Abstract
The Arabidopsis thaliana L. genome contains 58 membrane proteins belonging to the mitochondrial carrier family. Two mitochondrial carrier family members, here named AtNDT1 and AtNDT2, exhibit high structural similarities to the mitochondrial nicotinamide adenine dinucleotide (NAD+) carrier ScNDT1 from bakers' yeast. Expression of AtNDT1 or AtNDT2 restores mitochondrial NAD+ transport activity in a yeast mutant lacking ScNDT. Localization studies with green fluorescent protein fusion proteins provided evidence that AtNDT1 resides in chloroplasts, whereas only AtNDT2 locates to mitochondria. Heterologous expression in Escherichia coli followed by purification, reconstitution in proteoliposomes, and uptake experiments revealed that both carriers exhibit a submillimolar affinity for NAD+ and transport this compound in a counter-exchange mode. Among various substrates ADP and AMP are the most efficient counter-exchange substrates for NAD+. Atndt1- and Atndt2-promoter-GUS plants demonstrate that both genes are strongly expressed in developing tissues and in particular in highly metabolically active cells. The presence of both carriers is discussed with respect to the subcellular localization of de novo NAD+ biosynthesis in plants and with respect to both the NAD+-dependent metabolic pathways and the redox balance of chloroplasts and mitochondria.
Introduction
Nucleotides are metabolites of enormous importance for all living cells. They are the essential building blocks for DNA and RNA synthesis, energize most anabolic and many catabolic reactions, and fulfill critical functions in intracellular signal transduction (1, 2). Moreover, nucleotides serve as cofactors for a wide number of enzymes and are, with water, the most highly connected compounds within the metabolic network (3). Among these co-factors nicotinamide adenine dinucleotides are widely used for reductive/oxidative processes, playing important roles in the operation and control of a wide range of dehydrogenase activities. Accordingly, nucleotides are essential in nearly all cell organelles, and transport of these solutes into mitochondria, plastids, the endoplasmic reticulum, the Golgi apparatus, and peroxisomes has been observed (4–7).
Two types of nucleotide transport proteins have been identified to date at the molecular level: nucleotide transporter (NTT)2 type carriers and members of the mitochondrial carrier family. The former transporters occur in plastids from all plants (8) and in a limited number of intracellular pathogenic bacteria (9). Most NTT-type carrier proteins catalyze an ATP/ADP+Pi counter-exchange mode of transport (10–13), but several bacterial NTT proteins mediate either H+/nucleotide transport or NAD+/ADP counter-exchange (12, 14, 15). With the exception of the bacterial NAD+/ADP carrier (14), all NTT proteins exhibit 12 predicted trans-membrane domains, whereas none of the NTT proteins share structural or domain similarities to members of the mitochondrial carrier family (11).
Carriers belonging to the mitochondrial carrier family (MCF) represent the second group of nucleotide transporters (16, 17). The most prominent member is the mitochondrial ADP/ATP carrier AAC (4), but MCF-type nucleotide transporters have also been identified in peroxisomes (7, 18), in plastids (19, 20), and in the endoplasmic reticulum of higher plants (21). The transport modes catalyzed by MCF-type adenylate nucleotide transporters range from typical ADP/ATP counter-exchange in mitochondria (4) to ATP/AMP exchange in peroxisomes (7, 18) and in Arabidopsis mitochondria (22) range from ADP-glucose/ADP exchange in maize endosperm amyloplasts (23) to unidirectional adenylate export from plastids (20, 23).
Recently, the first NAD+ transporting MCF type carrier was identified in bakers' yeast (24). This carrier, named ScNDT, resides in the mitochondrial envelope, exhibits a high affinity for NAD+, and presumably provides the mitochondrial matrix with NAD+ to meet the requirements of several luminal enzymes (24). The bakers' yeast genome harbors two NDT isoforms, but to date transport specificity was only determined for ScNDT1 (24). Interestingly, although NAD+ transport into purified plant mitochondria has long been observed (25–27), the corresponding carrier protein has not yet been identified at the molecular level. Moreover, plant plastids also harbor NAD+-dependent enzymes (28), and similarly to the situation for plant mitochondria, no corresponding transport protein has been defined that would be capable of delivering NAD+ to these organelles.
The Arabidopsis genome harbors as many as 58 genes coding for MCF-type carrier proteins and three genes coding for NTT type proteins (29–31). In recent years several of these transporters have been cloned and investigated at the molecular and biochemical level (22, 31–34). These studies revealed that some members of the MCF in plants can be present in the plastid membranes (35–37). However, despite the research efforts of many laboratories, the transport functions of most members of the family remain, as yet, unknown.
The three Arabidopsis NTT type carriers have been documented to catalyze adenylate transport but do not accept NAD+ as substrate (Ref. 38 and data not shown). For this reason, we screened the Arabidopsis genome for the presence of homologues to the ScNDT proteins and performed localization using GFP fusions to two candidate genes as well as characterization of their biochemical properties after proteoliposome reconstitution. To the best of our knowledge, this report documents the first molecular description of the plant proteins proposed to be responsible for NAD+ uptake into both mitochondria and plastids and as such identify enigmatic transporters of vast importance for both metabolic and redox-mediated control of cellular processes.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
Arabidopsis plants were grown on soil in a greenhouse for a relatively short photoperiod (10 h light at 23 °C/14 h dark at 20 °C) under low light (100 μmol of photons m−2 s−1) and at 40–65% relative humidity. A short photoperiod and low light intensities are mandatory for large Arabidopsis plants.
Sequence Analysis
Multiple alignments of amino acid sequences from ScNDT1 and ScNDT2 and the plant homologues available at ARAMEMNON (30) were obtained using ClustalX2 (39).
Cloning and Transient Expression of GFP Fusion Constructs
The green fluorescent protein fusion constructs were prepared by amplification of the complete coding region of Atndt1 and Atndt2 using forward primer SK43-XbaI 5′- CGTTCAGATTCTAGAGATGTCCG-3′ (Atndt2, SK45-XbaI 5′-TTCTAGGGTCTAGAGATGATTGAA-3′) and reverse primer SK44-XhoI 5′-GAGCTTTGCTCGAGAGGTATATG-3′ (Atndt2, SK46-XhoI 5′-TTTATTTGCTCTCGAGAGGGATAT-3′), respectively. Both primers included restriction sites for an “in-frame” insertion into pGFP2 (40).
Protoplasts were prepared from tobacco plants (Nicotiana tabacum cv. W38) grown under sterile conditions as described previously (40). Protoplasts were transformed with column-purified plasmid DNA (30 μg/0.5 × 106 cells). After 18 or 36 h of incubation in the dark at 22 °C, protoplasts were analyzed for green fluorescence using a Zeiss Axiovert 200M fluorescence microscope (Carl Zeiss AG, Jena, Germany). GFP was excited at 488 nm, and emission was detected using a Zeiss digital camera AxioCam-MRm equipped with a 505–530-nm bandpass filter and a Plan Neofluar 40×/1.3 oil objective.
Generation of Promoter-β-glucuronidase (GUS) Plants and Staining for GUS Activity
For generation of promoter-GUS constructs, the promoter regions of about 1100 bp of the Atndt1 and Atndt2 genes (including 15 bp of the coding region) were amplified by PCR from genomic DNA, and the obtained PCR products were subcloned in T7 orientation into SmaI restricted pBSK (Stratagene). Both pBSK constructs were restricted with HindIII and SmaI, and the promoter inserts were further introduced in-frame into the binary vector pGPTV (41). For the Atndt1 construct, the following primers were used: 5′-ATGGTTATCGATGTCAAAGTTGTGATATG-3′ (forward) and 5′-GGAGGATGAGAATCCCGGGACATCTCTTGG-3 (reverse); for the Atndt2 construct, primers 5′-GAATCGAGTGAAGCTATTTCCATAAGC-3′ (forward) and 5′-ATACTCCGGTAATCCCGGGTAGAGTTCCCA-3′ (reverse) were used. The Atndt1- and Atndt2-promoter-GUS plasmids were used for Agrobacterium tumefaciens transformation. Transformation of Arabidopsis thaliana (ecotype Columbia) was conducted according to the floral dip method (42).
Seedlings or tissues from transgenic Atndt1- or Atndt2-promoter-GUS plants were collected in glass scintillation vials filled with ice-cold 90% acetone and incubated for 20 min at room temperature. For histochemical localization of GUS activity, the plant material was infiltrated for 30 min with 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid in staining buffer medium and subsequently stained according to a standard protocol (43). Images were taken using a Leica MZ10F stereo microscope (Leica Microsystems, Wetzlar, Germany) equipped with a Leica digital camera DFC 420 C.
Bacterial Expression and Purification of AtNDT1 and AtNDT2
The coding sequences of At2g47490 for AtNDT1 and At1g25380 for AtNDT2 (accession numbers NM_130317 and NM_102349, NCBT RefSeq, respectively) were amplified by PCR from A. thaliana root cDNA preparation. The oligonucleotide primers were synthesized corresponding to the extremities of the coding sequences, with additional BamHI and HindIII (for AtNDT1) or NdeI and EcoRI (for AtNDT2) restriction sites as linkers (AtNDT1, TAGGGATCCATGTCCGCTAATTCTCATCCTCC (forward) and CGAAAGCTTTTAAAGTATAGAGCTTTGCTCAGAAGGTATAT (reverse); AtNDT2, TGAGGATCCCATATGATTGAACATGGGAACTCTACC (forward) and CGAGAATTCTTATTTGCTTCCAAGAGGGATATG (reverse)). The amplified products were cloned into the Escherichia coli pRUN expression vector and transformed into E. coli DH5α cells. Transformants were selected on 2× YT plates containing ampicillin (100 μg/ml) and screened by direct colony PCR. The sequences of inserts were verified.
The expression of recombinant proteins was carried out at 37 °C in E. coli strain C0214(DE3) (44). Control cultures with the empty vector were processed in parallel. Inclusion bodies were purified on a sucrose density gradient (45) and washed at 4 °C, first with TE buffer (10 mm Tris/HCl, 1 mm EDTA, pH 7.0), then twice with a buffer containing Triton X-114 (3%, w/v), 1 mm EDTA, and 10 mm PIPES, pH 7.0, and finally with 10 mm PIPES, pH 7.0. The proteins were solubilized in 1.6% sarkosyl (w/v). Small residues were removed by centrifugation (258,000 × g for 20 min at 4 °C). Proteins were separated by SDS-PAGE and stained with Coomassie Blue dye. The N termini were sequenced, and the yield of purified proteins was estimated by laser densitometry of stained samples (44).
Reconstitution of AtNDT1 and AtNDT2 into Liposomes and Transport Measurements
The recombinant proteins in sarkosyl were reconstituted into liposomes in the presence of substrates, as described previously (46). External substrate was removed from proteoliposomes on Sephadex G-75 columns, pre-equilibrated with 50 mm NaCl and 10 mm PIPES at pH 7.0 (buffer A). The amount of protein incorporated into liposomes was measured as described previously (44). Transport at 25 °C was started by adding [3H]NAD (PerkinElmer Life Sciences) or [14C]AMP (Amersham Biosciences) to substrate-loaded proteoliposomes and terminated after the desired time by the addition of 20 mm pyridoxal 5′-phosphate and 20 mm bathophenanthroline. In controls, the inhibitors were added at the beginning together with the labeled substrate. Entrapped radioactivity was quantified (46). The experimental values were corrected by subtracting control values.
The initial transport rate was calculated from the radioactivity taken up by proteoliposomes in the linear range of substrate uptake. For efflux measurements, proteoliposomes containing 2 mm NAD+ or AMP were labeled with 5 μm [3H]NAD+ or [14C]AMP by carrier-mediated exchange equilibration (46). After 30 min for AtNDT1 and 10 min for AtNDT2, the external radioactivity was removed by passing the proteoliposomes through Sephadex G-75 columns. Efflux was started by adding unlabeled external substrate or buffer A alone and terminated by the addition of the inhibitors indicated above.
Complementation of a Yeast Mutant Lacking Mitochondrial NAD+ Transport Capacity (Δndt1Δndt2) by AtNDT1 and AtNDT2
The pYES2-AtNDT1 and pYES2-AtNDT2-short (lacking the last 47 C-terminal amino acids) plasmids were constructed by cloning the coding sequences of both carriers into the yeast pYES2 expression vector (Invitrogen) under the control of the inducible Gal1 promoter. Δndt1Δndt2 Saccharomyces cerevisiae cells (24) were transformed with the above plasmids and grown in liquid synthetic minimal medium or synthetic complete medium supplemented with 2% ethanol and with auxotrophic nutrients. Mitochondria were isolated as described previously (24) from cells grown until an optical density of 1.0 was reached, and the intramitochondrial NAD+ was assayed by a standardized method (47).
RESULTS
Identification of Two Homologues to ScNDT in A. thaliana
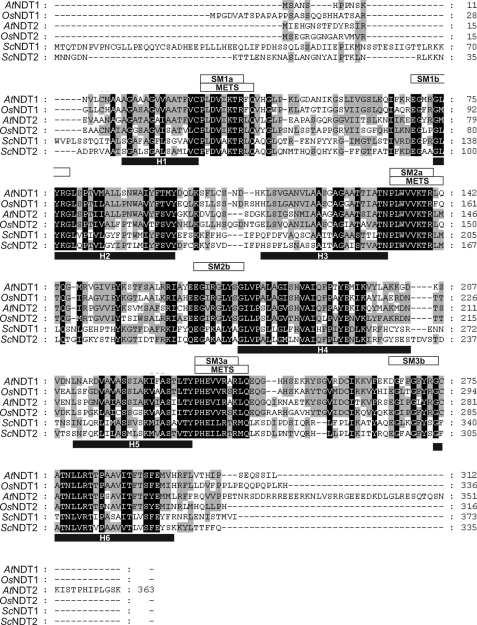
The Arabidopsis genome harbors 58 genes coding for MCF-type carrier proteins (29, 31). A detailed search for proteins in Arabidopsis exhibiting structural homology to the NDT type carriers previously identified in bakers' yeast revealed that two transporters show a substantial degree of similarity. AtNDT1 (At2g47490) comprises 312 amino acids in length leading to a calculated molecular mass of 33.9 kDa. AtNDT1 exhibits 52% similar- and 34% identical amino acids when compared with the NAD+ carrier NDT1 form yeast (Fig. 1). The isoform AtNDT2 (At1g25380) comprises 363 amino acids in length, leading to a calculated molecular mass of 39.5 kDa. AtNDT2 exhibits 46% similar and 28% identical amino acids when compared with ScNDT1 (Fig. 1).
FIGURE 1.
Alignment of the predicted amino acid sequences of AtNDT1 and AtNDT2 with NDT homologues. The residues identical or similar among all family members are indicated by black shading, and the residues conserved by three proteins are indicated by gray shading. Solid black bars underline six putative membrane-spanning helices (H1–H6). Three conserved mitochondrial energy transfer signatures (mitochondrial energy transfer signature = PX(DE)XX(RK)X(RK)) after each odd membrane-spanning helix are marked by white bars. Boxes above the sequences indicate part a and part b of the 3-fold repeated signature motive (synthetic minimal medium) characteristic of the MCF proteins. The numbers indicate the amino acid positions. AtNDT1, nicotinamide adenine dinucleotide transporter 1 from A. thaliana (NCB accession no. AAC62861); AtNDT2, nicotinamide adenine dinucleotide transporter2 from A. thaliana (NCB accession no. AAP42759); OsNDT1, nicotinamide adenine dinucleotide transporter1 from O. sativa (NCB accession no. AAV43947.1); OsNDT2, nicotinamide adenine dinucleotide transporter2 from O. sativa (NCB accession no. BAD73272.1); ScNDT1, nicotinamide adenine dinucleotide transporter1 from S. cerevisiae (NCB accession no. NP_012260); ScNDT2, nicotinamide adenine dinucleotide transporter2 from S. cerevisiae (NCB accession no. NP_010910).
Bioinformatics analysis revealed that the amino acid sequences of the two novel Arabidopsis proteins show the main characteristics of all members of the mitochondrial carrier family, namely a hydropathy profile of a six transmembrane protein (according to transmembrane prediction programs, TmMulticon, (30)) and the presence of a 3-fold repeated METS domain representing mitochondrial energy transfer signatures (assessed by Interpro analysis) (Fig. 1). These signature motifs are also present in the two NDT homologues in bakers' yeast (24). Moreover, the occurrence of NDT carriers is not limited to the dicotyledonous species A. thaliana as homologous proteins are also present in the monocotyledonous species Oryza sativa (Fig. 1).
Before the first predicted transmembrane domains, both AtNDT1 and AtNDT2 exhibited comparable short N-terminal extensions (Fig. 1). The N-terminal extension of AtNDT2 is, according to the ChloroP_V1.1 prediction server, proposed to represent a putative mitochondrial transit peptide, whereas in the case of AtNDT1, a localization in the plant endomembrane system may be assumed (30). The C-terminal extension of AtNDT1 is 45 amino acids shorter than that of AtNDT2 (Fig. 1). Without considering these 45 residues, AtNDT1 and AtNDT2 share 61% identical amino acids.
Bacterial Expression of AtNDT1 and AtNDT2
Reconstitution of recombinant proteins in proteoliposomes is a method frequently used to identify transport properties of uncharacterized carrier proteins. Thus, we expressed the open reading frames of At2g47490 and At1g25380 in E. coli CO214(DE3) cells (see supplemental Fig. 1, lanes 4 and 7, respectively). The gene products of At2g47490 and At1g25380 accumulated as inclusion bodies and were purified by centrifugation and washing. The apparent molecular masses of the purified proteins were about 35.0 and 40.5 kDa for AtNDT1 and AtNDT2 (supplemental Fig. 1, lanes 5 and 8; yield 60–80 mg/liter), in good agreement with their respective molecular masses. The identities of both recombinant proteins were further confirmed by N-terminal sequencing. The recombinant proteins were not detected in bacteria harvested immediately before induction of expression (supplemental Fig. 1, lane 3 for AtNDT1 and lane 6 for AtNDT2) nor in cells harvested after induction but lacking the coding sequence in the corresponding expression vector (supplemental Fig. 1, lane 2 for AtNDT1, and data not shown for AtNDT2).
Identification of Counter-exchange Substrates for AtNDT1 and AtNDT2
In the search for potential substrates of AtNDT1 and AtNDT2, we based our choice of metabolites on the fact that these proteins are related to ScNDT1, which has been demonstrated to be the NAD+ transporter in mitochondria from bakers' yeast (24). Proteoliposomes reconstituted with recombinant AtNDT1 and AtNDT2 catalyzed an [3H]NAD+/NAD+ homo-exchange that was completely inhibited by a mixture of pyridoxal 5′-phosphate and bathophenanthroline (data not shown). However, they did not catalyze homo-exchanges of pyruvate, malate, oxoglutarate, glutamate, or carnitine (data not shown). Importantly, no [3H]NAD+/NAD+ exchange activity was detected when AtNDT1 or AtNDT2 had been inactivated by boiling before incorporation into liposomes or when proteoliposomes were reconstituted with sarkosyl-solubilized protein from bacterial cells either lacking the expression vector for AtNDT1 or AtNDT2 or harvested immediately before induction of expression.
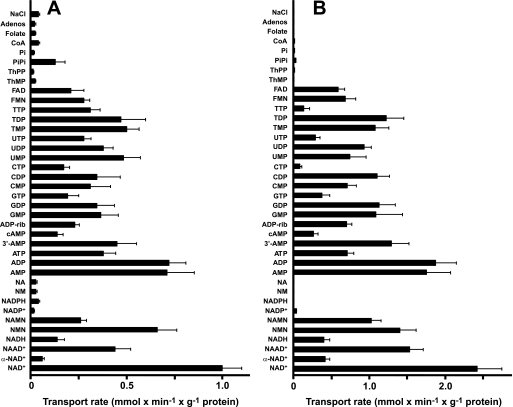
The substrate specificities of reconstituted AtNDT1 and AtNDT2 were examined in depth by measuring the rates of [3H]NAD+ uptake into proteoliposomes that had been preloaded with various potential substrates (Fig. 2). With both proteins, [3H]NAD+ exchanged not only with itself (homo-exchange) but also with some intraliposomal NAD+ analogues (i.e. nicotinic acid adenine dinucleotide, nicotinamide mononucleotide, and nicotinic acid mononucleotide; hetero-exchange) and several nucleotides of the bases A, G, C, U, and T (Fig. 2). In contrast, the uptake of radioactively labeled NAD+ was low in the presence of internal α-NAD+, NADH, and cAMP. Both carriers accept FAD and FMN as poor counter-exchange substrates (Fig. 2), but pyrophosphate is solely used by AtNDT1 and not by AtNDT2 (Fig. 2). Negligible NAD+ uptake rates were measured with internal NADP+, NADPH, nicotinamide, nicotinic acid, adenosine, thiamine mono- or diphosphate, inorganic phosphate, coenzyme A, folate, NaCl (Fig. 2), and (not shown) malate, malonate, citrate, fumarate, aspartate, glutamate, S-adenosylmethionine, lysine, arginine, and ornithine did not serve as suitable counter-exchange substrates. Among intraliposomal nucleotides, adenine nucleotides were exchanged more effectively than those of the other bases. For each type of nucleotide, nucleoside mono- and diphosphates were more effective than nucleoside triphosphates, especially in the case of AtNDT2 (Fig. 2B). It is worth mentioning that similar results were obtained by measuring the uptake of [14C]AMP instead of [3H]NAD+ under the same experimental conditions of Fig. 2 (data not shown).
FIGURE 2.
Dependence on internal substrate of the transport properties of proteoliposomes reconstituted with recombinant AtNDT1 (A) and AtNDT2 (B). Proteoliposomes were preloaded internally with various substrates (each concentration 10 mm). Transport was started by adding 0.6 mm and 0.15 mm [3H]NAD+ for AtNDT1 and AtNDT2, respectively. The reaction time was 2 min (AtNDT1) and 45 s (AtNDT2). Data are the means ± S.D. of at least three independent experiments. Adenos, adenosine; ADP-rib, ADP-ribose; NA, nicotinic acid; ThMP, thiamine monophosphate; ThPP, thiamine pyrophosphate; NM, nicotinamide.
Effect of Inhibitors on AtNDT1- and AtNDT2-mediated NAD+ Transport
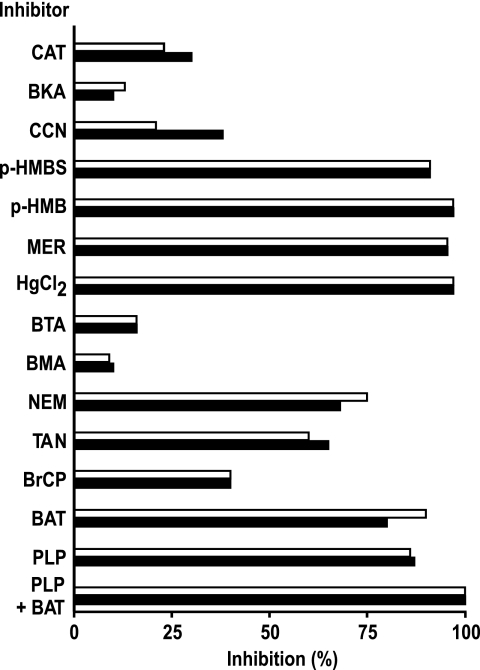
The [3H]NAD+/NAD+ exchange reactions catalyzed by reconstituted AtNDT1 and AtNDT2 were inhibited strongly by pyridoxal 5′-phosphate, bathophenanthroline, and mercurials (HgCl2, mersalyl, p-hydroxymercuribenzoate, and p-hydroxymercuribenzoate sulfonate), inhibitors of several mitochondrial carriers, and partially by bromcresol purple and tannic acid (inhibitors of the glutamate carrier (48)) as well as by N-ethylmaleimide (Fig. 3). In contrast, little inhibition was observed with butylmalonate, 1,2,3-benzenetricarboxylate, and bongkrekate (powerful inhibitors of other mitochondrial carriers). It is notable that carboxyatractyloside and α-cyano-4-hydroxycinnamate, at concentrations that completely inhibit the mitochondrial ADP/ATP- and the pyruvate carrier, respectively, had a small effect on the activity of AtNDT1 and a slightly greater effect on the activity of AtNDT2 (Fig. 3). The inhibitor sensitivity of AtNDT1 and AtNDT2, therefore, resembles that of ScNDT1 but is not identical.
FIGURE 3.
Effect of inhibitors on the [3H]NAD+/NAD+ exchange by reconstituted AtNDT1 and AtNDT2. Proteoliposomes were preloaded internally with 10 mm NAD+. Transport was initiated by adding 0.6 and 0.15 mm [3H]NAD+ for AtNDT1 (white bars) or AtNDT2 (black bars), respectively. The reaction time was 2 min (AtNDT1) and 45 s (AtNDT2), respectively. Thiol reagents were added 2 min before the labeled substrate; the other inhibitors were added together with the labeled substrate. The final concentrations of the inhibitors were 20 mm (PLP, pyridoxal 5′-phosphate; BAT, bathophenanthroline), 0.2 mm (MER, mersalyl; p-HMB, p-hydroxylmercuribenzoate; p-HMBS, p-hydroxymercuribenzensulfonate), 25 μm HgCl2, 2 mm (BM, butylmalonate; BTA, 1,2,3-benzenetricarboxylate), 0.3 mm (BrCP, bromcresol purple), 1 mm (NEM, N-ethylmaleimide; CCN, α-cyano-4-hydroxycinnamate), 0.2% (TAN, tannic acid), and 10 μm (BKA, bongkrekic acid; CAT, carboxyatractyloside). The extents of inhibition (%) for each carrier from a representative experiment are given.
Kinetic Characteristics of Recombinant AtNDT1- and AtNDT2 Proteins
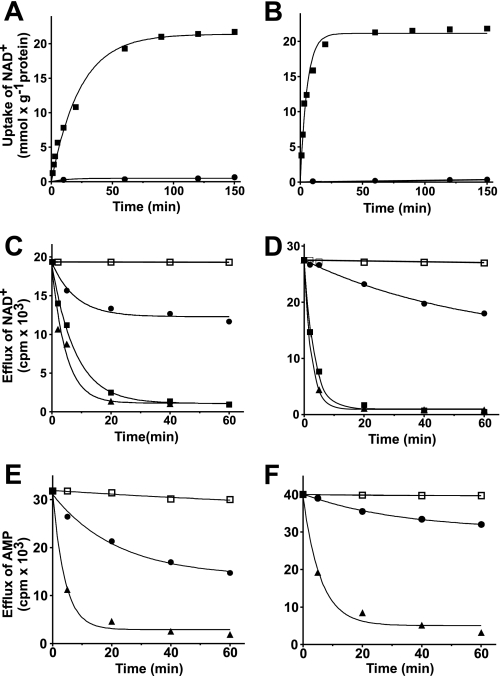
In Fig. 4, A and B, the [3H]NAD+ transport kinetics are compared for proteoliposomes measured either as uniport (in the absence of internal NAD+) or as exchange (in the presence of 10 mm internal NAD+). The exchange reactions catalyzed by both AtNDT1 and AtNDT2 followed first-order kinetics, isotopic equilibrium being approached exponentially (Fig. 4, A and B). The rate constants and the initial rates of NAD+ exchange deduced from the time-courses (46) were 0.04 and 0.18 min−1 and 0.90 and 3.78 mmol/min × g protein for AtNDT1 and for AtNDT2, respectively. In contrast, the uniport uptake of NAD+ by both carriers was negligible (Fig. 4, A and B).
FIGURE 4.
Kinetics of exchange reactions catalyzed by AtNDT1 and AtNDT2. Proteoliposomes were reconstituted with the recombinant AtNDT1 (A, C, and E) or AtNDT2 (B, D, and F). A and B, 0.6 mm [3H]NAD was added to proteoliposomes containing 10 mm NAD+ (■) or 10 mm NaCl (●). Similar results were obtained in three independent experiments. C, D, E, and F, the internal substrate of proteoliposomes (2 mm NAD+ in C and D or 2 mm AMP in E and F) was labeled with [3H]NAD (C and D) or [14C]AMP (E and F) by carrier-mediated exchange equilibration. After removal of the external substrate by Sephadex G-75, the efflux of [3H]NAD (C and D) or [14C]AMP (E and F) was started by adding buffer A alone (●), 5 mm NAD+ (▴), 5 mm AMP (■) or 5 mm NAD+, 20 mm pyridoxal 5′-phosphate, and 20 mm bathophenanthroline in buffer A (□). Similar results were obtained in three independent experiments. S.E. was always below 8% of the given value.
The uniport mode of transport was further investigated by measuring the efflux of [3H]NAD+ or [14C]AMP from proteoliposomes preloaded with these compounds. This experimental approach provides a more sensitive assay for unidirectional transport (46). With both reconstituted carriers, AtNDT1 and AtNDT2, little efflux of [3H]NAD+ or [14C]AMP was observed in the absence of external substrate, whereas substantial efflux occurred upon the addition of external NAD+ or AMP (Fig. 4, C–F). Both efflux processes, i.e. those with and without external substrate, were prevented completely by the presence of the inhibitors bathophenanthroline and pyridoxal 5′-phosphate (Fig. 4, C–F).
The kinetic constants of recombinant purified AtNDT1 and AtNDT2 were determined from the initial transport rates at various external [3H]NAD+ concentrations in the presence of a constant saturating internal NAD+ concentration of 10 mm. The half-saturation constant (Km) and specific activity (Vmax) values for NAD+/NAD+ exchange at 25 °C were 0.24 ± 0.04 mm and 1.41 ± 0.18 mmol/min × g protein for AtNDT1 and 0.15 ± 0.01 mm and 4.76 ± 0.75 mmol/min × g protein for AtNDT2, respectively (mean values of more than 60 experiments). The Vmax of AtNDT2 was, therefore, 3.4-fold greater than that of AtNDT1, and the Km of AtNDT2 for external NAD+ was nearly half that of AtNDT1.
Several external substrates were competitive inhibitors of AtNDT1 and AtNDT2 (Table 1), as they increased the apparent Km without changing the Vmax of the [3H]NAD+/NAD exchange (not shown). These results demonstrate that AtNDT1 has a greater affinity than AtNDT2 for all nucleotides of the purine and pyrimidine bases investigated herein. The inhibition constants (Ki) of AMP and ADP for both AtNDT1 and AtNDT2 were lower than those of the other purine and pyrimidine nucleotides. Moreover, the Ki of AtNDT1 and AtNDT2 for FMN and FAD (about 0.6 and 1.2 mm, respectively) were lower than those expected on the basis of their modest ability (at 10 mm concentration) to exchange with NAD+ (see Fig. 2). The latter findings suggest that these flavin adenine dinucleotides either have a rather high affinity for AtNDT1 and AtNDT2, although they are poorly transported, or exert an inhibitory effect on these proteins at high concentrations. In addition, in agreement with the results of Table 1, no inhibition was observed by the simultaneous addition of 5 mm CTP, UTP, TTP, or pyrophosphate with the labeled substrate on the AtNDT2-reconstituted [3H]NAD+/NAD+ exchange activity measured under the conditions described in Table 1.
TABLE 1.
Competitive inhibition by various substrates of [3H]NAD+ uptake into proteoliposomes reconstituted with AtNDT1 and AtNDT2
The Ki values were calculated from Lineweaver-Burk plots of the rate of [3H]NAD+ versus substrate concentrations. The competing substrates at appropriate constant concentrations were added together with 45–2000 μm [3H]NAD+ to proteoliposomes containing 10 mm NAD+. The values are the means of at least three independent experiments. ND, not determined.
| Substrate |
Ki |
|||
|---|---|---|---|---|
|
AtNDT1 |
AtNDT2 |
|||
| mm | ||||
| AMP | 0.12 | 0.02 | 0.38 | 0.04 |
| ADP | 0.36 | 0.05 | 1.50 | 0.10 |
| ATP | 0.70 | 0.10 | 6.80 | 0.80 |
| 3′-AMP | ND | ND | 2.10 | 0.30 |
| GMP | 0.80 | 0.20 | 3.00 | 0.50 |
| GDP | 1.20 | 0.10 | 3.10 | 0.40 |
| GTP | 3.10 | 0.50 | 9.20 | 1.70 |
| CMP | 1.10 | 0.20 | 3.20 | 0.50 |
| CDP | 2.00 | 0.30 | 3.90 | 0.50 |
| CTP | 3.40 | 0.50 | >10.00 | |
| UMP | 0.90 | 0.10 | 7.80 | 1.20 |
| UDP | 1.90 | 0.20 | 7.20 | 1.00 |
| UTP | 3.20 | 0.40 | >10.00 | |
| TMP | 0.28 | 0.04 | 4.70 | 0.60 |
| TDP | 0.60 | 0.10 | 4.90 | 0.70 |
| TTP | 2.50 | 0.50 | >10.00 | |
| FMN | 0.60 | 0.10 | 0.70 | 0.10 |
| FAD | 1.20 | 0.20 | 1.10 | 0.20 |
| NAAD | 0.50 | 0.10 | 1.80 | 0.30 |
| NMN | 1.30 | 0.30 | 3.30 | 0.40 |
| Pyrophosphate | 2.80 | 0.40 | >10.00 | |
Complementation of the Yeast Δndt1Δndt2 Double Mutant with AtNDT1 and AtNDT2
The S. cerevisiae ndt1Δndt2 strain lacks mitochondrial NAD+ uptake capacity and showed a growth delay on nonfermentable substrates which was more pronounced in the synthetic minimal medium than in the yeast-peptone medium (24). We, therefore, checked whether the introduction of each of the two Arabidopsis genes, At2g47490 (AtNDT1) or At1g25380 (AtNDT2), in S. cerevisiae Δndt1Δndt2 double mutant strain reversed the growth defect.
The Δndt1Δndt2 cells transformed with the empty pYES2 vector and grown on synthetic minimal medium supplemented with 2% ethanol exhibited a lower exponential growth rate (resulting in a 3-fold increase in doubling time) and a lower growth plateau at the stationary phase as compared with wild-type cells transformed with the same empty vector (data not shown). The growth parameters were completely recovered when the double mutant cells were transformed with the vector carrying the Arabidopsis carrier AtNDT1 or AtNDT2-short (without the 47 C-terminal amino acids) (data not shown). With full-length AtNDT2 only a partial recovery of the growth rate and the doubling time of the yeast mutant were obtained. The different effect between the full-length and truncated version of AtNDT2 is probably due to a toxic effect of the corresponding C-terminal extension. This possibility is substantiated by the finding that Δndt1Δndt2 cells transformed with AtNDT2 (but not with AtNDT2-short) did not grow on synthetic complete medium supplemented with 2% glucose or 2% galactose (data not shown).
Subsequently, we checked whether AtNDT1 or AtNDT2 short expression was able to increase the mitochondrial NAD+ content of the Δndt1Δndt2 strain, which is much lower than that of the wild-type yeast strain (24). The mitochondrial NAD+ content of the double mutant strain increased severalfold upon expression of Arabidopsis AtNDT1 or AtNDT2 short (Fig. 5). Taken together these results indicate that both Arabidopsis carriers are able to complement the phenotype of the S. cerevisiae cells devoid of their NAD+ mitochondrial transporters.
FIGURE 5.
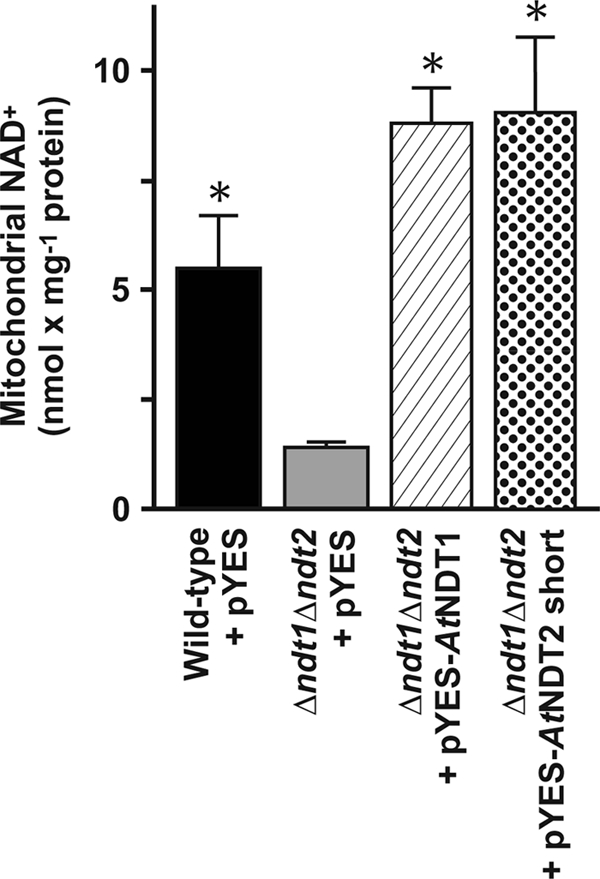
Effect of AtNDT1 and AtNDT2 expression on the mitochondrial NAD+ content of the Δndt1Δndt2 double mutant yeast cells. Mitochondria were isolated from wild-type + pYES2 (black bar), Δndt1Δndt2 + pYES2 (gray bar), Δndt1Δndt2 + pYES-AtNDT1 (hatched bar), and Δndt1Δndt2 + pYES-AtNDt2 short (dotted bar) cells grown on synthetic complete medium supplemented with 2% ethanol and 0.4% galactose. Data are the means ± S.D. of at least three independent experiments. The asterisk indicates significant differences in the mitochondrial NAD+ content as compared with that of Δndt1Δndt2 + pYES cells.
Subcellular Localization of AtNDT1-GFP and AtNDT2-GFP
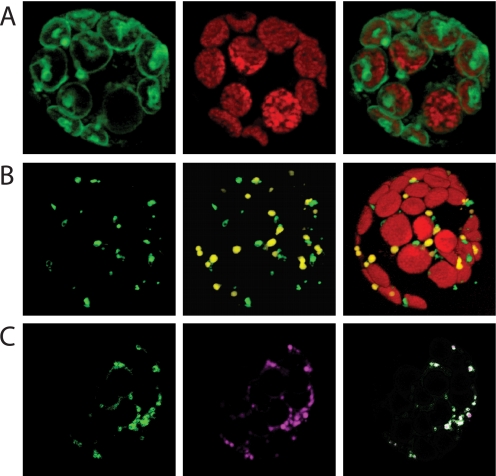
To provide experimental evidence on the subcellular localization of both carrier proteins, we generated corresponding GFP fusions and expressed these recombinant proteins in tobacco mesophyll protoplasts. The AtNDT1-GFP protein locates to the chloroplast membrane (Fig. 6A, left panel). This interpretation is based on the observation that the large green fluorescing organelle is fully congruent with the red autofluorescence of the chloroplasts (Fig. 6A, middle panel) and is further confirmed by the merge (Fig. 6A, right panel).
FIGURE 6.
Localization of AtNDT1- and AtNDT2-GFP proteins in tobacco leaf protoplasts. Tobacco protoplasts were prepared as given under “Experimental Procedures” and transformed using polyethylene glycol. Fluorescence was visualized after 18 h of incubation by fluorescence microscopy. Chloroplasts were visualized by their autofluorescence. A, shown is localization of AtNDT1-GFP. B, shown are localization of AtNDT2-GFP and cotransformation with SKL-DSred. C, shown is the localization of AtNDT2-GFP and coincubation with the Mito Tracker dye.
In the case of AtNDT2-GFP, the green fluorescence appears in organelles clearly smaller than the large red fluorescing chloroplasts (Fig. 6B, left panel). By use of further GFP fusion proteins we moreover excluded that AtNDT2-GFP resides in either Golgi vesicles or the endoplasmic reticulum (data not shown). To verify whether these small organelles represent either mitochondria or peroxisomes, we additionally expressed a GFP derivate carrying the peroxisomal targeting signal SKL (SKL22::DsRed, kindly provided by Dr. Ian Small) and used the Mito Tracker dye. A comparison between the AtNDT2-GFP fluorescence (Fig. 6B, left) and the GFP-SKL fluorescence in peroxisomes (Fig. 6B, middle panel) shows that AtNDT2-GFP does not reside in peroxisomes (Fig. 6B, middle and right panels). However, AtNDT2-GFP caused fluorescence (Fig. 6C, left), and the Mito Tracker labeled organelles (Fig. 6C, middle) merged perfectly (Fig. 6C, right) further underlining the mitochondrial localization of this carrier.
Expression Pattern of the Atndt1 and Atndt2 Gene
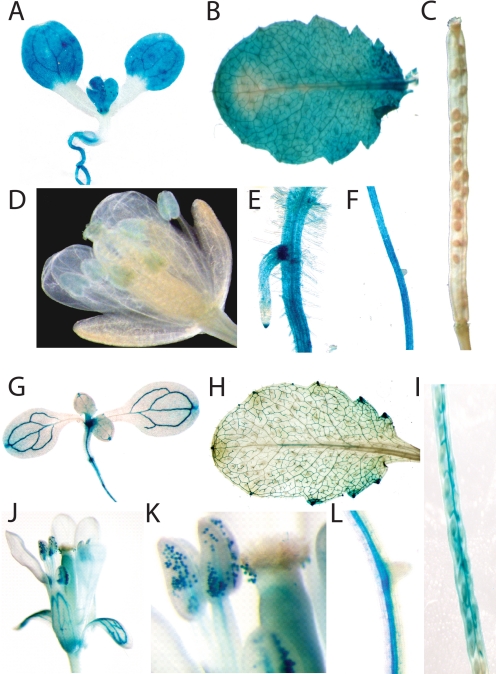
To evaluate the expression pattern of the Atndt1 and Atndt2 genes, we constructed corresponding promoter-GUS reporter plants. For each construct we generated 25 independent transgenic lines; representative data are shown in Fig. 7A–L. Atndt1-promoter-GUS activity is comparably high in young leaf mesophyll cells (Fig. 7, A and B), absent in developing siliques and seeds (Fig. 7C), low in all flower tissues (Fig. 7D), and high in root tips and at the branches of adventitious roots (Fig. 7E). Atndt2-promoter-GUS activity is remarkably high in the rapidly developing young meristematic shoot area (Fig. 7G), in vascular bundles (veins) from young and old leaves (Fig. 7, G and H), in developing siliques including the funiculi structures (Fig. 7I), in petal veins (Fig. 7J), in developing pollen (Fig. 7K), and in the central cylinder of Arabidopsis roots (Fig. 7L).
FIGURE 7.
Histochemical localization of GUS expression under control of the Atndt1 and Atndt2 promoter in A. thaliana. Plants were grown as described under “Experimental Procedures.” A, shown is Atndt1-promoter-GUS activity in a 10-day-old seedling. B, shown is Atndt1-promoter-GUS activity in a juvenile leaf from a 7-week-old plant. C, shown is Atndt1-promoter-GUS activity in developing siliques. D, shown is Atndt1-promoter-GUS activity in flowers. E and F, shown are Atndt1-promoter-GUS activity in root tips and root branches. G, shown is Atndt2-promoter-GUS activity in 2-week-old seedlings. H, shown is Atndt2-promoter-GUS activity in a mature leaf. I, shown is Atndt2-promoter-GUS activity in developing siliques. J, shown is Atndt2-promoter-GUS activity in flowers. K, shown is Atndt2-promoter-GUS activity in anthers. L, shown is Atndt2-promoter-GUS activity in roots.
DISCUSSION
Two different types of NAD+ transporting carrier proteins have been identified to date; the are the bacterial NTT-type carrier NTT4 from the bacterium Protochlamydia amoebophila and the MCF type carriers ScNDT1/2 from bakers' yeast cells (14, 24). NTT4-type carriers represent a subgroup of NTT proteins residing in intracellular bacteria or plant plastids, and they all exhibit between 10 to 12 predicted transmembrane domains. By contrast, the Arabidopsis carriers AtNDT1 and AtNDT2 clearly belong to the large MCF proteins. The reason for this conclusion lies not only in the fact that they show a high degree of overall similarity to the MCF carriers ScNDT1 and ScNDT2 (Fig. 1) but also because they exhibit a 3-fold repeated signature motif (Fig. 1). The presence of the 3-fold repeated signature motif, each representing the mitochondrial energy transfer signature (METS, Fig. 1), is a typical feature of transport proteins belonging to the mitochondrial carrier family (29, 49).
The name mitochondrial carrier family proteins already indicates that it was assumed that transporters belonging to this protein group reside in the mitochondrial envelope (50). However, today we know that the presence of MCF proteins is by no means restricted to mitochondrial membranes (51). The recent discovery of MCF-type nucleotide transporters in peroxisomes (7, 18, 52), plant plastids (19, 20, 23), and the endoplasmic reticulum of Arabidopsis (21) strikingly demonstrates that several organelles, especially in plant cells, exploit the large pool of MCF proteins to facilitate cross-membrane nucleotide transport. Thus, with our observation that AtNDT1-GFP resides in the chloroplast envelope (Fig. 6), we provide a further example of a MCF protein that does not locate to mitochondria.
In this context it is somewhat surprising that AtNDT1 does not exhibit an N-terminal-located transit peptide (Fig. 1) usually required for insertion into the plastid envelope (53). However, in recent years multiple examples of chloroplast envelope-located membrane proteins lacking N-terminal-located transit peptides have been reported (54), and about 15% of all proteins, which clearly locate to chloroplasts, lack such N-terminal transit peptide (55). Moreover, truncation experiments showed that the N-terminal-located extension of the plastid triose-phosphate transporter TPT is not required for proper direction into the inner envelope membrane as the required target information is located in the first hydrophobic domain (56).
Clearly, the experimentally conducted subcellular localization of AtNDT1 in chloroplasts (Fig. 6A) contradicts the observation that recombinant expression of this carrier, similar to AtNDT2, complements the absence of NAD+ transport activity in the yeast mutant Δndt1Δndt2 (Fig. 5). In fact, there are several examples that expression of recombinant carriers in yeast might lead to import into a membrane that differs to the authentic situation; e.g. the barley carrier protein HvSUT2 and the Arabidopsis carrier AtSUT4 are located in the plant vacuolar membrane (57), but both carriers mediate sucrose transport across the plasma membrane when heterologously synthesized in bakers' yeast (58, 59). Alternatively, the expression of the plastid triose-phosphate carrier from spinach leads to accumulation of the recombinant protein in yeast endomembranes (60). Obviously, the expression of carriers residing in plant membranes which are absent in yeast cells may provide false information on the subcellular localization. Moreover, it is not totally without precedence that an experimentally confirmed subcellular location of a MCF type carrier differs from that predicted bioinformatically. For example the Brittle1 carrier from Arabidopsis was predicted to reside to mitochondria but locates, similar to the homologue in maize, in the plastid envelope (19, 23).
Most, but not all (61), mitochondrial solute carriers in yeast and humans lack N-terminal-located transit peptides. In contrast, MCF homologues in plants show such extensions to their amino acid sequences (62). Although the N-terminal extension of AtNDT2, located in front of the first proposed transmembrane domain, is quite short (Fig. 1); the prediction programs locate this protein with high probability to the mitochondrial envelope (consensus prediction: 5.4 mitochondria to 0.5 endomembranes, no chloroplast prediction (30)). Thus, our experimental data suggesting that AtNDT2-GFP resides in mitochondria (Fig. 6, B and C) confirms the predicted subcellular localization of the authentic protein. In sum, we cannot as yet explain how the plant ensures correct targeting of AtNDT1 and AtNDT2 into their final cellular destinations, but is seems likely that internal domains in the structural parts of the carriers influence this process.
Without considering the 45-amino acid extension at the C-terminal end of AtNDT2 (Fig. 1), both carriers share 61% identical amino acids. However, it is not possible to make reliable assumptions on the substrate specificity or on the transport modes on basis of the amino acid similarity. Therefore, we decided to analyze the biochemical properties of both proteins in a reconstituted system. Similar to a range of other MCF carriers, AtNDT1 and AtNDT2 appear as inclusion bodies after recombinant synthesis in E. coli (supplemental Fig. 1, lanes 4 and 7). Such inclusions, however, are advantageous because they allow the enrichment of corresponding carriers to apparent homogeneity by centrifugation and washing (supplemental Fig. 1, lanes 5 and 8). Both AtNDT1 and AtNDT2 transport NAD+ in a counter-exchange mode across the liposomal membrane (Figs. 2 and 4, A–F). This counter-exchange mode of transport, which prefers NAD+ to its structural homologue α-NAD+ (Fig. 2), resembles that observed for the yeast homologue ScNDT1 (24). Given that NADH appeared as a low efficient counter-exchange substrate (Fig. 2) and knowing that cellular NAD+ levels exceed those of NADH several-hundredfold (63–65), binding of NADH to both types of carriers will only occur very rarely, excluding the importance of NADH transport under physiological conditions.
AtNDT1 and AtNDT2 share a number of similar transport properties; for example, both proteins accept AMP and ADP as highly efficient counter-exchange substrates for NAD+ as well as, although at a lower extent, all other RNA nucleotides tested but not NADP+, NADPH+, nicotinamide (NM), or nicotinic acid (NA) (Fig. 2), and they both respond similarly to all inhibitors tested (Fig. 3). Their transport affinities (Km) for NAD+ are lower than that of ScNDT1, and their specific activities (Vmax) values are similar or higher than those displayed by most mitochondrial carriers characterized so far (16, 17). However, AtNDT1 and AtNDT2 differ for their kinetic constants, AtNDT2 being more active and exhibiting a lower affinity for purine and pyrimidine nucleotides (Table 1). A major individual feature of the chloroplast carrier AtNDT1 is the acceptance of pyrophosphate as a counter-exchange substrate for NAD+ (Fig. 2). In the case of chloroplasts, a slow import of pyrophosphate has been observed (66) which is supposed to provide net phosphate into young plastids during increase of their volume. The Ki value (2.8 mm) of AtNDT1 for pyrophosphate (Table 1) is in line with a substantial cytosolic pyrophosphate level found in plant tissue (67). It remains to be shown whether AtNDT1 imports pyrophosphate during plastid development.
The close biochemical similarities between AtNDT1 and AtNDT2 are understandable given the commonality of their gene structures; both genes (Atndt1 and Atndt2) share a highly similar exon/intron structure (30). Therefore, we assume that they derive from a common molecular ancestor which would additionally explain similarities in their biochemical properties. However, it appears likely that after gene duplication, independent cellular evolution took place allowing the development of individual properties such as the different subcellular localization (Fig. 6, A and B) and different affinities for pyrophosphate and nucleotides (Fig. 2 and Table 1).
In plants, de novo synthesis of NAD+ takes place in the cytosol (68). Thus, newly synthesized NAD+ must enter cell organelles to supply NAD+ required for many enzymatic reactions. The need for NAD+ availability in mitochondria is clear from the presence of dehydrogenase-containing pathways like the tricarboxylic acid cycle or glycine oxidation as well as the highly active formate dehydrogenase (1). However, the requirement for the presence of NAD+ is not limited to mitochondria as many NAD+-dependent reactions are located in the chloroplast. As just one example, chloroplasts from both C3 and CAM plants possess an active NAD+-malate dehydrogenase (28) that together with the NAD+-dependent chloroplastic glyceraldehyde dehydrogenase (69) forms a highly active enzyme couple during the dark period (70). The subcellular location of AtNDT1 and AtNDT2 (Fig. 6, A and B) observed here and the demonstrated ability of both carriers to transport NAD+ (Fig. 2, Table 1), thus, fits with the requirement for net NAD+ import in both types of organelles.
AtNDT1and AtNDT2 are both able to catalyze a low unidirectional transport (uniport) of NAD+ (Fig. 4, C, and D) in addition to a fast counter-exchange reactions with a number of substrates (Figs. 4, A and B, and 2). However, we assume that in planta a counter-exchange, as opposed to a unidirectional mode of transport, takes place. In the chloroplast stroma, AMP or ADP is available as a counter-exchange substrate because de novo biosynthesis of adenylates is located in this compartment (71). The efflux of AMP or ADP via AtNDT1 would allow NAD+ import into the stroma. In mitochondria, adenylates can be imported unidirectionally by the recently identified carrier ADNT1 (22) and then be exchanged for NAD+ via AtNDT2 (Fig. 2). In this context it appears worth mentioning that the bacterial NAD+ transporter NTT4 from P. amoebophila, although structurally totally unrelated to AtNDT1 and AtNDT2, also transports NAD+ in counter-exchange with ADP (14). It will be interesting in the future to identify the micro-domains in both types of carriers (NTT and NDT) responsible for substrate binding.
Utilizing NAD+/NAD+ homo exchange as an experimental approach to study biochemical properties of both carriers in more detail, we were able to estimate apparent NAD+ affinities of 0.24 mm for AtNDT1 and 0.15 mm for AtNDT2. These affinity values are similar to the yeast carrier ScNDT1 (24) and are almost identical to the NAD+ affinity measured in isolated potato tuber mitochondria (26). In yeast cells NAD+ and NADH sum up to concentrations of between 1 and 3 mm (72), but it should be kept in mind that it is assumed that 90% of all nicotinamide dinucleotides are protein-bound (63). Thus, the apparent Km values measured for AtNDT1 and AtNDT2 can be predicted to allow a substantial influx into the corresponding organelles under in vivo conditions.
According to the subcellular location (in either chloroplasts or mitochondria) and to the proposed function of NDT proteins in Arabidopsis, we have to propose that both carriers must be active in cells exhibiting either high metabolic activities or exhibiting high rates of division. The promoter-GUS reporter plants clearly indicate that the Atndt1 gene expression is high throughout the whole leaf (Fig. 7, A and B). Photosynthetically active leaves represent a tissue with an extraordinary high number of chloroplasts which is in line with the subcellular localization of AtNDT1 (Fig. 6A). However, as yet we do not have an explanation as to why Atndt1 gene expression is additionally remarkably high at the branches of adventitious roots (Fig. 7E). The Atndt2 gene is highly active in young cells and in cells belonging to the vascular bundles. The need for high AtNDT2 activity in young cells is obvious, as in these cells organelles divide rapidly followed by a regain of organelle volume, processes that require vast quantities of NAD+ to support biosynthetic reactions. Vascular bundles are mainly comprised of phloem cells, companion cells, phloem parenchyma cells, and xylem tubes (73). Xylem tubes represent dead cells, and living phloem (sieve) cells do not possess functional mitochondria. Therefore, we speculate that Atndt2 gene expression is most likely high in companion cells. These cells exhibit a surprisingly high metabolic activity to allow active import of assimilates proposed for long distance transport (73), and it is long known that plant mitochondria must import NAD+ to exhibit high metabolic activities (26).
This speculation is strongly supported when the tissue-specific expression of the NDTs is compared with that of genes encoding enzymes of the de novo and salvage pathways of pyridine biosynthesis (Genevestigator). Recent studies of Arabidopsis knock-out mutants of the constituent enzymes of these pathway as well as of the external NAD(P)H dehydrogenases have highlighted the importance of pyridine nucleotides in a broad range of processes including germination (74), bolting (75), adaptation to osmotic stress (76), and senescence (77), suggesting that the NDTs may also play an important role in these processes.
Supplementary Material
This work was supported by a Deutsche Forschungsgemeinschaft Reinhard Koselleck grant (to E. N.), the Ministero dell'Università e della Ricerca, the Center of Excellence in Genomics, and the Italian Human ProteomeNet RBRN07BMCT_009 (to F. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- NTT
- nucleotide transporter
- GFP
- green fluorescent protein
- GUS
- -β-glucuronidase
- MCF
- mitochondrial carrier family
- PIPES
- 1,4-piperazinediethanesulfonic acid
- NDT
- nicotineamide adenine dinucleotide transporter.
REFERENCES
- 1.Heldt H. W. (2005) Plant Biochemistry, Elsevier Academic Press, Burlington, MA [Google Scholar]
- 2.Roux S. J., Steinebrunner I. (2007) Trends Plant Sci. 12, 522–527 [DOI] [PubMed] [Google Scholar]
- 3.Lehninger A. L., Nelson D. L., Cox M. M. (1994) Prinzipien der Biochemie, Spektrum, Akad. Verlag, Heidelberg, Berlin, Oxford [Google Scholar]
- 4.Klingenberg M. (2008) Biochim. Biophys. Acta 1778, 1978–2021 [DOI] [PubMed] [Google Scholar]
- 5.Strotmann H., Berger S. (1969) Biochem. Biophys. Res. Com. 35, 20–26 [DOI] [PubMed] [Google Scholar]
- 6.Abeijon C., Mandon E. C., Hirschberg C. B. (1997) Trends Biochem. Sci. 22, 203–207 [DOI] [PubMed] [Google Scholar]
- 7.Palmieri L., Rottensteiner H., Girzalsky W., Scarcia P., Palmieri F., Erdmann R. (2001) EMBO J. 20, 5049–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linka N., Hurka H., Lang B. F., Burger G., Winkler H. H., Stamme C., Urbany C., Seil I., Kusch J., Neuhaus H. E. (2003) Gene 306, 27–35 [DOI] [PubMed] [Google Scholar]
- 9.Schmitz-Esser S., Linka N., Collingro A., Beier C. L., Neuhaus H. E., Wagner M., Horn M. (2004) J. Bacteriol. 186, 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neuhaus H. E., Emes M. J. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 111–140 [DOI] [PubMed] [Google Scholar]
- 11.Winkler H. H., Neuhaus H. E. (1999) Trends Biochem. Sci. 24, 64–68 [DOI] [PubMed] [Google Scholar]
- 12.Trentmann O., Horn M., van Scheltinga A. C., Neuhaus H. E., Haferkamp I. (2007) PLoS Biol. 5, e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trentmann O., Jung B., Neuhaus H. E., Haferkamp I. (2008) J. Biol. Chem. 283, 36486–36493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haferkamp I., Schmitz-Esser S., Linka N., Urbany C., Collingro A., Wagner M., Horn M., Neuhaus H. E. (2004) Nature 432, 622–625 [DOI] [PubMed] [Google Scholar]
- 15.Haferkamp I., Schmitz-Esser S., Wagner M., Neigel N., Horn M., Neuhaus H. E. (2006) Mol. Microbiol. 60, 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmieri F. (2004) Pflügers Arch. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 17.Palmieri F., Agrimi G., Blanco E., Castegna A., Di Noia M. A., Iacobazzi V., Lasorsa F. M., Marobbio C. M., Palmieri L., Scarcia P., Todisco S., Vozza A., Walker J. (2006) Biochim. Biophys. Acta 1757, 1249–1262 [DOI] [PubMed] [Google Scholar]
- 18.Linka N., Theodoulou F. L., Haslam R. P., Linka M., Napier J. A., Neuhaus H. E., Weber A. P. (2008) Plant Cell 20, 3241–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchberger S., Tjaden J., Neuhaus H. E. (2008) Plant J. 56, 51–63 [DOI] [PubMed] [Google Scholar]
- 20.Leroch M., Kirchberger S., Haferkamp I., Wahl M., Neuhaus H. E., Tjaden J. (2005) J. Biol. Chem. 280, 17992–18000 [DOI] [PubMed] [Google Scholar]
- 21.Leroch M., Neuhaus H. E., Kirchberger S., Zimmermann S., Melzer M., Gerhold J., Tjaden J. (2008) Plant Cell 20, 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmieri L., Santoro A., Carrari F., Blanco E., Nunes-Nesi A., Arrigoni R., Genchi F., Fernie A. R., Palmieri F. (2008) Plant Physiol. 148, 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchberger S., Leroch M., Huynen M. A., Wahl M., Neuhaus H. E., Tjaden J. (2007) J. Biol. Chem. 282, 22481–22491 [DOI] [PubMed] [Google Scholar]
- 24.Todisco S., Agrimi G., Castegna A., Palmieri F. (2006) J. Biol. Chem. 281, 1524–1531 [DOI] [PubMed] [Google Scholar]
- 25.Neuburger M., Douce R. (1983) Biochem. J. 216, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobin A., Djerdjour B., Journet E., Neuburger M., Douce R. (1980) Plant Physiol. 66, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuburger M., Day D. A., Douce R. (1985) Plant Physiol. 78, 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berkemeyer M., Scheibe R., Ocheretina O. (1998) J. Biol. Chem. 273, 27927–27933 [DOI] [PubMed] [Google Scholar]
- 29.Millar A. H., Heazlewood J. L. (2003) Plant Physiol. 131, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W. B., Flügge U. I., Kunze R. (2003) Plant Physiol. 131, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picault N., Hodges M., Palmieri L., Palmieri F. (2004) Trends Plant Sci. 9, 138–146 [DOI] [PubMed] [Google Scholar]
- 32.Catoni E., Schwab R., Hilpert M., Desimone M., Schwacke R., Flügge U. I., Schumacher K., Frommer W. B. (2003) FEBS Lett. 534, 87–92 [DOI] [PubMed] [Google Scholar]
- 33.Palmieri L., Arrigoni R., Blanco E., Carrari F., Zanor M. I., Studart-Guimaraes C., Fernie A. R., Palmieri F. (2006) Plant Physiol. 142, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmieri L., Picault N., Arrigoni R., Besin E., Palmieri F., Hodges M. (2008) Biochem. J. 410, 621–629 [DOI] [PubMed] [Google Scholar]
- 35.Bedhomme M., Hoffmann M., McCarthy E. A., Gambonnet B., Moran R. G., Rébeillé F., Ravanel S. (2005) J. Biol. Chem. 280, 34823–34831 [DOI] [PubMed] [Google Scholar]
- 36.Bouvier F., Linka N., Isner J. C., Mutterer J., Weber A. P., Camara B. (2006) Plant Cell 18, 3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thuswaldner S., Lagerstedt J. O., Rojas-Stütz M., Bouhidel K., Der C., Leborgne-Castel N., Mishra A., Marty F., Schoefs B., Adamska I., Persson B. L., Spetea C. (2007) J. Biol. Chem. 282, 8848–8859 [DOI] [PubMed] [Google Scholar]
- 38.Möhlmann T., Tjaden J., Schwöppe C., Winkler H. H., Kampfenkel K., Neuhaus H. E. (1998) Eur. J. Biochem. 252, 353–359 [DOI] [PubMed] [Google Scholar]
- 39.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wendt U. K., Wenderoth I., Tegeler A., Von Schaewen A. (2000) Plant J. 23, 723–733 [DOI] [PubMed] [Google Scholar]
- 41.Becker D., Kemper E., Schell J., Masterson R. (1992) Plant Mol. Biol. 20, 1195–1197 [DOI] [PubMed] [Google Scholar]
- 42.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 43.Weigel D., Glazebrook J. (2002) Arabidopsis. A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 44.Fiermonte G., Palmieri L., Dolce V., Lasorsa F. M., Palmieri F., Runswick M. J., Walker J. E. (1998) J. Biol. Chem. 273, 24754–24759 [DOI] [PubMed] [Google Scholar]
- 45.Fiermonte G., Walker J. E., Palmieri F. (1993) Biochem. J. 294, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmieri F., Indiveri C., Bisaccia F., Iacobazzi V. (1995) Methods Enzymol. 260, 349–369 [DOI] [PubMed] [Google Scholar]
- 47.Gibon Y., Larher F. (1997) Anal. Biochem. 251, 153–157 [DOI] [PubMed] [Google Scholar]
- 48.Fiermonte G., Palmieri L., Todisco S., Agrimi G., Palmieri F., Walker J. E. (2002) J. Biol. Chem. 277, 19289–19294 [DOI] [PubMed] [Google Scholar]
- 49.Saraste M., Walker J. E. (1982) FEBS Lett. 144, 250–254 [DOI] [PubMed] [Google Scholar]
- 50.Aquila H., Link T. A., Klingenberg M. (1987) FEBS Lett. 212, 1–9 [DOI] [PubMed] [Google Scholar]
- 51.Haferkamp I. (2007) FEBS Lett. 581, 2375–2379 [DOI] [PubMed] [Google Scholar]
- 52.Arai Y., Hayashi M., Nishimura M. (2008) Plant Cell 20, 3227–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Heijne G., Steppuhn J., Herrmann R. G. (1989) Eur. J. Biochem. 180, 535–545 [DOI] [PubMed] [Google Scholar]
- 54.Stengel A., Soll J., Bölter B. (2007) Biol. Chem. 388, 765–772 [DOI] [PubMed] [Google Scholar]
- 55.Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K. J. (2008) PLoS ONE. 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knight J. S., Gray J. C. (1995) Plant Cell 7, 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Endler A., Meyer S., Schelbert S., Schneider T., Weschke W., Peters S. W., Keller F., Baginsky S., Martinoia E., Schmidt U. G. (2006) Plant Physiol. 141, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weschke W., Panitz R., Sauer N., Wang Q., Neubohn B., Weber H., Wobus U. (2000) Plant J. 21, 455–467 [DOI] [PubMed] [Google Scholar]
- 59.Weise A., Barker L., Kühn C., Lalonde S., Buschmann H., Frommer W. B., Ward J. M. (2000) Plant Cell 12, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loddenkötter B., Kammerer B., Fischer K., Flügge U. I. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2155–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zara V., Rassow J., Wachter E., Tropschug M., Palmieri F., Neupert W., Pfanner N. (1991) Eur. J. Biochem. 198, 405–410 [DOI] [PubMed] [Google Scholar]
- 62.Glaser E., Sjöling S., Tanudji M., Whelan J. (1998) Plant Mol. Biol. 38, 311–338 [DOI] [PubMed] [Google Scholar]
- 63.Zhang Q., Piston D. W., Goodman R. H. (2002) Science 295, 1895–1897 [DOI] [PubMed] [Google Scholar]
- 64.Coffe V., Carbajal R. C., Salceda R. (2004) J. Neurochem. 88, 885–890 [DOI] [PubMed] [Google Scholar]
- 65.Schwartz J. P., Passonneau J. V., Johnson G. S., Pastan I. (1974) J. Biol. Chem. 249, 4138–4143 [PubMed] [Google Scholar]
- 66.Lunn J. E., Douce R. (1993) Biochem. J. 290, 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farré E. M., Geigenberger P., Willmitzer L., Trethewey R. N. (2000) Plant Physiol. 123, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noctor G., Queval G., Gakière B. (2006) J. Exp. Bot. 57, 1603–1620 [DOI] [PubMed] [Google Scholar]
- 69.Yonuschot G. R., Ortwerth B. J., Koeppe O. J. (1970) J. Biol. Chem. 245, 4193–4198 [PubMed] [Google Scholar]
- 70.Neuhaus H. E., Schulte N. (1996) Biochem. J. 318, 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zrenner R., Stitt M., Sonnewald U., Boldt R. (2006) Annu. Rev. Plant Biol. 57, 805–836 [DOI] [PubMed] [Google Scholar]
- 72.Lin S. J., Ford E., Haigis M., Liszt G., Guarente L. (2004) Genes Dev. 18, 12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruiz-Medrano R., Xoconostle-Cázares B., Lucas W. J. (2001) Curr. Opin. Plant Biol. 4, 202–209 [DOI] [PubMed] [Google Scholar]
- 74.Hunt L., Holdsworth M. J., Gray J. E. (2007) Plant J. 51, 341–351 [DOI] [PubMed] [Google Scholar]
- 75.Liu Y. J., Nunes-Nesi A., Wallström S. V., Lager I., Michalecka A. M., Norberg F. E., Widell S., Fredlund K. M., Fernie A. R., Rasmusson A. G. (2009) Plant Physiol. 150, 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang G., Pichersky E. (2007) Plant J. 49, 1020–1029 [DOI] [PubMed] [Google Scholar]
- 77.Schippers J. H., Nunes-Nesi A., Apetrei R., Hille J., Fernie A. R., Dijkwel P. P. (2008) Plant Cell 20, 2909–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.