Abstract
An extracellular microenvironment, including an extracellular matrix (ECM), is an important factor in regulating stem cell differentiation. During tissue development, the ECM is dynamically remodeled to regulate stem cell functions. Here, we developed matrices mimicking ECM remodeling during the osteogenesis of mesenchymal stem cells (MSCs). The matrices were prepared from cultured MSCs controlled at different stages of osteogenesis and referred to as “stepwise osteogenesis-mimicking matrices.” The matrices supported the adhesion and proliferation of MSCs and showed different effects on the osteogenesis of MSCs. On the matrices mimicking the early stage of osteogenesis (early stage matrices), the osteogenesis occurred more rapidly than did that on the matrices mimicking undifferentiated stem cells (stem cell matrices) and the late stage of osteogenesis (late stage matrices). RUNX2 was similarly expressed when MSCs were cultured on both the early stage and late stage matrices but decreased on the stem cell matrices. PPARG expression in the MSCs cultured on the late stage matrices was higher than for those cultured on the stem cell and early stage matrices. This increase of PPARG expression was caused by the suppression of the amount of β-catenin and downstream signal transduction. These results demonstrate that the osteogenesis-mimicking matrices had different effects on the osteogenesis of MSCs, and the early stage matrices provided a favorable microenvironment for the osteogenesis.
Introduction
Stem cells pass through stepwise maturational stages for differentiation into somatic cells. During the stepwise differentiation of stem cells into somatic cells, the expression pattern of transcription factors changes, depending on their maturational stages (1). The expression of the transcription factors is regulated by the extracellular environment through the modulation of intracellular signaling. To alter the expression of the transcription factors, the extracellular microenvironment surrounding the stem cells changes through the differentiation processes. In particular, the extracellular matrix (ECM)2 is dynamically remodeled to activate the intracellular signaling to regulate the differentiation of stem cells into somatic cells (2, 3).
Mesenchymal stem cells (MSCs) can differentiate into chondrocytes, adipocytes, and osteoblasts (4). When osteoblasts originate from MSCs, the stepwise maturational stages are passed. At the early stage of MSC osteogenesis, the cells express RUNX2 (also known as CBFA1), an osteogenic transcription factor, and the genes controlled by RUNX2, such as ALP (liver/bone/kidney alkaline phosphatase) and SPP1 (osteopontin) (5, 6). However, at the early stage of osteogenesis, the cells cannot deposit calcium to form mineralized bone. In order to deposit calcium, the cells must enter the late stage of osteogenesis and express SP7 (also known as osterix), whose expression is regulated by RUNX2 (7, 8), and IBSP (bone sialoprotein 2), whose expression is controlled by Sp7 (7). During the osteogenesis processes of MSCs in vivo, the ECM surrounding the cells is dynamically remodeled (9–11). Fibronectin and versican are observed in the region of mesenchymal condensation (early stage of osteogenesis) and then disappear in mature mineralized bone (9, 10). Decorin is present in unmineralized bone matrix but disappears in mineralized bone (11). Biglycan is strongly detected in bone marrow surrounding MSCs but not in unmineralized and mineralized bone matrices (11). Similar to in vivo ECM remodeling, the ECM gene expression pattern also changes during the osteogenesis of MSCs in vitro (12). The effects of these ECM proteins on osteogenesis have been studied in vivo and in vitro using gene-deficient animals and cells, gene-overexpressed cells, and surfaces coated with isolated ECM proteins (13–17). However, ECM is formed from complex components to regulate proper cell functioning in vivo (18). It is expected that the matrices that formed from components similar to in vivo components will give us more precise and detailed insights into the role of ECM remodeling in the osteogenesis of MSCs.
There are many reports of the development of acellular matrices from tissues using various decellularization treatments (19, 20). In acellular matrices, it is difficult to identify and isolate the matrices at the different maturational stages of stem cells. In contrast, the cells cultured in vitro can secrete ECM proteins and deposit them beneath the cells. Similar to the acellular matrices from the tissue, these deposited ECM proteins can be used as matrices after decellularization (21–23). Mikos and co-workers (21, 22) made a matrix from cultured MSC-derived osteoblasts in vitro. This matrix enhanced the osteogenesis of MSCs compared with a Ti surface (21). Chen et al. (23) reported an undifferentiated MSC-derived matrix in vitro and also reported that this matrix suppressed the spontaneous differentiation of MSCs during culture. Therefore, it seems that the matrices formed by cultured cells can serve as an alternative to the acellular matrices of tissue.
In this study, we developed matrices mimicking in vivo ECM remodeling during the osteogenesis of human MSCs. To make the matrices mimicking in vivo ECM remodeling during the osteogenesis of MSCs, we prepared the matrices from cultured MSCs controlled at different stages of osteogenesis and referred to them as “stepwise osteogenesis-mimicking matrices.” In addition, the effects of stepwise osteogenesis-mimicking matrices on MSC functions, such as proliferation and osteogenesis, were also evaluated.
EXPERIMENTAL PROCEDURES
Osteogenesis of MSCs
Human MSCs were obtained from Osiris Therapeutics (Columbia, MD) and subcultured twice in Dulbecco's modified Eagle's medium (DMEM; Sigma) containing 10% fetal bovine serum (FBS; Equitech-Bio, Kerrville, TX), 4500 mg/liter glucose, 584 mg/liter glutamine, 100 units/ml penicillin, 100 mg/ml streptomycin, 0.1 mm nonessential amino acids, 1 mm sodium pyruvate, 0.4 mm proline, and 50 mg/liter ascorbic acid under an atmosphere of 5% CO2 at 37 °C. At passage 4, MSCs were cultured on tissue culture polystyrene (TCPS) plates for 1 and 3 weeks in osteogenic medium: DMEM containing 10% FBS, 1000 mg/liter glucose, 584 mg/liter glutamine, 100 units/ml penicillin, 100 mg/ml streptomycin, 0.1 mm nonessential amino acids, 1 mm sodium pyruvate, 0.4 mm proline, 50 mg/liter ascorbic acid, 10 nm dexamethasone, and 10 mm β-glycerophosphate. To create an undifferentiated condition, cells were cultured in osteogenic medium without dexamethasone and β-glycerophosphate for 1 week. The medium was changed every 3–4 days.
ALP (Alkaline Phosphatase) and Alizarin Red S Stainings
Cells were washed with phosphate-buffered saline (PBS) twice and were fixed with 10% formaldehyde in PBS for 30 min at room temperature. The cells were incubated with ALP substrate solution (0.1% naphthol AS-MX phosphate (Sigma) and 0.1% fast red violet LB salt (Sigma) in 56 mm 2-amino-2-methyl-1,3-propanediol (Sigma) or 0.5% alizarin red S (Acros Organics, Geel, Belgium) solution at room temperature for 10 or 30 min, respectively. After staining, the cells were washed with PBS and were observed under a light microscope. The positive areas of ALP staining were measured using Adobe Photoshop 6.0 and ImageJ (24).
Real-time PCR Analysis
Total RNA was extracted from the MSCs using ISOGEN reagent according to the manufacturer's instructions (Nippon Gene, Toyama, Japan). Total RNA (0.66 μg) was used as a first strand reaction that included random hexamer primers and murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA). Real-time PCR was amplified for GAPDH, 18 S rRNA, ALP, IBSP, SPP1, RUNX2, SP7, HOXA2, SOX9, and PPARG. The reaction was performed with 10 ng of cDNA, 300 nm PCR primers, 150 nm PCR probe, and TaqMan Universal PCR Master Mix (Applied Biosystems). The expression levels of 18 S rRNA were used as an endogenous control, and gene expression levels relative to GAPDH were calculated using the comparative Ct method. The sequences of primers and probes are listed in Table 1. All primers and probes were obtained from Applied Biosystems. For SOX9 expression analysis, cDNA synthesized from mRNA of human chondrocytes (at passage 3; Lonza, Basel, Switzerland) was used as a positive control.
TABLE 1.
Primers and probes for real-time PCR analysis
GAPDH, ALP, IBSP, and SPP1 were designed according to Martin et al. (25). SOX9 were designed according to Schaefer et al. (26). RUNX2, SP7, HOXA2, and PPARG were assay-on demand gene expression products (Applied Biosystems).
| mRNA | Oligonucleotide |
|---|---|
| 18 S rRNA | Forward 5′-GCCGCTAGAGGTGAAATTCTTG-3′ |
| Reverse 5′-CATTCTTGGCAAATGCTTTCG-3′ | |
| Probe 5′CCGGCGCAAGACGGACCAGA-3′ | |
| GAPDH | Forward 5′-ATGGGGAAGGTGAAGGTCG-3′ |
| Reverse 5′-TAAAAGCAGCCCTGGTGACC-3′ | |
| Probe 5′-CGCCCAATACGACCAAATCCGTTGAC-3′ | |
| ALP | Forward 5′-GACCCTTGACCCCCACAAT-3′ |
| Reverse 5′-GCTCGTACTGCATGTCCCCT-3′ | |
| Probe 5′-TGGACTACCTATTGGGTCTCTTCGAGCCA-3′ | |
| IBSP | Forward 5′-TGCCTTGAGCCTGCTTCC-3′ |
| Reverse 5′-GCAAAATTAAAGCAGTCTTCATTTTG-3′ | |
| Probe 5′-CTCCAGGACTGCCAGAGGAAGCAATCA-3′ | |
| SPP1 | Forward 5′-CTCAGGCCAGTTGCAGCC-3′ |
| Reverse 5′-CAAAAGCAAATCACTGCAATTCTC -3′ | |
| Probe 5′-AAACGCCGACCAAGGAAAACTCACTACC-3′ | |
| RUNX2 | Hs00231692_m1 |
| SP7 | Hs00541729_m1 |
| HOXA2 | Hs00534579_m1 |
| SOX9 | Forward 5′-CACACAGCTCACTCGACCTTG-3′ |
| Reverse 5′-TTCGGTTATTTTTAGGATCATCTCG-3′ | |
| Probe 5′-CCCACGAAGGGCGACGTTGG-3′ | |
| PPARG | Hs01115510_m1 |
Semiquantitative reverse transcription-PCR was also performed using rTaq DNA polymerase (TaKaRa, Tokyo, Japan) with specific human primer sets, as shown in supplemental Table 1. All primers were obtained from Hokkaido System Science (Sapporo, Japan). For each experiment, GAPDH was amplified to normalize the expression of other genes in the sample. The PCR products were analyzed by 1% agarose gel electrophoresis.
Immunocytochemical Analysis
Cells were fixed with glutaraldehyde for 6 h at 4 °C and then treated with 0.1 m glycine for 3 h. To block nonspecific interaction of the antibodies, cells were treated with 1% bovine serum albumin (BSA) in PBS at room temperature for 2 h. After the BSA blocking, cells were incubated with specific primary antibodies (Abs) at room temperature for 2 h and then treated with peroxidase-conjugated anti-rabbit Ab (Dako, Carpinteria, CA) or peroxidase-conjugated anti-mouse Ab (Dako) at room temperature for 1 h. The specific Abs used in this study were anti-fibronectin Ab (Sigma), anti-type I collagen Ab (Abcam, Cambridge, UK), anti-versican Ab (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-decorin Ab (Santa Cruz Biotechnology, Inc.), and anti-biglycan Ab (Santa Cruz Biotechnology, Inc.). Finally, the cells were incubated with 3,3′-diaminobenzidine solution (Dako) as a colorimetric substrate to visualize peroxidase-labeled proteins. In addition, cell nuclei were counterstained with methyl green (Wako, Osaka, Japan). The positive areas relative to the cell areas were measured using Adobe Photoshop 6.0 and ImageJ after tracing the cell shapes with a graphics tablet (Bamboo Fun, Wacom, Saitama, Japan).
Preparation of Stepwise Osteogenesis-mimicking Matrices
MSCs were cultured on TCPS plates in DMEM with or without osteogenic induction factors. After culture for specific periods, the cell-cultured TCPS was washed twice with PBS. The cellular components were removed from the matrices by incubation with PBS containing 0.5% Triton X-100 and 20 mm NH4OH for 5 min at 37 °C, similar to the reported method of decellularization (23). The matrices were then treated with 100 μg/ml DNase I (Roche Applied Science) and 100 μg/ml RNase A (Nacalai Tesque, Kyoto, Japan) for 1 h at 37 °C. After decellularization, the matrices were treated with 0.1% glutaraldehyde in PBS for 6 h at 4 °C to stabilize the structure and then treated with 0.1 m glycine in PBS. The obtained matrices were frozen in PBS and stored at −80 °C until use.
Confirmation of Decellularization and Protein Deposition in the Matrices
Cells were fixed with 0.1% glutaraldehyde for 6 h at 4 °C and then treated with 0.1 m glycine in PBS. After fixation, the cells were permeabilized by treatment with 0.2% Triton X-100 in PBS for 2 min. To visualize the cell nuclei, the cells and stepwise osteogenesis-mimicking matrices were stained with 10 μg/ml Hoechst 33258 (Wako) for 15 min at room temperature. To visualize the actin fibers, the cells and stepwise osteogenesis-mimicking matrices were incubated with Alexa 488-conjugated phalloidin (Invitrogen) for 1 h at room temperature. The cells and matrices were then observed under a fluorescence microscope. The results were shown in pseudocolor mode using Adobe Photoshop 6.0. To visualize whole proteins in the matrices, Coomassie Brilliant Blue (CBB) staining (Nacalai Tesque) was done.
Cell Attachment and Proliferation of MSCs on the Stepwise Osteogenesis-mimicking Matrices
For an attachment assay of MSCs, MSCs were seeded on the stepwise osteogenesis-mimicking matrices and TCPS at a density of 50,000 cells/cm2 and were incubated in FBS-free DMEM. After 1 or 4 h of incubation, non-adherent cells were removed by washing once with PBS containing 0.5 mm CaCl2 and 0.5 mm MgCl2. TCPS coated with 1% BSA was used as a negative control. Adherent cells were quantified by a colorimetric WST-1 assay (Roche Applied Science). The results were expressed as a percentage normalized to the MSCs attached to TCPS after 4 h of incubation.
For a proliferation assay of MSCs, MSCs were seeded on the stepwise osteogenesis-mimicking matrices and TCPS at a density of 5,000 cells/cm2 in DMEM with 10% FBS. After 1, 2, and 4 days of culture, the cell number was quantified by WST-1 assay.
Detection of DNA-synthesizing Cells
MSCs were seeded on the stepwise osteogenesis-mimicking matrices at a density of 5,000 cells/cm2. After 1 day of culture, the culture medium was changed to DMEM with 10% FBS in the presence of 10 μm BrdUrd, and the cells were incubated for 2 h. The DNA-synthesizing cells were labeled using the BrdUrd labeling and detection kit II (Roche Applied Science) according to the manufacturer's instructions. The BrdUrd-incorporated cells were visualized with 3,3′-diaminobenzidine (Dako). In addition, the cell nuclei were counterstained with hematoxylin (Muto Pure Chemicals, Tokyo, Japan). The percentage of BrdUrd-incorporated cells was calculated by counting the positively stained cells. Over 300 cells were counted in each sample.
Osteogenic Differentiation of MSCs on Stepwise Osteogenesis-mimicking Matrices
To investigate the effect of stepwise osteogenesis-mimicking matrices on the osteogenesis of MSCs, MSCs (at passage 4) were seeded on the matrices at a density of 5,000 cells/cm2 and cultured in osteogenic medium. To investigate the effect of exogenous BMP-2 on RUNX2 expression, 10 or 100 ng/ml BMP-2 (Sigma) was added to the osteogenic medium. For inhibition of the translocation of β-catenin into nuclei, 50 μm quercetin (Sigma) was added to the osteogenic medium.
Western Blot Analysis of Total β-Catenin
After 2 weeks of MSC culture on stepwise osteogenesis-mimicking matrices in the osteogenic medium, cells were lysed on ice for 1 h with lysis buffer (10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 0.1% SDS, 1% sodium deoxycholate, 1% Triton X-100, 1 μg/ml leupeptin, 2 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride, 0.1 mm Na3VO4, 0.5 mm NaP2O7, and 5 mm NaF). Cell lysates were collected as supernatant after centrifugation at 20,400 × g for 30 min and separated on 7.5% SDS-PAGE. After being transferred to a polyvinylidene difluoride membrane (Bio-Rad), proteins were reacted with antibodies and detected with the Immobilon Western system (Millipore, Bedford, MA) following treatment with 5% BSA containing PBS to prevent nonspecific reactions. The specific antibodies for this experiment were anti-β-catenin (Cell Signaling Technology, Beverly, MA) and β-actin (Sigma).
Statistical Analysis
All statistical analyses were performed using R, a language and environment for statistical computing. Statistical differences were determined by unpaired Student's t test when the difference between two samples was to be determined. When the differences among three or more samples were to be determined, statistical differences were determined by analysis of variance. The Tukey multiple comparison test was applied as a post hoc test. p values of <0.05 were considered statistically significant.
RESULTS
Stepwise Osteogenesis of MSCs
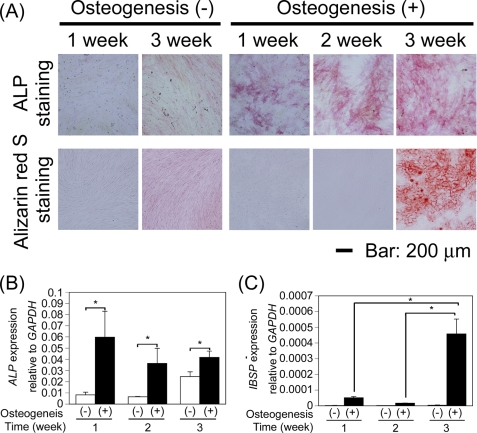
We confirmed the time schedule of the MSC osteogenesis using ALP and alizarin red S stainings (Fig. 1A). When MSCs were cultured without osteogenic induction factors, both ALP and alizarin red S stainings were negative after 3 weeks of culture. In contrast to the culture without the osteogenic induction factors, stepwise osteogenesis was achieved depending on the period of differentiation. After 1 week of osteogenic induction culture, ALP activity was positively stained by ALP staining. However, alizarin red S staining was negative until 2 weeks of osteogenic induction culture, indicating that calcium deposition did not occur for the first 2 weeks of osteogenic induction culture. After 3 weeks of osteogenic induction culture, alizarin red S staining was positive, indicating that calcium deposition started after 3 weeks of osteogenic induction culture.
FIGURE 1.
Stepwise osteogenic differentiation of MSCs. A, MSCs were differentiated into osteoblasts on TCPS. ALP and alizarin red S stainings were performed after culture for the indicated period. Scale bar, 200 μm. B and C, expressions of ALP (B) and IBSP (C) during the osteogenesis of the MSCs were investigated by real-time PCR analysis. Data represent means ± S.D. (n = 3). *, p < 0.05.
To further confirm the stepwise osteogenesis of the MSCs, the expressions of osteogenic genes, ALP and IBSP, were measured by real-time PCR analysis (Fig. 1, B and C). The osteogenic induction culture resulted in a 7.3-fold increase of ALP expression in the MSCs (Fig. 1B). The ALP expression level in the MSCs in the osteogenic culture maintained a higher level than did that in the non-osteogenic culture for the first 3 weeks. IBSP expression in the MSCs in the osteogenic induction culture was higher than was that in the non-osteogenic induction culture throughout the culture period. After 3 weeks of culture, the expression level of IBSP in the osteogenic induction culture increased 8.9-fold compared with that in the osteogenic induction culture after 1 week of culture (Fig. 1C). These results confirmed stepwise osteogenesis of the MSCs. Consequently, we defined the MSCs after 1 and 3 weeks of osteogenic induction culture as cells at the early and late stages of osteogenesis, respectively, and the MSCs cultured without osteogenic induction factors as being in an undifferentiated stem cell stage.
Components of ECM Change during the Osteogenesis of MSCs in Vitro
To investigate whether the components of ECM changed depending on the stages of osteogenesis of the MSCs in vitro, we first checked the expression pattern of ECM genes in the cells by the semiquantitative reverse transcription-PCR method (supplemental Fig. 1). The expression level of FN1 (fibronectin) was maintained through the progression of osteogenesis. However, the expression levels of VCAN (versican), BGN (biglycan), and DCN (decorin) changed according to the progression of osteogenesis. VCAN was expressed in the cells in the non-osteogenic induction culture and in the osteogenic induction culture after only 1 week. Its expression then disappeared after 2 and 3 weeks in the osteogenic induction culture. BGN was expressed only in the undifferentiated MSCs. The expression of BGN disappeared in the cells in the osteogenic induction culture. In contrast to the BGN expression, DCN was weakly expressed in the cells in the non-osteogenic induction culture, and its expression increased in the cells in the osteogenic induction culture.
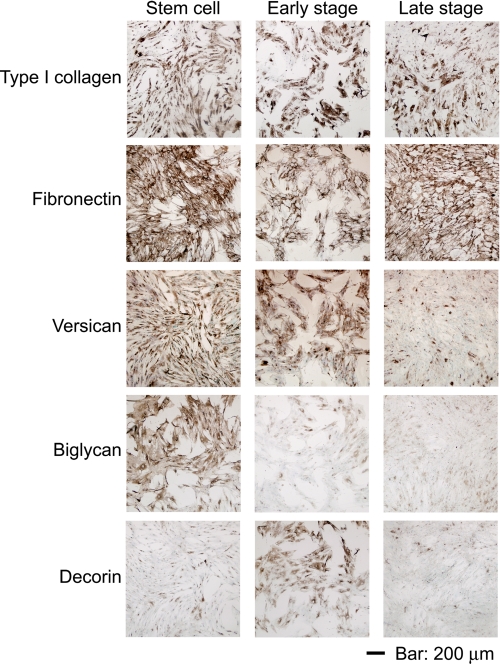
To further investigate the components of ECMs, the ECM proteins were detected by immunocytochemical analysis (Fig. 2 and supplemental Figs. 2 and 3). Fibronectin and type I collagen were positively stained in all of the matrices produced by the undifferentiated and osteogenic differentiated cells in both the early and late stages. Versican was strongly stained in the matrices of early stage osteogenic cells and undifferentiated MSCs but only weakly stained in the matrices of late stage osteogenic MSCs. Biglycan was detected in the matrices of undifferentiated MSCs but only weakly detected in the matrices of early and late stage osteogenic MSCs. Decorin was strongly detected in the matrices of early stage osteogenic cells and weakly detected in the matrices of late stage osteogenic cells and undifferentiated MSCs. These results indicate that the components of ECM changed during the osteogenesis of MSCs in vitro.
FIGURE 2.
ECM alteration during the osteogenesis of MSCs in vitro. ECM proteins were investigated by immunocytochemical analysis. Cell nuclei were counterstained with methyl green. Stem cell, Early stage, and Late stage, undifferentiated MSCs and differentiating cells at the early and late stages of osteogenesis, respectively. The results of negative control are shown in supplemental Fig. 2. Scale bar, 200 μm.
Stepwise Osteogenesis-mimicking Matrices and Their Cell Attachment Activities
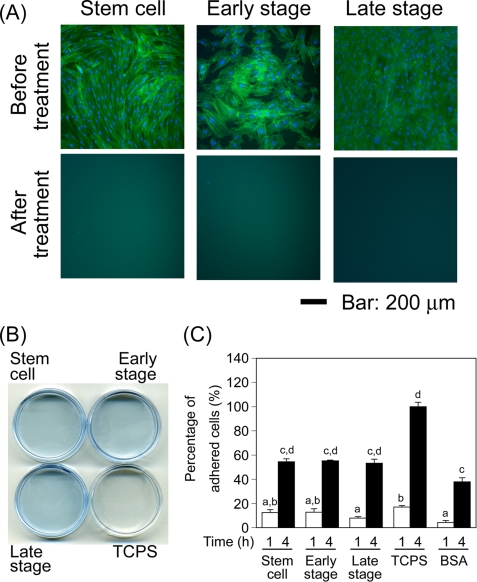
After the MSCs were cultured under the osteogenic conditions for 1 or 3 weeks or under the non-osteogenic condition for 1 week, we tried to remove the cellular components selectively from the matrices. To confirm whether the cellular components were removed from the matrices, the nuclei and actin filaments were stained after decellularization (Fig. 3A). Obvious nuclei and actin filaments were observed in the cells at the undifferentiated stem cell stage, early stage, and late stage of osteogenesis before decellularization. In contrast, no cell nuclei and actin filaments were observed in the matrices after decellularization, indicating that the cellular components were removed from the matrices. The matrices derived from the MSCs after 1 week of undifferentiated culture, and 1 and 3 weeks of osteogenic differentiation culture were referred to as stem cell matrices, early stage matrices, and late stage matrices, respectively.
FIGURE 3.
Preparation of the stepwise osteogenesis-mimicking matrices. A, the removal of cellular components from the stepwise osteogenesis-mimicking matrices was confirmed by cell nuclei and actin stainings. Cell nuclei and actin were stained in the samples before and after decellularization. Blue pseudocolor and green pseudocolor indicate cell nuclei and actin, respectively. B, remaining proteins after decellularization were confirmed by CBB staining. C, cell attachment activity on the stepwise osteogenesis-mimicking matrices was measured after 1 or 4 h of incubation. Data represent mean ± S.D. (n = 3). a, p < 0.01 versus TCPS (1 h); b, p < 0.01 versus BSA (1 h); c, p < 0.001 versus TCPS (4 h); d, p < 0.001 versus BSA (4 h). The differences among cell attachment activity of stepwise osteogenesis-mimicking matrices were not significant.
To check the existence of extracellular proteins after decellularization treatment, whole proteins were stained by CBB staining (Fig. 3B). The matrices after decellularization were obviously stained with CBB, whereas TCPS was not stained. This result indicates that the extracellular proteins (i.e. extracellular matrix proteins) remained after decellularization.
To check whether these matrices possess the attachment activity of MSCs, a cell attachment assay was performed after 1 and 4 h of incubation. More cells adhered to the stepwise osteogenesis-mimicking matrices than did to the BSA-coated surfaces after 1 h of culture. The number of MSCs attached to the stepwise osteogenesis-mimicking matrices was significantly greater than was that attached to the BSA-coated surfaces after 4 h of culture, indicating that these matrices had cell attachment activity (Fig. 3C). No significant difference in cell attachment activity was observed among the stepwise osteogenesis-mimicking matrices.
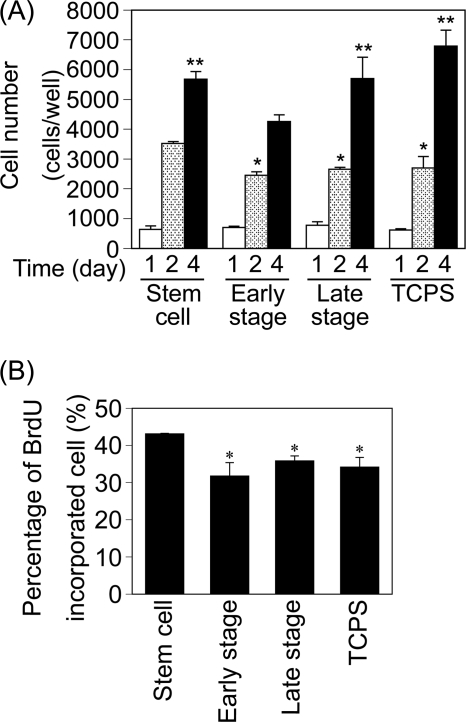
Stem Cell Matrices Enhanced the Proliferation of MSCs
To investigate the effect of stepwise osteogenesis-mimicking matrices on the proliferation of MSCs, cell proliferation assays were performed after 1, 2, and 4 days of culture. The MSCs proliferated with an increase in culture time on the three stepwise osteogenesis-mimicking matrices (Fig. 4A). After 2 days of culture, the cell number was significantly higher on the stem cell matrices than it was on the early stage and late stage matrices and TCPS. After 4 days of culture on the early stage matrices, the cell number was significantly lower than it was on the other matrices.
FIGURE 4.
Proliferation of MSCs on the stepwise osteogenesis-mimicking matrices. A, MSC proliferation was measured by a WST-1 assay. Data represent means ± S.D. (n = 3). *, p < 0.05 versus stem cell matrices (2 days). **, p < 0.01 versus early stage matrices (4 days). B, MSCs were cultured for 1 day and then were allowed to incorporate BrdUrd for 2 h. The results are expressed as a percentage of BrdUrd-incorporated cells to the total cell number. Data represent mean ± S.D. (n = 3). *, p < 0.05 versus stem cell matrices.
To further investigate the effect of stepwise osteogenesis-mimicking matrices on the proliferation of MSCs, DNA synthesis activity was also evaluated by measuring the rate of BrdUrd incorporation. The rate was highest when the MSCs were cultured on the stem cell matrix (Fig. 4B). These results suggest that the stepwise osteogenesis-mimicking matrices supported MSC proliferation, and their effects were in the order of stem cell matrices > late stage matrices > early stage matrices.
Early Stage Matrices Enhance the Osteogenesis of MSCs
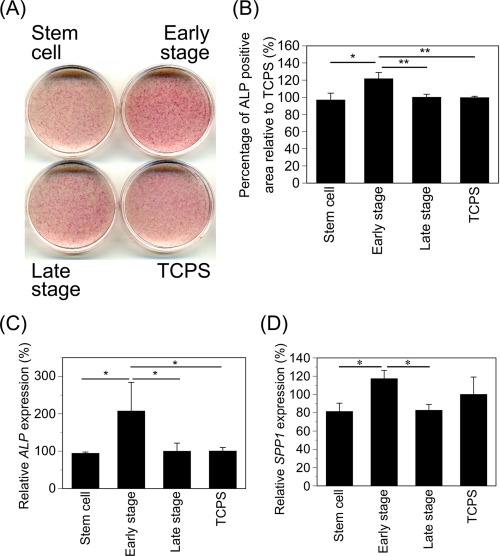
To investigate the effect of stepwise osteogenesis-mimicking matrices on the osteogenic differentiation of MSCs, ALP staining was performed after MSCs were cultured on the stepwise osteogenesis-mimicking matrices and TCPS in the presence of osteogenic induction factors for 2 weeks. MSCs that were cultured on the matrices in the presence of osteogenic induction factors showed osteogenesis, as evidenced by ALP staining. However, the MSCs on the early stage matrices were more strongly stained by ALP than were those on the stem cell matrices, late stage matrices, and TCPS after 2 weeks of culture (Fig. 5A). The positive ALP staining area was measured to quantify the results of ALP staining. The positive ALP staining areas on the early stage matrices were significantly higher than were those on the stem cell matrices, late stage matrices, and TCPS (Fig. 5B).
FIGURE 5.
Osteogenesis of MSCs on the stepwise osteogenesis-mimicking matrices. A, MSCs were cultured on the stepwise osteogenesis-mimicking matrices for 2 weeks in osteogenic medium. After the osteogenic culture, ALP staining of MSCs was done. B, positive areas of ALP staining on the stepwise osteogenesis-mimicking matrices measured after 2 weeks of osteogenic culture. The results are expressed as a percentage normalized to the positive area on TCPS. Data represent means ± S.D. (n = 3). *, p < 0.005; **, p < 0.01. C and D, MSCs were cultured on stepwise osteogenesis-mimicking matrices for 2 weeks in osteogenic medium. Expressions of ALP (C) and SPP1 (D) genes were investigated by real-time PCR analysis. The results are expressed as a percentage normalized to the expression level on TCPS. Data represent means ± S.D. (n = 3). *, p < 0.05.
To further confirm whether the early stage matrices enhanced the osteogenesis of MSCs, the expression levels of osteogenic genes, such as ALP and SPP1, were also measured by real-time PCR analysis after 2 weeks of culture with osteogenic induction factors. The expression level of ALP in the MSCs on the early stage matrices was ∼2 times greater than were those in the MSCs on other matrices (Fig. 5C). Furthermore, the expression level of SPP1 in the MSCs on the early stage matrices was ∼1.4 times greater than were those in the MSCs on stem cell matrices and late stage matrices and showed a higher level than did that in the MSCs on TCPS (Fig. 5D). These results indicate that the early stage matrices were more favorable to the osteogenesis of MSCs compared with the stem cell and late stage matrices. The MSCs cultured on the stepwise osteogenesis-mimicking matrices and TCPS in the medium without osteogenic induction factors were not positively stained by ALP staining (data not shown). The osteogenic differentiation of MSCs on the stepwise osteogenesis-mimicking matrices and TCPS requires the synergistic effect of osteogenic induction factors.
Different Expression Patterns of Osteogenic Transcription Factors among the Stepwise Osteogenesis-mimicking Matrices
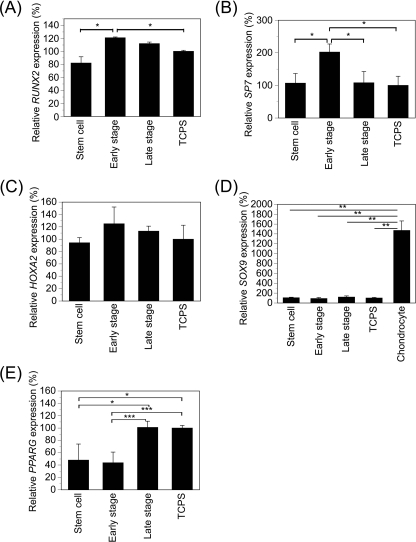
The osteogenesis effect of the osteogenesis-mimicking matrices may be explained by three different mechanisms: up-regulation of genes of osteogenesis-promoting transcription factors, up-regulation of genes encoding osteogenesis-inhibitory transcription factors, or suppression of differentiation to other cell types. To explore the mechanism of differentiation of the MSCs on the stepwise osteogenesis-mimicking matrices, the expression of transcriptional genes relative to osteogenesis, chondrogenesis, and adipogenesis was investigated. RUNX2 and SP7 are two of the main osteogenesis-promoting transcription factors (6, 7). HOXA2 is an inhibitory transcription factor of osteogenesis (27). SOX9 and PPARG are the transcription factors for chondrogenic (28) and adipogenic differentiation (29), respectively. The expression levels of genes encoding RUNX2, SP7, HOXA2, SOX9, and PPARG were measured by real-time PCR analysis after MSCs were cultured on the three stepwise osteogenesis-mimicking matrices and TCPS for 2 weeks. RUNX2 expression was high in the MSCs cultured on the early stage and late stage matrices. It decreased most obviously in the MSCs cultured on the stem cell matrices (33% lower than that on the early stage matrices) and decreased slightly on the TCPS (17% lower than that on the early stage matrices) (Fig. 6A), suggesting that the osteogenesis of the MSCs was inhibited on the stem cell matrices. In contrast, the expression level of SP7 in the MSCs cultured on the early stage matrices was ∼2 times greater than were those in the MSCs cultured on the other matrices (Fig. 6B). HOXA2 expression showed similar levels among the stem cell matrices, early stage matrices, late stage matrices, and TCPS, suggesting that any difference in osteogenesis was not caused by a difference of HOXA2 expression (Fig. 6C). The expression level of SOX9 in the MSCs cultured on the stepwise osteogenesis-mimicking matrices was significantly lower than was that in human chondrocytes as a positive control (Fig. 6D). The expression levels of PPARG in the MSCs cultured on the late stage matrices and TCPS were 2.0–2.4-fold higher than were those in the MSCs cultured on stem cell matrices and early stage matrices (Fig. 6E). This result suggests that insufficient specific differentiation into osteoblasts occurred on the late stage matrices and TCPS.
FIGURE 6.
Expressions of transcription factors related to osteogenesis of MSCs. MSCs were cultured on stepwise osteogenesis-mimicking matrices for 2 weeks in osteogenic medium. The expression levels of RUNX2 (A), SP7 (B), HOXA2 (C), SOX9 (D), and PPARG (E) were measured by real-time PCR analysis. The results are expressed as a percentage normalized to the expression level on TCPS. Data represent means ± S.D. (n = 3). *, p < 0.05; **, p < 0.001; ***, p < 0.01.
Exogenous BMP-2 Enhances Osteogenesis on Stem Cell Matrices but Not on Early Stage Matrices, Late Stage Matrices, and TCPS
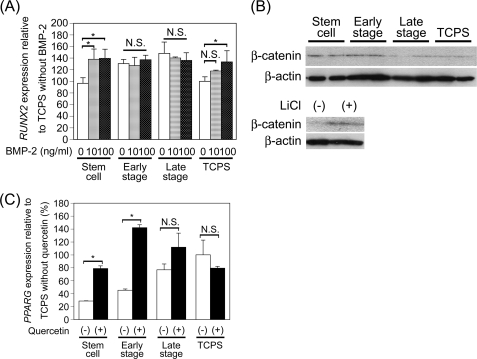
During osteogenesis, RUNX2 expression is mostly regulated by Smad; the activation of Smad is regulated by BMPs (30, 31). It has been reported that BMP-2 and BMP-4 that are produced by the cells themselves are associated with ECM and enhance osteogenesis (32, 33). In addition, ECM can suppress the osteogenesis of MSCs via the interaction of BMPs that were secreted from the cells themselves (24). It is possible that stepwise osteogenesis-mimicking matrices can interact with BMPs to regulate their activities and affect the osteogenesis of MSCs. If the activities of the endogenous BMPs were regulated by the stepwise osteogenesis-mimicking matrices, the responses to exogenous BMP-2 should be changed. To investigate the effect of exogenous BMP2, we examined the ability of MSCs to express RUNX2 in response to exogenous BMP-2 (Fig. 7A). RUNX2 expression level was high when MSCs were cultured on the early stage matrices and late stage matrices in the absence of exogenous BMP-2. When exogenous BMP-2 was added to the culture medium during the culture of MSCs on the early stage and late stage matrices, RUNX2 expression levels did not change significantly even at 100 ng/ml of BMP-2, suggesting that the cells on the early stage matrices and late stage matrices might already be stimulated by endogenous BMP-2 to express RUNX2. On the other hand, RUNX2 expression levels in the cells cultured on the stem cell matrices were lower than were those on the early stage and late stage matrices. When exogenous BMP-2 was added to the MSCs cultured on the stem cell matrices, the RUNX2 expression level was significantly elevated at 10 ng/ml BMP-2. This result suggests that the endogenous BMP signal may be suppressed in the cells cultured on the stem cell matrices. A large amount of exogenous BMP-2 (100 ng/ml) was required to increase the RUNX2 expression level beyond that already stimulated by the endogenous BMPs when the MSCs were cultured on TCPS.
FIGURE 7.
BMP-2 and Wnt signals regulated on the matrices. A, MSCs were cultured on stepwise osteogenesis-mimicking matrices for 2 weeks in osteogenic medium with or without exogenous BMP-2. The expression level of RUNX2 was measured by real-time PCR analysis. Data represent means ± S.D. (n = 3). *, p < 0.05. N.S., no significant difference. B, MSCs were cultured on stepwise osteogenesis-mimicking matrices for 2 weeks in osteogenic medium. The amount of total β-catenin was measured by Western blot analysis. The lower panel shows the control experiment. The sample from the MSCs cultured on TCPS with 25 mm of LiCl for 1 week was used as positive control of β-catenin. C, MSCs cultured on osteogenesis-mimicking matrices on stepwise osteogenesis-mimicking matrices for 2 weeks in osteogenic medium with 50 μm quercetin or DMSO. The expression level of PPARG was measured by real-time PCR analysis. Data represent means ± S.D. (n = 3). *, p < 0.05. N.S., no significant difference.
β-Catenin Increases on Stem Cell and Early Stage Matrices to Suppress PPARG Expression
It has been well reported that the increase of β-catenin in the cells suppresses PPARG expression through the expression of COUP-TFII (chicken ovalbumin upstream promoter-transcription factor II gene) following the translocation of β-catenin into the nuclei (34). To check the amount of β-catenin in the cells cultured on the stepwise osteogenesis-mimicking matrices, the total amount of β-catenin was measured by Western blot analysis. When MSCs were cultured on the stem cell matrices and early stage matrices, the total amount of β-catenin was higher than that cultured on the late stage matrices and TCPS (Fig. 7B). To confirm whether this increase in the amount of β-catenin causes the suppression of PPARG expression in the cells cultured on the stem cell matrices and early stage matrices, the effect of quercetin, an inhibitor of β-catenin translocation into the nuclei (35), on PPARG expression was investigated (Fig. 7C). PPARG expression was elevated by the addition of quercetin to the cells cultured on the stem cell matrices and early stage matrices but not on the late stage matrices and TCPS. These results suggest that PPARG expression was inhibited through the increase of the β-catenin amount on the stem cell matrices and early stage matrices.
DISCUSSION
Characterization of Stepwise Osteogenesis-mimicking Matrices
To investigate the components of the stepwise osteogenesis-mimicking matrices, we checked the ECM gene expression and ECM deposition in the matrices by semiquantitative reverse transcription-PCR and immunocytochemical analysis, respectively (Fig. 2 and supplemental Figs. 1–3). The immunocytochemical staining results of fibronectin, versican, and biglycan were coincident with their gene expression patterns. However, the immunocytochemical staining result for decorin was different from that of its gene expression. Its expression was strongly detected in the matrices at the early stage of osteogenesis and weakly detected in the matrices of undifferentiated MSCs but disappeared during the late stage of osteogenesis. It has been reported that ADAMTS-4 and -5 are also expressed at the late stage of osteogenesis to form lamellar (mature) bone (9). Since it is known that ADAMTS-4 and -5 can degrade decorin (36), the disappearance of decorin in the late stage matrices may be due to degradation by ADAMTS-4 and -5.
The components of ECM changed during the osteogenesis of MSCs in vitro (Fig. 2B). According to previous reports of ECM composition in vivo, expressions of versican are observed in the matrix at the early stage of osteogenesis (9). Decorin also exists in the matrix at the early stage of osteogenesis (11). In contrast, biglycan is strongly observed in the matrix surrounding the MSCs (11). Type I collagen increases in the matrix according to the progression of osteogenesis (10). Therefore, the results of immunocytochemical analysis revealed that the stepwise osteogenesis-mimicking matrices reflected to some degree the composition pattern of in vivo ECM during development. In particular, the compositions of proteoglycan, versican, biglycan, and decorin were similar to the composition pattern of in vivo ECM during development.
After decellularization, the matrices were positively stained by CBB staining, suggesting that ECM proteins remained. Although ECM proteins remained after decellularization, it is unclear whether the specific ECM proteins remained. Chen et al. reported that type I collagen, fibronectin, versican, biglycan, and decorin remained after decellularization treatment with 0.5% Triton X-100 and 20 mm NH4OH for 5 min at 37 °C (23). Therefore, it is likely that these proteins remained in our stepwise osteogenesis-mimicking matrices.
Different Proliferation Rates on the Stepwise Osteogenesis-mimicking Matrices
On the stepwise osteogenesis-mimicking matrices, different cell proliferation rates were observed (Fig. 4). At present, it is unknown how the matrices regulate the proliferation of MSCs. It has been shown that the ECM modulates the activity of growth factors by controlling the proteolytic activation of latent factors, as occurs in the case of transforming growth factor-β (37). In addition, ECM molecules, such as decorin, can interact with cell surface receptors so as to prevent binding of the cognate ligand, as occurs in the case of the epidermal growth factor receptor (38). The ECM may also bind growth-promoting factors from the serum for optimal presentation to MSCs. Finally, the ECM may modulate the proliferation of MSC.
Possible Mechanism of the Enhancement of Osteogenesis on Early Stage Matrices
We investigated the effect of the stepwise osteogenesis-mimicking matrices on the osteogenesis of MSCs (Fig. 5). The results of ALP staining and gene expression analysis revealed that early stage matrices enhanced the osteogenesis of MSCs. To explain the mechanism of the enhancement on early stage matrices, we checked the expression levels of transcription factors (Fig. 6). The results of the transcription factor expression levels revealed that the mechanism to enhance the expression of osteogenic phenotype of MSCs by stepwise osteogenesis-mimicking matrices and TCPS may be different. When MSCs were cultured on the early stage matrices, the expression of RUNX2 and its downstream transcription factor, SP7, were high, which indicates that the early stage matrices could directly enhance osteogenic differentiation. On the other hand, the expressions of RUNX2 and SP7 were suppressed by the stem cell matrices, which suggests that the stem cell matrices may directly suppress osteogenic differentiation. The cases of the late stage matrices and TCPS were slightly complicated. The late stage matrices and TCPS increased the expressions of both the RUNX2 and PPARG genes, meaning that the late stage matrices and TCPS may not directly suppress osteogenic differentiation. However, it has been reported that PPARγ can interact with RUNX2 to inhibit binding to the osteoblast-specific cis-acting element (OSE2) (39) and the expression of the downstream genes of RUNX2 that are regulated by OSE2, such as ALP and SPP1 (6). RUNX2 regulates SP7 expression through binding with the promoter region of SP7 (8). The SP7 expression decreased on the late stage matrices. Therefore, expression of the osteogenic phenotype might have been suppressed by the potential of unexpected adipogenic differentiation when the MSCs were cultured on the late stage matrices and TCPS.
On only the stem cell matrices did the cells retain the ability to respond to exogenous BMP-2 at a low concentration, suggesting that the BMP signal was suppressed (Fig. 7A). Our result is coincident with the report of Chen et al. (23). In addition, Chen et al. have recently reported that matrices derived from cultured mouse MSCs suppress the spontaneous osteogenesis of MSCs by binding between the matrices and BMP-2. It has been reported that ECM modulates intracellular signals activated by cytokines, such as BMP-2 and transforming growth factor-β, by binding with several proteoglycans (15, 16, 40, 41). Biglycan was abundant in the stem cell matrices. It has been reported that biglycan binds with BMP-2 to inhibit activation of the following signaling pathway (40, 41). In addition, biglycan increases the inhibitory activity of chordin, an antagonist of BMP-2 and -4 (41). Therefore, biglycan in the stem cell matrices may be one of the factors that suppresses the osteogenesis of MSCs.
When MSCs were cultured on stem cell matrices and early stage matrices, the amount of β-catenin increased to suppress PPARG expression (Fig. 7, B and C). There are many reports that PPARG expression is suppressed by Wnt/β-catenin signal during osteogenesis (42, 43). The Wnt/β-catenin signal may be strongly activated in the cells cultured on the stem cell matrices and early stage matrices to inhibit PPARG expression. Similar to the regulation of BMPs by the interaction of proteoglycan, the wingless protein, the Drosophila homologue of WNT1, can bind to glycosaminoglycans to promote signal transduction (44). In the stem cell matrices and early stage matrices, versican, biglycan, or decorin were detected. It seems that a high level of β-catenin and the suppression of PPARG expression in the MSCs may result from the interaction between these proteoglycans and Wnt ligands to activate Wnt/β-catenin signaling. On the other hand, in the late stage matrices, biglycan, versican, and decorin were barely detected. It seems that the low level of β-catenin and the unexpected PPARG expression in the MSCs cultured on the late stage matrices may result from the shortage of proteoglycans, which can bind to Wnt proteins. However, we cannot exclude other possible mechanisms responsible for suppressing the amount of β-catenin in the cells cultured on stem cell matrices and early stage matrices.
Stepwise Osteogenesis-mimicking Matrices as in Vitro ECM Models during Osteogenesis in Vivo
The effects of the ECM on osteogenesis have been studied in vivo and in vitro using gene-deficient animals and cells and surfaces coated with isolated ECM proteins (13–17). These studies have reported the individual roles of isolated ECM proteins. However, ECM is formed from complex components in vivo (18). Therefore, usage of matrices that form from components similar to the in vivo components is necessary to understand the role of ECM in osteogenesis precisely. To obtain matrices similar to in vivo components, the use of acellular matrices from the tissue after decellularization is one approach (19, 20). In these matrices, it is difficult to identify and isolate the matrices at different maturational stages of stem cells even using fetal tissue. Furthermore, obtained acellular matrices are too small to use for analyzing the roles in the differentiation of stem cells. Therefore, we tried to develop the matrices whose components are similar to in vivo components by cultured stem cells that were differentiating into osteoblasts. Such matrices can be prepared in a large amount from cultured cells and be used for analyzing the roles of ECM in osteogenesis. Therefore, these stepwise osteogenesis-mimicking matrices would be good in vitro ECM models for analyzing the roles of ECM in osteogenesis. Similar to these stepwise osteogenesis-mimicking matrices, the matrices formed by differentiating stem cells, precursor cells of other somatic cells (stepwise tissue development-mimicking matrices), may be good in vitro ECM models for analyzing the roles of ECM in tissue development and stem cell differentiation.
Application of the Stepwise Osteogenesis-mimicking Matrices for Tissue Engineering and Regenerative Medicine
The early stage matrices of stepwise osteogenesis-mimicking matrices enhanced osteogenesis more strongly than did stem cell matrices, late stage matrices, and TCPS, indicating that early stage matrices can provide a more favorable microenvironment for the osteogenesis of MSCs. Recently, Takeuchi et al. (45) reported that the rate of hepatic differentiation of embryonic stem cells increased when embryonic stem cells were cultured on frozen sectioned regenerating liver. By using an approach similar to the use of stepwise osteogenesis-mimicking matrices, it is possible to prepare matrices that mimic the in vivo ECM formed during the development of other tissue types. The matrices (stepwise tissue development-mimicking matrices) may provide a favorable microenvironment for the differentiation of stem cells. Furthermore, these matrices may provide us with novel insights into the design of new culture systems for the differentiation of stem cells.
In this study, we developed stepwise osteogenesis-mimicking matrices. The matrices mimicking the ECM at the early stage of osteogenesis enhanced the osteogenesis of MSCs. These stepwise osteogenesis-mimicking matrices may be good in vitro models for analyzing the roles of ECM in osteogenesis. Moreover, stepwise osteogenesis-mimicking matrices and the matrices mimicking ECM during tissue development may provide a favorable microenvironment for the differentiation of stem cells and also provide novel insights into the design of new culture systems for the differentiation of stem cells.
Supplementary Material
This work was supported in part by World Premier International Research Center Initiative on Materials Nanoarchitectonics and Grant-in-Aid for Young Scientists (Start-up) 20800073; Japan Society for the Promotion of Science Fellowship 21-10589 from the Ministry of Education, Culture, Sports, Science and Technology, Japan; and the New Energy and Industrial Technology Development Organization of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- ECM
- extracellular matrix
- MSC
- mesenchymal stem cell
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- TCPS
- tissue culture polystyrene
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- CBB
- Coomassie Brilliant Blue.
REFERENCES
- 1.D'Amour K. A., Bang A. G., Eliazer S., Kelly O. G., Agulnick A. D., Smart N. G., Moorman M. A., Kroon E., Carpenter M. K., Baetge E. E. (2006) Nat. Biotechnol. 24, 1392–1401 [DOI] [PubMed] [Google Scholar]
- 2.Daley W. P., Peters S. B., Larsen M. (2008) J. Cell Sci. 121, 255–264 [DOI] [PubMed] [Google Scholar]
- 3.Page-McCaw A., Ewald A. J., Werb Z. (2007) Nat. Rev. Mol. Cell. Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. (1999) Science 284, 143–147 [DOI] [PubMed] [Google Scholar]
- 5.Malaval L., Modrowski D., Gupta A. K., Aubin J. E. (1994) J. Cell. Physiol. 158, 555–572 [DOI] [PubMed] [Google Scholar]
- 6.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 7.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. (2002) Cell 108, 17–29 [DOI] [PubMed] [Google Scholar]
- 8.Nishio Y., Dong Y., Paris M., O'Keefe R. J., Schwarz E. M., Drissi H. (2006) Gene 372, 62–70 [DOI] [PubMed] [Google Scholar]
- 9.Nakamura M., Sone S., Takahashi I., Mizoguchi I., Echigo S., Sasano Y. (2005) J. Histochem. Cytochem. 53, 1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasano Y., Li H. C., Zhu J. X, Imanaka-Yoshida K., Mizoguchi I., Kagayama M. (2000) Histochem. J. 32, 591–598 [DOI] [PubMed] [Google Scholar]
- 11.Kamiya N., Shigemasa K., Takagi M. (2001) J. Oral Sci. 43, 179–188 [DOI] [PubMed] [Google Scholar]
- 12.Pham Q. P., Kasper F. K., Scott Baggett L., Raphael R. M., Jansen J. A., Mikos A. G. (2008) Biomaterials 29, 2729–2739 [DOI] [PubMed] [Google Scholar]
- 13.Xu T., Bianco P., Fisher L. W., Longenecker G., Smith E., Goldstein S., Bonadio J., Boskey A., Heegaard A. M., Sommer B., Satomura K., Dominguez P., Zhao C., Kulkarni A. B., Robey P. G., Young M. F. (1998) Nat. Genet. 20, 78–82 [DOI] [PubMed] [Google Scholar]
- 14.Chen X. D., Shi S., Xu T., Robey P. G., Young M. F. (2002) J. Bone Miner. Res. 17, 331–340 [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez J., Osses N., Brandan E. (2006) J. Cell. Physiol. 206, 58–67 [DOI] [PubMed] [Google Scholar]
- 16.Riquelme C., Larrain J., Schonherr E., Henriquez J. P., Kresse H., Brandan E. (2001) J. Biol. Chem. 276, 3589–3596 [DOI] [PubMed] [Google Scholar]
- 17.Moursi A. M., Damsky C. H., Lull J., Zimmerman D., Doty S. B., Aota S., Globus R. K. (1996) J. Cell Sci. 109, 1369–1380 [DOI] [PubMed] [Google Scholar]
- 18.Adachi E., Hopkinson I., Hayashi T. (1997) Int. Rev. Cytol. 173, 73–156 [DOI] [PubMed] [Google Scholar]
- 19.Gilbert T. W., Sellaro T. L., Badylak S. F. (2006) Biomaterials 27, 3675–3683 [DOI] [PubMed] [Google Scholar]
- 20.Ott H. C., Matthiesen T. S., Goh S. K., Black L. D., Kren S. M., Netoff T. I., Taylor D. A. (2008) Nat. Med. 14, 213–221 [DOI] [PubMed] [Google Scholar]
- 21.Datta N., Holtorf H. L., Sikavitsas V. I., Jansen J. A., Mikos A. G. (2005) Biomaterials 26, 971–977 [DOI] [PubMed] [Google Scholar]
- 22.Datta N., Pham Q. P., Sharma U., Sikavitsas V. I., Jansen J. A., Mikos A. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2488–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X. D., Dusevich V., Feng J. Q., Manolagas S. C., Jilka R. L. (2007) J. Bone Miner. Res. 22, 1943–1956 [DOI] [PubMed] [Google Scholar]
- 24.Lehr H. A., Mankoff D. A., Corwin D., Santeusanio G., Gown A. M. (1997) J. Histochem. Cytochem. 45, 1559–1565 [DOI] [PubMed] [Google Scholar]
- 25.Martin I., Jakob M., Schäfer D., Dick W., Spagnoli G., Heberer M. (2001) Osteoarthritis Cartilage 9, 112–118 [DOI] [PubMed] [Google Scholar]
- 26.Schaefer J. F., Millham M. L., de Crombrugghe B., Buckbinder L. (2003) Osteoarthritis Cartilage 11, 233–241 [DOI] [PubMed] [Google Scholar]
- 27.Kanzler B., Kuschert S. J., Liu Y. H., Mallo M. (1998) Development 125, 2587–2597 [DOI] [PubMed] [Google Scholar]
- 28.Bi W., Deng J. M., Zhang Z, Behringer R. R., de Crombrugghe B. (1999) Nat. Genet. 22, 85–89 [DOI] [PubMed] [Google Scholar]
- 29.Spiegelman B. M., Flier J. S. (1996) Cell 87, 377–389 [DOI] [PubMed] [Google Scholar]
- 30.Lee K. S., Kim H. J., Li Q. L., Chi X. Z., Ueta C., Komori T., Wozney J. M., Kim E. G., Choi J. Y., Ryoo H. M., Bae S. C. (2000) Mol. Cell. Biol. 20, 8783–8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lian J. B., Stein G. S., Javed A., van Wijnen A. J., Stein J. L., Montecino M., Hassan M. Q., Gaur T., Lengner C. J., Young D. W. (2006) Rev. Endocr. Metab. Disord. 7, 1–16 [DOI] [PubMed] [Google Scholar]
- 32.Suzawa M., Takeuchi Y., Fukumoto S., Kato S., Ueno N., Miyazono K., Matsumoto T., Fujita T. (1999) Endocrinology 140, 2125–2133 [DOI] [PubMed] [Google Scholar]
- 33.Abe E., Yamamoto M., Taguchi Y., Lecka-Czernik B., O'Brien C. A., Economides A. N., Stahl N., Jilka R. L., Manolagas S. C. (2000) J. Bone Miner. Res. 15, 663–673 [DOI] [PubMed] [Google Scholar]
- 34.Okamura M., Kudo H., Wakabayashi K., Tanaka T., Nonaka A., Uchida A., Tsutsumi S., Sakakibara I., Naito M., Osborne T. F., Hamakubo T., Ito S., Aburatani H., Yanagisawa M., Kodama T., Sakai J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5819–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park C. H., Chang J. Y., Hahm E. R., Park S., Kim H. K., Yang C. H. (2005) Biochem. Biophys. Res. Commun. 328, 227–234 [DOI] [PubMed] [Google Scholar]
- 36.Gendron C., Kashiwagi M., Lim N. H., Enghild J. J., Thøgersen I. B., Hughes C., Caterson B., Nagase H. (2007) J. Biol. Chem. 282, 18294–18306 [DOI] [PubMed] [Google Scholar]
- 37.Dallas S. L., Rosser J. L., Mundy G. R., Bonewald L. F. (2002) J. Biol. Chem. 277, 21352–21360 [DOI] [PubMed] [Google Scholar]
- 38.Santra M., Reed C. C., Iozzo R. V. (2002) J. Biol. Chem. 277, 35671–35681 [DOI] [PubMed] [Google Scholar]
- 39.Jeon M. J., Kim J. A., Kwon S. H., Kim S. W., Park K. S., Park S. W., Kim S. Y., Shin C. S. (2003) J. Biol. Chem. 278, 23270–23277 [DOI] [PubMed] [Google Scholar]
- 40.Hocking A. M., Shinomura T., McQuillan D. J. (1998) Matrix Biol. 17, 1–19 [DOI] [PubMed] [Google Scholar]
- 41.Moreno M., Muñoz R., Aroca F., Labarca M., Brandan E., Larraín J. (2005) EMBO J. 24, 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang S., Bennett C. N., Gerin I., Rapp L. A., Hankenson K. D., Macdougald O. A. (2007) J. Biol. Chem. 282, 14515–14524 [DOI] [PubMed] [Google Scholar]
- 43.Liu G., Vijayakumar S., Grumolato L., Arroyave R., Qiao H., Akiri G., Aaronson S. A. (2009) J. Cell Biol. 185, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reichsman F., Smith L., Cumberledge S. (1996) J. Cell Biol. 135, 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi T., Ochiya T., Takezawa T. (2008) Tissue Eng. Part A 14, 267–274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.