Abstract
Collapsin response mediator proteins (CRMPs) specify axon/dendrite fate and axonal growth of neurons through protein-protein interactions. Their functions in presynaptic biology remain unknown. Here, we identify the presynaptic N-type Ca2+ channel (CaV2.2) as a CRMP-2-interacting protein. CRMP-2 binds directly to CaV2.2 in two regions: the channel domain I-II intracellular loop and the distal C terminus. Both proteins co-localize within presynaptic sites in hippocampal neurons. Overexpression in hippocampal neurons of a CRMP-2 protein fused to enhanced green fluorescent protein caused a significant increase in Ca2+ channel current density, whereas lentivirus-mediated CRMP-2 knockdown abolished this effect. Interestingly, the increase in Ca2+ current density was not due to a change in channel gating. Rather, cell surface biotinylation studies showed an increased number of CaV2.2 at the cell surface in CRMP-2-overexpressing neurons. These neurons also exhibited a significant increase in vesicular release in response to a depolarizing stimulus. Depolarization of CRMP-2-enhanced green fluorescent protein-overexpressing neurons elicited a significant increase in release of glutamate compared with control neurons. Toxin block of Ca2+ entry via CaV2.2 abolished this stimulated release. Thus, the CRMP-2-Ca2+ channel interaction represents a novel mechanism for modulation of Ca2+ influx into nerve terminals and, hence, of synaptic strength.
Introduction
The N-type presynaptic Ca2+ channel (CaV2.2)2 is the predominant source of Ca2+ influx required for neurotransmitter release at immature CNS synapses (1). Enriched at functional release sites, CaV2.2 forms part of a large macromolecular complex, which facilitates efficient neurotransmitter release. Identification and analyses of protein-protein interactions within the nerve terminal have demonstrated a functional coupling between presynaptic Ca2+ channels and the transmitter release machinery (1–4). Because transmitter release is proportional to the third or fourth power of Ca2+ entry (5–7), even small changes in intracellular Ca2+ can elicit large changes in synaptic transmission. Since expression of N-type-mediated Ca2+ influx coincides with synapse formation, we sought to identify proteins that might modulate Ca2+ channel behavior and transmitter release during synaptogenesis. A proteomic screen of growth cone-associated proteins (see supplemental Fig. S1) and proteins interacting with the α1B subunit of CaV2.2 (8) identified collapsin response mediator protein-2 (CRMP-2) as a candidate CaV2.2-interacting protein.
CRMPs are intracellular phosphoproteins implicated in neuronal growth cone advance and migration (9–13). In neurons, CRMPs are expressed in the lamellipodia and filopodia of growth cones, in the shafts of axons, and in cell bodies. They contribute to axon formation by binding to tubulin heterodimers and promoting assembly of microtubules (14). CRMP phosphorylation causes dissociation from microtubules, leading to axonal outgrowth arrest (10, 15, 16). Overexpression of CRMP-2 in cultured hippocampal neurons induces extra axons (12), whereas CRMP-3/4/5 have been shown to be necessary for neurite extension (10), branching (17), and growth cone formation (18). The breadth of interactions of CRMP proteins with motor proteins, kinases, enzymes, and endocytosis-exocytosis-related proteins suggests that CRMPs may serve as adaptors/scaffold molecules (10). Although neurotransmission was not affected in CRMP-1 knock-out mice (19), long term potentiation, spatial learning, and memory were affected in both CRMP-1 and CRMP-3 knock-out mice (19, 20), suggesting a role for CRMPs in neurotransmission as well as synaptogenesis. As potential binding partners, CRMPs could affect neurotransmission by a physical interaction with either the synaptic machinery or CaV2.2 itself.
We tested this hypothesis by examining the molecular and mechanistic basis of CRMP-2 interaction with CaV2.2. We identify a direct binding between the proteins and show that CRMP-2 mediates an increase in Ca2+ current density, which, together with the effects of CRMP-2 on synapse size and synaptic vesicle loading, results in increased transmitter release. Thus, the CRMP-2-CaV2.2 interaction represents a novel mechanism for modulation of Ca2+ influx into nerve terminals and hence of synaptic strength.
EXPERIMENTAL PROCEDURES
Synaptosome Isolation
Synaptosomes were prepared as described (21, 22) with modifications. Briefly, brains from neonatal postnatal day 1 (PN1) Sprague-Dawley rats (Harlan Laboratories, Indianapolis, IN) were dissected into 10 volumes of ice-cold homogenization buffer A (0.32 m sucrose, 10 mm HEPES, pH 7.4, 2 mm EDTA, supplemented with protease inhibitors) and homogenized using 10–15 strokes of a glass Teflon hand-held homogenizer. The homogenate (H) was spun for 10 min at 1400 × g at 4 °C, and the supernatant was saved. The pellet (P1) from this spin was rehomogenized in buffer A and spun as before. Both low speed spin supernatants were combined and spun for 10 min at 13,800 × g at 4 °C. The pellet from this spin (P2) was homogenized in buffer B (0.32 m sucrose, 0.5 mm HEPES/KOH, pH 7.4, and protease inhibitors), layered onto a sequential gradient of 1 and 1.4 m sucrose, and centrifuged at 82,500 × g (4 °C) for 90 min in a swinging bucket rotor with slow acceleration and deceleration. The gradient interphase contained intact synaptosomes (P2′), which were used for depolarization experiments within 2 h of isolation or lysed in modified RIPA buffer (50 mm Tris-HCl, pH 8, 1% Nonidet P-40, 150 mm NaCl, 0.5% sodium deoxycholate, and 1 mm EDTA, supplemented with freshly added protease inhibitors: 1 μg/ml leupeptin, 2 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride (Sigma) together with a protease inhibitor mixture (Roche Applied Science)) and filtered through a 0.22-μm syringe filter before their protein concentration was determined using a BCA assay (Thermo Fisher Scientific, Shelbyville, IN). Fractions stored at −80 °C until use.
Co-immunoprecipitation and Western Blotting
Co-immunoprecipitation and Western blotting were performed as described previously (8, 23–25). Rat brain synaptosomes were precleared by a 1-h incubation with 20 μl of a 50% slurry of protein A/G beads (Pierce). The cleared lysate was then incubated overnight with various primary antibodies (see supplemental Table 2) or rabbit or mouse isotype-specific IgGs (Sigma) as controls. The antibody-captured complexes were recovered with fresh protein A-agarose (for rabbit polyclonal antibodies) or protein A/G-agarose (for mouse monoclonal antibodies) beads (20 μl of original bead slurry/sample) by incubation with lysate/antibody mixture at 4 °C for 2 h. The beads were then washed three times with lysis buffer. Prior to electrophoresis on SDS-polyacrylamide gels, protein samples were boiled in Laemmli sample buffer for 5 min. Proteins were fractionated on 5, 7.5, 10, or 4–15% separating gels with 4% stacking gels. Apparent molecular weights were determined using broad range standards (Fermentas). Following electrophoresis, proteins were transferred to polyvinylidene difluoride membranes (Invitrogen) for immunoblotting and stained with Amido Black (Bio-Rad) to monitor transfer efficiency. The membranes were blocked for 1 h in 5% skim milk powder in TBST (25 mm Tris-Cl, pH 8.0, 125 mm NaCl, 0.1–2% polyoxyethylene sorbitan monolaurate (Tween 20)) at room temperature. Primary antibody incubations were for 2 h at room temperature or overnight at 4 °C. Following incubations with primary antibody and secondary antibody (goat anti-rabbit or anti-mouse IgG horseradish peroxidase (1:10,000; Stressgen, Ann Arbor, MI)), blots were washed extensively in TBST and probed with enhanced chemiluminescence Western blotting substrate (Thermo Scientific) before exposure to photographic film. Blots were exposed for a range of durations to ensure the generation of a print in which the film is not saturated. Films were then scanned, digitized, and quantified using Un-Scan-It gel version 6.1 scanning software (Silk Scientific Inc., Orem, UT), limiting our analysis to the linear range.
Sucrose Density Gradient Centrifugation
To assess the subcellular distribution of CRMP-2 and CaV2.2 proteins, sucrose density gradients were prepared from PN1 rat brains as described (26). Rat brains (∼2 mg) were solubilized in 2 ml of 1% Triton X-100 (Sigma) in MBS (25 mm MES, pH 6.5, 150 mm NaCl). Following homogenization using 10–15 strokes of a glass Teflon hand-held homogenizer, the synaptosomes were adjusted to 45% with sucrose and overlaid with 4 ml of 30% sucrose in MBS and 4 ml of 5% sucrose in MBS. The sucrose gradient was formed by centrifuging at 200,000 × g for 18 h at 4 °C using a Beckman SW41 rotor (Beckman Coulter, Mississauga, Canada). Twelve 1-ml aliquots were removed, beginning at the top. The high density membrane fractions (fractions 9–12) at the bottom of the gradient contain proteins like the Na+/K+ ATPase and voltage-gated ion channels (26), and fractions 5 and 6 are rich in cholesterol and sphingolipids, confirming the ability of this centrifugation method in separating membrane and cellular components.
Cloning CRMPs and CaV2.2 Channel Fragments into pGEX-Glu Vectors
Regions of CRMP-2 were cloned into pGEX3X-GST, whereas the intracellular loops of the N-type Ca2+ channel were cloned into the dually tagged pGEX3X-GST-Glu vector for expression of recombinant proteins. Accession numbers for the clones used for this study are NM_147141 for CaV2.2 and NM_009955.2 for CRMP-2. Further details of the molecular cloning are provided in the supplemental material.
Recombinant Protein Production
Recombinant protein production was as previously described (27). DNA encoding sequence-verified pGex-Glu-CaV2.2-type channel constructs were transformed into BL21 (DE3) Escherichia coli for protein expression. CRMP-2-GST fusion constructs were generously provided by Dr. Akihiro Kurimasa (Tottori, Japan) (28), and a Munc18-1-GST fusion vector was provided by Dr. Debbie Thurmond (Indiana University School of Medicine). Expression of fusion proteins was induced with 1 mm isopropyl-β-d-thiogalactopyranoside. For purification, following overnight growth at 16 °C, transformed bacteria were pelleted and lysed in buffer containing 20 mm Tris, pH 7.5, 200 mm NaCl, 0.1 mm EDTA, 1 mm dithiothreitol, and protease inhibitors using an M-110L microfluidizer (Microfluidics Corp., Newton, MA). The microfluidized lysate was adjusted with Triton X-100 (1% final), and the cells were incubated on ice for 30 min followed by centrifugation at 30,000 × g for 45 min at 4 °C. The high speed supernatant was then incubated with either glutathione-Sepharose beads or protein G-agarose beads conjugated with mouse monoclonal Glu primary antibody (a gift from Dr. Clark Wells, Indiana University School of Medicine). Immunoblot of a representative purification is shown in Fig. 4B.
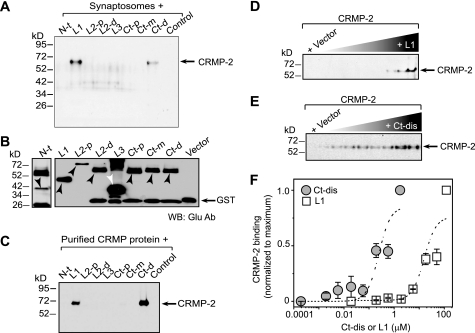
FIGURE 4.
CRMP-2 binds directly to two cytoplasmic regions within CaV2.2. A, pull-down assays of CRMP-2 with GST-Glu fusion CaV2.2 constructs. Synaptosomes (A) or purified CRMP-2 protein (C) were incubated with equimolar amounts of glutathione beads saturated with GST-CaV2.2-Glu fusion proteins or GST protein alone. Bound proteins were analyzed by immunoblotting with CRMP-2 antibody. CRMP-2 bound to L1 and the Ct-d. The N terminus (N-t), intracellular loop 2 proximal and distal parts (L2-p and L2-d), the third intracellular loop (L3), and the proximal and medial parts of the C terminus (Ct-p and Ct-m) as well as the GST-Glu protein (control) showed no binding in these assays. B, Western blot with Glu antibody of GST- and Glu-tagged calcium channel fragments used in the binding experiments. GST-Glu-L1 (D) or GST-Glu-Ct-dis (E) proteins at increasing concentrations, incubated with CRMP-2 (∼500 nm), were captured by protein A-Sepharose beads. Captured CRMP-2 proteins were identified by immunoblotting with CRMP-2 antibodies. Quantitative densitometry was performed on the bands shown in D and E. F, data representing the normalized average band intensity ± S.E. (n = 3) were subjected to a Sigmoidal curve-fitting method to evaluate the apparent dissociation constant (Kd). The CaV2.2-CRMP-2 Kd values were as follows: L1, ∼24.1 ± 7.1 μm; Ct-dis, ∼0.31 ± 0.09 μm.
CRMP-2 Purification
CRMP-2 purification was performed as described previously with slight modifications (29). Briefly, adult rat brains (∼10 g) were homogenized in 3 volumes of DEAE buffer (25 mm sodium phosphate (pH 7.8) containing 10 mm 2-mercaptoethanol and 1 mm phenylmethylsulfonyl fluoride) with a Teflon/glass homogenizer. The homogenate was centrifuged at 200,000 × g for 40 min at 4 °C, and the supernatant was applied to a 1-ml DEAE-Sephacel column (Sigma) equilibrated with DEAE buffer. The column was washed thoroughly with DEAE buffer, and bound proteins were eluted with 100, 200, and 300 mm NaCl step gradients in DEAE buffer. Two-ml fractions were collected at each salt concentration. Peak CRMP-containing fractions (as monitored by Western blot with a polyclonal CRMP-2 antibody) in the 200 mm NaCl elution was diluted 10-fold with S buffer (25 mm sodium phosphate (pH 6.0) containing 10 mm 2-mercaptoethanol) and applied to a 2-ml S-Sepharose (GE Healthcare) column equilibrated with S buffer. After washing, proteins were eluted in 150 mm NaCl in S buffer. One 3-ml fraction was collected, diluted 5-fold with HA buffer (10 mm potassium phosphate (pH 7.0) containing 10 mm 2-mercaptoethanol). The diluted sample was applied to a 1-ml hydroxyapatite column (Bio-gel HTP; Bio-Rad) and eluted with 100 mm potassium phosphate (pH 7.0) containing 10 mm 2-mercaptoethanol. Six fractions of 0.5 ml each were collected. At this step, Coomassie Blue staining and Western blot analysis was performed to verify CRMP purification at the predicted molecular mass of 65 kDa. Coomassie and immunoblot analysis of a representative purification is shown in supplemental Fig. S8.
In Vitro Protein-binding Assay
For binding studies, the cytoplasmic loops of CaV2.2 and full-length (as well as deletional) CRMP-2 proteins were purified as GST or GST-Glu-tagged proteins, respectively. Binding reactions were performed in binding buffer (20 mm HEPES, 150 mm NaCl, 2 mm MgCl2, 1 mm dithiothreitol, 0.5% (v/v) Triton X-100, pH 7.4). Monoclonal Glu antibody-saturated Protein G beads (GE Healthcare) carrying various CaV2.2 cytoplasmic loop constructs or GST-Glu were incubated with rat brain purified CRMP-2 protein in a total reaction volume of 100 μl. Reactions were incubated end-over-end for 4 h at 4 °C and washed three times with a 400-fold excess of binding buffer, and the proteins were eluted in 60 μl of SDS gel buffer and boiled for 5 min, after which 20 μl of each assay was run on SDS-PAGE and analyzed by immunoblotting with CRMP-2. To determine binding affinity (Kd), the experiments were performed using a fixed amount of rat brain purified CRMP-2 protein and increasing amounts of CaV2.2-GST-Glu fusion proteins.
Isolation and Culture of Hippocampal Neurons
Rat hippocampal neuron cultures were prepared from hippocampi dissected from PN1 rats as described (30). Briefly, rat hippocampi were dissected out of PN1 rats, and cells were dissociated enzymatically and mechanically (trituration through Pasteur pipette) in a Papain solution (12 units/ml; Worthington) containing Leibovitz's L-15 medium (Invitrogen), 0.42 mg/ml cysteine (Sigma), 250 units/ml DNase I (type IV; Sigma), 25 mm NaHCO3, penicillin (50 units/ml)/streptomycin (50 μg/ml), 1 mm sodium pyruvate, and 1 mg/ml glucose (Invitrogen). After dissociation, the cells were gently washed by sequential centrifugation in Neurobasal medium containing either 2 or 20 mg/ml bovine serum albumin and penicillin/streptomycin, glucose, pyruvate, and DNase I (as above) and then plated on poly-d-lysine-coated Aclar coverslips at high density (∼2000 cells/mm2). Growth media (1 ml/well) consisted of Neurobasal medium containing 2% NuSerum, 2% B27, supplemented with penicillin/streptomycin (100 units/ml; 50 μg/ml), 0.1 mm l-glutamine, and 0.4 mm l-Glutamax (Invitrogen). Cytosine β-d-arabinofuranoside (5 μm; Sigma) was added 24 h after plating to reduce the number of non-neuronal cells. After 4 days in culture and two times each week thereafter, half of the growth medium was replaced with medium without cytosine β-d-arabinofuranoside.
Immunocytochemistry, Confocal Microscopy, and Iterative Deconvolution Deblurring
24–48 h after transfection, hippocampal cultures were fixed with 4% paraformaldehyde (diluted in 0.1 mm PBS) for 10 min at room temperature, permeabilized with 0.2% Triton X-100 for 10 min, and then washed three times with 0.01 mm PBS. The neurons were then preincubated with 10% bovine serum albumin (diluted in 0.1 mm PBS) for 1 h at room temperature to block nonspecific binding with the primary antibody. Primary antibodies for monoclonal CRMP-2 (Chemicon International, Billerica, MA) or β-III tubulin (Abcam Inc., Cambridge, MA) and rabbit polyclonal N-type/CaV2.2 (Calbiochem) or synaptophysin (Abcam Inc.) were diluted (in 0.1 mm PBS) to 1:100 and applied to the cells. After incubation at 4 °C overnight, the neurons were washed again with PBS, and secondary antibodies (goat anti-mouse Alexa 488 or anti-rabbit Alexa 594, 1:1000; Molecular Probes, Inc., Eugene, OR) were incubated in blocking solution for 45 min at room temperature. Coverslips were mounted in Prolong Gold Antifade mounting media (Molecular Probes, Inc.). The hippocampal neurons were imaged on a Nikon Ti swept-field confocal microscope using a ×60, 1.4 numerical aperture lens and standard fluorescein isothiocyanate/Texas Red fluorescence cubes with a cooled Cascade 512B digital camera (Photometrics, Tucson, AZ). Z stack image pairs were captured at an interplane distance of 200 nm through the sample. Images were deblurred off-line by an iterative deconvolution protocol (Nikon Elements version 3.0) using a theoretical point spread function and pseudocolored for presentation.
Intensity Correlation Analysis (ICA)/Intensity Correlation Quotient (ICQ)
Developed by Dr. E. F. Stanley (Toronto Western Research Institute, Toronto, Canada), this analysis has been described in detail (23). ICA/ICQ analysis was carried out by means of an automated graphic plugin (23) for the public domain image analysis software ImageJ (Wayne Rasband, National Institutes of Health, Bethesda, MD). For the ICA, we calculated the function (Ai − a)(Bi − b), where a and b are the means of the pixel staining intensity values Ai and Bi. Ai or Bi was graphed in separate scatter plots against its respective (Ai − a)(Bi − b) value. Distributions that skew to the right reflect dependent staining patterns (where the two pixel staining intensity values vary in synchrony); distributions that are symmetrical about the 0 axis indicate random staining, whereas those that skew to the left reflect independent staining patterns, where the pixel staining intensity values vary inversely. Analysis for each stain was performed separately so that a dependence of stain A on B but a lack of dependence of B on A can be identified, and, further, these plots permit detection of complex or mixed staining relations. The ICQ reflects the ratio of the number of positive (Ai − a)(Bi − b) values to the total number of pixels in the region of interest, corrected to a −0.5 (independent staining) to +0.5 (dependent staining) range by subtracting 0.5. The ICQ provides a single value parameter that is used for statistical comparisons. Typically, with n > 6 regions of interest, a mean ICQ value of −0.05 to +0.05 indicates random staining, +0.05 to +0.10, indicates a moderate covariance, and >0.1 indicates a strong covariance. Images shown in Figs. 2, 8, and 10 were enhanced using the smart sharpen mask filter, high pass, and brightness-contrast adjustment functions in Adobe Photoshop. Images were processed with Nikon Elements version 3.0, and all figures were constructed in Adobe Illustrator 9 (Adobe Systems).
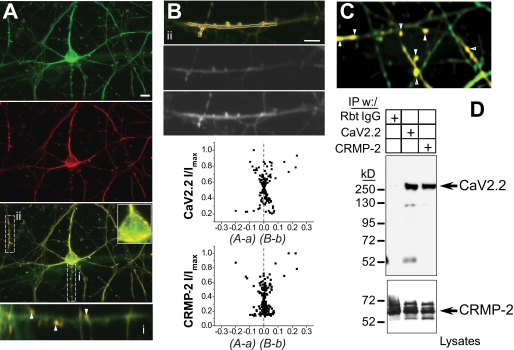
FIGURE 2.
CRMP-2 colocalizes and associates with CaV2.2 in hippocampal neurons. A, double labeling of endogenous CRMP-2 (green) and CaV2.2 (red) in cultured hippocampal neurons. The enlarged regions show co-localization of both proteins at the cell surface of the soma and in punctate structures along neurites (i and ii). B, merged and black and white confocal images of region ii from A. Quantitative co-localization analysis shows a moderate to strong skew in the ICA plots for CaV2.2 and CRMP-2, indicative of staining pairs that vary in synchrony. The ICQ of the region in ii was 0.178. C, confocal image of hippocampal neurons double-labeled with CRMP-2 (green) and CaV2.2 (red). The arrows show boutons and dendritic spines with strong colocalization. D, immunoblot with CaV2.2 antibody showing that CRMP-2 and CaV2.2 immunoprecipitate (IP) from purified hippocampal neurons grown for 14 DIV. The bottom blot, probed with CRMP-2 antibody, represents 10% of the lysates used in the immunoprecipitations. Rbt IgG, isotype-specific rabbit IgG (control) antibody.
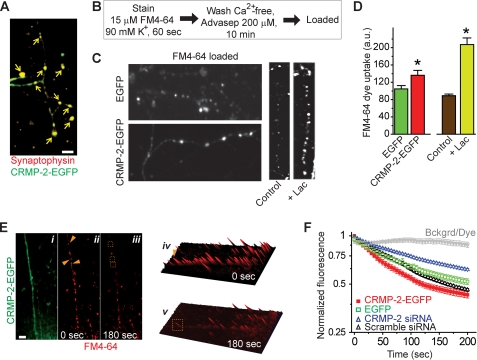
FIGURE 8.
CRMP-2 increases synaptic vesicle recycling. A, hippocampal neurons at 10 DIV were transfected with CRMP-2-EGFP (green) and immunostained with an antibody against synaptophysin (red), a marker of synapses. The arrows show that CRMP-2-EGFP is targeted to synapses. B, schematic of FM4-64 experiments to measure FM4-64 uptake. C, representative images of cultured hippocampal neurons, transfected with EGFP or CRMP-2-EGFP, taken at the end of the 10-min wash period in Ca2+-free solution containing the membrane-bound dye quencher Advasep-7. Also shown are images from untransfected cells treated with 0.01% DMSO (control) or 10 μm clastolactacystin-β-lactone (Lac) added 2 h prior to the start of the experiment. Treatment with the proteasome inhibitor Lac increased the amount of initial dye uptake, as reported previously (34). D, average FM4-64 dye uptake, in arbitrary units (a.u.), is plotted as mean ± S.E. Synapses in CRMP-2-EGFP-expressing neurons had a ∼30% higher initial dye uptake than EGFP neurons (p < 0.05). Consistent with previously published data (34), we observed an ∼2-fold increase in dye uptake in cells exposed to the proteasome inhibitor Lac. E, representative pseudocolor fluorescence images of synapses loaded with FM4-64 at time 0 (ii and iv) and 3 min after destaining with 90 mm KCl (iii and v). The corresponding green fluorescent protein image of this neuron is also shown (i). Images in iv and v represent three-dimensional intensity projections of FM4-64 at the indicated times. The arrows and boxes illustrate examples of synapses that destain during the course of the experiment. F, the destaining kinetic curves of FM4-64 taken up by presynaptic terminals expressing EGFP (green squares) and CRMP-2-EGFP (red squares) in the presence of 90 mm KCl. Normalized fluorescence intensities are plotted against time (s). Also shown are curves for neurons transfected with CRMP-2 siRNA (blue triangles) or scrambled siRNA controls (black triangles). As a control, fluorescence intensities from regions of background or styryl dye under repetitive laser scanning have been plotted (Bckgrd/Dye, gray triangles). Scale bar, 10 μm (all panels).
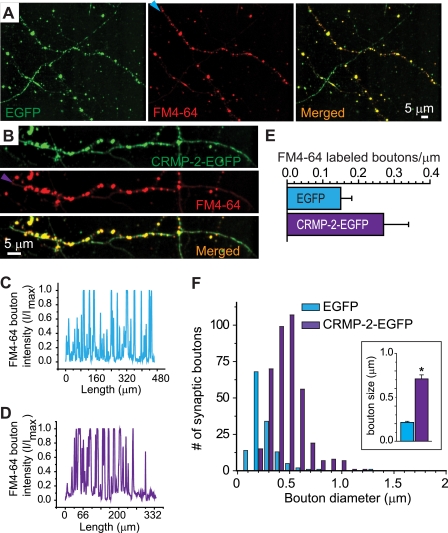
FIGURE 10.
CRMP-2 increases bouton size. Shown are representative pseudocolor images of EGFP (green) and FM4-64 (red) from neurons transfected with EGFP (A) or CRMP-2-EGFP (B). The arrowheads in A and B represent axons along which a line scan was performed to quantify the number of boutons (labeled with FM4-64) in EGFP (C) and CRMP-2-EGFP (D) neurons. Boutons were identified by counting intensity peaks that traversed the 50% maximum intensity level. Representative line intensity staining profiles of the FM4-64 panels in A and B are shown in C and D, respectively. E, summary of the average number of boutons/μm of axon. The density of boutons was not different between EGFP- and CRMP-2-EGFP-expressing neurons (p > 0.05). F, histogram of diameters from boutons of EGFP- and CRMP-2-EGFP-expressing neurons. The largest diameter of each bouton was measured in a double-blind manner; the resulting diameters were grouped into bins of 0.1 μm. Bouton diameters were distributed unimodally in both EGFP- and CRMP-2-EGFP-expressing neurons. Inset, mean bouton size of EGFP and CRMP-2-EGFP showed the average diameter of boutons to be ∼3.3-fold larger in CRMP-2-EGFP-expressing neurons than in EGFP-expressing neurons (p < 0.05, Student's t test).
Transfection/Infection of Hippocampal Neurons
Adherent hippocampal cultures were transfected with cDNAs using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions or infected with lentivirus expressing CRMP-2 or scrambled short hairpin RNAs (shRNAs). We routinely achieve about 10–15% transfection efficiencies in hippocampal neurons transfected with this method. With lentiviral infections, we have observed up to 95% infection in ≤7 days. Typically, cultured neurons were transfected with equal amounts of different cDNA constructs at 12 days in vitro (DIV), and electrophysiology experiments were performed at 14 DIV.
Cell Surface Biotinylation
Biotinylation was performed as described (27). Hippocampal neurons grown for 10 DIV were transfected with EGFP or CRMP-2-EGFP for 2 days and then processed for biotinylation experiments. Live cells were incubated with sulfosuccinimidyl 2-(biotinamido) ethyl-1,3′dithiopropionate (1 mg/mg protein; Pierce) for 30 min at 4 °C in cold PBS, pH 8.0. Excess biotin was quenched with PBS containing 100 mm glycine and washed three times with ice-cold PBS, and the pellet was resuspended in RIPA lysis buffer. The resuspended pellet was triturated 10 times (25-gauge needle) and centrifuged at 100,000 × g for 20 min. The biotinylated proteins were separated from clear solubilizate by adsorption onto streptavidin-agarose beads (Novagen) for 2–4 h at 4 °C. Beads were washed 3–5 times with RIPA buffer, and bound biotinylated proteins were gently eluted off of the beads with RIPA buffer containing 2% Triton X-100 and 650 mm NaCl by end-over-end incubation for 1 h at 30 °C. The biotinylated fraction was subjected to immunoblotting with CRMP-2 and CaV2.2 antibodies. To ensure that the cells remained intact throughout the surface labeling, biotinylation of β-tubulin, a cytosolic protein, was analyzed by immunoblotting (data not shown).
Electrophysiology
Whole-cell Ca2+ currents were recorded as described previously (31). Whole-cell calcium currents were isolated using the following solutions: external (bath) medium (128 mm NaCl, 5 mm KCl, 1 mm MgCl2, 10 mm BaCl2, 10 mm d-glucose, 10 mm HEPES-Na, and 10 mm tetraethylammonium-Cl, with an osmolarity of 300 mosmol/liter and pH 7.3) and patch electrode intracellular solution (110 mm CsCl2, 10 mm EGTA, 1 mm MgSO4, 25 mm HEPES(Cs), 2 mm ATP, and 0.2 mm GTP, with an osmolarity of 305 mosmol/liter and pH 7.4). Tetrodotoxin (1 μm; Alomone Laboratories) and nifedipine (1 μm; Calbiochem) were added in the bath solution to block Na+ and L-type Ca2+ channels. Under these conditions, the primary Ca2+ current component should be of the N- and P/Q-type with some R-type Ca2+ current (32).
Voltage Protocols and Data Analysis
Cells were held at a holding potential of −80 mV. Immediately after seal rupture, −80 to +80 mV ramp depolarizations were given until recording current stability was established. Current to voltage relations were assessed using incrementing 10-mV step depolarizing pulses at 1-s intervals. Leak currents were subtracted on-line using a −P/5 procedure. Data were acquired using an EPC-10 amplifier (HEKA Electronics) and analyzed using Pulsefit (version 8.5; HEKA) and Origin 7.0 software (Microcal, Northampton, MA).
FM4-64 Imaging
This was completed exactly as described (33). FM dye imaging was performed on hippocampal cultures 48 h post-transfection. Presynaptic terminals were loaded with the fluorescent styryl dye N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide (FM4-64; 15 μm) by incubation of the cells for 1 min in high K+ solution containing 58 mm NaCl, 90 mm KCl, 10 mm HEPES, 3 mm CaCl2·2H2O, 8 mm glucose, and 2 mm MgCl2·6H2O (pH 7.3). The cells were then washed in Ca2+-free solution for 10 min to reduce nonspecific staining. In some experiments, advasep-7 (200 μm; Biotium, Inc., Hayward, CA) was used to quench any FM4-64 dye not taken up by endocytosis (34, 35). The nerve terminals of optically distinguishable axons of neurons expressing EGFP or CRMP-2-EGFP were identified under a confocal microscope (Nikon Livescan SFC inverted microscope) using an oil immersion CFI Plan APO VC ×60 objective lens. EGFP-labeled terminals capable of dye uptake were considered to be functional release sites. Fluorescence of EGFP and the FM dye were excited at 448 and 543 nm, respectively. Quantitative measurements of FM4-64 fluorescence intensity at individual synapses were obtained by averaging a 4 × 4 square area of pixel intensities centered on the optical center of a given fluorescent punctum. The individual punctum was selected by hand, and the optical center of mass used to center the measurement box was calculated over a slightly larger area (typically 10 × 10 pixels). Large puncta, typically representative of clusters of smaller synapses, were not analyzed during the selection procedure, as were any puncta that were not clearly discernible during the time course of the experiment. Areas for imaging contained networks of well defined dendrites and no cell bodies. At least 100 boutons from each transfection condition were imaged in each of these experiments. During the time lapse imaging of fluorescence change, consecutive frames were acquired at 2-s intervals. After ∼20 s of steady-state recording, high K+ solution was locally perfused to destain FM4-64 dye. All data were acquired and analyzed with Nikon NIS-Elements version 3.0 software.
Construction of pLL3.7-CRMP-2 and Control shRNA Lentiviral Shuttle Vectors
Published target sequence against rat CRMP-2 (5′-GTAAACTCCTTCCTCGTGT-3′ (36) and 5′-GCCTATTGGCAGCCTTTGA-3′) harboring HpaI and XhoI restriction sites were cloned into pLentilox3.7 (pLL3.7; a generous gift of Dr. Michael R. Vasko, Indiana University School of Medicine) to create the pLL3.7-CRMP-2-v1 and pLL3.7-CRMP-2-v2 expression vectors. Scrambled shRNA sequences with approximately the same percentage of GC but no sequence homology were used to create control lentiviral vectors.
Recombinant Lentivirus Production
Self-inactivating LV vectors were packaged as described (37). Briefly, 293T cells were transfected with a four-plasmid vector system (i.e. the shuttle plasmid (LV-CRMP-2 shRNA-v1-CMV-EGFP, LV-CRMP-2 shRNA-v2-CMV-EGFP, or LV-CMV-EGFP) and the packaging plasmids pMDMg/pRRE, pRSV-Rev, and pCMV-VSV-G, to provide the VSV-G envelope. Two days post-transfection, the virus-containing medium was collected, and debris was removed by centrifugation. Virus was concentrated by ultracentrifugation, and the viral pellet was suspended in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, passed through a 0.45-μm filter, and frozen at −80 °C. The viral titer, as determined by analysis of the virus-associated p24 core protein (QuickTiter lentivirus quantification kit (Cell Biolabs, Inc., San Diego, CA)), was above 2 × 108 transducing units/ml for all viruses used in this study. Virus particles were produced in the Stark Neurosciences Research Institute Viral Production Core Facility.
Knockdown of Gene Expression by siRNA
Validated siRNAs against the rat CRMP-2 (5′-ACTCCTTCCTCGTGTACAT-3′) sequence (36) and controls (scrambled sequence with approximately the same percentage of GC but no sequence homology) were used for CRMP-2 knockdown (Invitrogen). Cells were incubated for 2–4 days with CRMP-2-siRNA or scrambled siRNA (50–1000 nm optimized for each condition), and assessment of knockdown was by quantitative reverse transcription-PCR, Western blot (see supplemental Fig. S6), immunocytochemistry, and electrophysiology.
Glutamate Release Assay, HPLC
Glutamate released into the cytosol in CRMP-2-overexpressing or ablated hippocampal neurons was measured by HPLC separation and electrochemical detection of the o-phthaldialdehyde-mercaptoethanol derivative using modifications of the methods of Donzanti and Yamamoto (38). Using this method, the minimal detectable amount of glutamate is ∼2.5 pmol. Briefly, following a series of washes in phosphate-buffered saline and a 1-h incubation in normal Tyrode's buffer, 400-μl samples were sequentially collected every 10 min from the cytosol: 1) bathed in Tyrode's buffer, 2) bathed in Tyrode's buffer in the absence or presence of drug, 3) bathed in high K+ Tyrode's buffer, and 4) rebathed in Tyrode's buffer. These samples were collected on ice, filtered with 0.22-μm filters (Millipore), and processed for HPLC analyses. The resulting four values represent basal 1, +drug, stimulated, and basal end for each sample. Samples were processed in triplicate and averaged to yield a single value.
Several measures were used to ensure that comparable numbers of viable cells were present in all wells within a dish. Cells in representative wells were counted immediately before the release experiment, and trypan blue exclusion was used to assess the viability of cells before and after the release experiment. In addition, the average basal releases of neurotransmitter from each treatment group (in the absence of drug) in an experiment were within 20% of each other. The cells exhibited robust KCl-stimulated neurotransmitter release at least 3-fold over the basal levels. Post-treatment basal release (in the absence of drug) was measured in some glutamate assays and was not more than 50% higher than initial basal levels, indicating that the release assay did not damage the cells or induce cell lysis (data not shown). These observations, when taken together, indicate that similar numbers of viable cells were present in all wells within an experiment.
Statistical Analyses
Differences between means were compared by either paired or unpaired two-tailed Student's t tests or an analysis of variance when comparing multiple groups (repeated measures whenever possible). With the analysis of variance, if a significant difference is determined, then Tukey's post hoc test was performed. Data are expressed as mean ± S.E., with p < 0.05 considered as the level of significance. Imaging (ICA/ICQ) values were tested either for two means between conditions (P) or for one mean for a difference from 0 (P=0). Each hippocampal ICQ value reflects the mean of two determinations from image planes at least 400 nm apart (two z axis increments). The normal approximation to the sign test was used for statistical tests of ICQ values for single experiments.
RESULTS
Identification of CRMPs as a Novel Ca2+ Channel Target
To identify Ca2+ channel binding proteins that might contribute to cytoskeletal interactions in nascent synapses, we performed tandem mass spectrometric analysis of growth cone particles. The purity of the biochemically fractionated growth cone particle fraction was evaluated using immunoblot analysis for proteins reported to be prevalent at the growth cone, such as GAP-43 (growth cone-associated protein 43) and those absent, such as GFAP (glial fibrillary acidic protein); both CaV2.2 and CRMP-2 were present in synaptosomes as well as in growth cone particle fractions (supplemental Fig. S1) of PN1 and PN8 neonatal rats. Liquid chromatography-tandem mass spectrometry analyses identified 148 and 464 proteins in the PN1 and PN8 growth cone particle fractions, respectively. The 612 identified proteins were classified within a number of broad functional classes (supplemental Fig. S1). Prominent among these were growth cone guidance and migration proteins, including CRMPs. CRMPs were also found in a screen for binding partners of the presynaptic Ca2+ channel from purified synaptosomes (8).
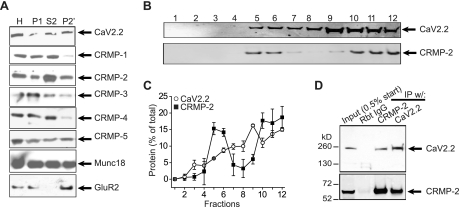
A CRMP-2-CaV2.2 Complex
To test for a biochemical complex between CaV2.2 and CRMPs, we prepared synaptosomes (P2′) from neonatal PN1 rat brains and determined that all five CRMP proteins were present in synaptosomes; CRMP-2 protein was relatively more abundant compared with its homogenate levels than the other CRMPs (Fig. 1A). Synaptic proteins Munc18 and GluR2 (glutamate receptor 2 subunit) were used as loading controls as well as for assessment of the fractions (39). Next, sucrose density gradient fractionation of PN1 rat brains revealed co-sedimentation of CRMP-2 and CaV2.2 in both cholesterol-rich/cytosolic (Fig. 1, B (lanes 5–8) and C) and membrane fractions (Fig. 1, B (lanes 9–12) and C), consistent with cytosolic and membrane distribution of both proteins. Finally, to confirm that CRMP-2 had a bona fide interaction with CaV2.2, we performed standard co-immunoprecipitations and immunoblotting. Precipitation of CaV2.2 with a polyclonal CaV2.2 antibody (Calbiochem; n = 11) or a polyclonal CRMP-2 antibody (Cell Signaling; n = 8) from PN1 rat brain synaptosomes reliably precipitated the associated protein (Fig. 1D). No proteins were observed in control immunoprecipitations using rabbit isotype-specific IgG antibodies (Fig. 1D). The interaction of CaV2.2 and CRMP-2 was also observed in hippocampal neurons cultured for 14 DIV (see Fig. 2D) and with chick and mouse brain synaptosomes (data not shown). Thus, biochemical evidence clearly shows that CRMP-2 and CaV2.2 co-exist in a complex.
FIGURE 1.
CRMP-2 association with CaV2.2. A, Western blots of CaV2.2, CRMP1 to -5, and synaptic proteins Munc18 and GluR2 in homogenate (H), pellet (P1), supernatant (S2), and synaptosomes (P2′) of PN1 rats. B and C, CRMP-2 and CaV2.2 are present in both cytosolic (lanes 5–8) and membrane (lanes 9–12) fractions in sucrose density gradients from PN1 rat brains. The summary of fractionation data represents mean ± S.E. of four experiments. D, CRMP-2 and CaV2.2 immunoprecipitate (IP) reciprocally from purified synaptosomes. Rbt IgG, isotype-specific rabbit IgG (control) antibody.
Immunocytochemistry on PN1 rat hippocampal neurons revealed the presence of CRMP-2 in soma, dendrites, neurite shafts, and punctate structures defined to be in presynaptic boutons by co-labeling with the synaptic vesicle marker protein synaptophysin (supplemental Fig. S2, A and B). Co-staining with growth cone-enriched protein F-actin showed strong colocalization within growth cones (supplemental Fig. S2C). We observed an overlap in the staining patterns for CRMP-2 and CaV2.2 in the soma as well as in presynaptic varicosities (Fig. 2). ICA was used to quantitatively assess the colocalization (23–25, 40). This method tests if staining for two proteins co-varies, as they should if the two proteins are part of the same complex. This test generates an ICQ, which ranges from −0.5 to +0.5 and can be used for statistical comparison. ICA plots of CaV2.2 and CRMP-2 staining of the soma cell surface, in the axons, and in boutons/dendritic spines (see examples in Fig. 2, B and C) exhibited varying degrees of positive skews, suggesting strong to moderate levels of covariance. The mean ICQ values were 0.078 ± 0.017 (axons, n = 12, P=0 < 0.001), 0.038 ± 0.052 (surface of soma, n = 7, P=0 < 0.001), and 0.245 ± 0.073 (boutons/dendritic spines, n = 9, P=0 < 0.001). We have previously shown that staining for a membrane protein, the Na+/K+ ATPase, does not co-vary with that for CaV2.2 (40). Thus, these results indicate that CaV2.2 and CRMP-2 exist in a complex.
Binding Requirements between CRMP-2 and N-type Ca2+ Channels
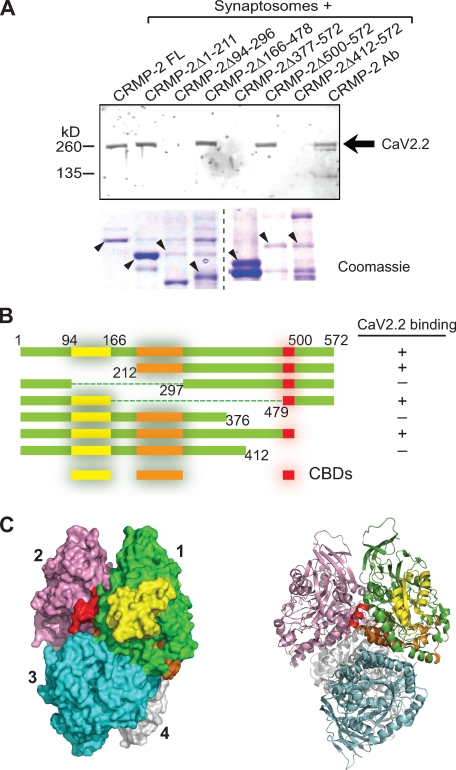
To examine the possibility that CRMP-2 and CaV2.2 proteins may bind directly to each other, in vitro pull-down assays were performed to assess binding between the two proteins. First, we purified bacterially expressed GST fusion proteins of full-length CRMP-2 or fragments and incubated them with PN1 rat synaptosomes. To ensure that the synaptosomes contained a sufficient amount of CaV2.2, the channels were partially purified with wheat germ agglutinin as described by Catterall and co-workers (41), followed by enrichment with heparin-agarose. The in vitro complexes were recovered by incubation with glutathione-Sepharose beads, washed extensively, and immunoblotted with a CaV2.2 antibody. CaV2.2 bound to full-length CRMP-2 (Fig. 3, A and B). Within CRMP-2, three regions of interactions were determined: a region in the CRMP-2 N terminus (residues 94–166), a region in the middle of the protein (residues 212–297), and a region near the C terminus of the CRMP-2 protein (residues 479–500), proximal to the microtubule binding domain (Fig. 3B). These regions, designated CaV binding domains 1–3 (CBD1 to -3) are identified in the crystal structure of CRMP-2 tetramer (Protein Data Bank code 2GSE; Fig. 3C) (42); for clarity, the CBDs are identified only on monomer 1. Interestingly, the CBD3 interaction lies within the oligomerization domain of the tetrameric CRMP-2 protein (Fig. 3C). A large portion of CBD2 appears to be buried within the CRMP-2 tetrameric structure and may interfere with binding of the other CBDs by an as yet unclear mechanism.3 This is consistent with the demonstration that large segments of the CRMP protein are involved in oligomerization (29).
FIGURE 3.
Mapping binding sites between CRMP-2 and CaV2.2. A, pull-down assay of CaV2.2 with GST fusion CRMP-2 mutants. GST fusion proteins immobilized on glutathione-Sepharose beads were incubated with Ca2+ channel enriched fractions from rat brain synaptosomes. Bound proteins were analyzed by Western blotting with an antibody for CaV2.2. Coomassie-stained SDS-polyacrylamide gel of GST- and Glu-tagged purified CRMP-2 constructs. B, summary of binding of CaV2.2 onto CRMP-2. Three CBDs on CRMP-2 were observed: a region in the CRMP-2 N terminus (residues 94–166, designated CBD1; yellow), a region (residues 212–297, CBD2; orange), and one near the C terminus of the CRMP-2 protein (residues 479–500, CBD3; red). The second region, CBD2, if deleted, may destabilize CBD3 binding and/or impact CRMP-2 tetramerization. These regions (CBD1, CBD2, and CDB3) are identified in the crystal structure of the CRMP-2 tetramer (Protein Data Bank code 2GSE). The crystal structure of CRMP-2 is truncated at residue 490. C, for clarity, the binding sites are identified only on monomer 1. Surface (left) and ribbon (right) views of the CRMP-2 tetrameric structure were rendered using PyMol (version 1.1).
To determine the site of the CRMP-2 interaction with CaV2.2, we constructed fusion constructs encoding the intracellular cytoplasmic loops of CaV2.2 in a dual tag pGEX-Glu bacterial expression vector harboring GST and Glu (amino acids EYMPME) tags. Synaptosomes or CRMP-2 protein purified from rat brains using a three-step affinity chromatography method (29) were used for the binding assay. Synaptosomes were incubated with equimolar amounts of glutathione beads saturated with GST-CaV2.2-Glu fusion proteins or GST protein alone for 2 h at 4 °C with gentle agitation. After washing, the in vitro complexes were eluted from the beads, resolved by SDS-PAGE, and immunoblotted for CRMP-2 protein. As shown in Fig. 4, A and C, CRMP-2 bound two regions on CaV2.2: loop 1 (L1) and the distal part of the C terminus (Ct-dis). Next, we measured the strength of the binding of the CaV2.2 L1 and Ct-dis regions using an in vitro binding assay. A fixed amount of CRMP-2 protein was incubated overnight at 4 °C with increasing amounts of protein A beads saturated with L1 or Ct-dis fusion proteins. The bound complexes were washed, resolved by SDS-PAGE, and subjected to immunoblotting with CRMP-2 antibody as above (Fig. 4, D and E). The intensities of the CRMP-2 band were quantified, normalized to the maximum intensity within an experiment (n = 3), and plotted against the concentration of the fusion protein. The CaV2.2-CRMP-2 binding affinity (Kd) values were as follows: L1 = 24.1 ± 7.1 μm (n = 3); Ct-dis = 0.31 ± 0.09 μm (n = 3) (Fig. 4F). As a control, the Kd of interaction between the presynaptic protein Munc 18 (27) and loop 2 (L2) of CaV2.2 was found to be ∼32 ± 8.5 nm (n = 3; data not shown), consistent with the value of ∼13.8 ± 4 nm we reported previously (27). Thus, CRMP-2 binds directly to CaV2.2 via a moderate affinity (Ct-dis) and a low affinity (L1) binding site.
Activity-dependent Regulation of CRMP-2-CaV2.2 Interaction
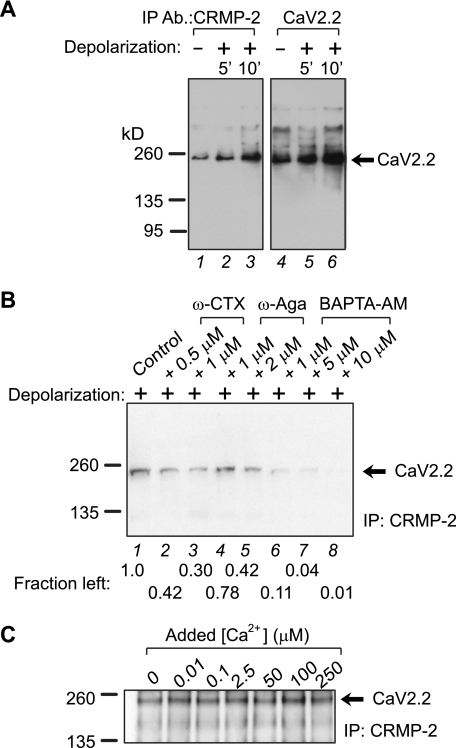
Having established that CRMP-2 binds to CaV2.2 and that CRMP-2 colocalizes with CaV2.2 in synapses, we next asked if changes in synaptic activity could affect the interaction. To determine if the CRMP-2-CaV2.2 interaction was dynamic, neonatal synaptosomes were used. These were chosen because a majority of the Ca2+ uptake in neonatal synaptosomes is dependent on CaV2.2 activity (43); this differs dramatically from adult synaptosomes in which CaV2.2 blockade has little to no effect on uptake. In order to study how synaptic activity may affect the interaction between CaV2.2 and CRMP-2, freshly prepared, unsolubilized synaptosomes were depolarized with high KCl for 5 or 10 min and then lysed in RIPA buffer and immunoprecipitated with CRMP-2 or CaV2.2 antibodies and immunoblotted with either antibody. The interaction between CRMP-2 and CaV2.2 was increased upon depolarization (Fig. 5A). Interestingly, there was relatively more CaV2.2 and CRMP-2 in the depolarized versus non-depolarized immunoprecipitates (Fig. 5A, compare lane 3 with lane 1 and lane 6 with lane 4). This result suggests that depolarization may cause changes in the conformations of the interacting proteins, which could lead to unmasking of the antigenic sites for either antibody, thereby facilitating greater recovery in the immunoprecipitates and thus greater association. This increase in CRMP-2-CaV2.2 interaction was dependent on Ca2+ influx via N-type Ca2+ channels, because incubation of synaptosomes with ω-CTX for 15 min prior to depolarization decreased the association (Fig. 5B, compare lane 2 or 3 with lane 1). Incubation of the synaptosomes with the P/Q-type blocker ω-agatoxin also partially affected the CRMP-2-CaV2.2 interaction (Fig. 5B). Incubation with increasing amounts of the membrane-permeable Ca2+ chelator BAPTA-AM (44) caused a dose-dependent decrease in CRMP-2-CaV2.2 association (Fig. 5B, lanes 6–8). The interaction exhibited maximal binding at around 100 μm added Ca2+ and declined at higher concentrations (Fig. 5C). Collectively, these results suggest an activity-dependent regulation of the interaction between CRMP-2 and CaV2.2.
FIGURE 5.
Activity-dependent regulation of CRMP-2-CaV2.2 interaction. A, equal amounts of freshly prepared, unsolubilized synaptosomes were depolarized with high 90 mm KCl for 5 or 10 min and then lysed in RIPA buffer and immunoprecipitated (IP) with CRMP-2 or CaV2.2 antibodies and immunoblotted with CaV2.2. B, to assess the importance of Ca2+ in the activity-dependent enhancement of the interactions, synaptosomes were depolarized for 5 or 10 min in the absence (control) or presence of the followings drugs, which were added 15 min before the addition of the KCl-depolarizing stimulus: ω-CTX, ω-agatoxin (ω-Aga), and the Ca2+ chelator BAPTA-AM. Following lysis, the synaptosomes were immunoprecipitated with CRMP-2 antibody and immunoblotted with CaV2.2. Fraction left, density of the CaV2.2 protein relative to control. C, to assess the importance of Ca2+ influx in the CRMP-2-CaV2.2 interaction, synaptosomes were incubated with increasing amounts of Ca2+ and immunoprecipitated with CRMP-2 antibody, followed by immunoblotting with CaV2.2. Maximal binding was observed at ∼100 μm added Ca2+. Representative blots from 2–4 experiments are shown.
Overexpression of CRMP-2 Increases Ca2+ Current Density
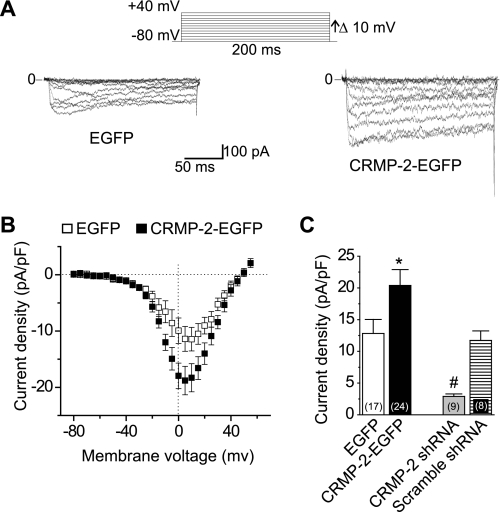
Using whole-cell voltage clamp electrophysiology, we tested if CRMP-2 expression had any effects on Ca2+ current properties in primary hippocampal neurons. Whole-cell Ca2+ currents were recorded from EGFP- and CRMP-2-EGFP-overexpressing cells. Whole-cell Ca2+ currents were elicited from a holding potential of −80 mV to depolarizing test potentials (ranging from −80 to +70 mV) for 200 ms using Ba2+ as the charge carrier. Fig. 6A shows representative voltage-dependent inward currents (IBa) from an EGFP- and a CRMP-2-EGFP-expressing neuron. Nifedipine (1 μm) was included in the bath solution to block L-type Ca2+ channels. Forty-eight h after transfection, the Ca2+ current density was significantly higher in CRMP-2-EGFP-transfected neurons than in EGFP neurons (Fig. 6). On average, the Ca2+ current density was increased by about 60%, from 12.8 ± 2.2 (n = 17) in EGFP neurons to 20.4 ± 2.5 (n = 24) pA pF−1 (Vt = 0 mV, p < 0.001, one-way analysis of variance) in CRMP-2-EGFP neurons, consistent with the values of 14.6 ± 1.0 pA pF−1 reported previously for control hippocampal neurons (45). Expression of CRMP-2-EGFP did not alter the size of the transfected neurons, because the cell capacitance was identical in CRMP-2-EGFP-transfected (21.8 ± 1.8 pF, n = 24) and EGFP-transfected (20.2 ± 1.7 pF, n = 17, p > 0.56) neurons. This result rules out changes in cell size as a potential contributing factor for the increased Ca2+ current density.
FIGURE 6.
CRMP-2 enhances Ca2+ current density in hippocampal neurons. A, representative current traces obtained from a cell transfected with EGFP and CRMP-2-EGFP evoked by 200-ms steps in 5-mV increments applied from a holding potential of −80 mV, as shown in the voltage protocol above the traces. For clarity, current traces at every 10 mV are shown. Bath solutions contained 1 μm tetrodotoxin, 10 mm tetraethylammonium-Cl, and 1 μm nifedipine to block Na+, K+, and L-type voltage-gated Ca2+ channels. Lines labeled 0 indicate the zero current level. B, I-V relationships for the currents shown in A. Peak currents were normalized to the cell capacitance. C, peak current density (pA/pF) measured at +10 mV for EGFP, CRMP-2-EGFP-transfected neurons, and CRMP-2 shRNA and scrambled shRNA-infected neurons. The numbers in parentheses represent numbers of cells tested. *, p < 0.05 versus all other conditions; #, p < 0.05 versus all other conditions (one-way analysis of variance).
To further confirm that CRMP-2 manipulation affected Ca2+ current density, we next asked if knockdown of CRMP-2 using lentiviral CRMP-2 shRNA would decrease Ca2+ current density. We confirmed efficient and specific knockdown of CRMP-2 using these knockdown strategies (supplemental Fig. S3). Interestingly, we observed that whole-cell recordings of 12 DIV hippocampal neurons infected with CRMP-2 shRNA lentivirus for 7 days in culture reduced Ca2+ currents to 2.9 ± 0.4 pA/pF (n = 9), an ∼80% reduction compared with CRMP-2-EGFP-overexpressing neurons (Fig. 6C). A scramble shRNA lentivirus did not affect Ca2+ currents; currents were 11.7 ± 1.5 pA/pF (n = 8; Fig. 6C). The specificity of the knockdown was further confirmed by measuring peak inward Na+ currents, which were not changed between the two groups: 126.04 ± 15.2 (n = 6; scramble shRNA lentivirus) and 112.25 ± 8.8 (n = 11; CRMP-2 shRNA lentivirus) pA pF−1 (Vt = 0 mV, p > 0.5, Student's t test; supplemental Fig. S4). Collectively, these results confirm that CRMP-2 has a specific effect on Ca2+ current density.
Current density can be affected by a change in channel gating (i.e. open probability) or changes in the number of CaV2.2 at the cell surface. To explore the first possibility, voltage-dependent properties of activation and inactivation of Ca2+ currents were examined in transfected neurons. There were no differences in the half-maximal activation (V½) and slope factors (k) of steady-state activation or inactivation between EGFP- and CRMP-2-EGFP-overexpressing neurons (supplemental Fig. S5). Inactivation time constants were also similar between the two groups (data not shown). Thus, changes in the Ca2+ channel gating properties are unlikely to account for the observed increase in Ca2+ current density.
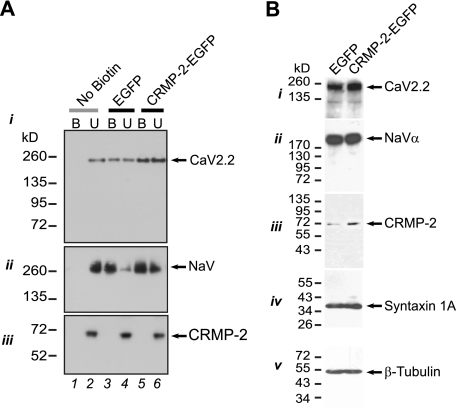
To test if there is a change in cell surface CaV2.2 following manipulation of CRMP-2 levels, biotinylation experiments were performed as previously described (27, 46). Immunoblotting with CaV2.2 for streptavidin-enriched complexes from biotinylated hippocampal neurons showed increased cell surface expression of CaV2.2 in CRMP-2-EGFP-expressing neurons compared with EGFP neurons (Fig. 7A, i, biotinylated (B) fractions). The surface levels of an unrelated membrane protein, the voltage-gated Na+ channel, were not affected by CRMP-2 overexpression (Fig. 7A, ii, compare lane 3 with lane 5). As a control for the biotinylation assay, no surface proteins were detected in samples in which the biotinylation reagent was omitted prior to the streptavidin precipitation (Fig. 7A, i and ii, lane 1). Thus, these results suggest that the increased Ca2+ current density in CRMP-2-overexpressing neurons probably occurs via increased cell surface trafficking of CaV2.2.
FIGURE 7.
CRMP-2 enhances surface CaV2.2 levels. A, cell surface expression of CaV2.2 and CRMP-2 was monitored using a biotinylation assay. Hippocampal neurons expressing EGFP or CRMP-2-EGFP were biotinylated, the cell-surface proteins were harvested from the cell lysates, and the precipitates were analyzed by immunoblotting with CaV2.2 (i), Na+ channel (ii), and CRMP-2 (iii) antibodies. Equal amounts of samples were used for the precipitation of biotinylated proteins. Of these samples, the entire biotinylated fraction (B) and 10% of the non-biotinylated (U) fraction were loaded. CRMP-2-EGFP overexpression increased the amount of CaV2.2 at the cell surface compared with EGFP-transfected cells. In the example shown, the cell surface CaV2.2 was increased by 2-fold. No change was observed in surface levels of the Na+ channel. Omission of the biotinylation reagent did not yield any surface CaV2.2 or NaV proteins. Representative blots from three experiments are shown. B, equal amounts of total proteins from EGFP- and CRMP-2-EGFP-transfected hippocampal neurons were immunoblotted with various antibodies as indicated. Except for the CRMP-2 protein level, which increased ∼3-fold, none of the other proteins (CaV2.2, Na+ channel, the SNARE protein syntaxin 1A, or the neuronal marker protein β-tubulin) were affected by CRMP-2 overexpression. See also supplemental Fig. S7B for additional proteins tested.
Western blotting of lysates from EGFP- and CRMP-2-EGFP-transfected cells revealed that CRMP-2 overexpression did not cause a change in the total levels of CaV2.2 and other ion channels (CaV1.3, voltage-gated Na+ channel) as well as other proteins, including SNARE proteins (syntaxin, RIM), SV2 (synaptic vesicle protein 2), scaffold protein (PSD-95), kinase (CaMKIIα), and cytoskeletal proteins (MAP2, β-tubulin) (Fig. 7B and supplemental Fig. S7). As expected, CRMP-2 protein levels were increased (∼3-fold) upon overexpression (Fig. 7B, iii). Quantitative reverse transcription-PCR data corroborated these findings, ruling out a transcriptional effect of CRMP-2 overexpression on CaV2.2 mRNA (see supplemental Fig. S7).
CRMP-2 Facilitates Increased Synaptic Vesicle Recycling
Because Ca2+ influx via CaV2.2 is the trigger for transmitter release in hippocampal synapses (1, 47–49) and CRMP-2 increases Ca2+ current density, we next investigated if the increased Ca2+ could contribute to changes in synaptic vesicle release and recycling. Synaptic vesicle fusion was measured by the unloading of the styryl dye FM4-64, using live cell confocal microscopy (33). Styryl dyes (FM dyes) are used as endocytic markers that enable detection of presynaptic vesicle recycling and release (50, 51). As shown in Fig. 8A, we first confirmed that CRMP-2-EGFP is present in the synapses of cultured hippocampal neurons by co-labeling with the synaptic vesicle marker protein synaptophysin. FM dye imaging was then performed on hippocampal neurons at 14 DIV, 48 h after transfection of CRMP-2-EGFP or EGFP. Functional presynaptic terminals were loaded with FM4-64 (15 μm) under stimulation with 90 mm KCl, followed by unloading in a Ca2+-free solution (Fig. 8B). This paradigm is believed to recruit the entire pool of actively recycling vesicles (52). Synaptic boutons of green fluorescent neurons that had taken up the FM dye were identified, and numerical fluorescence values were determined from defined regions of interest within boutons using imaging software (Image J) and graphed as the average amount of dye uptake or plotted as a change in fluorescence values (ΔF) as compared with base line. CRMP-2-EGFP neurons had slightly higher initial FM4-64 dye uptake than EGFP neurons (Fig. 8, C and D; p < 0.01; EGFP, 107.5 ± 8.95 arbitrary units, n = 5 (300 boutons); CRMP-2-EGFP, 138.8 ± 9.5 arbitrary units, n = 5 (365 boutons). The decrease in FM4-64 fluorescence intensity of the synaptic boutons over time in response to the second 1-min stimulation represents loss of dye from the fusion of vesicles that have previously taken up the dye. Representative examples of FM4-64 boutons stained and destained are shown in Fig. 8E, ii–v. The destaining kinetic curves of FM4-64 taken up by the presynaptic terminals expressing EGFP or CRMP-2-EGFP in the presence of 90 mm KCl are shown in Fig. 8F. Functional boutons from CRMP-2-EGFP-expressing neurons destained more than EGFP boutons. The extent of destaining (at t = 3 min) was ∼21% less in EGFP boutons (0.52 ± 0.02; n = 100) compared with CRMP-2-EGFP boutons (0.43 ± 0.02; n = 100; Fig. 8F). As a control, the styryl dye did not bleach under repetitive laser scanning without high stimulation (Fig. 8F). Knockdown of CRMP-2 changed the extent of destaining by ∼44% from 0.61 ± 0.03 in CRMP-2 siRNA boutons (n = 69) to 0.43 ± 0.02 in CRMP-2-EGFP-overexpressing boutons (n = 100). The extent of destaining in boutons from neurons treated with scrambled siRNA (0.46 ± 0.03, n = 79) was similar to control EGFP levels (0.43 ± 0.02, n = 100). These results suggest that loading of the initial pool of synaptic vesicles, as well as vesicle fusion, is influenced by CRMP-2 overexpression. Since CRMP-2 manipulation affects Ca2+ influx, it is likely that the extra Ca2+ could contribute to changes in transmitter release. However, we cannot rule out the possibility that CRMP-2 impacts transmitter release independently of CaV2.2.
CRMP-2 Regulates Transmitter Release
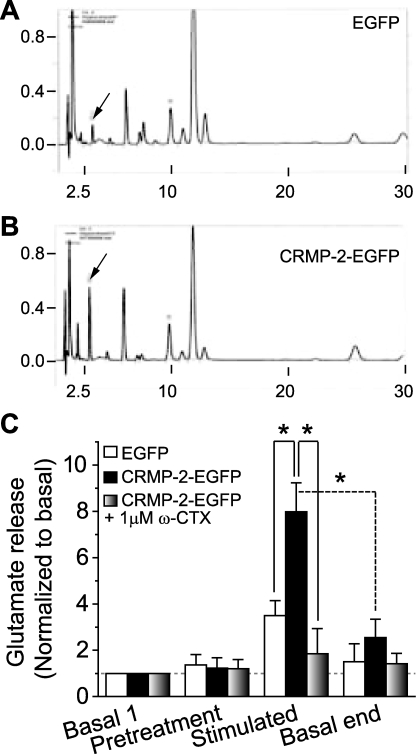
So far, we have shown that CRMP-2 can bind directly to the CaV2.2 channel and can enhance Ca2+ current density in hippocampal neurons, but do these events translate into an increase in transmitter release? To answer this question, we measured glutamate release from hippocampal neurons overexpressing EGFP or CRMP-2-EGFP using HPLC. At 10 DIV, hippocampal neurons were transfected with EGFP or CRMP-2-EGFP, and release experiments were performed 48 h later. Representative chromatographs are shown in Fig. 9, A and B, with a typical retention time of glutamate at about 3 min. Exposing hippocampal neurons transfected with EGFP to 30 mm KCl stimulated glutamate release by about 3.5-fold from a basal level of 3.8 ± 1.0 μm/well/10 min to 13.3 ± 0.9 μm/well/10 min. In CRMP-2-EGFP-overexpressing neurons, 30 mm KCl stimulated glutamate release by about 8-fold from a basal level of 3.7 ± 0.9 μm/well/10 min to 29.5 ± 1.9 (p < 0.001 versus EGFP). This shows that CRMP-2 can augment glutamate release by 2.3-fold over control EGFP-transfected cells. Although the CRMP-2-mediated glutamate release is smaller than the 2.7–12-fold increase in glutamate release that is predicted based on a ∼65% increase in Ca2+ current density, it is important to note that not every neuron overexpresses CRMP-2 due to incomplete transfection efficiency (∼10–15%).
FIGURE 9.
CRMP-2 modulates transmitter release. Shown are representative HPLC chromatographs of glutamate (arrows) from hippocampal neurons transfected with EGFP (A) or CRMP-2-EGFP (B) in response to a depolarizing (30 mm KCl) stimulus. The neurons were incubated with HEPES buffer at 37 °C for 1 h and then exposed to successive 10-min incubations in the absence or presence of drugs. Likewise, 10-min incubations with the addition of 30 mm KCl to the HEPES buffer were utilized to evoke neurotransmitter release in the presence or absence of drugs. After stimulation, the cells were exposed to HEPES buffer alone again to reestablish basal release. The supernatants were collected following each 10-min interval for glutamate assay by HPLC. C, stimulated glutamate release is increased in CRMP-2-overexpressing neurons versus controls (EGFP; p < 0.05, Student's t test; n = 9). The addition of 1 μm ω-conotoxin GVIA, the N-type Ca2+ channel blocker, completely attenuates this CRMP-2-mediated increase in glutamate release. Basal release is not affected by toxin block.
Importantly, the addition of 1 μm ω-CTX GVIA, the N-type-selective CaV2.2 blocker, decreased the magnitude of the potassium-stimulated glutamate release from CRMP-2-EGFP-expressing neurons by ∼77% (to 8.9 ± 1.9 μm/well/10 min) compared with potassium-stimulated glutamate release in the absence of blocker (Fig. 9C, p < 0.05). Block of Ca2+ channels did not affect the basal release of glutamate. These results suggest that, in hippocampal neurons at 12 DIV, CaV2.2 supports most, if not all, of the CRMP-2-enhanced, depolarization-associated glutamate release from cultured hippocampal neurons.
CRMP-2 Regulates Bouton Size but Not Bouton Number
To address the possibility that changes in levels of CRMP-2 could induce changes in the number and/or size of synaptic boutons, an analysis of bouton density and size was carried out as described (33). As illustrated in Fig. 10, A and B, live EGFP- and CRMP-2-EGFP-transfected neurons were labeled with FM4-64. A line scan of synapses labeled with FM4-64 revealed a similar density of boutons between the two conditions: 0.15 ± 0.04/μm (EGFP; n = 598 boutons from nine neurons) and 0.27 ± 0.07 (CRMP-2-EGFP; n = 615 boutons from nine neurons; p > 0.05; Fig. 10, C–E). A double-blind analysis of the bouton size was performed in which only the largest diameter of each bouton was measured. Bouton diameters were distributed unimodally in both EGFP- and CRMP-2-EGFP-expressing neurons (Fig. 10F). The average diameter of boutons was ∼3.3-fold larger in CRMP-2-EGFP-expressing neurons (0.71 ± 0.046 μm; 435 boutons from five images) than in EGFP-expressing neurons (0.21 ± 0.012 μm; 139 boutons from five images of EGFP neurons; p < 0.05, Student's t test; Fig. 10F, inset). These results suggest that CRMP-2 expression increases the average diameter of synaptic boutons.
DISCUSSION
The rapidity with which transmitter release is initiated following influx of Ca2+ through a single Ca2+ channel (2) suggests close spatial proximity between presynaptic Ca2+ channels and the release machinery (1). Major progress has been made toward identifying proteins of the release machinery at the transmitter release site and characterizing the effects of these proteins on synaptic transmission (1, 2, 4, 33, 53–60). Here we identify CRMP-2 as a novel “neuromodulator” of N-type Ca2+ channels. Our results demonstrate that an interaction between CRMP-2 and N-type Ca2+ channels (Figs. 3 and 4) increases calcium current density via increased cell surface CaV2.2 expression (Fig. 7), which, together with CRMP-2-mediated increase in SV loading (Fig. 8) and synapse size (Fig. 10), translates into an increased release of the neurotransmitter glutamate (Fig. 9). The CRMP-2-Ca2+ channel association may serve to sustain Ca2+ influx through functional regulation of Ca2+ channels and to recruit synaptic vesicles to Ca2+ channels. Thus, our results identify CRMP-2 as a novel “neuromodulator” of Ca2+ channels and of synaptic strength.
CRMP-2 overexpression leads to an increase in current density, suggesting a direct functional coupling between the two proteins. Changes in current amplitude could be explained by increased cell surface expression of Ca2+ channels, changes in channel availability, and/or changes in open probability (Po). Interestingly, cell-surface biotinylation experiments showed a significant increase in surface-expressed CaV2.2 in CRMP-2-EGFP-overexpressing neurons compared with those expressing EGFP (Fig. 7A). Analyses of the biophysical parameters of the measured currents showed that the gating properties of the channel were not affected by CRMP-2 manipulation, despite the finding that CRMP-2 binds to the same regions of the Ca2+ channel α subunit as the calcium channel β subunit (61–63). Although changes in the open probability of CaV2.2 cannot be ruled out, the most straightforward explanation for a change in current density is an increase in cell surface expression of Ca2+ channels; additional studies are needed to determine the precise mechanism(s) involved. Because CaV2.2 Ca2+ channels determine the amount, timing, and location of the Ca2+ influx, evoked transmitter release can be dramatically affected by changes in channel biophysical properties, activation and inactivation. Thus, a CRMP-2-mediated increase in Ca2+ current amplitude has significant implications because modulation of Ca2+ currents is critical for regulation of neurotransmission in the nervous system.
A mixed population of N- and P/Q-type Ca2+ channels can coexist at a single release site and contribute jointly to the local Ca2+ transient that triggers transmitter release (47, 48, 64, 65). There is a growing body of evidence that suggests that N- and P/Q-type Ca2+ channels undergo a developmental switch (66–68), although the mechanism underlying this is not understood. In a recent study on hippocampal neurons cultured for 10 DIV, N-type Ca2+ channels accounted for ∼50% of the total Ca2+ current, with P/Q-type supporting ∼25% (69). At 10–15 DIV, N-type Ca2+ channels accounted for >80% of synaptic transmission, as assessed by block with ω-CTX (66). In the CRMP-2-EGFP-overexpressing neurons, ∼35% of the cadmium-sensitive barium current was ω-conotoxin (1 μm)-sensitive, and ∼37% was ω-agatoxin (0.4 μm)-sensitive (n = 5; data not shown), at least in the cell soma. Importantly, block of N-type channels inhibits transmitter release by ∼80% in our experiments (Fig. 9), consistent with published data (66). Taken together, these results would argue against a significant involvement of P/Q-type Ca2+ channels in transmitter release, although they contribute to a similar fraction of the Ca2+ current as N-type Ca2+ channels in the cell soma. Remarkably, knockdown of CRMP-2 reduced Ca2+ current by ∼80%, suggesting that CRMP-2 modulates non-N-type (i.e. P/Q-type) Ca2+ channels. Furthermore, it appears that N-type Ca2+ channel expression dominates over other Ca2+ channels at active release sites because N-type Ca2+ channels were responsible for a majority of transmitter release but only ∼30% of the Ca2+ current. This may be due to the selective expression of N-type Ca2+ channels at nascent immature terminals or in a reserve pool of terminals held in an immature state, whereas P/Q-type channels might dominate at fully mature terminals. The finding of a CRMP-2-CaV2.2 complex in adult rat brain synaptosomes4 despite the low expression of CaV2.2 in adult brain (68) suggests that this interaction may play roles in mature nerve terminals as well. Differential expression, sorting, trafficking and targeting, postsynaptic cues, protein-protein interactions, and other factors may account for this developmental switch in Ca2+ channels. Thus, interactions with and/or functional regulation by CRMP-2 may be required for N-type, but not P/Q-type, Ca2+ channel expression at presynaptic terminals. Consequently, N-type Ca2+ channels may be of central importance to synaptic plasticity simply because of their predominant role in juvenile transmitter release at most systems studied to date (66, 67).
CRMPs have been implicated in synaptic function, where the CRMP-2-related CRMP-4b variant localizes to synaptic vesicles in growth cones and interacts with the Src homology 3A domain of intersectin, an endocytic-exocytic adaptor protein (17); intersectin also has been shown to associate with the N-type Ca2+ channel (24). Further support for CRMP involvement in the synaptic vesicle endocytosis and exocytosis pathway comes from studies showing that the other Src homology 3 domains of intersectin bind to SNAP-25 (synaptosome-associated protein of 25 kDa) (70), dynamin, and synaptojanin (71), key endocytic proteins. Both SNAP-25 and dynamin associate with the N-type Ca2+ channel (24); however, dynamin is also part of an endocytosis-related clathrin-containing subcomplex (24). These interactions with the Ca2+ channel and exocytosis-endocytosis proteins position CRMPs ideally for facilitating synaptic vesicle recycling and release. This could account for the increased synaptic vesicle recycling observed in CRMP-2-overexpressing neurons (Fig. 7B).
Another possibility may be that CRMP-2 affects synaptic vesicle recycling independent of its interaction with CaV2.2. In support of this idea, CRMP-2-mediated increase in synapse size (Fig. 10F) as well as changes in the numbers of vesicles loaded (Fig. 8C) may contribute to the observed increase in transmitter release (Fig. 9). Larger synapses have higher release probabilities because they have been shown to contain greater release site areas and more docked vesicles and have a correspondingly larger readily releasable pool of vesicles (72–74). Interestingly, although mRNAs of synaptic vesicle genes SV2a and SV2b are up-regulated in CRMP-2-EGFP-expressing neurons, the protein levels of SV2 are not affected (supplemental Fig. S7, A and B). In addition, because expression levels of synaptic proteins, such as syntaxin and synaptophysin, were unchanged, both CaV2.2-dependent and -independent effects on SV recycling remain possible.
The ability of the Ca2+ chelator BAPTA-AM and N-type blocker ω-CTX to prevent the increase in the activity-induced CRMP-2-CaV2.2 binding suggests that Ca2+ influx, not CaV2.2 activity, regulates the strength of the interaction in synaptosomes (Fig. 5). This is further supported by our findings that enhanced Ca2+ levels trigger CRMP-2-CaV2.2 binding (Fig. 5C), entirely consistent with previous reports of Ca2+ dependence of interactions between N-type Ca2+ channels and presynaptic core complex proteins (56, 75, 76). It is possible that depolarization-induced Ca2+ influx through voltage-gated Ca2+ channels mobilizes CRMP-2 protein to traffic channels into the presynaptic membrane. This could also explain why depolarization leads to an enhanced recovery of CRMP-2 and CaV2.2 in the immunoprecipitates (Fig. 5A). A 5–10-min depolarization may position both CRMP-2 and CaV2.2 in a conformation that favors increased association. Examples of such conformation-specific interactions have been reported previously (77–79). Finally, if CRMP-2 is a component of the CaV2.2 transmitter release site core complex (25), then increased CRMP-2 association is likely to facilitate synaptic vesicle recycling. Alternatively, activity-dependent CRMP-2 recruitment to the release site core complex may enable it to interact with other vesicular release proteins.
Several lines of evidence point to a putative link between CRMPs and transmitter release. The CRMP-1 mutant mice showed a reduction in long term potentiation in hippocampal CA1 region and impaired performance in hippocampus-dependent spatial learning and memory tests (19). CRMP-3 knock-out mice were found to have abnormal dendritic arborization, altered spine morphology, and abnormal neuronal orientation, leading to impaired long term potentiation and impaired spatial learning and memory (19, 20). Abnormalities in the dendritic morphologies may have impeded correct formation of synaptic connections and could be responsible for the long term potentiation deficit observed in the CRMP-3 knock-out mice. Second, a cleaved form of CRMP-2 regulated Ca2+ influx by down-regulating the NR2B subunit of the N-methyl-d-aspartate receptor. Third, activation of the neuropilin-1 receptor by the CRMP ligand, semaphorin-3A, in acute hippocampal slices caused synaptic depression (80); however, the probability of transmitter release was not affected (80). Fourth, the recruitment of kinases, such as Cdk5 (cyclin-dependent kinase 5), ROCK (RhoA (Ras homolog family A) kinase), and GSK3β (glycogen synthase kinase 3β) by semaphorin3A (10, 36, 81, 82) could lead to post-translational modifications of CRMP-2, leading to reduced binding between CRMP-2 and the N-type Ca2+ channel, thus accounting for the reduction in synaptic transmission. Collectively, the results from these studies and our findings of increased transmitter release show that CRMPs are important for synaptogenesis and neurotransmission.
CRMP proteins have been implicated in both developmental and adult neurological diseases (10). A common feature of several of these diseases is alteration of Ca2+ homeostasis, which appears to play a key role in the mechanisms of the neuronal/axonal injury underlying these diseases. These “calciumopathies” include Alzheimer disease (83), paraneoplastic neurological syndromes (84), cerebral ischemia and stroke (85), neuroinflammation (85), and Down syndrome (86). CRMP-2 levels are altered in epilepsy patients and in animal models of epilepsy (87). CRMP-2 levels were found to correlate with increased depression in an animal model of chronic mild stress (88). Calcium-dependent CRMP-2 proteolysis has been observed in traumatic brain injury and cerebral ischemia (89) and may be a limiting factor for postinjury axonal regeneration. Excitotoxicity and oxidative stress induce Ca2+ influx via voltage-gated Ca2+ channels (90), which elicits CRMP cleavage, thus providing an additional functional link between CRMPs and Ca2+ channels.
As described by Catterall and Few (1), the presynaptic Ca2+ channel signaling complex, composed of soluble N-ethylmaleimide-sensitive factor attachment protein receptor, Ca2+-binding, scaffolding, and endocytosis-exocytosis proteins, forms a regulatory node controlling synaptic transmission, presynaptic plasticity, and feedback regulation of Ca2+ entry. Our results position CRMP-2 as a novel regulator of presynaptic Ca2+ channels and identify the CRMP-2-Ca2+ channel interaction as a new node for integration of Ca2+ signals to synaptic transmission and plasticity. Dysregulation of CRMP-2 expression and the CRMP-2-CaV2.2 interaction may contribute to diseases of synaptic function or “synaptopathies.”
Supplementary Material
Acknowledgments
We thank Chunlu Guo and Dr. Michael R. Vasko (Indiana University School of Medicine) for assistance with HPLC analysis of glutamate release and Kashif Z. Kirmani (Stark Neurosciences Research Institute Viral Core Facility) for production of the lentiviral particles. We are grateful to Dr. Clark Wells for providing the Glu antibody, Dr. Akihiro Kurimasa (Tottori University, Yonago, Japan) for several CRMP-2 constructs, and Dr. Debbie Thurmond for the Munc18-1-GST construct. We thank Dr. Patrick L. Sheets (Northwestern University, Chicago) for earlier electrophysiology experiments that are not included here, Sarah Marie Walls (Indiana University School of Medicine) for bouton analysis, and Drs. May Khanna (University of Toronto) and Samy Meroueh (Indiana University School of Medicine) for helpful discussions regarding the CRMP-2 structure. We also thank Drs. Gerry Oxford, Andy Hudmon, and Grant Nicol and members of the Paul and Carole Stark Neurosciences Research Institute for helpful comments and constructive discussions regarding this study.
This work was supported in part by National Institutes of Health Grant NS053422 (to T. R. C.). This work was also supported by grants from the Indiana State Department of Health Spinal Cord and Brain Injury Fund (to R. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S8.
J. M. Brittain and R. Khanna, unpublished data.
J. M. Brittain, unpublished data.
- CaV2.2
- N-type voltage-gated Ca2+ channel
- CRMP
- collapsin response mediator protein
- CBD
- CaV binding domain on CRMP-2
- ICA
- intensity correlation analysis
- ICQ
- intensity correlation quotient
- EGFP
- enhanced green fluorescent protein
- DIV
- days in vitro
- ω-CTX
- ω-conotoxin GVIA
- siRNA
- short interfering RNA
- shRNA
- short hairpin RNA
- MES
- 2-morpholinoethanesulfonic acid
- BAPTA-AM
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester
- RIPA
- radioimmunoprecipitation assay
- GST
- glutathione S-transferase
- PN1 and -8
- postnatal day 1 and 8, respectively
- PBS
- phosphate-buffered saline
- HPLC
- high pressure liquid chromatography
- L1
- loop 1
- Ct-dis
- distal part of the C terminus
- pF
- picofarads
- FM4-64
- N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide.
REFERENCES
- 1.Catterall W. A., Few A. P. (2008) Neuron 59, 882–901 [DOI] [PubMed] [Google Scholar]
- 2.Stanley E. F. (1997) Trends Neurosci. 20, 404–409 [DOI] [PubMed] [Google Scholar]
- 3.Davies J. N., Zamponi G. W. (2008) Channels 2, 130–138 [DOI] [PubMed] [Google Scholar]
- 4.Kisilevsky A. E., Zamponi G. W. (2008) Handb. Exp. Pharmacol. 184, 45–75 [DOI] [PubMed] [Google Scholar]
- 5.Dodge F. A., Jr., Rahamimoff R. (1967) J. Physiol. 193, 419–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz B., Miledi R. (1970) J. Physiol. 207, 789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Augustine G. J., Charlton M. P., Smith S. J. (1987) Annu. Rev. Neurosci. 10, 633–693 [DOI] [PubMed] [Google Scholar]
- 8.Khanna R., Zougman A., Stanley E. F. (2007) J. Biochem. Mol. Biol. 40, 302–314 [DOI] [PubMed] [Google Scholar]
- 9.Goshima Y., Nakamura F., Strittmatter P., Strittmatter S. M. (1995) Nature 376, 509–514 [DOI] [PubMed] [Google Scholar]
- 10.Schmidt E. F., Strittmatter S. M. (2007) Adv. Exp. Med. Biol. 600, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arimura N., Inagaki N., Chihara K., Ménager C., Nakamura N., Amano M., Iwamatsu A., Goshima Y., Kaibuchi K. (2000) J. Biol. Chem. 275, 23973–23980 [DOI] [PubMed] [Google Scholar]
- 12.Inagaki N., Chihara K., Arimura N., Ménager C., Kawano Y., Matsuo N., Nishimura T., Amano M., Kaibuchi K. (2001) Nat. Neurosci. 4, 781–782 [DOI] [PubMed] [Google Scholar]
- 13.Arimura N., Menager C., Fukata Y., Kaibuchi K. (2004) J. Neurobiol. 58, 34–47 [DOI] [PubMed] [Google Scholar]
- 14.Fukata Y., Itoh T. J., Kimura T., Ménager C., Nishimura T., Shiromizu T., Watanabe H., Inagaki N., Iwamatsu A., Hotani H., Kaibuchi K. (2002) Nat. Cell Biol. 4, 583–591 [DOI] [PubMed] [Google Scholar]
- 15.Goshima Y., Hori H., Sasaki Y., Yang T., Kagoshima-Maezono M., Li C., Takenaka T., Nakamura F., Takahashi T., Strittmatter S. M., Misu Y., Kawakami T. (1999) J. Neurobiol. 39, 579–589 [DOI] [PubMed] [Google Scholar]
- 16.Uchida Y., Ohshima T., Sasaki Y., Suzuki H., Yanai S., Yamashita N., Nakamura F., Takei K., Ihara Y., Mikoshiba K., Kolattukudy P., Honnorat J., Goshima Y. (2005) Genes Cells 10, 165–179 [DOI] [PubMed] [Google Scholar]
- 17.Quinn C. C., Chen E., Kinjo T. G., Kelly G., Bell A. W., Elliott R. C., McPherson P. S., Hockfield S. (2003) J. Neurosci. 23, 2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hotta A., Inatome R., Yuasa-Kawada J., Qin Q., Yamamura H., Yanagi S. (2005) Mol. Biol. Cell 16, 32–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su K. Y., Chien W. L., Fu W. M., Yu I. S., Huang H. P., Huang P. H., Lin S. R., Shih J. Y., Lin Y. L., Hsueh Y. P., Yang P. C., Lin S. W. (2007) J. Neurosci. 27, 2513–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quach T. T., Massicotte G., Belin M. F., Honnorat J., Glasper E. R., Devries A. C., Jakeman L. B., Baudry M., Duchemin A. M., Kolattukudy P. E. (2008) FASEB J. 22, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huttner W. B., Schiebler W., Greengard P., De Camilli P. (1983) J. Cell Biol. 96, 1374–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlin R. K., Grab D. J., Cohen R. S., Siekevitz P. (1980) J. Cell Biol. 86, 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khanna R., Li Q., Sun L., Collins T. J., Stanley E. F. (2006) Neuroscience 140, 1201–1208 [DOI] [PubMed] [Google Scholar]
- 24.Khanna R., Li Q., Schlichter L. C., Stanley E. F. (2007) Eur. J. Neurosci. 26, 560–574 [DOI] [PubMed] [Google Scholar]
- 25.Khanna R., Li Q., Bewersdorf J., Stanley E. F. (2007) Eur. J. Neurosci. 26, 547–559 [DOI] [PubMed] [Google Scholar]
- 26.Wong W., Newell E. W., Jugloff D. G., Jones O. T., Schlichter L. C. (2002) J. Biol. Chem. 277, 20423–20430 [DOI] [PubMed] [Google Scholar]
- 27.Chan A. W., Khanna R., Li Q., Stanley E. F. (2007) Channels 1, 11–20 [PubMed] [Google Scholar]
- 28.Tahimic C. G., Tomimatsu N., Nishigaki R., Fukuhara A., Toda T., Kaibuchi K., Shiota G., Oshimura M., Kurimasa A. (2006) Biochem. Biophys. Res. Commun. 340, 1244–1250 [DOI] [PubMed] [Google Scholar]
- 29.Wang L. H., Strittmatter S. M. (1997) J. Neurochem. 69, 2261–2269 [DOI] [PubMed] [Google Scholar]
- 30.Goslin K., Banker G. (1989) J. Cell Biol. 108, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D., Wang F., Lai M., Chen Y., Zhang J. F. (2005) J. Neurosci. 25, 1914–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J. F., Randall A. D., Ellinor P. T., Horne W. A., Sather W. A., Tanabe T., Schwarz T. L., Tsien R. W. (1993) Neuropharmacology 32, 1075–1088 [DOI] [PubMed] [Google Scholar]
- 33.Pan P. Y., Cai Q., Lin L., Lu P. H., Duan S., Sheng Z. H. (2005) J. Biol. Chem. 280, 25769–25779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willeumier K., Pulst S. M., Schweizer F. E. (2006) J. Neurosci. 26, 11333–11341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kay A. R., Alfonso A., Alford S., Cline H. T., Holgado A. M., Sakmann B., Snitsarev V. A., Stricker T. P., Takahashi M., Wu L. G. (1999) Neuron 24, 809–817 [DOI] [PubMed] [Google Scholar]
- 36.Nishimura T., Fukata Y., Kato K., Yamaguchi T., Matsuura Y., Kamiguchi H., Kaibuchi K. (2003) Nat. Cell Biol. 5, 819–826 [DOI] [PubMed] [Google Scholar]
- 37.Zufferey R. (2002) Curr. Top. Microbiol. Immunol. 261, 107–121 [DOI] [PubMed] [Google Scholar]
- 38.Donzanti B. A., Yamamoto B. K. (1988) Life Sci. 43, 913–922 [DOI] [PubMed] [Google Scholar]
- 39.Takamori S., Holt M., Stenius K., Lemke E. A., Grønborg M., Riedel D., Urlaub H., Schenck S., Brügger B., Ringler P., Müller S. A., Rammner B., Gräter F., Hub J. S., De Groot B. L., Mieskes G., Moriyama Y., Klingauf J., Grubmüller H., Heuser J., Wieland F., Jahn R. (2006) Cell 127, 831–846 [DOI] [PubMed] [Google Scholar]
- 40.Khanna R., Sun L., Li Q., Guo L., Stanley E. F. (2006) Neuroscience 138, 1115–1125 [DOI] [PubMed] [Google Scholar]
- 41.Westenbroek R. E., Hell J. W., Warner C., Dubel S. J., Snutch T. P., Catterall W. A. (1992) Neuron 9, 1099–1115 [DOI] [PubMed] [Google Scholar]
- 42.Deo R. C., Schmidt E. F., Elhabazi A., Togashi H., Burley S. K., Strittmatter S. M. (2004) EMBO J. 23, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi K., Tatsuno M., Kiuchi Y. (1998) Brain Dev. 20, 234–238 [DOI] [PubMed] [Google Scholar]
- 44.Nichols R. A., Suplick G. R. (1996) Neurosci. Lett. 211, 135–137 [DOI] [PubMed] [Google Scholar]
- 45.Lai M., Wang F., Rohan J. G., Maeno-Hikichi Y., Chen Y., Zhou Y., Gao G., Sather W. A., Zhang J. F. (2005) Nat. Neurosci. 8, 435–442 [DOI] [PubMed] [Google Scholar]
- 46.Joiner W. J., Khanna R., Schlichter L. C., Kaczmarek L. K. (2001) J. Biol. Chem. 276, 37980–37985 [DOI] [PubMed] [Google Scholar]
- 47.Reid C. A., Clements J. D., Bekkers J. M. (1997) J. Neurosci. 17, 2738–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid C. A., Bekkers J. M., Clements J. D. (1998) J. Neurosci. 18, 2849–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reid B., Slater C. R., Bewick G. S. (1999) J. Neurosci. 19, 2511–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cochilla A. J., Angleson J. K., Betz W. J. (1999) Annu. Rev. Neurosci. 22, 1–10 [DOI] [PubMed] [Google Scholar]
- 51.Henkel A. W., Simpson L. L., Ridge R. M., Betz W. J. (1996) J. Neurosci. 16, 3960–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryan T. A., Reuter H., Wendland B., Schweizer F. E., Tsien R. W., Smith S. J. (1993) Neuron 11, 713–724 [DOI] [PubMed] [Google Scholar]
- 53.Altier C., Zamponi G. W. (2008) J. Recept. Signal. Transduct. Res. 28, 71–81 [DOI] [PubMed] [Google Scholar]
- 54.Sheng Z. H., Rettig J., Takahashi M., Catterall W. A. (1994) Neuron 13, 1303–1313 [DOI] [PubMed] [Google Scholar]
- 55.Mochida S., Sheng Z. H., Baker C., Kobayashi H., Catterall W. A. (1996) Neuron 17, 781–788 [DOI] [PubMed] [Google Scholar]
- 56.Sheng Z. H., Rettig J., Cook T., Catterall W. A. (1996) Nature 379, 451–454 [DOI] [PubMed] [Google Scholar]
- 57.Ilardi J. M., Mochida S., Sheng Z. H. (1999) Nat. Neurosci. 2, 119–124 [DOI] [PubMed] [Google Scholar]
- 58.Leenders M., Gerwin C., Sheng Z. H. (2004) in Current Protocols in Neuroscience (Taylor G. ed) pp. 2.7.1–2.7.18, John Wiley & Sons, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 59.Leenders A. G., Lin L., Huang L. D., Gerwin C., Lu P. H., Sheng Z. H. (2008) J. Neurosci. 28, 11333–11346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang J. S., Tian J. H., Pan P. Y., Zald P., Li C., Deng C., Sheng Z. H. (2008) Cell 132, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pragnell M., De Waard M., Mori Y., Tanabe T., Snutch T. P., Campbell K. P. (1994) Nature 368, 67–70 [DOI] [PubMed] [Google Scholar]
- 62.De Waard M., Scott V. E., Pragnell M., Campbell K. P. (1996) FEBS Lett. 380, 272–276 [DOI] [PubMed] [Google Scholar]
- 63.Walker D., Bichet D., Campbell K. P., De Waard M. (1998) J. Biol. Chem. 273, 2361–2367 [DOI] [PubMed] [Google Scholar]
- 64.Cao Y. Q., Piedras-Rentería E. S., Smith G. B., Chen G., Harata N. C., Tsien R. W. (2004) Neuron 43, 387–400 [DOI] [PubMed] [Google Scholar]
- 65.Reid C. A., Bekkers J. M., Clements J. D. (2003) Trends Neurosci. 26, 683–687 [DOI] [PubMed] [Google Scholar]
- 66.Scholz K. P., Miller R. J. (1995) J. Neurosci. 15, 4612–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iwasaki S., Momiyama A., Uchitel O. D., Takahashi T. (2000) J. Neurosci. 20, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones O. T., Bernstein G. M., Jones E. J., Jugloff D. G., Law M., Wong W., Mills L. R. (1997) J. Neurosci. 17, 6152–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grimm C., Holter N. I., Draguhn A., Bruehl C. (2008) Neurosci. Lett. 442, 44–49 [DOI] [PubMed] [Google Scholar]
- 70.Okamoto M., Schoch S., Südhof T. C. (1999) J. Biol. Chem. 274, 18446–18454 [DOI] [PubMed] [Google Scholar]
- 71.Yamabhai M., Hoffman N. G., Hardison N. L., McPherson P. S., Castagnoli L., Cesareni G., Kay B. K. (1998) J. Biol. Chem. 273, 31401–31407 [DOI] [PubMed] [Google Scholar]
- 72.Murthy V. N., Schikorski T., Stevens C. F., Zhu Y. (2001) Neuron 20, 673–682 [DOI] [PubMed] [Google Scholar]
- 73.Schikorski T., Stevens C. F. (1997) J. Neurosci. 17, 5858–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prange O., Murphy T. H. (1999) J. Neurophysiol. 81, 1810–1817 [DOI] [PubMed] [Google Scholar]
- 75.Chapman E. R., Hanson P. I., An S., Jahn R. (1995) J. Biol. Chem. 270, 23667–23671 [DOI] [PubMed] [Google Scholar]
- 76.Sheng Z. H., Yokoyama C. T., Catterall W. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5405–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu S. M., Hodges R. S. (2002) J. Biol. Chem. 277, 23515–23524 [DOI] [PubMed] [Google Scholar]
- 78.Panneerselvam K., Reitz H., Khan S. A., Bishayee S. (1995) J. Biol. Chem. 270, 7975–7979 [DOI] [PubMed] [Google Scholar]
- 79.Gupta A., Décaillot F. M., Gomes I., Tkalych O., Heimann A. S., Ferro E. S., Devi L. A. (2007) J. Biol. Chem. 282, 5116–5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bouzioukh F., Daoudal G., Falk J., Debanne D., Rougon G., Castellani V. (2006) Eur. J. Neurosci. 23, 2247–2254 [DOI] [PubMed] [Google Scholar]
- 81.Yamashita N., Morita A., Uchida Y., Nakamura F., Usui H., Ohshima T., Taniguchi M., Honnorat J., Thomasset N., Takei K., Takahashi T., Kolattukudy P., Goshima Y. (2007) J. Neurosci. 27, 12546–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arimura N., Ménager C., Kawano Y., Yoshimura T., Kawabata S., Hattori A., Fukata Y., Amano M., Goshima Y., Inagaki M., Morone N., Usukura J., Kaibuchi K. (2005) Mol. Cell. Biol. 25, 9973–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshida H., Watanabe A., Ihara Y. (1998) J. Biol. Chem. 273, 9761–9768 [DOI] [PubMed] [Google Scholar]
- 84.Honnorat J., Byk T., Kusters I., Aguera M., Ricard D., Rogemond V., Quach T., Aunis D., Sobel A., Mattei M. G., Kolattukudy P., Belin M. F., Antoine J. C. (1999) Eur. J. Neurosci. 11, 4226–4232 [DOI] [PubMed] [Google Scholar]
- 85.Chen A., Liao W. P., Lu Q., Wong W. S., Wong P. T. (2007) Neurochem. Int. 50, 1078–1086 [DOI] [PubMed] [Google Scholar]
- 86.Weitzdoerfer R., Fountoulakis M., Lubec G. (2001) J. Neural Transm. Suppl.61, 95–107 [DOI] [PubMed] [Google Scholar]
- 87.Ryu M. J., Lee C., Kim J., Shin H. S., Yu M. H. (2008) J. Neurochem. 104, 1260–1270 [DOI] [PubMed] [Google Scholar]
- 88.Bisgaard C. F., Jayatissa M. N., Enghild J. J., Sanchéz C., Artemychyn R., Wiborg O. (2007) J. Mol. Neurosci. 32, 132–144 [DOI] [PubMed] [Google Scholar]
- 89.Hou S. T., Jiang S. X., Desbois A., Huang D., Kelly J., Tessier L., Karchewski L., Kappler J. (2006) J. Neurosci. 26, 2241–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kowara R., Chen Q., Milliken M., Chakravarthy B. (2005) J. Neurochem. 95, 466–474 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.