Abstract
The 5′-untranslated region (UTR) of serine hydroxymethyltransferase 1 (SHMT1) contains an internal ribosome entry site (IRES) that regulates SHMT1 expression, a rate-limiting enzyme in de novo thymidylate biosynthesis. In this study, we show that the SHMT1 IRES is the first example of a cellular IRES that is poly(A) tail-independent. Interactions between the 5′-UTR and 3′-UTR functionally replaced interactions between the poly(A) tail and the poly(A)-binding protein (PABP) to achieve maximal IRES-mediated translational efficiency. Depletion of the SHMT1 IRES-specific trans-acting factor (ITAF) CUG-binding protein 1 (CUGBP1) from in vitro translation extracts or deletion of the CUGBP1 binding site on the 3′-UTR of the SHMT1 transcript decreased the IRES activity of non-polyadenylylated biscistronic mRNAs relative to polyadenylylated biscistronic mRNAs and resulted in a requirement for PABP. We also identified a novel ITAF, heterogeneous nuclear ribonucleoprotein H2 (hnRNP H2), that stimulates SHMT1 IRES activity by binding to the 5′-UTR of the transcript and interacting with CUGBP1. Collectively, these data support a model for the IRES-mediated translation of SHMT1 whereby the circularization of the mRNA typically provided by the eukaryotic initiation factor (eIF) 4G/PABP/poly(A) tail interaction is achieved instead through the hnRNP H2/CUGBP1-mediated interaction of the 5′- and 3′-UTRs of the SHMT1 transcript. This circularization enhances the IRES activity of SHMT1 by facilitating the recruitment and/or recycling of ribosomal subunits, which bind to the transcript in the middle of the 5′-UTR and migrate to the initiation codon via eIF4A-mediated scanning.
Introduction
Internal ribosome entry sites (IRES)2 are cis-acting elements that enable the cap-independent recruitment of 40 S ribosomal subunits to the 5′-untranslated region (UTR) of an mRNA transcript. Although originally identified in viruses (1, 2), IRESes have recently been discovered in many cellular mRNAs, particularly those encoding proteins involved in development, differentiation, cell cycle progression, cell growth, apoptosis, and stress response (reviewed in Refs. 3 and 4). It is now estimated that as many as 3–5% of cellular mRNAs can be translated by a cap-independent mechanism (5). Although the list of cellular IRESes continues to grow, little is known about their mechanism of action. However, there is increasing evidence that the poly(A) tail of the transcript and IRES-specific trans-acting factors (ITAFs) play major roles during cap-independent translation.
Almost all eukaryotic mRNAs possess a 3′ poly(A) tail that can enhance both cap-dependent and IRES-mediated translation initiation (6, 7). The interaction of poly(A)-binding protein (PABP) with eukaryotic initiation factor 4G (eIF4G) results in the formation of a “closed loop” by linking the poly(A) tail and either the 5′-cap (in cap-dependent translation) or the 5′-UTR (in IRES-mediated translation). Looping through the 5′- and 3′-ends of the transcript is thought to increase translation rates by facilitating the recycling of 40 S ribosomal subunits, promoting 40 S recruitment, and/or stimulating the formation of the 80 S ribosome (8–16). It has also recently been reported that the poly(A) tail can enhance 48S complex assembly through a process that is independent of PABP (17).
ITAFs are RNA-binding proteins that interact functionally with IRES elements to positively or negatively regulate internal initiation. Many of the ITAFs identified to date belong to the group of heterogeneous nuclear ribonucleoproteins (hnRNPs). These include hnRNP A1, C1/C2, I, E1/E2, K, and L (18–21). hnRNPs are located primarily in the nucleus but are known to translocate to the cytoplasm in specific cell or tissue types and as part of specific stress responses. It is hypothesized that hnRNPs and other ITAFs exert their effect on IRES activity by aiding in the recruitment of the 40 S ribosomal subunit through their interactions with the canonical initiation factors or ribosomal components or by acting as RNA chaperones to control the configuration of the IRES (reviewed in Refs. 4 and 22). Studies of viral IRESes have suggested that ITAFs can also establish a RNA/protein bridge between the IRES and the 3′-end of the transcript (23).
In this study, we investigated the role of the poly(A) tail, ITAFs, and the 40 S ribosomal subunit in the IRES-mediated translation of serine hydroxymethyltransferase 1 (SHMT1), an enzyme that regulates folate-dependent de novo thymidylate biosynthesis during S-phase (24, 25) and in response to UV exposure (see accompanying article (49)). The data presented here suggest a model for the IRES-mediated translation of SHMT1 and provide a mechanism that accounts for the previously reported finding that SHMT1 IRES activity is stimulated by the SHMT1 3′-UTR (25).
EXPERIMENTAL PROCEDURES
Cell Culture and Preparation of Extracts
Mammary adenocarcinoma (MCF-7) cells were obtained from ATCC (HTB22) and were cultured in α-MEM (Hyclone Laboratories) containing 11% fetal bovine serum (Hyclone Laboratories) at 37 °C and 5% CO2. When the cells reached ∼95% confluence, they were harvested by trypsinization and washed in phosphate-buffered saline. To obtain whole cell extract, cells were resuspended in lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 5 mm EDTA, 1 mm PMSF, 1:100 dilution of protease inhibitor mixture (Sigma)) and lysed on ice for 30 min. When necessary, the cell extract was treated with γ-phosphatase (Sigma) in the presence of 2 mm MnCl2 for 30 min at 30 °C. γ-Phosphatase activity was then inhibited by the addition of EDTA to a final concentration of 50 mm followed by heating at 65 °C for 1 h. The protein concentration of the extract was determined using the Lowry assay as modified by Bensadoun and Weinstein (26).
Vectors
The generation of bicistronic DNA templates containing the immunoglobulin heavy chain-binding protein (BiP) IRES, the SHMT1 5′-UTR, the full-length SHMT1 3′-UTR, and the reverse complement of the SHMT1 5′-UTR is described elsewhere (25). The poly(A)-interacting protein 2 (Paip2) coding sequence (27) was subcloned into the pGEX4T-2 vector (GE Healthcare) using the primers 5′-TAGGATCCATGAAAGATCCAAGTCGCAG-3′ and 5′-TAGTCGACTCAAATATTTCCGTACTTCAC-3′, where the BamHI and SalI restriction sites are shown in bold. The CUGBP1-pMAL vector was a gift from Lubov Timchenko (Baylor College of Medicine). The hnRNP H2 cDNA (a gift from Jeffrey Wilusz, Colorado State University) was subcloned into the pMAL-c2E vector (New England Biolabs) using the primers 5′-TCGGATCCATGATGCTGAGCACGGAAG-3′ and 5′-TAGTCGACCTAAGCAAGGTTTGACTG-3′, where the BamHI and SalI restriction sites are shown in bold. The SHMT1 3′-UTR truncations were cloned using the following primers: 3′-UTR Fwd = 5′-AGGAGCGGGCCCACTCTGGAC-3′, 3′-UTR (157) Rev = 5′-GTGAAGAAAACATGAAAAAAG-3′, 3′-UTR (200) Rev = 5′-GTCCCAGAATTACTAACAATGAG-3′, 3′-UTR (236) Rev = 5′-GAAAGCCAGGTTCAAATTTAAATCC-3′, 3′-UTR (317) Rev = 5′-TTGCCCTACACCACCATCTA-3′, 3′-UTR (477) Rev = 5′-AGCCTCAGAAGCTAATTCAG-3′, 3′-UTR (637) Rev = 5′-CTGGTTGCTTCTCACACCAG-3′. The SHMT1 5′-UTR truncations were cloned using the following primers: 5′-UTR Fwd = 5′-GCCTGGCGCGCAGAGTGCACCTTCC-3′, 5′-UTR Rev = 5′-TGCACTGGTTCGAAGCTGCCTAGCGAC-3′, 5′-UTR (104) Rev = 5′-GCGCACCGCCGCGGGCCAGCCACG-3′, 5′-UTR (105) Fwd = 5′-GGGGCGTTGGGTCAGCGGGTCTGGG-3′, 5′-UTR (50) Fwd = 5′-TTCGGGGTTTGGGGTTGGAGCGGCTG-3′, 5′-UTR (150) Rev = 5′-GCCGCCGCCGGTGCCACCAGTCCC-3′, 5′-UTR (114) Rev = 5′-CCAACGCCCCGCGCACCGCCGCGG-3′, 5′-UTR (131) Rev = 5′-GTCCCAGACCCGCTGACCCAACGCC-3′. The pcDNA 3 template containing the hepatitis C virus IRES 3′ of the Renilla luciferase reporter gene and 5′ of the firefly luciferase reporter gene was a gift from the laboratory of Partho Ray.
In Vitro Transcription
DNA templates were linearized and purified using the Roche PCR clean-up column. The templates were transcribed using the Ambion MEGAscript kit (for uncapped mRNA) or mMESSAGE mMACHINE kit (for capped mRNA) according to the manufacturer's protocol. For preparation of radiolabeled mRNA for electrophoretic gel mobility shift assays, 50 μCi of α-32P-labeled rUTP (800 Ci/mm, PerkinElmer Life Sciences) was included in in vitro transcription reactions. The crude mRNA was treated with DNase I (Ambion) for 15 min at 40 °C and precipitated in 2 m LiCl at −80 °C. All RNA procedures were conducted under RNase-free conditions, and all mRNA was stored with recombinant RNasin® ribonuclease inhibitor (Promega). The mRNA was quantified by spectrophotometry and its quality verified by electrophoresis.
siRNA Transfections
MCF-7 cells were grown to ∼40% confluence in 6-well plates. The cells were transfected with either 5 nm negative control siRNA (Ambion) or CUGBP1 siRNA (Qiagen; sense, r(GGAACUCUUCGAACAGUAU)dTdT; antisense, r(AUACUGUUCGAAGAGUUCC)dCdG) using HiPerFect transfection reagent (Qiagen) according to the manufacturer's instructions. Following incubation with siRNA for 50 h at 37 °C, the cells were either lysed and subjected to SDS-PAGE/immunoblot analysis to determine knockdown efficiency or used in mRNA transfection experiments.
RNA Affinity Chromatography
1.0 nmol of the indicated in vitro transcribed mRNA (uncapped and polyadenylylated) was incubated with 1.0 nmol of biotinylated oligo(dT) probe (Promega) and 200 μl of packed streptavidin-agarose (Novagen) in TMK buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 150 mm KCl) for 1 h at 4 °C. After extensive washing with TMK buffer, the agarose was resuspended in TMK buffer containing 1.0 nmol of the in vitro transcribed reverse complement of SHMT1 5′-UTR (RevUTR) mRNA (competitor mRNA, uncapped and non-polyadenylylated), 3.0 mg of whole cell extract, 150 μg of yeast tRNA (Ambion), and 600 units of recombinant RNasin® Ribonuclease inhibitor (Promega). Following incubation for 1 h at 4 °C, the agarose was washed extensively with TMK buffer, and bound proteins were eluted in 2× SDS-PAGE sample buffer (160 mm Tris, pH 6.8, 20 mm DTT, 4% SDS, 20% glycerol) at 95 °C for 5 min. The eluted proteins were separated by SDS-PAGE and either stained with Coomassie Blue or subjected to Western blot analysis. Sequencing was performed at the Harvard Microchemistry Facility by microcapillary reverse-phase high pressure liquid chromatography nanoelectrospray tandem mass spectrometry (μLC/MS/MS).
RNA Immunoprecipitation
MCF-7 cells were grown to ∼95% confluence and then treated with 10,000 μJ/cm2 UVC (254 nm) using the Stratagene UV Stratalinker 2400. 22 h after UV treatment, the cells were treated with formaldehyde at a final concentration of 1%. After a 20-min incubation at room temperature, the cross-linking reaction was quenched by the addition of glycine to a final concentration of 125 mm. Following a 10-min incubation at room temperature, the cells were washed in cold phosphate-buffered saline and resuspended in lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 5 mm EDTA, 1 mm PMSF, 1:100 dilution of protease inhibitor mixture (Sigma), 1 unit/ml recombinant RNasin® ribonuclease inhibitor (Promega)). Following a 30-min incubation on ice and centrifugation at 13,000 rpm for 10 min at 4 °C, the supernatant was removed and diluted 1:10 in immunoprecipitation buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris, pH 8, 167 mm NaCl, 1 mm PMSF, 1:100 dilution of protease inhibitor mixture (Sigma), 1 unit/ml recombinant RNasin® ribonuclease inhibitor (Promega)). 1-ml aliquots of the diluted lysate were incubated with 10 μg of antibody overnight at 4 °C. The antibodies used include ImmunoPure goat IgG (Pierce), ImmunoPure mouse IgG (Pierce), goat anti-hnRNP H (N-16, Santa Cruz Biotechnology), and mouse anti-CUGBP1 (3B1, Santa Cruz Biotechnology). 100 μl of packed immobilized protein G (Pierce) was then added, and the lysate was incubated for 2 h at 4 °C in the presence of 0.1 mg/ml bovine serum albumin. After washing the beads once in low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, pH 8, 150 mm NaCl), once in high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, pH 8, 500 mm NaCl), once in LiCl wash buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris, pH 8), and twice in TE (10 mm Tris, pH 8, 1 mm EDTA), bound proteins were eluted in elution buffer (1% SDS, 0.1 m NaHCO3, 1 unit/ml recombinant RNasin® ribonuclease inhibitor (Promega)) at room temperature for 15 min. To reverse the cross-linking, NaCl was added to a final concentration of 200 mm, and the samples were heated at 65 °C for 3 h. Following treatment with Proteinase K for 45 min at 42 °C, the nucleic acids were purified by phenol/chloroform extraction followed by ethanol precipitation. DNA was then removed by treatment with DNase I (Promega) according to the manufacturer's protocol. Reverse transcription was carried out on the remaining RNA using the SuperScript III first-strand synthesis system for reverse transcription-PCR (Invitrogen) according to the manufacturer's protocol. The resulting cDNA was amplified using the 5′-UTR (50) Fwd, 5′-UTR (150) Rev, 3′-UTR Fwd, and 3′-UTR (236) Rev primers described above.
Western Blotting
Proteins were separated by SDS-PAGE and transferred to a PVDF membrane. The membrane was blocked with 5% nonfat dry milk in phosphate-buffered saline containing 1% Nonidet P-40 for 1 h at room temperature, incubated with primary antibody overnight at 4 °C, and incubated with horseradish peroxidase-conjugated anti-IgG for 1–3 h at room temperature. After each incubation, the membrane was washed with phosphate-buffered saline, 0.1% Tween 20. Proteins were visualized using SuperSignal® substrate (Pierce) followed by autoradiography. When necessary, membranes were stripped with 0.2 m NaOH. Mouse anti-CUGBP1 (3B1, Santa Cruz Biotechnology) was used at a 1:10,000 dilution, sheep anti-human SHMT1 was used at a 1:40,000 dilution, goat anti-hnRNP H (N-16, Santa Cruz Biotechnology) was used at a 1:500 dilution, mouse anti-PABP (10E10, Santa Cruz Biotechnology) was used at a 1:1000 dilution, mouse anti-phosphoserine/threonine (BD Transduction Laboratories) was used at a 1:1000 dilution, and mouse anti-GAPDH (Novus Biologicals) was used at a 1:40,000 dilution. Goat anti-mouse IgG, rabbit anti-sheep IgG, and rabbit anti-goat IgG were all purchased from Pierce and used at a 1:5,000 dilution.
Purification of Recombinant Proteins
BL21* cells were transformed with pMAL, CUGBP1-pMAL, hnRNP H2-pMAL, Paip2-pGEX, or pGEX vector and grown to mid-log phase. Protein synthesis was induced with isopropyl β-d-thiogalactopyranoside (final concentration 0.1 mm) for 16 h at 18 °C. The cells were lysed in B-PER (Pierce) followed by sonication with a Branson digital sonifier at 50% amplitude with intermittent icing. After removal of the insoluble material, the clarified supernatant was applied directly to an amylose resin (New England Biolabs) for purification of MBP-tagged proteins or GST-Bind resin (Novagen) for purification of GST-tagged proteins, and the protein was purified according to the manufacturer's protocol. The purity of the protein was determined by SDS-PAGE, and its concentration was determined using the Lowry assay as modified by Bensadoun and Weinstein (26).
RNA Electrophoretic Gel Mobility Shift Assays
20 nm α-32P-labeled RNA and the indicated amount of recombinant protein were added to the binding buffer (20 mm Tris, pH 7.5, 5 mm MgCl2, 1 mm EDTA, 8 mm DTT, 2 μg bovine serum albumin, 1.25 μg of yeast tRNA (Ambion), 10% glycerol, 40 units of recombinant RNasin® ribonuclease inhibitor (Promega)) for a total volume of 10 μl. The binding reaction was incubated at 37 °C for 15 min and then run on a 4% native polyacrylamide gel at 32 mA. Electrophoresis buffer contained 25 mm Tris base, 0.2 m glycine, and 1 mm EDTA. For competition experiments, 200 nm, 1 μm, or 2 μm unlabeled RNA was preincubated with the protein for 5 min prior to its addition to the binding reaction.
Depletion of Proteins from Rabbit Reticulocyte Lysate and in Vitro Translation Reactions
To deplete PABP, Flexi® rabbit reticulocyte lysate (Promega) was incubated with GST-Paip2-bound resin for 1 h at 4 °C. The resin was then collected by centrifugation, and the supernatant was subjected to a second incubation with GST-Paip2-bound resin for 1 h at 4 °C. The resin was then collected by centrifugation, and the supernatant was used in in vitro translation reactions. To immunodeplete CUGBP1 or hnRNP H, Flexi® rabbit reticulocyte lysate was incubated with mouse anti-CUGBP1 (3B1, Santa Cruz Biotechnology) or goat anti-hnRNP H (N-16, Santa Cruz Biotechnology) for 1 h at 4 °C. Immobilized protein A/G beads (Pierce) were then added. Following a 1-h incubation at 4 °C, the beads were collected by centrifugation, and the supernatant was used in in vitro translation reactions. In vitro translation reactions (25 μl) contained 12.5 μl ofFlexi® rabbit reticulocyte lysate, 20 μm amino acids, 2 mm DTT, 100 ng of yeast tRNA (Ambion), 80 mm KCl, 0.5 mm magnesium acetate, 20 units of recombinant RNasin® ribonuclease inhibitor (Promega), and 125 ng of in vitro transcribed mRNA. When necessary, the mRNA was preincubated with recombinant protein for 10 min at room temperature prior to its addition to the in vitro translation reaction. Reactions were carried out at 30 °C for 20 min. Renilla and firefly luciferase expression was quantified on a Veritas microplate luminometer (Turner Biosystems) using the Dual-Glo luciferase assay system (Promega) according to the manufacturer's protocol.
mRNA Transfections
MCF-7 cells at ∼95% confluence were incubated in Opti-MEM (Invitrogen) containing a 1:100 dilution of DMRIE-C transfection reagent (Invitrogen) and 5 μg/ml mRNA (capped and polyadenylylated) for 4 h at 37 °C and 5% CO2. The Opti-MEM was then replaced with α-MEM and the cells incubated for an additional 6 h at 37 °C and 5% CO2. Renilla and firefly luciferase expression was quantified on a Veritas microplate luminometer (Turner Biosystems) using the Dual-Glo luciferase assay system (Promega) according to the manufacturer's protocol. Where indicated, cells were treated with 10,000 μJ/cm2 UVC (254 nm) using the Stratagene UV Stratalinker 2400 12 h prior to the mRNA transfection.
Yeast Two-hybrid Assay
CUGBP1 cDNA was amplified and cloned into the pGBK plasmid (Clontech) using the following primers: 5′-TCGAATTCATGAACGGCACCCTGGA-3′ and 5′-TCGGATCCTCAGTAGGGCTTGCTGT-3′ (EcoRI and BamHI sites are shown in bold). hnRNP H2 cDNA was amplified and cloned into the pGAD plasmid (Clontech) using the following primers: 5′-TCCATATGATGATGCTGAGCACGGAAG-3′ and 5′-TCCTCGAGCTAAGCAAGGTTTGACTG-3′. The NdeI and XhoI sites are shown in bold. The pGBK-CUGBP1 vector was transformed into yeast strain AH109, and stable clones were maintained in Trp−dropout medium. The pGAD-hnRNP H2 vector was transformed into yeast strain Y187, and stable clones were maintained in Leu− dropout medium. The transformed yeast were then mated following the Clontech Matchmaker protocol. After a 24-h mating, cells were plated on His−, Leu−, Trp− dropout medium containing X-α-gal and incubated at 30 °C for 4 days. Clones were validated against negative controls according to the Matchmaker protocol.
Coimmunoprecipitation
MCF-7 cells were grown to ∼95% confluence, and formaldehyde was added to a final concentration of 1%. After a 20-min incubation at 37 °C, the cross-linking reaction was quenched by the addition of glycine to a final concentration of 125 mm. After a 10-min incubation at 37 °C, the cells were washed in cold Tris-buffered saline, harvested, and resuspended in lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 5 mm EDTA, 1 mm PMSF, 1:100 dilution of protease inhibitor mixture (Sigma)). The cells were sonicated five times for 30 s with a Branson digital sonifier at 25% amplitude with intermittent icing. Following centrifugation at 13,000 rpm for 32 min at 4 °C, the supernatant was removed and incubated overnight with 10 μg of antibody at 4 °C. The antibodies used include ImmunoPure goat IgG (Pierce), mouse anti-HA (Santa Cruz Biotechnology), goat anti-hnRNP H (N-16, Santa Cruz Biotechnology), and mouse anti-CUGBP1 (3B1, Santa Cruz Biotechnology). 100 μl of packed immobilized protein G (Pierce) was then added, and the lysate was incubated for 3 h at 4 °C. After washing the beads extensively with lysis buffer, bound proteins were eluted in elution buffer (50 mm Tris, pH 7.5, 10 mm EDTA, 1%SDS) at 95 °C for 10 min. To reverse the cross-linking, 6× SDS-PAGE sample buffer (480 mm Tris, pH 6.8, 60 mm DTT, 12% SDS, 60% glycerol) was added to a final concentration of 1×, and the samples were heated at 95 °C for 20 min. For the recombinant protein immunoprecipitation, 1 μg of MBP-CUGBP1 was combined with 1 μg of MBP-hnRNP H2 and 4 μg of antibody in a final volume of 1 ml. Following an overnight incubation at 4 °C, 100 μl of packed immobilized protein G (Pierce) was added, and the mixture was incubated for 3 h at 4 °C. After washing the beads extensively with lysis buffer, bound proteins were eluted in 2× SDS-PAGE sample buffer (160 mm Tris, pH 6.8, 20 mm DTT, 4% SDS, 20% glycerol) at 95 °C for 10 min.
Hippuristanol Treatment
MCF-7 cells were transfected with in vitro transcribed mRNA according to the protocol above. 10 h after transfection, the indicated amount of hippuristanol (a gift from Junichi Tanaka, University of the Ryukyus, Japan) or an equal volume of vehicle (DMSO) was added to the culture medium, and the cells were and incubated at 37 °C and 5% CO2 for an additional 11 h. Luciferase activity was then quantified as stated above.
Introduction of Open Reading Frames (ORFs) into the SHMT1 5′-UTR
A stop codon was introduced into the 5′-UTR of the bicistronic construct lacking the 3′-UTR according to the QuikChange II site-directed mutagenesis protocol (Stratagene) using the following primers: Stop Fwd = 5′-GTCGCTAGGCAGCTTCGAACTAGTGCAATG-3′ and Stop Rev = 5′-CATTGCACTAGTTCGAAGCTGCCTAGCGAC-3′. Start codons were then introduced into the resulting construct via QuikChange II site-directed mutagenesis using the following primers: 52 AUG Fwd = 5′-GGTCCAGCGCCAAGTATGGGGTTTGGGGTTGG-3′, 52 AUG Rev = 5′-CCAACCCCAAACCCCATACTTGGCGCTGGACC-3′, 70 AUG Fwd = 5′-GGGTTTGGGGTTGGAATGGCTGGTCACGTGGC-3′, 70 AUG Rev = 5′-GCCACGTGACCAGCCATTCCAACCCCAAACCC-3′, 82 AUG Fwd = 5′-GGAGCGGCTGGTAACATGGCTGGCCCGC-3′, 82 AUG Rev = 5′- GCGGGCCAGCCATGTTACCAGCCGCTCC-3′, 103 AUG Fwd = 5′-GGCTGGCCCGCGGCGGAGCATGGGGCGTTGGGTCAGC-3′, 103 AUG Rev = 5′-GCTGACCCAACGCCCCATGCTCCGCCGCGGGCCAGCC-3′, 118 AUG Fwd = 5′-GGGCGTTGGGTCATGGGGTCTGGGACTGG-3′, 118 AUG Rev = 5′-CCAGTCCCAGACCCCATGACCCAACGCCC-3′, 139 AUG Fwd = 5′-GGGACTGGTGGCATGGGCGGCGGCGTAG-3′, 139 AUG Rev = 5′-CTACGCCGCCGCCCATGCCACCAGTCCC-3′, 151 AUG Fwd = 5′-GCACCGGCGGCGGCATGGGACGGAGGCGTCG-3′, 151 AUG Rev = 5′-CGACGCCTCCGTCCCATGCCGCCGCCGGTGC-3′, 169 AUG Fwd = 5′-GGACGGAGGCGTGGCATGGCAGCTTCGAAC-3′, and 169 AUG Rev = 5′-GTTCGAAGCTGCCATGCCACGCCTCCGTCC-3′. The stop and start codons are shown in bold. To determine whether the introduction of the start codon affected the secondary structure of the mRNA, each start codon was mutated via QuikChange II site-directed mutagenesis using the following primers: 52 GUG Fwd = 5′-GGTCCAGCGCCAAGTGTGGGGTTTGGGGTTGG-3′, 52 GUG Rev = 5′-CCAACCCCAAACCCCACACTTGGCGCTGGACC-3′, 70 UUG Fwd = 5′-GGGTTTGGGGTTGGATTGGCTGGTCACGTGGC-3′, 70 UUG Rev = 5′-GCCACGTGACCAGCCAATCCAACCCCAAACCC-3′, 82 UUG Fwd = 5′-GGAGCGGCTGGTAACTTGGCTGGCCCGC-3′, 82 UUG Rev = 5′-GCGGGCCAGCCAAGTTACCAGCCGCTCC-3′, 103 UUG Fwd = 5′-GGCTGGCCCGCGGCGGAGCTTGGGGCGTTGGGTCAGC-3′, 103 UUG Rev = 5′-GCTGACCCAACGCCCCAAGCTCCGCCGCGGGCCAGCC-3′, 118 AUA Fwd = 5′-GGGCGTTGGGTCATAGGGTCTGGGACTGG-3′, 118 AUA Rev = 5′-CCAGTCCCAGACCCTATGACCCAACGCCC-3′, 139 AUA Fwd = 5′- GGGACTGGTGGCATAGGCGGCGGCGTAGG-3′, 139 AUA Rev = 5′-CCTACGCCGCCGCCTATGCCACCAGTCCC-3′, 151 UUG Fwd = 5′-GCACCGGCGGCGGCTTGGGACGGAGGCGTCG-3′, 151 UUG Rev = 5′-CGACGCCTCCGTCCCAAGCCGCCGCCGGTGC-3′, 169 UUG Fwd = 5′- GGACGGAGGCGTGGCTTGGCAGCTTCGAAC-3′, and 169 UUG Rev = 5′-GTTCGAAGCTGCCAAGCCACGCCTCCGTCC-3′. The mutated start codons are shown in bold. All mutations were verified by sequencing at the Cornell Biotechnology Resource Center.
RESULTS
The poly(A) Tail and PABP Are Not Required for Maximal SHMT1 IRES Activity
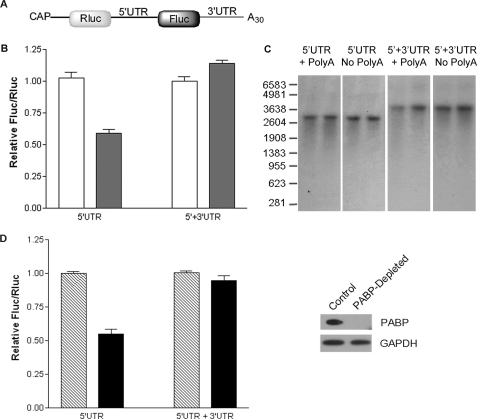
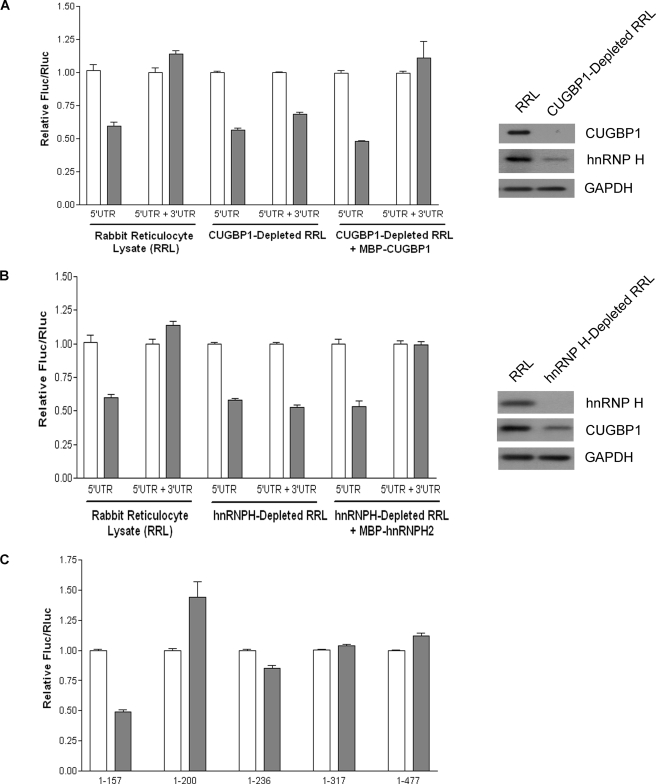
To investigate the influence of the poly(A) tail on the cap-independent translation of SHMT1, the IRES activity of bicistronic mRNAs with and without the stimulatory SHMT1 3′-UTR (25) and with and without an A30 tail (Fig. 1A) was determined in vitro using nuclease-treated rabbit reticulocyte lysate. This cell-free system was selected over a cell culture model, as it eliminates the potential for artificially reduced translation efficiency of the non-polyadenylylated bicistronic mRNA resulting from decreased competition with endogenous polyadenylylated mRNAs. IRES-mediated translation, as measured by the ratio of firefly luciferase (Fluc) to Renilla luciferase (Rluc) activity, was independent of the poly(A) tail when the SHMT1 3′-UTR was present in the transcript (Fig. 1B). However, removal of the 3′-UTR resulted in a 40% decrease in the IRES-mediated translation of non-polyadenylylated RNA relative to RNA containing a poly(A) tail (Fig. 1B). Changes in the stability of the bicistronic mRNA lacking the 3′-UTR were not responsible for the observed stimulatory effect of the poly(A) tail (Fig. 1C).
FIGURE 1.
The poly(A) tail and PABP are not required for maximal SHMT1 IRES activity. A, the bicistronic mRNA used to quantify SHMT1 IRES activity. It consists of (in the 5′ to 3′ direction) a cap analog, the Rluc reporter gene, the alternatively spliced form of the human SHMT1 5′-UTR lacking exon 2 (25), the Fluc reporter gene, and where indicated, the full-length human SHMT1 3′-UTR and a 30-nucleotide poly(A) tail (25). B, in vitro translation assays were carried out using rabbit reticulocyte lysate and in vitro transcribed bicistronic mRNAs with (5′-UTR + 3′-UTR) and without (5′-UTR) the SHMT1 3′-UTR. The white bars represent the ratio of IRES-mediated translation (Fluc) to cap-dependent translation (Rluc) of bicistronic mRNA containing a 30-nucleotide poly(A) tail, and the dark bars represent the Fluc/Rluc of bicistronic mRNA lacking a poly(A) tail. The relative ratio for each bicistronic mRNA containing a poly(A) tail was given a value of 1.0. The data represent the average of three independent experiments ± S.E. C, the bicistronic mRNAs described in A were labeled with 32P, and in vitro translation assays were performed as described under ”Experimental Procedures.“ The RNAs were resolved on an agarose gel and transferred to a positively charged nylon membrane. For each transcript, the left lane represents the mRNA before the in vitro translation reaction, and the right lane represents the transcript after the in vitro translation reaction. D, rabbit reticulocyte lysate was incubated with either GST (Control) or GST-Paip2 (PABP-depleted). The depletion of PABP by GST-Paip2 but not GST alone was confirmed by immunoblotting (right) using an antibody against PABP. GAPDH served as a control for equal protein loading. The graph on the left shows the relative IRES activity (as measured by Fluc/Rluc) of the bicistronic mRNAs in control (striped bars) and PABP-depleted (black bars) rabbit reticulocyte lysate. The relative luminosity for each bicistronic mRNA in the control reaction was given a value of 1.0. The data represent the average of three independent experiments ± S.E.
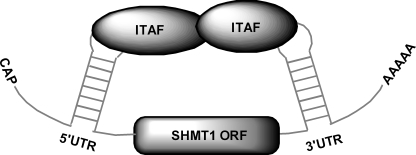
The poly(A) tail affects IRES activity through its interaction with PABP (8–15). Depletion of PABP from rabbit reticulocyte lysate with immobilized Paip2 (27) significantly reduced PABP protein levels (Fig. 1D). Depletion of PABP by this procedure has been shown not to affect the concentration of other initiation factors (7). PABP depletion had no significant effect on the IRES-mediated translation of the polyadenylylated bicistronic mRNA containing the SHMT1 3′-UTR but reduced the IRES activity of the polyadenylylated bicistronic mRNA lacking the 3′-UTR by 45% (Fig. 1D). These results led to the formulation of a model for the IRES-mediated translation of SHMT1 in which interactions between ITAFs bound to the 5′-UTR of the transcript and ITAFs bound to the 3′-UTR serve to circularize the transcript, thereby eliminating the need for both the poly(A) tail and PABP (Fig. 2).
FIGURE 2.
Proposed model for the IRES-mediated translation of SHMT1. In this model, an interaction between an ITAF bound to the 5′-UTR of the SHMT1 transcript and another ITAF bound to the 3′-UTR of the SHMT1 transcript serves to circularize the mRNA. This results in the formation of a closed loop similar to the one that is typically formed by the eIF4G/PABP/poly(A) tail interaction.
CUGBP1 Binds to the SHMT1 3′-UTR
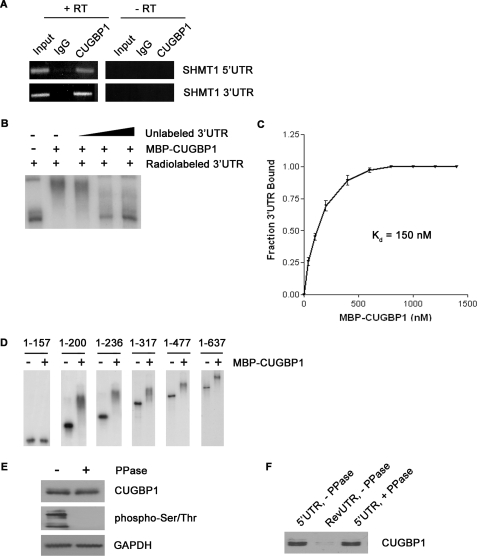
A promising candidate for the 3′-UTR-binding protein in our model was CUG-binding protein 1 (CUGBP1), an isoform of the hnRNP hNab50 (28) that has been shown to stimulate the IRES-mediated translation of SHMT1 (25). The effect of CUGBP1 on IRES activity is significant only when the 3′-UTR of SHMT1 is included in the transcript (25), indicating that the protein might act by binding to the 3′-UTR of the mRNA. The binding of CUGBP1 to the SHMT1 3′-UTR was confirmed in mammary adenocarcinoma (MCF-7) cells by RNA immunoprecipitation using an antibody against CUGBP1 and amplification of the immunoprecipitated RNA with PCR primers specific to the SHMT1 3′-UTR (Fig. 3A).
FIGURE 3.
The interaction of CUGBP1 with the SHMT1 UTRs. A, RNA was immunoprecipitated from UV-treated MCF-7 whole cell extract using an antibody against IgG (control for nonspecific binding) or CUGBP1. The RNA in lanes 1–3 (+RT) was reverse-transcribed into cDNA and then analyzed by PCR using primers specific to either the SHMT1 5′-UTR or the SHMT1 3′-UTR. The RNA in lanes 4–6 (−RT) did not undergo the reverse transcription step. Rather, they were analyzed directly by PCR to control for DNA contamination in the immunoprecipitates. Lanes 1 and 4 (Input) represent 1% of the RNA used in the immunoprecipitation. B, electrophoretic mobility shift assays were carried out in the presence of excess yeast tRNA to eliminate nonspecific binding of the probe, radiolabeled SHMT1 3′-UTR, and recombinant MBP-CUGBP1. A 10×, 50×, and 100× molar excess of unlabeled SHMT1 3′-UTR was also added in lanes 3, 4, and 5, respectively, to determine binding specificity. C, electrophoretic mobility shift assays were carried out using radiolabeled SHMT1 3′-UTR and increasing concentrations of MBP-CUGBP1. The fraction of the 3′-UTR bound by the recombinant protein was quantified using ChemiImager 4400 from Alpha Innotech Corp. The dissociation constant (Kd) was determined using GraphPad Prism. D, electrophoretic mobility shift assays were carried out using radiolabeled SHMT1 3′-UTR truncation mutants in the absence and presence of MBP-CUGBP1. The nucleotides of the 3′-UTR that comprise the truncation mutant are listed above each gel. The nucleotide at the 5′-end of the 3′-UTR is labeled 1. The nucleotide at the 3′-end of the 3′-UTR is labeled 637. E, MCF-7 whole cell extract was either not treated or treated with λ-phosphatase (PPase) and analyzed by Western blot using antibodies against CUGBP1 and phosphorylated serine and threonine residues. GAPDH served as a control for equal protein loading. F, the extracts from E were applied to RNA affinity columns to which either the SHMT1 5′-UTR or the RevUTR had been attached. Proteins that bound to the UTRs were eluted and analyzed by Western blotting using an antibody against CUGBP1.
The binding of CUGBP1 to the SHMT1 3′-UTR was also confirmed in vitro by RNA electrophoretic mobility shift assays (EMSAs) using recombinant MBP-CUGBP1 and an excess of nonspecific competitor tRNA (Fig. 3B). The addition of MBP alone had no effect on the mobility of the SHMT1 3′-UTR (data not shown), and MBP-CUGBP1 binding was abolished upon the addition of a molar excess of unlabeled 3′-UTR (Fig. 3B), indicating that the interaction was specific. As the EMSA was performed in the absence of other proteins, the results of the experiment indicate that CUGBP1 is capable of binding to the 3′-UTR of SHMT1 independent of any auxiliary factors. The dissociation constant (Kd) of MBP-CUGBP1 binding to the 3′-UTR was determined to be 150 nm (Fig. 3C).
CUGBP1 has been reported to bind several consensus sequences including (CUG)n triplet repeats (28), GU-rich elements (UGUUUGUUUGU) (29), and Bruno response elements (AAUGUAUGUUAAUUGUAUGUAUUA) (30). The 3′-UTR of SHMT1 contains a partial Bruno response element (UGUAUGUU) at nucleotide positions 496–503. However, mutation of this sequence did not affect the Kd of CUGBP1 binding in EMSAs (data not shown). To map the CUGBP1 binding site on the SHMT1 3′-UTR, the ability of recombinant protein to bind a series of 3′-UTR truncation mutants was determined by EMSA. The addition of MBP-CUGBP1 had no effect on the mobility of 3′-UTR(1–157), but it did decrease the mobility of 3′-UTR-(1–200), 3′-UTR-(1–236), 3′-UTR-(1–317), 3′-UTR-(1–477), and the full-length 3′-UTR (3′-UTR-(1–637)) (Fig. 3D). These results indicate that CUGBP1 binds to the 5′-end of the SHMT1 3′-UTR between nucleotides 157 and 200. This region contains a partial GU-rich element (UUUGUUU) located at nucleotide positions 167–173.
CUGBP1 Binding to the SHMT1 5′-UTR Requires an Auxiliary Factor
MBP-CUGBP1 did not bind to the SHMT1 5′-UTR in EMSAs (data not shown). However, the results from RNA immunoprecipitation experiments using an antibody against CUGBP1 revealed that the endogenous protein is associated with the SHMT1 5′-UTR in MCF-7 cells (Fig. 3A). Taken together, these data suggest that CUGBP1 binding to the SHMT1 5′-UTR requires either a post-translational modification that is absent from the bacterially-expressed protein or an auxiliary factor that binds to both the SHMT1 5′-UTR and to CUGBP1.
CUGBP1 is known to be phosphorylated in vivo (31, 32). To determine whether the phosphorylation status of CUGBP1 affects its interaction with the SHMT1 5′-UTR, RNA affinity chromatography was carried out using in vitro transcribed RNA and either untreated or phosphatase-treated MCF-7 whole cell extract (Fig. 3E). Phosphatase treatment did not interfere with the ability of the SHMT1 5′-UTR to pull down CUGBP1 (Fig. 3F), making it unlikely that a CUGBP1 post-translational modification is required for the RNA/protein interaction. We therefore hypothesized that CUGBP1 interacts with the 5′-UTR through its association with a novel SHMT1 ITAF that binds directly to the 5′-UTR.
Identification of hnRNP H2 as a SHMT1 5′-UTR-binding Protein
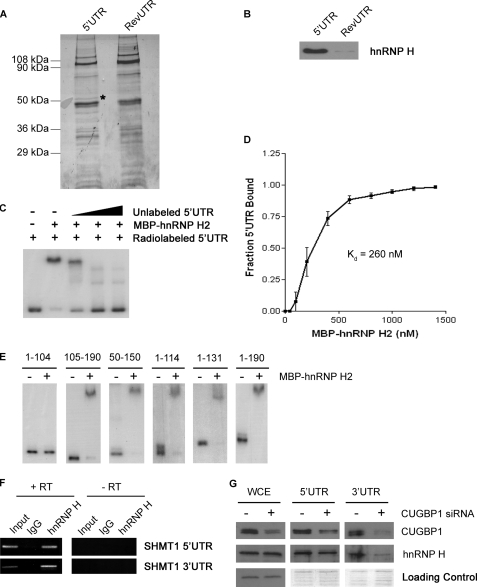
To identify the SHMT1 5′-UTR binding protein, RNA affinity chromatography was carried out using in vitro transcribed mRNA. The RevUTR, which lacks IRES activity (25), was used as a control for nonspecific binding. Analysis of the eluate revealed that a protein of ∼50 kDa bound to the SHMT1 5′-UTR but not the RevUTR (Fig. 4A). μLC/MS/MS identified this protein as hnRNP H2. Immunoblotting using an antibody against hnRNP H confirmed these results (Fig. 4B), although the antibody cannot distinguish between hnRNP H1 and hnRNP H2, which are 96% identical (33).
FIGURE 4.
Interaction of hnRNP H2 with the SHMT1 UTRs. A, MCF-7 whole cell extract was applied to RNA affinity columns to which either the SHMT1 5′-UTR or the RevUTR had been attached. Proteins that bound to the UTRs were eluted, separated by SDS-PAGE, and visualized by Coomassie Blue staining. The protein marked with an asterisk was excised from the gel and analyzed by μLC/MS/MS. B, the SDS-polyacrylamide gel from A was transferred to a PVDF membrane and analyzed by Western blotting using an antibody against hnRNP H. C, electrophoretic mobility shift assays were carried out in the presence of excess yeast tRNA to eliminate nonspecific binding of the probe, radiolabeled SHMT1 5′-UTR, and recombinant MBP-hnRNP H2. A 10×, 50×, and 100× molar excess of unlabeled SHMT1 5′-UTR was also added in lanes 3, 4, and 5, respectively, to determine binding specificity. D, electrophoretic mobility shift assays were carried out using radiolabeled SHMT1 5′-UTR and increasing concentrations of MBP-hnRNP H2. The fraction of the 5′-UTR bound by the recombinant protein was quantified using ChemiImager 4400 from Alpha Innotech Corp. The dissociation constant (Kd) was determined using GraphPad Prism. E, electrophoretic mobility shift assays were carried out using radiolabeled SHMT1 5′-UTR truncation mutants in the absence and presence of MBP-hnRNP H2. The nucleotides of the 5′-UTR that comprise the truncation mutant are listed above each gel. The nucleotide at the 5′-end of the 5′-UTR is labeled 1. The nucleotide at the 3′-end of the 5′-UTR is labeled 190. F, RNA was immunoprecipitated from UV-treated MCF-7 whole cell extract using an antibody against IgG (control for nonspecific binding) or hnRNP H. The RNA in lanes 1–3 (+RT) was reverse-transcribed into cDNA and then analyzed by PCR using primers specific to either the SHMT1 5′-UTR or the SHMT1 3′-UTR. The RNA in lanes 4–6 (−RT) did not undergo the reverse transcription step. Rather, they were analyzed directly by PCR to control for DNA contamination in the immunoprecipitates. Lanes 1 and 4 (Input) represent 1% of the RNA used in the immunoprecipitation. G, whole cell extract (WCE) from cells that were treated with either negative control siRNA (−CUGBP1 siRNA) or siRNA directed against CUGBP1 were applied to RNA affinity columns to which either the cSHMT 5′-UTR or the cSHMT 3′-UTR had been attached. Proteins that bound to the UTRs were eluted, separated by SDS-PAGE, and visualized by Western blotting using antibodies against CUGBP1 or hnRNP H. For the whole cell extract, equal protein loading was confirmed by using an antibody against GAPDH. For the affinity column elutions, equal protein loading was confirmed by staining the gel with Coomassie blue after transfer.
To further investigate the binding of hnRNP H2 to the SHMT1 5′-UTR, EMSAs were carried out using recombinant MBP-hnRNP H2 and an excess of nonspecific competitor tRNA. The results revealed that MBP-hnRNP H2 (but not MBP alone; data not shown) binds directly and specifically to the 5′-UTR (Fig. 4C) with a Kd of 260 nm (Fig. 4D). To map the hnRNP H2 binding site, the ability of the recombinant protein to bind several different 5′-UTR truncation mutants was determined by EMSA. MBP-hnRNP H2 bound to 5′-UTR-(105–190), 5′-UTR-(50–150), 5′-UTR-(1–114), 5′-UTR-(1–131), and the full-length 5′-UTR (5′-UTR-(1–190)) but not to 5′-UTR-(1–104) (Fig. 4E), indicating that nucleotides 105–114 comprise the hnRNP H2 binding site on the SHMT1 5′-UTR. This region contains a run of G nucleotides followed by a C (GGGGC), a sequence that has been shown to promote hnRNP H1 and hnRNP H2 binding specifically (34).
RNA immunoprecipitation using an antibody against hnRNP H verified that the association between hnRNP H1/H2 and the SHMT1 5′-UTR occurs in vivo (Fig. 4F). It also revealed that hnRNP H2 interacts with the 3′-UTR of SHMT1 (Fig. 4F), although it did not bind to the 3′-UTR directly in EMSAs (data not shown). In vitro the binding of hnRNP H2 to the 3′-UTR decreased significantly upon CUGBP1 depletion; in contrast, CUGBP1 depletion had little effect on the binding of hnRNP H2 to the 5′-UTR (Fig. 4G). These results are consistent with a role for hnRNP H2 as the auxiliary factor that enables CUGBP1 to interact with the SHMT1 5′-UTR.
hnRNP H2 Interacts with CUGBP1
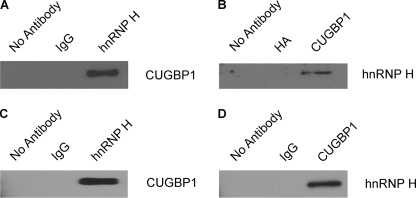
If hnRNP H2 indeed serves as the auxiliary factor for the interaction between CUGBP1 and the SHMT1 5′-UTR, hnRNP H2 and CUGBP1 must interact physically. To test this hypothesis, a yeast two-hybrid analysis was performed. An hnRNP H2-GAL4-activating domain fusion and a CUGBP1-GAL4 DNA-binding domain fusion were expressed in the Saccharomyces cerevisiae strains Y187 and AH109, respectively. The mating of these strains activated the HIS3 and MEL1 reporter genes (as evidenced by the growth of blue colonies on SD/Leu−/Trp−/His− medium containing X-α-gal; data not shown), providing evidence that the two proteins interact. The results of the yeast two-hybrid assay were confirmed by both the coimmunoprecipitation of CUGBP1 with the hnRNP H antibody (Fig. 5A) and the coimmunoprecipitation of hnRNP H with the CUGBP1 antibody (Fig. 5B) from MCF-7 cell extracts. Although it has been reported previously that hnRNP H and CUGBP1 interact in an RNA-dependent manner in myoblasts (35), our data suggest that the interaction is RNA-independent, as the two proteins coimmunoprecipitated from a mixture containing only recombinant hnRNP H2 and recombinant CUGBP1 (Fig. 5, C and D).
FIGURE 5.
hnRNP H2 binds to CUGBP1 in an RNA-independent manner. CUGBP1 (A) and hnRNP H (B) were coimmunoprecipitated from MCF-7 whole cell extracts using antibodies against hnRNP H and CUGBP1, respectively. The no antibody, IgG, and hemagglutinin (HA) coimmunoprecipitations serve as controls for nonspecific binding. CUGBP1 (C) and hnRNP H (D) were coimmunoprecipitated from a mixture of recombinant hnRNP H2 and recombinant CUGBP1 using antibodies against hnRNP H and CUGBP1, respectively. The no antibody and IgG coimmunoprecipitations serve as controls for nonspecific binding.
Depletion of CUGBP1 or hnRNP H2 Results in a Decrease in the IRES-mediated Translation of Non-polyadenylylated SHMT1 mRNA
If an interaction between hnRNP H2 bound to the 5′-UTR of the mRNA and CUGBP1 bound to the 3′-UTR functions to circularize the SHMT1 transcript, then removal of these factors from nuclease-treated rabbit reticulocyte lysate should reveal a requirement for the poly(A) tail. Indeed, the immunodepletion of CUGBP1 (Fig. 6A) and hnRNP H2 (Fig. 6B) reduced the IRES activity of the non-polyadenylylated bicistronic mRNA containing the SHMT1 3′-UTR to levels similar to those obtained when the IRES activity of the non-polyadenylylated bicistronic mRNA lacking the 3′-UTR was measured in rabbit reticulocyte lysate containing both factors. The immunodepletion of CUGBP1 and hnRNP H2 had no effect on the IRES activity of the non-polyadenylylated bicistronic mRNA lacking the SHMT1 3′-UTR. The addition of MBP-tagged recombinant protein (Fig. 6, A and B), but not MBP alone (data not shown), to the immunodepleted extracts restored the IRES activity of the non-polyadenylylated bicistronic mRNA containing the SHMT1 3′-UTR to the levels of its polyadenylylated counterpart, demonstrating that the immunodepletion procedure did not completely remove any other factor required for cap-independent translation. However, consistent with an RNA-independent interaction between CUGBP1 and hnRNP H2, the immunodepletion of CUGBP1 did reduce the levels of hnRNP H present in the nuclease-treated rabbit reticulocyte lysate (Fig. 6A) and vice versa (Fig. 6B).
FIGURE 6.
CUGBP1 and hnRNP H2 depletion result in a dependence on the poly(A) tail. CUGBP1 (A) and hnRNP H (B) were immunodepleted from rabbit reticulocyte lysate as described under ”Experimental Procedures.“ The depletion of these proteins was confirmed by immunoblotting (right) using antibodies against CUGBP1 (A) and hnRNP H (B). GAPDH served as a control for equal protein loading. The graphs on the left show the IRES activities of the bicistronic mRNAs containing a 30-nucleotide poly(A) tail (white bars) or lacking a poly(A) tail (dark bars) as measured in control rabbit reticulocyte, CUBBP1 (A)- or hnRNP H (B)-depleted rabbit reticulocyte lysate, or immunodepleted lysate supplemented with recombinant CUGBP1 (A) or hnRNP H2 (B). The relative ratio of Fluc/Rluc for each bicistronic mRNA containing a poly(A) tail was given a value of 1.0. The data represent the average of three independent experiments ± S.E. C, the 3′-UTR of the bicistronic mRNA was truncated by removal of nucleotides from the 3′-end, and the IRES activity of these truncation mutants was measured in rabbit reticulocyte lysate. The white bars represent the IRES activity of the truncated bicistronic mRNA containing a 300nucleotide poly(A) tail, and the shaded bars represent the IRES activity of the truncated bicistronic mRNA lacking a poly(A) tail. The relative ratio for each truncated bicistronic mRNA containing a poly(A) tail was given a value of 1.0. The data represent the average of three independent experiments ± S.E.
Like the immunodepletion of CUGBP1 from rabbit reticulocyte lysate, the deletion of all but the first 157 nucleotides from the SHMT1 3′-UTR in the bicistronic mRNA resulted in a 50% decrease in the IRES activity of non-polyadenylylated mRNA relative to that of its polyadenylylated counterpart (Fig. 6C). This requirement for the poly(A) tail was not observed when the SHMT1 3′-UTR contained nucleotides 1–200, 1–236, 1–317, and 1–477 (Fig. 6C). As only the 1–157 3′-UTR truncation mutant is incapable of binding CUGBP1 (Fig. 3D), these results further support a role for CUGBP1 in circularizing the SHMT1 transcript.
IRES-mediated Translation of SHMT1 Involves Ribosome Scanning
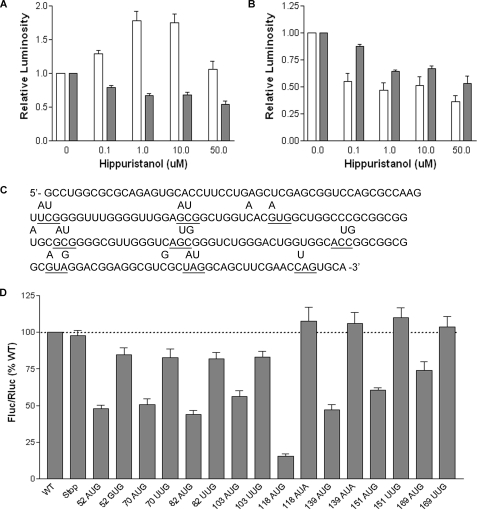
Internal initiation has been shown previously to proceed by a “land and scan” mechanism, whereby the 40 S ribosomal subunit binds upstream of the initiation codon and then migrates in a 5′-3′ direction until AUG recognition occurs (36–38). Consistent with this mechanism, most cellular IRESes have been found to require eIF4A (7), a DEAD box helicase that unwinds mRNA secondary structure and thereby facilitates ribosome binding and scanning (39, 40). To determine whether ribosome scanning is involved in the IRES-mediated translation of SHMT1, cells were treated with hippuristanol, a small molecule inhibitor of the eIF4A helicase (41). As in previous reports (41), hippuristanol treatment did not reduce the IRES activity of hepatitis C virus (Fig. 7A), which is known to be eIF4A-independent (42). However, it did result in a significant decrease in the IRES-mediated translation of SHMT1 (Fig. 7B).
FIGURE 7.
Ribosome scanning occurs between nucleotides 103 and 118 of the SHMT1 5′-UTR. MCF-7 cells were transiently transfected with polyadenylylated bicistronic mRNAs containing the hepatitis C virus IRES (A) or the SHMT1 5′-UTR (B) and then treated with the indicated amount of hippuristanol. Following 11 h of treatment, Fluc (white bars) and Rluc (dark bars) expression was quantified as described under ”Experimental Procedures.“ The relative luminosity in untreated cells was given a value of 1.0. The data represent the average of three independent experiments ± S.E. C, the sequence of the SHMT1 5′-UTR indicating the positions of the inserted open reading frames. The location of each start and stop codon is underlined, and the letters above the underlined nucleotides indicate changes to the wild-type sequence that were made by site-directed mutagenesis. D, MCF-7 cells were transiently transfected with bicistronic mRNAs containing the mutated SHMT1 5′-UTRs. The number of each mutant represents the position of the A in the AUG or AUA, or the U in the UUG. The relative ratio of Fluc/Rluc for each the wild-type (WT) bicistronic mRNA was given a value of 100%. The data represent the average of at least three independent experiments ± S.E.
To locate the ”landing“ point of the ribosome within the SHMT1 5′-UTR, a series of ORFs was introduced throughout the IRES by site-directed mutagenesis of the bicistronic construct. A common stop codon (UAG) was placed at the 3′-end of the IRES, but the locations of the start codon (AUG) varied. The ORFs were engineered within the 5′-UTR so that they were out of frame with the downstream ORF of Fluc. All AUG codons were placed in a sequence context that is highly favorable for initiation (with a purine three nucleotides upstream of the AUG and a G in position +4) (43) to eliminate leaky scanning by the 40 S ribosomal subunit (Fig. 7C). It was anticipated that if the inserted ORFs were located downstream of the ribosome entry site, IRES activity would decrease. However, if the ORFs were located upstream of the ribosome entry site, IRES activity would be unaffected.
The mRNA produced from each mutated bicistronic construct was transiently transfected into MCF-7 cells, and IRES activity was measured and compared with the wild type and stop codon only controls (Fig. 7D). The introduction of start codons at positions 52, 70, 82, and 103 resulted in similar reductions in IRES activity. However, the mutation of these AUGs to GUG or UUG also resulted in reduced IRES activity, indicating that mutations at these positions affect luciferase expression simply by perturbing the RNA structure. In contrast, the introduction of start codons at positions 118, 139, 151, and 169 resulted in reduced IRES activity that could be completely rescued by mutation of the AUGs to AUA or UUG, indicating that the 40 S ribosomal subunit begins scanning between nucleotides 103 and 118 of the SHMT1 5′-UTR. The gradual increase in IRES activity of the 118, 139, 151, and 169 AUG mutants is consistent with reinitiation, a process by which ribosomes remain associated with the mRNA following the translation of an upstream ORF and then reinitiate at a downstream ORF. The shorter the upstream ORF, the greater the efficiency of reinitiation (44).
DISCUSSION
There are few conserved properties shared among cellular IRESes in terms of their sequence, size, or structure, indicating that a universal mechanism for their mode of action is unlikely. This is evidenced by the growing list of ITAFs, many of which have only been implicated in the cap-independent translation of a small percentage of the known IRES-containing mRNA transcripts. In this study, we have provided evidence that the IRES-mediated translation of SHMT1 proceeds by a unique mechanism in that it involves two novel ITAFs, CUGBP1 and hnRNP H2, and does not require either the poly(A) tail or PABP.
The direct binding of hnRNP H2 to the SHMT1 5′-UTR, the direct binding of CUGBP1 to the SHMT1 3′-UTR, and the RNA-independent interaction between hnRNP H2 and CUGBP1 are all consistent with a model for the IRES-mediated translation of SHMT1 whereby an hnRNP H2/CUGBP1 bridge results in the circularization of the mRNA. Formation of this bridge would replicate a function of the eIF4G/PABP/poly(A) tail interaction and would account for the finding that both the poly(A) tail and PABP are dispensable when the SHMT1 3′-UTR, CUGBP1, and hnRNP H2 are present but are required for maximal SHMT1 IRES activity in the absence of any one of these factors. It also accounts for the previously observed stimulatory effect of the SHMT1 3′-UTR on IRES activity (25). Such a model of IRES-mediated translation is not unprecedented. The finding that several non-polyadenylylated viral RNAs contain sequences in their 3′-UTRs that are required for efficient IRES-mediated translation led to the hypothesis that an RNA/RNA or RNA/protein bridge can be established between the IRES and the 3′-UTR (23, 45–47). However, this is the first report, to our knowledge, that demonstrates a cellular IRES/protein bridge experimentally.
Because endogenous SHMT1 mRNA is polyadenylylated and therefore can presumably bind PABP, why would it utilize an alternative set of factors to form a closed loop? One hypothesis comes from examining the CUGBP1 binding site on the 3′-UTR. The data from this study indicate that CUGBP1 binds between nucleotides 157 and 200, which is more than 400 nucleotides upstream of the start of the poly(A) tail. Consequently, the interaction of CUGBP1 with the IRES would result in the formation of a much smaller closed loop than an interaction between PABP and the IRES. A smaller closed loop would likely increase the efficiency of 40 S ribosomal subunit recycling.
It remains to be determined whether hnRNP H2 and CUGBP1 serve any additional role(s) in the IRES-mediated translation of SHMT1 aside from circularizing the transcript. The proximity of the hnRNP H2 binding site (between nucleotides 105 and 114) and the 40 S ribosomal subunit binding site (between nucleotides 103 and 118) on the SHMT1 5′-UTR, combined with the previously reported association between CUGBP1 and the α and β subunits of eIF2 (48), raises the possibility that the ITAFs may physically recruit the translation initiation machinery to the 5′-UTR. In support of this model, preliminary results from our laboratory suggest that hnRNP H2 and CUGBP1 interact with eIF4A (data not shown). Further work is needed to investigate the requirement of the other canonical initiation factors in the IRES-mediated translation of SHMT1 and to determine whether the essential initiation factors bind to the SHMT1 ITAFs.
The role of ferritin in the IRES-mediated translation of SHMT1 also remains to be determined (25). Heavy chain ferritin (H ferritin) is not present in polysomes, but ongoing studies indicate that H ferritin stimulates SHMT1 IRES activity by enhancing the interaction of hnRNPH2 and CUGBP1 during the assembly of the initiation complex.3
This work was supported, in whole or in part, by National Institutes of Health Grant DK58144 from the Public Health Service.
P. J. Stover and J. T. Fox, unpublished results.
- IRES
- internal ribosome entry site
- SHMT1
- serine hydroxymethyltransferase 1
- UTR
- untranslated region
- ITAF
- IRES-specific trans-acting factor
- PABP
- poly(A)-binding protein
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- CUGBP1
- CUG-binding protein 1
- eIF
- eukaryotic initiation factor
- Rluc
- Renilla luciferase
- Fluc
- firefly luciferase
- siRNA
- small interference RNA
- EMSA
- electrophoretic mobility shift assay
- RevUTR
- reverse complement of the SHMT1 5′-UTR
- ORF
- open reading frame
- μLC/MS/MS
- microcapillary reverse-phase high pressure liquid chromatography nanoelectrospray tandem mass spectrometry
- PMSF
- phenylmethylsulfonyl fluoride
- DTT
- dithiothreitol
- Fwd
- forward
- Rev
- reverse
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MPB
- maltose-binding protein
- GST
- glutathione S-transferase
- X-α-gal
- 5-bromo- 4-chloro-3-indoyl-α-d-galactopyranoside.
REFERENCES
- 1.Jang S. K., Kräusslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. (1988) J. Virol. 62, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelletier J., Sonenberg N. (1988) Nature 334, 320–325 [DOI] [PubMed] [Google Scholar]
- 3.Hellen C. U., Sarnow P. (2001) Genes Dev. 15, 1593–1612 [DOI] [PubMed] [Google Scholar]
- 4.Stoneley M., Willis A. E. (2004) Oncogene 23, 3200–3207 [DOI] [PubMed] [Google Scholar]
- 5.Johannes G., Carter M. S., Eisen M. B., Brown P. O., Sarnow P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13118–13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallie D. R. (1991) Genes Dev. 5, 2108–2116 [DOI] [PubMed] [Google Scholar]
- 7.Thoma C., Bergamini G., Galy B., Hundsdoerfer P., Hentze M. W. (2004) Mol. Cell 15, 925–935 [DOI] [PubMed] [Google Scholar]
- 8.Jacobson A. (1996) in Translational Control (Hershey J. W. B., Mathews B. M., Sonenberg N. eds), pp. 451–480, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 9.Munroe D., Jacobson A. (1990) Mol. Cell. Biol. 10, 3441–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachs A. B., Davis R. W. (1989) Cell 58, 857–867 [DOI] [PubMed] [Google Scholar]
- 11.Sachs A. B., Varani G. (2000) Nat. Struct. Biol. 7, 356–361 [DOI] [PubMed] [Google Scholar]
- 12.Svitkin Y. V., Imataka H., Khaleghpour K., Kahvejian A., Liebig H. D., Sonenberg N. (2001) RNA 7, 1743–1752 [PMC free article] [PubMed] [Google Scholar]
- 13.Tarun S. Z., Jr., Sachs A. B. (1995) Genes Dev. 9, 2997–3007 [DOI] [PubMed] [Google Scholar]
- 14.Tarun S. Z., Jr., Wells S. E., Deardorff J. A., Sachs A. B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9046–9051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchida N., Hoshino S., Imataka H., Sonenberg N., Katada T. (2002) J. Biol. Chem. 277, 50286–50292 [DOI] [PubMed] [Google Scholar]
- 16.Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. (1998) Mol. Cell 2, 135–140 [DOI] [PubMed] [Google Scholar]
- 17.Thoma C., Fraterman S., Gentzel M., Wilm M., Hentze M. W. (2008) RNA 14, 1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonnal S., Pileur F., Orsini C., Parker F., Pujol F., Prats A. C., Vagner S. (2005) J. Biol. Chem. 280, 4144–4153 [DOI] [PubMed] [Google Scholar]
- 19.Giraud S., Greco A., Brink M., Diaz J. J., Delafontaine P. (2001) J. Biol. Chem. 276, 5668–5675 [DOI] [PubMed] [Google Scholar]
- 20.Hahm B., Kim Y. K., Kim J. H., Kim T. Y., Jang S. K. (1998) J. Virol. 72, 8782–8788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holcík M., Gordon B. W., Korneluk R. G. (2003) Mol. Cell. Biol. 23, 280–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komar A. A., Hatzoglou M. (2005) J. Biol. Chem. 280, 23425–23428 [DOI] [PubMed] [Google Scholar]
- 23.Ito T., Tahara S. M., Lai M. M. (1998) J. Virol. 72, 8789–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbig K., Chiang E. P., Lee L. R., Hills J., Shane B., Stover P. J. (2002) J. Biol. Chem. 277, 38381–38389 [DOI] [PubMed] [Google Scholar]
- 25.Woeller C. F., Fox J. T., Perry C., Stover P. J. (2007) J. Biol. Chem. 282, 29927–29935 [DOI] [PubMed] [Google Scholar]
- 26.Bensadoun A., Weinstein D. (1976) Anal. Biochem. 70, 241–250 [DOI] [PubMed] [Google Scholar]
- 27.Khaleghpour K., Svitkin Y. V., Craig A. W., DeMaria C. T., Deo R. C., Burley S. K., Sonenberg N. (2001) Mol. Cell 7, 205–216 [DOI] [PubMed] [Google Scholar]
- 28.Timchenko L. T., Miller J. W., Timchenko N. A., DeVore D. R., Datar K. V., Lin L., Roberts R., Caskey C. T., Swanson M. S. (1996) Nucleic Acids Res. 24, 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlasova I. A., Tahoe N. M., Fan D., Larsson O., Rattenbacher B., Sternjohn J. R., Vasdewani J., Karypis G., Reilly C. S., Bitterman P. B., Bohjanen P. R. (2008) Mol. Cell 29, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Good P. J., Chen Q., Warner S. J., Herring D. C. (2000) J. Biol. Chem. 275, 28583–28592 [DOI] [PubMed] [Google Scholar]
- 31.Kuyumcu-Martinez N. M., Wang G. S., Cooper T. A. (2007) Mol. Cell 28, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts R., Timchenko N. A., Miller J. W., Reddy S., Caskey C. T., Swanson M. S., Timchenko L. T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13221–13226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honoré B., Baandrup U., Vorum H. (2004) Exp. Cell Res. 294, 199–209 [DOI] [PubMed] [Google Scholar]
- 34.Caputi M., Zahler A. M. (2001) J. Biol. Chem. 276, 43850–43859 [DOI] [PubMed] [Google Scholar]
- 35.Paul S., Dansithong W., Kim D., Rossi J., Webster N. J., Comai L., Reddy S. (2006) EMBO J. 25, 4271–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jopling C. L., Spriggs K. A., Mitchell S. A., Stoneley M., Willis A. E. (2004) RNA 10, 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Quesne J. P., Stoneley M., Fraser G. A., Willis A. E. (2001) J. Mol. Biol. 310, 111–126 [DOI] [PubMed] [Google Scholar]
- 38.Mitchell S. A., Spriggs K. A., Coldwell M. J., Jackson R. J., Willis A. E. (2003) Mol. Cell 11, 757–771 [DOI] [PubMed] [Google Scholar]
- 39.Rogers G. W., Jr., Richter N. J., Merrick W. C. (1999) J. Biol. Chem. 274, 12236–12244 [DOI] [PubMed] [Google Scholar]
- 40.Svitkin Y. V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G. J., Sonenberg N. (2001) RNA 7, 382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bordeleau M. E., Mori A., Oberer M., Lindqvist L., Chard L. S., Higa T., Belsham G. J., Wagner G., Tanaka J., Pelletier J. (2006) Nat. Chem. Biol. 2, 213–220 [DOI] [PubMed] [Google Scholar]
- 42.Pestova T. V., Shatsky I. N., Fletcher S. P., Jackson R. J., Hellen C. U. (1998) Genes Dev. 12, 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozak M. (1986) Cell 44, 283–292 [DOI] [PubMed] [Google Scholar]
- 44.Luukkonen B. G., Tan W., Schwartz S. (1995) J. Virol. 69, 4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danthinne X., Seurinck J., Meulewaeter F., Van Montagu M., Cornelissen M. (1993) Mol. Cell. Biol. 13, 3340–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meulewaeter F., van Lipzig R., Gultyaev A. P., Pleij C. W., Van Damme D., Cornelissen M., van Eldik G. (2004) Nucleic Acids Res. 32, 1721–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S., Browning K. S., Miller W. A. (1997) EMBO J. 16, 4107–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timchenko N. A., Wang G. L., Timchenko L. T. (2005) J. Biol. Chem. 280, 20549–20557 [DOI] [PubMed] [Google Scholar]
- 49.Fox J. T., Shin W. K., Caudill M. A., Stover P. J. (2009) J. Biol. Chem. 284, 31097–31108 [DOI] [PMC free article] [PubMed] [Google Scholar]