Abstract
Adenosine deaminases acting on RNA (ADARs) catalyze the deamination of adenosine to inosine in double-stranded RNA templates, a process known as RNA editing. In Drosophila, multiple ADAR isoforms are generated from a single locus (dAdar) via post-transcriptional modifications. Collectively, these isoforms act to edit a wide range of transcripts involved in neuronal signaling, as well as the precursors of endogenous small interfering RNAs. The phenotypic consequences of a loss of dADAR activity have been well characterized and consist of profound behavioral defects manifested at the adult stage, including extreme uncoordination, seizures, and temperature-sensitive paralysis. However, the spatio-temporal requirements of adenosine to inosine editing for correct behavior are unclear. Using transgenic RNA interference, we show that network-wide editing in the nervous system is required for normal adult locomotion. Regulated restoration of editing activity demonstrates that the neuronal requirement of dADAR activity has a significant adult stage component. Furthermore we show that in relation to behavior there are no observable genetic interactions between dAdar and several loci encoding RNA interference components, suggesting that editing of neuronal transcripts is the key mode of ADAR activity for normal behavior in Drosophila.
Introduction
The catalytic deamination of adenosine to inosine in mRNAs of metazoan organisms is carried out by a highly conserved family of enzymes, the adenosine deaminases acting on RNA (ADARs)2 (1). A unifying feature of all ADAR substrates is a degree of double-stranded RNA (dsRNA) structure (2). However, ADARs may act on multiple distinct classes of RNAs. A-to-I editing can inhibit small interfering RNA (siRNA) production through disruption of dsRNA formation following the replacement of A-U Watson-Crick base-pairs with I·U wobble pairs or via competing with Dicers for shared dsRNA substrates (3, 4). ADARs have also been recently shown to modulate both micro RNA (miRNA) production and target recognition (5–7).
In addition, ADARs deaminate adenosines in a wide range of coding mRNAs. Intriguingly, in Drosophila, mice, and humans, transcripts encoding proteins involved in neuronal signaling comprise the vast majority of coding mRNAs known to undergo editing (8, 9). In these cases, editing is mediated by the formation of an imperfect dsRNA generated via base pairing between the region surrounding the edited adenosine and an adjacent complementary sequence, often found in neighboring intronic regions (10, 11). Because inosine is recognized by the cellular machinery as guanosine (12), adenosine deamination can act to “recode” the amino acid sequences of key regulators of neuronal firing properties and the release of neurotransmitter from synaptic vesicles. Indeed, A-to-I editing generally alters residues that are both highly conserved (or invariant) and functionally important (8, 11, 13–15).
The genome of Drosophila contains a single adar locus (termed dAdar) (16). Multiple dADAR isoforms are generated through alternative splicing and an auto-regulatory event in which dADAR edits its own transcript (16). Loss of dADAR activity results in extreme adult stage behavioral defects, including profound uncoordination, temperature-sensitive paralysis, seizures, progressive adult stage neuro-degeneration, and a complete lack of courtship displays in dAdar null males (17). Bio-informatic and comparative genomic approaches have identified a multitude of editing sites in mRNAs associated with electrical signaling in the nervous system (8, 18–21). In addition, recent evidence suggests that dADAR intersects with the RNAi pathway. Of the subpopulation of endogenous siRNAs that exhibit single base pair mismatches when compared with their genomic loci, adenosine-to-guanosine mismatches are vastly over-represented, suggesting A-to-I conversions (22).
Although the targets of dADAR are becoming increasingly well characterized, the spatio-temporal requirements of dADAR activity for wild-type behavior are less clear. Furthermore, no study has yet to delineate the relative importance of dADAR interactions with neuronal mRNA transcripts versus the various small RNA pathways. Here, we address these issues through two strategies. First, we test for epistatic interactions between dAdar and components of the RNAi pathway by generating fly lines null for both dAdar and genes involved in the production of a variety of small RNAs. Second, we use transgenic RNAi and dADAR-expressing transgenes coupled with the Gene-Switch system (23) to manipulate levels of dADAR activity in both time and space and investigate the corresponding effects on coordinated behavior in adult Drosophila.
EXPERIMENTAL PROCEDURES
Fly Stocks, Gene-Switch Expression Studies, and Data Analysis
The Drosophila melanogaster stocks Canton-S and w1118 were used as controls, depending on the genetic background of the experimental stocks. For all of the experiments involving dAdar null flies, we used the dAdar5g1 allele, which lacks all detectable editing activity (17). Progeny from all crosses were raised at a constant 25 °C and under 12-h day/night cycles. Both dAdar RNAi transgenes were obtained from the Vienna Drosophila RNAi Center stock center. Using the two individual lines, we subsequently generated a doubly balanced line (w1118; adr-IR1/CyO; adr-IR2/TM3) and picked double-homozygote males to cross to females carrying Gal4 insertions. Tissue-specific Gal4 lines were obtained from the Bloomington stock center. For Gene-Switch rescue experiments, we used the UAS-dADAR transgene line w1118;+; TM3::dADARwt5 (3/4 dADAR). Males from the transgene line were crossed to dAdar5g1/FM7;+; elav-Switch females, and dAdar5g1 males positive for both driver and transgene were collected and aged for 3–5 days. Gene-Switch activation was subsequently induced by placing flies on food containing 200 μm RU486 (Sigma) for 1 week before locomotor experiments or harvesting for preparation of RNA or protein. The original yw;+; elav-Switch stock was a kind gift from Stephen Helfand. The following lines were used to generate dAdar; RNAi double mutants: dicer-2 (dcr-2L811fsX), argonaute-2 (ago-251b), and lines with P-element insertions in the loquacious (loqs), r2d2, and piwi loci. dcr-2, loqs, piwi, and r2d2 mutations were balanced over CyO and crossed into a dAdar5g1/ FM7a::GFP; CyO/Sco background. The dAdar5g1/FM7:: GFP;+; ago-251b/TM3::GFP and UAS-dFMR+ stocks were kind gifts from Balpreet Bhogal and Thomas Jongens. Unpaired two-tailed Student's t tests were used for tests of statistical significance unless otherwise stated.
RNA Editing Analysis
RNA extractions from Drosophila heads (10–15/sample) were performed using TRIzol reagent (Invitrogen). At least three independent RNA samples were used as a template for each genotype. To measure editing at various sites, three methods were used. First, A and G peak heights from electropherogram traces were measured, and the percentage of editing was expressed at G/(A+G) × 100. For quantitative measurement of editing at site 7 of the Dα6 acetylcholine receptor and site 6 of eag, we used the EcoRI and Xmn-1 endonucleases, respectively (New England Biolabs), to digest the bulk RT-PCR product of each mRNA. In both cases, editing abolishes the respective endonuclease consensus site. Editing was calculated by measuring the relative fluorescent signal of cut and uncut products on an ethidium bromide gel, normalized to the size of the product. To control for incomplete cutting, the same reaction was performed using cDNA from dAdar null flies, and the intensity of the uncut product was subtracted from experimental samples. Finally, for measurement of edited isoform frequencies, we cloned bulk RT-PCR products into the TOPO vector (Invitrogen) and sequenced individual clones using gene-specific primers.
Western Blotting
Protein samples were prepared from ∼10–15 fly heads in buffer containing SDS and β-mercaptoethanol and run out on a 12.5% gel (Amresco). Anti-HA antibody (Covance) was used at 1:1000, whereas anti-actin (Chemicon International) was used at 1:10,000. Band intensities were quantified on a Kodak Image Station following background subtraction.
Behavioral Analysis
For locomotor assays, total activity was measured using population activity monitors (TriKinetics). Each monitor consists of a vertical glass vial surrounded by three concentric rings of infrared beams, covering the bottom, middle, and top of each vial. Locomotion is quantified as the total number of beam breaks over a 24-h period starting at 1 a.m. This timing allowed us to observe both morning and night peaks of activity. The flies were raised under 12-h light/day conditions and aged for 3–5 days post-eclosion prior to measurement (unless placed on RU486; see above). Nonanesthetized flies (n = 5 flies/vial) were used in all cases. For all genotypes except those harboring the dAdar5g1 allele, locomotion was measured following an acclimatization period of 36 h. For flies with the dAdar5g1 allele, a 12-h acclimatization period was used to avoid flies becoming stuck in the fly food. Temperature-sensitive paralysis was tested for by placing flies (1–5) in a glass vial into a 39 °C water bath for 2 min and scoring for subsequent paralysis. The vials were preheated for at least 30 min to ensure correct temperature equilibration.
Confocal Microscopy
All of the microscopy was performed on a Zeiss LSM 510 meta confocal microscope. The samples were prepared as previously described (24). Primary antibodies were used at the following concentrations: nc82 (anti-Bruchpilot, Developmental Studies Hybridoma Bank), 1:40; and anti-HA (Santa Cruz Biotech), 1:50. Alexa-fluor secondary antibodies (goat anti-mouse Cy3 and goat anti-rabbit fluorescein isothiocyanate (Invitrogen)) were used at 1:200. Confocal images were obtained at subsaturation levels of fluorescent intensity. The images were contrast-enhanced in Adobe Photoshop.
RESULTS
Inhibition of Small RNA Production or Targeting Fails to Rescue the dAdar Null Phenotype
Across a range of metazoan species, ADAR enzymes exhibit a dichotomous pattern of activity, editing both the precursors of small RNAs destined to enter the RNAi pathway (3–7) and imperfect duplexes in the coding regions of mRNAs (8, 9). A critical step in understanding how A-to-I editing is related to behavioral outputs is the determination of which of these two modes of ADAR activity is of greater relative importance.
In the invertebrate nematode Caenorhabditis elegans, loss of ADAR activity results in defects in chemotaxis (25), and wild-type chemotactic behavior can be restored by the introduction of mutations in genes involved in RNAi (26), implying that a major function of ADARs in C. elegans is to inhibit the action of specific small RNAs. Because Drosophila is also an invertebrate model organism, we asked whether the inhibition of small RNA production similarly rescued the behavioral defects observed in dAdar null flies.
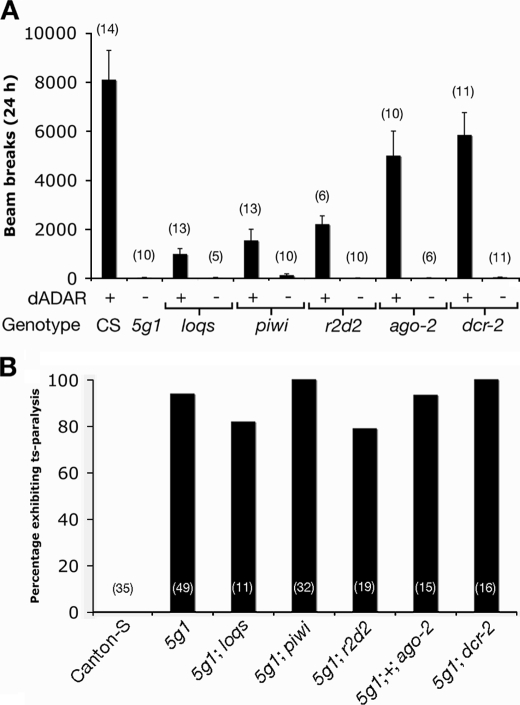
The primary phenotype of flies lacking editing activity is extreme adult stage uncoordination (17) (Fig. 1A). We used an automated monitoring system (see “Experimental Procedures”) to assess levels of locomotor activity in dAdar null flies over a 24-h time period. In agreement with previous observations (17, 27), flies lacking dAdar exhibited almost no detectable locomotion, in contrast to the robust levels of activity observed in wild-type controls (Fig. 1A). The introduction of mutations in several loci required for the production or targeting of a variety of small RNAs (28) failed to rescue locomotion in dAdar null flies to either wild-type levels or to levels observed in flies harboring the same mutations but wild type for dADAR activity (Fig. 1A).
FIGURE 1.
Lack of negative epistasis between dAdar and the RNAi pathway. A, graph of average locomotor activity for flies wild type for RNAi components (Canton-S, CS) or harboring mutations in loci involved in the production of siRNAs, miRNAs, or piwi-interacting RNAs, in a genetic background containing or lacking dADAR activity (5g1). The number of vials/genotype is indicated in parentheses. Error bars, S.E. values. B, graph showing proportion of flies/genotype exhibiting temperature-sensitive (ts) paralysis following incubation at 39 °C for 2 min. The number of flies tested per genotype is indicated in parentheses. The flies lacking editing activity are referred to by the dAdar allele used (5g1; see “Experimental Procedures”).
Furthermore, 78–100% of all of the double mutants tested showed clear temperature-sensitive paralysis at 39 °C, a level similar to that seen in dAdar nulls (93%) and in contrast to wild-type controls (0%) (Fig. 1B). These data suggest that the severe behavioral abnormalities observed in dAdar null flies do not result from the unrestrained action of small RNAs in the absence of dADAR but rather from a lack of specific editing in mRNA coding regions.
Down-regulation of dADAR Activity in Vivo via Transgenic RNAi
We next sought to determine the spatial requirements of dADAR activity for coordinated adult behavior in Drosophila. Previous studies have approached this issue through ectopic expression of individual dADAR isoforms in a dAdar null background and subsequently testing for rescue of locomotor activity (27). An obvious drawback of this method is the inability to recapitulate the diverse variety of dADAR isoforms expressed in the adult nervous system.
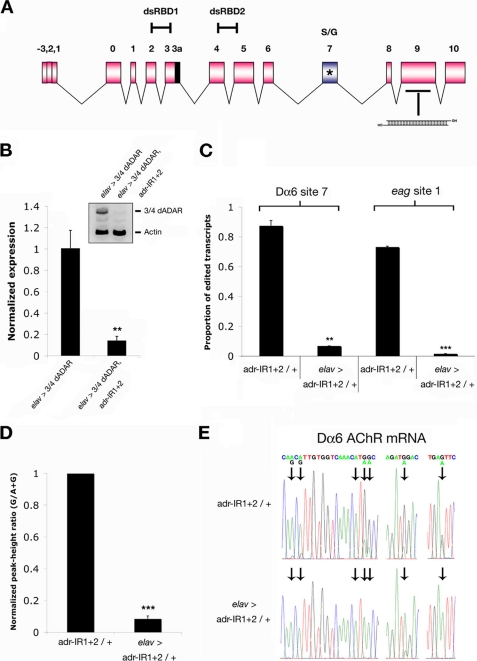
We therefore initially sought to adopt the converse strategy: to knock down expression of all dADAR isoforms simultaneously in an otherwise wild-type genetic background and to do this in a cell-specific manner. To achieve this end, we made use of two transgenic RNAi lines (termed adr-IR1 and adr-IR2, inserted on chromosomes 2 and 3, respectively) under the control of the UAS-Gal4 binary expression system (29), that generate perfect dsRNAs complementary to the 3′ end of the dAdar locus (30) (Fig. 2A). All of the dADAR isoforms are predicted to be down-regulated via the RNAi pathway in cells expressing these constructs. To maximize dADAR knock down, we generated a line containing both RNAi constructs and used this line for all subsequent experiments, because we found that the combined expression of two independent RNAi constructs was more efficient at reducing dADAR activity than either transgene line alone (supplemental Fig. S1, A and B).
FIGURE 2.
Knockdown of dADAR activity using transgenic RNAi. A, schematic diagram of the dAdar locus. Auto-editing occurs close to motif II of the catalytic domain in exon 7 (blue). Transgenic RNAi constructs align to exon 9, downstream of all post-transcriptional modifications. B, knockdown of dADAR protein levels demonstrated by co-expression of transgenic RNAi constructs with a HA-tagged dADAR transgene (3/4 dADAR; see text), driven with elav-Gal4. Inset is a representative example of n = 8 westerns from three independent samples. **, p < 0.005. C, effect of pan-neuronal expression of transgenic RNAi constructs on two editing sites, assayed by restriction digest analysis. The results are compared with transgenes/+ controls (n = 3 independent digests in each case; **, p < 0.005; ***, p < 0.0005). D, effects of dADAR knockdown on 33 editing sites in five mRNAs (Dα6, DopEcR, eag, shab, and Caα1D) measured by alteration in A/G peak-height ratio in control and experimental electropherograms (n = 3 independent PCRs/site). The data are shown normalized to transgenes/+ controls; ***, p < 0.0005, paired t test. E, representative electropherograms of transcripts encoding the Dα6 AChR amplified from control (transgenes/+) and experimental head cDNA. Editing at sites 1, 2, 4, 5, and 7 is abolished following knockdown. Editing at site 6 is greatly reduced but still present. Editing at site 3 is not detectable in control heads. Error bars, S.E. values.
Pan-neuronal expression of both RNAi constructs robustly reduced levels of a co-expressed HA-tagged dADAR transgene (Fig. 2B) and almost abolished detectable endogenous editing levels at two sites assayed by restriction digest analysis (Fig. 2C), as well as in the majority of a further 33 adenosines measured semi-quantitatively by comparison of A-G peak-height ratios in RT-PCR electropherograms (Fig. 2, D and E, and supplemental Fig. S1, C and D). Thus, expression of both RNAi constructs robustly diminished both dADAR protein levels and the editing activity at a wide range of editing sites.
Pan-neuronal Expression of dADAR Is Required for Normal Locomotion
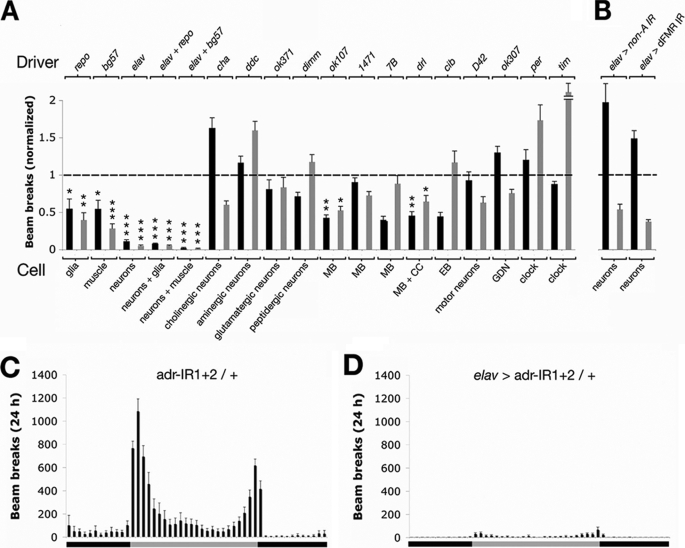
Using the above methodology, we knocked down dADAR expression in a variety of physiologically important cell types and examined the subsequent impact on locomotor activity. We initially examined the effects of dADAR knockdown in three cell types: neurons, glia, and muscle. Knockdown of dADAR in all of these excitable cell types correlated with a significant reduction in locomotion relative to both control genotypes (Fig. 3A). However, only pan-neuronal dADAR knockdown reduced locomotion close to levels seen in dAdar null flies (89 and 95% reductions compared with driver/+ and transgenes/+, respectively), completely abolishing both diurnal peaks of activity (Fig. 3, C and D). Simultaneous expression of both RNAi constructs in neurons and muscle (but not neurons and glia) yielded a significant additive effect (p < 0.05). Locomotion in these flies was further reduced compared with pan-neuronal knockdown alone and was only 4-fold higher than that exhibited by flies lacking dAdar (supplemental Fig. S2).
FIGURE 3.
Effects of cell-specific dADAR knockdown on adult locomotion. A, graph showing mean locomotor activity of experimental populations (n = 9–12 vials/genotype) normalized to driver/+ (black) or transgenes/+ control lines (gray), measured over 24 h. The cell type and driver are indicated in each case. Error bars, S.E. values. MB, mushroom body; CC, central complex; EB, ellipsoid body; GDN, giant descending neuron. *, p < 0.05; **, p < 0.005; ***, p < 0.0005, relative to controls (dotted line illustrates deviation from control means, normalized to 1). B, pan-neuronal expression of RNAi constructs targeted to non-A or dFMR does not phenocopy the effect of pan-neuronal dADAR knockdown (n = 9–11 vials/genotype). C and D, effects of dADAR knockdown on temporal patterns of locomotor activity. Flies harboring dAdar RNAi constructs with no driver exhibit robust peaks of activity at lights-on (morning; gray bars) and lights-off (night; black bars) (C). Pan-neuronal dADAR knockdown results in complete suppression of both peaks of activity (D).
In contrast, pan-neuronal expression of UAS-RNAi lines corresponding to two RNA-binding proteins, non-A and dFMR, failed to illicit a significant decrease in locomotion relative to both controls, indicating that the reduction in locomotor activity observed following dADAR knockdown in the nervous system is not a nonspecific effect of RNAi induction (Fig. 3B).
We next asked whether elimination of dADAR activity in discrete neuronal subtypes also conferred locomotor defects. Knockdown of dADAR in a range of spatially restricted neuronal subsets failed to phenocopy the effect of pan-neuronal dADAR knockdown (Fig. 3A), although small but significant reductions in locomotion were observed in two lines driving the RNAi constructs in the mushroom bodies, which in addition to their well known role in learning and memory also act to modulate locomotor activity (31, 32).
Cholinergic neurons have previously been suggested to be the primary locus for A-to-I editing in Drosophila. In a 2-min manual two-dimensional planar locomotor assay, ectopic expression of an uneditable dADAR transgene in cholinergic neurons was reported to rescue the locomotor deficits of dAdar null flies (27). However, in our automated system, transgenic flies expressing dAdar RNAi constructs in cholinergic neurons showed no reduction in activity compared with driver/+ controls when measured over a 24-h period and only a 40% reduction relative to transgenes/+ controls. To rule out the possibility that dADAR knockdown is inefficient in this cell type because of particularly high levels of dADAR expression or other properties that make these neurons refractory to RNAi, we took two complementary approaches. First, because cholinergic neurons are widely distributed throughout the adult brain (33), we reasoned that successful knockdown of dADAR might still leave an observable signature in bulk RT-PCR products derived from the entire nervous system. Indeed, we observed substantial reductions in editing of three pan-neuronal transcripts (lap, dUnc-13, and stn-B) following knockdown of dADAR in cholinergic neurons (supplemental Fig. S3). In contrast, editing of syt-1, which also exhibits pan-neuronal expression, was unaffected.
Negative results in this bulk assay are difficult to interpret, because the editing status of syt-1 in cholinergic neurons is unknown. It also remains possible that homeostatic mechanisms may increase editing in other neuronal populations in response to a reduction of editing activity in cholinergic neurons. Thus, we developed a method to directly analyze A-to-I editing in defined neurons using a molecular reporter of dADAR activity based on the syt-1 mRNA. We used this particular template because the intronic cis-elements that direct editing of sites C and D of syt-1 have been previously defined, residing in the intron directly downstream of the edited exon (11).
We expressed a transgene using the UAS-Gal4 system containing the edited exon of syt-1, flanked by the neighboring intronic regions as well as the downstream and upstream exons (supplemental Fig. S4A). When ectopically expressed using elav-Gal4 or cha-Gal4, the pattern of editing at sites C and D in the syt-1 reporter transgene (which we term syt-T), mimicked that of the endogenous syt-1 mRNA, with site D exhibiting a substantially higher level of editing relative to site C (supplemental Fig. S4, B and C). For both sites C and D, expression of syt-T using cha-Gal4 did not yield higher levels of editing relative to elav-Gal4 (supplemental Fig. S4C). We next examined the effect of co-expressing the syt-T reporter along with both dAdar RNAi constructs in cholinergic neurons. Under these conditions, editing at site C in the syt-T reporter was abolished, and editing at sites D was reduced by >20% (supplemental Fig. S4, B and C). From the above data we conclude that driving both RNAi constructs using cha-Gal4 is sufficient to dramatically reduce editing at some, though perhaps not all, editing sites. Thus, although editing may not be completely abolished in cholinergic neurons following dADAR knockdown (perhaps because of low expression of Gal4 from the cha promoter), it appears that the maintenance of wild-type levels of editing in this cell type is not essential for normal locomotor activity.
Controlled Restoration of Editing Activity in Time and Space Using the Gene-Switch System
How is neuronal dADAR activity linked to adult stage behavior? Hypothetically, two (mutually nonexclusive) paradigms can be envisioned. A-to-I editing may be required early in development for the establishment or maintenance of appropriate neuronal connections. Alternatively, fine-tuning of neuro-transmission via dADAR-mediated coding changes may be solely required at the adult stage. We sought to delineate between these two conceptual schemes. Given that (i) dAdar null larvae do not exhibit any obvious locomotor defects (17); (ii) expression of dAdar mRNA is highest at the pupal and adult stages (16); and (iii) editing at many adenosines is restricted to later stages of Drosophila development (34–36), we favored the hypothesis of a strong adult stage requirement for dADAR activity. This hypothesis makes a clear prediction that restoration of A-to-I editing specifically in the adult stage nervous system should partially rescue the dAdar null phenotype. To test this we utilized the Gene-Switch system (23), which allows temporal control of transgene expression through the use of a modified Gal4 (“switch”) that is active only in the presence of the ligand RU486.
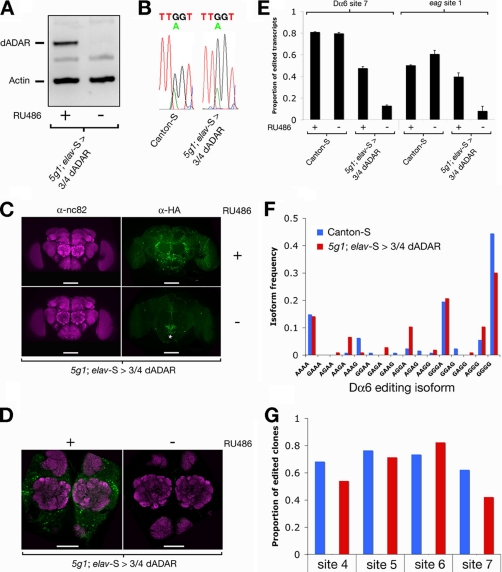
We used a pan-neuronal Gene-Switch driver (elav-S), to induce expression of a HA-tagged dADAR transgene corresponding to the most abundant adult stage isoform of dADAR (termed 3/4 dADAR) (16) specifically in the adult stage nervous system of dAdar null flies. Switch activation was initiated by transferring 3–5-day-old dAdar null males onto food containing 200 μm RU486 for 1 week. This concentration of RU486 elicited robust expression of the 3/4 dADAR transgene (Fig. 4A). Examination of the transgene mRNA revealed high levels of auto-editing, above that observed in endogenous dAdar transcripts isolated from total head mRNA in Canton-S controls (Fig. 4B). Confocal microscopy demonstrated that 3/4 dADAR was expressed throughout the nervous system when driven by elav-S and was localized to neuronal nuclei and cell bodies (Fig. 4, C and D). We did not detect any 3/4 dADAR expression in the absence of RU486 using both Western blotting and microscopy, illustrating that the Gene-Switch system allows stringent regulation over dADAR transgene expression. Furthermore, induction of 3/4 dADAR restored editing at two sites (site 7 of the Dα6 acetylcholine receptor (AChR) and site 1 of the eag potassium channel) to ∼60 and 80%, respectively, of levels observed in wild-type flies placed on RU486 (Fig. 4E).
FIGURE 4.
Restoration of adult stage editing using the Gene-Switch system. A, temporal control of transgene expression. In dAdar null flies (5g1) carrying the elav-S driver and a HA-tagged dADAR transgene (3/4 dADAR), transgene expression is only observed following placement of flies on food containing 200 μm RU486 (representative example of n = 3 Western blots). B, transgene auto-editing occurs at high levels following induction with RU486. Levels of editing are higher in the ectopically expressed transgene than dAdar mRNA isolated from whole Canton-S heads (n = 3 electropherograms for each genotype). C, contrast-enhanced confocal z-stack of 3/4 dADAR localization. In dAdar null flies expressing 3/4 dADAR under control of elav-S, punctate transgene expression (green) can be observed throughout the nervous system (neuropil is stained with anti-bruchpilot, nc82, purple) and is absent in flies of the same genotype fed on food lacking RU486 (n = 5, ± RU486). Bar, 100 μm. The staining in the subesophageal ganglion in the RU486 controls (asterisk) is an antibody artifact, as it is also present in w1118 control flies (data not shown). D, contrast-enhanced confocal slice showing 3/4 dADAR expression in nuclei/cell bodies surrounding the antennal lobes. Transgene expression is not observed in the neuropil. Bar, 50 μm. E, rescue of editing at Dα6 site 7 and eag site 1 by expression of 3/4 dADAR (n = 3 independent restriction digests/population). Error bars, S.E. values. F, frequency distribution of edited isoforms of Dα6 mRNAs is substantially recapitulated by expression of 3/4 dADAR in a dAdar null background. The frequencies were calculated by sequencing individual Dα6 cDNA clones derived from total head cDNA (Canton-S: n = 133; 5g1; elav-S > 3/4 dADAR: n = 109). G, total levels of editing at four sites in Dα6 mRNA in control and rescue backgrounds.
In the Drosophila nervous system, many mRNAs targeted for deamination by dADAR are edited at multiple adenosines, creating a combinatorial spectrum of edited isoforms. We investigated whether expression of the 3/4 dADAR transgene in a dAdar null background was able to phenocopy the wild-type distribution of editing in mRNAs encoding the Dα6 AChR subunit. The Dα6 transcript is edited at seven adenosines (19), leading to coding changes in the ligand-binding domain of the receptor. We focused on the four most 3′ editing sites (sites 4–7), which exhibit robust editing in wild-type head cDNA (supplemental Table S2).
We sequenced individual clones from bulk RT-PCR products derived from the heads of wild-type Canton-S controls and dAdar null flies expressing 3/4 dADAR driven by elav-S (Fig. 4F). Of the 16 possible combinations of editing sites, 12 were detected in Dα6 cDNA from Canton-S heads. However, the frequency distribution of edited isoforms was highly nonuniform, with three isoforms (fully edited, GGGG; fully unedited, AAAA; and edited at sites 4–6 but not site 7, GGGA) accounting for almost 80% of the 133 clones sequenced. Transcripts edited at all sites were the most abundant isoform (45%). Remarkably, expression of 3/4 dADAR in a dAdar null background using elav-S generated a strikingly similar frequency distribution to that of Canton-S, with the fully edited isoform found at a frequency of 30% (Fig. 4F). However, we also observed novel editing combinations (AAGA and AGGA) in the experimental genotype at frequencies of 6–13% that are extremely rare in the wild-type nervous system (1 of 133 and 3 of 133 clones, respectively). Examination of total editing at all four sites in wild-type and rescue genotypes further demonstrated the robust restoration of editing achieved following induction of 3/4 dADAR. Editing at sites 4–7 was rescued to 67–112% of wild-type levels, with site 7 rescued to the lowest degree, and site 6 rescued to the highest degree (Fig. 4G).
Broad Substrate Recognition by a 3/4 dADAR Transgene
Mammalian genomes encode three ADAR loci (ADAR1–3), which often exhibit specificity for particular dsRNA duplexes. For example, within the serotonin-2C receptor mRNA, site A is edited by ADAR1, but site D is not (37). Similarly, editing of the Gln/Arg site of the GluR-2 AMPA receptor is solely mediated by ADAR2, whereas the Gln/Arg site of the paralogous GluR-6 kainate receptor is an exclusive substrate of ADAR1 (38).
Whether post-transcriptional modifications of dAdar mRNAs affect substrate specificity is unknown. If the 3/4 dADAR transgene used in our Gene-Switch experiments was unable to edit a wide range of target adenosines, this would make the interpretation of any subsequent behavioral assays difficult. Therefore, we investigated the ability of the 3/4 dADAR transgene to mimic global editing levels in wild-type Drosophila.
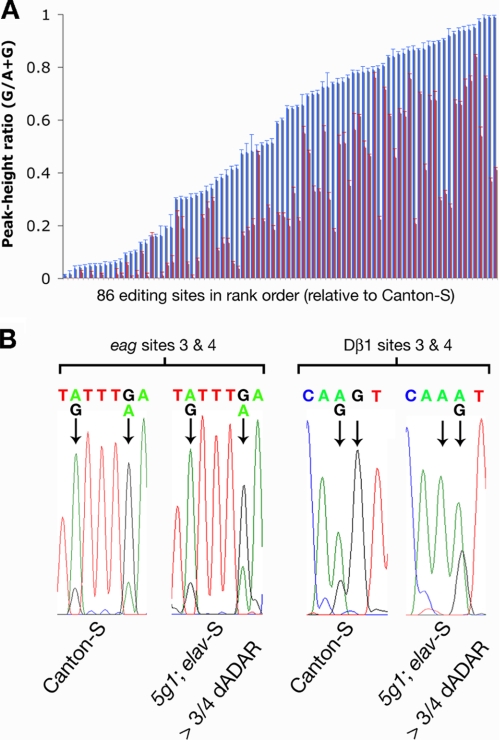
To do so, we assayed editing at a wide range of target adenosines (n = 86) in 19 neuronal mRNAs derived from the heads of Canton-S controls and dAdar null flies expressing 3/4 dADAR under control of elav-S. The mRNAs studied encompass three classes of protein translated from dADAR mRNA targets: voltage-gated ion channels (VGICs), ligand-gated ion channels (LGICs), and presynaptic release proteins, and exhibit diverse editing levels ranging from <5 to >90% (Fig. 5A).
FIGURE 5.
Global rescue of A-to-I editing via ectopic expression of a 3/4 dADAR transgene. A, peak-height ratios measured for 86 neuronal editing sites in Canton-S controls (blue) or dAdar null flies (red) expressing 3/4 dADAR driven by elav-S. Editing sites are presented in rank order of levels observed in Canton-S controls. The data are from 3–12 electropherograms from at least three independent total cDNA samples. The number of electropherogram peaks analyzed for each genotype: Canton-S, 382; 5g1; elav-S > 3/4 dADAR, 423. Error bars, S.E. values. B, examples of site-specific sensitivity to the expression of 3/4 dADAR. Editing at sites 3 and 4 in the eag potassium channel (edited to ∼16 and 80%, respectively, in Canton-S heads) is rescued by >85% via ectopic expression of 3/4 dADAR. In contrast, editing at sites 3 and 4 of the Dβ1 AChR mRNA (edited to ∼30 and 100% in Canton-S) is only rescued by 2 and 37%, respectively.
All of the sites with editing levels above 6% in wild-type heads (n = 76) exhibited detectable levels of editing following expression of 3/4 dADAR in a dAdar null background, illustrating that this single dADAR isoform can edit an extremely diverse range of target adenosines. Levels of editing induced by expression of 3/4 dADAR in dAdar null flies broadly correlate with the levels of endogenous editing over the 86 sites tested (R2 = 0.722 compared with Canton-S; supplemental Fig. S5A). However, in 83 of the 86 editing sites tested, expression of 3/4 dADAR was unable to fully restore wild-type levels of editing. The inability of 3/4 dADAR to fully rescue editing at many sites is not due to low expression levels, because increasing expression severalfold using a stronger global driver (tubulin-Gal4) failed to further restore editing at 36 adenosines tested and did not shift the spectrum of editing in Dα6 AChR mRNAs toward more highly edited isoforms (supplemental Fig. S6).
To delineate between sites with high and low affinities for the 3/4 dADAR isoform, we examined the identities of transcripts containing adenosines edited above 10% in Canton-S that were either highly (>75%) or weakly (<25%) rescued in dAdar null flies expressing 3/4 dADAR, examples of which are shown in Fig. 5B. Intriguingly, transcripts encoding VGICs dominated the population of mRNAs with adenosines rescued by >75% relative to Canton-S controls (14 of 17 adenosines) (supplemental Table S3). In contrast, 8 of 9 adenosines rescued by <25% were found in transcripts encoding LGICs. This skewed distribution occurs despite the fact that an equal proportion of the editing sites tested reside in mRNAs encoding VGICs and LGICs (39 of 86 and 38 of 86 sites, respectively). The mean percentage rescue relative to wild type is significantly higher for VGICs (61 ± 4%) compared with LGICs (44 ± 4%; p < 0.01). VGIC editing in the rescue background also exhibits a greater correlation with wild-type levels than that shown by LGICs (supplemental Fig. S5, B and C).
In addition to the above demarcation, high and low affinity sites for 3/4 dADAR tended to cluster within particular mRNAs. Just three transcripts (encoding the Caα1D calcium channel and the eag and shab potassium channels) account for 12 of 17 high affinity adenosines. Similarly, 8 of 9 low affinity adenosines in LGICs are clustered in three mRNAs (the Dα5 and Dβ2 AChR subunits and the rdl GABAA receptor α-subunit) (supplemental Table S3).
These results first indicate that the 3/4 dADAR isoform exhibits a very low degree of absolute substrate specificity and second suggest that post-transcriptional modifications that lead to the inclusion of alternative exons not present in the 3/4 dADAR transgene may act to modulate the affinity of dADAR for dsRNA duplexes in mRNAs encoding distinct classes of neuronal proteins.
Adult Stage Restoration of A-to-I Editing Partially Rescues the dAdar Null Locomotor Defect
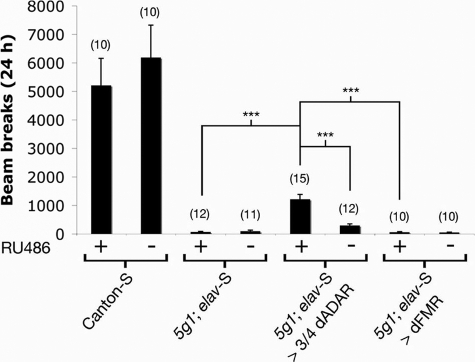
Given that expression of 3/4 dADAR was able to restore editing at almost all of the sites tested (although in the majority of cases, not to wild-type levels), we next examined the effect of adult stage restoration of A-to-I editing on coordinated locomotion using the same automated locomotor assay described above (Figs. 1A and 3). A week-long exposure to RU486-containing fly food did not induce an alteration in locomotor activity in either wild-type or dAdar null flies heterozygous for elav-S nor in dAdar null flies overexpressing a dFMR transgene under the control of elav-S (Fig. 6). In contrast, adult stage neuronal expression of 3/4 dADAR increased locomotor activity in dAdar null flies 18-fold relative to dAdar nulls carrying the elav-S driver but lacking the 3/4 dADAR transgene (1214 ± 166 versus 68 ± 22 beam breaks over 24 h). This increase represents a rescue to ∼25% of locomotor activity exhibited by Canton-S controls fed RU486. These results demonstrate a clear adult stage requirement for dADAR activity in Drosophila.
FIGURE 6.
Induction of editing in the adult nervous system partially restores locomotion in dAdar null flies. Mean activity in control and experimental genotypes with or without a 1-week exposure to RU486. The presence of RU486 neither alters locomotor activity over 24 h in wild-type controls nor rescues locomotion in dAdar null flies heterozygous for elav-S alone or overexpressing dFMR via elav-S. In contrast, expression of 3/4 dADAR in the adult nervous system significantly restores locomotion relative to the above controls (***, p < 0.0005) to ∼25% of wild type. Error bars, S.E. values.
DISCUSSION
Understanding how gene function is related to ethology requires a detailed knowledge of not only the molecular pathways but also the cell types and developmental stages in which gene activity is most crucial. A-to-I editing is essential for correct adult stage behavior in Drosophila (17). It is becoming increasingly clear that dADAR is likely to exhibit extreme functional pleiotropy, deaminating adenosines within a multitude of coding mRNAs as well as the precursors of endogenous siRNAs (8, 22). In this study we have sought to untangle this pleiotropy by (i) defining the primary substrates of dADAR for correct behavior, and (ii) investigating the spatial and temporal loci in which A-to-I editing is most crucial.
If the predominant function of A-to-I editing in Drosophila was to antagonize small RNA production or targeting, then an overabundance of small RNA activity would be expected to substantially contribute to the dAdar null phenotype. If so, reducing small RNA production should rescue wild-type behavior in flies lacking dADAR activity. Although such negative epistasis has been documented in another invertebrate model organism, the nematode C. elegans (26), we were unable to observe any genetic interactions between dAdar and several loci involved in the production of a range of small RNAs (Fig. 1). Positive interactions with the RNAi pathway (for example, the requirement of edited endogenous si- or miRNAs) are also unlikely to significantly contribute to the dAdar null phenotype, because dcr-2, ago-2, and loqs mutants do not phenocopy flies lacking editing activity (Fig. 1A).3
A number of caveats should be made clear when interpreting this data. First, we cannot fully rule out the possibility of negative epistasis with the miRNA pathway, because miRNA production is not fully abolished in loqs null flies (39), and the generation of dAdar; dicer-1, or argonaute-1 double nulls (which would completely lack miRNA activity) was impossible given the lethality of dicer-1 and argonaute-1 mutants. Second, it has previously been reported that certain siRNAs may be initially generated by Dicer-1 and subsequently loaded into Ago-2 complexes (40). Therefore, a small degree of residual siRNA activity may still be present in dcr-2 null flies. Third, in the double-null mutants we generated, we did not investigate the full spectrum of abnormalities observed in dAdar null flies, such as seizure frequency, disorganization of the retina, or progressive adult stage neuro-degeneration (17).
Nonetheless, while keeping the above caveats in mind, given that extreme uncoordination and temperature-sensitive paralysis are still present in all the double mutants generated, we suggest that the key function of dADAR in relation to behavior is likely to be the alteration of the protein landscape through deamination of adenosines in the coding regions of mRNAs. This conclusion is parsimonious with the ontology of mRNA targets of dADAR (which are almost exclusively involved in controlling the electrophysiological and synaptic release properties of excitable cells) and the phenotype of dAdar null males (severe behavioral abnormalities without any gross morphological defects) (8, 17–21).
We present two additional pieces of evidence that genetic recoding of neuronal transcripts is the crucial mode of dADAR activity. First, although our locomotor assays combined with cell-specific dADAR knockdown indicate a role for A-to-I editing in non-neuronal tissues, they strongly illustrate that the primary locus for editing is the nervous system. Loss of neuronal editing activity correlates with a 90% reduction in locomotor activity, close to levels seen in dAdar null flies (Fig. 3, A and D, and supplemental Fig. S2). The requirement of dADAR in the Drosophila brain appears to be “network-wide” rather than limited to a restricted population of neurons, because knockdown of editing in a wide range of neuronal subtypes failed to phenocopy the effect of pan-neuronal loss of dADAR activity (Fig. 3A).
Second, restoration of editing specifically in the adult stage nervous system of dAdar null flies rescued locomotion to ∼25% of wild type (Figs. 4–6). Several constraints are likely to limit the level of rescue achieved by this method. dADAR activity is not solely required in the nervous system for locomotor activity (Fig. 3). Knockdown of dADAR in the muscle reduced locomotion from ∼7200 beam breaks in 24 h (transgenes/+ controls) to ∼2900 (driver + transgenes) (supplemental Fig. S2). Thus, activity levels in our rescued dAdar mutants (∼1200 beam breaks in 24 h) are likely to be approaching the maximum level achievable by transgene expression solely in the nervous system, particularly when considering the possibility of any additional requirements of editing in glial or other cell types.
In addition, although we were able to restore editing at almost all of the adenosines tested (Fig. 5A) and substantially recapitulate the spectrum of edited isoforms of one transcript (the Dα6 AChR mRNA; Fig. 4F), we were unable to restore wild-type levels of editing at the majority of editing sites (Fig. 5A). Our data indicate that the most prevalent adult stage dADAR isoform (termed 3/4 dADAR) is capable of editing almost all target adenosines but not to wild-type levels. Surprisingly, mRNAs encoding ligand-gated ion channels appeared significantly more resistant to editing by 3/4 dADAR relative to those encoding voltage-gated ion channels (supplemental Fig. S5 and supplemental Table S3). It will be interesting to test whether varying auto-editing levels, expressing dADAR transgenes that include alternative exons not present in 3/4 dADAR, or co-expressing multiple dADAR isoforms can further rescue editing in this class of mRNAs.
The lack of a complete restoration of editing is likely to act as a further limit to the level of rescue attainable by expressing a single dADAR isoform. Functional analysis has demonstrated strong epistatic interactions between editing sites in both the Shaker potassium channel and the Rdl GABAA receptor (35, 36), suggesting that regulation of the spectrum of edited isoforms of a given protein is of adaptive importance. The inability to correctly mimic wild-type levels of editing by expression of 3/4 dADAR may lead to the generation of novel edited isoforms (Fig. 4F) and consequent downstream effects on neuronal physiology that may mitigate a more complete rescue. Therefore, our rescue experiments are likely to significantly under-estimate the actual adult stage requirement for neuronal A-to-I editing.
In conclusion, although our data does not rule out the hypothesis that as yet unknown neuro-developmental defects contribute to the dAdar null phenotype, we demonstrate that a major role of editing is to fine-tune neuronal physiology at the adult stage through genetic recoding. Altering the expression pattern and biophysical properties of several dADAR targets has been shown to modulate complex behaviors such as learning and memory, sleep, and the production of the mating song (41–45). The generation of tools to robustly knock down dADAR in a cell-specific manner will now facilitate experiments to determine how A-to-I editing molds the transcriptome in discrete neuronal subpopulations, contributing to evolved and adaptive ethological outputs in Drosophila.
Supplementary Material
Acknowledgments
We acknowledge Selena Gell and Leila Rieder for constructive criticism of the manuscript. Cynthia Staber provided technical assistance, and Yiannis Savva generated the syt-T reporter construct used in supplemental Fig. S4.
This work was supported by an Ellison Medical Foundation Senior Scholar award.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S6.
J. E. C. Jepson, personal observations.
- ADAR
- adenosine deaminase acting on RNA
- A-to-I
- adenosine to inosine
- dsRNA
- double-stranded RNA
- siRNA
- small interfering RNA
- miRNA
- microRNA
- VGIC
- voltage-gated ion channel
- LGIC
- ligand-gated ion channel
- AChR
- acetylcholine receptor
- syt-1
- synaptotagmin-1
- RNAi
- RNA interference
- RT
- reverse transcription
- HA
- hemagglutinin
- GABAA
- γ-aminobutyric acid, type A.
REFERENCES
- 1.Keegan L. P., Gallo A., O'Connell M. A. (2001) Nat. Rev. Genet. 2, 869–878 [DOI] [PubMed] [Google Scholar]
- 2.Bass B. L. (2001) RNA Editing, pp. 94–95, Oxford University Press, Oxford [Google Scholar]
- 3.Scadden A. D., Smith C. W. (2001) EMBO Rep. 2, 1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W., Wang Q., Howell K. L., Lee J. T., Cho D. S., Murray J. M., Nishikura K. (2005) J. Biol. Chem. 280, 3946–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W., Chendrimada T. P., Wang Q., Higuchi M., Seeburg P. H., Shiekhattar R., Nishikura K. (2006) Nat. Struct. Mol. Biol. 13, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawahara Y., Zinshteyn B., Chendrimada T. P., Shiekhattar R., Nishikura K. (2007) EMBO Rep. 8, 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A. G., Nishikura K. (2007) Science 315, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoopengardner B., Bhalla T., Staber C., Reenan R. (2003) Science 301, 832–836 [DOI] [PubMed] [Google Scholar]
- 9.Seeburg P. H., Hartner J. (2003) Curr. Opin. Neurobiol. 13, 279–283 [DOI] [PubMed] [Google Scholar]
- 10.Higuchi M., Single F. N., Köhler M., Sommer B., Sprengel R., Seeburg P. H. (1993) Cell 75, 1361–1370 [DOI] [PubMed] [Google Scholar]
- 11.Reenan R. A. (2005) Nature 434, 409–413 [DOI] [PubMed] [Google Scholar]
- 12.Basilio C., Wahba A. J., Lengyel P., Speyer J. F., Ochoa S. (1962) Proc. Natl. Acad. Sci. U.S.A. 48, 613–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhalla T., Rosenthal J. J., Holmgren M., Reenan R. (2004) Nat. Struct. Mol. Biol. 11, 950–956 [DOI] [PubMed] [Google Scholar]
- 14.Jin Y., Tian N., Cao J., Liang J., Yang Z., Lv J. (2007) BMC Evol. Biol. 7, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verdoorn T. A., Burnashev N., Monyer H., Seeburg P. H., Sakmann B. (1991) Science 252, 1715–1718 [DOI] [PubMed] [Google Scholar]
- 16.Palladino M. J., Keegan L. P., O'Connell M. A., Reenan R. A. (2000) RNA 6, 1004–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palladino M. J., Keegan L. P., O'Connell M. A., Reenan R. A. (2000) Cell 102, 437–449 [DOI] [PubMed] [Google Scholar]
- 18.Semenov E. P., Pak W. L. (1999) J. Neurochem. 72, 66–72 [DOI] [PubMed] [Google Scholar]
- 19.Grauso M., Reenan R. A., Culetto E., Sattelle D. B. (2002) Genetics 160, 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stapleton M., Carlson J. W., Celniker S. E. (2006) RNA 12, 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith L. A., Peixoto A. A., Hall J. C. (1998) J. Neurogenet. 12, 227–240 [DOI] [PubMed] [Google Scholar]
- 22.Kawamura Y., Saito K., Kin T., Ono Y., Asai K., Sunohara T., Okada T. N., Siomi M. C., Siomi H. (2008) Nature 453, 793–797 [DOI] [PubMed] [Google Scholar]
- 23.Roman G., Endo K., Zong L., Davis R. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12602–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J. S., Luo L. (2006) Nat. Protoc. 1, 2110–2115 [DOI] [PubMed] [Google Scholar]
- 25.Tonkin L. A., Saccomanno L., Morse D. P., Brodigan T., Krause M., Bass B. L. (2002) EMBO J. 21, 6025–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tonkin L. A., Bass B. L. (2003) Science 302, 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keegan L. P., Brindle J., Gallo A., Leroy A., Reenan R. A., O'Connell M. A. (2005) EMBO J. 24, 2183–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamura K., Lai E. C. (2008) Nat. Rev. Mol. Cell Biol. 9, 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand A. H., Perrimon N. (1993) Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 30.Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., Couto A., Marra V., Keleman K., Dickson B. J. (2007) Nature 448, 151–156 [DOI] [PubMed] [Google Scholar]
- 31.Helfrich-Förster C., Wulf J., de Belle J. S. (2002) J. Neurogenet. 16, 73–109 [DOI] [PubMed] [Google Scholar]
- 32.Martin J. R., Ernst R., Heisenberg M. (1998) Learn. Mem. 5, 179–191 [PMC free article] [PubMed] [Google Scholar]
- 33.Salvaterra P. M., Kitamoto T. (2001) Brain Res. Gene Expr. Patterns 1, 73–82 [DOI] [PubMed] [Google Scholar]
- 34.Hanrahan C. J., Palladino M. J., Ganetzky B., Reenan R. A. (2000) Genetics 155, 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones A. K., Buckingham S. D., Papadaki M., Yokota M., Sattelle B. M., Matsuda K., Sattelle D. B. (2009) J. Neurosci. 29, 4287–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingleby L., Maloney R., Jepson J., Horn R., Reenan R. (2009) J. Gen. Physiol. 133, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y., Emeson R. B., Samuel C. E. (1999) J. Biol. Chem. 274, 18351–18358 [DOI] [PubMed] [Google Scholar]
- 38.Maas S., Melcher T., Herb A., Seeburg P. H., Keller W., Krause S., Higuchi M., O'Connell M. A. (1996) J. Biol. Chem. 271, 12221–12226 [DOI] [PubMed] [Google Scholar]
- 39.Förstemann K., Tomari Y., Du T., Vagin V. V., Denli A. M., Bratu D. P., Klattenhoff C., Theurkauf W. E., Zamore P. D. (2005) PLoS Biol. 3, e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Förstemann K., Horwich M. D., Wee L., Tomari Y., Zamore P. D. (2007) Cell 130, 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agosto J., Choi J. C., Parisky K. M., Stilwell G., Rosbash M., Griffith L. C. (2008) Nat. Neurosci. 11, 354–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parisky K. M., Agosto J., Pulver S. R., Shang Y., Kuklin E., Hodge J. J., Kang K., Liu X., Garrity P. A., Rosbash M., Griffith L. C. (2008) Neuron 60, 672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cirelli C., Bushey D., Hill S., Huber R., Kreber R., Ganetzky B., Tononi G. (2005) Nature 434, 1087–1092 [DOI] [PubMed] [Google Scholar]
- 44.Liu X., Krause W. C., Davis R. L. (2007) Neuron 56, 1090–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peixoto A. A., Hall J. C. (1998) Genetics 148, 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.