Abstract
Calumenin is a multiple EF-hand Ca2+-binding protein localized in the sarcoplasmic reticulum (SR) with C-terminal SR retention signal HDEF. Recently, we showed evidence that calumenin interacts with SERCA2 in rat cardiac SR (Sahoo, S. K., and Kim, D. H. (2008) Mol. Cells 26, 265–269). The present study was undertaken to further characterize the association of calumenin with SERCA2 in mouse heart by various gene manipulation approaches. Immunocytochemical analysis showed that calumenin and SERCA2 were partially co-localized in HL-1 cells. Knockdown (KD) of calumenin was conducted in HL-1 cells and 80% reduction of calumenin did not induce any expressional changes of other Ca2+-cycling proteins. But it enhanced Ca2+ transient amplitude and showed shortened time to reach peak and decreased time to reach 50% of baseline. Oxalate-supported Ca2+ uptake showed increased Ca2+ sensitivity of SERCA2 in calumenin KD HL-1 cells. Calumenin and SERCA2 interaction was significantly lower in the presence of thapsigargin, vanadate, or ATP, as compared with 1.3 μm Ca2+, suggesting that the interaction is favored in the E1 state of SERCA2. A glutathione S-transferase-pulldown assay of calumenin deletion fragments and SERCA2 luminal domains suggested that regions of 132–222 amino acids of calumenin and 853–892 amino acids of SERCA2-L4 are the major binding partners. On the basis of our in vitro binding data and available information on three-dimensional structure of Ca2+-ATPases, a molecular model was proposed for the interaction between calumenin and SERCA2. Taken together, the present results suggest that calumenin is a novel regulator of SERCA2, and its expressional changes are tightly coupled with Ca2+-cycling of cardiomyocytes.
Introduction
Sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA)2 is a major player in muscle relaxation of mammalian heart (2). Tight regulation of SERCA activity is important for normal Ca2+ homeostasis in heart, and altered activity could lead to impaired excitation-contraction (E-C) coupling and cardiac diseases (3). SERCA2a is the predominant isoform in mouse heart, compared with SERCA2b (4). SERCA2a and SERCA2b are identical up to 994 amino acids. However, the C terminus of SERCA2b contains additional 50 amino acids, as compared with only 4 amino acids in SERCA2a (5). Structural and biochemical studies have shown that SERCA2 contains 10 transmembrane segments (M1–10), and the large globular cytoplasmic part is composed of three domains: the β strand domain between M2 and M3, the phosphorylation domain attached to M4 at one end, and the nucleotide binding domain at the other end. The nucleotide binding domain contains a hinge domain, which is connected to the M5 segment (6). The luminal side contains five luminal domains connecting the following segments M1 and M2, M3 and M4, M5 and M6, M7 and M8, and M9 and M10, respectively (6–8).
Recent studies have shown that a number of proteins interact with SERCA2 and regulate its stability and activity. Among them phospholamban (PLN) is the most extensively studied molecule (9). PLN interacts with SERCA2 and decreases apparent affinity of SERCA2 for Ca2+, and this inhibition can be disrupted by phosphorylation of PLN or by elevation of cytosolic Ca2+, which leads to reversal of the inhibition (10). PLN is the important regulator of SERCA activity and contractility in heart (11). Other proteins related to the apoptotic pathway such as Bcl2 (12) and Hax1(13) interact with SERCA2 in the cytosolic side of SERCA2 and regulate SERCA2 protein level and stability. EF-hand protein S100A1 interacts with SERCA2 in the cytosolic side and regulates contractility in heart (14). Recent studies have suggested that SR luminal proteins such as calreticulin (15), ER protein 57 (16), sarcalumenin (17), histidine-rich Ca2+-binding protein (HRC) (18), and calumenin (1) interact with SERCA2. HRC binds to SERCA2 in a Ca2+-dependent manner, and its overexpression could inhibit SERCA2 activity and Ca2+ cycling in cardiomyocytes (18, 19). Sarcalumenin also interacts with SERCA2, which may consequently increase the tendency of its retention in the SR lumen and increase the SERCA2 protein stability (17).
Calumenin is a multiple EF-hand Ca2+-binding protein and is found to have unique C-terminal SR retention signal HDEF (20, 21). Calumenin is associated with the ryanodine receptor (RyR) in rabbit skeletal muscle, and its overexpression shows decreased depolarization-induced Ca2+ release in C2C12 myotubes (22). Recently we showed that calumenin interacts with SERCA2 in rat SR, and its overexpression in rat neonatal cells showed decreased SR Ca2+ uptake and decreased fractional Ca2+ release (1). However, the detailed biochemical nature of the interaction between calumenin and SERCA2 and precise role of calumenin in E-C coupling remains to be clarified.
In the present study, both biochemical and physiological experiments were conducted to identify the nature of interaction between calumenin and SERCA2. In this study, we showed evidence that the interaction between calumenin and SERCA2 could dynamically change during the Ca2+-cycling process, suggesting calumenin as an important regulator of SERCA2 during E-C coupling in mouse heart.
EXPERIMENTAL PROCEDURES
Materials
All enzymes for DNA restriction digestion and ligation were purchased from New England Biolabs. All enzymes for PCR amplification was purchased from Stratagene, and protein-A Sepharose and GST-Sepharose 4B beads were purchased from Amersham Biosciences. HL-1 cells derived from the AT-1 murine atrial cardiomyocytes tumor lineage (23) were obtained as a kind gift from Dr. W. Claycomb (Louisiana State University Medical Center). All chemicals were purchased from Sigma-Aldrich Co. unless otherwise mentioned.
Cell Culture of HL-1 Cells
HL-1 cells were maintained as described previously (23). Briefly, cells were cultured on gelatin (0.02%, w/v)/fibronectin (10 μg/ml)-coated plates. The cells were maintained in Claycomb medium (JRH Biosciences) supplemented with 10% fetal bovine serum (JRH Biosciences), 2 mm l-glutamine, 0.1 mm norepinephrine, 100 unit/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). The culture medium was changed with fresh medium every 24 h. The cells were grown at 37 °C in an atmosphere of 5% CO2 and 95% air in an incubator.
Immunofluorescence Studies
HL-1 cells were grown on glass coverslips coated with gelatin and fibronectin at low confluence. After 24 h, cells were fixed with 4% paraformaldehyde for 10 min at room temperature, and washed three times with PBS. Cells were permeabilized with 0.1% Triton X-100 for 10 min at room temperature followed by washing three times with PBS. Cells were blocked with 10% bovine serum albumin in PBS for 30 min at 37 °C. The cells were incubated with respective primary antibodies in 3% bovine serum albumin in PBS overnight at 4 °C. The following day cells were washed six times in PBS and incubated with respective secondary antibodies in 3% bovine serum albumin in PBS for 1 h at 37 °C. After incubation cells were washed six times and mounted on slides using Vectashield mounting medium (Vector Laboratories). Mouse anti-SERCA2 and rabbit anti-calumenin primary antibodies were used for staining SERCA2 and calumenin, respectively. Alexa Flour 488-conjugated anti-mouse and Alexa Fluor 594-conjugated anti-rabbit IgG (Molecular Probes) secondary antibodies were used for detection of SERCA2 and calumenin, respectively. The slides were examined with an LSM 700 confocal laser scanning microscope (Carl Zeiss, Jena, Germany).
siRNA Design and Transfection
The siRNA oligonucleotides and transfection reagents were purchased from Dharmacon, Inc. (Lafayette, CO). The predesigned ON-TARGETplus SMARTpool for mouse calumenin gene, containing a mixture of four targeting siRNA oligonucleotides, was used for knockdown. An ON-TARGETplus Non-Targeting pool, containing four nonspecific siRNA oligonucleotides, was used as control. For detection of successfully siRNA-transfected cells, siGLO green transfection indicator from Dharmacon was used. The fluorescence signal was specifically localized to the nucleus as a sign of siRNA transfection in the cells. For siRNA transfection, HL-1 cells were cultured in culture plates overnight and transfected with control or calumenin siRNA oligonucleotides with siGLO green transfection indicator using DharmaFECT transfection reagent according to the manufacturer's instruction. The medium was changed with fresh culture medium in every 24 h, and cells were used for physiological and biochemical experiments after 72 h of siRNA transfection.
SDS-PAGE and Western Blot Analysis
For Western blot analysis, siRNA-transfected HL-1 cells were washed in cold PBS and lysed in radioimmune precipitation assay buffer with protease inhibitor mixture (Roche Applied Science). Protein concentration was measured by Bradford assay kit (Bio-Rad). Cell lysates solubilized in 2× sample buffer (24) were separated by electrophoresis on 6–15% gels and transferred to nitrocellulose membranes. The membranes were incubated with 5% skim milk in Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% Tween 20 (TBST) for 1 h at room temperature. After washing with TBST the membranes were incubated with one of the following antibodies overnight at 4 °C: anti-calumenin rabbit polyclonal antibody as described previously (1); SERCA2, calsequestrin (CSQ), PLN, and RyR2 (Affinity Bioreagents); Na+-Ca2+ exchanger and dihydropyridine receptor (Abcam); phospho-2809-RyR2 (Badrilla); Ser16-phosphorylated PLN (Upstate), and glyceraldehyde-3-phosphate dehydrogenase (Abfrontier, Korea). After primary antibody incubation, membranes were washed with TBST and further incubated with appropriate peroxidase-conjugated secondary antibody. Western blot band signal was detected by using a SuperSignal West Pico chemiluminescence kit (Pierce). Western blot band intensities were measured by using ImageJ software.
Co-immunoprecipitation of Calumenin from Cardiac Homogenates after Treatment with ATP, Vanadate, Thapsigargin, and/or Ca2+
Co-immunoprecipitation with anti-calumenin antibody was performed after addition of different ligands to mouse cardiac samples, as described previously with some modifications (10). Briefly, mouse cardiac homogenates (0.3 mg) in 50 μl of 0.25 m sucrose, 10 mm Tris-HCl, pH 7.5, 20 μm CaCl2, 3 mm 2-mercaptoethanol, and 150 mm KCl were mixed with 50 μl of reaction mixture containing 280 mm KCl, 10 mm EGTA, 50 mm PIPES-NaOH, pH 7.0, and 300 mm sucrose in the presence or absence of 10 mm ATP, 800 μm sodium vanadate, 20 μm thapsigargin, and 8.6 mm CaCl2 ([Ca2+]free = 1.3 μm after mixing). After 5-min incubation at room temperature, 100 μl of lysis buffer containing 40 mm HEPES-NaOH, pH 7.5, 300 mm NaCl, 4 mm phenylmethylsulfonyl fluoride, and 1% Tween 20 was added to the reaction mixture and vortexed for 30 s followed by centrifugation at 16,000 × g for 30 min at 4 °C. The supernatant was pre-cleared with protein-A-Sepharose beads and incubated overnight with anti-calumenin antibody. After incubation, protein-A-Sepharose beads were added for 4 h to make the immune complex and washed in the wash buffer containing 20 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 1 mm EDTA, and 0.5% Tween 20 for three times at 4 °C. Samples were solubilized in 2× sample buffer and analyzed by SDS-PAGE. Protein was transferred into nitrocellulose membrane, and probing was done with anti-SERCA2 and anti-calumenin antibody.
Generation and Purification of Recombinant Proteins
GST fusion protein constructs were generated from mouse cardiac cDNA using PCR. PCR-amplified products were digested with EcoRI/XhoI and subcloned into EcoRI/XhoI cloning sites of pGEX 4T-1 vector (Amersham Biosciences). The following primer sets with attached restriction enzyme sites at the 5′-end were used for generation of fusion proteins: 5′-GAA TTC ATG GAC CTG CGT CAG TTT-3′ and 5′-CTC GAG AAC GTA GCC GTA GGT GGC-3′ for calumenin-A (1–137 aa), 5′-GAA TTC GCC ACC TAC GGC TAC GTT-3′ and 5′-CTC GAG CCC ATC ATG ACT GTA CAT-3′ for calumenin-B (132–222 aa), 5′-GAA TTC ATG TAC AGT CAT GAT GGG AAT-3′ and 5′-CTC GAG TCA GAA CTC ATC ATG TCG-3′ for calumenin-C (217–315 aa), 5′-GAA TTC ATG GAC CTG CGT CAG TTT-3 and 5′-CTC GAG CCC ATC ATG ACT GTA CAT-3′ for calumenin-AB (1–222 aa), 5′-GAA TTC GCC ACC TAC GGC TAC GTT-3′ and 5′-CTC GAG TCA GAA CTC ATC ATG TCG-3′ for calumenin-BC (132–315 aa), and 5′-GAA TTC ATG GAC CTG CGT CAG TTT-3′ and 5′-CTC GAG TCA GAA CTC ATC ATG TCG-3′ for calumenin full (1–315 aa). The following luminal regions of SERCA2 were amplified and subcloned into pGEX 4T-1 vector: SERCA2-L1 (74–90 aa) connecting M1 and M2, SERCA2-L2 (275–295 aa) connecting M3 and M4, SERCA2-L3 (779–786 aa) connecting M5 and M6, SERCA2-L4 (853–892 aa) connecting M7 and M8, and SERCA2-L5 (951–960 aa) connecting M9 and M10. The following primer sets with attached restriction enzyme sites at the 5′-end were used: 5′-GAA TTC GTT TTG GCT TGG TTC GA-3′ and 5′-CTC GAG CTC TAC AAA GGC TGT A-3′ for SERCA2-L1 (74–90 aa), 5′-GAA TTC AAC ATT GGG CAT TTC AA-3′ and 5′-CTC GAG GTA GTA GAT GGC ACC CC-3′ for SERCA2-L2 (275–295 aa), 5′-AAT TCG CCC TTG GGT TTC CTG AGG CTT TAC-3′ and 5′-TCG AGT AAA GCC TCA GGA AAC CCA AGG GCG-3′ for SERCA2-L3 (779–786 aa), 5′-GAA TTC TGG TGG TTC ATC GCT GCT-3′ and 5′-CTC GAG GGA CTC AAA GAT TGC AC-3′ for SERCA2-L4 (853–892 aa), and 5′-AAT TCC CTT TGC CGC TCA TTT TCC AGA TCA CAC CGC-3′ and 5′-TCG AGC GGT GTG ATC TGG AAA ATG AGC GGC AAA GGG-3′ for SERCA2-L5 (951–960 aa). For the generation of SERCA2-L3 and SERCA2-L5, sense and antisense primers were allowed to anneal at 90 °C for 3 min followed by incubation for 1 h at 37 °C in 1× annealing solution (Ambion Inc.). After annealing the DNA fragments were ligated to the EcoRI- and XhoI-digested pGEX 4T-1 vectors. In other constructs, PCR-amplified segments were digested with EcoRI and XhoI and subcloned into EcoRI- and XhoI-digested pGEX 4T-1 vectors. Correct clones with desired inserts were confirmed by direct sequencing and fusion protein expression. GST fusion proteins were expressed in Escherichia coli (BL21) cells after induction with 0.5 mm isopropyl-β-d-thiogalactopyranoside for 2 h at 30 °C. Cells were harvested and washed with cold STE buffer composed of 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1 mm EDTA. Fusion proteins were purified following an Amersham Biosciences fusion protein purification procedure (25).
Site-directed Mutagenesis
Point mutations (F866A, Y867A, L869A, and L873A) within the SERCA2-L4 region were generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instruction. The following primer sets were used for mutagenesis: 5′-GCG GTC CAA GAG TCT CCG CCT ACC AGC TGA GTC ATT-3′ and 5′-AAT GAC TCA GCT GGT AGG CGG AGA CTC TTG GAC CGC-3′ for SERCA2-L4-F866A, 5′-GGT CCA AGA GTC TCC TTC GCC CAG CTG AGT CAT TTC CT-3′ and 5′-AGG AAA TGA CTC AGC TGG GCG AAG GAG ACT CTT GGA CC-3′ for SERCA2-L4-Y867A, 5′-AAG AGT CTC CTT CTA CCA GGC GAG TCA TTT CCT ACA GTG T-3′ and 5′-ACA CTG TAG GAA ATG ACT CGC CTG GTA GAA GGA GAC TCT T-3′ for SERCA2-L4-L869A, and 5′-CTA CCA GCT GAG TCA TTT CGC ACA GTG TAA GGA GGA CAA C-3′ and 5′-GTT GTC CTC CTT ACA CTG TGC GAA ATG ACT CAG CTG GTA G-3′ for SERCA2-L4-L873A. The presence of all mutations was confirmed by direct sequencing, and fusion proteins were expressed as described previously (25).
GST Pulldown Assay
GST pulldown assays were performed as described previously with minor modifications (26). Briefly, mouse hearts were homogenized in protein extraction buffer containing 50 mm potassium phosphate, pH 7.4, 10 mm sodium fluoride (NaF), 1 mm EDTA, 300 mm sucrose, and 0.5 mm DTT, and protease inhibitor mixture. The homogenates were solubilized for 30 min on ice in buffer containing 3% CHAPS, 1 m NaCl, 1 mm DTT, 20 mm Tris-HCl, pH 7.4, and protease inhibitor mixture. Solubilized proteins were obtained by centrifugation and diluted with 20 mm Tris-HCl, pH 7.4, 1 mm DTT, and protease inhibitor mixture to reduce high salt and detergent concentration. Solubilized proteins were pre-cleared with 50% slurry of glutathione-Sepharose 4B beads for 2 h at 4 °C. The pre-cleared supernatant was incubated with equivalent amount of GST fusion proteins coupled to glutathione-Sepharose 4B beads for 16 h at 4 °C. After the incubation, the fusion protein-Sepharose 4B complexes were collected by centrifugation and washed five times with wash buffer containing 20 mm Tris-HCl, pH 7.4, 0.15 m NaCl, and 0.3% CHAPS. The bound proteins were solubilized in 2× SDS-PAGE sample buffer, and the solubilized proteins were analyzed by SDS-PAGE and immunoblotting with anti-calumenin or anti-SERCA2 antibody, as described previously.
Measurement of Ca2+ Transient
Ca2+ transients in HL-1 cells were measured as described previously (1). Briefly, transfected HL-1 cells on glass coverslips were incubated with Fura-2 AM (Molecular Probes) in Tyrode solution (10 mm HEPES-NaOH, pH 7.4, 135 mm NaCl, 4.0 mm KCl, 1.0 mm MgCl2, 1.8 mm CaCl2, and 10 mm glucose) for 30 min and washed in dye-free Tyrode solution. The cells were placed in a circulating bath with Tyrode solution held at 37 ± 1 °C under an inverted microscope. Cells exhibiting visible unresponsiveness or showing ectopic beats during 1-Hz pulsing were excluded from analysis. A dual-beam excitation spectrofluorometer setup (IONOPTIX) was used to record fluorescence emissions (505 nm) elicited from exciting wavelengths of 340 and 380 nm. Ca2+ transient amplitude measured as fluorescence ratio (340:380 nm), time required to reach 50% of baseline (T50), and time to reach peak of Ca2+ transients were acquired. Total SR Ca2+ content was estimated by rapid application of 20 mm caffeine in Ca2+-free Tyrode solution. Data were analyzed using Ion Wizard software (IONOPTIX).
Ca2+ Uptake Measurement
Ca2+ uptake rate was measured in HL-1 cells using a Millipore filtration method as described previously (27). Briefly, cell lysates were prepared in protein extraction buffer containing 50 mm potassium phosphate, pH 7.0, 10 mm NaF, 1 mm EDTA, 300 mm sucrose, 0.5 mm DTT, 0.3 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture, and Ca2+ uptake was measured by the Millipore filtration technique. HL-1 cell lysates (80 μg/ml) were incubated in Ca2+ uptake medium containing 40 mm imidazole, pH 7.0, 100 mm KCl, 5 mm MgCl2, 5 mm NaN3, 5 mm potassium oxalate, 0.5 mm EGTA, and various concentrations of CaCl2 to yield 0.03–10 μm free Ca2+. To inhibit Ca2+ release by RyR2, 1 μm Ruthenium Red was added immediately prior to the addition of samples. The reaction was started by the addition of 5 mm ATP and terminated at different time intervals by filtration. The membrane was washed two times, and radioactivity was measured by scintillation counting. The Ca2+ concentration required for half-maximal velocity for Ca2+ uptake (EC50) was determined by non-linear curve fitting using Origin 6.0 software.
Structural Model Building
The comparative modeling program MODELLER v8 (28) was used to generate the model for calumenin-B and SERCA2-L4 complex structure. Among 12 EF-hand domain structures complexed with their ligands in the Protein Data Bank, troponin C (pdb id 1OZS, 99–161 aa) showed highest sequence homology (53%, 30% identity) to calumenin-B domain (158–220 aa), and therefore used as a template for the calumenin-B domain modeling. The structure of SERCA2-L4 (853–892 aa) was modeled based on the SERCA1-L4 structure in E1 state (pdb id 1SU4, 854–893 aa) having 86% sequence homology (58% sequence identity). Docking of calumenin-B and SERCA2-L4 was performed based on the interaction of hydrophobic residues (Phe866, Tyr867, Leu869, and Leu873) in SERCA2-L4 and hydrophobic surface of calumenin-B. The HADDOCK program (29) was used for calumenin-B and SERCA2-L4 complex docking. The best model was kept and further refined by energy minimization using the program CNS (30).
Statistics
The experimental values are represented as means ± S.E. Significance was determined by using Student's t test or analysis of variance. A value of p < 0.05 was used as criteria for statistical significance.
RESULTS
Co-localization of SERCA2 and Calumenin in Cardiomyocytes Examined by Immunofluorescence Image Analysis
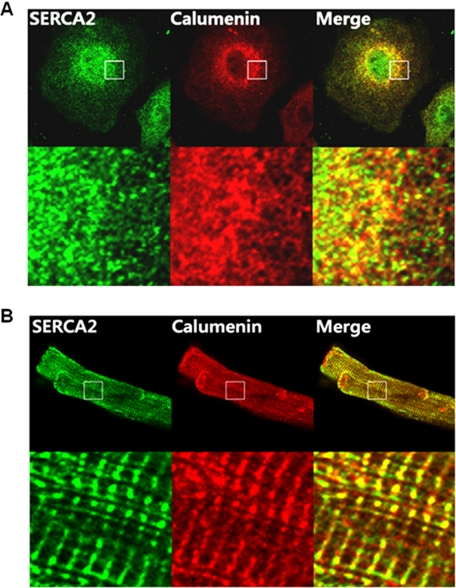
HL-1 is the only available mouse cardiac cell line showing the functional properties of adult cardiomyocytes (23, 31). In the present study, we used HL-1 cells to examine the role of calumenin in mouse cardiomyocytes. The possible in vivo localization of SERCA2 and calumenin was determined by dual confocal imaging analysis in HL-1 cells and adult rat ventricular cardiomyocytes using mouse anti-SERCA2 and rabbit anti-calumenin antibodies (Fig. 1). The results showed that both SERCA2 (green) and calumenin (red) molecules were widely expressed in the lateral areas of HL-1 cells (first and second images, Fig. 1A). Their merged images (yellow, third image) showed significant co-localization of SERCA2 and calumenin molecules especially in the lateral region of the cells (Fig. 1A). Adult rat ventricular cardiomyocytes were also stained for visualization of SERCA2 and calumenin molecules (Fig. 1B). SERCA2 (green) was stained in both junctional and longitudinal regions of cardiomyocytes (first image), whereas calumenin (red) showed clear localization only in the junctional region of SR along with the Z-line (second image, Fig. 1B). Their merged images (yellow) showed co-localization mainly along the Z-line (third image, Fig. 1B). Our immunofluorescence results also support the previous evidence of luminal localization of calumenin and its association with SR (20).
FIGURE 1.
Localization of SERCA2 and calumenin in cardiomyocytes. HL-1 cells (A) and adult rat ventricular myocytes (B) were stained with mouse anti-SERCA2 and rabbit anti-calumenin antibodies. SERCA2 was detected using Alexa Fluor 488-conjugated anti-mouse secondary antibody (first panels), and calumenin was detected using Alexa Fluor 594-conjugated anti-rabbit secondary antibody (second panels). The merged figure of SERCA2 and calumenin staining is also shown (third panels). The lower panels show the magnified views of the marked region in each upper panel.
Calumenin KD in HL-1 Cells Does Not Alter the Expression of Ca2+-handling Proteins
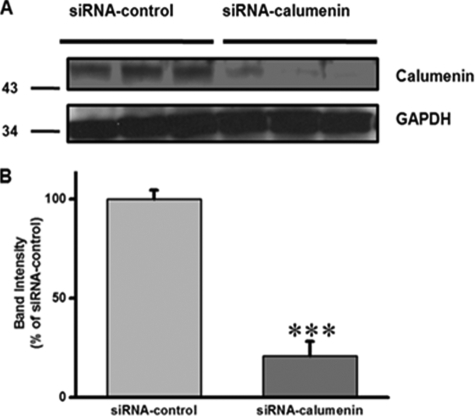
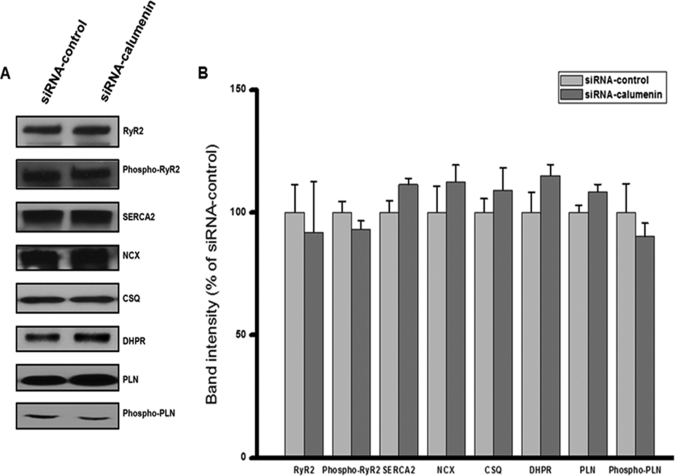
We previously reported that overexpression of calumenin led to prolonged relaxation time, increased SR Ca2+ load, and decreased fractional Ca2+ release in rat neonatal cells (1). In the present study, we knocked down calumenin in HL-1 cells using siRNA oligonucleotides targeted to calumenin. Cells transfected with calumenin siRNA showed 80% reduction of calumenin (Fig. 2). We further examined whether calumenin knockdown (KD) affected other Ca2+-handling proteins (Fig. 3). As shown in Fig. 3, the expression levels of RyR2, phospho-RyR2, dihydropyridine receptor, Na+-Ca2+ exchanger, SERCA2, CSQ, PLN, and Ser16 phospho-PLN did not change significantly between the two groups. These results suggest that there is no compensatory effect on the expression of above Ca2+-handling proteins. Also it does not affect the phosphorylation levels of RyR2 and PLN, which could alter the Ca2+ release and uptake activity of Ca2+ transients, respectively (9, 32).
FIGURE 2.
Calumenin knockdown in HL-1 cells. HL-1 cells were transfected with siRNA-control or siRNA-calumenin oligonucleotides. A, 72 h after transfection, protein sample was prepared, and immunoblotting was done using anti-calumenin and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies. B, band intensities were measured and presented as a percentage of siRNA-control cells. Data represent means ± S.E., n = 3 and ***, p < 0.001 versus siRNA-control.
FIGURE 3.
Effect of calumenin knockdown on expression of Ca2+-cycling proteins in HL-1 cells. Western blot analysis for Ca2+-cycling proteins in HL-1 cells transfected with siRNA-control or siRNA-calumenin oligonucleotides. A, cells transfected with oligonucleotides were harvested 72 h after transfection, and cell lysates was prepared as described under “Experimental Procedures.” After immunoblotting, the blots were probed with RyR2, phospho-RyR2, SERCA2, Na+-Ca2+ exchanger, CSQ, dihydropyridine receptor, PLN, and Ser16 phospho-PLN antibodies. B, band intensities of proteins were normalized to the siRNA-control Western bands. Data represent means ± S.E., n = 3–4.
Altered Ca2+ Transients in Calumenin KD HL-1 Cells
We further examined whether Ca2+ transients were affected by calumenin KD in HL-1 cells. Fura-2 AM-loaded cells were stimulated at 1-Hz electrical stimulation, and Ca2+ transients were recorded (Fig. 4). The transient peak height was significantly larger in calumenin KD cells than the control cells (Fura-2 fluorescence ratio (340/380 nm): siRNA-control cells, 0.81 ± 0.03, n = 44 versus siRNA-calumenin cells, 1.00 ± 0.02, n = 58; p < 0.001) (Fig. 4B), whereas time to peak of Ca2+ transients decreased significantly (time to peak (seconds): siRNA-control cells, 0.107 ± 0 .003, n = 44 versus siRNA-calumenin cells, 0.093 ± 0.001, n = 58; p < 0.001) (Fig. 4C). These results suggest that the sensitivity of RyR2 for luminal Ca2+ was increased in calumenin KD cells (33, 34). Time to reach 50% baseline of Ca2+ transient, which shows the relaxation phase, decreased significantly in calumenin KD cells (T50 (seconds): siRNA-control cells, 0.235 ± 0.006, n = 44 versus siRNA-calumenin cells, 0.172 ± 0.005, n = 58; p < 0.001) (Fig. 4D). It appears that calumenin KD significantly enhances SERCA2 activity which is an important player for relaxation (35). The SR Ca2+ load in the lumen was examined by applying 20 mm caffeine (Fig. 5A). The SR Ca2+ load did not change significantly by calumenin KD (Fura-2 fluorescence ratio (340/380 nm): siRNA-control cells, 2.0 ± 0.1, n = 17 versus siRNA-calumenin cells, 2.1 ± 0.1, n = 19; p value is not significant) (Fig. 5B). This result suggests that calumenin KD has no significant effect on the storage function of the SR. However, the fractional Ca2+ release was increased in calumenin KD cells, due to the increased transient amplitude and unaltered SR Ca2+ load (data not shown) (36).
FIGURE 4.
Electrically evoked Ca2+ transients in calumenin knockdown HL-1 cells. 72 h post transfection, HL-1 cells were incubated with Fura-2 AM for 30 min, and Ca2+ transients were measured as described under “Experimental Procedures” at 37 °C. A, representative traces of Ca2+ transients in siRNA-control and siRNA-calumenin-transfected HL-1 cells. B, Ca2+ transient amplitude in siRNA-control and siRNA-calumenin-transfected cells (Fura-2 ratio, 340/380 nm). C, time to reach peak of Ca2+ transient in siRNA-control and siRNA-calumenin-transfected cells. D, T50 (time to 50% of baseline) of Ca2+ transient in siRNA-control and siRNA-calumenin-transfected cells. n = 44 for siRNA-control cells and n = 58 for siRNA-calumenin cells. Data represents means ± S.E. and ***, p < 0.001 versus siRNA-control.
FIGURE 5.
Caffeine-evoked Ca2+ transients in calumenin knockdown HL-1 cells. 72 h post-transfection with oligonucleotides, HL-1 cells were incubated with Fura-2 AM for 30 min, and Ca2+ transients were measured at 37 °C. Cells were stimulated at 1 Hz for 1 min, the electrical stimulation was stopped, and 20 mm caffeine was added rapidly. A, representative tracing of caffeine-evoked transient in siRNA-control and siRNA-calumenin transfected cells. B, peak amplitude of caffeine-evoked Ca2+ transient in siRNA-control and siRNA-calumenin-transfected cells (Fura-2 ratio, 340/380 nm). n = 17 for siRNA-control cells and n = 19 for siRNA-calumenin cells.
Enhanced Ca2+ Uptake Activity in Calumenin KD HL-1 Cells
To determine whether the reduction in calumenin protein levels led to altered SR Ca2+ uptake function, the initial rates of oxalate-supported Ca2+ uptake were assayed in HL-1 cell lysates after treatment with siRNA-control and siRNA-calumenin oligonucleotides. Ca2+ uptake assay results showed significant leftward shift in the sigmoid curve measuring the Ca2+ dependence of Ca2+ uptake in KD cells (Fig. 6). The EC50 of SERCA2 for Ca2+ was significantly decreased in calumenin KD cells (EC50 (micromoles of Ca2+): siRNA-control cells, 0.30 ± 0.02, n = 3 versus siRNA-calumenin cells, 0.24 ± 0.01, n = 4; p < 0.05). However, the maximum velocity of Ca2+ uptake (Vmax) of SERCA2 did not change between the two groups (Vmax (nanomoles of Ca2+/mg of protein/min): siRNA-control cells, 65.5 ± 6.3, n = 3 versus siRNA-calumenin cells, 58.4 ± 3.6, n = 4; p value is not significant). This result suggests that calumenin KD enhances Ca2+ sensitivity of SERCA2 supporting the faster relaxation of the Ca2+ transients (Fig. 4D).
FIGURE 6.
Initial rates of ATP-dependent, oxalate-supported Ca2+ uptake rates at various Ca2+ concentrations in calumenin knockdown HL-1 cells. HL-1 cell lysates transfected with siRNA-control or siRNA-calumenin oligonucleotides were prepared as described under “Experimental Procedures.” Ca2+ uptake rates were measured by using the samples derived from three or four independent transfections. Vertical lines show the point of calculated EC50 values, and siRNA-calumenin-treated cells show leftward shift of curve in comparison to siRNA-control cells. Values are expressed as percentage of maximum uptake rates in each group. n = 3 for siRNA-control cells and n = 4 for siRNA-calumenin cells.
Interaction between Calumenin and SERCA2 Is Higher in the E1 State of SERCA2
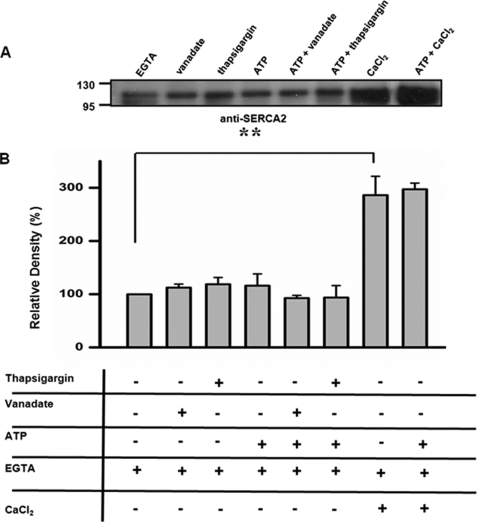
Our study shows that calumenin KD significantly increased SERCA2 activity in siRNA-calumenin-treated cells, and this supports our previous observation of a direct interaction between SERCA2 and calumenin in rat heart and its inhibitory effect on Ca2+ cycling in calumenin-overexpressing rat neonatal cardiomyocytes (1). The interaction between SERCA2 and calumenin was further studied considering that SERCA2 undergoes kinetic cycles for its active Ca2+ transport activity. A previous report by Asahi et al. (10) showed that SERCA2 interaction with PLN was affected by the presence of ATP, vanadate, and thapsigargin. Thapsigargin and vanadate are the compounds that stabilize SERCA2 into the E2 state, and ATP is known to induce a conformational change of SERCA2 in the absence of Ca2+ (10, 37–39). To determine the conformational state of SERCA2, which is more favorable for interaction with calumenin, the cardiac whole homogenates were treated with 400 μm vanadate, 10 μm thapsigargin, or 1.3 μm free Ca2+ in the presence or absence of 5 mm ATP and subjected to a co-immunoprecipitation assay (Fig. 7). The co-immunoprecipitation assay by anti-calumenin antibody carried out in the presence of 400 μm vanadate, 10 μm thapsigargin, or 5 mm ATP did not show any alterations in the interaction between calumenin and SERCA2 (Fig. 7). On the other hand, the level of interaction between calumenin and SERCA2 was significantly increased when 1.3 μm Ca2+ in the absence or presence of ATP was present in the reactions. This result suggests that the presence of Ca2+, which makes SERCA2 into the E1 state, shows stronger association with calumenin in comparison to the E2 state.
FIGURE 7.
The effects of thapsigargin, ATP, and vanadate on interaction between calumenin and SERCA2. Mouse cardiac homogenates were treated with either 400 μm vanadate, 10 μm thapsigargin, or 1.3 μm free Ca2+ in the presence or absence of 5 mm ATP, as described under “Experimental Procedures.” After the addition of Tween 20, solubilized supernatant was used for coimmunoprecipitation for calumenin and SERCA2 using anti-calumenin antibody. A, SERCA2 band was detected using anti-SERCA2 antibody. B, SERCA2 band intensity was plotted as percentage of EGTA-treated band. Data represents means ± S.E., n = 3 and **, p < 0.01 versus EGTA treated sample.
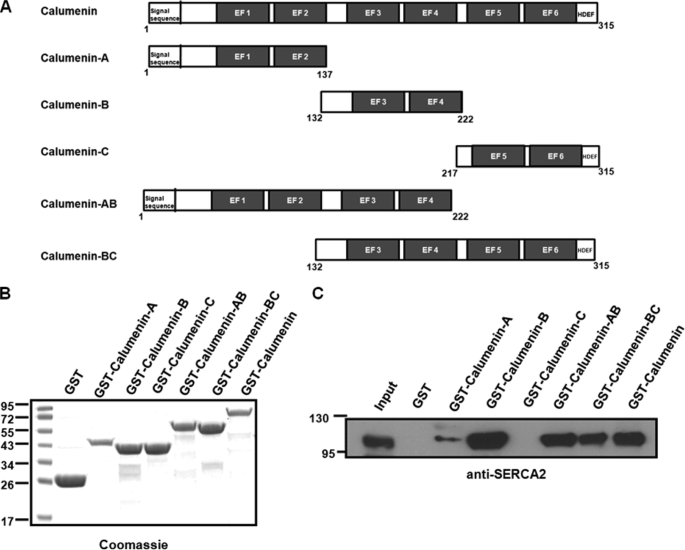
Calumenin Containing EF-hand 3 and 4 Region Mainly Interacts with SERCA2
To determine the region of calumenin that interacts with SERCA2, a pulldown assay was performed using GST fusion proteins, containing different regions of calumenin (Fig. 8A). Calumenin contains six EF-hands (21, 22), and we prepared different GST fusion proteins of the following regions, calumenin-A (1–137 aa) containing EF-hands 1 and 2, calumenin-B (132–222 aa) containing EF-hands 3 and 4, calumenin-C (217–315 aa) containing EF-hands 5 and 6, calumenin-AB (1–222 aa) containing EF-hands 1–4, calumenin-BC (132–315 aa) containing EF-hands 3–6, and full calumenin (1–315 aa). The estimated approximate molecular sizes of the proteins were ∼42.0, ∼37.0, ∼38.0, ∼52.0, ∼48.0, and ∼63.0 kDa, respectively (Fig. 8B). Equivalent amounts of the different GST fusion proteins bound to glutathione-Sepharose 4B beads were incubated with solubilized mouse cardiac whole homogenates. Western blotting of the pulldown samples with anti-SERCA2 antibody showed that mainly calumenin-B, calumenin-AB, calumenin-BC, and full-length calumenin protein interacted with SERCA2, whereas calumenin-A, calumenin-C, or control GST did not show major interaction with SERCA2 (Fig. 8C). This result suggests that the calumenin-B region contains the binding site for interaction with SERCA2.
FIGURE 8.
Region of calumenin interacting with SERCA2. A, schematic representation of mouse calumenin and the series of calumenin deletion construct. B, GST, recombinant GST-calumenin-A (1–137 aa, ∼42.0 kDa), GST-calumenin-B (132–222 aa, ∼37.0 kDa), GST-calumenin-C (217–315 aa, ∼38.0 kDa), GST-calumenin-AB (1–222 aa, ∼52.0 kDa), GST-calumenin-BC (132–315 aa, ∼48.0 kDa), and GST-calumenin (1–315 aa, ∼63.0 kDa) proteins were subjected to SDS-PAGE and stained with Coomassie Blue. C, GST pulldown assays were performed using control GST, GST-calumenin-A, GST-calumenin-B, GST-calumenin-C, GST-calumenin-AB, GST-calumenin-BC, and GST-calumenin fusion proteins bound to Sepharose 4B by incubating with cardiac homogenates. The pulldown samples were separated by SDS-PAGE and immunoblotted with anti-SERCA2 antibody.
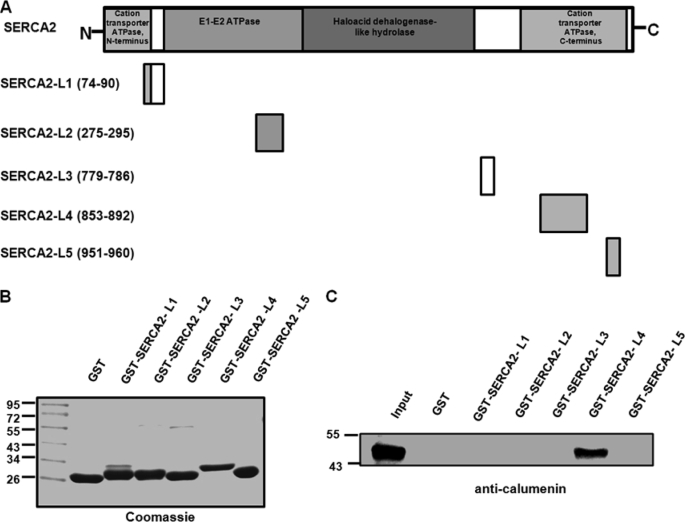
SERCA2-L4 Region Interacts with Calumenin
Calumenin is localized to the SR lumen and interacts with SERCA2 through the calumenin-B region (132–222 aa) (Fig. 8C). We further examined the region of SERCA2, which predominantly interacts with the calumenin molecule. It has been reported that SERCA2 molecule has five luminal loops (Fig. 9A) (5, 18). The GST fusion peptides of the five SERCA2 luminal loops were purified and used for pulldown assay with cardiac homogenates. The predicted approximate sizes of the GST fusion peptides of L1–L5 were ∼28.0, ∼28.3, ∼26.8, ∼30.6, and ∼27.0 kDa, respectively (Fig. 9B). Equivalent amounts of different GST fusion proteins bound to glutathione-Sepharose 4B were incubated with solubilized mouse cardiac homogenates. Western blotting with anti-calumenin antibody showed that SERCA2-L4 was the only site for calumenin interaction (Fig. 9C).
FIGURE 9.
Region of SERCA2 interacting with calumenin. A, schematic representation of mouse SERCA2 and its five luminal loop region constructs. The loops are marked by their amino acid positions, L1–L5 corresponding to mouse SERCA2 domains that face the luminal side of SR. B, GST and GST-SERCA2 fusion proteins L1–L5 (predicted approximate molecular sizes ∼28.0, ∼28.3, ∼26.8, ∼30.6, and ∼27.0 kDa, respectively) were analyzed by SDS-PAGE and stained with Coomassie Blue. C, pulldown assay was performed using equivalent amounts of control GST protein and GST-SERCA2 fusion peptides L1–L5 bound to Sepharose 4B by incubating with cardiac homogenates. The pulldown samples were separated by SDS-PAGE and immunoblotted with anti-calumenin antibody.
Prediction of Amino Acids in SERCA2-L4 Important for Interaction with Calumenin
GST pulldown study using cardiac homogenates showed that calumenin-B region was critical for binding with the SERCA2-L4 region (Figs. 8C and 9C). Hydrophobic residues of the SERCA2-L4 region were well conserved among all SERCA isoforms (Fig. 10A), and alanine mutations of Phe866, Tyr867, Leu869, and Leu873 abolished calumenin-B binding (Fig. 10, B and C). To have a structural insight for the calumenin-B and SERCA2-L4 interaction, we generated two structures using homology modeling method. The structure of calumenin-B domain (158–220 aa) was modeled based on the structure of the C-terminal EF-hand domain of troponin C (pdb id 1OZS), and the structure of the SERCA2-L4 region (853–892 aa) was modeled based on the structure of SERCA1-L4 in E1 state (pdb id 1SU4). Docking of calumenin-B domain and SERCA2-L4 was performed based on the interaction of hydrophobic residues (Phe866, Tyr867, Leu869, and Leu873) of the SERCA2-L4 structure and the hydrophobic surface of calumenin-B (Fig. 10, D and E). Consistent with the conventional hydrophobic interaction between EF-hand domain and ligand, α-helices 1, 2, and 4 in calumenin-B create hydrophobic surface to accommodate hydrophobic residues in the SERCA2-L4 region. Collectively, these results suggest that the four hydrophobic residues (Phe866, Tyr867, Leu869, and Leu873) in SERCA2-L4 are critical for the interaction with calumenin.
FIGURE 10.
Molecular modeling and characterization of the interaction between calumenin-B and SERCA2-L4. A, the multiple sequence alignment of SERCA isoforms using the ClustalX program (53). SERCA2-L4 region is shown in blue. Four hydrophobic residues of SERCA2-L4 involved in the interaction with calumenin-B are indicated by arrows. The dashed line indicates the disulfide bond (Cys875–Cys887). B, Coomassie Blue staining of the purified GST control, GST-SERCA2-L4, GST-SERCA2-L4-F866A, GST-SERCA2-L4-Y867A, GST-SERCA2-L4-L869A, and GST-SERCA2-L4-L873A fusion proteins. C, pulldown assay was performed using equivalent amounts of GST control protein and different GST-SERCA2-L4 fusion peptides bound to Sepharose 4B by incubating with cardiac homogenates. The pulldown samples were separated by SDS-PAGE and immunoblotted with anti-calumenin antibody. D, molecular modeling of SERCA2 structure. The model for SERCA2 structure was built using the structure of SERCA1 in E1 state (pdb id 1SU4). The luminal region of SERCA2 model structure contains five loops (L1–L5). L4 is colored blue. L1, L2, L3, and L5 are colored orange. More open conformational state of SERCA2-L4 may be required upon calumenin binding (arrow). E, molecular surface representation of the SERCA2-L4 binding site in calumenin-B. The residues (Phe866, Tyr867, Leu869, and Leu873) of SERCA2-L4 involved in the hydrophobic interaction with calumenin-B are shown as ball-and-stick models. The surface of calumenin-B binding to SERCA2-L4 is shown in yellow.
DISCUSSION
Our previous study in rat neonatal cardiac cells showed decreased SR Ca2+ uptake in calumenin-overexpressed cells (1). On the basis of the above observation, we hypothesized that calumenin could inhibit Ca2+ cycling in murine cardiomyocytes through inhibition of SERCA2. In the present study, we further characterized the hypothesis by siRNA-mediated calumenin knockdown using HL-1 cells. We found that the electrically evoked Ca2+ transients showed faster Ca2+-cycling in KD HL-1 cells. Furthermore calumenin KD in HL-1 cells led to increased Ca2+ affinity of SERCA2 without any significant change in the maximal Ca2+ uptake velocity. Further biochemical study suggests that calumenin interaction with SERCA2 is enhanced in the E1 conformation of SERCA2.
The unique ER/SR retention signal HDEF in calumenin should help its retention in the lumen of mouse cardiac SR. The partial co-localization of calumenin and SERCA2 (Fig. 1A) in HL-1 cells may be due to the irregular morphology of HL-1 cells. In HL-1 cells, it is difficult to examine the sarcomere structure and the difference between the junctional and longitudinal regions of SR. On the other hand, staining of adult rat ventricular cardiomyocytes showed clearer localization of SERCA2 and calumenin along the Z-line of sarcomere (Fig. 1B). This result suggests that calumenin is co-localized with SERCA2 primarily in the junctional region of SR.
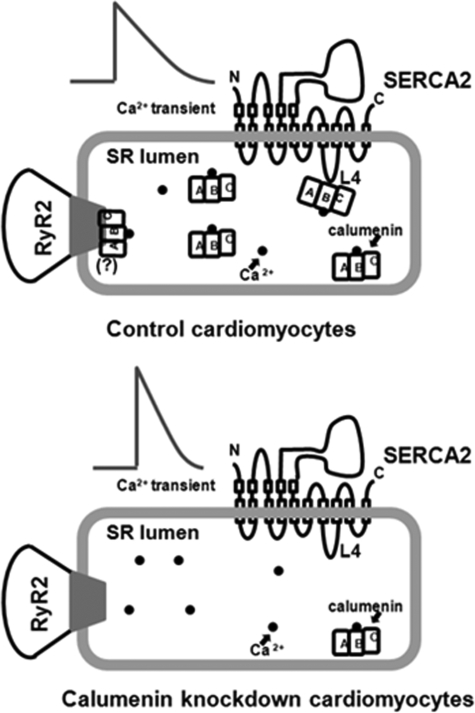
To characterize the physiological role of calumenin in cardiomyocytes, we knocked down calumenin protein in HL-1 cells. Calumenin KD showed ∼80% decrease in calumenin protein level in HL-1 cardiomyocytes (Fig. 2). There was no significant change in protein expression of other Ca2+-cycling proteins suggesting that calumenin KD does not affect the expressional balance of other Ca2+-handling proteins in HL-1 cells (Fig. 3). Electrically stimulated Ca2+ transient amplitude was increased without change in the SR Ca2+ load (Figs. 4 and 5). The negligible effect of calumenin KD on SR Ca2+ loading could be due to its lower amount in the SR than that of CSQ (3:100)3 and its lower Ca2+ binding capacity than that of CSQ (7:45) (40, 41). The increased amplitude for Ca2+ transients and the decreased time to reach peak of Ca2+ transient upon calumenin KD (Fig. 4) suggest that calumenin inhibits the function of RyR2. In a similar context, we previously found that calumenin overexpression decreased the size of fractional Ca2+ release in rat neonatal cells (1). Our preliminary results showing a physical interaction between calumenin and RyR2 in mouse heart suggest a dual functional role of calumenin during excitation-contraction coupling (Fig. 11).
FIGURE 11.
Schematic representation of functional role of calumenin on Ca2+ cycling in cardiac SR. Regulation of SERCA2 and cardiac ryanodine receptor (RyR2) function by calumenin in cardiomyocytes. In control cardiomyocytes (upper panel), Ca2+ (black dot)-binding protein calumenin is associated with SERCA2 and RyR2 (possible interaction between RyR2 and calumenin is shown by “(?)” and is based on our previous report about interaction between RyR1 and calumenin in rabbit skeletal muscle (22)). The calumenin-B region interacts with SERCA2-L4 in the SR lumen. Calumenin knockdown (lower panel), shows decreased interaction in SERCA2 and RyR2. This leads to increased Ca2+ transient amplitude, faster Ca2+ release, and decreased relaxation time of Ca2+ transients, without change in the SR Ca2+ content. This suggests that calumenin knockdown enhances SERCA2 and RyR2 activity in cardiomyocytes.
The time to reach 50% baseline was significantly decreased (27%), possibly due to the increased SERCA2 activity in calumenin KD cells. SERCA2 is the primary transporter of Ca2+ during relaxation, and the unchanged expression levels of Na+-Ca2+ exchanger protein suggest that calumenin KD enhanced SERCA2 Ca2+ uptake activity (42). The observed decrease in EC50 value of SERCA2 Ca2+ uptake in calumenin KD cells (20%) without change in maximal velocity suggests that calumenin KD enhances SERCA2 affinity for Ca2+ as seen in PLN KD cells (43). Our present data supports the previous finding that calumenin overexpression in rat neonatal cells led to prolonged relaxation time (1). Our data also suggest that the functional role of calumenin is somewhat similar to that of another luminal Ca2+-binding protein, HRC, in cardiomyocytes (19, 44).
The reaction cycle of SERCA2 is generally classified into two major conformations, the E1 and E2 states (6, 8, 45, 46). The E1 state has high affinity for Ca2+ binding sites, which faces the cytoplasmic side, whereas the E2 state has low affinity for Ca2+ binding and releases Ca2+ to the luminal side. Micromolar Ca2+ stabilizes the E1 conformation of the enzyme, whereas vanadate or thapsigargin in the absence of Ca2+ stabilizes the E2 conformation (10). SERCA2-mediated Ca2+ uptake in the SR is therefore closely related with cytosolic free Ca2+ concentration. It has been shown that SERCA2 interaction with other proteins (e.g. PLN) is also dependent on the intracellular Ca2+ level (10, 11, 46, 47). For the present study, the relationship between the enzyme conformation and the binding between SERCA2 and calumenin was tested under the various conditions as shown in Fig. 7. The results show that the interaction between the two proteins was maximal near pCa 6.0, which favors the E1 conformation. However, under the conditions where the E2 conformation is favored (vanadate or thapsigargin in the absence of Ca2+) the degree of interaction was significantly less. Considering the much higher Kd of calumenin EF-hands for Ca2+ (∼600 μm) than that of SERCA2, the effects of cytosolic Ca2+ on the interaction between the two proteins must be on the SERCA2 side (40). In the present study, however, we did not find any major changes in the interaction between calumenin and SERCA2 after addition of ATP in the presence or absence of either vanadate or thapsigargin (Fig. 7), suggesting that ATP-induced structural changes of SERCA2 do not affect calumenin binding (48). The altered interaction between the two proteins by the physiological Ca2+ level could further determine the possible role of calumenin in cardiac muscle.
Calumenin interaction with SERCA2 was mapped to 132–222 aa of calumenin, which contains the EF-hands 3 and 4 (Fig. 8). The minimal domain in SERCA2 required for interaction with calumenin lies between the 853 and 892 amino acids, which form the luminal loop L4 (Fig. 9). Because our study used the SERCA2 luminal loops in the form of GST fusion proteins, the possibility that other luminal loops are also involved in the interactions cannot be excluded completely. The SERCA2-L4 is the longest luminal loop in SERCA2 connecting transmembrane loops M7 and M8 (7). The L4 loop of SERCA2 contains two cysteine residues, which form a disulfide bond and regulate the luminal Ca2+ balance by modulating SERCA2 activity (49). A previous study in Xenopus oocytes shows that ER chaperone ER protein 57 interacts with SERCA2b in a Ca2+-dependent manner and regulates SERCA Ca2+ uptake activity (16). It has been also reported that calreticulin is associated with SERCA2a, in response to oxidative stress (50). Other luminal proteins like sarcalumenin and HRC interact with SERCA2 and regulate its activity (17, 18). HRC interacts with the luminal loop L1 of SERCA2 and inhibits SERCA2 activity in cardiomyocytes, whereas the interaction site of SERCA2 with sarcalumenin is not known yet (17). The mechanism of interaction between the L4 loop region and the counterpart in calumenin (132–222 aa) may be important for understanding the role of calumenin as a regulator of SERCA2 activity. However, because our study used the SERCA2 luminal loops in the form of GST fusion proteins, the possibility that other luminal loops are also involved in the interactions cannot be completely ruled out.
To elucidate a structural aspect of SERCA2-L4 and calumenin interaction, we modeled calumenin-B and SERCA2-L4 complex structure (Fig. 10) based on the GST pulldown assay (Fig. 10C) showing that the hydrophobic residues in SERCA2-L4 were critical for the calumenin-B binding. In the complex structure, four hydrophobic residues (Phe866, Tyr867, Leu869, and Leu873) in SERCA2-L4 are involved in the hydrophobic interaction with calumenin-B, consistent with conventional EF-hand domain-ligand interaction. In addition, we modeled full-length SERCA2 structure based on the SERCA1 crystal structure in E1 state (pdb id 1SU4), and the docked complex structure with calumenin-B domain (data not shown). In this complex structure, minimal structural crashes were observed between calumenin-B domain (helix2-loop2 region, 178–186 aa) and SERCA2 luminal regions (especially, the SERCA2-L3 region), suggesting that the structural rearrangement of luminal loop regions of SERCA2 into a more open conformation would be necessary upon calumenin binding. Collectively, these results suggest that conserved hydrophobic residues in the SERCA2-L4 region are critical for its interaction with calumenin.
Analysis of SERCA2 Ca2+ uptake activity showed that there was no significant change in the maximal Ca2+ uptake, but the affinity of SERCA2 for Ca2+ was significantly increased in KD cells (Fig. 6). The increased affinity for Ca2+ suggests that calumenin association with SERCA2 in the luminal loop L4 region regulates Ca2+ binding. The Ca2+ binding sites are present in the SERCA2 transmembrane regions M4, M6, and M8. So the loop L4 may regulate the Ca2+ binding affinity of SERCA2 through the modulation of the binding pocket in M8 (8).
The expression of some ER luminal chaperone proteins is regulated during development in the heart (51). In our study we found that the calumenin protein level was decreased in the adult heart in comparison to the embryonic and neonatal stages.3 A previous report showed that calumenin is expressed ubiquitously, and its RNA expression is higher in the embryonic stages than the adult heart (20). The decreased calumenin protein in the adult heart is similar to the trend seen in other ER chaperone proteins such as calreticulin, protein disulfide isomerase, ER protein 57, and glucose-regulated proteins, which are highly expressed during the early embryonic stages but decreased in the adult heart (51). Calumenin has been designated as an ER chaperone protein, and this phenomenon supports the hypothesis that the chaperonic effects of calumenin are essential during the early stages of development similar to other ER chaperon proteins like calreticulin, protein disulfide isomerase, ER protein 57, and glucose-regulated protein (51). Upon further development, SERCA2 activity is increased (52), whereas calumenin expression is decreased gradually until it is stabilized in the adult heart. Then calumenin molecules remain as an important regulator of SERCA2 activity during the adulthood.
Fig. 11 summarizes a hypothetical model describing the interaction of calumenin-B region with SERCA2-L4 and possible interaction with RyR2 in cardiomyocytes. KD of calumenin results in enhanced Ca2+ transient amplitude and shorter relaxation time without any change in the luminal Ca2+ concentration. This shows that calumenin inhibits the affinity of SERCA2 for Ca2+. Therefore, KD of calumenin enhances its Ca2+ affinity. Calumenin KD may also enhance the Ca2+ sensitivity of RyR2; however this phenomenon still remains to be elucidated in the future. The detailed mechanism concerning the effect of calumenin on Ca2+ affinity of SERCA2 in vivo requires future studies perhaps using a genetically altered mice model.
This work was supported by Korea Ministry of Science and Technology Systems Biology Research Grant M1050301001-6N0301-0110, Brain Korea 21 Project, and the Gwangju Institute of Science and Technology Systems Biology Infrastructure Establishment Grant (2009).
S. K. Sahoo, T. Kim, G. B. Kang, J. G. Lee, S. H. Eom, and D. H. Kim, unpublished data.
- SERCA2
- sarco(endo)plasmic reticulum Ca2+-ATPase
- RyR
- ryanodine receptor
- PLN
- phospholamban
- PBS
- phosphate-buffered saline
- GST
- glutathione S-transferase
- DTT
- dithiothreitol
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammino]-1-propanesulfonic acid
- siRNA
- small interfering RNA
- aa
- amino acid(s)
- KD
- knockdown
- E-C
- excitation-contraction coupling
- SR
- sarcoplasmic reticulum
- ER
- endoplasmic reticulum
- HRC
- histidine-rich Ca2+-binding protein
- CSQ
- calsequestrin.
REFERENCES
- 1.Sahoo S. K., Kim D. H. (2008) Mol. Cells 26, 265–269 [PubMed] [Google Scholar]
- 2.Periasamy M., Huke S. (2001) J. Mol. Cell Cardiol. 33, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 3.Hasenfuss G., Pieske B. (2002) J. Mol. Cell Cardiol. 34, 951–969 [DOI] [PubMed] [Google Scholar]
- 4.Vangheluwe P., Louch W. E., Ver Heyen M., Sipido K., Raeymaekers L., Wuytack F. (2003) Cell Calcium 34, 457–464 [DOI] [PubMed] [Google Scholar]
- 5.Campbell A. M., Kessler P. D., Sagara Y., Inesi G., Fambrough D. M. (1991) J. Biol. Chem. 266, 16050–16055 [PubMed] [Google Scholar]
- 6.Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 7.Dode L., Andersen J. P., Leslie N., Dhitavat J., Vilsen B., Hovnanian A. (2003) J. Biol. Chem. 278, 47877–47889 [DOI] [PubMed] [Google Scholar]
- 8.Toyoshima C., Inesi G. (2004) Annu. Rev. Biochem. 73, 269–292 [DOI] [PubMed] [Google Scholar]
- 9.Koss K. L., Kranias E. G. (1996) Circ. Res. 79, 1059–1063 [DOI] [PubMed] [Google Scholar]
- 10.Asahi M., McKenna E., Kurzydlowski K., Tada M., MacLennan D. H. (2000) J. Biol. Chem. 275, 15034–15038 [DOI] [PubMed] [Google Scholar]
- 11.Bluhm W. F., Kranias E. G., Dillmann W. H., Meyer M. (2000) Am. J. Physiol. Heart Circ. Physiol. 278, H249–H255 [DOI] [PubMed] [Google Scholar]
- 12.Dremina E. S., Sharov V. S., Kumar K., Zaidi A., Michaelis E. K., Schöneich C. (2004) Biochem. J. 383, 361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vafiadaki E., Arvanitis D. A., Pagakis S. N., Papalouka V., Sanoudou D., Kontrogianni-Konstantopoulos A., Kranias E. G. (2009) Mol. Biol. Cell 20, 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Most P., Pleger S. T., Völkers M., Heidt B., Boerries M., Weichenhan D., Löffler E., Janssen P. M., Eckhart A. D., Martini J., Williams M. L., Katus H. A., Remppis A., Koch W. J. (2004) J. Clin. Investig. 114, 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John L. M., Lechleiter J. D., Camacho P. (1998) J. Cell Biol. 142, 963–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Camacho P. (2004) J. Cell Biol. 164, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimura M., Minamisawa S., Takeshima H., Jiao Q., Bai Y., Umemura S., Ishikawa Y. (2008) Cardiovasc. Res. 77, 362–370 [DOI] [PubMed] [Google Scholar]
- 18.Arvanitis D. A., Vafiadaki E., Fan G. C., Mitton B. A., Gregory K. N., Del Monte F., Kontrogianni-Konstantopoulos A., Sanoudou D., Kranias E. G. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H1581–H1589 [DOI] [PubMed] [Google Scholar]
- 19.Fan G. C., Gregory K. N., Zhao W., Park W. J., Kranias E. G. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H1705–H1711 [DOI] [PubMed] [Google Scholar]
- 20.Yabe D., Nakamura T., Kanazawa N., Tashiro K., Honjo T. (1997) J. Biol. Chem. 272, 18232–18239 [DOI] [PubMed] [Google Scholar]
- 21.Jung D. H., Kim D. H. (2004) Gene 327, 185–194 [DOI] [PubMed] [Google Scholar]
- 22.Jung D. H., Mo S. H., Kim D. H. (2006) Biochem. Biophys. Res. Commun. 343, 34–42 [DOI] [PubMed] [Google Scholar]
- 23.Claycomb W. C., Lanson N. A., Jr., Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., Izzo N. J., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25.Frangioni J. V., Neel B. G. (1993) Anal. Biochem. 210, 179–187 [DOI] [PubMed] [Google Scholar]
- 26.Lee H. G., Kang H., Kim D. H., Park W. J. (2001) J. Biol. Chem. 276, 39533–39538 [DOI] [PubMed] [Google Scholar]
- 27.Babu G. J., Bhupathy P., Petrashevskaya N. N., Wang H., Raman S., Wheeler D., Jagatheesan G., Wieczorek D., Schwartz A., Janssen P. M., Ziolo M. T., Periasamy M. (2006) J. Biol. Chem. 281, 3972–3979 [DOI] [PubMed] [Google Scholar]
- 28.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 29.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 30.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. S. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 31.White S. M., Constantin P. E., Claycomb W. C. (2004) Am. J. Physiol. Heart Circ. Physiol. 286, H823–H829 [DOI] [PubMed] [Google Scholar]
- 32.Wehrens X. H., Lehnart S. E., Marks A. R. (2005) Annu. Rev. Physiol. 67, 69–98 [DOI] [PubMed] [Google Scholar]
- 33.Györke I., Hester N., Jones L. R., Györke S. (2004) Biophys. J. 86, 2121–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Györke S., Terentyev D. (2008) Cardiovasc. Res. 77, 245–255 [DOI] [PubMed] [Google Scholar]
- 35.Frank K. F., Bölck B., Erdmann E., Schwinger R. H. G. (2003) Cardiovasc. Res. 57, 20–27 [DOI] [PubMed] [Google Scholar]
- 36.Bassani J. W. M., Yuan W., Bers D. M. (1995) Am. J. Physiol. 268, C1313–C1319 [DOI] [PubMed] [Google Scholar]
- 37.Mueller B., Zhao M., Negrashov, Bennett R., Thomas D. D. (2004) Biochemistry 43, 12846–12854 [DOI] [PubMed] [Google Scholar]
- 38.Pick U. (1982) J. Biol. Chem. 257, 6111–6119 [PubMed] [Google Scholar]
- 39.Sagara Y., Inesi G. (1991) J. Biol. Chem. 266, 13503–13506 [PubMed] [Google Scholar]
- 40.Vorum H., Liu X., Madsen P., Rasmussen H. H., Honoré B. (1998) Biochim. Biophys. Acta 1386, 121–131 [DOI] [PubMed] [Google Scholar]
- 41.MacLennan D. H., Wong P. T. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 1231–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terracciano C. M., Souza A. I., Philipson K. D., MacLeod K. T. (1998) J. Physiol. 512, 651–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe A., Arai M., Yamazaki M., Koitabashi N., Wuytack F., Kurabayashi M. (2004) J. Mol. Cell Cardiol. 37, 691–698 [DOI] [PubMed] [Google Scholar]
- 44.Gregory K. N., Ginsburg K. S., Bodi I., Hahn H., Marreez Y. M., Song Q., Padmanabhan P. A., Mitton B. A., Waggoner J. R., Del Monte F., Park W. J., Dorn G. W., Bers D. M., Kranias E. G. (2006) J. Mol. Cell Cardiol. 40, 653–665 [DOI] [PubMed] [Google Scholar]
- 45.Toyoshima C., Asahi M., Sugita Y., Khanna R., Tsuda T., MacLennan D. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asahi M., Nakayama H., Tada M., Otsu K. (2003) Trends Cardiovasc. Med. 13, 152–157 [DOI] [PubMed] [Google Scholar]
- 47.Asahi M., Sugita Y., Kurzydlowski K., De Leon S., Tada M., Toyoshima C., MacLennan D. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5040–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahaney J. E., Froehlich J. P., Thomas D. D. (1995) Biochemistry 34, 4864–4879 [DOI] [PubMed] [Google Scholar]
- 49.Daiho T., Yamasaki K., Saino T., Kamidochi M., Satoh K., Iizuka H., Suzuki H. (2001) J. Biol. Chem. 276, 32771–32778 [DOI] [PubMed] [Google Scholar]
- 50.Ihara Y., Kageyama K., Kondo T. (2005) Biochem. Biophys. Res. Commun. 329, 1343–1349 [DOI] [PubMed] [Google Scholar]
- 51.Papp S., Zhang X., Szabo E., Michalak M., Opas M. (2008) Open Cardiovasc. Med. J. 2, 31–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramesh V., Kresch M. J., Park W. J., Kim D. H. (2001) J. Biochem. Mol. Biol. 34, 573–577 [Google Scholar]
- 53.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]