Abstract
12/15-Lipoxygenase (12/15LO) plays a role in the pathogenesis of atherosclerosis and diabetes and has been implicated in low density lipoprotein oxidation. Murine macrophages express high levels of 12/15LO and are key cells involved in the accumulation and efflux of oxidized low density lipoprotein in the arterial wall. During this process, macrophages up-regulate scavenger receptors that regulate lipid uptake, and ATP-binding cassette (ABC) transporters, that regulate lipid efflux. We have previously demonstrated that 12/15LO enhances the turnover and serine phosphorylation of ABCG1. In the current study, we further elucidate the mechanisms by which 12/15LO regulates ABCG1. Proteasomal inhibitors blocked the down-regulation of ABCG1 expression and resulted in accumulation of phosphorylated ABCG1. Macrophages that lack 12/15LO have enhanced transporter expression, reduced ABCG1 phosphorylation, and increased cholesterol efflux. Conversely, macrophages that overexpress 12/15LO have reduced ABCG1 expression, increased transporter phosphorylation, and reduced cholesterol efflux. 12/15LO plays a key role in activating the MAPK pathway. Inhibition of the p38 or JNK pathways with pharmacological inhibitors or dominant negative constructs blocked 12S-hydroxyeicosatetranoic acid-mediated degradation of ABCG1. Moreover, we isolated macrophages from JNK1-, JNK2-, and MKK3-deficient mice to analyze the involvement of specific MAPK pathways. JNK2- and MKK3-, but not JNK1-deficient macrophages were resistant to the down-regulation of ABCG1 protein, reduction in efflux, and increase in serine phosphorylation by 12S-hydroxyeicosatetranoic acid. These findings provide evidence that 12/15LO regulates ABCG1 expression and function through p38- and JNK2-dependent mechanisms, and that targeting these pathways may provide novel approaches for regulating cholesterol homeostasis.
Introduction
Lipoxygenases comprise a family of enzymes capable of mediating selective lipid oxidation. 12/15-lipoxygenase (12/15LO)2 catalyzes the conversion of arachidonic or linoleic acid to produce 12S- and 15S-hydroxyeicosatetranoic acids (12SHETE and 15SHETE) and 13S-hydroxyoctadecadienoic acid, respectively, and oxidizes esterified fatty acids within cholesteryl esters and phospholipids (1, 2). Mouse leukocyte 12/15LO is highly related to the 15LO enzyme in that they are ∼74% identical in primary structure, and both are dual-specificity lipoxygenases (1). Mouse 12/15LO likely represents the orthologue of human 15LO (2–4). Murine macrophages express high levels of 12/15LO. Human or mouse peripheral blood monocytes do not express 12/15LO in circulation, but 12/15LO expression in these cells can be elicited by IL-4 or IL-13 (5–7).
The role of 12/15LO in atherogenesis has been controversial, with studies attributing both pro-atherogenic (8–10) and anti-atherogenic properties to the enzyme (11). We and others have shown that mice overexpressing 12/15LO are more susceptible to developing spontaneous aortic fatty streak lesions on a rodent chow diet (9), whereas mice deficient in the enzyme are protected from atherosclerosis (10). Conversely, Chan and colleagues have recently demonstrated that 12/15LO overexpression protects mice from atherosclerosis through production of local lipid mediators, including lipoxin A4, resolvin D1, and protectin D1 (12). 12/15LO has been detected in atherosclerotic lesions and colocalizes with macrophages (13). However, Habenicht and colleagues have reported that the expression of 15LO in human atherosclerotic lesions was either undetectable or several orders of magnitude lower than 5LO (14). Products of the 12/15LO pathway are potent signal transducers and can modify the inflammatory responses in cells. 12/15LO products have been demonstrated to activate specific isoforms of protein kinase C (PKC) (15), activate extracellular signal-regulated kinase (ERK) (16), c-Jun NH2-terminal kinase (JNK) (17), p38 MAPK pathways (18), and p21-activated kinase (19), and regulate transcription factor activation (20).
The ATP-binding cassette transporters G1 (ABCG1) and A1 (ABCA1) are up-regulated in differentiated macrophages to mediate the efflux of cholesterol to high density lipoproteins (HDLs) (21). A role for 12/15LO activity in the vessel wall in regulating the clearance of excess cholesterol from macrophages and contributing to the development of macrophage foam cell formation has not been fully elucidated. We have previously demonstrated that 12/15LO activity in macrophages enhances the degradation of ABCG1, resulting in reduced cholesterol efflux (22). Correlating with increased turnover of the transporter, the 12/15LO eicosanoid product 12SHETE enhanced the serine phosphorylation of ABCG1 (22). The current study provides further understanding of the mechanisms by which 12/15LO regulates ABCG1 expression and function. We have found that 12/15LO activates JNK2 and p38 MAPK pathways to enhance transporter phosphorylation and degradation. These results define a novel pathway for the regulation of ABCG1 and cholesterol homeostasis in macrophages.
MATERIALS AND METHODS
Chemicals and Reagents
FBS was obtained from HyClone (Logan, UT). HDL was from Intracel (Frederick, MD). NuPAGE 4–12% denaturing gels, MOPS running and transfer buffers, and nitrocellulose were from Invitrogen. Mouse anti-ABCG1 antibody was from Novus Biologicals, Inc. Mouse anti-ABCA1 antibody and human lipid-free apoA-I were kind gifts from John Parks, Ph.D., at Wake Forest University. Mouse anti-β-actin antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). RNeasy Mini kit was from Qiagen. An adenovirus expressing murine 12/15LO was prepared by our laboratory with the assistance of the University of Iowa Gene Transfer Vector Core. 12SHETE, SB203580, SP600125, PD98059, anti-phosphoserine, cinnamyl-3,4-dihydroxy-α-cyanocinnamate, PKC inhibitors, H89, and MG132 were purchased from Biomol (Plymouth Meeting, PA). Lactacystin and casein kinase II inhibitor DMAT were from Calbiochem. Phorbol 12-myristate 13-acetate was purchased from Sigma- Aldrich. Anti-phospho-p38 was from R&D Systems. Anti-phospho-JNK, anti-phospho-MKK3/6, anti-p38, and anti-JNK were from Cell Signaling Technology.
Mice and Bone Marrow-derived Macrophage Isolation
12/15LO transgenic (LOTG) mice on the C57BL6/J background were generated as previously described (9). Ten-week-old female B6.129S2-Alox15tm1Fun/J (LOKO) mice (stock no. 002778), B6.129S1-Mapk8tm1Flv/J (JNK1 KO) mice (stock no. 004319), B6.129S2-Mapk9tm1Flv/J (JNK2 KO) mice (stock no. 004321), B6.129S1-Map2k3tm1Flv/J (MKK3 KO) mice (stock no. 006416), and their controls, C57BL6/J (stock no. 000664), were purchased from Jackson Laboratories (Bar Harbor, ME). Mice were fed a standard rodent chow diet and housed in microisolator cages in a pathogen-free facility. All experiments followed University of Virginia Animal Care and Use Committee guidelines, and approval for use of rodents was obtained from the University of Virginia. Cells were obtained from the tibia and femur bone marrow of aged matched mice and were cultured in the presence of L-929 conditioned medium as described in detail previously (23).
Recombinant Adenoviral Delivery
We generated a recombinant adenovirus to express murine 12/15LO. Briefly, the full-length cDNA for murine 12/15LO (GenBankTM accession number U04331.1) was subcloned into pACCMVpLpA. Recombination, adenoviral production, and titering were performed by the University of Iowa Gene Transfer Vector Core. Bone marrow-derived macrophages were infected at a multiplicity of infection of 50 for 48 h with the recombinant adenoviral vectors AdLO (to overexpress 12/15LO) or control AdLacZ.
Transient Transfection of Bone Marrow-derived Macrophages
Dominant-negative MAPK kinase-4 (dnMKK4) was generously provided by Dr. Michael Karin (University of California, San Diego, CA), and dnMKK7 was generously provided by Dr. Tse-Hua Tan (Baylor College of Medicine, Houston, TX). Dominant-negative p38 (dnp38) contains two phosphorylation mutations of the isoform p38α, Thr-180 to Ala and Tyr-182 to Phe (24). Bone marrow-derived macrophages were isolated from mice as described above. Four million cells were transfected with 1 μg of dnp38, dnMKK4, dnMKK7, or pcDNA control constructs by nucleofection using a mouse macrophage nucleofector kit (Amaxa, Gaithersburg, MD). Transfection efficiency was ∼65% as measured by green fluorescent protein transfection. Transfected cells were incubated for 24 h in Dulbecco's modified Eagle's medium supplemented with 10% FBS. After 24 h, cells were treated with 500 nm 12SHETE for 30 min, and protein was collected for analysis as described.
Cellular Cholesterol Efflux Measurements
Cholesterol efflux assays were performed as described, with minor modification (25). Bone marrow-derived macrophages were plated in 12-well plates at a density of 6 × 105 cells/well. To measure effects on cholesterol efflux, cells were radiolabeled with 2 μCi/ml [3H]cholesterol for 16 h in Dulbecco's modified Eagle's medium containing 10% lipoprotein-depleted FBS. After radiolabeling, cells were washed three times with phosphate-buffered saline. Cells were allowed to equilibrate for 2 h in 0.2% fatty acid free bovine serum albumin (FAFBSA). In some experiments, macrophages were treated with 500 nm 12SHETE in 0.2% FAFBSA for 2 h. Cholesterol efflux was conducted for 4 or 8 h at 37 °C in media containing: 1) 0.2% FAFBSA, 2) 0.2% FAFBSA plus 15 μg/ml lipid-free human apolipoprotein A-I (apoA-I), or 3) 0.2% FAFBSA plus 50 μg of protein/ml of human HDL. Human apoA-I and HDL were isolated as described previously (26, 27). The efflux medium was removed and a 100-μl aliquot was taken for 3H radioactivity determination. Adherent cells were rinsed three times with cold phosphate-buffered saline, cells were dried, and isopropanol was added for overnight extraction at room temperature. A 100-μl aliquot of the extract was taken for 3H radioactivity determination. Results are expressed as [3H]cholesterol in medium/([3H]cholesterol in medium + cell) × 100%. Specific efflux to apoA-I or HDL was calculated by subtracting nonspecific efflux in the presence of 0.2% FAFBSA only.
Immunoblotting for ABCA1, ABCG1, and 12/15LO
RIPA buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Igepal, 10 mm NaF, 2 mm Na3VO4 containing Sigma protease inhibitor mixture) was added to macrophages to generate whole cell lysates. Lysates were sonicated, and protein was quantified via a protein assay kit (Bio-Rad). Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and blocked for 1 h with 2.5% milk-Tris-buffered saline plus 1% Tween 20 (TBST) at room temperature. Fifty micrograms of whole cell lysate was used to detect all proteins. ABCA1 and ABCG1 antibodies (1:500 dilution) were incubated with the blot at 4 °C overnight in 2.5% milk-TBST. The 12/15LO antibody (1:1000 dilution) was incubated with the blot at 4 °C overnight in 2.5% Milk-TBST. ABCA1, ABCG1, and 12/15LO blots were then incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:2000 dilution) in 2.5% milk-TBST for 1 h at room temperature. The β-actin antibody (1:10000 dilution) was incubated with the blot for 1 h at room temperature in 2.5% milk-TBST. The β-actin blots were then incubated with horseradish peroxidase-conjugated anti-mouse secondary antibody (1:5000 dilution) in 2.5% milk-TBST for 1 h at room temperature. Proteins were visualized using chemiluminescence and normalized to β-actin as gel loading control. Densitometry was performed using Stratagene Zero D-Scan densitometry software.
Quantitative Real-time PCR
Total cellular RNA was collected from macrophages using the RNeasy Micro kit (Qiagen) following the manufacturer's protocol. One microgram of cDNA was synthesized using the Iscript cDNA synthesis kit (Bio-Rad). Total cDNA was diluted 1:10 in H2O, and 4 μl was used for each real-time condition using a Bio-Rad MyIQ Single Color Real-Time PCR Detection System and iQ SYBR Green Supermix (Bio-Rad). Primer sequences were as follows: ABCA1 (forward 5′-GGTTTGGAGATGGTTATACAATAGTTGT-3′ and reverse 5′-CCCGGAAACGCAAGTCC-3′), ABCG1 (forward 5′-TTCCCCTGGAGATGAGTGTC-3′ and reverse 5′-CAGTAGGCCACAGGGAACAT-3′), LXR alpha (forward 5′-GGATAGGGTTGGAGTCAGCA-3′ and reverse 5′-GGAGCGCCTGTTACACTGTT-3′), LXR beta (forward 5′-GCTCAGGAGCTGATGATCCA-3′ and reverse 5′-GCGCTTGATCCTCGTGTAG-3′), and cyclophilin (forward 5′-TGGAGAGCACCAAGACAGACA-3′ and reverse 5′-TGCCGGAGTCGACAATGAT-3′). Samples were normalized to cyclophilin using the ΔCt method.
Immunoprecipitation of ABCG1
Macrophage lysates were prepared with radioimmune precipitation assay buffer as described above. Lysates were incubated overnight at 4 °C with antibody against ABCG1 (Novus Biologicals). The antibody-antigen complex was isolated by protein A-Sepharose beads (Invitrogen) and resolved by SDS-PAGE. Each gel lane received equal amounts of immunoprecipitated protein. ABCG1 phosphorylation was assayed by immunoblot analysis using anti-phosphoserine antibody (Biomol) that recognizes a broad range of serine-phosphorylated proteins.
ABCG1 Site-directed Mutagenesis
Full-length FLAG-tagged human ABCG1 open reading frame 678 (ORF678) and truncated FLAG-tagged human ABCG1 ORF638 were generously provided by Dr. John Oram (University of Washington, Seattle, WA) (28). ABCG1 ORF638 mutation constructs on CKII consensus sites (Ser-45, Ser-205, and Ser-349) and N-terminal serines (Ser-65, Ser-70, Ser-119, Ser-141, and Ser-168) to alanines were generated by PCR using a QuikChange multisite-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions and confirmed by sequencing. HEK293 cells were grown in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum then transiently transfected with 1 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Statistical Analysis
Data for all experiment comparisons between groups was performed using analysis of variance (ANOVA) methods using the StatView 6.0 software program. Data is graphically represented as the mean ± S.E., in which the mean consists of a minimum of three experiments performed in triplicate. Comparisons between groups and tests of interactions were performed assuming a two-factor analysis with the interaction term testing each main effect with the residual error testing the interaction. All comparisons were made using Fisher's least standard difference procedure, so that multiple comparisons were performed at the alpha = 0.05 level only if the overall F-test from the ANOVA were significant at p < 0.05 (29).
RESULTS
Serine Phosphorylation Targets ABCG1 for Proteasomal Degradation
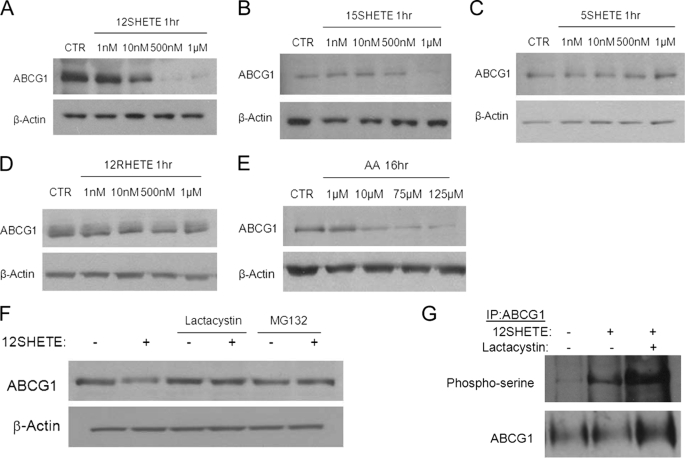
We have previously shown that the 12/15LO product 12SHETE enhances the serine phosphorylation of ABCG1 in macrophages (22). Increased serine phosphorylation correlated with enhanced turnover of the transporter (22). We performed dose-response experiments to examine whether this response was specific for the 12/15LO pathway. 12SHETE treatment reduced ABCG1 expression in a dose-dependent manner (Fig. 1A). Another product of the 12/15LO pathway, 15SHETE, also reduced ABCG1 expression, but at a higher concentration than that observed for 12SHETE (Fig. 1B). Further, we examined the effects of 5SHETE and 12RHETE, which are products of the 5(S)-lipoxygenase and 12(R)-lipoxygenase pathways, respectively. At concentrations ranging from 1 nm to 1 μm, we observed no change in ABCG1 expression by 5SHETE or 12RHETE (Fig. 1, C and D), indicating that this response is specific to products of the 12/15LO pathway. Additionally, we treated macrophages with the fatty acid substrate of the 12/15LO pathway, arachidonic acid. Arachidonic acid also reduced ABCG1 expression in a dose-dependent manner (Fig. 1E), but the reduction in ABCG1 occurred at higher concentrations and longer incubation times than for 12/15LO eicosanoids.
FIGURE 1.
ABCG1 is targeted for proteasomal degradation by 12SHETE. A–D, C57BL6/J bone marrow-derived macrophages were treated with vehicle control (CTR) or the indicated eicosanoids (HETEs) at the indicated concentrations for 1 h. Cell lysates were analyzed by immunoblotting for ABCG1 or β-actin. Blot is representative of three different experiments. E, C57BL6/J bone marrow-derived macrophages were treated with vehicle control (CTR) or arachidonic acid (AA) at the indicated concentrations for 16 h. Cell lysates were analyzed by immunoblotting for ABCG1 or β-actin. Blot is representative of three different experiments. F, C57BL6/J bone marrow-derived macrophages were pre-treated with 10 μm lactacystin, 10 μm MG132, or vehicle control DMSO for 30 min prior to the addition of 500 nm 12SHETE or vehicle control ethanol for an additional 30 min. Cell lysates were analyzed by immunoblotting for ABCG1 or β-actin. Blot is representative of five different experiments. G, ABCG1 was isolated by immunoprecipitation, and blots were probed with phosphoserine or ABCG1 antibodies. Blot is representative of three different experiments.
We hypothesized that 12SHETE targets ABCG1 for degradation by the proteasome. To evaluate this hypothesis, C57BL6/J (B6) macrophages were pre-treated with the proteasomal inhibitors lactacystin or MG132 for 30 min before addition of 12SHETE for 30 min. As shown previously (22), 12SHETE reduced ABCG1 protein levels (Fig. 1F). Both lactacystin and MG132 blocked the degradation of ABCG1 protein by 12SHETE (Fig. 1F). Moreover, lactacystin treatment enhanced the accumulation of phosphorylated ABCG1 (Fig. 1G), indicating that serine phosphorylation of ABCG1 by 12SHETE likely targets ABCG1 for proteasomal degradation.
12/15LO Regulates ABCG1 Expression and Function
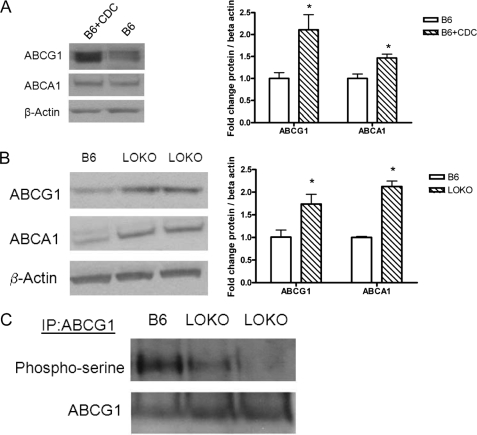
To examine whether 12/15LO activity in the macrophage can regulate ABCG1 expression and function, we treated B6 macrophages with the 12/15LO pharmacological inhibitor cinnamyl-3,4-dihydroxy-α-cyanocinnamate for 24 h. Treatment of B6 macrophages with cinnamyl-3,4-dihydroxy-α-cyanocinnamate resulted in a 2-fold increase in ABCG1 protein expression (Fig. 2A). Next we isolated bone marrow-derived macrophages from mice deficient for 12/15LO (LOKO). LOKO macrophages had a significant 2-fold increase in ABCG1 and ABCA1 protein expression (Fig. 2B) compared with wild-type B6 macrophages. Concomitant with these findings, we observed a reduction in serine phosphorylation of ABCG1 in LOKO macrophages (Fig. 2C). Moreover, we observed no changes in mRNA expression of ABCG1, ABCA1, LXRα, or LXRβ expression (data not shown).
FIGURE 2.
Inhibition of 12/15LO regulates ABCG1 protein expression and phosphorylation. In A: left, C57BL6/J (B6) bone marrow-derived macrophages were incubated 10 μm cinnamyl-3,4-dihydroxy-α-cyanocinnamate or vehicle control DMSO for 24 h. Cell lysates were analyzed by immunoblotting for ABCG1, ABCA1, or β-actin. Right, densitometry of immunoblots normalized to β-actin. Data represent the mean ± S.E. of three mice per group (*, significantly higher than B6 p < 0.03 by ANOVA). In B: left, cell lysates from 12/15LO-deficient (LOKO) or C57BL6/J (B6) bone marrow-derived macrophages were analyzed by immunoblotting for ABCG1, ABCA1, or β-actin. Right, densitometry of immunoblots normalized to β-actin. Data represent the mean ± S.E. of 5 mice per group (*, significantly higher than B6 p < 0.02 by ANOVA). C, ABCG1 was isolated by immunoprecipitation, and blots were probed with phospho-serine or ABCG1 antibodies. Blot is representative of three different experiments.
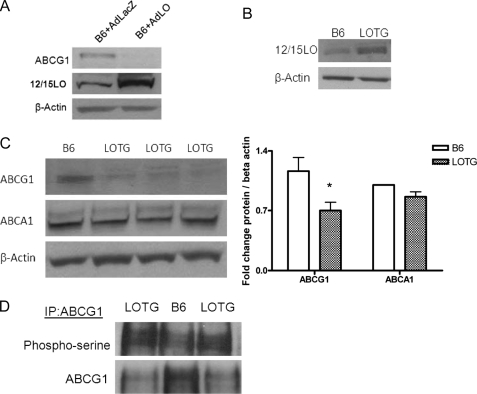
To further examine a role for 12/15LO in regulating ABCG1 protein levels, we overexpressed 12/15LO in B6 macrophages with an adenovirus (indicated AdLO) for 48 h. We confirmed that the adenovirus increased 12/15LO expression ∼2-fold compared with macrophages treated with a control adenovirus (indicated AdLacZ) (Fig. 3A). B6 macrophages treated with AdLO had almost no ABCG1 protein expression (Fig. 3A).
FIGURE 3.
Overexpression of 12/15LO regulates ABCG1 protein expression and phosphorylation. A, C57BL6/J (B6) bone marrow-derived macrophages were incubated with recombinant adenovirus expressing either 12/15LO (+AdLO) or control LacZ (+AdLacZ) for 48 h. Cell lysates were analyzed by immunoblotting for ABCG1, 12/15LO, or β-actin. Blot is representative of four different experiments. B, cell lysates from C57BL6/J (B6) or 12/15LO-overexpressing (LOTG) bone marrow-derived macrophages were analyzed by immunoblotting for 12/15LO or β-actin. Blot is representative of three different experiments. In C: left, Cell lysates from 12/15LO-overexpressing (LOTG) or C57BL6/J (B6) bone marrow-derived macrophages were analyzed by immunoblotting for ABCG1, ABCA1, or β-actin. Right, densitometry of immunoblots normalized to β-actin. Data represent the mean ± S.E. of seven mice per group (*, significantly lower than B6 p < 0.03 by ANOVA). D, ABCG1 was isolated by immunoprecipitation, and blots were probed with phosphoserine or ABCG1 antibodies. Blot is representative of three different experiments.
We previously generated transgenic mice on a C57BL/6J (B6) background that modestly overexpressed the murine 12/15LO gene (designated LOTG) (9). LOTG mice had significant elevations in levels of urinary 12SHETE and 13S-hydroxyoctadecadienoic acid, the primary eicosanoid products of the 12/15LO pathway, and a 2-fold increase in 12/15LO protein expression in vivo (9). We confirmed transgene overexpression of 12/15LO in bone marrow-derived macrophages isolated from these mice (Fig. 3B). Bone marrow-derived macrophages isolated from LOTG mice had a significant 40% reduction in ABCG1 protein expression compared with B6 (Fig. 3C). No change in ABCA1 protein expression was observed (Fig. 3C). Moreover, we observed an increase in serine phosphorylation of ABCG1 by immunoprecipitation in macrophages from LOTG mice (Fig. 3D). Additionally, ABCG1 and ABCA1 mRNA expression was unchanged (data not shown), indicating that the inhibitory effects of 12/15LO on ABCG1 expression are not attributable to reduced ABCG1 mRNA expression.
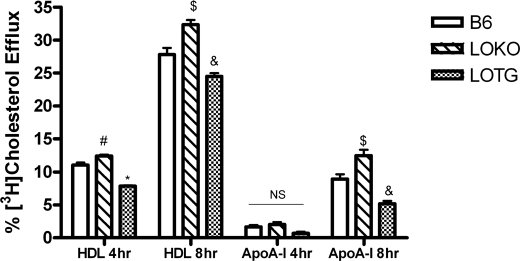
Because ABCG1 is known to regulate cholesterol efflux, we next assayed the effects of 12/15LO activity on this process. B6, LOKO, or LOTG macrophages were labeled with [3H]cholesterol, and HDL- and apoA-I-mediated cholesterol efflux was measured. Tall and colleagues have reported that HDL is the preferred cholesterol acceptor from ABCG1, whereas lipid-free apoA-I is the preferred cholesterol acceptor from ABCA1 (30). LOKO macrophages had a significant increase in cholesterol efflux measured for 4 or 8 h to HDL compared with B6 macrophages (Fig. 4). Conversely, LOTG macrophages had a significant reduction in cholesterol efflux measured for 4 or 8 h to HDL (Fig. 4). No change in cholesterol efflux to apoA-I was measured in either genotype (Fig. 4) at 4 h; however, after 8 h we observed a significant increase in cholesterol efflux from LOKO bone marrow-derived macrophages and a significant reduction in efflux from LOTG bone marrow-derived macrophages to lipid-free apoA-I. ABCG1 degradation is not regulated by PKA- or PKC-dependent pathways.
FIGURE 4.
12/15LO regulates macrophage cholesterol efflux. Control B6, LOKO, and LOTG bone marrow-derived macrophages were isolated and utilized in a cholesterol efflux assay as described under “Materials and Methods.” Cholesterol efflux to lipid-free apoA-I (apoA-I) and to HDL (HDL) for 4 or 8 h was assessed. Data represent the mean ± S.E. of 6 experiments (*, significantly lower than B6 HDL 4 h p < 0.0001; #, significantly higher than B6 HDL 4 h p < 0.05; $, significantly higher than B6 HDL 8 h p < 0.02; and &, significantly lower than B6 HDL 8 h p < 0.03 by ANOVA).
There is evidence that phosphorylation plays an important role in the function of ABCA1. Hayden and colleagues have reported that site-specific serine phosphorylation of ABCA1 by PKA regulates apoA-I-dependent phospholipid efflux (31). Oram and colleagues have shown that phosphorylation of ABCA1 through a PKCδ-dependent pathway is regulated by unsaturated fatty acids and results in transporter degradation (32). Although no link between PKA and 12/15LO has been found to date, we and others have previously demonstrated that the 12/15LO product 12SHETE is a potent activator of the PKC pathway (15). However, using inhibitors of both PKA and PKC isoforms, we were unable to block the degradation of ABCG1 by 12SHETE (supplemental Fig. 1 and data not shown), indicating that 12SHETE does not appear to regulate ABCG1 through either PKA- or PKC-dependent pathways. Thus, kinase regulation of ABCG1 expression and function is through a mechanism that is distinct from ABCA1.
12/15LO Regulates Activation of the MAPK Pathway
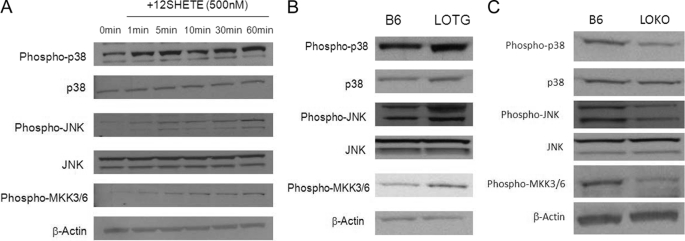
Studies by our laboratory and others have indicated a role for 12/15LO in the activation of members of the MAPK pathway. 12SHETE has been demonstrated to be an activator of ERK, JNK, and p38 MAPK. To examine the regulation of the MAPK pathway by 12/15LO, we treated B6 macrophages with 12SHETE for 1–60 min. Within 5 min of treatment, the p38 and JNK pathways were activated, indicated by increased phosphorylation of p38, MKK3/6, and JNK, respectively (Fig. 5A). We have previously found that serine phosphorylation of ABCG1 is significantly up-regulated within 30 min of treatment (22), indicating that the p38 and JNK pathways are activated in advance of the observed increase in ABCG1 phosphorylation. Adenoviral overexpression of 12/15LO in B6 bone marrow-derived macrophages resulted in activation of p38 and JNK MAPK pathways compared with control adenovirus (data not shown). Macrophages from LOTG also displayed activation of both the p38 and JNK pathways (Fig. 5B). Conversely, macrophages from LOKO macrophages had reduced activation of both the p38 and JNK MAPK pathways, as indicated by decreased phosphorylation (Fig. 5C).
FIGURE 5.
12/15LO regulates activation of p38 and JNK MAPK pathways. Cell lysates were analyzed by immunoblotting using antibodies specific for the phosphorylated forms of p38, JNK, or MKK3/6. Bands were normalized to total p38, total JNK, or β-actin. A, C57BL6/J bone marrow-derived macrophages were incubated with 500 nm 12SHETE for 1–60 min. Cell lysates were analyzed for MAPK pathway activation by immunoblot analysis. Blot is representative of three different experiments. B, cell lysates from B6 or LOTG bone marrow-derived macrophages were analyzed for MAPK pathway activation by immunoblot analysis. Blot is representative of four different experiments. C, cell lysates from B6 or LOKO bone marrow-macrophages were analyzed for MAPK pathway activation by immunoblot analysis. Blot is representative of four different experiments.
Inhibition of p38 and JNK Pathways Blocks 12SHETE-mediated Degradation and Phosphorylation of ABCG1
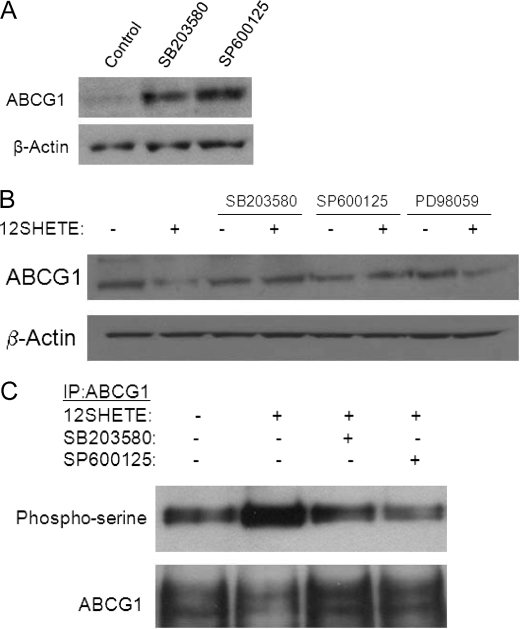
To determine if activation of the MAPK pathway is responsible for 12/15LO-regulated degradation of ABCG1, we used pharmacological inhibitors of p38 (SB203580), JNK (SP600125), and MEK1 (PD98059). Initially, we treated B6 bone marrow-derived macrophages with SB203580 or SP600125 for 24 h and observed a 2-fold increase in ABCG1 expression (Fig. 6A). To further examine a role for 12/15LO in the regulation of ABCG1 through the MAPK pathway, B6 macrophages were pre-treated for 30 min with inhibitors of the MAPK pathway, prior to the addition of 12SHETE for 30 min. The inhibitors SB203580 and SP600125, but not PD98059, were able to block the 12SHETE-mediated degradation of ABCG1 (Fig. 6B). Moreover, SB203580 and SP600125 blocked serine phosphorylation of ABCG1 induced by 12SHETE (Fig. 6C). Thus, 12SHETE appears to specifically target the p38 and JNK, but not ERK, pathways for regulation of ABCG1.
FIGURE 6.
Inhibition of p38 and JNK MAPK pathways blocks 12SHETE-mediated ABCG1 degradation and phosphorylation. A, C57BL6/J bone marrow-derived macrophages were treated with 10 μm SB203580, 10 μm SP600125, or vehicle control DMSO for 24 h. Cell lysates were analyzed by immunoblotting for ABCG1 or β-actin. Blot is representative of three different experiments. B, C57BL6/J bone marrow-derived macrophages were pretreated with 10 μm SB203580, SP600125, PD98059, or vehicle control DMSO for 30 min prior to the addition of 500 nm 12SHETE or vehicle control ethanol for 30 min. Cell lysates were analyzed by immunoblotting for ABCG1 or β-actin. Blot is representative of five different experiments. C, C57BL6/J bone marrow-derived macrophages were pretreated with 10 μm SB203580, SP600125, or vehicle control DMSO for 30 min prior to the addition of 12SHETE or vehicle control ethanol for 30 min. ABCG1 was isolated by immunoprecipitation, and blots were probed with phosphoserine or ABCG1 antibodies. Blot is representative of three different experiments.
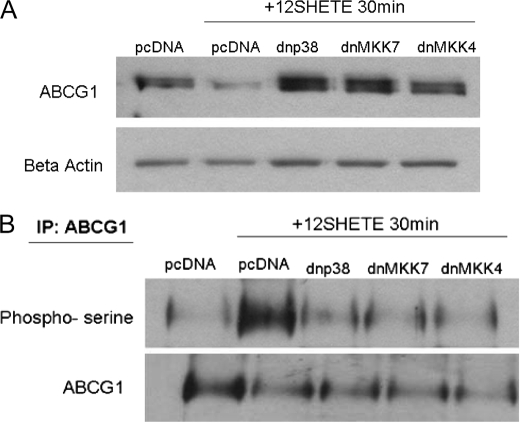
To further determine if the p38 and JNK MAPK pathways were involved in the regulation of ABCG1 by 12/15LO, we transfected dominant-negative MAPK constructs into B6 macrophages. B6 bone marrow-derived macrophages were transfected with dominant negative p38 (indicated dnp38) or the upstream activating kinases of JNK, MKK4 (dnMKK4) or MMK7 (dnMKK7), for 24 h prior to the addition of 12SHETE. We have previously described these constructs (24, 33). B6 macrophages transfected with dnp38, dnMKK4, or dnMKK7 were resistant to degradation of ABCG1 by 12SHETE compared with pcDNA-transfected control macrophages (Fig. 7A). Additionally, transfection of B6 macrophages with dnp38, dnMKK4, or dnMKK7 blocked serine phosphorylation of ABCG1 by 12SHETE (Fig. 7B).
FIGURE 7.
Dominant-negative constructs of p38 and JNK MAPK pathways block the degradation of ABCG1 by 12SHETE. C57BL6/J bone marrow-derived macrophages were transfected with dominant negative p38 (dnp38), MKK4 (dnMKK4), MKK7 (dnMKK7) or pcDNA control vectors (pcDNA). After 24 h, transfected macrophages were treated with 12SHETE or vehicle control ethanol for 30 min. A, cell lysates were analyzed by immunoblotting for ABCG1 or β-actin. Blot is representative of three different experiments. B, ABCG1 was isolated by immunoprecipitation, and blots were probed with phosphoserine or ABCG1 antibodies. Blot is representative of three different experiments.
JNK2- and MKK3-deficient Macrophages Are Resistant to 12SHETE-mediated Degradation and Phosphorylation of ABCG1
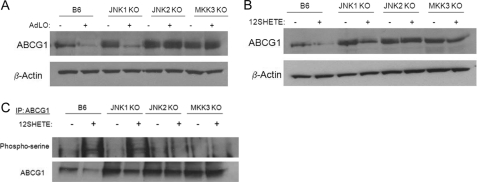
To further delineate a role for the MAPK pathway in mediating the regulation of ABCG1 by 12/15LO, we utilized macrophages deficient in specific MAPK pathways. We isolated bone marrow-derived macrophages from JNK1-, JNK2-, and MKK3-deficient mice. JNK1- and JNK2-deficient macrophages have deficiencies in activation of JNK1 and JNK2 pathways, respectively, whereas a deficiency in one of the upstream kinases of p38, MKK3, results in reduced p38 activation in MKK3-deficient mice. Basal, unstimulated expression of ABCG1 was significantly enhanced in JNK1-, JNK2-, and MKK3-deficient macrophages compared with B6 control macrophages (Fig. 8, A and B). We overexpressed 12/15LO in these macrophages using an adenovirus construct. Overexpression of 12/15LO in B6 or JNK1-deficient macrophages significantly reduced ABCG1 protein expression compared with control LacZ-expressing adenovirus (Fig. 8A). This reduction in ABCG1 protein expression by 12/15LO was blocked in JNK2- and MKK3-deficient macrophages (Fig. 8A). Additionally we treated bone marrow-derived macrophages from these mice with 12SHETE for 30 min. Degradation of ABCG1 protein by 12SHETE was blocked in JNK2 - and MKK3-deficient macrophages, but not in JNK1-deficient macrophages (Fig. 8B). Additionally, 12SHETE-mediated serine phosphorylation of ABCG1 was blocked in JNK2- and MKK3-deficient macrophages, but not in JNK1-deficient macrophages (Fig. 8C). Thus, 12/15LO regulates ABCG1 protein expression through JNK2- and p38-dependent pathways.
FIGURE 8.
The MAPK pathway regulates 12/15LO-mediated down-regulation of ABCG1. A, JNK1-deficient (JNK1 KO), JNK2-deficient (JNK2 KO), MKK3-deficient (MKK3KO), or C57BL6/J (B6) control bone marrow-derived macrophages were incubated with recombinant adenovirus expressing either 12/15LO (+AdLO) or control LacZ (+AdLacZ) for 48 h. Cell lysates were analyzed by immunoblotting for ABCG1 or β-actin. Blot is representative of four different experiments. B, JNK1-deficient (JNK1 KO), JNK2-deficient (JNK2 KO), MKK3-deficient (MKK3 KO), or C57BL6/J (B6) control macrophages were treated with 500 nm 12SHETE or vehicle control ethanol for 30 min. Cell lysates were analyzed by immunoblotting for ABCG1 or β-actin. Blot is representative of four different experiments. C, ABCG1 was isolated by immunoprecipitation, and blots were probed with phosphoserine or ABCG1 antibodies. Blot is representative of three different experiments.
MAPKs target serines and threonines followed by prolines (Ser/Thr-Pro) for phosphorylation. The sequence of ABCG1 contains no serine or threonine residues immediately preceding a proline residue. Thus, we hypothesized that a downstream kinase such as MK2/3, MNK1/2, PRAK, or casein kinase may be activated to target ABCG1 for degradation. We pretreated B6 bone marrow-derived macrophages with the casein kinase II (CKII) inhibitor DMAT for 2 or 16 h prior to the addition of 12SHETE for 30 min. DMAT blocked the degradation of ABCG1 by 12SHETE (supplemental Fig. 2A), indicating that casein kinase may be activated downstream of the MAPK pathway to target ABCG1.
To examine the effects of serine residue phosphorylation on ABCG1 degradation by 12/15LO, we were generously provided an N-terminal FLAG-tagged human ABCG1 cDNA coding for a full-length 678-amino acid protein (GenBankTM accession number NP_004906.3) and an N-terminal truncated 638-amino acid protein (GenBankTM accession number NP_004906.1, indicated ORF638) from Dr. John Oram at the University of Washington (28). We cotransfected a murine 12/15LO full-length construct (GenBankTM accession number U04331.1) or control pcDNA vector into HEK293 cells. We confirmed overexpression of 12/15LO in these cells. Both human ABCG1 constructs were degraded and phosphorylated by 12/15LO (data not shown), indicating that serine phosphorylation of ABCG1 is not occurring within the first 40 amino acids of the N-terminal tail. This eliminated eight potential serine residues in the N terminus of ABCG1 that might be phosphorylated.
We have identified three potential consensus sequences for CKII (SXX(D/E)) located in the N terminus of human ABCG1 ORF638. We performed multisite-directed mutagenesis of those residues converting serine residues 45, 205, and 349 to alanine residues. We cotransfected HEK293 cells expressing either human ABCG1 or the CKII mutant with full-length murine 12/15LO or pcDNA control. 12/15LO overexpression enhanced the degradation of both human ABCG1 and the CKII mutant (supplemental Fig. 2B), indicating that it is unlikely that CKII is activated to target ABCG1 for degradation, at least at the serine residues 45, 205, or 349.
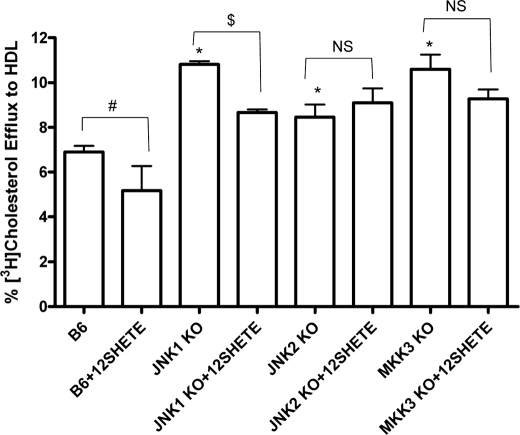
To examine a role for the MAPK pathway in regulating cholesterol efflux, bone marrow-derived macrophages from B6, JNK1-, JNK2-, and MKK3-deficient mice were labeled with [3H]cholesterol. Subsequently, macrophages were treated with 12SHETE, and HDL-mediated cholesterol efflux was measured. Basal cholesterol efflux was significantly increased in JNK1-, JNK2-, and MKK3-deficient macrophages compared with control B6 macrophages (Fig. 9). 12SHETE treatment significantly reduced cholesterol efflux to HDL by ∼20% in B6 and JNK1 KO macrophages (Fig. 9). Interestingly, the reduction in cholesterol efflux by 12SHETE was completely blocked in JNK2- and MKK3-deficient macrophages (Fig. 9). Thus, 12SHETE preferentially regulates the expression of ABCG1 and transporter function as measured by cholesterol efflux through JNK2- and p38-dependent pathways.
FIGURE 9.
JNK2- and MKK3-deficient macrophages are resistant to 12SHETE-mediated reduction in cholesterol efflux. B6, JNK1 KO, JNK2 KO, and MKK3 KO bone marrow-derived macrophages were isolated and utilized in a cholesterol efflux assay as described under “Materials and Methods.” Macrophages were treated with 500 nm 12SHETE or vehicle control ethanol for 2 h and efflux to HDL was assessed. Data represent the mean ± S.E. of three experiments (*, significantly higher than B6 p < 0.04; #, significantly lower than B6 p < 0.05; and $, significantly lower than JNK1 KO p < 0.02 by ANOVA).
N-terminal Serine Residues Regulate ABCG1 Degradation by 12/15LO
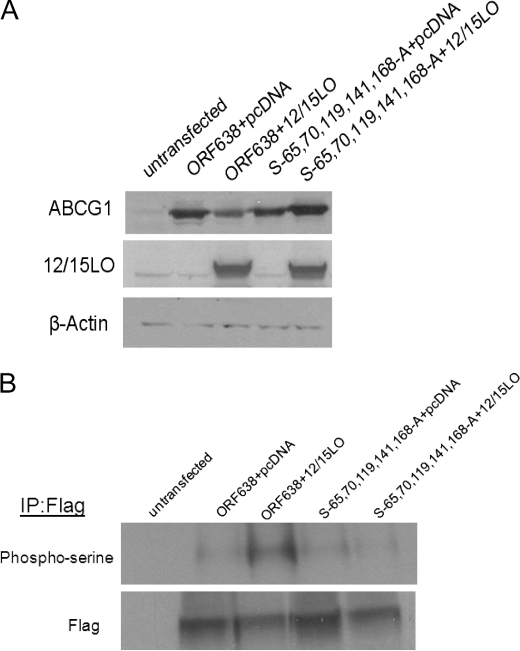
We performed site-directed mutagenesis of serine residues in the truncated ABCG1 construct (termed ORF638). All mutations were confirmed by sequencing. Sequentially, beginning at the N terminus, we changed serine residues Ser-27, Ser-28, Ser-43, Ser-45, Ser-80, and Ser-85 to alanine residues. All of these mutants were degraded by 12/15LO (data not shown). We continued multisite-directed mutagenesis of Ser-65, Ser-70, Ser-119, Ser-141, and Ser-168 (termed S-65,70,119,141,168-A). This mutant with multiple serine residues changed to alanine residues was resistant to degradation by 12/15LO (Fig. 10). Additionally, we examined serine phosphorylation of FLAG-tagged ABCG1. ABCG1 ORF638 was phosphorylated on serine residues when 12/15LO was co-overexpressed. However, the ABCG1 mutant with serines 65, 70, 119, 141, and 168 changed to alanines was resistant to serine phosphorylation by 12/15LO, indicating that phosphorylation of ABCG1 is likely occurring within this region of the N terminus.
FIGURE 10.
N-terminal serines regulate the degradation of ABCG1 by 12/15LO. A, multisite-directed mutagenesis was performed to sequentially mutate N-terminal serine residues in human ABCG1 ORF638. Human ABCG1 ORF638 or the serine mutant (indicated as S-65,70,119,141,168-A) were co-overexpressed in HEK293 cells with full-length 12/15LO or pcDNA control. Cell lysates were analyzed by immunoblotting for ABCG1, 12/15LO, or β-actin. Blot is representative of three different experiments. B, ABCG1 was isolated by immunoprecipitation for FLAG, and blots were probed with phosphoserine or FLAG antibodies. Blot is representative of three different experiments.
DISCUSSION
The ATP-binding cassette transporter ABCG1 is essential for macrophage cholesterol homeostasis and the process of reverse cholesterol transport. We previously demonstrated a role for the 12/15LO pathway in regulating cholesterol efflux and the degradation of ABCG1 (22). In the current study, we demonstrate for the first time that 12/15LO targets ABCG1 for degradation through enhanced serine phosphorylation. Moreover, we define a novel critical role for the JNK2 and p38 MAPK pathways in regulating ABCG1 expression and function.
12/15LO has been localized to macrophages in both early and advanced lesions; however, the pro- versus anti-atherogenic role of this enzyme in atherogenesis is debatable. Recently, Belkner et al. demonstrated that overexpression of 12/15LO in macrophages attenuates intracellular lipid accumulation and foam cell formation through a reduction in SR-A expression and accelerated lipid metabolism (34). Weibel et al. have indicated a role for human 15LO in enhancing cholesterol ester hydrolysis and reverse cholesterol transport (35). These authors determined that overexpression of human 15LO in RAW macrophages increases cholesterol efflux, ABC transporter expression, cholesteryl ester hydrolysis, and reverse cholesterol transport. Additionally they determined that metabolites of the 12/15LO pathway had no effect on cholesterol efflux or transporter expression. Conversely, we have found that 12/15LO overexpression and 12/15LO eicosanoid products reduce cholesterol efflux and enhance the turnover of ABCG1 protein. In the study by Weibel et al., overexpression of human 15LO primarily produces 15SHETE as the major product (35). Previously we determined that 15SHETE reduced ABCG1 expression but to a lesser extent than the product 12SHETE (22). We also determined that 15SHETE had no significant effect on cholesterol efflux, whereas 12SHETE significantly reduced efflux from macrophages (22). Weibel et al. utilized a mouse macrophage cell line for their studies and treated overexpressing macrophages with 12/15LO eicosanoid products, whereas in the current study we utilized primary macrophages and analyzed the effects of the eicosanoid product 12SHETE on basal ABCG1 expression. Additionally, in performing cholesterol efflux assays, Weibel et al. utilized an acylcoA:cholesterolacyltransferase inhibitor to prevent esterification, treated macrophages with 12/15LO products for longer time points than our assays (4 versus 2 h), and measured efflux to acceptor particle for shorter time points than our assays (2 versus 4 h). The conflicting results between the two studies may be attributable to LO species differences, preferential production of specific eicosanoid species, varied assay conditions, or varied expression patterns.
Leukocyte 12/15LO is expressed primarily in macrophages and is not expressed in monocytes without IL-4 or IL-13 induction. The mouse leukocyte-type 12/15LO produces more 12SHETE than 15SHETE (3:1 ratio) (36), whereas human reticulocyte-type 15LO produces more 15SHETE than 12SHETE (12:1 ratio) (37). Indeed, we have observed that these eicosanoids have different effects on ABCG1 degradation and cholesterol efflux (22). In Fig. 1, higher concentrations of 15SHETE were required to decrease ABCG1 expression than 12SHETE. Wen et al. reported that 15SHETE can activate the JNK pathway, but activation was observed over a shorter time course than 12SHETE (17). Thus, eicosanoid products of the 12/15LO pathway may have functional differences in regards to cholesterol metabolism. It is still unresolved how 12SHETE regulates activity of the MAPK pathway. Studies using pertussis toxin have indicated that 12SHETE induces JNK activation through Gi protein signaling (17) and that pertussis toxin blocks the degradation of ABCG1 by 12HETE (data not shown). Other eicosanoids have been observed to signal through G-protein-coupled receptors (38, 39); however the molecular basis for activation of the MAPK pathway requires further extensive study.
Foam cells are responsible for the majority of lipid accumulation within atherosclerotic lesions. Mobilization of lipid accumulation in foam cells is essential for plaque regression. The MAPK pathway has been shown to play a role in regulating macrophage cholesterol homeostasis. Ricci and colleagues found that the MAPK pathway can regulate macrophage cholesterol accumulation, ultimately resulting in enhanced foam cell formation and in vivo plaque formation (40). Specifically, these authors showed that JNK2 phosphorylates SR-A, leading to enhanced internalization of lipids (40). Thus, taken together with our data, inhibition of the JNK2 pathway may provide a therapeutic approach to combat atherogenesis. First, inhibition of the JNK2 pathway results in decreased macrophage lipid uptake. Second, inhibition of the JNK2 pathway enhances cholesterol efflux through increased ABCG1 expression and cholesterol efflux (Figs. 8 and 9). Because foam cell formation is a balance between lipid accumulation and removal, JNK2-dependent phosphorylation of SR-A and ABCG1 would shift the paradigm toward a “pro-atherogenic” phenotype.
The 12/15LO pathway has been shown to regulate MAPK activation. Lipoxygenase products have been shown to activate JNK (17), p38 (18), and ERK MAPKs (16). Concurrently, we found that the 12/15LO eicosanoid product 12SHETE was able to rapidly activate (within 1–5 min) the p38 and JNK MAPK pathways (Fig. 5). This activation occurs prior to the down-regulation and phosphorylation of ABCG1 by 12SHETE (22). Additionally, macrophages that overexpressed (LOTG) or were deficient in 12/15LO (LOKO) had corresponding up-regulation or down-regulation of MAPK activation, respectively (Fig. 5). By inhibiting the p38 or JNK pathways with pharmacological inhibitors (Fig. 6) or dominant-negative constructs (Fig. 7), we were able to block the down-regulation and phosphorylation of ABCG1 by 12SHETE. Interestingly, pharmacological inhibition of the p38 and JNK pathways for 24 h increased ABCG1 protein expression (Fig. 6A), suggesting that these pathways regulate basal expression of ABCG1.
Phosphorylation of ATP-binding cassette transporters has been shown to play a key role in transporter expression and function. PKA site-specific phosphorylation of ABCA1 enhances cholesterol and phospholipid efflux (31). Oram and colleagues have demonstrated that activation of a PKC δ pathway by polyunsaturated fatty acids results in enhanced serine phosphorylation of ABCA1 and targets the transporter for degradation (32). However, our data indicate that 12/15LO regulation of ABCG1 is not through a PKA- or PKC-dependent pathway (supplemental Fig. 1 and data not shown). Tall and colleagues have reported that phosphorylation of a PEST sequence in ABCA1 regulates degradation of the transporter by calpain protease (41). Phosphorylation of the ATP-binding cassette transporter G family member, ABCG2/BCRP, has also been demonstrated to play an essential role in the multidrug resistance capabilities of this transporter. The anti-apoptotic kinase Pim-1 has been shown to phosphorylate ABCG2 to promote transporter dimerization and drug-resistant activity (42). Moreover, epidermal growth factor has been shown to increase ABCG2 expression through ERK1/2 and JNK pathways (43). Based on our findings in the current study, we hypothesize that serine phosphorylation of ABCG1 mediates the reduction in transporter expression by 12/15LO by sensitizing ABCG1 to the action of the proteasome. Accordingly, the proteasomal inhibitors lactacystin or MG132 inhibited ABCG1 degradation by 12SHETE (Fig. 1). Phosphorylation serves as a positive signal for the targeting of several proteins to the 26 S proteasome, including IRS-1 (44), cyclin E (45), cyclin D1 (46), progesterone receptor (47), and IκBα (48). Numerous studies have implicated the MAPK pathway in regulation of protein stability, including promoting the phosphorylation and proteasomal-dependent degradation of the proapoptotic protein BimEL (49), MAPK phosphatase 3 (50), cyclin D2 (51), and the adaptor protein cFLIP (52).
The protein sequence of ABCG1 contains no serines or threonines followed by a proline residue. MAPKs almost invariably phosphorylate serine or threonine residues that are followed by a proline (Ser/Thr-Pro) (53). There are reports of proteins phosphorylated by MAPKs on residues that are not proline-directed (54, 55). However, it is likely that JNK2 or p38 may not directly phosphorylate ABCG1. Instead, activation of a downstream kinase such as MK2/3, MNK1/2, PRAK, or casein kinase, may mediate the phosphorylation of ABCG1 and target the transporter for degradation. Roosbeek et al. have reported that site-specific phosphorylation of ABCA1 by CKII down-regulates ABCA1-mediated cholesterol and phospholipid efflux and reduces apoprotein binding (56). Indeed, the CKII inhibitor DMAT blocked the down-regulation of ABCG1 by 12SHETE (supplemental Fig. 2). We identified three potential consensus sequences for CKII (SXX(D/E)) located in the N terminus of human ABCG1. However, 12/15LO was able to degrade the CKII mutant ABCG1 (supplemental Fig. 2), indicating that it is unlikely that the MAPK pathway activates CKII to target ABCG1.
ABCG1 has an N-terminal intracellular domain that contains the Walker motifs for ATP binding and a short intracellular C-terminal tail. The C-terminal tail contains no serine residues; however, the N-terminal region contains 40 possible target serine residues. We have performed sequential mutational analysis of each serine located with the first 208 residues in the N-terminal region of human ABCG1. There are 19 serine residues located within this region. Mutational analysis of serines 65, 70, 119, 141, and 168 in human ABCG1 indicated that 12/15LO targets these residues for degradation and phosphorylation (Fig. 10). We are currently in the process of mutating each of these residues singularly; however, it is also likely that a combination of serine residues is required for degradation of the transporter.
Elevated levels of plasma free fatty acids are prevalent among subjects with metabolic syndrome and Type 2 diabetes, and these free fatty acids can accumulate in atherosclerotic lesions. We have previously shown that the lipoxygenase pathway is up-regulated in Type 2 diabetic mice (57) and humans (58) and that diabetic mice have reduced ABCG1 expression and function (59). Thus, 12/15LO-mediated regulation of ABCG1 may contribute to the accelerated cardiovascular disease observed in patients with Type 2 diabetes. The involvement of a signaling pathway indicates that 12/15LO-induced down-regulation of ABCG1 has a biological function. The 12/15LO product 12SHETE destabilizes ABCG1 at concentrations that fall within the physiological range for eicosanoid products (60), and the destabilization is rapid within 30 min. Although the reason for the suppression of ABCG1 by fatty acids of the 12/15LO pathway is unknown, it is consistent with other studies demonstrating cross-regulation of the fatty acid and sterol metabolic pathways (61, 62). 12/15LO metabolites have been demonstrated to have mitogenic effects (63). Thus, the suppression of ABCG1 might retain a pool of cholesterol and phospholipids within the cell in preparation for new membrane synthesis. It is also possible that the accumulation of excess cholesterol and phospholipids is needed for the formation of triglyceride-rich lipid droplets formed by the influx of fatty acids, as is found in adipocytes (64).
In summary, 12/15LO targets ABCG1 for serine phosphorylation and destabilization through p38- and JNK2-dependent pathways. Ultimately, this results in a reduction in cholesterol efflux, impairing reverse cholesterol transport. Regulation of the 12/15LO and/or the MAPK pathways may provide important therapeutic targets for combating foam cell formation and regulating the reverse cholesterol transport pathway.
Supplementary Material
Acknowledgments
We thank Dr. John S. Parks (Wake Forest University) for recombinant apoA-I and helpful advice regarding cholesterol efflux, and Dr. David L. Brautigan (University of Virginia) for providing invaluable advice regarding immunoprecipitation studies.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 HL55798 (to C. C. H. and to J. L. N.) and R01 HL085790 (to C. C. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- 12/15LO
- 12/15-lipoxygenase
- 12SHETE
- 12S-hydroxyeicosatetranoic acid
- 15SHETE
- 15S-hydroxyeicosatetranoic acid
- IL-4
- interleukin-4
- PKC
- protein kinase C
- PKA
- protein kinase A
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- ABCG1
- ATP-binding cassette transporters G1
- ABCA1
- ATP-binding cassette transporters A1
- HDL
- high density lipoprotein
- FBS
- fetal bovine serum
- MOPS
- 4-morpholinepropanesulfonic acid
- LOTG
- 12/15LO transgenic mice
- LOKO
- B6.129S2-Alox15tm1Fun/J mice
- dnMKK4
- dominant-negative MAPK kinase-4
- FAFBSA
- fatty acid free bovine serum albumin
- apoA-I
- apolipoprotein A-I
- ORF
- open reading frame
- CKII
- casein kinase II
- ANOVA
- analysis of variance
- DMAT
- 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole.
REFERENCES
- 1.Yamamoto S., Takahashi Y., Hada T., Hagiya H., Suzuki H., Reddy G. R., Ueda N., Arakawa T., Nakamura M., Matsuda S., Taketani Y., Yoshimoto T., Azekawa T., Morita Y., Ishimura K., Arase S., Glasgow W. C., Brash A. R., Anton M., Kuhn H. (1997) Adv. Exp. Med. Biol. 400A, 127–131 [DOI] [PubMed] [Google Scholar]
- 2.Funk C. D. (1996) Biochim. Biophys. Acta 1304, 65–84 [DOI] [PubMed] [Google Scholar]
- 3.Kuhn H., Belkner J., Suzuki H., Yamamoto S. (1994) J. Lipid Res. 35, 1749–1759 [PubMed] [Google Scholar]
- 4.Yoshimoto T., Takahashi Y. (2002) Prostaglandins Other Lipid Mediat. 68–69, 245–262 [DOI] [PubMed] [Google Scholar]
- 5.Conrad D. J., Kuhn H., Mulkins M., Highland E., Sigal E. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heydeck D., Thomas L., Schnurr K., Trebus F., Thierfelder W. E., Ihle J. N., Kuhn H. (1998) Blood 92, 2503–2510 [PubMed] [Google Scholar]
- 7.Brinckmann R., Topp M. S., Zalan I., Heydeck D., Ludwig P., Kuhn H., Berdel W. E., Habenicht J. R. (1996) Biochem. J. 318, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cyrus T., Pratico D., Zhao L., Witztum J. L., Rader D. J., Rokach J., Fitzgerald G. A., Funk C. D. (2001) Circulation 103, 2277–2282 [DOI] [PubMed] [Google Scholar]
- 9.Reilly K. B., Srinivasan S., Hatley M. E., Patricia M. K., Lannigan J., Bolick D. T., Vandenhoff G., Pei H., Natarajan R., Nadler J. L., Hedrick C. C. (2004) J. Biol. Chem. 279, 9440–9450 [DOI] [PubMed] [Google Scholar]
- 10.Huo Y., Zhao L., Hyman M. C., Shashkin P., Harry B. L., Burcin T., Forlow S. B., Stark M. A., Smith D. F., Clarke S., Srinivasan S., Hedrick C. C., Pratico D., Witztum J. L., Nadler J. L., Funk C. D., Ley K. (2004) Circulation 110, 2024–2031 [DOI] [PubMed] [Google Scholar]
- 11.Shen J., Herderick E., Cornhill J. F., Zsigmond E., Kim H. S., Kuhn H., Guevara N. V., Chan L. (1996) J. Clin. Invest. 98, 2201–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merched A. J., Ko K., Gotlinger K. H., Serhan C. N., Chan L. (2008) FASEB J. 22, 3595–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yla-Herttuala S., Rosenfeld M. E., Parthasarathy S., Glass C. K., Sigal E., Witztum J. L., Steinberg D. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6959–6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spanbroek R., Grabner R., Lotzer K., Hildner M., Urbach A., Ruhling K., Moos M. P., Kaiser B., Cohnert T. U., Wahlers T., Zieske A., Plenz G., Robenek H., Salbach P., Kuhn H., Radmark O., Samuelsson B., Habenicht A. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natarajan R., Lanting L., Xu L., Nadler J. (1994) Mol. Cell Endocrinol. 101, 59–66 [DOI] [PubMed] [Google Scholar]
- 16.Wen Y., Nadler J. L., Gonzales N., Scott S., Clauser E., Natarajan R. (1996) Am. J. Physiol. 271, C1212–C1220 [DOI] [PubMed] [Google Scholar]
- 17.Wen Y., Scott S., Liu Y., Gonzales N., Nadler J. L. (1997) Circ. Res. 81, 651–655 [DOI] [PubMed] [Google Scholar]
- 18.Reddy M. A., Thimmalapura P. R., Lanting L., Nadler J. L., Fatima S., Natarajan R. (2002) J. Biol. Chem. 277, 9920–9928 [DOI] [PubMed] [Google Scholar]
- 19.Wen Y., Gu J., Knaus U. G., Thomas L., Gonzales N., Nadler J. L. (2000) Biochem. J. 349, 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao G. N., Glasgow W. C., Eling T. E., Runge M. S. (1996) J. Biol. Chem. 271, 27760–27764 [DOI] [PubMed] [Google Scholar]
- 21.Vaughan A. M., Oram J. F. (2006) J. Lipid Res. 47, 2433–2443 [DOI] [PubMed] [Google Scholar]
- 22.Nagelin M. H., Srinivasan S., Lee J., Nadler J. L., Hedrick C. C. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1811–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bruijn M. F., Slieker W. A., van der Loo J. C., Voerman J. S., van Ewijk W., Leenen P. J. (1994) Eur. J. Immunol. 24, 2279–2284 [DOI] [PubMed] [Google Scholar]
- 24.Srinivasan S., Bolick D. T., Hatley M. E., Natarajan R., Reilly K. B., Yeh M., Chrestensen C., Sturgill T. W., Hedrick C. C. (2004) J. Biol. Chem. 279, 31930–31936 [DOI] [PubMed] [Google Scholar]
- 25.Wang N., Silver D. L., Thiele C., Tall A. R. (2001) J. Biol. Chem. 276, 23742–23747 [DOI] [PubMed] [Google Scholar]
- 26.Nichols A. B., Ravenscroft C., Lamphiear D. E., Ostrander L. D., Jr. (1976) JAMA 236, 1948–1953 [PubMed] [Google Scholar]
- 27.Parks J. S., Rudel L. L. (1979) J. Biol. Chem. 254, 6716–6723 [PubMed] [Google Scholar]
- 28.Vaughan A. M., Oram J. F. (2005) J. Biol. Chem. 280, 30150–30157 [DOI] [PubMed] [Google Scholar]
- 29.Milliken G. A. (1995) Stat. Med. 14, 701–702 [DOI] [PubMed] [Google Scholar]
- 30.Wang N., Lan D., Chen W., Matsuura F., Tall A. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9774–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.See R. H., Caday-Malcolm R. A., Singaraja R. R., Zhou S., Silverston A., Huber M. T., Moran J., James E. R., Janoo R., Savill J. M., Rigot V., Zhang L. H., Wang M., Chimini G., Wellington C. L., Tafuri S. R., Hayden M. R. (2002) J. Biol. Chem. 277, 41835–41842 [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Oram J. F. (2007) J. Lipid Res. 48, 1062–1068 [DOI] [PubMed] [Google Scholar]
- 33.Wen Y., Gu J., Chakrabarti S. K., Aylor K., Marshall J., Takahashi Y., Yoshimoto T., Nadler J. L. (2007) Endocrinology 148, 1313–1322 [DOI] [PubMed] [Google Scholar]
- 34.Belkner J., Chaitidis P., Stender H., Gerth C., Kuban R. J., Yoshimoto T., Kuhn H. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 797–802 [DOI] [PubMed] [Google Scholar]
- 35.Weibel G. L., Joshi M. R., Alexander E. T., Zhu P., Blair I. A., Rothblat G. H. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X. S., Kurre U., Jenkins N. A., Copeland N. G., Funk C. D. (1994) J. Biol. Chem. 269, 13979–13987 [PubMed] [Google Scholar]
- 37.Kuhn H., Thiele B. J., Ostareck-Lederer A., Stender H., Suzuki H., Yoshimoto T., Yamamoto S. (1993) Biochim. Biophys. Acta 1168, 73–78 [DOI] [PubMed] [Google Scholar]
- 38.Obinata H., Hattori T., Nakane S., Tatei K., Izumi T. (2005) J. Biol. Chem. 280, 40676–40683 [DOI] [PubMed] [Google Scholar]
- 39.Toda A., Yokomizo T., Shimizu T. (2002) Prostaglandins Other Lipid Mediat. 68–69, 575–585 [DOI] [PubMed] [Google Scholar]
- 40.Ricci R., Sumara G., Sumara I., Rozenberg I., Kurrer M., Akhmedov A., Hersberger M., Eriksson U., Eberli F. R., Becher B., Boren J., Chen M., Cybulsky M. I., Moore K. J., Freeman M. W., Wagner E. F., Matter C. M., Luscher T. F. (2004) Science 306, 1558–1561 [DOI] [PubMed] [Google Scholar]
- 41.Martinez L. O., Agerholm-Larsen B., Wang N., Chen W., Tall A. R. (2003) J. Biol. Chem. 278, 37368–37374 [DOI] [PubMed] [Google Scholar]
- 42.Xie Y., Xu K., Linn D. E., Yang X., Guo Z., Shimelis H., Nakanishi T., Ross D. D., Chen H., Fazli L., Gleave M. E., Qiu Y. (2008) J. Biol. Chem. 283, 3349–3356 [DOI] [PubMed] [Google Scholar]
- 43.Meyer zu Schwabedissen H. E., Grube M., Dreisbach A., Jedlitschky G., Meissner K., Linnemann K., Fusch C., Ritter C. A., Volker U., Kroemer H. K. (2006) Drug Metab. Dispos. 34, 524–533 [DOI] [PubMed] [Google Scholar]
- 44.Pederson T. M., Kramer D. L., Rondinone C. M. (2001) Diabetes 50, 24–31 [DOI] [PubMed] [Google Scholar]
- 45.Won K. A., Reed S. I. (1996) EMBO J. 15, 4182–4193 [PMC free article] [PubMed] [Google Scholar]
- 46.Diehl J. A., Zindy F., Sherr C. J. (1997) Genes Dev. 11, 957–972 [DOI] [PubMed] [Google Scholar]
- 47.Lange C. A., Shen T., Horwitz K. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1032–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Z., Hagler J., Palombella V. J., Melandri F., Scherer D., Ballard D., Maniatis T. (1995) Genes Dev. 9, 1586–1597 [DOI] [PubMed] [Google Scholar]
- 49.Ley R., Balmanno K., Hadfield K., Weston C., Cook S. J. (2003) J. Biol. Chem. 278, 18811–18816 [DOI] [PubMed] [Google Scholar]
- 50.Marchetti S., Gimond C., Chambard J. C., Touboul T., Roux D., Pouyssegur J., Pages G. (2005) Mol. Cell Biol. 25, 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kida A., Kakihana K., Kotani S., Kurosu T., Miura O. (2007) Oncogene 26, 6630–6640 [DOI] [PubMed] [Google Scholar]
- 52.Chang L., Kamata H., Solinas G., Luo J. L., Maeda S., Venuprasad K., Liu Y. C., Karin M. (2006) Cell 124, 601–613 [DOI] [PubMed] [Google Scholar]
- 53.Alvarez E., Northwood I. C., Gonzalez F. A., Latour D. A., Seth A., Abate C., Curran T., Davis R. J. (1991) J. Biol. Chem. 266, 15277–15285 [PubMed] [Google Scholar]
- 54.Cheung P. C., Campbell D. G., Nebreda A. R., Cohen P. (2003) EMBO J. 22, 5793–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuma Y., Campbell D. G., Cuenda A. (2004) Biochem. J. 379, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roosbeek S., Peelman F., Verhee A., Labeur C., Caster H., Lensink M. F., Cirulli C., Grooten J., Cochet C., Vandekerckhove J., Amoresano A., Chimini G., Tavernier J., Rosseneu M. (2004) J. Biol. Chem. 279, 37779–37788 [DOI] [PubMed] [Google Scholar]
- 57.Hatley M. E., Srinivasan S., Reilly K. B., Bolick D. T., Hedrick C. C. (2003) J. Biol. Chem. 278, 25369–25375 [DOI] [PubMed] [Google Scholar]
- 58.Antonipillai I., Nadler J., Vu E. J., Bughi S., Natarajan R., Horton R. (1996) J. Clin. Endocrinol. Metab. 81, 1940–1945 [DOI] [PubMed] [Google Scholar]
- 59.Mauldin J. P., Srinivasan S., Mulya A., Gebre A., Parks J. S., Daugherty A., Hedrick C. C. (2006) J. Biol. Chem. 281, 21216–21224 [DOI] [PubMed] [Google Scholar]
- 60.Spector A. A., Gordon J. A., Moore S. A. (1988) Prog. Lipid Res. 27, 271–323 [DOI] [PubMed] [Google Scholar]
- 61.Brown M. S., Goldstein J. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osborne T. F. (2000) J. Biol. Chem. 275, 32379–32382 [DOI] [PubMed] [Google Scholar]
- 63.Setty B. N., Graeber J. E., Stuart M. J. (1987) J. Biol. Chem. 262, 17613–17622 [PubMed] [Google Scholar]
- 64.Angel A., Farkas J. (1974) J. Lipid Res. 15, 491–499 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.