Abstract
NeuN (neuronal nuclei) is a neuron-specific nuclear protein which is identified by immunoreactivity with a monoclonal antibody, anti-NeuN. Anti-NeuN has been used widely as a reliable tool to detect most postmitotic neuronal cell types in neuroscience, developmental biology, and stem cell research fields as well as diagnostic histopathology. To date, however, the identity of its antigen, NeuN itself, has been unknown. Here, we identify NeuN as the Fox-3 gene product by providing the following evidence: 1) Mass spectrometry analysis of anti-NeuN immunoreactive protein yields the Fox-3 amino acid sequence. 2) Recombinant Fox-3 is recognized by anti-NeuN. 3) Short hairpin RNAs targeting Fox-3 mRNA down-regulate NeuN expression. 4) Fox-3 expression is restricted to neural tissues. 5) Anti-Fox-3 immunostaining and anti-NeuN immunostaining overlap completely in neuronal nuclei. We also show that a protein cross-reactive with anti-NeuN is the synaptic vesicle protein, synapsin I. Anti-NeuN recognizes synapsin I in immunoblots with one order of magnitude lower affinity than Fox-3, and does not recognize synapsin I using immunohistology. Fox-3 (also called hexaribonucleotide-binding protein 3 and D11Bwg0517e) contains an RNA recognition motif and is classified as a member of the Fox-1 gene family that binds specifically to an RNA element, UGCAUG. We demonstrate that Fox-3 functions as a splicing regulator using neural cell-specific alternative splicing of the non-muscle myosin heavy chain II-B pre-mRNA as a model. Identification of NeuN as Fox-3 clarifies an important element of neurobiology research.
Introduction
Mullen et al. (1) have reported a monoclonal antibody (mAb)2, which was generated using brain cell nuclei as antigens. This mAb recognizes 2–3 protein bands with apparent molecular masses of 46–48 kDa following SDS-PAGE that are expressed in neuronal tissues. Intensive immunohistochemical analyses using embryonic and adult murine tissues have demonstrated that this mAb stains exclusively neuronal cells in the central and peripheral nervous systems, especially postmitotic and differentiating neurons, as well as terminally differentiated neurons. The mAb staining is localized primarily to nuclei. Thus, the authors named the antigen recognized by this mAb NeuN for “Neuronal Nuclei.” The original study and subsequent studies by others have shown that anti-NeuN stains most types of neurons throughout the nervous system with few exceptions (1, 2). In no case has NeuN expression been observed in glial cells. This mAb can detect the NeuN antigen in a wide range of vertebrate species, including mammals, birds, and amphibians. Because of its high specificity for postmitotic neurons, its broad specificity for most types of neuronal cells, and its cross-reactivity with multiple species, anti-NeuN has gained widespread acceptance as a reliable tool to detect neuronal cells in neuroscience and developmental biology research and in diagnostic histopathology for neural diseases and tumors (3–10). More recently, it has also been used to monitor neuronal differentiation of stem cells (11–15).
Despite widespread use of NeuN as a general neuronal marker for over 15 years, only a few studies have addressed the biochemical properties and the potential function of NeuN (1, 16). To date, there is still no definitive characterization for NeuN, and in this post-genome era, molecular identification of NeuN is anticipated. In this study, we determine the gene that encodes NeuN using the following criteria: 1) Its protein product reacts with anti-NeuN. 2) Its protein products have apparent molecular masses similar to 46–48 kDa in SDS-PAGE. 3) Its expression is restricted to neuronal tissues. 4) Its protein products localize predominantly to the nuclei of neurons. We present evidence that NeuN is the Fox-3 gene product, a member of the RNA-binding protein Fox-1 gene family, and that Fox-3 functions as a splicing regulator. We also address the identity of the ∼70-kDa protein(s) which cross-react with anti-NeuN.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Short Hairpin (shRNA) RNA
P19 cells were obtained from ATCC and maintained in MEMα (Invitrogen) containing 7.5% bovine calf serum and 2.5% fetal bovine serum (FBS). To induce neural differentiation of P19 cells, 1 × 106 cells were cultured in a bacteria grade Petri dish in 10 ml of medium containing 5 × 10−7 m all-trans retinoic acid (Sigma) for 4 days. The cell aggregates were resuspended by mild pipetting and trypsin/EDTA treatment. The resuspended cells were then transferred to a poly-d-lysine-coated tissue culture dish and cultured in retinoic acid-free medium for an additional 4 days. To establish shRNA-expressing P19 cells, the cells were transfected with pRS-shRNA constructs encoding shRNA against mouse Fox-3 or green fluorescent protein (GFP, OriGene) using the Effectene transfection reagent (Qiagen). The stably transfected clonal cells were selected with 2 μg/ml puromycin and maintained in 1 μg/ml puromycin. The pRS-shRNA constructs ID TI543802 (target CGCACAGACTCATCCTGAGCAGCCAGGCA), TI543803 (target TTGAAACTAGCTCAGATGCTGACCGAGCC), and TR30003 (non-effective negative control) are shown as shRNA T-2, T-3, and GFP, respectively, in Fig. 1E. The human embryonic kidney cell line HEK-293 and the human neuroblastoma cell line SK-N-SH cells were maintained in Dulbecco's modified Eagle's medium containing 10% FBS and MEMα containing 10% FBS, respectively. Effectene transfection reagent was used for transfection of the plasmid constructs.
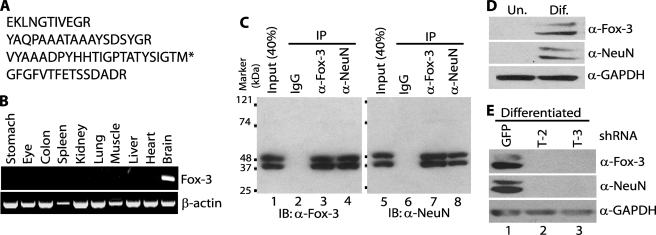
FIGURE 1.
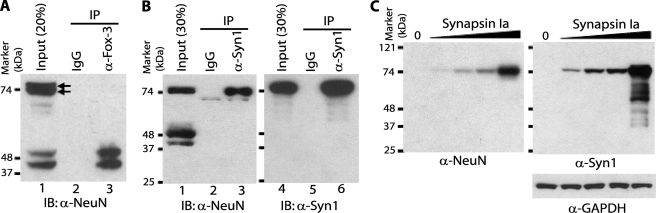
Identification of the protein recognized by anti-NeuN as Fox-3. A, tryptic peptide sequences of the 40-kDa protein identified as Fox-3 by MS analysis. The 40-kDa protein immunoprecipitated by anti-NeuN was subjected to MS analysis. * indicates the C-terminal end peptide. B, brain-restricted expression of Fox-3 mRNA. Expression of Fox-3 mRNAs in the indicated tissues from adult mice was analyzed by RT-PCR. β-Actin mRNA is used as a positive control. Ethidium bromide-stained agarose gels are shown. C, detection by anti-Fox-3 of proteins immunoprecipitated by anti-NeuN and detection by anti-NeuN of proteins immunoprecipitated by anti-Fox-3. Nuclear extract of brain and spinal cord was immunoprecipitated with nonspecific mouse immunoglobulin (IgG), anti-Fox-3 (α-Fox-3), and anti-NeuN (α-NeuN). Immunoprecipitation (IP) was followed by immunoblot (IB) analysis with anti-Fox-3 (lanes 1–4) or anti-NeuN (lanes 5–8). D, neural differentiation results in expression of protein(s) detected by anti-NeuN and anti-Fox-3. Whole cell extracts of P19 cells in the undifferentiated (Un.) and neurally differentiated (Dif.) stages were analyzed by immunoblots using the indicated Abs. GAPDH serves as a loading control. E, down-regulation of NeuN expression by the Fox-3-targeting shRNAs. P19 cells expressing the indicated shRNAs were treated with retinoic acid. Whole cell extracts were analyzed by immunoblots using the indicated Abs. GFP shRNA is used as a non-effective negative control.
RNA Preparation and RT-PCR
Total RNA was isolated from adult mouse tissues using an RNeasy mini kit (Qiagen). RT-PCR was performed using Superscript III RNase H− reverse transcriptase (Invitrogen) and Pfu Turbo DNA polymerase (Stratagene). The PCR primers used for Fig. 1B were as follows: for Fox-3 5′-CCAGGCACTGAGGCCAGCACACAGC-3′ and 5′-CTCCGTGGGGTCGGAAGGGTGG-3′; for β-actin 5′-ATCGTGGGCCGCCCTAGGCA-3′ and 5′-TGGCCTTACCCTTCAGAGGGG-3′. The PCR primers to obtain the full-length coding region of Fox-3 cDNAs were 5′-ctcaggcctccactagtgATGGCCCAGCCCTACCCCCCTG-3′ and 5′-ctcaggcctcctctagaaGTAGGGGGTGAAGCGGCTGTA-3′. Lowercase letters represent adapter sequences including restriction enzyme sites. RT-PCR for the minigene-derived mRNAs was carried out as previously described (17).
Construction of Expression Plasmids and Minigenes
The full-length coding regions of the Fox-3 cDNAs were introduced into a plasmid pCS3+MT, which contained 6 copies of a Myc epitope (Figs. 2A, 6, and 8). Defined cDNA fragments encoding various domains of Fox-3 were generated by PCR with appropriate sets of primers and cloned in the pEGFP-C3 vector (Clontech) or the pFN2A(GST)Flexi vector (Promega) which had been modified to delete the Barnase sequence (Fig. 2, B–D). For preparation of antigen, the Fox-3 cDNA fragment encoding amino acids 1–97 was generated by PCR and cloned in the pGEX-5X-1 vector (GE Healthcare). The expression plasmids for Fox-1 and Fox-2 (Fig. 2A) have been described previously as A016 and F011, respectively (17). The expression plasmids for synapsin Ia (Figs. 5C and 7, B and C) and IIa were obtained from Open Biosystems and OriGene, respectively. The synapsin Ia cDNA insert was cut out from the pCMV-SPORT6 vector using SmaI and XbaI and was transferred to the pCS3+MT vector between the StuI and XbaI sites (Fig. 6). Minigene constructs (Fig. 8) used to analyze splicing activity were described previously (17).
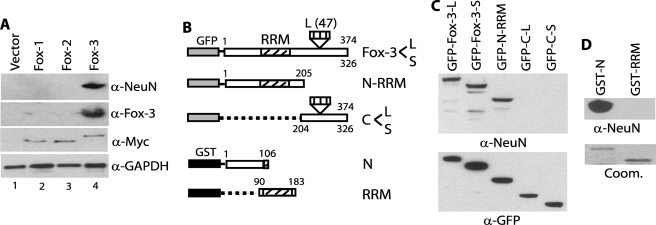
FIGURE 2.
Recognition and epitope mapping of recombinant Fox-3 by anti-NeuN. A, recognition of recombinant Fox-3 by anti-NeuN. The expression constructs encoding Myc-tagged Fox-1, -2, and -3 and the empty vector were transfected in SK-N-SH cells. Total cell lysates were analyzed by immunoblots using the indicated Abs. B, schematic representation of full-length and deletion constructs of Fox-3 fused to GFP or GST. Each construct includes the region indicated by the boxes and deletions are indicated by the broken lines or the empty spaces. Numbers represent amino acids. GenBankTM accession numbers for mouse Fox-3-L and Fox-3-S are FJ958310 and FJ958311, respectively. C, detection of the N-terminal 1–205-amino acid fragment of Fox-3 by anti-NeuN. The indicated GFP fusion constructs were transfected into HEK-293 cells. Total cell lysates were analyzed by immunoblots using the indicated Abs. Anti-GFP (α-GFP) verifies the expression of each protein. D, detection of the bacterially expressed N-terminal 1–106-amino acid fragment of Fox-3 by anti-NeuN. The indicated GST fusion constructs were expressed in bacteria. Purified proteins were analyzed by immunoblot using anti-NeuN. Approximately 0.1% of the protein amounts shown in the Coomassie Blue-stained gel (Coom.) were loaded for the immunoblot analysis.
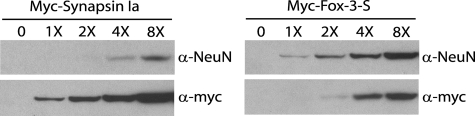
FIGURE 6.
Different immunoreactivities of anti-NeuN with Fox-3 and synapsin I. HEK-293 cells were transfected with either Myc-tagged Fox-3-S or Myc-tagged synapsin Ia expression constructs. Various amounts of total cell lysates indicated by 1X-8X (the numbers reflect relative amounts in each lane) were analyzed in parallel by immunoblotting using anti-Myc (α-myc) and anti-NeuN (α-NeuN). Immunoblots using anti-Myc show relative amounts of the expressed proteins in both panels. 0, untransfected cell lysates.
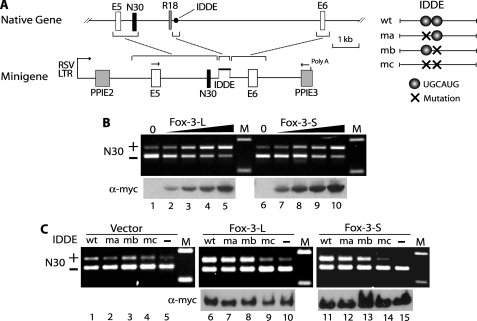
FIGURE 8.
Activation of N30 splicing by Fox-3. A, schematic diagrams of NMHC II-B gene, minigene constructs and IDDEs. Native gene shows only a part of the human NMHC II-B gene surrounding exon N30. E5 and E6 are constitutive exons, and N30 and R18 are alternative exons. The IDDE is an intronic region consisting of 201 nucleotides and is located ∼1.5-kb downstream of N30 in the native gene. Rectangles and horizontal lines in the diagrams indicate exons and introns, respectively. Exon size and the IDDE are not drawn to scale. Minigenes include the NMHC II-B genomic DNA fragments indicated by brackets, which are flanked by exons E2 and E3 of the rat preproinsulin gene (PPI). The wild-type (wt) and mutant (ma, mb, and mc) IDDEs are inserted at the indicated location in the minigene. Transcription of the minigene is driven by the Rous sarcoma virus long terminal repeat (RSVLTR). Arrows above E5 and PPIE3 indicate the location of the primers used for RT-PCR. B, dose-dependent activation of N30 splicing by Fox-3. Increasing amounts (0.03, 0.1, 0.3, and 1 μg, indicated by triangle) of the expression constructs encoding Myc-tagged Fox-3-L and -S were co-transfected into SK-N-SH cells with the wild-type minigene. The mRNAs derived from the minigene were analyzed by RT-PCR (upper panel). The upper band includes N30 (+) and the lower band excludes N30 (−). Immunoblots using anti-Myc (lower panel) show relative amounts of the expressed proteins. 0, the empty vector alone; M, molecular size marker. C, UGCAUG-dependent activation of N30 splicing by Fox-3. The minigenes containing wild-type (wt), mutations (ma, mb, and mc) and deletion (−) of the IDDE were co-transfected into SK-N-SH cells with the expression constructs encoding Myc-tagged Fox-3-L and -S. RT-PCR of the minigene mRNAs (upper panel) and immunoblots for the expressed proteins (lower panel) are shown.
FIGURE 5.
Cross-reactivity of anti-NeuN with synapsin I. A, failure of anti-Fox-3 to cross-react with the 70-kDa proteins detected by anti-NeuN. Nonspecific IgG or anti-Fox-3 were used to immunoprecipitate proteins from whole tissue extract of brain and spinal cord. Immunoprecipitation (IP) was followed by immunoblot (IB) analysis with anti-NeuN (α-NeuN). The arrows indicate doublet band. B, detection by anti-NeuN of proteins immunoprecipitated by anti-synapsin I. IgG or anti-synapsin I (α-Syn1) was used to immunoprecipitate from whole tissue extract of spinal cord. Immunoprecipitation (IP) was followed by immunoblot (IB) analysis using the indicated Abs. C, recognition of recombinant synapsin Ia by anti-NeuN. HEK-293 cells were transfected with increasing amounts (0.1, 0.3, 1, 3 μg, indicated by triangle) of the expression construct encoding mouse synapsin Ia. Total cell lysates were analyzed by immunoblots using the indicated Abs. 0, the empty vector alone.
FIGURE 7.
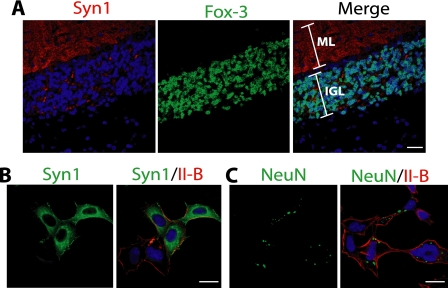
Failure of anti-NeuN to cross-react with synapsin I in fixed tissues and cells. A, different staining pattern of anti-synapsin I from that of anti-NeuN or anti-Fox-3. The mouse cerebellar section was co-stained with anti-synapsin I (red) and anti-Fox-3 (green). Compare this image with that in Fig. 3A. DAPI stains nuclei. ML, molecular layer; IGL, internal granular layer. B and C, failure of anti-NeuN to cross-react with synapsin I. HEK-293 cells were transfected with the expression construct encoding synapsin Ia. The fixed and permeabilized cells were stained with anti-synapsin I (B, green) or anti-NeuN (C, green) together with anti-NMHC II-B (red). Bars indicate 20 μm. Syn1, synapsin I: II-B, NMHC II-B.
Generation of Fox-3 Antisera and Other Antibodies (Abs)
The N-terminal 1–97-amino acid fragment of Fox-3 fused to glutathione S-transferase (GST) was expressed in BL21 bacteria and purified by a GSTrap FF column (GE Healthcare). The GST-Fox-3-(1–97) fusion protein was digested with Factor-Xa and the cleaved Fox-3-(1–97) protein fragment was purified using Benzamidine Sepharose 4 FF and GSTrap FF columns. Purified Fox-3-(1–97) protein was used to generate polyclonal anti-Fox-3 in rabbits (Biosynthesis, Inc). The primary Abs used in this study were mouse monoclonal anti-NeuN (Millipore), mouse monoclonal anti-Myc (Invitrogen), mouse monoclonal anti-GAPDH (Biodesign), mouse monoclonal anti-synapsin I (Synaptic Systems), rabbit polyclonal anti-GFP (Clontech), and rabbit polyclonal anti-non-muscle myosin heavy chain II-B (18).
Preparation of Extracts, Immunoprecipitation, and Immunoblot Analysis
Whole tissue or cell extracts were prepared using radioimmune precipitation assay buffer (Sigma) supplemented with a protease inhibitor mixture (Sigma). Nuclear extracts were prepared using a NE-PER kit (Pierce) from single cell suspensions, which were prepared by passing the tissue through a cell strainer. For immunoprecipitation, the extracts containing 0.5–1 mg of protein in 5–10 ml of Co-IP buffer (Pierce Biotechnology) were incubated with the primary Abs overnight at 4 °C. The immunocomplexes were collected by incubation with protein A/G PLUS-agarose (Santa Cruz Biotechnology) and washed in the Co-IP buffer. The complexes were solubilized in a SDS sample buffer and subjected to SDS-PAGE using a 4–20% gradient polyacrylamide NuPAGE Bis-Tris gel and a NuPAGE MOPS SDS running buffer (Invitrogen). ProSieve color protein markers (Lonza) were used as Mr standards. Total cell lysates from cultured cells were prepared by direct addition of SDS sample buffer to whole cells. For immunoblot analysis, the protein samples were transferred to a nitrocellulose membrane following SDS-PAGE. Binding of antibodies was detected by the SuperSignal system (Pierce).
Mass Spectrometry
Following SDS-PAGE, the Coomassie Blue-stained protein bands were digested in the gels with trypsin. The recovered peptides were analyzed using an Applied Biosystems 4700 MALDI-TOF/TOF to acquire tandem MS/MS spectra. A compiled protein data base was searched for the peptide sequences.
Flow Cytometry
Brain tissues were passed through a cell strainer to prepare a single cell suspension. The cells were washed in phosphate-buffered saline containing bovine serum albumin and then fixed and permeabilized in Fix/perm buffer (eBioscience). Nonspecific binding sites were blocked with 5% goat serum for 30 min at 4 °C, and then the cells were labeled with either anti-NeuN alone, anti-Fox-3 alone, or both Abs at a final dilution of 1:100 for 45 min at 4 °C. The cells were further incubated with Alexa-594 or Alexa-488-conjugated goat Abs against mouse IgG and rabbit IgG (Molecular Probes), respectively, at a 1:500 dilution for 15 min at 4 °C. The resulting cells were analyzed using FACSCalibur (BD Biosciences).
Immunofluorescence Microscopy
HEK-293 cells transfected with a synapsin Ia expression plasmid were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline. Mouse brains and spinal cords were processed as described previously (19). Briefly, paraformaldehyde-fixed tissues were embedded in paraffin and cut into 4-μm sections. Antigen retrieval was performed on hydrated sections by immersing them in 10 mm sodium citrate (pH 6.0) and microwaving for 10 min. The primary Abs are described above. The Alexa-488- and Alexa-594-conjugated goat Abs against mouse IgG and rabbit IgG were used as secondary Abs. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). The specimens were examined using a Zeiss LSM 510 Meta confocal laser-scanning microscope.
RESULTS
Identification of the 40–50-kDa NeuN Antigen as Fox-3
Because mouse brain cell nuclei were originally used as antigens to generate the mAb anti-NeuN and NeuN is enriched in nuclei, we first used mouse brain nuclear extracts as source materials to identify the NeuN protein. In agreement with the original report (1), anti-NeuN detects two major bands with approximate masses of 40 and 50 kDa by immunoblot analysis following SDS-PAGE (Fig. 1C, lane 5). These estimated masses are slightly different from those in the original report (46–48 kDa), presumably due to differences in the buffer conditions during SDS-PAGE and the Mr markers used. We then carried out immunoprecipitation using anti-NeuN to isolate these proteins from crude nuclear extracts. Although the 50-kDa band co-migrated with the immunoglobulin heavy chain following SDS-PAGE, the 40-kDa band separated from it and was subjected to mass spectrometry (MS) analysis. The largest numbers of peptide sequences obtained are derived from the product of a gene called D11Bwg0517e in mice, and its orthologs in other species called hexaribonucleotide-binding protein 3 and FOX3. Hereafter we designate it as Fox-3. The amino acid sequences of the tryptic peptides obtained for Fox-3 are listed in Fig. 1A. The full-length Fox-3 amino acid sequence is in supplemental Fig. S1. Fox-3 is predicted to be an RNA-binding protein. However its expression and its function have not been characterized. Therefore, we next analyzed the expression of Fox-3 mRNA in various mouse tissues by RT-PCR. Fox-3 expression is confined to the brain (Fig. 1B), consistent with the neuron-specific expression of NeuN.
In an effort to generate antibodies (Abs) against Fox-3, we cloned its cDNAs and bacterially expressed the N-terminal region of Fox-3 (amino acids 1–97), which was used to immunize rabbits. Using the rabbit polyclonal anti-Fox-3 that was generated, we analyzed immunoblots from mouse brain nuclear extracts or immunoprecipitates of extracts with anti-NeuN or anti-Fox-3. Anti-Fox-3 detects the same Mr proteins detected by anti-NeuN in all three samples (Fig. 1C, left panel), providing strong evidence that NeuN and Fox-3 are the same proteins. Of note, despite performing MS analysis using only the 40-kDa protein, anti-Fox-3 detects both the 40- and 50-kDa proteins. This result indicates that both the 40- and 50-kDa proteins are Fox-3 gene products, presumably generated by alternative splicing (see “Discussion”). Reciprocal immunoprecipitation and immunoblot analysis demonstrate that the proteins immunoprecipitated with anti-Fox-3 are also detected by anti-NeuN (Fig. 1C, right panel). Moreover the observation that the relative ratio of the band intensities in the input extracts versus the immunoprecipitates on the immunoblot using anti-Fox-3 is similar to that on the immunoblot using anti-NeuN (compare the ratio of lanes 1 versus 4 to that of lanes 5 versus 8) supports the notion that anti-NeuN and anti-Fox-3 are immunoprecipitating and detecting the same proteins.
To further verify the identity of 40–50-kDa NeuN as Fox-3 by manipulating NeuN expression or Fox-3 expression in cell culture, we made use of the mouse embryonic carcinoma cell line P19. It has been reported that NeuN expression is up-regulated in P19 cells during neural differentiation triggered by retinoic acid treatment (1). As shown in the immunoblot in Fig. 1D, undifferentiated and proliferating P19 cells do not express NeuN at a detectable level. Upon retinoic acid treatment, they withdraw from the cell cycle and extend dendrites. In this post-mitotic, neurally differentiated stage, P19 cells now express NeuN at 40 and 50 kDa. Anti-Fox-3 also detects the same Mr bands under the differentiated condition (Fig. 1D). Notably, both NeuN and Fox-3 have the same Mr isoforms with a similar ratio, suggesting that NeuN is Fox-3. Next, we attempted to down-regulate Fox-3 expression using shRNAs. P19 cells were stably transfected with the construct encoding shRNA T-2 or T-3, which target different regions of Fox-3 mRNA, and then these cells were treated with retinoic acid. For mock transfection, the construct which encodes shRNA targeting GFP mRNA was used. The GFP shRNA-expressing cells continue to express Fox-3 proteins with masses of 40 and 50 kDa (Fig. 1E, upper panel, lane 1). However, as expected, the Fox-3 protein expression is down-regulated in the T-2 and T-3 shRNA-expressing cells (Fig. 1E, upper panel, lanes 2 and 3). Importantly, in these cells anti-NeuN also fails to detect the same Mr proteins, whereas it detects them in the control GFP shRNA-expressing cells (Fig. 1E, middle panel). These data support the idea that NeuN is the product of Fox-3 mRNAs.
To definitively determine that NeuN is Fox-3, the full-length coding region of one of the Fox-3 isoforms was cloned and exogenously expressed in SK-N-SH cells, which do not express endogenous Fox-3. The Myc-tagged recombinant Fox-3 is indeed detected by anti-NeuN as well as by anti-Fox-3 and anti-Myc (Fig. 2A, lane 4), indicating that Fox-3 is bona fide NeuN.
Data base analysis has revealed that there are two genes, Fox-1 and Fox-2, which are closely related to Fox-3 in mammalian genomes (supplemental Fig. S1) (20). Fox-1, -2, and -3 contain an almost identical RNA recognition motif (RRM, 95–100% amino acid identity). These three genes are predicted to express proteins with a similar range of Mr and all are expressed in brain (17, 20–23). Therefore we checked the possibility that anti-NeuN might cross-react with Fox-1 or Fox-2. Myc-tagged Fox-1 and Fox-2 were also expressed in SK-N-SH cells. Anti-Myc verifies that similar levels of Fox-1, -2, and -3 are expressed (Fig. 2A, third panel). Neither Fox-1 nor Fox-2 is detected by anti-NeuN (Fig. 2A, top panel), demonstrating the specificity of anti-NeuN for Fox-3.
Epitope Mapping of Anti-NeuN to the N-terminal Region of Fox-3
Next, we determined which region of the Fox-3 protein is recognized by anti-NeuN. During the course of cDNA cloning for Fox-3, we found that there are at least two alternatively spliced mRNA variants, Fox-3-L and Fox-3-S, which include or exclude 141 nucleotides encoding 47 amino acids located at amino acid 252 (supplemental Fig. S1 and Fig. 2B). The full-length, N-terminal half of Fox-3 including the RRM (N-RRM) and the C-terminal half (C-L, C-S) were fused to GFP, as depicted in Fig. 2B, and the fusion proteins were expressed in HEK-293 cells. Anti-NeuN detects both the full-length Fox-3-L and -S and the N-RRM fragment, but not the C-terminal C-L and C-S fragments (Fig. 2C). To further define the region recognized by anti-NeuN and to examine whether a post-translational modification unique to mammalian cells is required for Ab recognition, the N-terminal one-third of Fox-3 (N) and the RRM were separately fused to GST and were expressed in bacteria. Anti-NeuN recognizes the N-terminal region but not the RRM (Fig. 2D), consistent with the lack of recognition of Fox-1 and -2 (see supplemental Fig. S1 for sequence comparison). The epitope recognized by anti-NeuN is mapped to the N-terminal 1–106-amino acid region of Fox-3. Mammalian post-translational modification appears not to be required for the Ab recognition. Taken together, all the above biochemical analyses establish that the 40–50-kDa proteins detected by anti-NeuN in immunoblots are Fox-3 gene products and that the epitope resides in the first 106 amino acids.
Restricted Localization of Fox-3 to Neuronal Nuclei, Identical to NeuN
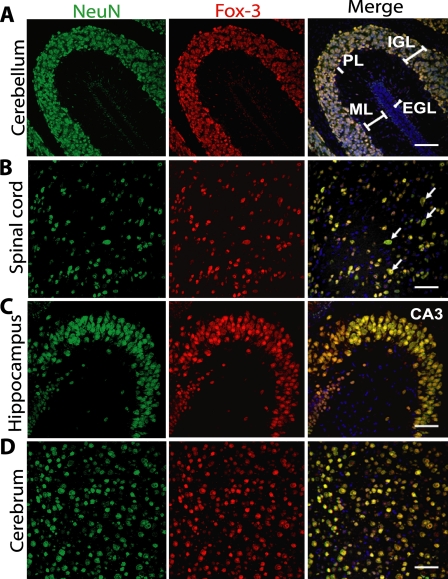
We next extended our analysis to determine whether Fox-3 met histological criteria for identity with NeuN. Anti-NeuN has been utilized extensively in histological and cell biological analyses and primarily stains nuclei of most neuronal cell types with a few exceptions, including cerebellar Purkinje cells (1, 2). We compared the staining patterns of anti-Fox-3 to those of anti-NeuN by co-staining with both Abs. Fig. 3 shows confocal immunofluorescence images of various regions of mouse brain and spinal cord at postnatal day 10. Careful inspection of these images as well as others reveals that anti-Fox-3 staining and anti-NeuN staining exactly overlap in every single cell. Both Abs stain nuclei of a majority of neural cells. Characteristically, however, the nuclei of Purkinje cells and external germinal cells in the cerebellum are devoid of staining with anti-Fox-3, similar to anti-NeuN (Fig. 3A, PL, EGL). In the case of some of the large neurons, such as motor neurons in the spinal cord, the cytoplasm of the soma is also diffusely stained with anti-NeuN, and this is also the case with anti-Fox-3 (Fig. 3B, arrows). The tissue distribution as well as subcellular localization of Fox-3 are the same as those of NeuN. Thus, Fox-3 fulfills the original histological definition for NeuN, Neuronal Nuclei.
FIGURE 3.
Co-localization of Fox-3 and NeuN in the mouse central nervous system. The sagittal section of the brain and the transverse section of the spinal cord (A, B, C, and D, as indicated) from postnatal day 10 mice were co-stained for NeuN (green) and Fox-3 (red). DAPI was used to stain nuclei (blue). Bars indicate 50 μm. Arrows in B point to motor neurons. EGL, external germinal layer; ML, molecular layer; PL, Purkinje cell layer; IGL, internal granular layer.
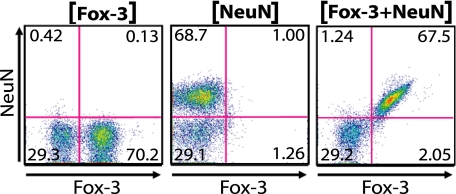
We also undertook a second approach to verify cytologically whether Fox-3-positive brain cells exactly equated to NeuN-positive cells. Dissociated cells from the adult mouse cerebrum were labeled with anti-NeuN and anti-Fox-3 and were subjected to fluorescence-activated cell sorting (FACS) analysis. As shown in Fig. 4, practically all double-labeled cerebral cells are sorted out either in the double-positive (67.5%) or double-negative (29.2%) quadrant (right panel). The left and center panels show the sorting profiles of cells labeled with either one of the two Abs to adjust the quadrants. These histological and cytological analyses further establish the identity of NeuN as Fox-3.
FIGURE 4.
Co-expression of Fox-3 and NeuN demonstrated by FACS analysis. Dissociated mouse cerebral cells were stained for either Fox-3 alone (left panel), NeuN alone (middle panel), or both Fox-3 and NeuN (right panel). The number in each quadrant represents % of cell numbers.
Cross-reactivity of Anti-NeuN Ab with Synapsin I in Immunoblot Analysis
For nearly two decades during the widespread use of anti-NeuN, a number of investigators have experienced cross-reactivity of this mAb to an ∼70-kDa protein(s) using immunoblot analysis (2, 24). Therefore we undertook to identify these proteins. Because detection of the 70-kDa proteins in our nuclear extracts was not consistent, we used total extracts of mouse brains and spinal cords without subcellular fractionation to avoid losing these proteins. As shown in Fig. 5A, anti-NeuN detects the 70-kDa proteins as doublet bands with similar intensity to the 40–50-kDa proteins in total extracts. We first asked whether these 70-kDa proteins could be additional Fox-3 gene products or not by immunoprecipitation using anti-Fox-3. Anti-Fox-3 fails to immunoprecipitate the 70-kDa proteins (Fig. 5A, lane 3). We then performed immunoprecipitation using anti-NeuN, followed by MS analysis. The majority of the peptide sequences from each of the doublets are products of synapsin I (supplemental Fig. S2). The synapsin I gene generates two alternatively spliced isoforms, synapsin Ia and Ib with masses of 74 and 70 kDa, respectively (25, 26). Only a small region of sequence at the C-terminal differs between Ia and Ib. The peptide sequences from the upper band included a sequence unique to synapsin Ia and those from the lower band are all common to both isoforms. Therefore, the lower band could be synapsin Ib or a proteolytic fragment of synapsin Ia. Synapsin I is relatively well characterized and its specific Ab and cDNA clone are readily available. As shown in Fig. 5B, the 70-kDa proteins immunoprecipitated with anti-synapsin I are recognized by anti-NeuN using immunoblotting (lane 3). The ratio of band intensity in the input extracts versus the immunoprecipitates on the immunoblot using anti-NeuN is similar to that obtained with anti-synapsin I (compare the ratio of lanes 1 versus 3 to that of lanes 4 versus 6 in Fig. 5B), suggesting that anti-NeuN and anti-synapsin I react with the same proteins, and not to co-precipitated proteins. Finally, recombinant synapsin Ia exogenously expressed in HEK-293 cells cross-reacts with anti-NeuN in a dose-dependent manner (Fig. 5C, left panel). However, as shown in Fig. 6, the immunoreactivity of anti-NeuN to synapsin I and that to Fox-3 differ in degree. Immunoblots of various amounts of cell lysates which contain the Myc-tagged synapsin Ia and the Myc-tagged Fox-3-S were analyzed in parallel using anti-NeuN and anti-Myc. Anti-Myc detects the same epitope of both proteins and determines the relative amounts of both Myc-tagged synapsin I and Fox-3. By quantifying immunoblot signals, we estimated that the reactivity of anti-NeuN to Fox-3 is ∼20-fold greater than that to synapsin I.
The finding that the 70-kDa anti-NeuN-reactive proteins are synapsin Is is consistent with the failure to detect them in nuclear extracts, since synapsin I is known to be localized to the cytoplasmic surface of synaptic vesicles in the presynaptic terminals (26). Immunofluorescence staining of mouse cerebellum with anti-synapsin I shows a punctate staining pattern in the molecular layer where dendrites from cerebellar neurons form extensive synapses (Fig. 7A, left panel). This staining pattern is quite different from that of anti-NeuN shown in Fig. 3A. The specimen shown in Fig. 7A has been co-stained with anti-Fox-3 rather than with anti-NeuN, since the staining patterns of two Abs completely overlap (Fig. 3). Unlike anti-NeuN and anti-synapsin I which were produced in mouse hybridomas, anti-Fox-3 was generated in rabbits, so use of Abs from different species was technically easier for double staining. Although anti-Fox-3 and anti-NeuN show extranuclear staining to some extent, their staining is diffuse in soma and not punctate in dendrites (Fig. 3). These observations indicate that anti-NeuN does not recognize synapsin I in tissue sections. The exogenously expressed synapsin Ia in HEK-293 cells was stained by either anti-synapsin I or anti-NeuN following fixation with paraformaldehyde and permeabilization with Triton X-100. Anti-synapsin I shows diffuse staining with strong punctate staining in the cytoplasm (Fig. 7B). On the other hand, anti-NeuN staining is hardly detected. We can only detect small amounts of punctate staining in the cytoplasm with an increased microscope gain (Fig. 7C). In addition to the lower affinity of anti-NeuN to synapsin I, it appears that paraformaldehyde fixation also contributes, at least in part, to the inability of anti-NeuN to detect synapsin I in tissue sections and cell culture.
Function of Fox-3/NeuN as Splicing Regulator
Although anti-NeuN shows cross-reactivity to synapsin I in immunoblots, the true antigen for anti-NeuN is Fox-3. Therefore we addressed a potential function of Fox-3. Because the Fox-3 amino acid sequence shows high homology to Fox-1 and Fox-2, which have recently been characterized as regulators of alternative pre-mRNA splicing (17, 20, 23, 27–30), we asked whether Fox-3 is capable of regulating alternative splicing of pre-mRNA. We used neural cell-specific alternative splicing of exon N30 of the non-muscle myosin heavy chain (NMHC) II-B gene as a model. The cassette type alternative exon N30 encoding 10 amino acids is included in the mRNAs from some neural cells, but is skipped in all other cells (19, 31). We have previously established a minigene reporter system which can be used to analyze RNA elements and protein factors that regulate alternative splicing of N30 (17, 32). The minigene which consists of exon N30 and the flanking exons and introns is outlined in Fig. 8A. Following transfection of the minigene in SK-N-SH cells, the mRNA derived from the minigene was analyzed by RT-PCR. Using endogenous splicing factors, most of the mRNAs from the wild-type minigene exclude N30 as shown in Fig. 8B (lanes 1 and 6). Exogenous expression of Fox-3-L or -S results in a dose-dependent increase of N30 inclusion in the minigene mRNAs with slightly higher activity using Fox-3-L compared with Fox-3-S (compare lanes 3 versus 8 and lanes 4 versus 9 in Fig. 8B). Previous studies have defined the intronic region downstream of N30, called the intronic distal downstream enhancer (IDDE), as the critical region for activation of N30 splicing (17, 32). Within the IDDE, there are two copies of UGCAUG elements. Mutation of one of two copies does not affect Fox-3-induced splicing of N30 (Fig. 8C, lanes 7, 8, 12, and 13). Mutation of both copies of this element abolishes the enhancement effects of Fox-3-L and -S on N30 splicing (Fig. 8C, lanes 9 and 14). Therefore, at least, one copy of the UGCAUG element is required for Fox-3 to exert its effect on N30 splicing. Consistent with the dependence of Fox-3 enhanced splicing on a UGCAUG element, deletion of the IDDE also abolishes the effects of Fox-3 (Fig. 8C, lanes 10 and 15). These results indicate that Fox-3 can function as a splicing activator for exon N30 of NMHC II-B via the intronic UGCAUG element. The nucleotide sequences for Fox-3-L and Fox-3-S have been deposited in the GenBankTM database under GenBankTM Accession Numbers FJ958310 and FJ958311, respectively.
DISCUSSION
The aim of this study was to determine the molecular identity of NeuN. We conclude that NeuN is Fox-3, based on the following findings: 1) MS analysis of the anti-NeuN immunoreactive protein yields the Fox-3 amino acid sequence. 2) The anti-Fox-3 immunoreactive protein is also recognized by anti-NeuN and vice versa. 3) NeuN protein expression is decreased by the shRNA for Fox-3. 4) Recombinant Fox-3 is recognized by anti-NeuN. 5) Anti-Fox-3 staining and anti-NeuN staining overlap completely at the cellular and subcellular levels. 6) Fox-3 is expressed only in neural tissues. Furthermore, we demonstrate that Fox-3 serves as a regulator of alternative splicing of pre-mRNA.
The Fox-3 gene was first recognized in a genome-wide search for RNA-binding proteins. Its mRNA expression has been shown to be restricted to the post-mitotic region of the developing brain at embryonic day 13.5 and postnatal day 0 by in situ hybridization (33). We have demonstrated that its expression is also confined to the brain among all major organs in adult mice. The Fox-3 expression pattern is consistent with NeuN expression reported previously (1). Our cDNA cloning and analysis of the EST and other sequence databases have revealed that the Fox-3 pre-mRNA undergoes alternative splicing in at least two regions. Alternative 3′ splice site usage of an internal exon and the last exon generates protein isoforms which include or exclude 47 and 13 amino acids, respectively (see supplemental Fig. S1). The predicted 4 isoforms would have masses ranging from 34 to 41 kDa. These masses are slightly smaller than those estimated by SDS-PAGE (40–50 kDa). The Myc-tagged recombinant Fox-3 expressed in cultured cells also migrates more slowly than the Mr calculated from the amino acid sequence. Moreover the migration of Fox-3 proteins is affected by the polyacrylamide concentration and the buffer system. Therefore, the discrepancy in Mrs estimated by the primary sequence and SDS-PAGE could be explained by protein conformation or post-translational modification. However, we cannot rule out the possibility that some of the isoforms might use a more upstream initiating codon that we have not yet identified, although the 5′ cDNA sequences obtained so far using rapid amplification of 5′ cDNA end analysis give the same initiating codon.3 This study also does not completely exclude the possibility that the multiple protein bands of Fox-3/NeuN could result from proteolysis despite our use of proteolytic inhibitors.
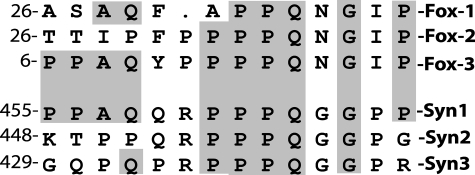
In this study, we have also provided evidence that the 70-kDa, anti-NeuN immunoreactive proteins include synapsin I gene products Ia (74 kDa) and/or Ib (70 kDa). Synapsins are a family of neuron-specific synaptic vesicle-associated phosphoproteins that play a role in synaptogenesis and modulation of neurotransmitter release (26). Three genes, synapsin I, II, and III make up the synapsin gene family. The predicted masses of synapsin IIa, IIb, and III isoforms are 63, 52, and 63 kDa, respectively. We have not detected the protein bands corresponding to these masses and recombinant synapsin IIa is not recognized by anti-NeuN (data not shown). It appears that anti-NeuN cross-reacts with only synapsin I but not other synapsins. The finding that the monoclonal anti-NeuN recognizes Fox-3 and synapsin I, which belong to quite different gene families is somewhat unexpected, so we have tried to estimate a potential epitope for anti-NeuN. Because the epitope recognized by anti-NeuN is located in the N-terminal 1–106-amino acid region of Fox-3, we have searched for a homologous sequence between Fox-3 amino acids 1–106 and the full-length synapsin Ia amino acids 1–586. A best alignment is a stretch of 14 amino acids and is shown, together with the sequences of other members of the two gene families, in Fig. 9. Several amino acids within this 14-amino acid segment from other members are also aligned to some extent with Fox-3 and synapsin I. However, since Fox-1 and -2 and synapsin II are not recognized by anti-NeuN, we speculate that the core sequence of the epitope for anti-NeuN might include PPAQ. There is a report suggesting that the phosphorylation of NeuN is required for Ab recognition (16). Although a tyrosine residue exists within the 14-amino acid segment of Fox-3, we do not know whether it is phosphorylated in mammalian cells or bacteria.
FIGURE 9.
Amino acid alignment of Fox-3 and synapsin I and their families. Fox-3 amino acids 1–106 and full-length synapsin Ia (Syn1) were subjected to a homology search using the ExPASy Proteomic Server program. Only one homology alignment composed of 14 amino acids was found. The sequences of Fox-1 and -2 homologous to Fox-3 and those of synapsin II and III (Syn2 and 3) homologous to Syn1 are also included. Amino acids with a gray background are identical to both Fox-3 and Syn1. The numbers represent the starting amino acids.
Another puzzling observation is that the anti-NeuN does not recognize synapsin I in tissue sections or cell culture, despite its ability to recognize it as an SDS-denatured form or a native form in extracts. This is presumably due to the inaccessibility of the epitope, resulting from the process of fixation and embedding for sectioning. Cross-linking of synapsin I intra-molecularly and inter-molecularly with associated proteins in situ by paraformaldehyde during the fixation process is likely one of the causes of a lack of synapsin I staining by anti-NeuN, as demonstrated in Fig. 7. Another reason is that anti-NeuN has only a low affinity for synapsin I, as shown in Fig. 6, and therefore it might not detect synapsin I in situ. For immunoblotting, however, the lower affinity of anti-NeuN for synapsin I would be compensated for by the high concentration of synapsin I in extracts, since synapsins are known to be the most abundant phosphoproteins in the brain (26). The fact that anti-NeuN can cross-react with synapsin I calls for caution in the use of this mAb and interpretation of results.
What might be the function of NeuN/Fox-3? Since NeuN is predominantly localized to nuclei, it has been thought to function in regulating gene expression such as transcription and RNA processing. An attempt to detect NeuN binding to DNA has been reported without addressing whether the binding is direct or indirect and whether the binding has any sequence specificity (1). Now, we have identified NeuN as Fox-3 and we provide the primary sequence of Fox-3. Fox-3 contains a single copy of an RRM at the middle part of the molecule. This RRM shows high homology (only 4 amino acids are substituted for out of 77 amino acids) to that of Fox-1 (also called A2BP1, Hrnbp1) and Fox-2 (also called Rbm9, Fxh, Hrnbp2, RTA) and these three genes constitute the Fox-1 gene family (see supplemental Fig. S1 for the sequence alignment). Fox-1 and -2 have an identical primary sequence for the RRM in humans and mice and this sequence is well conserved in vertebrates. The RNA sequences bound by zebrafish Fox-1 and human Fox-1 have been determined to be GCAUG and UGCAUG, respectively (23, 27). Further, the solution structure of the RRM of human Fox-1 in complex with UGCAUGU has been determined (34). The RRM of Fox-3 has the identical amino acid residues that are important for RNA binding, as well as those important for sequence specificity. Therefore, it is very likely that Fox-3 can bind specifically to UGCAUG. Since the discovery of the target RNA sequence of Fox-1, several laboratories, including ours, have reported that Fox-1 and Fox-2 are capable of regulating alternative splicing of pre-mRNAs in a UGCAUG sequence-specific manner in a number of model systems (17, 20, 23, 27–30). In addition to the RRM, the C-terminal region following the RRM, which seems to be important for regulation of alternative splicing, contains a number of segments highly conserved among Fox-1, -2, and -3. We demonstrated that Fox-3 can activate neural cell-specific splicing of a cassette exon, N30, of the NMHC II-B pre-mRNA. This activation is absolutely dependent on the downstream intronic element UGCAUG, consistent with the idea that Fox-3 binds to the UGCAUG element. These observations suggest that Fox-3 plays a role in neuron-specific alternative splicing.
The brain is the organ where alternative splicing occurs most frequently (35, 36), presumably to generate large numbers of neuronal cell types and to support their diverse functions. Global computational analysis of intronic sequences and genome-wide splicing microarrays have revealed that UGCAUG is over-represented in the introns in which splicing is regulated in a cell type- and developmental stage-specific manner, compared with constitutively spliced introns (37–40). Fox-1 is expressed in brain and striated muscles and Fox-2 is expressed ubiquitously (17, 20–23, 27). In contrast, Fox-3 expression is restricted to neurons. Therefore, Fox-3 is an attractive candidate to act as a determinant factor of neural specificity during splicing. It is tempting to speculate that alternative splicing regulated by Fox-3 would play a key role in regulation of neural cell differentiation and development of the nervous system. Future studies will determine whether Fox-3 expression plays a causal role in neurogenesis or is instead only a convenient marker for neural differentiation. This study identifying NeuN as Fox-3 affords an opportunity to re-interpret previous studies of NeuN. Our findings should promote future work on neural development and disease in relation to Fox-3 function.
Supplementary Material
Acknowledgments
We thank Xuefei Ma, Mary Anne Conti, and Jong H. Kim for reagents and helpful discussions. We thank Christian A. Combs and Daniela Malide (Light Microscope Core Facility, NHLBI), Eric Billings (Proteomics Core Facility, NHLBI), and J. Philip McCoy Jr. (Flow Cytometry Core Facility, NHLBI) for professional skills and advice. We also thank Rebecca Nichols (Anne Arundel Community College) and Antoine F. Smith for technical assistance and Mary Anne Conti for critical reading of the manuscript.
This work was supported, in whole or in part, by the Division of Intramural Research, NHLBI, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) FJ958310 and FJ958311.
K. K. Kim and S. Kawamoto, unpublished observation.
- mAb
- monoclonal antibody
- FBS
- fetal bovine serum
- GFP
- green fluorescent protein
- Ab(s)
- antibody(ies)
- GST
- glutathione S-transferase
- DAPI
- 4,6-diamidino-2-phenylindole
- MS
- mass spectrometry
- FACS
- fluorescence-activated cell sorting
- RRM
- RNA recognition motif
- NMHC
- non-muscle myosin heavy chain
- IDDE
- intronic distal downstream enhancer
- NeuN
- neuronal nuclei
- shRNA
- short hairpin RNA.
REFERENCES
- 1.Mullen R. J., Buck C. R., Smith A. M. (1992) Development 116, 201–211 [DOI] [PubMed] [Google Scholar]
- 2.Weyer A., Schilling K. (2003) J. Neurosci. Res. 73, 400–409 [DOI] [PubMed] [Google Scholar]
- 3.Magavi S. S., Leavitt B. R., Macklis J. D. (2000) Nature 405, 951–955 [DOI] [PubMed] [Google Scholar]
- 4.Cummings B. J., Uchida N., Tamaki S. J., Salazar D. L., Hooshmand M., Summers R., Gage F. H., Anderson A. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14069–14074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gauthier A. S., Furstoss O., Araki T., Chan R., Neel B. G., Kaplan D. R., Miller F. D. (2007) Neuron 54, 245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suh H., Consiglio A., Ray J., Sawai T., D'Amour K. A., Gage F. H. (2007) Cell Stem Cell 1, 515–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckingham B. P., Inman D. M., Lambert W., Oglesby E., Calkins D. J., Steele M. R., Vetter M. L., Marsh-Armstrong N., Horner P. J. (2008) J. Neurosci. 28, 2735–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagel D. M., Ganat Y., Cheng E., Silbereis J., Ohkubo Y., Ment L. R., Vaccarino F. M. (2009) J. Neurosci. 29, 1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf H. K., Buslei R., Schmidt-Kastner R., Schmidt-Kastner P. K., Pietsch T., Wiestler O. D., Blümcke I. (1996) J. Histochem. Cytochem. 44, 1167–1171 [DOI] [PubMed] [Google Scholar]
- 10.Preusser M., Laggner U., Haberler C., Heinzl H., Budka H., Hainfellner J. A. (2006) Histopathology 48, 438–444 [DOI] [PubMed] [Google Scholar]
- 11.Brazelton T. R., Rossi F. M., Keshet G. I., Blau H. M. (2000) Science 290, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 12.Mezey E., Chandross K. J., Harta G., Maki R. A., McKercher S. R. (2000) Science 290, 1779–1782 [DOI] [PubMed] [Google Scholar]
- 13.Sigurjonsson O. E., Perreault M. C., Egeland T., Glover J. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5227–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wynder C., Hakimi M. A., Epstein J. A., Shilatifard A., Shiekhattar R. (2005) Nat. Cell Biol. 7, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 15.Goetz A. K., Scheffler B., Chen H. X., Wang S., Suslov O., Xiang H., Brüstle O., Roper S. N., Steindler D. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11063–11068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lind D., Franken S., Kappler J., Jankowski J., Schilling K. (2005) J. Neurosci. Res. 79, 295–302 [DOI] [PubMed] [Google Scholar]
- 17.Nakahata S., Kawamoto S. (2005) Nucleic Acids Res. 33, 2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips C. L., Yamakawa K., Adelstein R. S. (1995) J. Muscle Res. Cell Motil. 16, 379–389 [DOI] [PubMed] [Google Scholar]
- 19.Ma X., Kawamoto S., Uribe J., Adelstein R. S. (2006) Mol. Biol. Cell 17, 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Underwood J. G., Boutz P. L., Dougherty J. D., Stoilov P., Black D. L. (2005) Mol. Cell. Biol. 25, 10005–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibata H., Huynh D. P., Pulst S. M. (2000) Hum. Mol. Genet. 9, 1303–1313 [DOI] [PubMed] [Google Scholar]
- 22.Lieberman A. P., Friedlich D. L., Harmison G., Howell B. W., Jordan C. L., Breedlove S. M., Fischbeck K. H. (2001) Biochem. Biophys. Res. Commun. 282, 499–506 [DOI] [PubMed] [Google Scholar]
- 23.Jin Y., Suzuki H., Maegawa S., Endo H., Sugano S., Hashimoto K., Yasuda K., Inoue K. (2003) EMBO J. 22, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unal-Cevik I., Kilinç M., Gürsoy-Ozdemir Y., Gurer G., Dalkara T. (2004) Brain Res. 1015, 169–174 [DOI] [PubMed] [Google Scholar]
- 25.Kao H. T., Porton B., Czernik A. J., Feng J., Yiu G., Häring M., Benfenati F., Greengard P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4667–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gitler D., Xu Y., Kao H. T., Lin D., Lim S., Feng J., Greengard P., Augustine G. J. (2004) J. Neurosci. 24, 3711–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponthier J. L., Schluepen C., Chen W., Lersch R. A., Gee S. L., Hou V. C., Lo A. J., Short S. A., Chasis J. A., Winkelmann J. C., Conboy J. G. (2006) J. Biol. Chem. 281, 12468–12474 [DOI] [PubMed] [Google Scholar]
- 28.Baraniak A. P., Chen J. R., Garcia-Blanco M. A. (2006) Mol. Cell. Biol. 26, 1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H. L., Baraniak A. P., Lou H. (2007) Mol. Cell. Biol. 27, 830–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang G., Huang S. C., Wu J. Y., Benz E. J., Jr. (2008) Blood 111, 392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh K., Adelstein R. S. (1995) J. Biol. Chem. 270, 14533–14540 [DOI] [PubMed] [Google Scholar]
- 32.Kawamoto S. (1996) J. Biol. Chem. 271, 17613–17616 [PubMed] [Google Scholar]
- 33.McKee A. E., Minet E., Stern C., Riahi S., Stiles C. D., Silver P. A. (2005) BMC Dev. Biol. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auweter S. D., Fasan R., Reymond L., Underwood J. G., Black D. L., Pitsch S., Allain F. H. (2006) EMBO J. 25, 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modrek B., Resch A., Grasso C., Lee C. (2001) Nucleic Acids Res. 29, 2850–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo G., Holste D., Kreiman G., Burge C. B. (2004) Genome Biol. 5, R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brudno M., Gelfand M. S., Spengler S., Zorn M., Dubchak I., Conboy J. G. (2001) Nucleic Acids Res. 29, 2338–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castle J. C., Zhang C., Shah J. K., Kulkarni A. V., Kalsotra A., Cooper T. A., Johnson J. M. (2008) Nat. Genet. 40, 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C., Zhang Z., Castle J., Sun S., Johnson J., Krainer A. R., Zhang M. Q. (2008) Genes Dev. 22, 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeo G. W., Coufal N. G., Liang T. Y., Peng G. E., Fu X. D., Gage F. H. (2009) Nat. Struct. Mol. Biol. 16, 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.