Abstract
α-Synuclein is an intrinsically unstructured protein that binds to membranes, forms fibrils, and is involved in neurodegeneration. We used a reconstituted in vitro system to show that the molecular chaperone Hsp90 influenced α-synuclein vesicle binding and amyloid fibril formation, two processes that are tightly coupled to α-synuclein folding. Binding of Hsp90 to monomeric α-synuclein occurred in the low micromolar range, involving regions of α-synuclein that are critical for vesicle binding and amyloidogenesis. As a consequence, both processes were affected. In the absence of ATP, the accumulation of non-amyloid α-synuclein oligomers prevailed over fibril formation, whereas ATP favored fibril growth. This suggests that Hsp90 modulates the assembly of α-synuclein in an ATP-dependent manner. We propose that Hsp90 affects these folding processes by restricting conformational fluctuations of α-synuclein.
Introduction
α-Synuclein (AS)2 is an intrinsically unstructured 14-kDa protein with the propensity to form proteinaceuos inclusions known as Lewy Bodies (LBs) in the substantia nigra of dopaminergic neurons (1). LBs have a spherical morphology and are surrounded by radially oriented amyloid fibrillary tangles generated by self-assembly of AS (2).
The physiological function of AS is still elusive (3, 4). Research is mainly focused on the etiology of a series of neurologic disorders known as synucleinopathies, first among them is Parkinson disease.
In vitro folding and/or aggregation of AS is influenced by a plethora of different factors, including (poly)cations, polyanions, alcohols, and lipids. Notably, AS adopts an α-helical-fold when bound to artificial lipid membranes (5), whereas it forms β-sheets in amyloid fibrils (6). By varying the experimental in vitro conditions, AS forms oligomeric species differing in size and shape (4), some of them displaying a pronounced cytotoxicity (7–9).
In the cell, such a high degree of conformational freedom requires a stringent control that counteracts the formation of undesired or toxic folding species. Most likely, the breakdown in folding control leads to the occurrence of synucleinopathies. This raises the question of how living organisms manage productive folding and assembly of AS, and avoid the accumulation of noxious folding intermediates. Yet, the factors that modulate this balance are unclear.
This issue provides a conceptual link to the protein class of molecular chaperones, which assist the formation of the native structure of proteins over unproductive and potentially dangerous folding populations (10, 11). These proteins usually act in concert creating a sophisticated cellular folding machinery. Several studies suggest that molecular chaperones are interlaced to AS folding and the insurgence of neurotoxic folding species. Torsin A, Hsp70, Hsp40, and α-crystallins were shown to co-localize and generally reduce AS amyloid spreading (12–15).
Among this class of proteins, Hsp90 has been recently identified as the predominant chaperone implicated in AS-evoked pathologies (16). Although the generally accepted role of Hsp90 is to stabilize functionally immature proteins, this chaperone seems also to possess decisional power on whether to direct a non-native folding substrate toward maturation or degradation (17, 18). Such an overlap might represent a pivotal mechanism for controlling the delicate folding balance of an intrinsically unstructured protein such as AS in a cell. In this context, Uryu et al. (16) showed Hsp90 together with ubiquitin to massively co-localize in vivo with soluble AS as well as amyloid filaments, suggesting a possible commitment of Hsp90 in modulating folding and the suppression of incorrectly folded AS. A proteomic survey identified Hsp90 associated with AS in oxidative damaged dopaminergic neurons (19). Hsp90 was further found to suppress AS toxicity in yeast genetic screenings (20). Finally, Hsp90 was shown to regulate rab11a-dependent secretion and recycling of AS (21).
To analyze the relationship between these two proteins, we investigated how Hsp90 influenced two folding-related processes of AS, namely aggregation and vesicle binding. Our analysis reveals that Hsp90 binds to AS and abolishes binding of this polypeptide to small unilamellar vesicles. Hsp90 further promotes fibril formation in an ATP-dependent manner via oligomeric intermediates. These results suggest that Hsp90 critically influences hallmark features of AS interaction and assembly.
EXPERIMENTAL PROCEDURES
Unless specified otherwise, all reagents were from Sigma.
Protein Expression and Purification
The expression vector containing the N-terminal His6-fused HSP90β gene (a kind gift of Sophie Jackson, Cambridge, UK) was transformed into Bl21DE3(Star) cells (Invitrogen). The protein was expressed in LB medium containing 100 μg/ml ampicillin. Cultures were grown at 37 °C to an A600 of 0.8. Protein expression was induced by addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. After 4–5 h, cells were harvested by centrifugation at 6,000 × g for 15 min at 4 °C. The cells were resuspended in 20 mm Tris-HCl, 300 mm NaCl, pH 7.4 (buffer A), containing an EDTA-free Complete® protease inhibitor mixture (Roche Applied Science), and disrupted on ice by sonification. After centrifugation, the supernatant was loaded on a Ni-CAM HC column equilibrated with buffer A, and Hsp90 was eluted with buffer A + 300 mm imidazole. The protein was dialyzed against 20 mm Tris, 50 mm NaCl, 5 mm dithiothreitol, pH 7.4, and loaded on a Resource Q column (GE Healthcare). Elution was carried out using a gradient from 50 mm to 1 m NaCl. Finally, the protein was loaded on a Superdex 200 gel filtration column (GE Healthcare) in 20 mm Tris, 150 mm NaCl, 5 mm dithiothreitol, pH 7.4. The purified protein was concentrated to 10 mg/ml and stored at −80 °C.
The pRSETB vector containing the AS gene was transformed into BL21(DE3)Star cells (Invitrogen). The unlabeled protein was expressed in LB medium containing 100 μg/ml ampicillin. Isotope-labeled 15N-AS and 15N,13C-AS were expressed in minimal medium (6.8 g/liter of Na2HPO4, 3 g/liter of KH2PO4, 0.5 g/liter of NaCl, 1.5 g/liter of (15NH4)2SO4, 2 g/liter of glucose (or [13C]glucose for doubly labeled protein), 1 μg/liter of biotin, 1 μg/liter of thiamin, 100 μg/ml of ampicillin, and 1 ml of ×1000 microsalts).
Cultures were grown at 37 °C to an A600 of approximately 0.8. Protein expression was induced by addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. After 5 h, cells were harvested by centrifugation at 6,000 × g for 15 min at 4 °C. The protein was purified according to Ref. 22 and extensively dialyzed against 50 mm NH4HCO3. Finally, both unlabeled and labeled AS were aliquoted, lyophilized, and stored at −80 °C. Each reconstituted AS solution was centrifuged and checked for aggregation (DLS).
The Hsp70 expression plasmid was a kind gift of Johannes Buchner (Munich, Germany). His6-fused Hsp70 was from Affinity Bioreagents (Golden, CO). Protein concentrations were determined by the BCA Protein Assay (Pierce).
Preparation of Fluorescent Small Unilamellar Vesicles (SUVs)
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPC and POPS, both from Avanti Polar Lipids, Alabaster, AL), and diphenyl-1,3,5-hexatriene (DPH, 20 mm stock solution in CHCl3) were mixed in CHCl3 at a ratio of 1:1:0.04 in a flat bottomed glass vial. CHCl3 was evaporated in a nitrogen stream; the lipid film was resuspended in 20 mm Na2HPO4, pH 7.4, incubated at 42 °C for 5 min, briefly vortexed, and sonicated in a sonicating water bath for 3 h. SUVs (16 mm final concentration) were stored in the dark at 4 °C and used within 24 h of preparation.
Fluorescence Titrations
Fluorescence emission spectra of a 100 nm Hsp90 or Hsp70 solution in 20 mm Tris-HCl, 150 mm NaCl, pH 7.4, were measured using a LB50 spectrofluorimeter (PerkinElmer Life Sciences) at 25 °C and an excitation wavelength of 295 nm. The slit widths were 10 and 15 nm for excitation and emission, respectively. Emission spectra were recorded between 300 and 400 nm.
Aliquots of a 600 μm AS stock solution in 20 mm Tris-HCl, 150 mm NaCl, pH 7.4, were added stepwise to the Hsp90 solution. An equilibration period of 3 min was allowed after each addition. The spectra were then background-corrected against the corresponding spectrum of AS alone. The normalized changes in fluorescence intensity resulting from three independent experiments were plotted against the AS concentration. Resulting binding isotherms were fitted by non-linear regression to supplemental Equation 1 (23).
Stoichiometric Fluorescence Titrations
The molar ratio of AS:Hsp90 was measured using the same experimental settings as for isothermal fluorescence titrations, except for the concentration of Hsp90, which was 8.5 μm.
Fluorescence Anisotropy
The fluorescence anisotropy of a 1 μm DPH-labeled SUV preparation in 20 mm Na2HPO4, pH 7.4, was measured at 25 °C using a LB50 spectrofluorimeter (PerkinElmer Life Sciences) at an emission wavelength of 430 nm upon excitation at 359 nm. Slits were set at 15 and 20 nm for excitation and emission, respectively. The fluorescence anisotropy is defined as r = (IVV − G × IVH)/(IVV + 2G × IVH), where IVV is the fluorescence intensity recorded with excitation and emission polarizers in vertical positions, and IVH is the fluorescence intensity recorded with the emission polarizer aligned in a horizontal position (24). The G factor is the ratio of the sensitivities of the detection system for vertically and horizontally polarized light G = IHV/IHH.
A 1 μm DPH-labeled SUV preparation was titrated against increasing amounts of AS in 20 mm Na2HPO4, pH 7.4. The same experiment was then repeated in the presence of 5 μm Hsp90. For each point in the experiments, the anisotropy was recorded over 30 s and the mean r values for each measurement were used. Anisotropy changes were fitted according to supplemental Equation 2.
ATPase Measurements
The ATPase activity of 2.5 μm Hsp90 in 50 mm Hepes/KOH, pH 7.5, 150 mm KCl, 10 mm MgCl2 was measured at 25 °C in a Hitachi U-2100 spectrophotometer (Hitachi, Krefeld, Germany) using a regenerative ATPase assay (25).
To calculate the apparent Kd, the rate of hydrolysis v of ATP was measured by varying the concentration of AS. Changes were plotted against the AS concentration and fitted to supplemental Equation 3.
ThioflavinT (ThioT) Measurements
A 60 μm AS solution in 20 mm Tris-HCl, 150 mm NaCl, pH 7.4, was agitated in a thermomixer (Eppendorf, Hamburg, Germany) at 37 °C and 900 rpm. 10-μl aliquots were taken at the time points indicated and diluted into a 5 μm ThioT solution prepared in the same buffer. Spectra of the samples were recorded in a Cary Eclipse fluorescence spectrometer (Varian, Palo Alto, CA) at 25 °C and 482 nm upon excitation at 442 nm. The slit widths were set to 10 and 20 nm for excitation and emission, respectively.
Turbidity
The turbidity of the sample solution was measured in a Cary Eclipse fluorescence spectrometer (Varian, Palo Alto, CA) at 25 °C with excitation and emission wavelengths set at 360 nm using a slit width of 5 nm for both excitation and emission.
Oligomer to Fibril Transition
An AS:Hsp90 solution (1:2) was agitated for 1 week at 37 °C, 1000 × g, until the oligomeric species were detectable (TEM, DLS, turbidity). Then, ATP was added and agitation was continued. The reaction was monitored by the ThioT assay.
Transmission Electron Microscopy (TEM)
Fibril growth was performed as for ThioT measurements. Samples were agitated for 2–3 weeks. 10-μl sample aliquots were applied onto carbon-coated grids (Plano, Wetzlar, Germany). After 1 min, the samples were washed with water and incubated with 10 μl of 2% (w/v) uranyl acetate for 1 min. Samples were air-dried and negative stained images were taken using a Philips CM12 electron microscope operating at 120 kV.
Dynamic Light Scattering (DLS)
Stokes radii were determined using a DynaPro instrument (Protein Solutions, Lakewood, NJ) in a 1.5-mm path length 12-μl quartz cuvette at 25 °C. Samples were centrifuged before measuring.
Pulldown Affinity Capture
10 μg of His6-Hsp90 was incubated with 25 μg of AS in 20 mm Tris, 50 mm NaCl, pH 7.4, for 15 min at 25 °C under gentle shaking. 25 μl of Ni-CAM HC resin was added, and the solution was shaken for a further 30 min. The resin was centrifuged and washed at least 5 times with 20 mm Tris, 150 mm NaCl, pH 7.4. Bound proteins were then eluted with 300 mm imidazole, resolved on a 4–12% NuPAGE denaturing gel (Invitrogen), and stained with Coomassie Blue stain.
NMR Spectroscopy
All spectra were acquired at 25 °C on a Varian Unity INOVA 600 MHz NMR spectrometer (Varian, Palo Alto, CA) using an HCN triple-resonance probe with single axis z-gradients. Data were processed using NMRPipe (26) and analyzed in NMRView (27). Backbone 1H and 15N,13C-resonance assignments of AS at the conditions used were confirmed by a three-dimensional HNCACB NMR experiment, which correlates backbone amide proton and nitrogen with Cα and Cβ resonances. Data were collected using samples consisting of 40 μm 15N-, or 13C,15N-labeled AS in 50 mm potassium Pi, 100 mm NaCl, pH 6.8. 10% D2O were added for field-frequency locking.
RESULTS
Hsp90 Binds to Monomeric AS
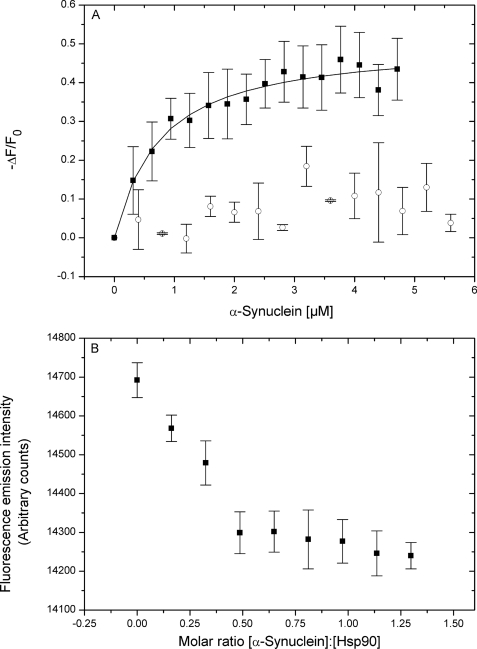
We first aimed to investigate if Hsp90 was able to bind to monomeric AS. To this end, fluorescence titrations were performed by measuring the intrinsic tryptophan fluorescence emission changes of Hsp90 upon the addition of increasing amounts of AS. Because AS lacks any tryptophan residues, it does not contribute to the fluorescence at the given excitation wavelength. Fig. 1A shows that fluorescence quenching of a Hsp90 solution was saturable by monomeric AS, yielding a Kd value of 1.2 ± 0.2 μm. As a control, AS did not quench the fluorescence of Hsp70, in accordance to Dedmon et al. (28), who demonstrated that this chaperone binds to pre-fibrillar, but not to monomeric AS. As no significant turbidity changes were observed during the titrations, we could exclude a possible influence of oligomerization on the fluorescence signal (supplemental Fig. S1).
FIGURE 1.
A, fluorescence binding isotherms of Hsp90 (■) and Hsp70 (□) interacting with AS. The changes in Hsp90 or Hsp70 fluorescence are plotted as the normalized change in fluorescence (−ΔF/F0, where F0 is the initial fluorescence intensity before addition of AS) versus the concentration of AS. B, stoichiometric titration curve for the AS-Hsp90 interaction. Measured values are averages including S.E. of three independent determinations.
Stoichiometric fluorescence titrations (Fig. 1B) yielded an AS:Hsp90 ratio of 0.5:1, suggesting one AS molecule bound per Hsp90 dimer. A similar asymmetric substrate binding stoichiometry was reported by ultrastructural analysis of the kinase Cdk4 in complex with Hsp90 and the co-chaperone Cdc37 (29).
A complex between Hsp90 and monomeric AS could also be isolated by pulldown affinity capture (Fig. 2, lane 3). We were not able to isolate AS using Hsp70 as bait (supplemental Fig. S2), confirming that Hsp90, but not Hsp70 bound to monomeric AS. The modest recovery is explainable as the pulldown procedure favors the dissociation of a pre-existing equilibrium in the direction of the free components (e.g. when transferring the complex from an originally saturated solution into an unsaturated wash solution), whereas titrations shift the equilibrium toward the complex by increasing the amount of one reaction partner.
FIGURE 2.
Pulldown affinity capture using His6-Hsp90 (bait) and AS (prey) at different stages of amyloidogenesis (M, marker; 1, background; 2, AS control; 3, after 0 h; 4, after 10 h; 5, after 25 h; 6, after 40 h; 7, after 0 h in the presence of 5 mm MgATP; 8, after 0 h in the presence of 5 mm MgATP+ 0.5 mm 17-AAG). The bands around 21 and 31 kDa correspond to partial degradation products of Hsp90. Fibrillogenesis was performed as described under “Experimental Procedures” (ThioT measurements), except that the AS solution was 200 μm and agitation was 1200 rpm.
In the presence of ATP, we could not detect any alteration of the pulldown recovery of the Hsp90·AS complex (Fig. 2, lane 7). Also, the small macrocylic compound 17-AAG (17-(allylamino)-17-demethoxygeldanamycin), which competes with ATP and inhibits the ATPase activity of Hsp90 (30), did not visibly alter the affinity of this interaction (Fig. 2, lane 8).
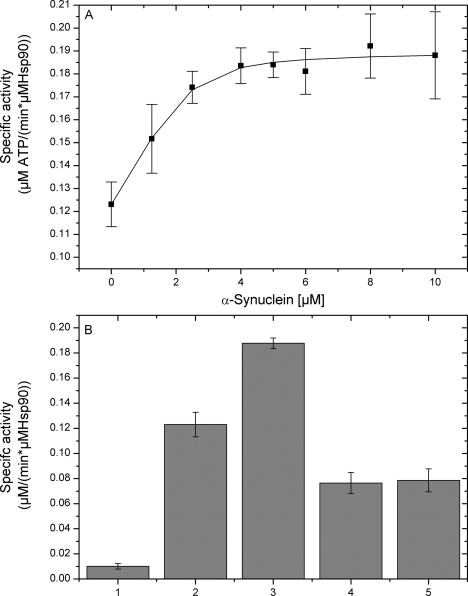
We then moved on to explore how AS might affect the ATPase activity of Hsp90. The measured slow turnover (0.12 min−1) is in accordance to previous reports (31, 32). As shown in Fig. 3A, the rate of ATP turnover of a Hsp90 solution was increased up to 1.6-fold by increasing the concentration of monomeric AS, which showed a negligible ATPase activity by itself (Fig. 3B). The stimulatory effect of AS was saturable and could be suppressed by 17-AAG (Fig. 3B). The resulting saturation curve yielded an apparent Kd value of 1.56 ± 0.40 μm, which was comparable with the value obtained by fluorescence titrations (Fig. 1A).
FIGURE 3.
A, specific activity of Hsp90 ATPase with increasing amounts of AS. B, specific ATPase activity of: 1) AS in the absence of Hsp90 (control); 2) Hsp90; 3) AS:Hsp90; 4) AS:Hsp90:17-AAG; and 5) Hsp90:17-AAG. Measured values are averages including S.E. of three independent determinations.
Hsp90 Influences Amphipatic and Hydrophobic Segments of AS
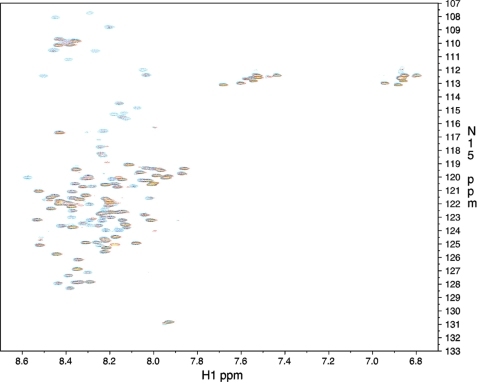
To trace the topology of the Hsp90-AS interaction, we exploited heteronuclear correlation NMR spectroscopy. The 1H-15N HSQC spectrum of AS recorded at 25 °C showed limited signal dispersion typical for intrinsically unstructured proteins (Fig. 4). 116 of the 135 non-prolyl backbone residues could be unambiguously assigned by a three-dimensional HNCACB (supplemental Fig. S3).
FIGURE 4.
15N-HSQC NMR spectra of AS in the absence (blue) and presence (yellow with red borders) of a 2.5-fold excess of Hsp90. See supplemental Fig. S1 for the assignments.
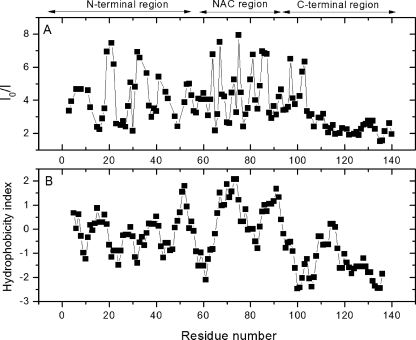
Upon the addition of a 2.5-fold molar excess of unlabeled Hsp90, we observed a pronounced decrease of several peak intensities (Fig. 5A). Affected residues were located along a segment of AS spanning from the amphipatic N-terminal (1–60), over the highly hydrophobic NAC (“non-Aβ component”) core region (61–95), up to residues 96–105 of the upper C-terminal region. The remaining residues (106–140) were not significantly affected.
FIGURE 5.
HSQC signal intensity ratio I0/I of 15N-AS residues (A) in relation to the residue hydrophobicity of AS (B) (50). I0 is the signal intensity of 15N-AS residues in the absence of Hsp90, I is the signal intensity of 15N-AS residues in the presence of Hsp90.
The most affected residues formed a pattern located around two basic residues on amphipatic repeats (AAXKTK) on the N-terminal domain (Lys21 and Lys32), a T/G-rich region in the NAC region (Thr64, Gly67, Thr75, Thr81, and Gly85), and two acidic residues in the upper C-terminal domain (Asp98 and Asn103). These findings suggest multiple contacts between AS and Hsp90 rather than a localized interaction occurring. The hydrophobicity plot of AS (Fig. 5B) further suggests that among these patterns, the primary binding site of Hsp90 might be the hydrophobic stretch constituting the NAC region, as it coincides with the most prominent perturbations.
An issue that has been correlated to the decrease in peak intensity is the occurrence of a disorder to order transition (5, 33, 34). Our observed perturbation pattern is reminiscent to what was previously observed for AS upon lipid binding, which induces an α helical conformation of the amphipatic N-terminal region (5, 33, 34). An analogous decrease in peak intensities was also observed upon folding of other intrinsically unstructured proteins, such as Salmonella typhimurium FlgM and the thyroid cancer-related protein TC-1 (35, 36). This raises the possibility of secondary structural rearrangements of the N-terminal and the middle domain taking place when Hsp90 binds to AS. Bona fide support for this notion comes from circular dichroism measurements (supplemental Fig. S4), which suggest that AS undergoes conformational rearrangements in complex with Hsp90.
Hsp90 Prevents AS from Vesicle Binding
The observation that Hsp90 affected regions that were shown to be critical for lipid binding (the N-terminal amphipatic region) (5) and for amyloid formation (the hydrophobic NAC core) (6) led us to investigate how this chaperone might influence the two processes.
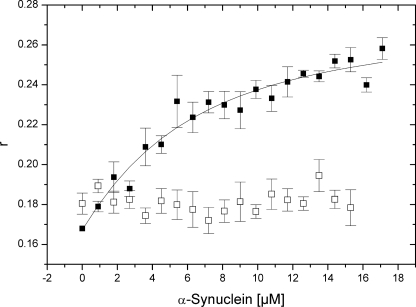
First, we tested the binding of AS to SUVs. We prepared POPC:POPS vesicles containing DPH, which is a fluorophore that shows a strong fluorescence emission at 430 nm when incorporated within lipid assemblies. We then measured the fluorescence anisotropy change of the vesicle probe upon the addition of AS. Fig. 6 shows that upon titration with increasing amounts of AS, the anisotropy of a fluorescent SUV preparation increased up to a saturable value (Kd = 4.56 ± 1.18 μm). In the presence of Hsp90, the addition of AS no longer caused significant anisotropy changes. Similarly, the anisotropy of fluorescence-labeled vesicles did not change in the presence of ATP or 17-AAG (see supplemental Fig. S6), implying that the Hsp90·AS complex was not altered in the presence of these molecules.
FIGURE 6.
Anisotropy changes of a 1 μm DPH-SUV preparation upon titration with AS in the absence (■) and presence (□) of 10 μm Hsp90. Measured values are averages including S.E. of three independent determinations.
As Hsp90 did not bind to vesicles (supplemental Fig. S5), we could exclude competition between Hsp90 and AS for binding to the vesicle surface. We also excluded that the increase in anisotropy is due to the binding of free fluorophore to the protein, as the fluorescence emission of DPH was not significant in the presence of proteins and in the absence of lipids (data not shown).
ATP Drives Hsp90-mediated AS Assembly
Next, we studied the influence of Hsp90 on AS fibril formation. We induced AS amyloidogenesis by agitation using a standard procedure (see “Experimental Procedures”) and assayed its progression in the absence and presence of Hsp90. Kinetics of fibril formation were monitored by ThioT, a dye that exhibits pronounced fluorescence emission intensity at 482 nm when bound to amyloids (37). In addition, the turbidity (i.e. the formation of light scattering particles) of the samples was measured, which mirrors the overall aggregation of a solution in relation to the size of the aggregating species, independently of their conformation.
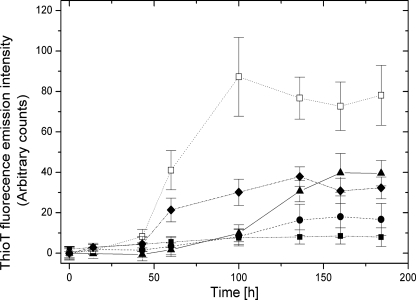
Under our experimental conditions, AS converted to a ThioT-binding species after a lag phase, emphasizing the accumulation of amyloids in solution (Fig. 7). Consistently, we observed a concomitant increase in turbidity (supplemental Fig. S8). The presence of Hsp90 caused a significant suppression of ThioT fluorescence intensity, attesting to the efficient inhibition of fibril formation by this heat shock protein over our experimental time interval.
FIGURE 7.
Kinetics of AS amyloid formation in the absence (▴) and presence of Hsp90 (●, AS:Hsp90 = 1:2; □, AS:Hsp90:MgATP = 1:2:10; ■, AS:Hsp90:AMPPNP = 1:2:10; ♦, AS:Hsp90:MgATP:17-AAG = 1:2:10:5). Measured values are averages including S.E. of three independent determinations.
Binding and hydrolysis of ATP are critical for the Hsp90 chaperone cycle (38–40). Intriguingly, Hsp90·ATP strongly accelerated fibril formation (see Fig. 7). This effect was not observed when the non-hydrolysable nucleotide analogue AMPPNP was used instead of ATP. Also, the Hsp90-specific ATPase inhibitor 17-AAG counteracted the effect of ATP. These results indicate that the observed effect was related to ATP hydrolysis. Thus, in the presence of ATP, Hsp90 promoted amyloidogenesis, whereas in the absence of ATP, fibril formation was suppressed. To reconcile these two opposites, we hypothesized that, in the absence of ATP, Hsp90 might stabilize soluble pre-fibrillar oligomeric AS intermediates, which might not accumulate in the presence of ATP due to their accelerated conversion into fibrils.
In agreement with this notion, partition analysis/SDS-PAGE showed that during the agitation, AS remained soluble exclusively in the presence of ATP-free Hsp90, whereas it accumulated into the unsoluble fraction in the presence of Hsp90·ATP and in the complete absence of Hsp90 (supplemental Fig. S7). Furthermore, in the absence of ATP we noticed an increase of AS turbidity despite the efficient attenuation of fibrillogenesis by Hsp90 (supplemental Fig. S8).
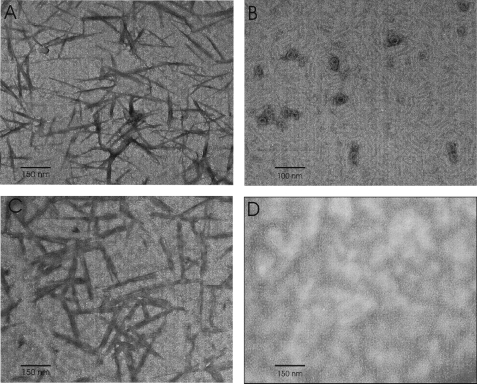
We addressed this issue in more detail by TEM imaging. Under our experimental fibril growth conditions, AS assembled into needle-like fibers with a size of 100–300 nm (Fig. 8 A). In accordance to the ThioT data, these were also obtained in the presence of Hsp90·ATP (Fig. 8C). In the presence of Hsp90 alone, fibrils were no longer detectable. Instead, granular species with a radius of 10–20 nm appeared (Fig. 8B). In agreement, oligomers with a mean Stokes radius of 14.8 nm were observed in solution by DLS (Table 1 and supplemental Fig. S9), whereas soluble species of similar size were not detectable in preparations of mature fibrils (data not shown).
FIGURE 8.
TEM images of AS (A), AS:Hsp90 = 1:2 (B), AS:Hsp90:MgATP = 1:2:10 (C), and Hsp90 (D) (×33,000 magnification).
TABLE 1.
Stokes radius r for 50 μm monomeric AS, 50 μm Hsp90, and soluble oligomers obtained by agitation of AS in the presence of Hsp90 (AS:Hsp90 = 1:2)
For preparations of mature fibrils, little residual monomeric AS and no higher sized species were detectable in solution after sample centrifugation (data not shown).
| Stokes radius r | |
|---|---|
| nm | |
| Monomeric AS | 3.0 ± 0.2a |
| Hsp90 | 7.1 ± 0.6 |
| AS agitated in the presence of Hsp90 | 14.8 ± 1.7 |
a Value reported in literature: 3.2 nm (41).
As the current opinion is that Hsp90 stabilizes protein folding intermediates pending ATP-driven maturation (42), we investigated the effect of ATP on the Hsp90-stabilized oligomers. To this end, we added ATP to an oligomeric solution generated by agitation of AS and Hsp90 (see “Experimental Procedures”). This provoked the conversion of oligomers into fibrils (Fig. 9). This effect was reduced in the presence of AMPPNP and 17-AAG (Fig. 9B). We conclude that, in the presence of ATP, Hsp90 restored the conversion of soluble intermediate oligomers into mature fibrils.
FIGURE 9.
A, kinetics of the oligomer to fibril transition of pre-fibrillar Hsp90-stabilized AS oligomers in the absence (■) and presence (●) of MgATP. B, effects of MgATP, AMPPNP, and 17-AAG on oligomer to fibril transition (t = 140 h). Measured values are averages including S.E. of three independent determinations. C, TEM image of pre-fibrillar AS oligomers generated by agitating an AS:Hsp90 solution for 1 week. D, amyloid fibrils obtained after MgATP was added to the same oligomeric solution.
We finally asked ourselves whether an interaction existed between Hsp90 and mature AS fibrils. For this purpose, we employed a pulldown affinity capture assay, in which Hsp90 was bound to Ni-CAM beads via its N-terminal His6 tag, and aliquots of AS withdrawn during different stages of fibril formation were added. Fig. 2 shows binding of Hsp90 to AS as fibril formation proceeds. The ability of Hsp90 to capture AS decreased as the amount of fibrillar fraction increased. AS was not recovered out of aliquots containing mature fibrils. In contrast, a modest amount was recovered from an aliquot drawn preceding fibrillary growth, suggesting an inverse correlation between Hsp90 binding and fibril progression. Therefore, Hsp90-binding AS species were present predominantly in the early phase preceding the onset of fibrillogenesis, then decrease as soon as AS interconverted into the fibrillar species. This suggests that mature fibrils do not bind to Hsp90 and is consistent with the addition of Hsp90 being ineffective to an advanced fibril solution (data not shown).
DISCUSSION
One major challenge in understanding Parkinson disease is to pinpoint the factors regulating the conformational flexibility and the assembly of its signature protein AS. The predisposition of AS to adopt different conformations leads to the key question of how the cell controls folding of this polypeptide.
Lipid binding and amyloid formation are two hallmark properties of AS, which are tightly coupled to folding of this polypeptide both in vivo and in vitro. Using a reconstituted in vitro system, we showed that Hsp90 affects regions in AS that are responsible for vesicle binding and amyloid formation, consequently interfering with both processes. We could further show that Hsp90 promotes fibril formation in an ATP-dependent manner via oligomeric intermediates, which we were able to resolve in vitro due to their accumulation in the absence of ATP.
It was shown, separately, for AS and Hsp90 that both proteins are related to vesicular trafficking, although with opposite roles. Whereas Hsp90 is an essential component of intracellular vesicle transport (43), AS has a deleterious effect by directly inhibiting lipid docking to Golgi membranes (44). Recently, Liu et al. (21) demonstrated that Hsp90 regulates the rab11a-mediated recycling of AS. Our results support a functional tie between these two proteins, whereupon Hsp90 binds to AS and thus suppresses vesicle binding.
The mechanisms controlling oligomerization and amyloid formation of AS in vivo are scarcely explored. Based on the present data, we propose a model (Fig. 10), where in a first step Hsp90 binds to and restricts conformational fluctuations of monomeric AS, most likely by stabilizing a partially folded ensemble that then favors the further assembly of AS. According to NMR data, Hsp90 seems to interact primarily with the highly hydrophobic fibril forming NAC region of AS (see Fig. 5), as the majority of the affected residues was situated in this particular region. Further changes observed by NMR around the charged NAC region might correspond to secondary binding events, in accordance to the high flexibility of this polypeptide, which, because of its intrinsically unstructured nature, might be reshaped to generate a Hsp90-binding surface patchwork.
FIGURE 10.
An in vitro model for the Hsp90 modulated AS assembly. In the presence of ATP, Hsp90 promotes the rapid conversion of monomeric AS into fibrils. Stable oligomeric intermediates do not accumulate (a). In the absence of ATP, Hsp90 stabilizes intermediate pre-fibrillar oligomers, which consequently accumulate in solution (b). The addition of ATP restores their conversion into unsoluble fibrils (c).
Our model further proposes that, as a consequence of AS binding, Hsp90 promotes fibril maturation in an ATP-dependent manner via an intermediate oligomeric pathway. In the presence of ATP, oligomers do not accumulate due to their rapid conversion into fibrils (Fig. 10, a), whereas in the absence of ATP, the oligomer to fibril transition (Fig. 10, c) is stalled. This leads to populating of the intermediate oligomeric state (Fig. 10, b), which consequently can be detected empirically (TEM and DLS). We speculate that in the cell this state might be evoked by ATPase modulating co-chaperones. Under given circumstances, inhibition (p23 or HOP) might promote soluble oligomers, whereas stimulation (Aha1) might trigger amyloid fibrils.
As the addition of ATP to Hsp90-stabilized AS oligomers restores their conversion into mature fibrils (Fig. 10, c), we conclude that, rather than “dead-end” aggregates, these oligomers represent pre-fibrillar intermediates located on the AS amyloid pathway. This supports the current view of Hsp90 to stabilize intermediately folded polypeptides until maturation (42).
Given that AS fibrils are a signature of Parkinson disease, one question emerging in the light of our results is whether Hsp90 favors Parkinson disease by promoting fibril formation. Against this hypothesis, growing indications show that the neurotoxic agents of the disease are not the amyloid fibrils. Instead, the toxicity is ascribed to soluble, intermediate AS-oligomers that accumulate in vivo (8, 9, 45). Under these premises, Hsp90 might accomplish a scavenging role by favoring the conversion of potentially dangerous intermediate oligomers into less noxious fibrils. Alternatively, Hsp90 might maintain a correct in vivo homeostasis of AS by discriminating “productive” from “non-productive” and potentially lethal AS intermediates in collaboration with other chaperones or degrading enzymes. Strong support comes from the observation that (a) the chaperone Hsp70 also influences fibril formation (28, 46, 47), and (b) the ubiquitin ligase CHIP acts for the disposal of toxic AS aggregates (48). Because both proteins cooperate with Hsp90, pathologic uncoupling of folding and degradation would contribute to the accumulation of a noxious AS phenotype, and finally to the outbreak of Parkinson disease. Although the in vivo validity of our model remains to be established, such an early acting chaperone identifies the particular susceptibility of AS to misfolding and misassembly, which implies a stringent requirement to prevent such derangements in the cell.
In vivo, substrate binding and maturation probably depends on conformational rearrangements that Hsp90 undergoes during the ATPase cycle (40). However, it remains elusive how these events are coupled mechanistically.
We could not discriminate an immediate influence of the nucleotide at the level of the AS·Hsp90 complex. This apparent indifference might be explained by the observation that conformational changes of Hsp90 are only weakly linked to ATP hydrolysis (49). This might impede the efficient coupling of ATP turnover and AS binding.
Supplementary Material
Acknowledgments
We thank Johannes Buchner for expert support, Bettina Richter for TEM imaging, Michael Uhl for DLS support, and Karl Gruber for constructive discussions.
This work was supported by Austrian Science Fund number P20020 (to K. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Equations 1–3 and Figs. S1–S11.
- AS
- α-synuclein
- Hsp90
- heat shock protein 90
- ThioT
- ThioflavinT
- SUVs
- small unilamellar vesicles
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- DPH
- diphenyl-1,3,5-hexatriene
- TEM
- transmission electron spectroscopy
- DLS
- dynamic light scattering
- 17-AAG
- 17-(allylamino)-17-demethoxygeldanamycin
- NAC region
- non-Aβ component region
- AMPPNP
- adenosine 5′-(β,γ-imido)triphosphate
- HSQC
- heteronuclear single quantrum coherence.
REFERENCES
- 1.Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M. (1997) Nature 388, 839–840 [DOI] [PubMed] [Google Scholar]
- 2.Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6469–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer K. (2006) Acta Neuropathol. 112, 237–251 [DOI] [PubMed] [Google Scholar]
- 4.Uversky V. N. (2007) J. Neurochem. 103, 17–37 [DOI] [PubMed] [Google Scholar]
- 5.Chandra S., Chen X., Rizo J., Jahn R., Südhof T. C. (2003) J. Biol. Chem. 278, 15313–15318 [DOI] [PubMed] [Google Scholar]
- 6.Vilar M., Chou H. T., Lührs T., Maji S. K., Riek-Loher D., Verel R., Manning G., Stahlberg H., Riek R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8637–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright J. A., Wang X., Brown D. R. (2009) FASEB J. 23, 2384–2393 [DOI] [PubMed] [Google Scholar]
- 8.Lashuel H. A., Hartley D., Petre B. M., Walz T., Lansbury P. T., Jr. (2002) Nature 418, 291. [DOI] [PubMed] [Google Scholar]
- 9.Pountney D. L., Voelcker N. H., Gai W. P. (2005) Neurotox. Res. 7, 59–67 [DOI] [PubMed] [Google Scholar]
- 10.Walter S., Buchner J. (2002) Angew. Chem. Int. Ed. Engl. 41, 1098–1113 [DOI] [PubMed] [Google Scholar]
- 11.Barral J. M., Broadley S. A., Schaffar G., Hartl F. U. (2004) Semin. Cell. Dev. Biol. 15, 17–29 [DOI] [PubMed] [Google Scholar]
- 12.Cantuti-Castelvetri I., Klucken J., Ingelsson M., Ramasamy K., McLean P. J., Frosch M. P., Hyman B. T., Standaert D. G. (2005) J. Neuropathol. Exp. Neurol. 64, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 13.McLean P. J., Kawamata H., Shariff S., Hewett J., Sharma N., Ueda K., Breakefield X. O., Hyman B. T. (2002) J. Neurochem. 83, 846–854 [DOI] [PubMed] [Google Scholar]
- 14.Klucken J., Shin Y., Masliah E., Hyman B. T., McLean P. J. (2004) J. Biol. Chem. 279, 25497–25502 [DOI] [PubMed] [Google Scholar]
- 15.Rekas A., Adda C. G., Andrew Aquilina J., Barnham K. J., Sunde M., Galatis D., Williamson N. A., Masters C. L., Anders R. F., Robinson C. V., Cappai R., Carver J. A. (2004) J. Mol. Biol. 340, 1167–1183 [DOI] [PubMed] [Google Scholar]
- 16.Uryu K., Richter-Landsberg C., Welch W., Sun E., Goldbaum O., Norris E. H., Pham C. T., Yazawa I., Hilburger K., Micsenyi M., Giasson B. I., Bonini N. M., Lee V. M., Trojanowski J. Q. (2006) Am. J. Pathol. 168, 947–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClellan A. J., Scott M. D., Frydman J. (2005) Cell 121, 739–748 [DOI] [PubMed] [Google Scholar]
- 18.Dickey C. A., Kamal A., Lundgren K., Klosak N., Bailey R. M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C. B., Patterson C., Dickson D. W., Nahman N. S., Jr., Hutton M., Burrows F., Petrucelli L. (2007) J. Clin. Invest. 117, 648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang J., Clark-Dixon C., Wang S., Flower T. R., Williams-Hart T., Zweig R., Robinson L. C., Tatchell K., Witt S. N. (2008) Hum. Mol. Genet. 17, 3784–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y., Gu G., Goodlett D. R., Zhang T., Pan C., Montine T. J., Montine K. S., Aebersold R. H., Zhang J. (2004) J. Biol. Chem. 279, 39155–39164 [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Zhang J. P., Shi M., Quinn T., Bradner J., Beyer R., Chen S., Zhang J. (2009) J. Neurosci. 29, 1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narhi L., Wood S. J., Steavenson S., Jiang Y., Wu G. M., Anafi D., Kaufman S. A., Martin F., Sitney K., Denis P., Louis J. C., Wypych J., Biere A. L., Citron M. (1999) J. Biol. Chem. 274, 9843–9846 [DOI] [PubMed] [Google Scholar]
- 23.Nomanbhoy T. K., Cerione R. (1996) J. Biol. Chem. 271, 10004–10009 [DOI] [PubMed] [Google Scholar]
- 24.Lakowicz J. R. (1999) Principles of Fluorescence Spectroscopy, 2nd Ed., Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 25.Grimminger V., Richter K., Imhof A., Buchner J., Walter S. (2004) J. Biol. Chem. 279, 7378–7383 [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 27.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 28.Dedmon M. M., Christodoulou J., Wilson M. R., Dobson C. M. (2005) J. Biol. Chem. 280, 14733–14740 [DOI] [PubMed] [Google Scholar]
- 29.Vaughan C. K., Gohlke U., Sobott F., Good V. M., Ali M. M., Prodromou C., Robinson C. V., Saibil H. R., Pearl L. H. (2006) Mol. Cell 23, 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulte T. W., Neckers L. M. (1998) Cancer Chemother. Pharmacol. 42, 273–279 [DOI] [PubMed] [Google Scholar]
- 31.Richter K., Soroka J., Skalniak L., Leskovar A., Hessling M., Reinstein J., Buchner J. (2008) J. Biol. Chem. 283, 17757–17765 [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin S. H., Smith H. W., Jackson S. E. (2002) J. Mol. Biol. 315, 787–798 [DOI] [PubMed] [Google Scholar]
- 33.Eliezer D., Kutluay E., Bussell R., Jr., Browne G. (2001) J. Mol. Biol. 307, 1061–1073 [DOI] [PubMed] [Google Scholar]
- 34.Dedmon M. M., Patel C. N., Young G. B., Pielak G. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12681–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodner C. R., Dobson C. M., Bax A. (2009) J. Mol. Biol. 390, 775–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gall C., Xu H., Brickenden A., Ai X., Choy W. Y. (2007) Protein Sci. 16, 2510–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeVine H., 3rd (1999) Methods Enzymol. 309, 274–284 [DOI] [PubMed] [Google Scholar]
- 38.Wandinger S. K., Richter K., Buchner J. (2008) J. Biol. Chem. 283, 18473–18477 [DOI] [PubMed] [Google Scholar]
- 39.Southworth D. R., Agard D. A. (2008) Mol. Cell 32, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hessling M., Richter K., Buchner J. (2009) Nat. Struct. Mol. Biol. 16, 287–293 [DOI] [PubMed] [Google Scholar]
- 41.Yamin G., Uversky V. N., Fink A. L. (2003) FEBS Lett. 542, 147–152 [DOI] [PubMed] [Google Scholar]
- 42.Pratt W. B., Morishima Y., Osawa Y. (2008) J. Biol. Chem. 283, 22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lotz G. P., Brychzy A., Heinz S., Obermann W. M. (2008) J. Cell Sci. 121, 717–723 [DOI] [PubMed] [Google Scholar]
- 44.Gitler A. D., Bevis B. J., Shorter J., Strathearn K. E., Hamamichi S., Su L. J., Caldwell K. A., Caldwell G. A., Rochet J. C., McCaffery J. M., Barlowe C., Lindquist S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Outeiro T. F., Putcha P., Tetzlaff J. E., Spoelgen R., Koker M., Carvalho F., Hyman B. T., McLean P. J. (2008) PLoS One 3, e1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang C., Cheng H., Hao S., Zhou H., Zhang X., Gao J., Sun Q. H., Hu H., Wang C. C. (2006) J. Mol. Biol. 364, 323–336 [DOI] [PubMed] [Google Scholar]
- 47.Luk K. C., Mills I. P., Trojanowski J. Q., Lee V. M. (2008) Biochemistry 47, 12614–12625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tetzlaff J. E., Putcha P., Outeiro T. F., Ivanov A., Berezovska O., Hyman B. T., McLean P. J. (2008) J. Biol. Chem. 283, 17962–17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mickler M., Hessling M., Ratzke C., Buchner J., Hugel T. (2009) Nat. Struct. Mol. Biol. 16, 281–286 [DOI] [PubMed] [Google Scholar]
- 50.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.