approved by the Animal Care and Use Committee.
Cell Purification
For γδ T cells purification, LN cells from TCRβ−/− mice were stained for 20 min on ice with a panel of phycoerythrin-labeled mAbs containing anti-B220, anti-TCRβ, anti-CD4, anti-CD8, and DX5 mAbs. Cells were washed and then negatively sorted by a FACSAria (BD Biosciences). Following separation, γδ T cells from TCRβ−/− mice were about 80% pure, with no detectable contamination from γδ TCR− CD3+ cells. For αβ T cell purification, LN cells from C57BL/6 mice were stained with anti-B220-FITC, pacific blue-CD4, and pacific orange anti-CD8α, washed, and then separated on a FACSAria. CD4 T cells or CD8 T cells were more than 90% pure upon reanalysis. N15 T cells were purified by removing I-Ab positive cells using anti-I-Ab and magnetic beads from N15TCRtgRag2−/− mice. All LN cells from C57BL/6 and N15TCRtg Rag2−/− mice were used immediately after purification and without prior stimulation (i.e. naïve T cells) unless specifically indicated otherwise.
Antibody and pMHC-coated Beads
For in vitro stimulation, 17A2, 2C11, or 500A2 were covalently immobilized on protein G-coupled dynabeads (Invitrogen). Antibody-coated beads were resuspended in freshly made 5 mm disuccinimidyl suberate (Pierce) for covalent cross-linking followed by removing free antibody. 17A2, 2C11, and H35 (anti-CD8β) mAbs labeled using Alexa 555 dye (Invitrogen) with an equivalent molar ratio ([Alexa 555]/[mAb] = ∼1) on protein G-coupled polystyrene beads (1 μm diameter, Polysciences) were immobilized as above. H-2Kb and β2-microglobulin protein expression, refolding, and purification were as described in detail (14). Preparation of H-2Kb with a biotinylation tag (GGGLNDIFEAQKIEWH) for birA enzymatic recognition at the C terminus was previously described (15). Each biotinylated H-2Kb complex was used for incubations with streptavidin-coupled polystyrene beads (1 μm diameter, Polysciences) after Alexa 555 labeling with an equivalent molar ratio ([Alexa 555]/[H-2Kb] = ∼1).
Calcium Flux
For calcium flux experiments, the T cells were loaded with Fluo-4AM (Molecular Probes) at 5 μm for 30 min in the presence of 2.5 mm probenecid. T cells were transferred to minimal imaging media (colorless RPMI with 5% fetal calf serum and 10 mm HEPES) and allowed to adhere to a 2% l-lysine-coated cover glass for 30 min at 37 °C before data collection. Recordings were made with a Nikon TE2000-U inverted microscope under temperature control. Fluo-4 was imaged at 30-s intervals for 10 min. All the samples were incubated in the imaging chambers at 37 °C for 5 min before data collection. We quantified Fluo-4 responses by determining the average intensity of a region within each cell as a function of time using the Image J program (NIH).
NMR Experiments
All NMR spectra were acquired on Bruker Avance750 or Avance600 spectrometers equipped with a 5-mm cryogenic probe. Unbound CD3ϵγ or the CD3ϵγ/17A2, 2C11 Fab complex was dissolved in 10 mm phosphate buffer, pH 6.8, containing 100 mm NaCl at a concentration of 0.5–1 mm. All NMR experiments are performed at 298 K. 15N-1H heteronuclear single quantum correlation experiments (HSQCs) using transverse relaxation optimization spectroscopy (TROSY) techniques were performed. Resonance assignments for unbound scCD3ϵγ were already established in previous studies (10) and were repeated for the new scCD3ϵγ construct. Native conformation was confirmed by its identity in long-range amide-amide patterns with the former construct. The bound CD3ϵγ assignments are determined by conventional three-dimensional NMR spectroscopy with TROSY techniques (16), for comparison with the free assignments. Data were analyzed with XWINNMR (Bruker) and SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco, CA). The binding surfaces of CD3ϵγ for Fabs were mapped with cross-saturation experiments following established protocols in the solvent (20% H2O, 80% D2O) (17). The saturation period was set to 0.5 s in all CD3ϵγ·Fab complexes.
Modeling Antibody Binding to the TCR Complex
We searched for plausible docking models for the CD3ϵγ·antibody complex using TREEDOCK software (18) while incorporating known experimental results. The coordinates were taken from the most representative NMR structures of scCD3ϵγ (PDB ID code 1JBJ). Structures of 17A2 and 2C11 were built by homology modeling in MODELER and refined in SCWRL3. To define the 17A2 sequence, we cloned the cDNAs of variable domains from 17A2 expressing hybridoma cells by PCR and obtained pre-existing 2C11 sequence data available in the Kabat data base (supplemental Fig. S5). Specifically, the target docking area in CD3ϵγ was centered at the mean position of the amide protons affected in cross-saturation experiments and residues within the 10 Å distance were used as potential docking anchors. The models were further selected from antibodies best accommodating scCD3ϵγ using their CDR loops and best satisfying the mutant analysis and former TCR complex model (11). The linker residues and flexible N terminus regions of CD3ϵγ were excluded from these models before docking.
Calcium Flux Experiment with External Mechanical Force Using Optical Tweezers
For calcium flux experiments, the purified N15 T cells from N15TCRtgRag2−/− mice were loaded with calcium orange (Invitrogen) at 5 μm for 30 min in the presence of 2.5 mm probenecid. These T cells were transferred to a 2% l-lysine-coated cover glass for 15 min at 37 °C to allow for cell attachment to the cover glass surface. After blocking the surface with 1 mg/ml casein solution for 5 min, antibody-coated or pMHC-coated beads (1 μm) were flowed into the sample for manipulation using a trapping beam (1064 nm). Single trapped beads were guided into contact with single surface-bound T cells using an automated piezoelectric stage, while monitoring the bead position relative to the trap so contact could be achieved with minimal force application. After T cell-bead contact was achieved, a continuous sinusoidal tangential (shear) or normal (vertical) force was applied to the bead by moving the trapping laser back and forth using acouto-optic deflectors at ∼0.25 Hz with an amplitude of up to ∼50 pN. The force on the bead was determined by measuring the trap stiffness (∼0.3 pN/nm) prior to cell-bead contact by the equipartition method (19), and by measuring the displacement of the bead from the trapping laser via post-processing of experiment videos. During the force application T cells were excited using a 532-nm laser to monitor calcium orange fluorescence. As an important control, similar experiments were performed with no force application. All of these experiments were done using a highly automated custom-built optical trap designed for simultaneous force and fluorescence experiments at the single molecule and single cell scale described previously (19).
RESULTS
Specificity of Anti-CD3 mAbs
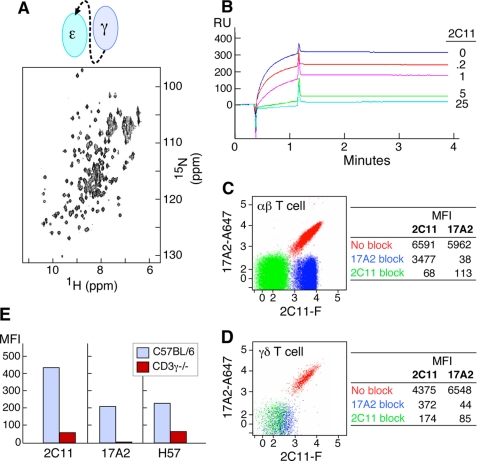
We designed and produced ectodomain fragments of CD3 heterodimers in E. coli for structural study. As shown by the 15N-1H two-dimensional HSQC NMR spectrum in Fig. 1A, this recombinant CD3ϵγ heterodimer, unlike an earlier protein (10), is stable under physiological conditions (pH 7.4, phosphate-buffered saline). A competitive binding assay shows that 2C11 preincubation with CD3ϵγ inhibits the ability of 17A2 to capture the heterodimer in a concentration-dependent manner (Fig. 1B) and vice versa (data not shown). Binding experiments on CD4 and CD8 T cells isolated from C57BL/6 mice LN as assessed by quantitative measurement of mean fluorescence intensity (MFI) indicate that 17A2 inhibits 2C11 binding by only 50%, whereas 2C11 blocks 17A2 binding completely in both CD4 (Fig. 1C) and CD8 (data not shown) populations. These results strongly support the notion that 17A2 recognizes only one of two CD3 heterodimers within the native αβ TCR complex on the T cell surface. In contrast, 17A2 and 2C11 have an equivalent number of binding sites on γδ T cells (numerically the minor T cell subset), and 17A2 blockade completely abrogates 2C11 binding (Fig. 1D). Because murine γδ T cells lack CD3ϵδ but express two CD3ϵγ heterodimers per TCR complex (20), this differential blocking activity on αβ versus γδ T cells is expected. Furthermore, as shown in Fig. 1E, 17A2 cannot bind to αβ T cells from CD3γ−/− mice, whereas residual 2C11 reactivity is still present. Serial cross-blocking results support that 17A2 competes with 2C11 on the TCRβ·CD3ϵγ side of the TCR complex (supplemental Fig. S1). Thus, 17A2 recognizes the CD3ϵγ heterodimer but not the CD3ϵδ heterodimer on the surface of T cells.
FIGURE 1.
Specificity of anti-CD3 mAbs. A, 15N-1H two-dimensional HSQC spectrum of recombinant CD3ϵγ (schematically represented) is stable under physiological conditions (pH 7.4, phosphate-buffered saline). B, SPR analysis of competitive scCD3ϵγ binding between 17A2 and 2C11. Direct binding between recombinant CD3ϵγ (1 μm) and surface-bound 17A2 was measured using a BIA3000 Biosensor. Sensorgrams without and with 2C11 pre-addition to the CD3ϵγ protein are shown (numbers represent antibody to CD3ϵγ protein molar ratios). C and D, binding competition between 17A2 and 2C11 on the T cell surface. LNs were isolated from C57BL/6 mice. mAbs against T cell surface markers including CD3 were used for flow cytometric analysis. Each dot represents 2C11-FITC (2C11-F) and 17A2-Alexa 647 (17A2-A647) binding after unlabeled 17A2 blocking (blue), 2C11 blocking (green), or no blocking (red). CD4 single-positive αβ TCR-positive or γδ TCR-positive populations were gated for analysis. Data are representative of three independent experiments and plotted on a 5 log scale (0–5). MFI values are given in the tables for binding of each fluorochrome-labeled antibody without or with the indicated unlabeled mAb blockade. E, differential binding of 17A2 to thymocytes of wild type (blue) and CD3γ−/− (red) mice. CD4 single-positive thymocytes were gated for analysis. H57 recognizes the FG loop in the constant region of TCRβ (24).
Differential Capacity of Anti-CD3 mAbs to Activate T Cells
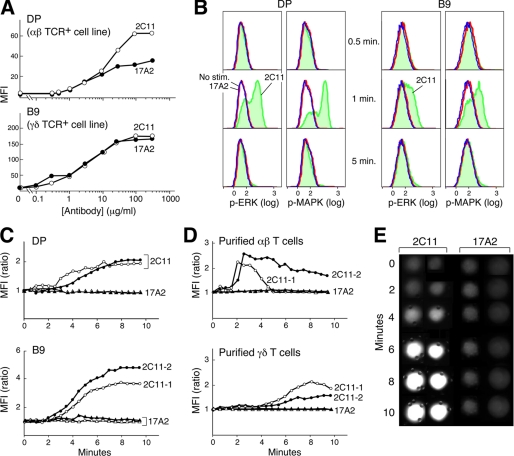
We next assessed how 17A2 versus 2C11 mAbs influence T cell activation. Flow cytometric analysis on the αβ TCR-expressing DP (257-20-109) cell line (21), and the γδ TCR expressing B9 cell line (22) indicates that 17A2 and 2C11 binding affinities are similar (Fig. 2A). Moreover, because the fluorescein labeling ratio of 2C11 and 17A2 mAbs are identical (supplemental Fig. S2), it is significant that 2C11 MFI is twice that of 17A2 on the DP cells, consistent with 2C11 binding to CD3ϵ in both CD3 heterodimers, whereas 17A2 only binds to CD3ϵγ. On the other hand, the 2C11 MFI is similar to that of 17A2 on B9 cells, consistent with the known γδ TCR stoichiometry (20). These results enabled us to then directly compare the functional consequence of 17A2 versus 2C11 mAb binding.
FIGURE 2.
Differential functional activation of T cells by anti-CD3 mAbs. A, flow cytometry analysis with MFI binding quantitation of 2C11-FITC and 17A2-FITC at varying concentrations to DP (αβ TCR+) and B9 (γδ TCR+) cell lines. B, phosphorylation states of ERK or MAPK after 2C11- or 17A2-coupled bead stimulation (blue = no stimulation; red = 17A2 stimulation; green = 2C11 stimulation). C and D, intracellular calcium dynamics upon interaction with 2C11- or 17A2-coupled beads. Circles represent cells stimulated by 2C11 beads and triangles represent cells stimulated by 17A2 beads (two independent measurements are shown for each with open and filled symbols). For calcium flux experiments, γδ and αβ T cells were purified from LNs of TCRβ−/− and CL57BL/6 mice, respectively, as described. Time 0 = time of bead addition to cells. E, microscopic images of calcium flux in individual cells using Fluo-4 and microscopy as described under “Experimental Procedures.” Fluo-4 responses were quantified by determining the average intensity of a region within each cell as a function of time using the Image J program (NIH). Results are representative of the 64 individual T cells analyzed. Two-tailed t test analysis was performed to compare MFI values from 64 T cells. The increase in intracellular free calcium levels induced by 2C11 beads was significantly different from those induced by 17A2 beads (i.e. p < 0.05 at 6 min).
To monitor initial T cell activation triggered by 17A2 and 2C11, we measured ERK phosphorylation at the single cell level using intracellular staining with anti-phospho-ERK antibody and flow cytometry. We immobilized equivalent amounts of 2C11 or 17A2 on protein G-coated magnetic beads (supplemental Fig. S2) and measured signaling dynamics in T cell-antibody immobilized bead conjugates. Phosphorylation of ERK was detected upon 2C11 bead stimulation in both αβ TCR+ and γδ TCR+ cells (Fig. 2B). These data regarding the ERK signaling response to 2C11 are consistent with the highly amplified, digital ERK responses induced by agonist pMHC (23). Similar results were found for MAPK phosphorylation stimulated by 2C11 beads. In contrast, neither phosphorylation of ERK nor MAPK was detected upon 17A2 bead stimulation (Fig. 2B). The number of antibody binding sites per TCR complex might account for such a functional difference in 2C11 versus 17A2 (2 versus 1, respectively) in αβ T cells. To test this possibility, comparative analysis was performed using γδ T cells that exclusively incorporate CD3ϵγ heterodimers into their TCR complex (20). However, as with αβ T cells, ERK/MAPK phosphorylation in γδ T cells is only induced by 2C11 beads. 500A2 beads, like 2C11 beads, induce phosphorylation of ERK and MAPK, consistent with their strikingly similar binding behavior on T cells (supplemental Fig. S3).
Because TCR signaling is associated with elevations in intracellular-free calcium, the ability of these anti-CD3 mAbs to induce a characteristic intracellular calcium flux was also tested. No detectable calcium flux was induced by 17A2-immobilized beads, but incubation with 2C11 beads triggered a response in αβ and γδ T cell lines as well as LN purified αβ and γδ T cells (Fig. 2, C and D); curves are representative of the multiple individual cells monitored. Calcium flux is detected after a delay of ∼2 min, related to the interval required for conjugates to form between a T cell and beads and evident in single cell microscopic analysis (Fig. 2E).
NMR Cross-saturation Analysis Maps Anti-CD3 mAbs to Membrane- and Interface-distal Corners of CD3ϵ
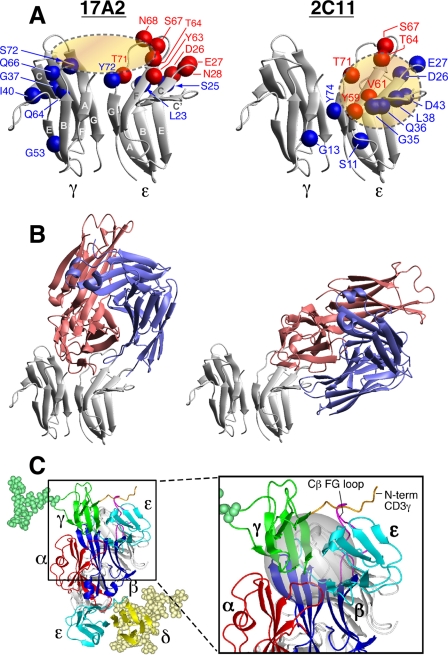
In the absence of differences in affinity or stoichiometry to explain the distinct functional effects of anti-CD3 mAbs, we performed structural comparison of 17A2 and 2C11 binding sites on the CD3ϵγ molecule. We established the bound assignment of these Fab·CD3ϵγ complexes by conventional three-dimensional NMR spectroscopy using TROSY techniques (16). Benefitting from line narrowing due to the TROSY effect and per-deuteration of the CD3ϵγ molecule, most of the resonances remain sharp and are easily detected even with the relatively large molecular mass of the 71-kDa complex. Exceptions are amide groups in close contact with other protons, as happens for certain interface residues juxtaposed to protonated Fabs. Red spheres in Fig. 3A indicate the position of those residues that are not observed in the complex due to this protonated Fab-induced relaxation. In addition, we performed cross-saturation NMR experiments (17) to map the complete binding sites of 17A2 and 2C11 on the CD3ϵγ molecule. In the cross-saturation experiment, irradiation of Fab resonances causes indirect saturation of CD3ϵγ amide resonances only for those located close in space (<7 Å) to the Fab fragment in a complex. The positions of amides (blue spheres) in the CD3ϵγ molecule that have greatly reduced intensity upon irradiation of Fab resonances (Isat/Iref < 0.5, see also the signal reductions for each residues in supplemental Fig. S4A) or disappeared (red spheres) upon Fab binding are shown. These residues are located around the cleft formed between CD3ϵ and CD3γ by 17A2 binding (shown as the brown ellipsoid in Fig. 3A, left), whereas 2C11 binding is centered on the CD3ϵ chain (brown circle in Fig. 3A, right). 17A2 and 2C11 have distinct binding sites on CD3ϵγ with some overlap of CD3ϵ footprint residues (Asp-26, Glu-27, Thr-64, Ser-67, and Thr-71).
FIGURE 3.
NMR cross-saturation experiments to map antibody binding surfaces on CD3ϵγ and TCR quaternary structure. A, the residues in the binding sites of CD3ϵγ for 17A2 (left) and 2C11 (right) Fabs were mapped using cross-saturation analysis. Blue and red spheres indicate residues that experience significant cross-saturation (signal reduction < 0.5) or disappear upon Fab binding, respectively, and thus mediate direct contact with the Fabs. B, representative models of CD3ϵγ/17A2 Fab and CD3ϵγ/2C11 Fab complexes as described in the text. Red represents the heavy chain fragments and blue the light chain of the Fab. C, proposed TCR quaternary structure model as viewed from the T cell membrane. Magnified boxed region in the complex model shows the location of the Cβ FG loop (magenta) and N-terminal segment of CD3γ (orange) with the Cβ/Cα cave (24) represented as a gray sphere. For simplicity, only CD3γ (green) and CD3δ (yellow) glycans are shown. Hypothetical distance constraints between nearby charged residues were employed with a number of residues, including the entire N-terminal segment of CD3ϵ, which was allowed to be flexible during annealing between the docking steps.
To further investigate their distinctive binding modes, we performed theoretical calculations using Treedock (18), after incorporating structural models of their respective Fab variable domains (supplemental Fig. S5). The residues proximal to the two Fabs in CD3ϵγ identified in the cross-saturation experiments were used as candidate anchors for the docked complexes. As illustrated in Fig. 3B, 17A2 Fab binds to CD3ϵγ in a more upright mode, largely between the CD3ϵ and CD3γ molecules, whereas 2C11 binds to the side top “corner” of CD3ϵ to assume a diagonal orientation. Consistent with the cross-saturation experiment, 17A2 contacts primarily the top end of the GFCC′ face of CD3ϵ, whereas it is also close to the N terminus of the A strand and BC loop, and the top end of the FG strand in CD3γ. On the other hand, 2C11 binding involves primarily the C′ strand and the beginning of the C′E loops in CD3ϵ. The complex models also clearly explain the direct competition between 2C11 and 17A2 for the CD3ϵγ binding site because they partially overlap with each other, sharing C and C′ strands as well as the end of the F strand in the same CD3ϵ molecule. The binding site of 500A2 (25), a second activating anti-CD3 mAb, is similar to that of 2C11, being restricted in its interaction with the same top corner of CD3ϵ as 2C11 (supplemental Fig. S6).
A Molecular Model of TCR Quaternary Topology
Prior models of the αβ TCR complex (11, 26) were improved upon by accommodating biochemical and functional data herein (Fig. 3C). This quaternary structure model, constructed by iterative rigid body docking using XPLOR-NIH (27), illustrates the relative orientation between the ectodomains of the N15TCRαβ heterodimer (PDB code 1NFD), CD3ϵγ (PDB code 1JBJ), and CD3ϵδ (PDB code 1XMW). As shown in Fig. 3C, the binding sites for 2C11 on CD3ϵ of the CD3ϵδ heterodimer and on CD3ϵ of the CD3ϵγ heterodimer are both exposed in the TCR complex consistent with our analyses. The CD3ϵ site of CD3ϵγ is located in proximity to the TCR Cβ FG loop (Fig. 3C, inset), compatible with the direct competition noted between 2C11 Fab and H57 Fab (28) (supplemental Fig. S1). Furthermore, binding competition between H57 and heterosera recognizing the N terminus of CD3γ (29) (supplemental Fig. S7) is also consistent with this model.
The TCR as a Mechanoreceptor: Optical Tweezer Analysis
The data amassed from NMR cross-saturation on murine CD3ϵγ show that activating anti-CD3 mAbs such as 2C11 and 500A2 bind to the membrane- and interface-distal corner of murine CD3ϵ in an orthogonal mode. Crystal structures of the human CD3ϵγ heterodimer complexed with OKT3 (30) and the human CD3ϵδ heterodimer complexed with UCHT-1 (31), two mitogenic anti-human CD3ϵ mAbs, likewise identify a similar binding architecture.
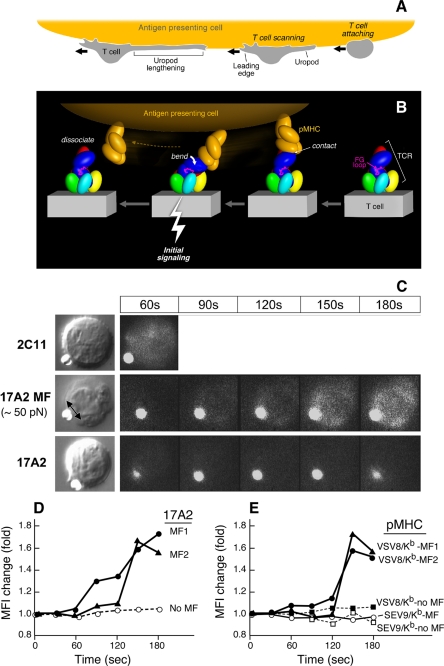
How might this particular binding orientation be stimulatory for human and mouse T cells? Fig. 4A schematically shows that scanning by T cells of APC surfaces in search of a specific pMHC can create a tensile mechanical force prior to transduction of a “stop movement” signal (Fig. 4A). Upon TCR-pMHC ligation, extracellular mechanical torque can result in quaternary structural changes within the TCR complex among the αβ and CD3 heterodimers (Fig. 4B), triggering downstream signaling via immunoreceptor tyrosine-based activation motifs in the CD3 cytoplasmic tails enhanced by Lck-linked CD4 and CD8 co-receptor function (not shown) during TCR/co-receptor approximation (32). Of note, the exposed TCR β chain constant domain FG loop, known to efficiently facilitate αβ T cell activation (33, 34), likely functions as a lever, contributing to antigen-specific signal transfer from the αβ heterodimer to the CD3ϵγ heterodimer (Fig. 4B, magenta loop). Impairing TCR complex quaternary structure and requisite conformational change, either by addition of the FG loop-specific H57 Fab to antigen-specific T cells (supplemental Fig. S8B) or mutations in the adjacent CD3ϵγ heterodimer ectodomain, blocks specific pMHC-stimulated cytokine production (data not shown).
FIGURE 4.
Mechanosensor model for TCR signaling. A, T cell scanning an APC surface in search of specific pMHC. Upon attachment, the T cell assumes a polarized morphology with a leading edge and lengthening uropod. B, one pMHC molecule is shown in orange on the APC, whereas the ectodomains of TCR subunits on the T cell membrane below are colored as described in the legend to Fig. 3. The view is from the CD3ϵγ side. Initial ligation of the TCR by pMHC (right) constitutes a detachable mechanosensor that, as a result of continued T cell scanning, transmits an external torque (bend) into initial signaling (bolt) via the rigid components of the TCR complex prior to dissociation. The β FG loop is shown in magenta. C, calcium flux in naïve T cells after application of external mechanical force using optical tweezers. Equivalent amounts of Alexa 555-labeled 17A2 or 2C11 were immobilized on protein G-coupled polystyrene beads (1 μm diameter). T cell-bead contact was manipulated via the trapping beam as shown in bright field images (left panel). The direction of the external mechanical force (MF) for a 17A2 bead is denoted by the double-headed arrow. Microscopic fluorescence images were recorded using a 532-nm laser for both Alexa 555 and calcium orange (cellular dye for detecting calcium flux) under a temperature control at 37 °C (right panels). For 2C11 beads, the 60-s point only is shown, given the rapidity of calcium flux and the absence of a requirement of any additional MF. The fluorescent antibody-bound bead is the smaller object next to the T cell in every frame. D, quantitative intracellular calcium dynamics upon 17A2 bead interaction. Results are representative of 112 cellular N15 T cell events quantitated in eight separate experiments. Two-tailed t test was performed to compare MFI values of all cells between the 150- and 180-s intervals. Only 17A2 bead with applied tangential mechanical force significantly induces the intracellular calcium flux (p < 0.05). Three representative T cells are shown; two receiving ∼50 pN mechanical force (MF1/2) and one without (No MF). E, calcium dynamics upon pMHC bead interaction. Equivalent numbers (∼1,000) of VSV8/Kb and SEV9/Kb complexes were immobilized on streptavidin-coupled polystyrene beads (1 μm diameter). Tangential mechanical force (∼50 pN) was provided for each of three pMHC beads (pMHC MF) but not two others (no MF). Five representative cells are shown among 190 cell determinations analyzed. Only VSV8/Kb beads upon MF application induced significant calcium flux (p < 0.05) when ∼10 and ∼1,000 pMHC complexes per bead were examined. The extent of calcium flux was not diminished at the lower pMHC complex density nor kinetics of activation lengthened.
In this process, the pMHC may behave as a detachable effort arm of a complex-lever system or “gaff”; its binding is determined by the specificity of an individual TCRαβ heterodimer but is not sufficient to mediate signal transduction per se. An external torque is additionally required to provide the energy for this signaling. Such an off-centered torque might be generated either via a shear force or pressure between opposing APC and T cell membranes. No additional energy costs would be incurred by the T cell during this process because energy expended on immune surveillance is productively channeled into signal transduction upon specific immune recognition. Note that in the view shown in Fig. 4B, scanning and attendant pMHC ligation results in a force on the membrane distal lobe of CD3ϵ akin to that contacted by activating anti-CD3ϵ mAbs.
If the distinctive antibody binding modes of 2C11 and 500A2 versus 17A2 account for their differential stimulatory activities, we reasoned that application of an external tangential force via 17A2 to T cells should convert 17A2 into a cellular activator. Thus, we exploited an optical trapping methodology to provide controlled external mechanical force to 17A2, thereby investigating how mAb binding and subsequent torque affect TCR signaling. Optical trapping combined with fluorescence imaging is established as a powerful tool for studying proteins and molecules (35). Here we use ligand, specifically 17A2 mAb-coated beads to probe cell membrane machinery. Strikingly, only after application of a tangential force (50 pN) to a 17A2 mAb-bound bead was calcium flux induced (Fig. 4C). Quantitation of changes in intracellular free calcium over time are shown for two representative T cells to which this 17A2 mAb-bound bead force was applied (MF) and, by comparison, that of a T cell only approximated to a 17A2 mAb bead (no MF) (Fig. 4D). In contrast, 2C11 mAb-bound bead binding itself triggers rapid calcium flux, without requirement for horizontal motion. This observation suggests that the intrinsic diagonal binding orientation of 2C11 provides the requisite activation force. Furthermore, neither an applied vertical (normal to the surface of the cell) movement of the 17A2 mAb bead nor a comparable tangential force on a bead coupled with anti-CD8 mAb (H35) binding to the CD8 co-receptor on T cells was able to induce a calcium flux (supplemental Fig. S9). These results show that an asymmetric force or torque applied to the CD3ϵγ ectodomain rather than a normal movement is essential for TCR signaling initiation by antibody binding.
To next assess if TCR ligation by pMHC also required application of external force to induce a rise in intracellular free calcium in T cells, we used either VSV8/Kb or SEV9/Kb pMHC complexes. These were expressed recombinantly, purified, and coupled to streptavidin beads at equivalent amounts as detailed under “Experimental Procedures” and in supplemental Fig. S10. VSV8/Kb is the specific cognate ligand for the N15 TCR, whereas SEV9/Kb is irrelevant for the vesicular stomatitis virus-directed N15 TCR. The Sendai virus nonamer binds to Kb with identical affinity to VSV8, but is not activating (36).
As shown in Fig. 4E, application of 50 pN of tangential mechanical force to each of two naïve N15 TCR transgenic lymph node T cells via VSV8/Kb-coupled beads induced a significant change in the MFI of the calcium sensing dye, calcium orange. In contrast, application of the same force to SEV9/Kb beads induced no intracellular calcium change when monitored in the same way. Approximation of VSV8/Kb bead to the N15 T cell but without force application induced no calcium change and was indistinguishable from the SEV9/Kb curve as depicted during a 5-min of continuous observation. In summary, TCR triggering by pMHC in this system is antigen specific and requires application of force during the pMHC interaction with the TCR.
DISCUSSION
The TCR complex projects less than 100 Å from the T cell membrane and is therefore resistant to nonspecific perturbations such as fluid shear in distinction to mechanosensors in the nervous system (37). The highly selective signaling torque generated through relevant pMHC ligation during T cell scanning of APC surfaces likely further precludes “nonspecific” activation to foster antigen-specific activation events including calcium flux and integrin up-regulation. This in turn can facilitate stop movement for the T cell to form the immunologic synapse. Mechanoreceptor function is most likely tunable by inducible actin cytoskeletal interactions with TCR and pMHC on T cells and APCs, respectively, because torque will be greatest in their presence. Within the synapse, force could be generated on the TCR via microcluster formation and actin-based trafficking (38). Mechanosensing can be further amplified at the synapse where intermembrane distances (∼150 Å) are optimal for TCR/pMHC ligation (supplemental Fig. S11A) in conjunction with other signaling molecules to sustain activation from both p-SMAC and c-SMAC components. Rapid transport of TCRs to the immunological synapse and TCR signaling after disengagement of pMHC likely gives rise to serial TCR triggering for high activation efficiency within the immunological synapse. The force required in this dynamic mechanosensor model could also be mediated by dimeric antibody or multimeric pMHC (i.e. tetramers) bound to two sets of TCR complexes (supplemental Fig. S11B) without a prerequisite for TCR or pMHC oligomerization via “permissive” geometry on the T cell membrane. This suggestion also explains why monomeric pMHC and anti-CD3 Fab are ineffective at T cell activation.
Our data suggest a model wherein tangential force is applied along the pseudo-2-fold symmetry axis of the TCR complex such that the αβ heterodimer exerts torque on CD3ϵγ and CD3ϵδ. There is a directionality to the TCR mechanosensor at the molecular level. However, given the rapid rotational motion of both TCR and pMHC, the assembly likely reorients relative to the direction of cell movement.
2C11 mAb binding exploits the most sensitive triggering direction of pMHC signaling (supplemental Fig. S11). Thus only minute force is required for T cell activation in contrast to the more vertically binding 17A2 and pMHC. Such lateral force may come from Brownian motion of the bead used in the optical tweezer experiments or diffusion/transportation of the TCR complex. Given the close proximity of the Cβ FG loop to the CD3ϵ of the CD3ϵγ heterodimer, we envision the FG loop as a lever to exert force on CD3ϵ upon pMHC ligation at the moving cell surface. This is similar to the torque mediated by 2C11 pulling on CD3ϵ. Obviously, physical force is required for the FG loop to impinge on the adjacent CD3ϵγ heterodimer (supplemental Fig. S11) in agreement with results depicted in Fig. 4E.
The application of 50 pN of force as used herein is a reasonable force to be anticipated under physiologic conditions. In this regard, Wulfing et al. (39) determined the minimum adhesion strength between activated T cells and APCs to be 90 dynes/cm2. A force of 50 pN results in an applied stress of 80 dynes/cm2, assuming the contact area between the cell and the bead is approximately half the bead surface area.
These data and our mechanosensor model need to be considered in the context of alternative signaling mechanisms proposed for the TCR. For example, earlier studies suggesting that pMHC applies an external force to push on or “deform” the TCR (10, 40) are not inconsistent with the current view, although the present model is more detailed, being guided by further structural and functional information. Moreover, a singularly vertical or piston-like motion previously suggested as a signaling basis seems excluded by the optical trap analysis herein. The notions of permissive geometry (41) involving dimers of TCRs and pMHC or pseudodimers (42) are unlikely given the extensive glycosylation of the TCR complex. However, such considerations do not exclude microclustering of TCR complexes in the absence of TCR and/or co-receptor protein ectodomain oligomerization, nor models of kinetic segregation. The latter is suggested to partition inhibitory phosphatases away from the activation machinery in the T cell-APC contact zone (43).
That the TCR is an evolved mechanosensor activated by direction-specific physical force has several immediate implications. First, because the total force applied to the T cell surface is essentially defined during movement of the T cell membrane relative to that of the APC, ligation of several TCRs by several cognate pMHCs on the opposing APC will exert a greater physical force on each individual TCR than multiple TCR ligations on the same T cell by a large number of TCR-pMHC interactions. Hence, specificity and sensitivity are built into TCR mechanosensor function. Second, in principle, shear forces can form catch bonds at the TCR-pMHC interface to enhance binding and/or confer additional ligand specificity. These bonds that are strengthened by tensile force have been described for cell adhesion molecules (44). Third, because the B cell receptor is made up of cell surface immunoglobulin associated with immunoreceptor tyrosine-based activation motif-containing Igα and Igβ invariant proteins (45), physical force may be important for B cell receptor-mediated B cell activation as well. Precedent for mechanoreceptors in the hematopoietic system is the von Willebrand factor receptor on platelets where tensile stress on bonds between the glycoprotein Ibα subunit and the von Willebrand factor A1 domain under fluid dynamic conditions triggers integrin αIIbβIII activation to support platelet adhesion (46). Fourth, this anisotropic (direction-dependent) non-equilibrium signaling mechanism implies that isotropic equilibrium constants may not be sufficient to describe mechanoreceptor signaling kinetics.
Supplementary Material
Acknowledgments
We thank Drs. Herman N. Eisen, Oreste Acuto, Darrell J. Irvine, Kristine Brazin, Linda Clayton, and Robert Mallis for thoughtful reviews of this manuscript. We acknowledge Dr. Haesook Kim and Dr. Guanglan Zhang for statistical analysis and Maris Handley for flow cytometry.
This work was supported, in whole or in part, by National Institutes of Health Grants AI19807 (to E. L. R.) and AI37581, GM47467, and EB002026 (to G. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S11.
- TCR
- T cell receptor
- pMHC
- antigenic peptide bound to a major histocompatibility complex molecule
- mAb
- monoclonal antibody
- HSQC
- hetero-nuclear single-quantum coherence
- MFI
- mean fluorescence intensity
- FITC
- fluorescein isothiocyanate
- MES
- 4-morpholineethanesulfonic acid
- APC
- antigen presenting cell
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- tg
- transgenic
- VSV
- vesicular stomatitis virus
- LN
- lymph node
- PDB
- Protein data bank
- TROSY
- transverse relaxation optimization spectroscopy
- N
- newton
- sc
- single-chain
- p-SMAC
- peripheral supramolecular activation cluster
- c-SMAC
- central supramolecular activation cluster.
REFERENCES
- 1.Rudolph M. G., Stanfield R. L., Wilson I. A. (2006) Annu. Rev. Immunol. 24, 419–466 [DOI] [PubMed] [Google Scholar]
- 2.Iwashima M., Irving B. A., van Oers N. S., Chan A. C., Weiss A. (1994) Science 263, 1136–1139 [DOI] [PubMed] [Google Scholar]
- 3.Shinkai Y., Ma A., Cheng H. L., Alt F. W. (1995) Immunity 2, 401–411 [DOI] [PubMed] [Google Scholar]
- 4.Sprent J., Surh C. D. (2002) Annu. Rev. Immunol. 20, 551–579 [DOI] [PubMed] [Google Scholar]
- 5.Teixeiro E., Daniels M. A., Hamilton S. E., Schrum A. G., Bragado R., Jameson S. C., Palmer E. (2009) Science 323, 502–505 [DOI] [PubMed] [Google Scholar]
- 6.Reth M. (1989) Nature 338, 383–3842927501 [Google Scholar]
- 7.Letourneur F., Klausner R. D. (1992) Science 255, 79–82 [DOI] [PubMed] [Google Scholar]
- 8.Crabtree G. R., Clipstone N. A. (1994) Annu. Rev. Biochem. 63, 1045–1083 [DOI] [PubMed] [Google Scholar]
- 9.Robb R. J., Munck A., Smith K. A. (1981) J. Exp. Med. 154, 1455–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Z. J., Kim K. S., Wagner G., Reinherz E. L. (2001) Cell 105, 913–923 [DOI] [PubMed] [Google Scholar]
- 11.Sun Z. Y., Kim S. T., Kim I. C., Fahmy A., Reinherz E. L., Wagner G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16867–16872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haks M. C., Krimpenfort P., Borst J., Kruisbeek A. M. (1998) EMBO J. 17, 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghendler Y., Hussey R. E., Witte T., Mizoguchi E., Clayton L. K., Bhan A. K., Koyasu S., Chang H. C., Reinherz E. L. (1997) Eur. J. Immunol. 27, 2279–2289 [DOI] [PubMed] [Google Scholar]
- 14.Kern P. S., Teng M. K., Smolyar A., Liu J. H., Liu J., Hussey R. E., Spoerl R., Chang H. C., Reinherz E. L., Wang J. H. (1998) Immunity 9, 519–530 [DOI] [PubMed] [Google Scholar]
- 15.Moody A. M., Xiong Y., Chang H. C., Reinherz E. L. (2001) Eur. J. Immunol. 31, 2791–2799 [DOI] [PubMed] [Google Scholar]
- 16.Ferentz A. E., Wagner G. (2000) Q. Rev. Biophys. 33, 29–65 [DOI] [PubMed] [Google Scholar]
- 17.Takahashi H., Nakanishi T., Kami K., Arata Y., Shimada I. (2000) Nat. Struct. Biol. 7, 220–223 [DOI] [PubMed] [Google Scholar]
- 18.Fahmy A., Wagner G. (2002) J. Am. Chem. Soc. 124, 1241–1250 [DOI] [PubMed] [Google Scholar]
- 19.Brau R. R., Tarsa P. B., Ferrer J. M., Lee P., Lang M. J. (2006) Biophys. J. 91, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes S. M., Love P. E. (2002) Immunity 16, 827–838 [DOI] [PubMed] [Google Scholar]
- 21.Nishida T., Matsuki Y., Ono T., Oguma T., Tsujimoto K., Sato M., Tadakuma T. (2004) J. Immunol. 172, 6634–6641 [DOI] [PubMed] [Google Scholar]
- 22.Mizoguchi A., Mizoguchi E., de Jong Y. P., Takedatsu H., Preffer F. I., Terhorst C., Bhan A. K. (2003) Int. Immunol. 15, 97–108 [DOI] [PubMed] [Google Scholar]
- 23.Altan-Bonnet G., Germain R. N. (2005) PLoS Biol. 3, e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Lim K., Smolyar A., Teng M., Liu J., Tse A. G., Liu J., Hussey R. E., Chishti Y., Thomson C. T., Sweet R. M., Nathenson S. G., Chang H. C., Sacchettini J. C., Reinherz E. L. (1998) EMBO J. 17, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Havran W. L., Poenie M., Kimura J., Tsien R., Weiss A., Allison J. P. (1987) Nature 330, 170–173 [DOI] [PubMed] [Google Scholar]
- 26.Kuhns M. S., Davis M. M. (2007) Immunity 26, 357–369 [DOI] [PubMed] [Google Scholar]
- 27.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 28.Ghendler Y., Smolyar A., Chang H. C., Reinherz E. L. (1998) J. Exp. Med. 187, 1529–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touma M., Sun Z. Y., Clayton L. K., Marissen W. E., Kruisbeek A. M., Wagner G., Reinherz E. L. (2007) J. Immunol. 178, 3668–3679 [DOI] [PubMed] [Google Scholar]
- 30.Kjer-Nielsen L., Dunstone M. A., Kostenko L., Ely L. K., Beddoe T., Mifsud N. A., Purcell A. W., Brooks A. G., McCluskey J., Rossjohn J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7675–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnett K. L., Harrison S. C., Wiley D. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16268–16273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer E., Naeher D. (2009) Nat. Rev. Immunol. 9, 207–213 [DOI] [PubMed] [Google Scholar]
- 33.Sasada T., Touma M., Chang H. C., Clayton L. K., Wang J. H., Reinherz E. L. (2002) J. Exp. Med. 195, 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Touma M., Chang H. C., Sasada T., Handley M., Clayton L. K., Reinherz E. L. (2006) J. Immunol. 176, 6812–6823 [DOI] [PubMed] [Google Scholar]
- 35.van Mameren J., Modesti M., Kanaar R., Wyman C., Peterman E. J., Wuite G. J. (2009) Nature 457, 745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghendler Y., Teng M. K., Liu J. H., Witte T., Liu J., Kim K. S., Kern P., Chang H. C., Wang J. H., Reinherz E. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10061–10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kung C. (2005) Nature 436, 647–654 [DOI] [PubMed] [Google Scholar]
- 38.Yokosuka T., Kobayashi W., Sakata-Sogawa K., Takamatsu M., Hashimoto-Tane A., Dustin M. L., Tokunaga M., Saito T. (2008) Immunity 29, 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wülfing C., Sjaastad M. D., Davis M. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6302–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Z., Janmey P. A., Finkel T. H. (2008) FASEB J. 22, 1002–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minguet S., Schamel W. W. (2008) Trends Biochem. Sci. 33, 51–57 [DOI] [PubMed] [Google Scholar]
- 42.Krogsgaard M., Li Q. J., Sumen C., Huppa J. B., Huse M., Davis M. M. (2005) Nature 434, 238–243 [DOI] [PubMed] [Google Scholar]
- 43.Davis S. J., van der Merwe P. A. (2006) Nat. Immunol. 7, 803–809 [DOI] [PubMed] [Google Scholar]
- 44.Marshall B. T., Long M., Piper J. W., Yago T., McEver R. P., Zhu C. (2003) Nature 423, 190–193 [DOI] [PubMed] [Google Scholar]
- 45.Kulathu Y., Hobeika E., Turchinovich G., Reth M. (2008) EMBO J. 27, 1333–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruggeri Z. M. (2007) Thromb. Haemost. 98, 55–62 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.