Abstract
Chemokine receptors constitute an attractive family of drug targets in the frame of inflammatory diseases. However, targeting specific chemokine receptors may be complicated by their ability to form dimers or higher order oligomers. Using a combination of luminescence complementation and bioluminescence resonance energy transfer assays, we demonstrate for the first time the existence of hetero-oligomeric complexes composed of at least three chemokine receptors (CCR2, CCR5, and CXCR4). We show in T cells and monocytes that negative binding cooperativity takes place between the binding pockets of these receptors, demonstrating their functional interaction in leukocytes. We also show that specific antagonists of one receptor (TAK-779 or AMD3100) lead to functional cross-inhibition of the others. Finally, using the air pouch model in mice, we show that the CCR2 and CCR5 antagonist TAK-779 inhibits cell recruitment promoted by the CXCR4 agonist SDF-1α, demonstrating that cross-inhibition by antagonists also occurs in vivo. Thus, antagonists of the therapeutically important chemokine receptors regulate the functional properties of other receptors to which they do not bind directly with important implications for the use of these agents in vivo.
Introduction
Chemokines are small chemoattractant cytokines that control a wide variety of biological and pathological processes, ranging from immunosurveillance to inflammation and from viral infection to cancer (1, 2). They mediate their effects by binding to cell surface receptors, which belong to the large family of G protein-coupled receptors (GPCRs).4 Receptor binding initiates a cascade of intracellular events mediated by the association of the receptor with a heterotrimeric G protein, promoting guanine nucleotide exchange in the α subunit and activation of the G protein (3). The G proteins trigger various effector enzymes, leading to chemotaxis and the regulation of a wide range of other functions, which vary in different cell populations. Because of their key role in inflammatory diseases, chemokines and their receptors constitute an attractive family of drug targets. However, the transfer of therapeutic compounds to clinical use has been hampered by the complexity and the functional redundancy of the chemokine system (4). The recent demonstration that chemokine receptors form both homo- and heteromers brings an additional layer of complexity to this system (reviewed in Ref. 5). There is therefore a need for a better understanding of how chemokine receptors are organized and regulated at the supramolecular level in the plasma membrane of primary leukocytes and how this organization affects the activity of agonists and antagonists of these receptors and their subsequent intracellular signaling network.
Homodimerization has been reported for four chemokine receptors: CCR2, CCR5, CXCR2, and CXCR4 (6–12). A recent report suggests that CXCR4 can also form constitutive homo-oligomers composed of at least three protomers (13). Heteromerization has been demonstrated between CCR2 and CCR5 or CXCR4 (6–12). CXCR4 was reported to heterodimerize with CCR2 but not with its closest homologue CCR5 (8), a surprising result given the high sequence similarity between the two CC-receptors, particularly within the transmembrane segments, which have been shown to constitute the dimer interface in other GPCR families (14–16). Indeed, interaction of CXCR4 and CCR5 in the immunological synapse of T cells was recently reported, but the pharmacological consequences of this interaction were not investigated (17). Because CCR2, CCR5, and CXCR4 are frequently expressed at the surface of leukocyte populations, their interactions might affect greatly the signaling of these receptors and the activity of antagonists acting on these important therapeutic targets. Receptor interactions might also impact the role of CCR5 and CXCR4 as HIV co-receptors. In previous studies, we investigated the functional consequences of chemokine receptor dimerization and demonstrated that interaction between CCR2 and CCR5 or CXCR4 resulted in a negative binding cooperativity (10, 18). The consequences of heteromerization on the chemokine binding properties of the receptors were also observed in native cells, demonstrating that such heteromers exist in cells naturally co-expressing these chemokine receptors.
In the present study, we build on these previous findings and show for the first time that CCR2, CCR5, and CXCR4 can form hetero-oligomers containing all three receptors. Consequences of heteromerization for the in vitro and in vivo properties of “selective” antagonists are illustrated.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Human and mouse recombinant chemokines were from R&D Systems. AMD3100 was obtained from Sigma, and TAK-779 was from the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. The anti-CCR5 (2D7) and anti-CXCR4 (12G5) antibodies were from BD Biosciences. The DOC-1 anti-CCR2 antibody was kindly provided by Matthias Mack (University of Regensburg, Germany). Expression of human chemokine receptors was analyzed by fluorescence-activated cell sorting using phycoerythrin-conjugated anti-hCCR2 (FAB151P), anti-hCCR5 (FAB1802P), and anti-hCXCR4 (MAB173) antibodies from R&D Systems. Mouse leukocyte populations were determined using fluorescein isothiocyanate-conjugated anti-mCD11c (HL3, 553801), anti-mCD3 (17A2, 555272), and phycoerythrin-conjugated anti-mCD4 (L3T4, 555308) or anti-mI-A/I-E (M5/114.15.2, 557000) from BD Biosciences. Cell surface expression of mCXCR4 was detected by incubation with an anti-mCXCR4 antibody (MAB21651, R&D Systems) followed by the addition of an allophycocyanin-conjugated anti-rat secondary antibody (112-136-071, ImmunoReseach). Expression of mCCR5 was detected using biotinylated anti-mCCR5 antibody (13-1951, eBioscience) and PerCP/Cy5.5-labeled streptavidin (551419, BD Biosciences).
Cell Lines and Leukocyte Populations
CHO-K1 cells were cultured in Ham's F-12 medium supplemented with 10% fetal bovine serum (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). The CCR5 coding sequence was cloned between the BamHI and XbaI sites of the bicistronic expression vector pEFIB3 (19), and the construct was transfected by FuGENE 6 (Roche Applied Science) into a CHO-K1 cell line expressing apoaequorin, Gα16, and wild-type CXCR4. Cells expressing CCR5 were selected by 10 μg/ml blasticidin (Invitrogen). Human peripheral blood lymphocytes were isolated from buffy coats of healthy blood donors (homozygotes for the wild type or Δ32 alleles of CCR5) by centrifugation on Ficoll. CD4+-T lymphocytes were isolated by negative selection by using a magnetic bead cell sorting kit (130-091-155; Millenyi Biotec, Sunnyvale, CA). After this procedure, CD4+ blasts were generated by incubating the lymphocytes with anti-CD3 (1:100; Janssen, Cilag) and anti-CD28 (1:1000; BD Biosciences) antibodies for 3 days. Cells were maintained in a medium supplemented with human IL-2 (2 ng/ml; R&D Systems) for an additional 7 days. Monocytes were isolated by positive selection using a CD14 magnetic bead cell sorting kit (130-050-201; Millenyi Biotec).
Bioluminescence Resonance Energy Transfer (BRET) Assays
The cDNAs encoding full-length EYFP, monomeric Venus, or Renilla luciferase (RLuc) were fused in frame to the 3′-end of CCR2, CCR5, and CXCR4 in the pcDNA3.1 vector. Similarly, the cDNAs encoding the L1 (amino acids 1–229) or L2 (amino acids 230–311) fragments of RLuc8 were fused in frame to the 3′-end of each receptor. The BRET assays were performed as described previously (10). Briefly, human embryonic kidney cells (HEK-293T) were transfected, using a constant amount of plasmid DNA but various ratios of plasmids encoding the fusion protein partners (29). A control corresponding to mock-transfected cells was included in order to subtract raw basal luminescence and fluorescence from the data. Expression of EYFP or monomeric Venus fusion proteins was estimated by measuring fluorescence at 535 nm following excitation at 485 or 510 nm, respectively. Expression of RLuc fusion proteins was estimated by measuring the luminescence of the cells after incubation with 5 μm coelenterazine H (Promega). Likewise, bimolecular luminescence complementation (BiLC) used for trimer experiments was measured (29). In parallel, BRET was measured as the fluorescence of the cells at 535 nm at the same time points using a Mithras LB940 reader (Berthold) (for the experiment regarding “dimers” or “two-way BRET”) or a Pherastar reader (BMG) for experiments with the trimers or “three-way BRET” because of its higher sensitivity.
Binding Assays
Competition binding experiments were performed as described (10). Membrane preparations were incubated in the assay buffer (50 mm Hepes, pH 7.4, 1 mm CaCl2, 5 mm MgCl2, 0.5% BSA) with 0.1 nm 125I-MCP-1/CCL2, 0.1 nm 125I-MIP-1β, or 0.1 nm 125I-SDF-1α as tracers and variable concentrations of unlabeled competitors. Samples were incubated for 1 h, and the bound tracer was separated by filtration through GF/B filters presoaked in 1% BSA. Filters were counted in a γ-scintillation counter, and binding parameters were determined with the PRISM software (Graphpad Software) using nonlinear regression applied to single-site or two-site binding models. The software compared the sum of square and the degree of freedom of each regression by using the F test and selected the most appropriate equation.
Dissociation Kinetics Experiments
Ligand dissociation experiments were performed as described (18). Membrane preparations were first incubated in assay buffer (50 mm Hepes, pH 7.4, 1 mm CaCl2, 5 mm MgCl2, 0.5% BSA) with 0.1 nm 125I-MIP-1β or 125I-SDF-1α in a final volume of 500 μl. After 1 h, the membranes were centrifuged, and the unbound radioligand was removed by aspiration. The membrane pellet was washed once with assay buffer and resuspended in 2.5 ml of assay buffer, with or without unlabeled ligands. At different time points, aliquots were collected, the bound tracer was separated by filtration through GF/B filters presoaked for 1 h in 1% BSA, and the filters were counted for 1 min in a γ-scintillation counter. In all experiments, the total binding and total tracer remaining at the initiation of the dissociation phase represented less than 10% of the amount of tracer engaged initially. The data are presented as the ratio between bound cpm at the various dissociation time points and total bound cpm at time zero of dissociation. The curves were fitted with the PRISM software (Graphpad Software) using nonlinear regression and a single-phase decay model.
Intracellular Calcium Mobilization Assay
Functional responses were analyzed with an aequorin-based assay as described previously (20). Briefly, cells were incubated for 4 h in the dark in the presence of 5 μm coelenterazine H (Promega Corp.). Variable concentrations of chemokines were added to cell suspension (25,000 cells/well), and luminescence was measured for 30 s in an EG&G Berthold luminometer (PerkinElmer Life Sciences). Half-maximal effective concentrations (EC50) were determined with the PRISM software (GraphPad Software) using nonlinear regression applied to a sigmoidal dose-response model.
Chemotaxis Assay
The migration of CD4+ T cells was performed in 96-well transwell chambers (5-μm pore size; Costar, Corning, NY). The lower compartment of the chambers was loaded with serial dilutions of MCP-1/CCL2, MIP-1β, or SDF-1α in Dulbecco's modified Eagle's medium/F-12 supplemented with 0.1% BSA. The upper compartment was loaded with 5 × 105 cells preincubated or not with specific antagonists (300 nm). Migration was stopped after 1 h, and cells in the lower chamber were counted by using the ATPlite luminescence assay kit (PerkinElmer Life Sciences). The results are expressed as chemotaxis index (i.e. the ratio of cells migrating in response to the chemoattractant over cells migrating toward the medium alone).
Air Pouch Assay
The air pouch assay was performed on 7-week-old BALB/c mice. On days 1 and 3, 3 ml of sterile air was injected under the back skin to create the pouch. On day 6, mice were divided in groups and received an intraperitoneal injection of 0.5 ml of PBS containing or not containing TAK-779 or AMD-3100 (10 mg/kg). Thirty minutes later, SDF-1α (2 μg) was injected into the pouch, alone or together with TAK-779 or AMD-3100 (10 mg/kg). Four hours later, the mice were killed in a CO2 chamber, and the air pouch was washed. The leukocytes were collected, counted, and characterized by flow cytometry. All experimental procedures with animals were conducted according to standard ethical guidelines and approved by the local ethical committee (number 222N- LA1230332).
RESULTS
Homo- and Hetero-oligomerization of CXCR4, CCR5, and CCR2
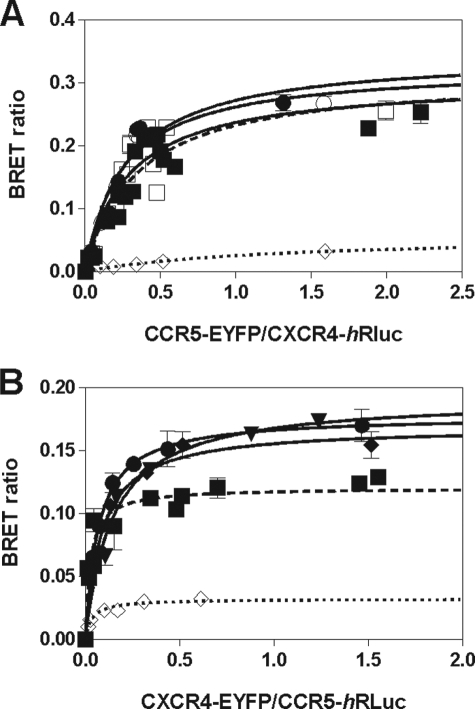
We investigated by BRET experiments the ability of CXCR4 to interact with CCR5 in living cells. A significant energy transfer was observed between CXCR4-hRLuc and CCR5-EYFP as well as between CCR5-hRLuc and CXCR4-EYFP (Fig. 1). As a control for its specificity, the GABAbR2 receptor fused to full-length EYFP was used, which led to a lower energy transfer. When considering the energy transfer between CCR5-hRLuc and CXCR4-EYFP, the addition of chemokine led to an increase of BRETmax of about 50% (p < 0.05) and to a 2–4-fold increase of BRET50 (p < 0.05 and p < 0.001, respectively). In contrast, no significant BRET changes were detected between CXCR4-hRLuc and CCR5-EYFP upon chemokine stimulation (supplemental Table S1). In keeping with our previous studies, these results show that CCR2, CCR5, and CXCR4 form homomers as well as heteromers with one another, raising the question of their natural organization at the surface of immune cells endogenously expressing these three receptors.
FIGURE 1.
Heterodimerization of CCR5 and CXCR4 as measured by BRET. HEK 293T cells were transfected with a constant amount of the CCR5-hRLuc fusion and increasing amounts of the CXCR4-EYFP fusion (A) or a constant amount of the CXCR4-hRLuc fusion and increasing amounts of the CCR2-EYFP fusion (B), and heterodimerization of CCR5 and CXCR4 was investigated by measuring the energy transfer between the two partners. Heterodimerization was investigated in the absence of agonist (■; dotted curve) or 5 min after the addition of 100 nm MIP-1β (□), 100 nm SDF-1α (○), or 50 nm each MIP-1β and SDF-1α (●) at room temperature. As a control, an increasing amount of GABAbR2-EYFP (◇) was used as a BRET acceptor. All data points were performed in triplicate (error bars indicate S.E.). The BRET50 and BRETmax values were calculated by nonlinear regression using a single-site saturation binding model.
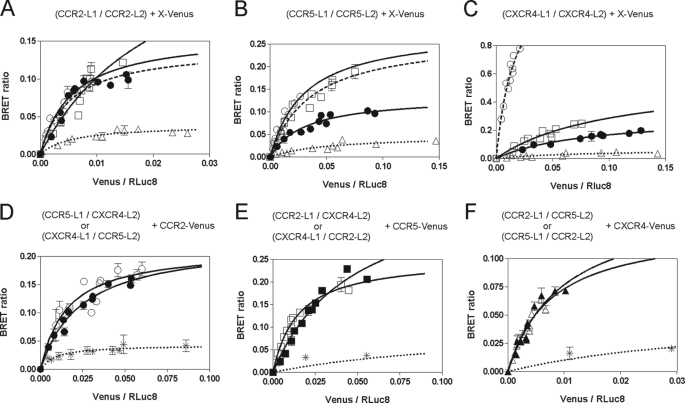
We next investigated the ability of these receptors to form higher order oligomeric structures by using a luminescence complementation BRET assay. The complementation assay is based on the principle that split bioluminescent proteins, such as RLuc, are not luminescent unless the two parts can reconstitute a functional enzyme, as a result of the interaction of the proteins to which they are fused (22). We recently used this assay to demonstrate homo-oligomerization of the dopamine D2 receptor (29). We generated CCR2, CCR5, and CXCR4 receptors fused at their C terminus with the L1 or L2 fragments of RLuc8 (23) (i.e. CCR2-L1 and CCR2-L2, CCR5-L1 and CCR5-L2, or CXCR4-L1 and CXCR4-L2). Coexpression of all of any pair of constructs containing an L1 and L2 fusion led to luminescence, consistent with interaction of the receptors (supplemental Fig. S1). Co-expressing a constant amount of the complementation constructs of each receptor with increasing amounts of the receptors fused to full-length monomeric Venus (24) led to specific and saturable BRET for each combination tested (Fig. 2, A–C). These results demonstrate that the chemokine receptor “homodimers” identified by BiLC can interact with at least one additional receptor of either the same or different nature. In order to better understand the nature of the oligomeric assemblies, we co-expressed constant amounts of heteromeric split RLuc8 constructs (i.e. CCR2-L1 with CCR5-L2, CCR2-L1 with CXCR4-L2, or CCR5-L1 with CXCR4-L2) with increasing amounts of each of the receptors fused to full-length monomeric Venus. Again, the results showed a saturable BRET signal that was independent from the nature of the receptor fused to the L1 and L2 fragments of RLuc (Fig. 2, D–F). As a control for its specificity, the metabotropic glutamate receptor mGluR1 fused to full-length monomeric Venus was used in combination with the various constructs, which led to a lower energy transfer (Fig. 2, A–F). Altogether, these results show that the chemokine receptors CCR2, CCR5, and CXCR4 form homo-oligomers as well as hetero-oligomeric structures containing all three receptors.
FIGURE 2.
Homo- and hetero-oligomerization of CCR2, CCR5, and CXCR4 as measured by a luminescence complementation BRET assay. A–C, the interaction of a chemokine receptor dimer with a third receptor was monitored in HEK 293T cells transfected with a constant amount of CCR2-L1 and CCR2-L2 (A), CCR5-L1 and CCR5-L2 (B), or CXCR4-L1 and CXCR4-L2 (C) as a BRET donor (complemented RLuc) and an increasing amount of either CCR2-Venus (●), CCR5-Venus (□), CXCR4-Venus (○), or metabotropic glutamate receptor mGluR1-Venus (▵) as the acceptor. The BRET signal resulting from the interaction between the partners was recorded. D–F, detection of oligomers involving three different chemokine receptors. HEK 293T cells were transfected with a constant amount of CCR5-L1 and CXCR4-L2 (○), CXCR4-L1 and CCR5-L2 (●), CCR2-L1 and CXCR4-L2 (□), CXCR4-L1 and CCR2-L2 (■), CCR5-L1 and CCR2-L2 (▵), or CCR2-L1 and CCR5-L2 (▴) as donor (complemented RLuc) and an increasing amount of CCR2-Venus (D), CCR5-Venus (E), or CXCR4-Venus (F) as a BRET acceptor, and the BRET signal was recorded. As a control, an increasing amount of metabotropic glutamate receptor mGluR1-Venus (*) was used as a BRET acceptor. All data points were performed in triplicate (error bars indicate S.E.). The BRET50 and BRETmax values were calculated by nonlinear regression using a single-site saturation binding model.
Pharmacological Characterization of CHO-K1 Cells Co-expressing CXCR4 and CCR5
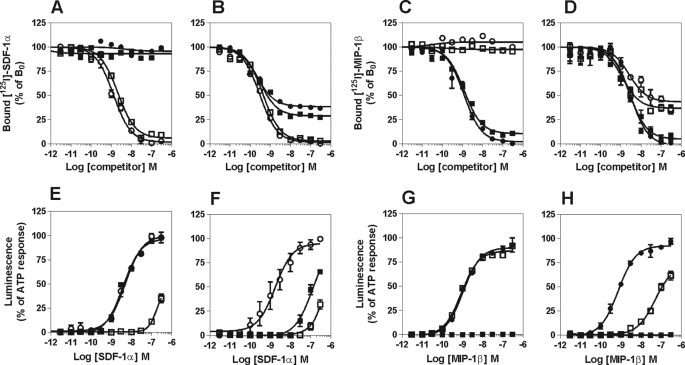
With the aim of investigating the pharmacological properties of CCR5/CXCR4 heteromers, we constructed a CHO-K1 cell line stably co-expressing CXCR4 and CCR5 at about equimolar amounts. The parental cell line expressing CXCR4 (Bmax, 3.2 ± 0.1 pmol/mg membrane proteins) was used as the recipient for CCR5 co-expression. In saturation binding assays, the Bmax values of the selected cell line were estimated to be 3.3 ± 0.4 pmol/mg proteins for CXCR4 and 3.6 ± 0.5 pmol/mg proteins for CCR5. Using membranes prepared from these cells co-expressing CCR5 and CXCR4, we observed a strong negative binding cooperativity between the two receptor binding pockets, very similar to what we reported previously for the CCR2/CXCR4 and CCR5/CCR2 heteromers (Fig. 3 and Table 1). The CXCR4 ligands (the chemokine agonist SDF-1α and the chemical antagonist AMD3100) were unable to inhibit 125I-MIP-1β binding to membranes prepared from cells expressing CCR5 alone, in agreement with their reported specificity (Fig. 3C). Both ligands were, however, able to inhibit 125I-MIP-1β binding with high apparent affinity when CXCR4 and CCR5 were coexpressed (Fig. 3D). Conversely, the CCR5-specific ligands (the chemokine MIP-1β and the chemical antagonist TAK-779) competed for 125I-SDF-1α binding only in cells coexpressing both receptors (Fig. 3, A versus B). The extent of the cross-competition (∼60%) is roughly compatible with the expected proportion of receptors involved in the formation of heteromers, provided the equal expression levels of the partners. Moreover, the cross-competition was observed both for agonist and antagonist molecules, suggesting that negative binding cooperativity across CXCR4/CCR5 hetero-oligomers does not require receptor signaling or the promotion of an active receptor conformation.
FIGURE 3.
Competition binding assays in cells co-expressing CCR5 and CXCR4. A–D, competition binding assays were performed on cells expressing CXCR4 (A), CCR5 (C), or both receptors (B and D). Membranes were incubated with 125I-SDF-1α or 125I-MIP-1β as tracer and unlabeled MIP-1β (●), SDF-1α (○), TAK-779 (■), or AMD3100 (□) as competitors. The data were normalized for nonspecific binding (0%), in the presence of 300 nm SDF-1α (A and B) or MIP-1β (C and D), and specific binding in the absence of competitor (100%). All points were run in triplicates (error bars indicate S.E.). The displayed data are representative of two independent experiments. E–H, aequorin-based functional assay in cells co-expressing CCR5 and CXCR4. The functional responses of CHO-K1 cells expressing CXCR4 (E), CCR5 (G), or both receptors (F and H) were measured using the aequorin-based functional assay. E and F, cells were stimulated with SDF-1α (○), SDF-1α + AMD3100 (□), or SDF-1α + TAK-779 (■). G and H, cells were stimulated with MIP-1β (●), MIP-1β + AMD3100 (□), or MIP-1β + TAK-779 (■). Luminescence was recorded for 30 s. The results were normalized for base-line activity (0%) and the maximal response obtained with 25 μm ATP (100%). The displayed data are representative of three independent experiments. All points were run in triplicates (error bars indicate S.E.).
TABLE 1.
Binding parameters of CHO-K1 cells expressing CCR5 and/or CXCR4
Binding parameters were measured on CHO-K1 cells expressing CCR5 and/or CXCR4. The IC50 and percentage of inhibition were obtained from competition binding experiments as displayed in Fig. 3. Values represent the mean ± S.E. of at least two independent experiments.
| Cells | Tracer | Competitor | IC50 | Inhibition |
|---|---|---|---|---|
| nm | % | |||
| CCR5 | 125I-MIP-1β | MIP-1β | 2.19 ± 0.57 | 100 |
| TAK-779 | 2.46 ± 1.77 | 94.8 ± 7.4 | ||
| CXCR4 | 125I-SDF-1α | SDF-1α | 1.46 ± 0.14 | 100 |
| AMD-3100 | 2.03 ± 0.71 | 90.6 ± 5.3 | ||
| CCR5 + CXCR4 | 125I-MIP-1β | MIP-1β | 2.21 ± 1.26 | 100 |
| TAK-779 | 2.54 ± 0.22 | 92.5 ± 1.5 | ||
| SDF-1α | 1,39 ± 1.71 | 51.9 ± 6.7 | ||
| AMD-3100 | 2.09 ± 1.26 | 36.7 ± 0.1 | ||
| CCR5 + CXCR4 | 125I-SDF-1α | MIP-1β | 0.80 ± 0.85 | 59.8 ± 2.5 |
| TAK-779 | 1.75 ± 2.05 | 65.0 ± 8.7 | ||
| SDF-1α | 0.35 ± 0.07 | 100 | ||
| AMD-3100 | 0.32 ± 0.16 | 92.3 ± 7.1 |
It is conceivable that the cross-competition might result from a steric impact at the extracellular surface of one receptor protomer on the other, although we showed this not to be the case previously with another pair of protomers (10, 12). Using a similar approach, we tested the impact on ligand binding of anti-CXCR4 and anti-CCR5 monoclonal antibodies targeted to the extracellular surfaces of the receptors. Although the anti-CXCR4 MAB173 prevented 125I-SDF-1α binding to membranes containing CXCR4 or the two receptors, it did not affect 125I-MIP-1β binding. Similarly, the anti-CCR5 2D7 prevented 125I-MIP-1β binding but not 125I-SDF-1α binding (supplemental Fig. S2). In order to characterize further the negative binding cooperativity between CXCR4 and CCR5, the rate of radioligand dissociation from heterodimers was assayed in “infinite” tracer dilution conditions as we have described previously (18). The co-expression of CCR5 and CXCR4 in CHO-K1 cells drastically increased the spontaneous dissociation rate of SDF-1α (t½ = 58.8 min versus t½ > 200 min) and MIP-1β (t½ = 44.6 min versus t½ > 200 min) (supplemental Fig. S3). Such changes in basal dissociation rates were also detected when CCR2 was co-expressed with CXCR4 but not following CCR2 and CCR5 co-expression (12, 18). This accelerated dissociation probably reflects conformational changes of the receptors according to the partners with which they interact. We also showed that specific CCR5 ligands (MIP-1β and TAK779) had no effect on the dissociation of bound tracer from membranes containing CXCR4 only. However, they slightly accelerated the dissociation of SDF-1α from membrane co-expressing CXCR4 and CCR5 (t½ = 41.7 and 43.3 min, respectively; supplemental Fig. S3B). Similarly, SDF-1α and AMD3100 promoted a faster dissociation of MIP-1β when CCR5 and CXCR4 were co-expressed (t½ = 38.3 and 26.7 min, respectively; supplemental Fig. S3D), indicating that ligand binding to one receptor in a heteromer modifies further the conformation of the other.
Functional Properties of CHO-K1 Cells Coexpressing CXCR4 and CCR5
We next compared the functional response of cells co-expressing CXCR4 and CCR5 to cell lines expressing only CCR5 or CXCR4. Calcium mobilization following stimulation by MIP-1β was similar for cells co-expressing both receptors or expressing only CCR5 (Fig. 3, G and H). Similarly, the functional response to SDF-1α was identical in CXCR4-expressing cells whether CCR5 was co-expressed or not (Fig. 3, E and F). Interestingly, co-stimulation by MIP-1β and SDF-1α at equimolar concentrations resulted in a functional response similar to that produced by the single chemokines, consistent with a lack of additivity. Finally, we investigated the effect of specific CCR5 or CXCR4 antagonists in cells coexpressing both receptors. In line with the binding data, we showed that the specific CXCR4 antagonist AMD3100 antagonized CXCR4 but also partially CCR5 signaling (Fig. 3, F and H), whereas TAK-779 inhibited CCR5 and also partially CXCR4 signaling (Fig. 3, F and H). The two antagonists, however, maintained their specificity when tested in cells expressing only CCR5 or CXCR4. These results demonstrate the ability of antagonists to inhibit the signaling of receptors on which they do not bind directly.
Functional Oligomerization of CCR2, CCR5, and CXCR4 in Native Cells
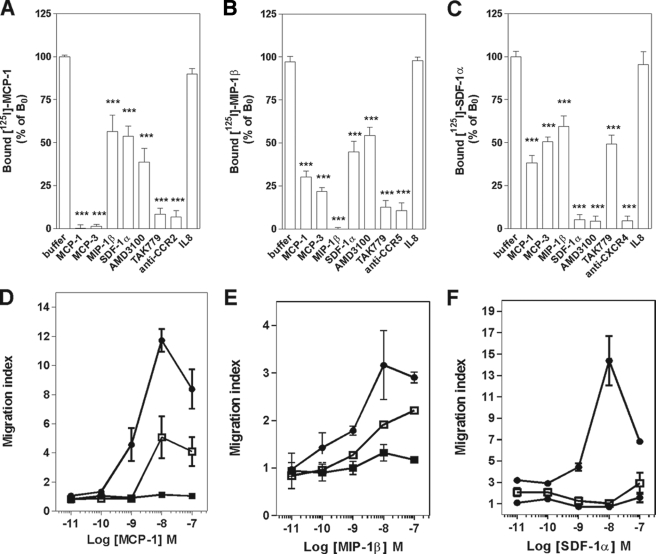
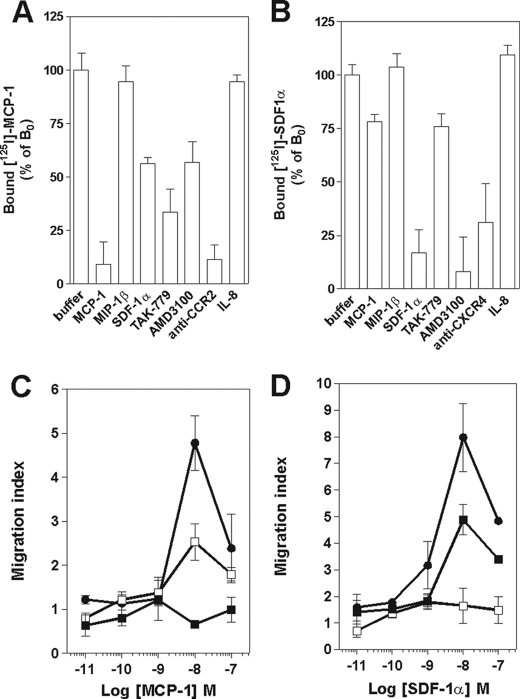
We next investigated whether the negative binding cooperativity could be demonstrated in cells endogenously co-expressing CCR2, CCR5, and CXCR4. For that purpose, human CD4+ T lymphocytes were isolated and activated with anti-CD3 antibodies (OKT3) and interleukin-2. Specific 125I-MCP-1, 125I-MIP-1β, and 125I-SDF-1α binding could be detected on these cells, as demonstrated by the competition achieved with the corresponding unlabeled chemokine, a chemical antagonist, or specific blocking antibodies for each receptor (Fig. 4, A–C). 125I-MCP-1 binding to CCR2 was inhibited by MCP-1, MCP-3, and TAK779 but also partially by MIP-1β, SDF-1α, and AMD-3100, which are CCR5- and CXCR4-specific ligands (Fig. 4A). Similarly, 125I-MIP-1β binding to CCR5 was fully inhibited by CCR5 ligands but also partially inhibited by CCR2 ligands (MCP-1 and MCP-3) or CXCR4 ligands (SDF-1α and AMD3100) (Fig. 4B). Finally, 125I-SDF-1α binding to CXCR4 was partially inhibited both by CCR2 and CCR5 ligands (Fig. 4C). Altogether, these data suggest that negative binding cooperativity takes place between the binding pockets of each receptor and that CXCR4/CCR5, CXCR4/CCR2, and CCR2/CCR5 heteromers and probably CCR2/CCR5/CXCR4 heteromers as well exist in native cells. We also performed those experiments in purified blood monocytes and reached similar conclusions regarding the negative binding cooperativity among CCR2, CCR5, and CXCR4 (supplemental Fig. S5, A–C).
FIGURE 4.
Competition binding and chemotaxis assays on CD4+ lymphoblasts. Competition binding assays were performed on CD4+ lymphoblasts by using 125I-MCP-1 (A), 125I- MIP-1β (B), or 125I-SDF-1α (C) as tracer and chemokines (300 nm), antagonists (300 nm), and monoclonal antibodies (10 μg/ml) as competitors. The data were normalized for nonspecific binding (0%) and specific binding in the absence of competitor (100%). Statistical significance as compared with the 100% values was tested by two-way analysis of variance followed by Tukey's test (***, p < 0.001). The displayed data are the mean of five independent experiments performed with lymphoblasts prepared from five different donors. All data points were performed in triplicate (error bars indicate S.E.). D–F, CD4+ lymphoblasts were either left untreated (■) or exposed to 300 nm TAK779 (□) or AMD3100 (●) for 30 min, and the chemotactic response of cells to MCP-1 (D), MIP-1β (E), or SDF-1α (F) was determined. The data are representative of three independent experiments. All points were run in duplicates (error bars indicate S.E.).
We next performed competition experiments with human CD4+ T lymphocytes prepared from a donor homozygous for the Δ32 allele of CCR5 (Fig. 5). The CCR5Δ32 allele encodes a truncated and nonfunctional CCR5 variant that is not transported to the cell surface (25, 26). Fluorescence-activated cell sorting analysis and 125I-MIP-1β binding confirmed that these cells failed to express cell surface CCR5 (data not shown). In CCR5Δ32 T cells, MIP-1β was unable to compete for 125I-MCP-1 or 125I-SDF-1α binding to CCR2 and CXCR4 receptors, respectively (Fig. 5, A and B). However, 125I-MCP-1 binding to CCR2 was still completely inhibited by CCR2 ligands and partially inhibited by the CXCR4 agonist and antagonist. Similarly, 125I-SDF-1α binding to CXCR4 was still inhibited both by CXCR4 and CCR2 ligands. These observations demonstrate that the absence of functional CCR5 did not impair the negative binding cooperativity between CCR2 and CXCR4 but completely abolished the cross-inhibition by CCR5-specific ligands.
FIGURE 5.
A and B, competition binding and chemotaxis assays on CCR5Δ32 CD4+ lymphoblasts. Competition binding assays were performed on CD4+ lymphoblasts by using 125I-MCP-1 (A) or 125I-SDF-1α (B) as tracer and chemokines (300 nm), antagonists (300 nm), and monoclonal antibodies (10 μg/ml) as competitors. The data were normalized for nonspecific binding (0%) and specific binding in the absence of competitor (100%). All data points were performed in triplicate (error bars indicate S.E.). C and D, CD4+ lymphoblasts were either left untreated (●) or exposed to 300 nm TAK779 (■) or AMD3100 (□) for 30 min, and the chemotactic response of cells to MCP-1 (C) or SDF-1α (D) was determined. The data are representative of three independent experiments. All points were run in duplicates (error bars indicate S.E.).
We also investigated whether in native cells antagonists could block the functional response of receptors to which they do not bind by using an ex vivo chemotaxis assay (Fig. 4, D–F). Cell migration of CD4+ T blasts toward MCP-1 or MIP-1β was totally inhibited by TAK-779 but also partially inhibited by the CXCR4-specific antagonist AMD3100 (Fig. 4, D and E). Similarly, migration toward SDF-1α was fully blocked by AMD3100 and partially blocked by TAK-779 (Fig. 4F). CCR5Δ32 T cell migration was observed toward SDF-1α and MCP-1 (Fig. 5, C and D) but not MIP-1β. The inhibition of SDF-1α-induced migration by TAK-779 was less significant than for cells expressing wild type CCR5 (Fig. 5F versus Fig. 4F). This is attributed to the exclusive effect of TAK-779 through CCR2 in the absence of functional CCR5. Functional cross-inhibition of chemotaxis was also shown in monocytes (supplemental Fig. S5, D–F). Altogether, these data indicate that CCR2, CCR5, and CXCR4 heteromers exist in native cells and that negative binding cooperativity takes place between each of the receptors. Importantly, these data also demonstrate that chemokine receptor heteromerization results in a cross-inhibitory effect by small molecule antagonists on the functional responses of receptors on which they do not bind.
Cross-inhibition of Cell Recruitment in Vivo
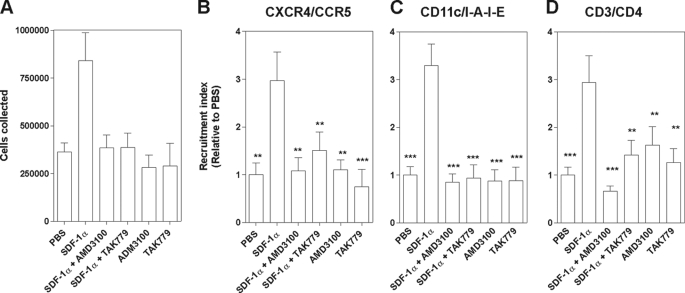
Finally, we investigated using the air pouch model in mice whether antagonists could also cross-inhibit cell migration in vivo. Air pouches were raised on the back of BALB/c mice, and an injection of PBS containing or not containing TAK-779 or AMD3100 was made into the air pouches. Thirty minutes later, SDF-1α was injected into the pouches, and recruitment was performed for 4 h. Leukocytes were collected, counted, and characterized by flow cytometry (Fig. 6). Injection of PBS only or PBS containing TAK-779 and AMD3100 induced a basal recruitment of cells co-expressing CXCR4 and CCR5, such as CD11c+/I-A-IE+ dendritic or CD3+/CD4+ T cells. The injection of SDF-1α induced a stronger cell recruitment that was completely inhibited by preinjection of the CXCR4-specific antagonist AMD3100. In line with the in vitro and ex vivo binding experiments and ex vivo cell recruitment assays, preinjection of the CCR2 and CCR5 antagonist TAK-779 also strongly blocked the recruitment of cells into the pouch. Altogether, these data show that TAK-779 inhibits the CXCR4-dependent cell recruitment in vivo.
FIGURE 6.
In vivo cell recruitment assay (air pouch). Air pouches were raised on the backs of 7-week-old BALB/c mice. Mice were given an injection into the pouches of 0.5 ml of PBS containing or not TAK-779 or AMD3100 (10 mg/kg). Thirty minutes later, SDF-1α (2 μg in 0.5 ml) was injected into the pouches. After 4 h, mice were killed, and the cells recruited to the pouches were collected and counted. A, graph showing the amount of cells collected from the pouch in a representative experiment. B–D, cell surface expression of CCR5 and CXCR4 and leukocyte populations were determined by fluorescence-activated cell sorting analysis. The recruitment index corresponds to the number of cells recruited divided by the number of cells collected after PBS injection alone. Statistical significance as compared to the SDF-1α condition was tested by two-way analysis of variance followed by Tukey's test (**, p < 0.01; ***, p < 0.001). The displayed data are the mean of three different experiments performed with a total of 10–17 mice (error bars indicate S.E.).
DISCUSSION
Chemokine receptors play a key role in the pathogenesis of autoimmune diseases, inflammation, and viral infection. Several approaches are currently being pursued with the aim of blocking the deleterious effects of specific chemokines, including the development of small molecule antagonists, the designing of N-terminally modified chemokines, and the generation of blocking antibodies (1, 2). However, the complexity and the functional redundancy of the chemokine system have represented significant hurdles in this development. Moreover, recent studies showing that the pharmacological properties of a given receptor subtype can be influenced by the array of its dimerization or oligomerization partners adds a layer of complexity to this system. There is, therefore, an urgent need for a better understanding of receptor organization at the surface of immune cells and how this organization influences receptor function.
We showed using BRET assays that the chemokine receptors CCR5 and CXCR4 form both homo- and heteromers in living cells. These results are in perfect agreement with two recent FRET studies (16, 21) but contradict a previous BRET study claiming that CXCR4 was not able to heteromerize with CCR5 (8).
The parameters of energy transfer (BRET50 and BRETmax) for the CCR5/CXCR4 interaction were in the same range as those reported for CCR2/CXCR4 and CCR2/CCR5 heteromers (Table S2) (10, 12). The BRETmax value for the CXCR4/CXCR4 interaction was 10-fold higher than that obtained for CXCR4/CCR2 or CXCR4/CCR5 interactions. Such a high BRET signal was previously reported in the study of Percherancier et al. (11) and might indicate either a higher propensity of CXCR4 to dimerize or a more efficient energy transfer between CXCR4-linked BRET probes, resulting from the relative positioning of the energy donor and acceptor.
Stimulation by specific chemokine agonist influences BRET energy transfer between CCR5 and CXCR4 in a manner that depends on the relative position of energy transfer donor and acceptor. This feature was reported for other chemokine receptor heteromers (11, 12) and attributed to conformational changes reflected differently according to the structural particularities of each receptor-probe chimera used in the assay. It is now widely accepted that BRET changes upon ligand stimulation more likely reflect conformation changes rather than dimer association or dissociation.
Furthermore, we showed, using luminescence complementation BRET, that these receptors can also form higher order homo- and hetero-oligomeric structures, containing at least three receptors at the same time. To our knowledge, this is the first demonstration of hetero-oligomerization of three different chemokine receptors. In line with the classical BRET assay, a much higher BRETmax value was obtained for CXCR4 homo-oligomers. Chemokine receptors might thus be part of large oligomeric complexes reminiscent of the rhodopsin arrays observed by atomic force microscopy in photoreceptors (27, 28) and recently demonstrated for the D2 dopamine receptor (29). Such arrays might also include other chemokine receptors as well as other GPCRs or even other classes of membrane proteins. The exact three-dimensional organization of chemokine receptor oligomers is not known precisely. It was proposed by Guo et al. (29) that the D2 dopamine receptor forms homo-oligomers through at least two distinct contact interfaces involving TM4 and TM1 plus helix 8. Oligomerization of GPCRs implies the existence of multiple receptor interfaces, but whether these interfaces are similar in different receptor or even in homo- and hetero-oligomers remains to be determined.
Using recombinant cells, we showed that co-expression of any two of these receptors results in strong negative binding cooperativity (i.e. the specific ligands of one receptor inhibit the binding of a radiolabeled chemokine to the other). We hypothesize that such negative binding cooperativity is a direct consequence of allosteric modulation across the receptor heteromer interface. Negative cooperativity was also observed in native leukocyte populations, such as CD4+ lymphoblasts and monocytes, which endogenously express the three receptors. Negative binding cooperativity was monitored for antagonists in recombinant systems and native cells, and in both intact cells and membrane fractions (supplemental Fig. S4). These observations support the view that negative binding cooperativity does not require classical intracellular signaling or the induction of a fully active conformation in the ligand-bound receptor. These experimental data do not support a recent model proposed by Chabre et al. (30) suggesting that cooperativity is dependent of protein-G availability. Given our similar pharmacological observations in recombinant systems expressing several receptors and in native cells, we infer that oligomerization of chemokine receptors also occurs in native cells. However, the formal demonstration of such organization in leukocytes will be much more difficult to achieve by biophysical means. Our results also showed that the cooperativity between the binding sites of chemokine receptors is not an artifactual consequence of their overexpression in recombinant systems and that recombinant cells constitute adequate models for studying the pharmacological consequences of receptor homo- and heteromerization.
We also showed that in leukocytes derived from a CCR5 Δ32/Δ32 donor, MIP-1β had no effect on the binding of MCP-1 and SDF-1α, demonstrating that the expression of functional CCR5 at the cell surface is essential for mediating the negative cooperative effects of CCR5 ligands. In contrast, the negative binding cooperativity between CCR2 and CXCR4 was still observed in these cells, demonstrating that the lack of CCR5 did not disrupt the oligomerization status of the two other receptors. These results support the existence of a complex organization of chemokine receptor oligomers at the surface of primary leukocytes, depending on their relative expression levels and their affinity for one another.
Importantly, we showed both in recombinant cell lines and in primary leukocytes that specific antagonists of one receptor inhibit the binding of chemokines to the others, apparently as a consequence of their heteromerization. This heterologous binding inhibition resulted in significant impairment of calcium mobilization and cell chemotaxis. These results illustrate how the pharmacological properties of a given molecule can be influenced by the range of dimers/oligomers in which its target receptor is involved, which will depend on the potential partners coexpressed in the same cells. Using the air pouch model in mice, we established that the cross-inhibition by antagonists has major consequences on the migration of cells in vivo. We showed that the CCR2 and CCR5 antagonist TAK-779 inhibits both lymphocyte and dendritic cell recruitment into the pouch in response to SDF-1α-induced stimulation of CXCR4. In line with the binding and the ex vivo chemotaxis experiments, this experiment demonstrates that TAK-779 inhibits CXCR4 receptor function in vivo although it does not bind to CXCR4 or inhibit its activity when this receptor is expressed alone. These results provide further support that small molecule antagonists can inhibit the function of receptors on which they do not bind directly, as the result of heteromerization, with important implications for the activities of chemokine receptor antagonists in vivo (31).
Although the exact molecular mechanisms underlying this regulation remain unknown, they probably involve a concerted conformational change of the antagonist target and the associated receptors. In this context, it should be noted that AMD3100 and TAK-779 were shown to display weak agonist and inverse agonist properties, respectively (32, 33), indicating that these molecules indeed affect the conformation of their respective targets. It is not yet known whether cross-inhibition will be a property shared by all antagonists of these receptors or restricted to some molecules only. Detailed functional testing of a larger range of antagonists will be required to address this issue. It seems, however, that antagonists might affect the conformation of heterodimers through different mechanisms. For instance, selective antagonists of the δ-opioid receptor were shown to modulate the pharmacology of the μ-opioid receptor by increasing the number of binding sites and enhancing the receptor signaling (34, 35). In another study, Ellis et al. (36) showed that antagonists of the cannabinoid CB1 and orexin-1 receptors regulate the cellular localization and the function of receptors.
It should be noted that both in recombinant systems and in primary leukocytes, the inhibition of the functional responses appears more important than the 50% inhibition of ligand binding. This observation suggests that the functional interaction between receptor units could extend beyond the direct allosteric interaction within heteromers. It could, for instance, involve the propagation of conformational changes to additional receptor dimers as a result of oligomerization or protomer exchange, thereby affecting the binding and functional properties of these receptors. This model could explain some of the data showing the ability of unlabeled ligands to promote the complete dissociation of prebound chemokines from cells co-expressing two receptors, whereas in competition binding assays at equilibrium, only a fraction of the specific binding is inhibited by the same (10, 18). However, we cannot exclude formally the possibility of cross-talk between downstream signaling events as contributing to the inhibition of the functional response of receptors.
The heteromerization of GPCRs in general and chemokine receptors in particular has important implications in the field of drug development and in understanding receptor function in native environments. Antagonist molecules characterized as selective for one receptor may in fact inhibit the functional response of other receptors in the same leukocyte. Cross-inhibition may imply either an increased therapeutic benefit, as a result of the partial blockade of other receptors contributing to an inflammatory process, or lead to development of unexpected and detrimental side effects. Future evaluation of the therapeutic benefit of targeting chemokine receptors or other GPCR classes will therefore have to consider the existence of oligomerization, which will greatly influence the practice of modern pharmacology.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DA022413 and MH54137 (to J. A. J.). This work was also supported by the Actions de Recherche Concertées of the Communauté Française de Belgique; the Interuniversity Attraction Poles Programme (P6–14), Belgian State, Belgian Science Policy, the Walloon region (Programme d'Excellence “CIBLES”); European Union Grant LSHB-CT-2005-518167/INNOCHEM; the French Agence Nationale de Recherche sur le SIDA; the Fonds de la Recherche Scientifique Médicale of Belgium; and the Fondation Médicale Reine Elisabeth.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- GPCR
- G protein-coupled receptor
- BRET
- bioluminescence resonance energy transfer
- EYFP
- enhanced yellow fluorescent protein
- RLuc
- Renilla luciferase
- hRLuc
- human RLuc
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Wells T. N., Power C. A., Shaw J. P., Proudfoot A. E. (2006) Trends Pharmacol. Sci. 27, 41–47 [DOI] [PubMed] [Google Scholar]
- 2.Pease J. E., Williams T. J. (2006) Br. J. Pharmacol. 147, S212–S221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gether U. (2000) Endocr. Rev. 21, 90–113 [DOI] [PubMed] [Google Scholar]
- 4.Horuk R. (2009) Nat. Rev. Drug Discov. 8, 23–33 [DOI] [PubMed] [Google Scholar]
- 5.Springael J. Y., Urizar E., Parmentier M. (2005) Cytokine Growth Factor Rev. 16, 611–623 [DOI] [PubMed] [Google Scholar]
- 6.Benkirane M., Jin D. Y., Chun R. F., Koup R. A., Jeang K. T. (1997) J. Biol. Chem. 272, 30603–30606 [DOI] [PubMed] [Google Scholar]
- 7.Mellado M., Rodríguez-Frade J. M., Vila-Coro A. J., Fernández S., Martín de Ana A., Jones D. R., Torán J. L., Martínez-A C. (2001) EMBO J. 20, 2497–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Issafras H., Angers S., Bulenger S., Blanpain C., Parmentier M., Labbé-Jullié C., Bouvier M., Marullo S. (2002) J. Biol. Chem. 277, 34666–34673 [DOI] [PubMed] [Google Scholar]
- 9.Hernanz-Falcón P., Rodríguez-Frade J. M., Serrano A., Juan D., del Sol A., Soriano S. F., Roncal F., Gómez L., Valencia A., Martínez-A C., Mellado M. (2004) Nat. Immunol. 5, 216–223 [DOI] [PubMed] [Google Scholar]
- 10.El-Asmar L., Springael J. Y., Ballet S., Andrieu E. U., Vassart G., Parmentier M. (2005) Mol. Pharmacol. 67, 460–469 [DOI] [PubMed] [Google Scholar]
- 11.Percherancier Y., Berchiche Y. A., Slight I., Volkmer-Engert R., Tamamura H., Fujii N., Bouvier M., Heveker N. (2005) J. Biol. Chem. 280, 9895–9903 [DOI] [PubMed] [Google Scholar]
- 12.Sohy D., Parmentier M., Springael J. Y. (2007) J. Biol. Chem. 282, 30062–30069 [DOI] [PubMed] [Google Scholar]
- 13.Hamatake M., Aoki T., Futahashi Y., Urano E., Yamamoto N., Komano J. (2009) Cancer Sci. 100, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reggio P. H. (2006) AAPS J. 8, E322–E336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filizola M., Weinstein H. (2005) FEBS J. 272, 2926–2938 [DOI] [PubMed] [Google Scholar]
- 16.Guo W., Shi L., Filizola M., Weinstein H., Javitch J. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17495–17500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contento R. L., Molon B., Boularan C., Pozzan T., Manes S., Marullo S., Viola A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Springael J. Y., Le Minh P. N., Urizar E., Costagliola S., Vassart G., Parmentier M. (2006) Mol. Pharmacol. 69, 1652–1661 [DOI] [PubMed] [Google Scholar]
- 19.Samson M., Labbe O., Mollereau C., Vassart G., Parmentier M. (1996) Biochemistry 35, 3362–3367 [DOI] [PubMed] [Google Scholar]
- 20.Blanpain C., Migeotte I., Lee B., Vakili J., Doranz B. J., Govaerts C., Vassart G., Doms R. W., Parmentier M. (1999) Blood 94, 1899–1905 [PubMed] [Google Scholar]
- 21.Isik N., Hereld D., Jin T. (2008) PLoS ONE 3, e3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulmurugan R., Massoud T. F., Huang J., Gambhir S. S. (2004) Cancer Res. 64, 2113–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 24.Loening A. M., Fenn T. D., Wu A. M., Gambhir S. S. (2006) Protein Eng. Des. Sel. 19, 391–400 [DOI] [PubMed] [Google Scholar]
- 25.Samson M., Libert F., Doranz B. J., Rucker J., Liesnard C., Farber C. M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C., Muyldermans G., Verhofstede C., Burtonboy G., Georges M., Imai T., Rana S., Yi Y., Smyth R. J., Collman R. G., Doms R. W., Vassart G., Parmentier M. (1996) Nature 382, 722–725 [DOI] [PubMed] [Google Scholar]
- 26.Huang Y., Paxton W. A., Wolinsky S. M., Neumann A. U., Zhang L., He T., Kang S., Ceradini D., Jin Z., Yazdanbakhsh K., Kunstman K., Erickson D., Dragon E., Landau N. R., Phair J., Ho D. D., Koup R. A. (1996) Nat. Med. 2, 1240–1243 [DOI] [PubMed] [Google Scholar]
- 27.Levitzki A. (1974) J. Theor. Biol. 44, 367–372 [DOI] [PubMed] [Google Scholar]
- 28.Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., Palczewski K. (2003) Nature 421, 127–128 [DOI] [PubMed] [Google Scholar]
- 29.Guo W., Urizar E., Kralikova M., Mobarec J. C., Shi L., Filizola M., Javitch J. A. (2008) EMBO J. 27, 2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chabre M., Deterre P., Antonny B. (2009) Trends Pharmacol. Sci. 30, 182–187 [DOI] [PubMed] [Google Scholar]
- 31.Springael J. Y., Urizar E., Costagliola S., Vassart G., Parmentier M. (2007) Pharmacol. Ther. 115, 410–418 [DOI] [PubMed] [Google Scholar]
- 32.Zhang W. B., Navenot J. M., Haribabu B., Tamamura H., Hiramatu K., Omagari A., Pei G., Manfredi J. P., Fujii N., Broach J. R., Peiper S. C. (2002) J. Biol. Chem. 277, 24515–24521 [DOI] [PubMed] [Google Scholar]
- 33.Lagane B., Ballet S., Planchenault T., Balabanian K., Le Poul E., Blanpain C., Percherancier Y., Staropoli I., Vassart G., Oppermann M., Parmentier M., Bachelerie F. (2005) Mol. Pharmacol. 67, 1966–1976 [DOI] [PubMed] [Google Scholar]
- 34.Gomes I., Jordan B. A., Gupta A., Trapaidze N., Nagy V., Devi L. A. (2000) J. Neurosci. 20, RC110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan B. A., Devi L. A. (1999) Nature 399, 697–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellis J., Pediani J. D., Canals M., Milasta S., Milligan G. (2006) J. Biol. Chem. 281, 38812–38824 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.