Abstract
8-Oxo-2′-deoxyguanosine (8-oxodG) is one of the most important oxidative DNA lesions, and G-rich telomeric DNA is especially susceptible to oxidative DNA damage. RecQ helicases WRN and BLM and telomere-binding protein POT1 are thought to play roles in telomere maintenance. This study examines the ability of WRN, BLM, and RecQ5 to unwind and POT1 to bind telomeric D-loops containing 8-oxodG. The results demonstrate that WRN and BLM preferentially unwind telomeric D-loops containing 8-oxodG and that POT1 binds with higher affinity to telomeric D-loops with 8-oxodG but shows no preference for telomeric single-stranded DNA with 8-oxodG. We speculate that telomeric D-loops with 8-oxodG may have a greater tendency to form G-quadruplex DNA structures than telomeric DNA lacking 8-oxodG.
Introduction
Telomeres are structures at the ends of the eukaryotic linear chromosomes that enhance chromosome stability by preventing DNA end-initiated recombination, exonucleolytic attack, and replication-associated terminal recession. Telomeres are composed of long tracts of short tandem repeated DNA sequences (5′-TTAGGG-3′ in human and mouse) and telomere-specific DNA-binding proteins. The length of telomeres is maintained by an active process, and defects in telomere maintenance lead to telomere attrition, genome instability, cell cycle arrest, and senescence or apoptosis. Telomere attrition is frequently associated with aging (1) and premature aging syndromes (2).
The unique structure of telomeres plays an important role in maintenance of telomeric DNA. Telomeres are composed of double-stranded tandem repeat sequences followed by a single-stranded short 3′-overhang (3, 4). The length of the double-stranded telomere tract varies from about 10 kbp in humans to up to 100 kbp in mice (5, 6). In mammals telomeres normally exist in a loop structure with the 3′-single-stranded overhang invading the telomeric double-stranded DNA (7). This so-called “D-loop” configuration is stabilized by telomere-binding proteins, known as shelterin, and associated proteins (8). Telomere repeat binding Factors 1 and 2 (TRF1 and -2) bind to duplex telomeric sequences (9, 10), whereas protection of telomeres 1 (POT1) binds to single-stranded telomeric DNA (11).
One family of proteins that is actively involved in maintaining genome stability is the RecQ helicase family, a highly conserved group of DNA helicases which functions in multiple DNA metabolic processes. Sgs1 is the sole RecQ homolog in Saccharomyces cerevisiae, and Rqh1 is the soleRecQ homolog in Schizosaccharomyces pombe (12). Curiously, five RecQ homologs have been identified in mammalian cells, RECQ1, BLM, WRN, RECQ4, and RECQ5. Three of the gene products have been shown to be associated with autosomal recessive disorders characterized by genomic instability and cancer predisposition. Bloom syndrome, Werner syndrome, and Rothmund Thomson syndrome are associated with defects in BLM, WRN, and RECQ4, respectively (13, 14). BLM and WRN are known to play important roles in DNA repair and replication (12, 15, 16) and have been implicated in telomere maintenance. However, none of the other helicases have been implicated in telomere functioning so far.
Reactive oxygen species, such as inorganic and organic oxygen ions, free radicals, and peroxides, are one of the most important contributors to DNA damage (17). These species are generated regularly in cells through various processes including endogenous and exogenous chemical exposures and ionizing radiation. The genome is frequently attacked by reactive oxygen species generating different lesions that essentially lead to DNA strand breaks (18). 8-Oxoguanine (8-oxodG)2 is one of the most well studied oxidative lesions (19, 20). This lesion can form Watson-Crick base pairs with 2′-deoxycytidine (dC) and also form Hoogsteen base pairs with 2′-deoxyadenosine (dA). In the Hoogsteen mode, 8-oxodG readily mispairs with dA during replication, and if left unrepaired, an 8oxodG:dA base pair can lead to GC → TA mutations (21).
Telomeric DNA is prone to oxidative damage due to the presence of easily oxidizable guanines in the TTAGGG telomeric repeats. Additionally, oxidative stress has been implicated in the telomeric shortening process (22). However, very little is known about the interaction of helicases with oxidatively damaged telomeric DNA.
Several lesions can be generated by oxidative damage, including 8-oxodG, thymine glycol, formamidopyrimidine-2′-deoxyguanosine, formamidopyrimidine-2′-deoxyadenosine, and many more. Although many of these lesions may have important biological roles, most of them are not well studied. It is also improbable to assess the effects of all the lesions due to the complications involved in the synthesis of DNA strands containing the damages. Among these lesions, 8-oxodG is an abundant and well characterized lesion and is of great biological significance. It is also a relatively stable lesion, making it a good candidate to study. We have constructed in vitro D-loop structures containing 8-oxodG lesions within telomeric repeat sequences to determine how the presence of DNA damage changes the helicase and DNA binding properties of WRN, BLM, and RecQ5. Additionally, we report on the POT1 DNA binding activity on D-loops containing 8-oxodG lesions. Our experiments clearly suggest that WRN and BLM efficiently unwind these damaged D-loops, but RecQ5 does not. Although DNA damage is present, POT1 still binds to the damaged D-loops. These findings indicate that these proteins have important roles in damaged telomeric DNA processing and repair.
EXPERIMENTAL PROCEDURES
Proteins
Recombinant histidine-tagged wild type WRN protein was purified using a baculovirus/insect cell expression system as described previously (23, 24). Recombinant histidine-tagged BLM was overexpressed in S. cerevisiae and purified as described previously (25). RecQ5 was purified using a protocol described elsewhere (26) Recombinant human POT1 protein was purified using a baculovirus/insect cell expression system as described previously (11) and contains a GST tag. Protein concentration was determined by the Bradford assay (Bio-Rad), and purity was determined by SDS-PAGE and Coomassie staining. Bacterial UvrD helicase was a generous gift from Dr. Steve Mattson (University of North Carolina, Chapel Hill, NC).
DNA Substrates
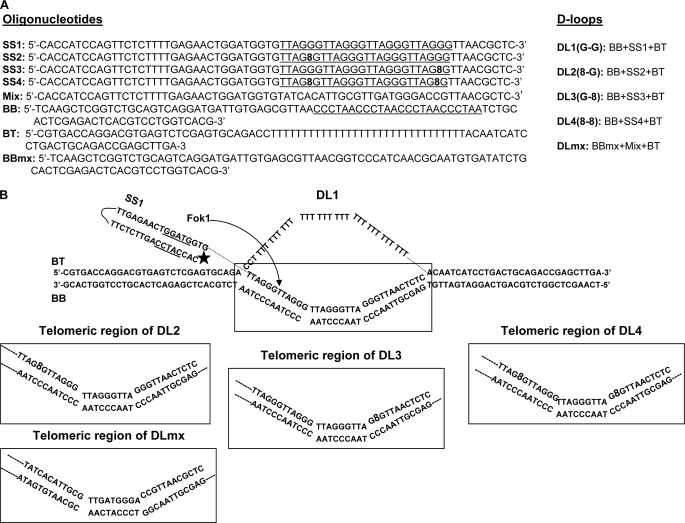
All the unmodified oligonucleotides were from Integrated DNA Technologies, Coralville, IA, and PAGE was purified by the manufacturer. The modified (8-oxodG-containing) oligonucleotides were synthesized and purified by The Midland Certified Reagent Co., Midland, TX. The D-loop DL1 substrate containing the (TTAGGG)4 sequence in the 33-bp duplex portion of the invading strand was constructed by annealing the BT, BBtel, and invading strand (SS1) oligonucleotides (Table 1) as described previously (23). 8-OxodG-containing D-loops (DL2–4) were prepared similarly, except SS2–4 oligonucleotides contained 8-oxodG instead of dG at specific positions (Table 1). The non-telomeric D-loop DLmx was constructed by mixing BT, BBmx, and non-telomeric invading strand (Mix). All the D-loops contain 5′-end-labeled invading strands and were constructed using the same amounts of invading strands. Oligonucleotides were 5′-end-labeled using [γ-32P]ATP (PerkinElmer Life Sciences) and T4-polynucleotide kinase (New England Biolabs) per the manufacturer's directions. However, the radiolabeled strands were not separated from the unlabeled strands. The complete formation of D-loops was established by running the D-loops in a non-denaturing gel side by side with the corresponding single-stranded oligonucleotides. The structure of D-loop DNA substrates was confirmed by Fok1 incision assays and mung bean nuclease (MBN) digestion. The Fok1 (New England Biolabs) incision reaction was carried out as described previously (23). Briefly, 210 fmol of the D-loops were incubated with Fok1 for 3 h at 37 °C. The reactions were stopped by the addition of stop buffer (80% formamide, 1× TBE, 0.25% bromphenol blue, 0.01% SDS), and the products were heat-denatured (95 °C for 5 min followed by rapid cooling on ice for 2 min) then resolved through a 12% TBE-urea-PAGE gel at 15 watts for 2 h in 1× TBE buffer. Radiolabeled oligos were run side by side as size marker. Mung bean nuclease was used to probe for any single-stranded character in the D-loops. The D-loops (14 ng) were treated with 1 unit of mung bean nuclease (New England Biolabs) for 3 h at 4 °C. The reactions were stopped by the addition of formamide loading buffer (80% formamide, 0.01% SDS, 0.1% bromphenol blue), heat-denatured (95 °C for 5 min, followed by rapid cooling on ice for 2 min), and then analyzed on a 12% denaturing (7 m urea) PAGE gel. Undamaged telomeric single strand (SS1) was treated similarly and used as positive control for this experiment.
TABLE 1.
Oligonucleotide substrates
Underline indicates telomeric region.
8 indicates 8-oxodG lesion.
Helicase Assay
Reactions were performed in standard reaction buffer (40 mm Tris, pH 8.0, 4 mm MgCl2, 5 mm DTT, 2 mm ATP, and 1 mg/ml BSA) unless otherwise indicated. DNA substrate and protein concentrations were as indicated in the figure legends. For WRN and BLM helicase reactions, the substrates were preincubated at 37 °C for 5 min before the addition of proteins. The reactions were initiated by the addition of WRN, BLM, or RecQ5 and incubated at 37 °C for 30 min. However, RecQ5 did not show any helicase activity in standard reaction buffer, and a different buffer (30 mm HEPES, pH 7.45, 5% glycerol, 40 mm KCl, 100 ng/μl BSA, 4 mm MgCl2, and 2 mm ATP) was used for RecQ5 helicase assays thereafter. For the helicase assays in the presence of POT1, the indicated amounts of POT1 were added together with WRN, BLM, or RecQ5 to initiate the reaction. After incubation, WRN and RecQ5 reactions were mixed with native stop dye (40% glycerol, 50 mm EDTA, 0.9% SDS, 0.1% bromphenol blue) and analyzed on an 8% native polyacrylamide (0.1% SDS) gel. Products of BLM helicase assays were treated with 10 μl of native stop dye supplemented with 75 μg/ml proteinase K and a 10 μm excess of unlabeled competitor oligonucleotide (27). The products were deproteinized for 30 min at 37 °C and were then run on 8% native polyacrylamide gels. Products were visualized using a PhosphorImager, and quantitation was performed using ImageQuant software (GE Healthcare).
WRN helicase assays were quantified by calculating the percent of total ssDNA product (all product species divided by sum of product and substrate, i.e. total radioactivity in the lane (Fig. 5, upper panel). For BLM and RecQ5 helicase assays, percent product was calculated as described previously (27) Values were corrected for background in negative control assays (i.e. no enzyme and heat-denatured substrate). UvrD was used as positive control (28).
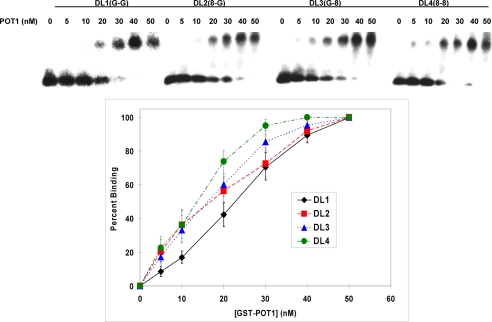
FIGURE 5.
GST-POT1 binding of telomeric D-loop DNA. Upper panel, shown is an autoradiogram of EMSA showing the appearance of slow moving POT1-DNA complex with increasing GST-POT1 concentration. Lower panel, shown is a plot of percentage formation of DNA-POT1complex (binding) against the corresponding GST-POT1 concentration. The error bars represent the S.E. calculated from four independent experiments.
Electrophoretic Mobility Shift Assay (EMSA)
Binding reactions were performed in EMSA buffer (40 mm Tris, pH 8.0, 12 mm MgCl2, 5 mm DTT, 10 mm ATP, 1 mg/ml BSA). For the kinetics study 50 mm LiCl was added to increase D-loop solubility. DNA substrates (1 nm SS1–4 or DL1–4 or DLmx) were preincubated at 25 °C for 5 min, and the reaction was started by the addition of GST-POT1 (concentrations are as shown in Figs. 4 and 5). The complete reaction mixture was incubated at 25 °C for 15 min, and then the reaction was stopped by the addition of EMSA loading buffer (0.25% bromphenol blue, 10% Ficoll, 0.5× TBE). The GST-POT1 binding reactions were resolved by electrophoresis on a 1% agarose gel (Seakem GTG, BioWhittaker Molecular Applications) in 0.5× TBE, 200 V for 2 h. Products were visualized using a PhosphorImager and quantified using ImageQuant software (GE Healthcare). Percent binding was determined by dividing bound DNA substrate (slow moving species) by bound plus unbound DNA substrate (total radioactivity in the lane). The dissociation constants (Kd) were obtained from the polynomial fit (Graphpad Prism 5.0) of the plots of percent binding against the GST-POT1 concentration. DNA binding assays in the presence of telomestatin (TMS) were conducted by preincubating D-loop DNA substrates with TMS at the indicated concentrations at 25 °C for 30 min before the addition of POT1. Telomestatin was kind gift from Dr. Robert Brosh (NIA, NIH).
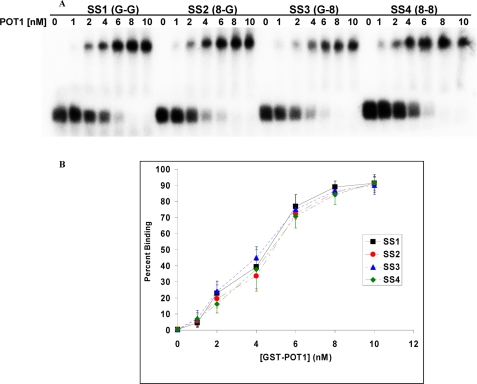
FIGURE 4.
GST-POT1 binding of telomeric single-stranded DNA. A, an autoradiogram of EMSA shows the appearance of slow moving POT1-DNA complex with increasing GST-POT1 concentration. B, shown is a plot of the percentage formation of DNA-POT1 complex (binding) against the corresponding GST-POT1 concentration. The error bars represent the S.E. calculated from four independent experiments.
Dimethyl Sulfate (DMS) Assay
The D-loops were kept at 0 °C for 15 min before incubation of the reaction. The reactions were performed in EMSA buffer (40 mm Tris, pH 8.0, 12 mm MgCl2, 5 mm DTT, 10 mm ATP, 1 mg/ml BSA) in the presence of 200 mm KCl. D-loop (1 nm) was treated with 2 μl 20% DMS (Sigma) in a 10-μl reaction volume. The reactions were carried out at 0 °C for 15 min and stopped by the addition of 50 μl of stop buffer (46 μl of H2O, 2 μl of BME, 2 μl of tRNA). The DNA was precipitated by adding 100 μl of ethanol and 10 μl of 0.5 m NaOAc and then treated with 1 m piperidine (10 μl) at 95 °C for 20 min. The samples were dried, resuspended in loading buffer (80% formamide, 0.01% SDS, 0.1% bromphenol blue), heat-denatured (95 °C for 5 min, followed by rapid cooling on ice for 2 min), and then resolved through a 12% TBE-urea-PAGE gel at 15 watts for 2.5 h in 1× TBE buffer.
8-OxodG Glycosylase Assay
0.5 nm DL2 was treated with either 10 nm WRN, 10 nm BLM, 50 nm POT1, or 1/16 unit α-OGG1 (New England Biolabs). The reactions with WRN and BLM were performed in helicase reaction buffer (40 mm Tris, pH 8.0, 4 mm MgCl2, 5 mm DTT, 2 mm ATP, and 1 mg/ml BSA), POT1 reactions were carried out in EMSA buffer (40 mm Tris, pH 8.0, 12 mm MgCl2, 5 mm DTT, 10 mm ATP, 1 mg/ml BSA), and OGG1 reactions were performed in NEB buffer 2. All the reactions were run for 30 min at 37 °C and stopped by the addition of formamide loading buffer (80% formamide, 1× TBE, 0.25% bromphenol blue, 0.01% SDS). The products were heat-denatured (95 °C for 5 min, followed by rapid cooling on ice for 2 min) then resolved through a 12% TBE-urea-PAGE gel for 15 watts for 2 h in 1× TBE buffer.
RESULTS
Construction of D-loops with and without the 8-OxodG Lesion
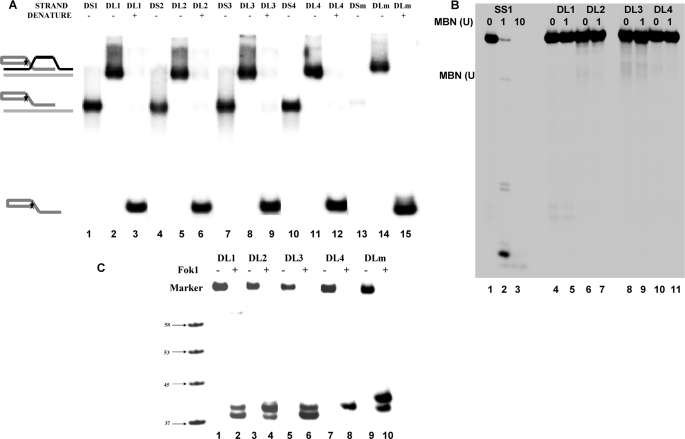
To investigate the interaction of WRN, BLM, RecQ5, and POT1 with damaged D-loops, we have crafted D-loop structures with 8-oxodG lesions at specific positions. Previous studies indicate that oxidative damage typically occurs at the middle and 5′ guanines of a GGG site (29–31). Thus, the damaged D-loops used here have been designed in such a way that the 8-oxodG lesion is positioned at the middle G of the telomeric GGG region (Table 1, Fig. 1). Three variations of damaged D-loops have been constructed (DL2–4). DL2 has the 8-oxodG (8) lesion in the first telomeric repeat, 5′-(TTAG8G)-3′, DL3 has the lesion in the last telomeric repeat in middle guanine, and DL4 has two lesions one at each site as shown in Fig. 1B. The D-loop DL1 does not have any lesion, and DLmx does not contain the telomeric repeat sequence. All the D-loops have been constructed using a method described previously (23). The D-loops were resolved on an 8% native PAGE, and the appearance of a single band confirmed the annealing of all three oligonucleotides to form the D-loops. Single- and double-stranded telomeric oligonucleotides were run side by side as size markers (Fig. 2A). This experiment also suggests that all the invading strands completely took part in the formation of D-loops. The invading strands were used in limiting amounts to prepare the D-loops, and the concentrations of these strands were considered as the concentrations of the corresponding D-loops. Proper alignment of the telomeric repeats was confirmed by Fok1 restriction enzyme digestion (Fig. 2C). DL2 and DL4 were less sensitive to Fok1 endonuclease, as these D-loops have 8-oxodG lesions next to the incision site. To obtain complete digestion, all the D-loops were treated with Fok1 for 3 h, which was more than usual (1–2 h). Fok1 digestion should produce 40-mer oligos from these D-loops. The autoradiogram shows two bands corresponding to 39- and 40-mer oligos for DL1, DL2, DL3, and DLmx, whereas DL4 shows only 40-mer band. The 39-mer product arises from the extended reaction, and this product was almost absent when DL1 and DL3 were treated with Fok1 for 2 h (supplemental Fig. 1A). Also, to verify the absence of any single-stranded regions in the radiolabeled telomeric strand, MBN digestion was performed on the D-loops. MBN was used for this experiment because under optimal conditions it shows a preference for single-stranded DNA over double-stranded DNA by a factor of 30,000:1 (32). Under these reaction conditions 90% of the single-stranded SS1 DNA (14 ng) was digested by 1 unit of MBN, and 10 units of MBN completely digested that strand (Fig. 2B, lanes 1–3). However, when 14 ng of the D-loops (DL1–4) were treated with 1 unit of MBN, all the strands with telomeric sequence remained intact (Fig. 2B, lanes 4–11). This result clearly suggests that there are no unhybridized single-stranded telomeric regions in the D-loops and that the damaged D-loops are as stable as the undamaged D-loops.
FIGURE 1.
Structures of the D-loops used in this study. The entire D-loop structure of DL1 is shown, and the telomeric region is boxed. Only the telomeric region of DL2, DL3, DL4, and DLmx is shown. 8 indicates 8-oxodG lesion. The Fok1 recognition site is underlined, and the incision site is shown by an arrow. An star indicates the position of 32P radiolabel. The Fok1 recognition site and position of the radiolabel are same for all the D-loops.
FIGURE 2.
Test of D-loop formation; DL indicates the D-loops described in Fig. 1, DS indicates corresponding double-stranded DNAs prepared by annealing of BB strand and corresponding single strands, and SS indicates only the single strand. A, for all the D-loops, corresponding double-stranded DNA, untreated D-loop, and denatured D-loop have been shown. The untreated D-loops show single band. B, MBN digestion of single-stranded (SS1) and D-loop DNAs (DL1–4) is shown. 0, 1, or 10 unit (U) of MBN was incubated with SS1 (lanes 1–3), and 0 or 1 unit of MBN was incubated with the D-loops (lanes 4–11). C, Fok1 endonuclease digestion of all the D-loops is shown. 10 nm D-loop was treated with 1 unit of Fok1 (lanes 2, 4, 6, 8, and 10) at 37 °C for 3 h. A radiolabeled marker indicates the position of 58-, 53-, 45-, and 37-mer oligos in the gel.
All the competitive experiments mentioned in this work were performed using the same amounts of D-loops. The radiolabeling efficiencies of different invading strands are different, and this leads to different amounts of radiolabeled substrates in the reaction mixture. However, as these substrates were constructed from the same amounts of invading strands and because these strands were not purified after labeling, they all contain the same amounts of D-loops (radiolabeled and unlabeled).
8-OxodG Glycosylase Activity of WRN, BLM, and POT1
To assess whether there was any 8-oxodG glycosylase activity in the WRN, BLM, and POT1 preparations used in this study, 8-oxodG containing D-loop DL2 was treated with these proteins, and the products were analyzed by denaturing PAGE (supplemental Fig. 1B). 8-OxodG glycosylase α-OGG1 was used as positive control to mark the position of incision product. WRN, BLM, and POT1 do not produce any incision products (lanes 2-4), whereasα-OGG1 incises at the 8-oxodG (lane 5). This result clearly demonstrates that there is no contaminating glycosylase activity in the proteins used in this study.
BLM, WRN, and RecQ5 Helicase Activity on Damaged and Undamaged D-loops
BLM and WRN helicases play important roles in telomere maintenance (33, 34) and have been implicated in DNA repair (12, 15, 16). On the other hand, the role of RecQ5 is not clear yet (35). To explore the possible role of these helicases in telomeric DNA repair, interactions of BLM, WRN, and RecQ5 helicases with the damaged and undamaged D-loops were studied. For these experiments, D-loops (0.5 nm) were treated with each helicase (either 5 or 10 nm) under reaction conditions described under “Experimental Procedures.” Standard helicase buffer has been used for BLM and WRN helicase reactions. RecQ5 did not show any helicase activity in this buffer and a different buffer was used to obtain a quantifiable helicase activity. Bacterial 3′ → 5′ helicase UvrD (10 nm) was used as a positive control in these experiments.
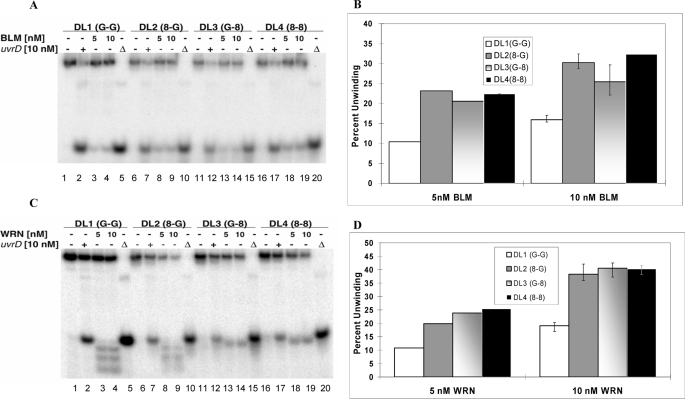
The helicase activities of BLM on undamaged and 8-oxodG-containing D-loops are shown in Fig. 3A. Unreacted DL1, DL2, DL3, and DL4 were run in lanes 1, 6, 11, and 16, respectively. Heat-denatured D-loops were used as markers for denatured, single-stranded DNA and run in lanes 5, 10, 15, and 20. The helicase activity of UvrD on DL1–4 is visible in lanes 2, 7, 12, and 17, respectively. From the gel it is evident that the unwinding ability of UvrD is similar on all four D-loops and that the presence of the 8-oxodG lesion in DL2–4 does not affect UvrD helicase activity. The percent unwinding was calculated as the relative density of the faster moving band to the total radiation present in the lane, and results from Fig. 3A are shown graphically in Fig. 3B.
FIGURE 3.
BLM and WRN helicase activity on telomeric D-loops with and without 8-oxodG lesion. A and C, an autoradiogram shows BLM and WRN helicase (5 and 10 nm) activity, respectively, on each of the telomeric D-loop substrates (0.5 nm) (lanes 3 and 4, 8 and 9, 13 and 14, and 18–19). The bacterial 3′-5′ helicase UvrD (10 nm) is used as a positive control (lanes 2, 7, 12, and 17). Each substrate is denatured (▵) at 95 °C (lanes 5, 10, 15, and 20). B and D, quantitative analysis of A and C, respectively, shows the percentage of unwinding of each D-loop in the absence of any helicase and in the presence of UvrD and BLM/WRN. The error bars represent the S.E. calculated from four independent experiments.
Results show that DL1–4 are unwound efficiently by 5 nm BLM and more efficiently by 10 nm BLM (Fig. 3A, compare lanes 3, 8, 13, and 18 with lanes 4, 9, 14, and 19). However, the 8-oxodG lesion does have a prominent effect on the D-loop unwinding ability of BLM. There is a 2-fold increase in the percent unwinding in case of DL2–4 as compared with DL1, and this effect is consistent for both dilutions of BLM used in this experiment. There is very little or no difference in unwinding activity of BLM on DL2, -3, and -4. This indicates that the position of the 8-oxodG lesion does not significantly affect the BLM helicase activity.
Similar experiments were done using WRN and RecQ5 helicases. The helicase activities of WRN on undamaged and damaged D-loops are summarized in Fig. 3C. Because WRN has a 3′ → 5′ exonuclease, all the products moving faster than the single-stranded DNA were considered as denatured products (lanes 3, 4, 8, and 9) (23). As has been previously observed, the presence of the 8-oxodG lesion in the GGG region close to the 3′-end of the strand, DL3, inhibits the exonuclease activity (36); thus, the shorter products are not visible in case of DL3 and DL4 (lanes 13, 14, 18, and 19). The quantification of the gels (Fig. 3D) shows that the helicase activity of WRN on D-loops is similar to those of BLM and that the presence of 8-oxodG lesions has approximately a 2-fold stimulatory effect on the unwinding ability of WRN. Although the position of the 8-oxodG served to inhibit the exonuclease function of WRN, there were no other discernible effects.
RecQ5 helicase unwinds D-loops much less efficiently than BLM and WRN (supplemental Figs. 2 and 3). The presence of 8-oxodG lesions in the D-loop enhances the RecQ5 helicase activity to some extent in DL3 and DL4. Unlike BLM and WRN, the enhancement is more pronounced in DL4 than in DL3. Additionally, there is a small decrease in RecQ5 helicase activity in DL2 as compared with DL1 (supplemental Fig. 2).
POT1 Binding to Damaged and Undamaged D-loops
POT1 is an important protein involved in the maintenance of telomeres and is known to interact with WRN and other proteins present in the telomeric region (23, 37). It has been shown in vitro that POT1 binds to the single-stranded telomeric DNA (11). Here, we tested its binding ability to bind to undamaged and damaged D-loops by EMSA. In the preliminary experiment the POT1 binding ability is studied on undamaged D-loop (DL1), single-stranded telomeric DNA (SS1), double-stranded DNA with telomeric region (DS1, prepared by annealing SS1 and BB), D-loop containing two 8-oxodG lesion (DL4) and corresponding single-stranded (SS4) and double-stranded (DS4) DNA and the non-telomeric D-loop (DLmx). Single-stranded, double-stranded, or DLmx DNA (1 nm) was tested for POT1 binding in the presence of 80 nm GST-POT1. For D-loops DL1 (1 nm) and DL4 (1 nm), a concentration gradient of POT1 (0 to 80 nm) was used. To prove that the GST tag present in POT1 does not affect the binding ability, both the D-loops were tested in the presence of GST only. The autoradiogram generated from this experiment (supplemental Fig. 4) shows that POT1 efficiently binds to the single-stranded telomeric DNA oligonucleotides with and without damage, whereas it does not bind to any of the double-stranded DNA. For the D-loops DL1 and DL4, slower-moving bands corresponding to the bound DNA showed up after the addition of 10 nm POT1. Intensity of that band increases with increase in POT1 concentration. However, there is a difference in POT1 binding ability between DL1 and DL4, discussed later. GST does not interact with any of the D-loops. The telomere-specific binding of POT1 is demonstrated by the fact that it does not bind to the non-telomeric D-loop, DLmx. Interestingly, the complex of POT1 with single-stranded DNA moves slower than the complex with D-loop DNA. Both the single-stranded telomeric DNA and D-loop have multiple POT1 binding sites. However, more than one GST-POT1 can easily bind to the flexible single-stranded DNA and run slowly in the gel, whereas the rigid structure of D-loop may prevent binding of multiple GST-POT1.
The apparent difference in POT1 binding between DL1 and DL4 led us to study the effect of the presence of 8-oxodG lesions on GST-POT1 binding to telomeric single-stranded and D-loop DNA substrates. EMSA was performed with all the single-stranded telomeric oligonucleotides (SS1–4) in the presence of different concentrations of GST-POT1 (Fig. 4). Each oligonucleotide (1 nm) was used in this experiment, and the GST-POT1 concentration ranged from 0 to 10 nm. Maximum binding was achieved after the addition of 10 nm GST-POT1, and it remained the same with higher concentrations of GST-POT1 (data not shown). The percent binding was calculated from the amount of radioactivity in the slow-moving bands relative to the total radioactivity in the corresponding lane and was plotted against the POT1 concentrations (Fig. 4B). The polynomial fit and calculation of binding parameters was performed for individual site-specific binding using Graphpad Prism 5.0 software. The polynomial fits were almost perfect for all the strands (R2 value almost 1). There is no significant difference in the dissociation constants (Kd) values between the strands (supplemental Table 1A). This implies that 8-oxodG lesions do not affect the GST-POT1 binding to single-stranded telomeric DNA. Also, the Hill coefficients for all the substrates are greater than 1 (supplemental Table 1A), implicating positive cooperative binding.
Similar EMSA experiments were performed with all the telomeric D-loops (DL1–4) in the presence of different concentrations of GST-POT1 (Fig. 5). Each D-loop (1 nm) was used in this experiment, and the GST-POT1 concentration ranged from 0 to 50 nm. Maximum binding was obtained in the presence of 50 nm GST-POT1, and the binding efficiency remained the same with higher concentrations of GST-POT1 (data not shown). The D-loops-containing lesions (especially DL4) were not completely soluble in the EMSA buffer; thus, the addition of 50 mm LiCl improved the solubility, and all the binding experiments (including the single-stranded ones) were done in the presence of 50 mm LiCl. We speculate that DL4 forms secondary structures such as G-quadruplexes and, thus, are poorly soluble in EMSA buffer. The addition of LiCl disrupts these structures and makes it soluble. The percent binding was calculated using the same method as in the previous experiment. The polynomial fits for one-site-specific binding fit almost perfectly (R2 values are almost 1). Hill coefficients (supplemental Table 1B) were also calculated using Graphpad Prism 5.0 software, and these are indicative of positive cooperative binding. Hill plots using a logarithmic scale were also analyzed and were also indicative of positive cooperative binding (supplemental Fig. 5). There are some differences in the dissociation constants (supplemental Table 1B) of GST-POT1 binding to these D-loops. The differences in POT1 binding to DL1, DL2, and DL3 are modest and are largely within the margin of error. However, the difference in POT1 binding between DL1 (no 8-oxodG lesion) and DL4 (two 8-oxodG lesion) is quite notable. In fact, there is more than a 2-fold decrease in the dissociation constant for DL4 compared with DL1. This result suggests that GST-POT1 binding is more facile to oxidatively damaged D-loops than the undamaged.
Involvement of G-quadruplex Structure
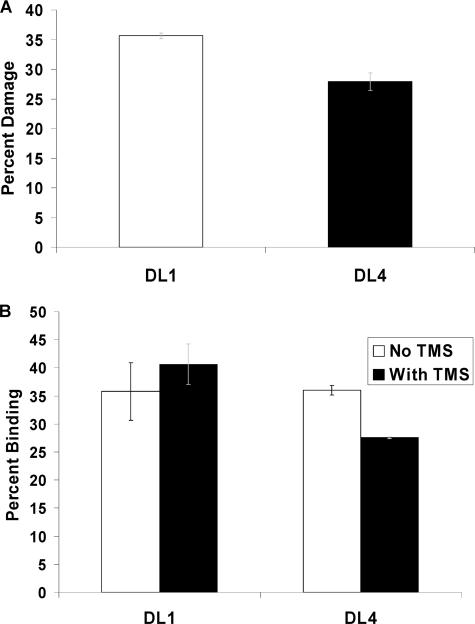
We also investigated whether there was any involvement of higher order structures (G-quadruplex etc) that could account for the enhancement of helicase activities of BLM and WRN on the damaged D-loop structures. The formation of G-quadruplex structures in telomeric DNA is well documented (38, 39). DMS has been used previously to map G-quadruplex formation in single-stranded telomeric DNA (40). We used this reagent to map the formation of G-quadruplex in the D-loops used in our study. DMS methylates the N7 of guanine and the generation of N7-methylguanine is easily detectable by piperidine treatment. However, if the guanine forms a G-quadruplex structure or any other secondary structures involving the N7, the methylation by DMS would be inhibited. This will result in less or no degradation upon piperidine treatment. DL1 (D-loop without any damage) and DL4 (D-loop with two 8-oxodG lesions) were treated with 20% DMS followed by piperidine treatment, and the amount of degradation was compared. The results derived from this experiment are shown in Fig. 6A. The percent damage is calculated as the intensity of the bands moving faster than the undamaged band relative to the total radiation present in the lane. The damage in DL4 is about 20% less than that in DL1. Although the difference is very minimal, the result indicates that the methylation is inhibited by DL4 in comparison to DL1 and suggests that secondary structure (G-quadruplex) formation is little more prominent in DL4 than in DL1.
FIGURE 6.
A, shown is the relative damage on DL1 (undamaged D-loop) and DL4 (D-loop with two 8-oxodG lesion) upon methylation by DMS and piperidine treatment. B, shown is GST-POT1 (10 nm) binding to DL1 (1 nm) and DL4 (1 nm) in the presence and absence of TMS (2 μm). The error bars represent the S.E. calculated from three independent experiments.
The G-quadruplex interacting agent TMS is a potent telomerase inhibitor (41). It also affects the POT1 binding in single-stranded telomeric DNA by stabilizing G-quadruplex structures (42). We investigated the effect of TMS on POT1 binding to damaged and undamaged D-loops. D-loops (1 nm, either DL1 or DL4) were treated with telomestatin at room temperature for 30 min before the addition of GST-POT1. Different concentrations of TMS and POT1 were used, and in the presence of 2 μm TMS, GST-POT1 (10 nm) binding to DL4 was affected by 20%; Fig. 6B. The binding of GST-POT1 to DL1 is hardly affected. Using a higher concentration of TMS did not affect the difference. When 2.5 nm GST-POT1 was used in the presence of 2 μm TMS, a 2-fold decrease in binding of GST-POT1 to DL4 (1 nm) was observed (supplemental Fig. 6), and there was no effect of TMS on GST-POT1 binding to DL1. However, the binding was very low under these conditions, making it difficult to evaluate the significance of the data. Nevertheless, these experiments suggest that telomestatin has a slightly more pronounced effect on GST-POT1 binding to the damaged D-loop (DL4) and indicates that there may be a possibility of preferential G-quadruplex formation in DL4 due to the presence of 8-oxodG.
Effect of POT1 on Helicase Activity
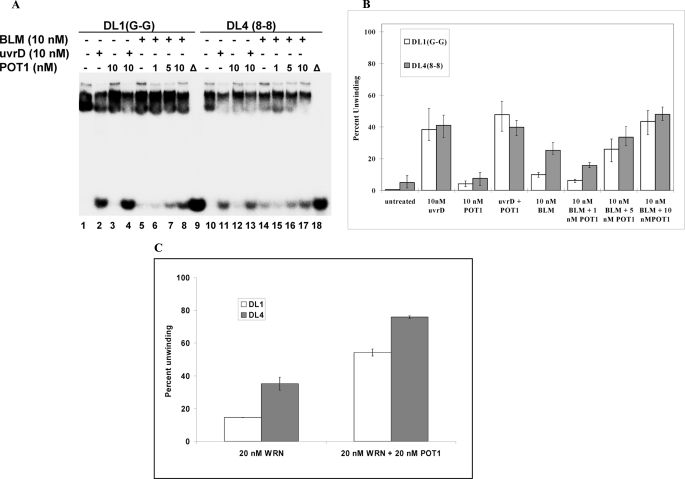
It was shown previously that GST-POT1 interacts with WRN and BLM to stimulate their helicase activities (28). Because GST-POT1 efficiently binds to undamaged and damaged D-loops, we investigated whether it could affect the helicase activities of BLM, WRN, and RecQ5 on undamaged and damaged D-loops. Fig. 7A summarizes the effect of GST-POT1 on BLM helicase activity. The D-loops without any damage (DL1) and with two 8-oxodG lesions (DL4) were used in this experiment. D-loops (0.5 nm) were treated with 10 nm BLM (lanes 5 and 14) or 10 nm BLM with different concentrations of GST-POT1 (lanes 6-8 and 15-17). Control experiments were performed to check helicase activity of 10 nm GST-POT1 alone (lanes 3 and 12). In another set of control experiments, the effect of 10 nm GST-POT1 was tested on helicase activity of 10 nm UvrD (lanes 2 and 4 and lanes 11 and 13). The quantitative analysis of this experiment (Fig. 7B) revealed that there is a small decrease in BLM helicase activity in the presence of 1 nm GST-POT1, and then the helicase activity increases consistently with increasing GST-POT1 concentration. The BLM helicase activity on DL1 was enhanced 4-fold, and there was an almost 2-fold increase in the helicase activity on DL4 in the presence of 10 nm GST-POT1. At low concentrations (1 nm), GST-POT1 cannot interact with BLM (10 nm) and inhibits its helicase activity by binding to the D-loops. At increasing concentrations (5 and 10 nm) it interacts with BLM and enhances its helicase activity. The UvrD helicase activity on both the strands remained unaffected by GST-POT1. This is in accord with our earlier finding that Pot1 does not stimulate UvrD helicase activity (28).
FIGURE 7.
Effect of GST-POT1 on helicase activity of BLM on telomeric D-loop DNAs with and without the 8-oxodG lesion. A, BLM helicase (10 nm) activity is measured by itself on each of the D-loop substrates (0.5 nm) (lanes 5 and 14). The bacterial 3′-5′ helicase UvrD (10 nm) was used as a positive control for both substrates both in the presence (lanes 4 and 13) and absence (lanes 2 and 11) of GST-POT1 (10 nm). Increasing amounts of GST-POT1 (1, 5, and 0 nm) were added to each substrate (POT1 +DL1(G-G) in lanes 6–8 and POT1 DL4 (8-8) in lanes 15–17.) Each substrate was denatured (Δ) at 95 °C. (lanes 9 and 18). B, shown is the quantitative analysis of helicase assay showing the stimulation of BLM helicase activity by GST-POT1 on damaged and undamaged telomeric D-loop structures. C, shown is the quantitative analysis of the effect of GST-POT1 (20 nm) on WRN (20 nm) helicase activity on undamaged (DL1, 0.5 nm) and oxidatively damaged (DL4, 0.5 nm) D-loops. The error bars represent the S.E. calculated from four independent experiments.
In addition, the effect of GST-POT1 on WRN helicase activity on D-loops was tested. A concentration of 20 nm WRN was used to generate ∼10% unwinding of 0.5 nm DL1. A different batch of WRN was used in this experiment, with less active helicase than the WRN used in experiments up to this point. When 0.5 nm DL1 was treated with 20 nm WRN and 20 nm GST-POT1 (Fig. 7C), an ∼4-fold increase in WRN helicase activity was observed. GST-POT1 also enhanced the WRN helicase function 2-fold on damaged D-loop (DL4). In contrast, the helicase activity of RecQ5 on both D-loops was unaltered by the presence of GST-POT1 (data not shown).
DISCUSSION
Oxidation of guanine to 8-oxoguanine is a common and well known oxidative DNA base modification, and G-rich DNA, including telomeric repeats, is susceptible to accumulation of this potentially mutagenic lesion (19, 20). Although there are several other important oxidative DNA base modifications, 8-oxoguanine is the most well characterized lesion with demonstrated biological significance. 8-Oxoguanine containing D-loops mimic oxidatively damaged telomeric DNA regions. WRN and BLM are thought to play roles in telomere maintenance, and it has been shown that WRN unwinds undamaged D-loop structures (23). The present study examines the ability of WRN, BLM, and RecQ5 to unwind telomeric D-loops containing 8-oxodG. The results demonstrate that WRN and BLM preferentially unwind telomeric D-loops containing 8-oxodG. It is important to point out that the 8-oxodG-containing D-loops used in this study, DL2–4, contain one (DL2) or two (DL3, DL4) 8-oxodG residues and are predicted to have the identical stability and melting temperature as DL1, which contains no damage, and to lack any gross structural perturbation in the vicinity of the 8-oxodG residues (43, 44). This is consistent with our observations that DL1–4 are equally resistant to MBN digestion (Fig. 2) and are unwound with identical efficiency by UvrD (Fig. 3). These results suggest that the difference in helicase activity does not arise from any instability in the damaged D-loops due to the 8-oxodG lesions but, rather, reflects a specific helicase/lesion interaction. Both BLM and WRN have been reported to be structure-specific proteins (45). Without affecting the overall structure of the D-loops, the 8-oxodG lesions in a telomeric region might be forming some specialized local structures. These structures could make the damaged D-loops the preferred substrates for BLM and WRN helicase activity. One of the possible structures could be G-quadruplex (G4) formation, as both BLM and WRN work particularly efficiently on these structures (33). Szalai et al. (46) reported the formation of G-quadruplex and other secondary structures upon conversion of guanine to 8-oxodG in telomeric DNA in vitro. Additionally, Gros et al. (47) tested the G-quadruplex-forming abilities of different modified bases in TG4T and TG5T structures. They found that 8-oxodG enhances G-quadruplex-forming ability of the strand and stabilizes the secondary structure. However, the involvement of 8-oxoguanine in formation of G4 structures is still speculative and needs further examination.
POT1, which plays an important role in telomere maintenance, binds to but does not show preference for 8-oxodG-containing telomeric ssDNA (Fig. 4). However, POT1 preferentially binds 8-oxodG-containing telomeric D-loops (Fig. 5) and it binds telomeric D-loops with or without 8-oxodG with lower affinity than telomeric ssDNA (Kd ∼ 14–35 nm for D-loop versus Kd ∼ 5 nm for ssDNA). This could reflect different levels of conformational flexibility in these two DNA substrates (11, 48) or that POT1 binds the two DNA substrates in a different manner. These results indicate that although POT1 is known to bind single-stranded telomeric DNA, it also can bind to a telomeric D-loop structure in vitro. Interaction with POT1 may transiently disrupt the D-loop conformation, resulting in a single-stranded telomeric region that promotes POT1 binding. This structural disruption could be elevated in the presence of 8-oxodG, resulting in a higher POT1 binding affinity. Analysis of POT1 binding shows that the cooperative nature of this POT1/DNA interaction is conserved independent of the structure of the telomeric DNA substrate.
Although POT1 binds relatively weakly to telomeric D-loops, our data suggest that the presence of 8-oxodG enhances the affinity of GST-POT1 to these DNA substrates. This may reflect an altered local DNA conformation in the vicinity of the DNA lesion; for example, the 8-oxodG-dC base pair could exist in a syn conformation instead of the anti conformation typical of a dG-dC base pair (21). Conversely, POT1 binding to telomeric ssDNA is independent of the presence or absence of 8-oxodG. It is possible that GST-POT1 binds readily to undamaged telomeric ssDNA to protect it from accumulating oxidative DNA lesions. Although previous studies indicate that POT1 may play a role in resolving G-quadruplex structures in telomeric double-stranded DNA (49), our data indicate that POT1 preferentially binds to 8-oxodG-containing telomeric D-loops, suggesting that POT1 could facilitate repair of oxidative DNA damage in telomeric D-loops.
The involvement of WRN and BLM in the dissociation of alternate/complex DNA intermediates during replication at telomeres is well documented (28). Association of both WRN and BLM with telomeres during S-phase in immortalized, telomerase-deficient cells has been reported, and these cells use recombination based ALT pathways (23). These two proteins also colocalize with nuclear foci of human ALT cells that contain telomeric DNA and telomere-associated proteins TRF1, TRF2, and POT1 (23, 28). Dissociation of telomeric t-loop/D-loop structures is required for progression of the ALT-based DNA elongation process at the chromosome ends, and resolution of these structures is also essential for complete repair of any damage (50). Our results suggest that WRN and BLM not only assist the replication process by unwinding telomeric D-loop structures but also facilitate repair of oxidatively damaged lesions by preferentially resolving them. It is also possible that accumulation of DNA damage at the telomeres increases WRN and BLM unwinding and leads to an increase in telomere length. This hypothesis is in agreement with studies of double knock-out mice for telomerase and WRN, which show signs of shortened telomeres, indicating that WRN is an important component of telomere maintenance in vivo (51, 52).
It is likely that the ability of POT1 to associate with telomeric D-loop structures enables it to play an important role in replication of telomeric DNA and repair of oxidative damage at telomeres. It may cooperate with WRN and BLM to resolve complex structures such as G-quadruplex to facilitate proper elongation of telomeric ends. POT1 may also help repair proteins to access the oxidatively damaged bases by resolving the complex secondary structures. Our observation of preferential binding of POT1 to the damaged D-loop supports this possibility.
In summary, we have successfully constructed 8-oxodG lesion-containing D-loops and demonstrated that the WRN and BLM helicases unwind the damaged D-loops more efficiently than undamaged D-loops. This supports a likely role of these helicases in telomeric DNA repair processes. It was also shown that the telomeric-binding protein GST-POT1 binds to the undamaged and damaged D-loops. This binding is less robust than its binding to single-stranded telomeric DNA, but it is a novel observation that the presence of lesions in the substrate enhances its binding ability. From these observations we propose that POT1 is involved in secondary structure disruption and, thus, participates in the DNA repair process at the telomere. Our study also indicates the possibility that a damaged D-loop might be a better candidate for forming complex structures like G-quadruplex than an undamaged D-loop. It has been shown that the quadruplex structure is preferred for the RecQ helicases. Thus, the formation of these structures in the D-loop region might explain our observed increased helicase activity.
Supplementary Material
Acknowledgments
We thank Dr. Penny Mason for help in the experimental setup and Dr. Patricia Opresko for comments.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Program of the National Institute on Aging.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6 and Table 1.
- 8-oxodG
- 8-oxo-2′-deoxyguanosine
- ssDNA
- single-stranded DNA
- MBN
- mung bean nuclease
- EMSA
- electrophoretic mobility shift assay
- DMS
- dimethyl sulfate
- TMS
- telomestatin
- GST
- glutathione S-transferase
- TBE
- Tris borate-EDTA
- DTT
- dithiothreitol
- BSA
- bovine serum albumin
- WRN
- Werner syndrome protein
- BLM
- Bloom syndrome protein.
REFERENCES
- 1.Harley C. B., Futcher A. B., Greider C. W. (1990) Nature 345, 458–460 [DOI] [PubMed] [Google Scholar]
- 2.Opresko P. L. (2008) Mech. Ageing Dev. 129, 79–90 [DOI] [PubMed] [Google Scholar]
- 3.Smogorzewska A., de Lange T. (2004) Annu. Rev. Biochem. 73, 177–208 [DOI] [PubMed] [Google Scholar]
- 4.Watson J. D. (1972) Nat. New Biol. 239, 197–201 [DOI] [PubMed] [Google Scholar]
- 5.Greider C. W., Blackburn E. H. (1996) Sci. Am. 274, 92–97 [DOI] [PubMed] [Google Scholar]
- 6.Blackburn E. H. (2001) Cell 106, 661–673 [DOI] [PubMed] [Google Scholar]
- 7.Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. (2002) Oncogene 21, 532–540 [DOI] [PubMed] [Google Scholar]
- 9.Bianchi A., Stansel R. M., Fairall L., Griffith J. D., Rhodes D., de Lange T. (1999) EMBO J. 18, 5735–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Steensel B., Smogorzewska A., de Lange T. (1998) Cell 92, 401–413 [DOI] [PubMed] [Google Scholar]
- 11.Lei M., Podell E. R., Cech T. R. (2004) Nat. Struct. Mol. Biol. 11, 1223–1229 [DOI] [PubMed] [Google Scholar]
- 12.Bohr V. A. (2008) Trends Biochem. Sci. 33, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickson I. D. (2003) Nat. Rev. Cancer 3, 169–178 [DOI] [PubMed] [Google Scholar]
- 14.Yu C. E., Oshima J., Fu Y. H., Wijsman E. M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., Martin G. M., Mulligan J., Schellenberg G. D. (1996) Science 272, 258–262 [DOI] [PubMed] [Google Scholar]
- 15.Brosh R. M., Jr., Karow J. K., White E. J., Shaw N. D., Hickson I. D., Bohr V. A. (2000) Nucleic Acids Res. 28, 2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walpita D., Plug A. W., Neff N. F., German J., Ashley T. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 5622–5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro L., Freeman B. A. (2001) Nutrition 17, 163–165 [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T. (1993) Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 19.Michaels M. L., Miller J. H. (1992) J. Bacteriol. 174, 6321–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grollman A. P., Moriya M. (1993) Trends Genet. 9, 246–249 [DOI] [PubMed] [Google Scholar]
- 21.Bruner S. D., Norman D. P., Verdine G. L. (2000) Nature 403, 859–866 [DOI] [PubMed] [Google Scholar]
- 22.von Zglinicki T. (2002) Trends Biochem. Sci. 27, 339–344 [DOI] [PubMed] [Google Scholar]
- 23.Opresko P. L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kølvraa S., May A., Seidman M. M., Bohr V. A. (2004) Molecular Cell 14, 763–774 [DOI] [PubMed] [Google Scholar]
- 24.Orren D. K., Brosh R. M., Jr., Nehlin J. O., Machwe A., Gray M. D., Bohr V. A. (1999) Nucleic Acids Res. 27, 3557–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karow J. K., Newman R. H., Freemont P. S., Hickson I. D. (1999) Curr. Biol. 9, 597–600 [DOI] [PubMed] [Google Scholar]
- 26.Peakman L. J., Antognozzi M., Bickle T. A., Janscak P., Szczelkun M. D. (2003) J. Mol. Biol. 333, 321–335 [DOI] [PubMed] [Google Scholar]
- 27.Opresko P. L., von Kobbe C., Laine J. P., Harrigan J., Hickson I. D., Bohr V. A. (2002) J. Biol. Chem. 277, 41110–41119 [DOI] [PubMed] [Google Scholar]
- 28.Opresko P. L., Mason P. A., Podell E. R., Lei M., Hickson I. D., Cech T. R., Bohr V. A. (2005) J. Biol. Chem. 280, 32069–32080 [DOI] [PubMed] [Google Scholar]
- 29.Henle E. S., Han Z., Tang N., Rai P., Luo Y., Linn S. (1999) J. Biol. Chem. 274, 962–971 [DOI] [PubMed] [Google Scholar]
- 30.Kawanishi S., Oikawa S., Murata M., Tsukitome H., Saito I. (1999) Biochemistry 38, 16733–16739 [DOI] [PubMed] [Google Scholar]
- 31.Oikawa S., Kawanishi S. (1999) FEBS Lett. 453, 365–368 [DOI] [PubMed] [Google Scholar]
- 32.Aldaye F. A., Sleiman H. F. (2006) Angew. Chem. Int. Ed. Engl. 45, 2204–2209 [DOI] [PubMed] [Google Scholar]
- 33.Li J. L., Harrison R. J., Reszka A. P., Brosh R. M., Jr., Bohr V. A., Neidle S., Hickson I. D. (2001) Biochemistry 40, 15194–15202 [DOI] [PubMed] [Google Scholar]
- 34.Shen J. C., Loeb L. A. (2000) Trends Genet 16, 213–220 [DOI] [PubMed] [Google Scholar]
- 35.Izumikawa K., Yanagida M., Hayano T., Tachikawa H., Komatsu W., Shimamoto A., Futami K., Furuichi Y., Shinkawa T., Yamauchi Y., Isobe T., Takahashi N. (2008) Biochem. J. 413, 505–516 [DOI] [PubMed] [Google Scholar]
- 36.Machwe A., Ganunis R., Bohr V. A., Orren D. K. (2000) Nucleic Acids Res. 28, 2762–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sowd G., Lei M., Opresko P. L. (2008) Nucleic Acids Res. 36, 4242–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han H., Langley D. R., Rangan A., Hurley L. H. (2001) J. Am. Chem. Soc. 123, 8902–8913 [DOI] [PubMed] [Google Scholar]
- 39.Parkinson G. N., Lee M. P., Neidle S. (2002) Nature 417, 876–880 [DOI] [PubMed] [Google Scholar]
- 40.Han H., Langley D. R., Rangan A., Hurley L. H. (2001) J. Am. Chem. Soc. 123, 8902–8913 [DOI] [PubMed] [Google Scholar]
- 41.Shin-ya K., Wierzba K., Matsuo K., Ohtani T., Yamada Y., Furihata K., Hayakawa Y., Seto H. (2001) J. Am. Chem. Soc. 123, 1262–1263 [DOI] [PubMed] [Google Scholar]
- 42.Gomez D., O'Donohue M. F., Wenner T., Douarre C., Macadré J., Koebel P., Giraud-Panis M. J., Kaplan H., Kolkes A., Shin-ya K., Riou J. F. (2006) Cancer Res. 66, 6908–6912 [DOI] [PubMed] [Google Scholar]
- 43.Lipscomb L. A., Peek M. E., Morningstar M. L., Verghis S. M., Miller E. M., Rich A., Essigmann J. M., Williams L. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92, 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanvah S., Schuster G. B. (2005) Nucleic Acids Res. 33, 5133–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohaghegh P., Karow J. K., Brosh R. M., Jr., Bohr V. A., Hickson I. D. (2001) Nucleic Acids Res. 29, 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szalai V. A., Singer M. J., Thorp H. H. (2002) J. Am. Chem. Soc. 124, 1625–1631 [DOI] [PubMed] [Google Scholar]
- 47.Gros J., Rosu F., Amrane S., De Cian A., Gabelica V., Lacroix L., Mergny J. L. (2007) Nucleic Acids Res. 35, 3064–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei M., Podell E. R., Baumann P., Cech T. R. (2003) Nature 426, 198–203 [DOI] [PubMed] [Google Scholar]
- 49.Zaug A. J., Podell E. R., Cech T. R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102, 10864–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henson J. D., Neumann A. A., Yeager T. R., Reddel R. R. (2002) Oncogene 21, 598–610 [DOI] [PubMed] [Google Scholar]
- 51.Chang S., Multani A. S., Cabrera N. G., Naylor M. L., Laud P., Lombard D., Pathak S., Guarente L., DePinho R. A. (2004) Nat. Genet. 36, 877–882 [DOI] [PubMed] [Google Scholar]
- 52.Du X., Shen J., Kugan N., Furth E. E., Lombard D. B., Cheung C., Pak S., Luo G., Pignolo R. J., DePinho R. A., Guarente L., Johnson F. B. (2004) Mol. Cell. Biol. 24, 8437–8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.