Abstract
ClpA is a ring-shaped hexameric chaperone that binds to both ends of the protease ClpP and catalyzes the ATP-dependent unfolding and translocation of substrate proteins through its central pore into the ClpP cylinder. Here we study the relevance of ATP hydrolysis in the two ATPase domains of ClpA. We designed ClpA Walker B variants lacking ATPase activity in the first (D1) or the second ATPase domain (D2) without impairing ATP binding. We found that the two ATPase domains of ClpA operate independently even in the presence of the protease ClpP or the adaptor protein ClpS. Notably, ATP hydrolysis in the first ATPase module is sufficient to process a small, single domain protein of low stability. Substrate proteins of moderate local stability were efficiently processed when D1 was inactivated. However, ATP hydrolysis in both domains was required for efficiently processing substrates of high local stability. Furthermore, we provide evidence for the ClpS-dependent directional translocation of N-end rule substrates from the N to C terminus and propose a mechanistic model for substrate handover from the adaptor protein to the chaperone.
Introduction
The chaperone ClpA is a member of the AAA+ protein family (ATPase-associated with various cellular activities) and catalyzes the energy-dependent degradation of proteins through interaction with the protease ClpP in Escherichia coli (1–3). Like many other AAA proteins (4), ClpA oligomerizes into a ring structure, shaping a central pore through which substrate proteins are routed into the proteolytic core ClpP. This process involves unfolding and translocation of the substrate protein and requires the consumption of ATP. AAA proteins can be grouped into class I (two ATPase domains) and class II (one ATPase domain) ATPases. The fact that some ATPases have only one AAA module whereas others seem to require two ATPase domains stimulated researchers to investigate the roles and interdependence of the two ATP-binding modules in class I AAA proteins like Hsp104 and ClpB (5–8). ClpA also features two ATPase domains, termed D1 and D2. They are highly homologous, but differences in the amino acid sequence around the conserved regions in D1 and D2 suggested that they might have a somewhat different function (9). As was shown for several other class I members, ClpA assembles into its oligomeric state only upon binding of nucleotide (10). Indeed, substituting the invariant lysine in the Walker A motif demonstrated that nucleotide binding to D1 triggers ClpA hexamerization, whereas ATP turnover is mainly catalyzed by D2 (11, 12). However, mutations in the Walker A motif also abolish or drastically decrease the affinity for ATP, making it impossible to distinguish between effects due to ATP binding and those due to ATP hydrolysis.
To study the role of ATP hydrolysis in both ATPase domains uncoupled from nucleotide binding events, we designed ClpA Walker B variants that lack the ability to hydrolyze ATP in either D1 (ClpAE286A), D2 (ClpAE565A), or both domains (ClpAE286A/E565A) but still bind ATP in both domains. This allowed us to probe the function of ATP turnover in D1 independent from ATP hydrolysis in D2, and vice versa, to reveal whether ATP hydrolysis is allosterically coupled or whether ATP consumption in D1 and D2 occurs nonconcomitantly. ClpA recognizes several classes of substrates. One example are the ssrA-tagged proteins that carry an 11-amino acid tag that is added to stalled nascent chains. This tag is recognized at the ClpA pore.
The substrate specificity of ClpA is modulated by an adaptor protein, ClpS, that inhibits degradation of ssrA-tagged substrates (13, 14) and simultaneously promotes processing of N-end rule substrates (15–18). These substrates are recognized due to their N-terminal amino acids followed by a flexible stretch of amino acids, for example the FR in the model substrate FRli-GFP. We studied the influence of various substrate proteins, ClpS-dependent or ClpS-independent, bearing an N- or C-terminal degradation tag, on the ATPase and degradation activity of ClpA harboring only one active ATPase domain. Furthermore, we addressed the directionality of ClpS-mediated N-end rule substrate processing. Taken together, this study provides new mechanistic insights on how the two ATPase domains of ClpA work together to unfold and translocate substrate proteins into ClpP.
EXPERIMENTAL PROCEDURES
Cloning and Protein Purification
The mutated genes for ClpAE286A, ClpAE565A and ClpAE286A/E565A, were generated by using the QuikChange protocol (Stratagene). The ssrA tag (AANDENYALAA) was C-terminally linked to GFP3 (GFP-ssrA) (19) and λ-repressor92C (λ-repressor92C-ssrA) (20). A truncated self-complemented FimA variant (FimAta) was designed for recombinant production of stable monomeric FimA. The N-terminal donor strand (AATTVNGGTVHFKGEVVNA) of FimA was deleted to prevent self-polymerization of FimA and C-terminally elongated by a short linker (Gly6) followed by the donor strand (AATTVNGGTVHFKGEVVNA). The plasmid containing fimAta was kindly provided by Prof. Rudolf Glockshuber. FimAta was fused N-terminally to a His6-TEV tag (His6-ENLYFQ, where TEV is tobacco etch virus protease cleavage site) and C-terminally extended by a linker followed by the ssrA tag (GITHGMDELYKAANDENYALAA), which yields His6-TEV-FimAta-li-ssrA. The His10-EK-FRli tag (His10-DDDDKFRSKGEELVTGVSSGHIEGRH, where EK is enterokinase cleavage site) was linked to the N terminus of GFP (15) and T4 lysozyme. The His6-TEV-FRli tag was added N-terminally to the eYFP-linker-GFPuv construct (His6-SGSENLYFQFRSKGEELVTGVSSGHIEGRH-eYFP-SPNGASESGSAPKTSSAPGS-GFPuv). All constructs were verified by DNA sequencing. ClpA, ClpP, ClpS, and all ClpA variants as well as all tagged model substrates (GFP-ssrA, His6-TEV-FimAta-li-ssrA, λ-repressor92C-ssrA, His10-EK-FRli-GFP and His10-EK-FRli-T4 lysozyme) were overproduced in E. coli strain Rosetta (DE3) (Invitrogen) from plasmids harboring the respective gene under control of the T7 promoter.
ClpP, ClpA, and all ClpA variants were purified as described previously (21). ClpS was purified as established by Dougan et al. (13). GFP-ssrA was purified by heat denaturation (30 min at 65 °C and GFP-ssrA remains soluble) followed by ion exchange chromatography (Q-Sepharose Fast Flow, Source30Q). λ-repressor92C-ssrA was purified by ion exchange chromatography (DEAE-Sephacel) before being applied to a heparin-Sepharose column followed by size exclusion chromatography (Superdex 75). His6-TEV-FimAta-li-ssrA was purified as described previously (22) using chelating Ni2+ chromatography instead of anion exchange chromatography as the last step of purification. Correct folding of purified His6-TEV-FimAta-li-ssrA was confirmed by circular dichroism spectroscopy. Both N-end rule substrates (His10-EK-FRli-GFP and His10-EK-FRli-T4 lysozyme) were purified by chelating Ni2+ chromatography. FRli substrate was released from purified His10-EK-FRli substrate by enterokinase digestion using the enterokinase cleavage capture kit (Novagen) according to the manufacturer's instructions. The His10 tag was subsequently removed by chelating Ni2+ chromatography. The His6-TEV-FRli-eYFP-li-GFPuv construct was purified by heat denaturation (30 min at 60 °C). The soluble fraction was dialyzed against 50 mm HEPES, pH 7.5, 0.3 m KCl, 15% glycerol (v/v), 2 mm EDTA, and a purity of ∼90% was confirmed by SDS-PAGE. FRli-eYFP-li-GFPuv was released by incubating with recombinant tobacco etch virus protease prior to the biochemical assay.
All proteins were stored in 50 mm HEPES, pH 7.5, 0.3 m KCl, 15% glycerol (v/v), and 2 mm EDTA. All purified protein constructs were verified by mass spectroscopy (Functional Genomics Center, Zurich, Switzerland). Chromatographic materials were purchased from GE Healthcare. Protein concentrations were determined spectroscopically by measuring the absorption at 280 nm. The concentrations of GFP-ssrA, FRli-GFP, and FRli-eYFP-li-GFPuv were determined by measuring the absorption at 447 nm in 0.1 m NaOH. All ClpA concentrations refer to hexamer and ClpP concentrations to 14-mer.
Analytical Gel Filtration
20 μl of 1.5 μm ClpAP complex (1.5 μm ClpP, 3 μm ClpA) preassembled for 1 h in 1 mm ATPγS was applied onto a Superose 6PC (2.4 ml) gel filtration column (Amersham Biosciences). The column was equilibrated in buffer R (50 mm HEPES, pH 7.5, 0.3 m KCl, 15% glycerol (v/v), 20 mm MgCl2) and completed with 1 mm ATPγS. Absorbance was monitored at 227 nm. Accordingly, 20 μl of 1.5 μm ClpP was applied to the gel filtration column equilibrated in buffer R. All experiments were performed at 23 °C.
Continuous Spectrophotometric ATPase Assay
The ATPase activity was determined using a continuous spectrophotometric assay coupled to the release of inorganic phosphate using 7-methylinosine and purine nucleoside phosphorylase (23). The depletion of 7-methylinosine was followed by monitoring the absorption at 291 nm. For all steady-state measurements the concentration of ClpA was 0.1 μm. Accordingly, 0.05 μm ClpP and 0.6 μm ClpS were used as indicated. Only for the ClpAE565A variant was 0.5 μm used to record the ATP saturation curve due to the low ATPase activity. The reaction was started by adding 5 mm ATP, 1 mm 7-methylinosine, and 1.5 units/ml purine nucleoside phosphorylase. All measurements were carried out in buffer R completed with 0.5 mm DTT at 23 °C in a Cary UV-visible spectrophotometer (Varian). The ATPase activity of the ClpAP complex (0.1 μm ClpA and 0.05 μm ClpP) was also measured in the presence of 3 μm GFP-ssrA, 3 μm FRli-GFP (with 0.6 μm ClpS), 30 μm FRli-GFP, 6 μm FRli-T4 lysozyme (with 0.6 μm ClpS), and 6 μm His6-TEV-FimAta-li-ssrA. All given ATPase rate constants k (min−1) refer to ClpA hexamer.
Labeling and Degradation of λ-Repressor92C-ssrA
The C-terminal cysteine of λ-repressor92C-ssrA was selectively labeled with the thiol-reactive fluorophore fluorescein 5-maleimide (Invitrogen) as described previously by Reid et al. (20). Degradation of λRep92C-ssrA-F was carried out at 23 °C in a fluorescence spectrophotometer (Photon Technology International). 0.5 μm ClpA, 0.25 μm ClpP, and 6 μm λ-repressor92C-ssrA-F were mixed with 5 mm ATP in buffer R completed with 80 mm phosphocreatine (Sigma), 10 units/ml creatine phosphokinase (Sigma), and 0.5 mm DTT. The excitation wavelength was set to 495 nm, and emission was recorded at 515 nm. The fluorescence intensity of the fluorescein moiety increases upon protein degradation (24). The relative fluorescence change (Frel = F(t)/Finitial) was plotted over time.
Degradation of GFP-ssrA and FRli-GFP
GFP degradation was followed by monitoring the fluorescence intensity at 507 nm (excitation at 397 nm). All steady-state measurements were carried out in a fluorescence spectrometer (Photon Technology International) in buffer R completed with 50 mm phosphocreatine (Sigma), 10 units/ml creatine phosphokinase (Sigma), and 0.5 mm DTT at 23 °C. Degradation of GFP was initiated by mixing 0.5 μm ClpA, 0.25 μm ClpP, and 6 μm GFP (FRli-GFP or GFP-ssrA) with 5 mm ATP. FRli-GFP degradation was measured in the presence of 3 μm ClpS. Single-turnover experiments were conducted in a stopped-flow apparatus (SX-20, Applied Photophysics) in buffer R completed with 50 mm phosphocreatine (Sigma), 10 units/ml creatine phosphokinase (Sigma), and 0.5 mm DTT at 23 °C. 2 μm ClpA and 1 μm ClpP were preassembled in 1 mm ATPγS and rapidly mixed with 0.4 μm GFP-ssrA and 5 mm ATP. Accordingly 2 μm ClpA, 1 μm ClpP, and 12 μm ClpS were preassembled in 1 mm ATPγS and rapidly mixed with 0.4 μm FRli-GFP and 5 mm ATP. The relative fluorescence change (Frel = F(t)/Finitial) was plotted over time.
Degradation of FRli-eYFP-li-GFPuv
In a stopped-flow fluorescence experiment, a solution containing 2 μm ClpA, 1 μm ClpP, and 12 μm ClpS (preassembled in 1 mm ATPγS) was rapidly mixed with 1.5 μm FRli-eYFP-li-GFPuv and 5 mm ATP in buffer R completed with 50 mm phosphocreatine (Sigma), 20 units/ml creatine phosphokinase (Sigma), and 0.5 mm DTT. The excitation wavelength was set to 514 nm, and fluorescence emission was measured at 527 nm to observe eYFP degradation. Degradation of GFPuv was measured by monitoring the fluorescence at 507 nm with excitation at 397 nm. All experiments were carried out at 23 °C in an SX-20 stopped-flow device (Applied Photophysics). The relative fluorescence change (Frel = F(t)/Finitial) was plotted over time.
Degradation of FRli-T4 Lysozyme
6 μm FRli-T4 lysozyme was mixed with 0.5 μm ClpA, 0.25 μm ClpP, and 3 μm ClpS in the presence of 50 mm phosphocreatine (Sigma), 10 units/ml creatine phosphokinase (Sigma), and 5 mm ATP in buffer R completed with 0.5 mm DTT at 23 °C. Aliquots were taken at the indicated time points, and further degradation was prevented by mixing with SDS sample buffer. Proteins were separated by SDS-PAGE followed by Coomassie staining. The remaining amount of FRli-T4 lysozyme was quantified using the spot analysis program AlphaEaseFC version 4.0 (Alpha Innotech Corp.).
RESULTS
ATPase Activity of the First and the Second AAA Module Can Be Selectively Disabled
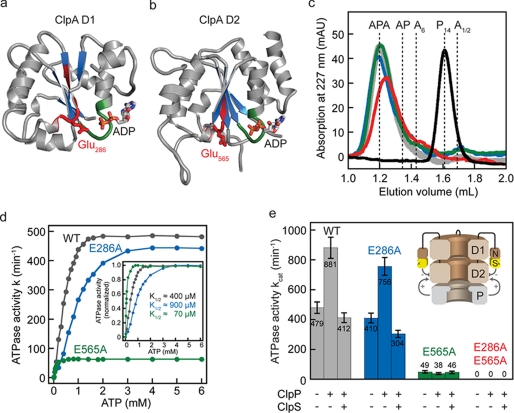
ClpA is composed of an N-terminal domain followed by two ATPase modules termed D1 and D2. Each ATPase module consists of an αβ-Rossmann fold domain, which contains the catalytic glutamate in the conserved Walker B motif (Fig. 1, a and b), and a small α-helical domain. The carboxyl moiety of the catalytic glutamate is thought to facilitate ATP hydrolysis by accepting a proton from a water molecule in the active site. We replaced the catalytic glutamate by alanine, thereby selectively abolishing ATP hydrolysis in D1 (E286A), D2 (E565A), or in both domains (E286A/E565A). Analogous Walker B protein variants were recently reported for ClpX, ClpB, and Hsp104 (8, 25, 26).
FIGURE 1.
Substitution of the catalytic glutamate abolishes ATP hydrolysis. The αβ-Rossmann fold of ClpA D1 (a) and D2 (b) consists of a central parallel β-sheet (blue) surrounded by α-helices (gray) (PDB 1KSF). The glutamate (shown as sticks) in the Walker B motif (red) activates the water molecule by abstracting one proton allowing a nucleophilic attack on the γ-phosphate of ATP. The nucleotide (ADP) is mainly bound by the Walker A motif (green). c, size exclusion chromatography confirms the correct assembly of the ClpA Walker B variants. A solution of 3 μm ClpA (ClpAwt in gray; ClpAE286A in blue; ClpAE565A in green; ClpAE286A/E565A in red) and 1.5 μm ClpP (black) was incubated for 1 h in the presence of 1 mm ATPγS to allow complete complex formation prior to gel filtration. All ClpA Walker B variants elute at a position similar to ClpAwt bound to ClpP indicating that they form the fully assembled APA complex. d, ATP saturation curves for wild type (WT) ClpA (0.1 μm, gray) and for the two ClpA Walker B variants (0.1 μm ClpAE286A, blue; 0.5 μm ClpAE565A, green). The rate constant (k in min−1) for the ATPase reaction refers to ClpA hexamer. e, ATPase activity of ClpA Walker B variants is affected by the protease ClpP and the adaptor ClpS. Catalytic amounts of ClpA (0.1 μm ClpA) were mixed with 5 mm ATP in the presence of 0.05 μm ClpP and 0.6 μm ClpS as indicated. The release of inorganic phosphate was measured in an enzyme-coupled photometric assay. The schematic representation of the ClpAP complex illustrates the opposing effects of ClpP and ClpS on the ATPase activity of ClpA.
ATP binding to the first ATPase domain of ClpA triggers the assembly into a hexameric ring structure, which is the prerequisite for activity and for binding to the protease ClpP (10, 21). Therefore, we investigated the assembly state of all ClpA variants and their ability to associate with the protease ClpP. ClpA was incubated with ClpP in the presence of ATPγS for 1 h to allow complete ClpAP complex formation prior to analytical size exclusion chromatography. All ClpA Walker B variants (E286A, E565A, and E286A/E565A) eluted at the position of wild type ClpA bound to ClpP, demonstrating binding of ClpA to both ends of the proteolytic core (Fig. 1c). This was confirmed by negative stain electron microscopy, visualizing ClpAP complexes for all ClpA Walker B variants (data not shown). Notably, the assembly process is slowed down considerably in ATPγS when Glu-286 was replaced (ClpAE286A and ClpAE286A/E565A), whereas the hexamerization time course of ClpAE565A is similar to ClpAwt. However, in the presence of ATP all ClpA variants assembled into the hexameric state with rates comparable with wild type ClpA (data not shown).
Once assembled, ClpA recruits ssrA-tagged substrate proteins through direct interaction of ClpA pore residues with the ssrA tag (27). We used fluorescein-labeled λ-repressor (λ-repressor92C-ssrA-F) to test whether ssrA-tagged substrates can be recognized by the ClpA Walker B variants used in this study (Fig. 2a). When λ-repressor92C-ssrA-F was mixed with hexameric ClpA (preassembled in ATPγS), we observed an increase in fluorescence anisotropy indicating that neither Glu-286 nor Glu-565 is crucial for binding ssrA-tagged substrates (data not shown). A recent study showed that ClpAE286A/E565A co-elutes with substrate (fluorescein isothiocyanate-casein) during gel filtration, substantiating the substrate binding competence of ClpA Walker B variants (28).
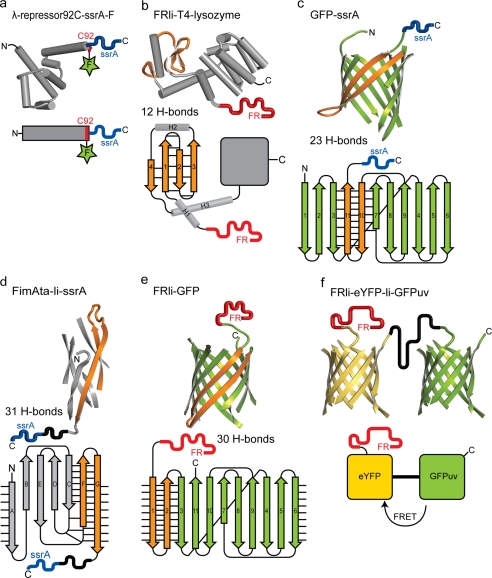
FIGURE 2.
Structure and topology of the model substrates containing the C-terminal ssrA tag (blue) or the N-terminal FRli tag (red). a, λ-repressor-ssrA (N-terminal domain of the cI repressor protein from bacteriophage λ, PDB 1LMB) was labeled with fluorescein (green star) at Cys-92 (red). The fluorescence intensity of fluorescein increases upon substrate degradation. b, T4 lysozyme (PDB 4LZM) was linked to the N-terminal degradation tag (FRli). Four antiparallel β-strands (orange) form a β-sheet stabilized by 12 hydrogen bonds. c and e, β-barrel protein GFP (PDB 1GFL) was either linked to the C-terminal ssrA tag (c) or to the N-terminal FRli tag (e). Unraveling GFP from its C terminus requires breakage of 23 hydrogen bonds (c), whereas 30 hydrogen bonds have to be resolved when mechanical unfolding is initiated from the N terminus (e). d, bacterial pilus subunit FimA (PDB 2JTY) was N-terminally truncated and C-terminally extended with the donor strand (strand G), which completes the immunoglobulin-like fold and yields FimAta. The β-strand G is embedded in a β-sheet that is stabilized by 31 hydrogen bonds. A linker (11 amino acids) followed by the ssrA tag was fused to the C terminus of FimAta. f, fusion protein eYFP-li-GFPuv (PDB 1YFP and 1B9C) was linked N-terminally to the FRli tag. The FRli-eYFP-GFPuv model substrate allows us to monitor the degradation of the eYFP barrel (λexc = 514 nm; λem = 527 nm) or the GFPuv barrel (λexc = 397 nm; λem = 507 nm) independently. FRET, fluorescence resonance energy transfer.
Activity of the Second but Not of the First ATPase Domain of ClpA Is Modulated by ClpP and ClpS
To gain a better understanding of how the two ATPase domains work together to unfold and translocate substrate proteins, we measured the ATPase activity of ClpA using an enzyme-coupled assay (23). As expected, ClpAE286A/E565A completely lost the ability to hydrolyze ATP (Fig. 1e, red). We measured the ATPase activity of ClpAwt, ClpAE286A, and ClpAE565A at increasing ATP concentrations to determine the K½ values for the ATPase reaction. All ClpA variants display saturation at 5 mm ATP. Thus, all further measurements were performed at that ATP concentration. Interestingly, D1 (K½ ≈ 70 μm) displays an ∼10-fold lower half-saturation point than D2 (K½ ≈ 900 μm) (Fig. 1d). In the wild type complex, the titration curve is dominated by D2 due to the low contribution of D1 to ATPase activity. The fact that ClpAwt displays a K½ of ∼400 μm suggests that the mutation in D1 mildly affects the ATP binding to D2.
The residual ATPase activity of ClpAE286A (kcat = 410 ± 33 min−1) and ClpAE565A (49 ± 9 min−1) adds up (96 ± 7%) to wild type activity (479 ± 38 min−1), demonstrating that ATP hydrolysis in D1 and D2 is independent of one another (Fig. 1e and Table 1). D2 hydrolyzed ATP approximately nine times faster than D1. Thus D1 only contributes 10% to the overall ATPase activity, as shown previously by Singh and Maurizi (11). Binding of ClpP to ClpA almost doubles (+84%) the ATPase activity of the chaperone (Fig. 1e, gray) as shown recently by Hinnerwisch et al. (29). ClpAE286A is stimulated to the same extent (+85%), whereas ATP hydrolysis of ClpAE565E is not accelerated when bound to ClpP (Fig. 1e, compare blue with green). Adding up the individual ATPase activities (E286A and E565E) yields 90 ± 7% of wild type ClpAP activity, which excludes a strong coupling of the two AAA modules within the ClpAP complex (Table 1). The adaptor protein ClpS that binds to the N-terminal domain of ClpA has an opposite, inhibitory effect (14). We observed a drop in ATPase activity by 53% for ClpAwt and 60% for ClpAE286A when ClpS was added to the ClpAP complex. ClpAE565A was not affected by ClpS.
TABLE 1.
ATPase activities of ClpA Walker B variants relative to wild type ClpA
| ClpA variant | ATPase activity relative to wild type ClpA in % |
|||||||
|---|---|---|---|---|---|---|---|---|
| ClpA | ClpAP | ClpAPS | ClpAP + GFP-ssrA | ClpAPS + FRli-GFP | ClpAP + FRli-GFPa | ClpAP + FimAta-ssrAb | ClpAPS + FRli-T4L | |
| E286A | 86 ± 7 | 86 ± 7 | 74 ± 6 | 79 ± 6 | 23 ± 5 | 20 ± 5 | 50 ± 6 | 83 ± 7 |
| E565A | 10 ± 2 | 4 ± 1 | 11 ± 2 | 5 ± 1 | 10 ± 2 | 7 ± 2 | 6 ± 2 | 15 ± 3 |
| E286A and E565A | 96 ± 7 | 90 ± 7 | 85 ± 6 | 84 ± 6 | 33 ± 5 | 27 ± 5 | 56 ± 6 | 98 ± 7 |
a30 μm FRli-GFP was used accounting for the low affinity in the absence of ClpS.
b His6 tag was linked to the N terminus of FimAta-li-ssrA for purification and anti-His6 immunoblot.
Our data clearly demonstrate that ClpP stimulates the ATP turnover in D2, the module that makes direct contact with the protease. Remarkably, ClpS also exerts its inhibitory effect on the second ATPase domain, although the adaptor binds to the ClpA N domain.
ATPase Activity of the First AAA Module Can Be Sufficient to Catalyze Substrate Unfolding and Translocation
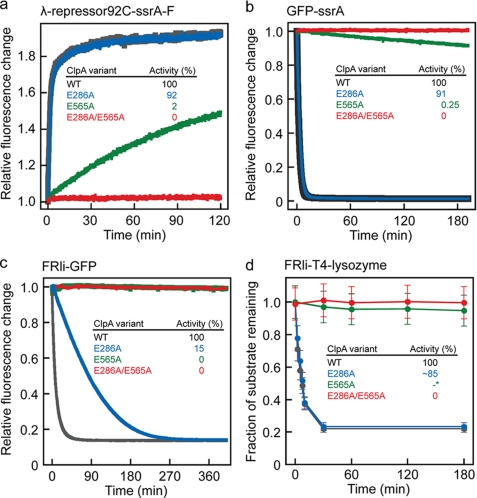
Because D1 not only binds but also hydrolyzes ATP, albeit at a slower rate than D2, we addressed the function of ATP consumption not only in D2 but also in D1. First, we used the N-terminal domain of the cI repressor protein from bacteriophage λ (λ-repressor-(1–92)) as a model substrate to probe the degradation activity of ClpAP complexes built from all three ClpA Walker B variants. λ-Repressor-(1–92) (Fig. 2a) is a small α-helical protein of low global stability but of sufficient size to require ClpA-assisted, energy-dependent unfolding prior to degradation in ClpP (30). To detect protein degradation spectroscopically, we used a cysteine variant covalently modified with fluorescein, λ-repressor92C-ssrA-F (Fig. 2a) (20). The Walker B double variant, ClpAE286A/E565A, could not promote degradation of λ-repressor92C-ssrA-F (Fig. 3a, red), whereas the ClpAE286A-ClpP complex is only mildly affected (Fig. 3a, blue). For this substrate, the second ATPase module alone can support an activity close to the wild type level. Interestingly, ClpAE565A is also capable of substrate translocation into ClpP (Fig. 3a, green) showing that D1 supports chaperone activity.
FIGURE 3.
ATPase activity of the first AAA module can be sufficient to catalyze substrate unfolding and translocation. Substrate degradation was assayed by incubating 6 μm λ-repressor92C-ssrA-F (a), 6 μm GFP-ssrA (b), 6 μm FRli-GFP (c), or 6 μm FRli-T4 lysozyme (d) with 0.5 μm ClpA, 0.25 μm ClpP, and if required 3 μm ClpS (ClpAwt is shown in gray, ClpAE286A in blue, ClpAE565A in green, and ClpA E286A/E565A in red). The insets show the relative degradation activities determined by linear fitting of the initial phase. *, note that the degradation rate for FRli-T4 lysozyme (d) cannot be determined reliably for ClpAE565A by SDS-PAGE and subsequent Coomassie staining. WT, wild type.
To challenge ClpA with a more stable protein, we used GFP C-terminally extended with the ssrA tag (19, 31) or N-terminally with the FRli tag (15, 32) (Fig. 2, c and e). Remarkably, D1 is still capable of unfolding the very stable β-barrel of GFP, as ClpAE565A catalyzes slow degradation of GFP-ssrA in the presence of ClpP (Fig. 3b, green). However, the low ATPase activity of D1 is not sufficient for unfolding the N-end rule substrate FRli-GFP in the presence of ClpP and ClpS (Fig. 3c, green). Interestingly, processing of FRli-GFP is strongly impaired when D1 is not active, although this module displays only low ATPase activity (Fig. 3c, blue). It can be assumed that GFP-ssrA and FRli-GFP exhibit similar global thermodynamic stabilities. However, it has been shown that local stabilities are more relevant for the ability of chaperone proteases to process substrates (33, 34). Hence, two active ATPase domains likely present an advantage when processing substrates displaying high local stability near the degradation tag.
Consequently, an N-end rule substrate of low local stability should be efficiently degraded by the ClpAE286A-ClpP complex. To test this, we linked the N-degron tag (FRli) to the N terminus of T4 lysozyme (FRli-T4 lysozyme). T4 lysozyme is a mainly α-helical protein containing a small N-terminal β-sheet stabilized by 12 hydrogen bonds (Fig. 2b). Indeed, analyzing FRli-T4 lysozyme degradation by SDS-PAGE revealed that ClpAE286A efficiently promotes substrate translocation into ClpP (Fig. 3d, blue). Because of the low sensitivity of the assay, it cannot be ruled out that also the ClpAE565A-ClpP complex is capable of very slow degradation of this substrate (Fig. 3d, green). Note that the FRli-T4 lysozyme was not degraded entirely because a fraction of the sample (∼20%) lost the first two amino acids (FR) during protein preparation (mass spectrometry data not shown). Our results indicate that although ATP hydrolysis in D1 is not generally required for substrate unfolding and translocation, it plays an important role when ClpA encounters a substrate of high local stability.
SsrA-tagged proteins are directionally processed from the C to N terminus (20). Thus, an ssrA-tagged substrate that is highly stabilized at its C terminus should also require two active ATPase domains to be efficiently unfolded and translocated. Therefore, we C-terminally extended FimAta, a bacterial pilus subunit variant, with a linker followed by the ssrA tag (Fig. 2d, FimAta-li-ssrA). FimA is the main subunit in E. coli type 1 pili that are formed by a mechanism known as “donor strand exchange” (35–38). Each FimA subunit has an unstructured N-terminal extension (= donor strand) that binds to and completes the fold of the adjacent FimA subunit. FimAta is an artificial FimA variant that is N-terminally truncated and C-terminally elongated by a short linker followed by the donor strand, leading to extensive stabilization of the C terminus through “self-complementation” (22) (Fig. 2d). The typical free energy of protein folding varies between −20 and −60 kJ/mol (39). Strikingly, FimG, a structurally related pilus subunit, is stabilizes by ∼70 kJ/mol when complemented with the FimF donor strand (40). Similar results were obtained for self-complemented FimA.4 Hence, FimAta is one of the most stable proteins known. Thus, maybe not too surprisingly, degradation was neither detectable in presence of the ClpA Walker B variants nor in the presence of ClpAwt. Instead, we found that ClpA autodegradation occurred within the observed time range. To exclude competition with autodegradation and to speed up the reaction, we mixed equimolar amounts of ClpAΔ9C (41) (ClpA variant without autodegradation signal) and FimAta-li-ssrA in presence of ClpP. Under these conditions, FimAta-li-ssrA is degraded slowly (t½ ≈ 25 min) in the presence of ATP (supplemental Fig. S1). When ATP is omitted, the substrate is not degraded, demonstrating that the activity is dependent on ATP hydrolysis. Although the degradation activity of the ClpAP complex cannot be assayed with this model substrate, we used FimAta-li-ssrA to probe the impact of this very stable substrate on the ATPase activity of the ClpA Walker B variants (see below).
ATP Hydrolysis in the First AAA Module Is Required for Efficient Processing of Very Stable Protein Structures
The slow processing of FRli-GFP by ClpAE286A implies that either ATP hydrolysis in D2 becomes extensively uncoupled from substrate unfolding or that FRli-GFP slows down the up and down movements of the D2 loop, responsible for substrate translocation, thereby decreasing ATP turnover in D2.
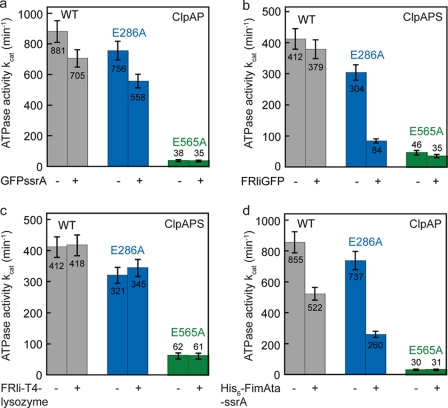
To test this, we measured the ATPase activity of ClpA in the absence and presence of various substrates (Fig. 4). The rate in ATP hydrolysis is reduced by ∼20% in ClpAwt when GFP-ssrA is being processed. We found a comparable decrease in ATPase activity for ClpAE286A (∼26%) when GFP-ssrA is present, whereas ClpAE565A is not affected, indicating that ATP turnover is decelerated mainly in the second AAA module during GFP-ssrA processing (Fig. 4a). Adding up the individual ATP hydrolysis rates (E286A and E565A) yielded 90 ± 7% (GFP-ssrA absent) and 84 ± 6% (GFP-ssrA present) of wild type ATPase activity, demonstrating that ATP hydrolysis in D1 and D2 occurs independently from each other irrespective of GFP-ssrA (Table 1).
FIGURE 4.
ATPase activity of the ClpA Walker B variants in the absence and presence of various substrates. The ATPase activity of 0.1 μm ClpA was measured in presence of 0.05 μm ClpP. 3 μm GFP-ssrA (a), 3 μm FRli-GFP (plus 0.6 μm ClpS) (b), 6 μm FRli-T4 lysozyme (plus 0.6 μm ClpS) (c), or 6 μm His6-TEV-FimAta-li-ssrA (d) was added where indicated (+). The error bars represent the standard deviation of at least three independent measurements. WT, wild type.
Next we determined the rate of ATP hydrolysis of the ClpAPS complex in the absence and presence of the N-terminally tagged model substrate FRli-GFP. Both, ClpAwt and ClpAE565A displayed similar ATPase activities (within error) irrespective of the absence or presence of FRli-GFP (Fig. 4b). Strikingly, the ATPase activity of ClpAE286A was reduced by 72% during FRli-GFP processing (Fig. 4b, blue). As shown in Table 1, ClpAE286A holds only 23% of wild type ATPase activity. Thus, the sum of the individual relative rates (E286A and E565A) yielded only 33% of wild type ATPase activity. Apparently FRli-GFP slows down ATP turnover in D2, when D1 is not active, underlining the importance of ATP hydrolysis in D1 under these conditions. GFP-ssrA degradation by the protease formed with ClpAE286A is still entirely inhibited by ClpS, ensuring that ClpS binding to ClpAE286A is not affected (data not shown).
To test whether the importance of ATP hydrolysis in D1 for unfolding of FRli-GFP is strictly ClpS-dependent, we measured the ATPase activity of ClpAE286A in the presence of FRli-GFP but without the adaptor protein. In this experiment, we employed a 10 times higher concentration of FRli-GFP (30 μm), accounting for the low affinity of the substrate to ClpA without the adaptor ClpS. Indeed, we found a decreased ATPase activity for ClpAE286A comparable with the deceleration observed in presence of ClpS, indicating that this inhibition is not caused by ClpS per se (Table 1). Furthermore, we could show that FRli-T4 lysozyme does not affect the ATPase activity of the ClpAPS complex (Fig. 4c) and that the individual ATP hydrolysis rates of the ClpA Walker B variants (E286A and E565A) add up to wild type level (Table 1).
Finally, we probed the ATPase activity of the ClpAP complex in the presence of FimAta-li-ssrA. Strikingly, when D1 is deactivated, FimAta-li-ssrA inhibits the second ATPase domain to a large extent (−65%) similar to what we observe for FRli-GFP (Fig. 4d, blue, and Table 1). Taken together, our data suggest that ATP hydrolysis in D1 is important for processing substrates displaying a high local stability near the degradation tag. Such substrates, upon interaction with the D2 loops, decrease the frequency of translocating D2 loop movements, which, in turn, reduces the rate of ATP hydrolysis.
N-end Rule Substrates Are Processed from N to C Terminus
The local protein stability near the degradation tag in the two model substrates GFP-ssrA and FRli-GFP is different. Consequently, the stability near the site of unfolding initiation is different, if we assume that unfolding is initiated at the tagged end. It was demonstrated that C-terminally tagged (ssrA tag) proteins are processed from the C to N terminus (20). However, N-end rule substrates are recruited with the help of the adaptor protein ClpS (14, 15, 18), and it is not clear whether they are translocated with the same directionality as ssrA-tagged substrates (C → N terminus) or the directionality of translocation is defined by the position of the tag (N → C terminus).
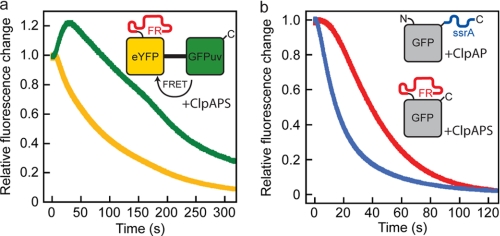
To test the directionality of N-end rule substrate processing, we designed a model substrate composed of eYFP C-terminally linked to GFPuv yielding FRli-eYFP-li-GFPuv (Fig. 2f). Degradation of the two linked fluorescent proteins can be observed independently because their spectral properties differ. Fluorescence resonance energy transfer occurs from GFPuv to eYFP. A single-turnover experiment is depicted in Fig. 5a. An excess of ClpAPS complex (preassembled in ATPγS) was rapidly mixed with FRli-eYFP-li-GFPuv in the presence of ATP. A brief lag phase of about 5 s was observed, indicative of a slower reaction step preceding the entry of the substrate into the ClpA pore (Fig. 5a). After that, we detected an instant drop in eYFP fluorescence, whereas the GFPuv fluorescence transiently increased because of the loss in fluorescence resonance energy transfer upon eYFP degradation. After a lag phase that is due to its delayed degradation, the fluorescence of GFPuv also decreases. This provides clear evidence that the N-terminal eYFP barrel is degraded first, demonstrating that the ClpS-mediated translocation of N-end rule substrates occurs directional from the N to C terminus. Consistently, eYFP-GFPuv bearing a C-terminal ssrA tag (eYFP-GFPuv-His6-ssrA) is degraded from the C to N terminus by ClpAP (supplemental Fig. S2).
FIGURE 5.
Directional processing of N-end rule substrates and handover from ClpS to ClpA. a, 2 μm ClpA, 1 μm ClpP, and 12 μm ClpS were preassembled in 1 mm ATPγS and rapidly mixed with 1.5 μm FRli-eYFP-li-GFPuv and 5 mm ATP. Degradation of the eYFP moiety was observed at 527 nm (λexc = 514 nm), and degradation of the GFPuv barrel was monitored at 507 nm (λexc = 397 nm). b, 2 μm ClpA and 1 μm ClpP were preassembled in 1 mm ATPγS and rapidly mixed with 0.4 μm GFP-ssrA and 5 mm ATP (blue). Accordingly, 2 μm ClpA, 1 μm ClpP, and 12 μm ClpS were preassembled in 1 mm ATPγS and rapidly mixed with 0.4 μm FRli-GFP and 5 mm ATP (red). Degradation of the GFP barrel was followed by monitoring the intrinsic GFP fluorescence (λexc = 397 nm and λem = 507 nm).
As the N-terminal tag is recognized by ClpS at a site remote from the ClpA unfolding pore, N-terminal processing must involve a handover step. Consequently, the initiation of translocation into ClpP is expected to be delayed for the N-end rule substrate in comparison with the ssrA substrate that binds directly to the ClpA pore. We performed a single-turnover experiment in which GFP-ssrA or FRli-GFP was rapidly mixed with ClpAP or ClpAPS, respectively (Fig. 5b). Indeed, degradation of the N-terminally tagged FRli-GFP is delayed by about 5 s, whereas GFP-ssrA degradation is initiated instantly (Fig. 5b). We observed the same kinetics when FRli-GFP was incubated with ClpS prior to mixing with ClpAP, ruling out that substrate binding to ClpS is limiting the degradation.
DISCUSSION
In this study we investigated the function of ATP turnover in the two ATPase domains of ClpA. We show that ATP hydrolysis in D1 and D2 can be selectively disabled by replacing the catalytic glutamate located in the Walker B motif (Glu-286 or Glu-565) (Fig. 1, a and b). The retained binding of nucleotide to the catalytically inactive ATP-binding sites allows correct and complete assembly of all ClpA Walker B variants as confirmed by analytical gel filtration. Thus, any observed change in ATPase activity can be ascribed to the replacement of the catalytic glutamate rather than to an assembly defect. We further found that the ability of ClpA to bind ssrA-tagged proteins is also not impaired by replacement of the catalytic glutamate.
Comparing ATPase activities of the two single Walker B variants (ClpAE286A and ClpAE565A) shows that the minor part (∼10%) of the overall ATPase activity is provided by the first AAA module as demonstrated previously by Singh and Maurizi (11). The ATP saturation curves for the two ClpA Walker B variants reveal that D1 has an ∼10-fold lower half-saturation point for ATP than D2. ClpAwt displays a K½ of ∼400 μm suggesting that the mutation in the D1 Walker B motif slightly affects ATP binding to D2. However, the individual ATPase activities of ClpAE286A and ClpA565A add up to ClpAwt level (Fig. 1e and Table 1) suggesting that ATP hydrolysis in D1 is not coupled to ATP hydrolysis in D2. The absence of allosteric interdependence of the two ATPase domains is surprising, because a strong coupling of the two ATPase domains was reported for Hsp104 (8), a related class I AAA protein. When D1 was deactivated in Hsp104 (E285Q), the ATPase activity increased approximately eight times, whereas abolishing ATP binding to D1 (K218T) led to a 45-fold decrease. Similar results were obtained for Hsp104 D2 variants proving inter-domain cooperativity.
The ATPase activity of ClpA, however, is modulated by its binding partners. For example, binding of ClpP stimulates ATP hydrolysis, whereas binding of ClpS on the other hand inhibits the overall ATPase activity (14, 29). By analyzing how the two variants are affected, i.e. whether ClpAE565A, ClpAE286A, or both variants are modulated by binding of ClpP or ClpS, we established that it is only the second AAA module that is stimulated by ClpP and inhibited by ClpS (Fig. 1e). Hence, ClpP and ClpS affect selectively the second ATPase domain of ClpA. Both the stimulatory and inhibitory effects are comparable with ClpAwt, and consequently, the individual ATPase activities of ClpAE286A and ClpA565A add up to the wild type level (Table 1). This demonstrates that neither ClpP nor ClpS causes a strict coupling of D1 and D2.
The stimulatory effect of ClpP on D2 is comprehensible, because in the hexameric ClpA ring the D2 domain makes contact with the ClpP cylinder. The inhibitory effect of ClpS cannot be explained by a stable domain interface, because ClpS has been shown to bind to the flexible N-terminal domain remote from D2 (13). The crystal structure of ClpS in complex with the ClpA N domain might offer an explanation (42, 43). Guo et al. (42) suggest two models of ClpS binding resulting in two species present, when concentrations of ClpS reach a certain threshold. At low ClpS concentrations, the model where ClpS fits into an empty space between D1 and D2 predominates, so that ClpS makes contact with a helix connecting D1 and D2 (42). In this position ClpS is in close proximity to D2 allowing ClpS to interact with the second AAA module.
It has been suggested previously that ATP hydrolysis occurs mainly in D2 providing the energy for substrate unfolding (11, 12). Because ClpAE565A retains a low ATPase activity, we tested whether D1 can actively unfold and translocate substrate proteins into the ClpP cavity when D2 has been deactivated. Using the fluorescently labeled model substrate λ-repressor92C-ssrA-F, we monitored the degradation activity of the ClpAP complex (24). Because substrate unfolding and translocation into ClpP are energy-consuming processes, the double Walker B variant (ClpAE286A/E565A) was not capable of catalyzing substrate degradation (Fig. 3a, red). Remarkably, despite the low ATPase activity, ClpAE565A could still promote substrate degradation (Fig. 3a, green), providing direct evidence for the functional integrity of the first AAA module capable of unfolding and translocation of substrate proteins (Fig. 3a).
The following mechanistic scenario is most likely. D1 converts the chemical energy of ATP into movements of D1 loops, thereby contributing to the overall chaperone activity of ClpA. Despite the low ATPase activity of D1 in comparison with D2, such movements could result in the generation and/or capture of partially unraveled C or N termini, preventing their refolding and dissociation from ClpA and facilitating their handover to the D2 loops. However, without the active motion of the D2 loops, the D1 activity supports only slow degradation, presumably because the further translocation steps into ClpP are unassisted.
To test whether the catalytic activity of D1 is substrate-dependent, we analyzed whether D1 could also process more stable substrate proteins. GFP-ssrA and FRli-GFP exhibit very similar free energies of protein folding. Although the global thermodynamic stability of a substrate protein was previously shown to play only a minor role for its susceptibility to degradation (44, 45), the local protein stability adjacent to the terminal degradation tag is important (33, 34). Both the N and C termini of GFP lead into strands that participate in the β-barrel and thus should be in a fairly stable local environment. Hence, GFP either bearing the C-terminal ssrA tag or the N-terminal FRli tag was used to test D1-supported degradation. Remarkably, D1 was also capable of unfolding the very stable β-barrel of GFP (GFP-ssrA) when unraveling occurred from the C terminus (Fig. 3b). In case of FRli-GFP, however, the first ATPase domain alone was now no longer sufficient to support even slow substrate degradation (Fig. 3c, green), indicating that the local thermodynamic stability of the substrate exceeds the energy provided by D1. Furthermore, the second AAA module alone was no longer as efficient in supporting degradation as was wild type ClpA (Fig. 3c, blue).
Because the C and N termini of GFP lead into different strands within the β-barrel structure, presumably their local thermodynamic stability also differs (Fig. 2, c and e, and supplemental Fig. S3). In agreement with this, loading the ClpAPS complex with FRli-GFP as a substrate greatly reduced the ATPase activity of ClpAE286A. This effect was not dependent on ClpS because an equivalent experiment without ClpS yielded the same result (Table 1). The individual ATPase activities of ClpAE286A and ClpAE565A no longer add up to wild type ATPase activity, indicating that D1 plays a significant role during FRli-GFP degradation (Table 1). For the C-terminally tagged GFP-ssrA, only a mild reduction in ATPase activity was observed, and the ATPase activities in the first and second module remained additive. We conclude that GFP is more resistant to unraveling from its N terminus.
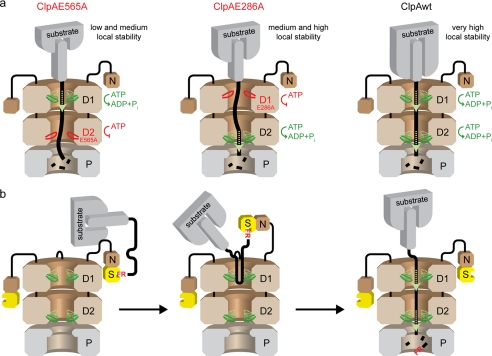
Proteins with the highest mechanical resistance are often all-β proteins, where the β-sheet hydrogen bonding is mostly responsible for opposing mechanical unfolding (46). Furthermore, there is a correlation between the observed change in energy and the number of methylene and methyl groups deleted from hydrophobic side chains in the hydrophobic core of a protein (39, 47–49). Hence, the local stability against mechanical unfolding is primarily determined by the amount of hydrogen bonds and, presumably to a lesser extent, by hydrophobic interactions. The N-terminal β-strands that are encountered first in the FRli-GFP model substrate are stabilized by a total number of 30 hydrogen bonds, and 27 methylene and methyl groups contribute to the hydrophobic core (Fig. 2e and supplemental Fig. S3). However, the C-terminal β-strands form only 23 hydrogen bonds with neighboring β-strands, and only 17 methylene and methyl groups make hydrophobic contacts, rendering GFP-ssrA less resistant against unraveling from that terminus (Fig. 2c and supplemental Fig. S3). These results suggest that ATP hydrolysis in D1 becomes important when ClpA encounters a very stable protein structure next to the terminal degradation tag. We propose a mechanistic model for ClpAP according to which the local protein stability of the substrate molecule determines whether D1, D2, or both AAA modules have to be hydrolytically active to promote efficient substrate unfolding and translocation (Fig. 6a). For those substrates requiring both ATPase domains to be active, the D1 ATPase activity is mandatory for efficient capture and handover to D2 of the tag-bearing end. This model is consistent with a previous study that states that methotrexate-stabilized E. coli dihydrofolate reductase with an N-terminal tag was not degraded by ClpAP, whereas a circular permutated variant of the same molecule (new N terminus at position 25) was degraded even in the presence of methotrexate (34). Interestingly, the first seven N-terminal residues of dihydrofolate reductase form a highly stabilized β-strand, whereas residue 25 is located at the N-terminal end of an α-helix rendering the circular permutated protein variant less resistant against mechanical unfolding (50).
FIGURE 6.
Suggested model of ClpAP/ClpAPS mechanism. a, local stability of the substrate molecule near the degradation tag determines whether D1 is required for efficient substrate processing by ClpA. The low ATPase activity of D1 is sufficient to catalyze slow translocation of substrates with a low and medium local stability (left) into ClpP. Substrates that display a somewhat higher local stability (middle) can only be processed by D2, which holds ∼90% of ClpA ATPase activity. However, both AAA modules are required for efficient processing of substrates displaying very high local protein stability near the degradation tag (right). b, N-end rule substrate degradation by ClpAPS. The N-terminal residues of the N-end rule substrate are bound by ClpS (yellow) tethered to the ClpA N domain (N). The flexible linker between N and D1 facilitates the approach of the substrate molecule toward the ClpA pore (left). The unstructured and highly flexible linker between the FR moiety and the folded substrate protein forms a loop that is bound by ClpA pore loops (middle). Eventually, the FR moiety is released from ClpS upon substrate unfolding occurring directional from the N to C terminus (right).
To exclude the possibility that the difference in the mode of recruitment (adaptor-mediated versus direct binding to the pore) could result in different requirements for contributions from the two ATP-binding sites, we designed two additional model substrates. One displays adaptor-mediated recruitment but features a lower local stability at the N terminus than FRli-GFP (Fig. 2b, FRli-T4-lysozyme), and the other is recruited by an ssrA tag directly to the pore without mediation through an adaptor and features a strongly stabilized C-terminal strand (Fig. 2d, FimAta-li-ssrA). In accordance with our proposed model (Fig. 6a), ClpAE286A processed FRli-T4 lysozyme only slightly slower than ClpAwt, and the ATP hydrolysis rate was not reduced (Fig. 3d and Fig. 4c). The C-terminally tagged variant FimAta-li-ssrA, however, induced a strong decrease in ATP turnover when the first AAA module was deactivated in ClpA (Fig. 4d, blue), indicating that D1 assists the second ATPase domain to unravel very stable protein structures. FimAta-li-ssrA indeed appears to be a very demanding substrate with respect to mechanical unfolding by the ClpAP complex. Degradation could not be observed in the presence of ClpAwt and ClpP due to ClpA autodegradation. However, when ClpAΔ9C was used, a ClpA variant lacking the autodegradation signal FimAta-li-ssrA was slowly degraded in an ATP-dependent manner (41) (supplemental Fig. S1).
We speculate that unfolding of a stable substrate by D2 alone decelerates the up and down movements of the D2 loop that was suggested to transfer the mechanical force to the substrate molecule (27, 51). Analogous loops were assigned to the same function in ClpB, HslU, and ClpX (52–55). Assuming a direct conversion of chemical into mechanical energy, slowing down D2 loop movements must result in a lower ATP turnover in D2. Hence, the decelerated ATP turnover in D2, when D1 is inactive, results directly from high local protein stability of the substrate. This is in good agreement with a previous study by Kenniston et al. (33) demonstrating that destabilizing the substrate structure near the degradation tag increases the rate of ATP hydrolysis in ClpX. However, unfolding supported by D1 alone appears to be affected also by the ClpS mode of recruitment, because neither FRli-GFP nor FRli-T4 lysozyme can be efficiently processed by D1 in presence of ClpS.
Because degradation of λ-repressor92C-ssrA-F, FRli-T4 lysozyme, and GFP-ssrA was also impaired, if only slightly, when D1 was deactivated, we conclude that the first ATPase domain generally contributes to the overall chaperone activity irrespective of ClpS binding to ClpA. It also appears that substrate processing is always assisted by ATP hydrolysis in D1 irrespective of the local substrate stability or location of the tag (Fig. 3, a, b, and d).
The considerations of local stability discussed above only apply if the degradation tag, which is recognized directly by ClpA or ClpS, is also the site of unfolding/translocation initiation. It was demonstrated that ssrA-tagged substrate proteins are translocated directionally such that the C terminus enters the ClpP cavity first (20). Thus, if the position of the tag defines the directionality of translocation, N-end rule substrates should be processed from the N to C terminus. However, the recruitment here is more complex due to the involvement of an adaptor protein, ClpS (15–18). To test the directionality in context of the adaptor protein, we designed an N-end rule substrate consisting of eYFP linked C-terminally to GFPuv (FRli-eYFP-GFPuv). Indeed, we found that the eYFP domain, which is preceded by the N-terminal FRli tag, was degraded before the GFPuv domain. This verifies the directional translocation of N-end rule substrates from N to C terminus by the ClpAPS complex. However, recruitment of N-end rule substrates via ClpS implies a handover step from ClpS to ClpA to initiate unfolding through the pore. We resolved the substrate handover step kinetically by comparing the degradation time courses of GFP-ssrA and FRli-GFP under single-turnover conditions. GFP-ssrA degradation by ClpAP commenced instantly, whereas FRli-GFP degradation by ClpAPS was delayed by ∼5 s. A straightforward handover of the very N terminus from ClpS to ClpA is unlikely, as it was previously shown that N-end rule substrates additionally require an unstructured N-terminal region to be degraded by the ClpAPS machinery (15, 16). Indeed, a hydrophobic motif within this region was recently suggested to serve as a ClpA-binding site (56). Therefore, it appears likely that ClpS-mediated substrate recruitment increases the local substrate concentration, which facilitates binding of the substrate to the ClpA pore. We suggest a mechanism that involves binding of the unstructured N-terminal region to the ClpA pore leading to substrate threading and thus eventually triggering the release of the substrate from ClpS. Hence, the 5-s delay can be assigned to a combination of events as follows: (i) the N domain-ClpS complex approaching the ClpA pore, (ii) the binding of a loop structure to the ClpA pore, and (iii) initiation of unfolding (Fig. 6b). The handover of N-end rule substrates is not strictly dependent on ATP hydrolysis in D1 as both model substrates (FRli-T4 lysozyme and FRli-GFP) can be processed by ClpA, even when D1 is not active.
In summary, the first AAA module, previously considered as only important for ClpA hexamerization, plays an important role in assisting the second AAA module to resolve very stable protein structures. ATP hydrolysis in D1 could cause loop movements that generate and/or capture the partially unraveled C or N terminus of the substrate protein. Thereby, refolding and dissociation from ClpA of the substrate is prevented and the handover to the D2 loops is facilitated.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
C. Puorger, personal communication.
- GFP
- green fluorescent protein
- PDB
- Protein Data Bank
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- eYFP
- enhanced yellow fluorescent protein
- DTT
- dithiothreitol.
REFERENCES
- 1.Hwang B. J., Woo K. M., Goldberg A. L., Chung C. H. (1988) J. Biol. Chem. 263, 8727–8734 [PubMed] [Google Scholar]
- 2.Katayama Y., Gottesman S., Pumphrey J., Rudikoff S., Clark W. P., Maurizi M. R. (1988) J. Biol. Chem. 263, 15226–15236 [PubMed] [Google Scholar]
- 3.Katayama-Fujimura Y., Gottesman S., Maurizi M. R. (1987) J. Biol. Chem. 262, 4477–4485 [PubMed] [Google Scholar]
- 4.Striebel F., Kress W., Weber-Ban E. (2009) Curr. Opin. Struct. Biol. 19, 209–217 [DOI] [PubMed] [Google Scholar]
- 5.Doyle S. M., Shorter J., Zolkiewski M., Hoskins J. R., Lindquist S., Wickner S. (2007) Nat. Struct. Mol. Biol. 14, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hattendorf D. A., Lindquist S. L. (2002) EMBO J. 21, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mogk A., Schlieker C., Strub C., Rist W., Weibezahn J., Bukau B. (2003) J. Biol. Chem. 278, 17615–17624 [DOI] [PubMed] [Google Scholar]
- 8.Schaupp A., Marcinowski M., Grimminger V., Bösl B., Walter S. (2007) J. Mol. Biol. 370, 674–686 [DOI] [PubMed] [Google Scholar]
- 9.Gottesman S., Squires C., Pichersky E., Carrington M., Hobbs M., Mattick J. S., Dalrymple B., Kuramitsu H., Shiroza T., Foster T., et al. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 3513–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurizi M. R., Singh S. K., Thompson M. W., Kessel M., Ginsburg A. (1998) Biochemistry 37, 7778–7786 [DOI] [PubMed] [Google Scholar]
- 11.Singh S. K., Maurizi M. R. (1994) J. Biol. Chem. 269, 29537–29545 [PubMed] [Google Scholar]
- 12.Seol J. H., Baek S. H., Kang M. S., Ha D. B., Chung C. H. (1995) J. Biol. Chem. 270, 8087–8092 [DOI] [PubMed] [Google Scholar]
- 13.Dougan D. A., Reid B. G., Horwich A. L., Bukau B. (2002) Mol. Cell 9, 673–683 [DOI] [PubMed] [Google Scholar]
- 14.Hou J. Y., Sauer R. T., Baker T. A. (2008) Nat. Struct. Mol. Biol. 15, 288–294 [DOI] [PubMed] [Google Scholar]
- 15.Erbse A., Schmidt R., Bornemann T., Schneider-Mergener J., Mogk A., Zahn R., Dougan D. A., Bukau B. (2006) Nature 439, 753–756 [DOI] [PubMed] [Google Scholar]
- 16.Wang K. H., Oakes E. S., Sauer R. T., Baker T. A. (2008) J. Biol. Chem. 283, 24600–24607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K. H., Roman-Hernandez G., Grant R. A., Sauer R. T., Baker T. A. (2008) Mol. Cell 32, 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K. H., Sauer R. T., Baker T. A. (2007) Genes Dev. 21, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber-Ban E. U., Reid B. G., Miranker A. D., Horwich A. L. (1999) Nature 401, 90–93 [DOI] [PubMed] [Google Scholar]
- 20.Reid B. G., Fenton W. A., Horwich A. L., Weber-Ban E. U. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3768–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kress W., Mutschler H., Weber-Ban E. (2007) Biochemistry 46, 6183–6193 [DOI] [PubMed] [Google Scholar]
- 22.Vetsch M., Erilov D., Molière N., Nishiyama M., Ignatov O., Glockshuber R. (2006) EMBO Rep. 7, 734–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rieger C. E., Lee J., Turnbull J. L. (1997) Anal. Biochem. 246, 86–95 [DOI] [PubMed] [Google Scholar]
- 24.Twining S. S. (1984) Anal. Biochem. 143, 30–34 [DOI] [PubMed] [Google Scholar]
- 25.Hersch G. L., Burton R. E., Bolon D. N., Baker T. A., Sauer R. T. (2005) Cell 121, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 26.Werbeck N. D., Schlee S., Reinstein J. (2008) J. Mol. Biol. 378, 178–190 [DOI] [PubMed] [Google Scholar]
- 27.Hinnerwisch J., Fenton W. A., Furtak K. J., Farr G. W., Horwich A. L. (2005) Cell 121, 1029–1041 [DOI] [PubMed] [Google Scholar]
- 28.Erbse A. H., Wagner J. N., Truscott K. N., Spall S. K., Kirstein J., Zeth K., Turgay K., Mogk A., Bukau B., Dougan D. A. (2008) FEBS J. 275, 1400–1410 [DOI] [PubMed] [Google Scholar]
- 29.Hinnerwisch J., Reid B. G., Fenton W. A., Horwich A. L. (2005) J. Biol. Chem. 280, 40838–40844 [DOI] [PubMed] [Google Scholar]
- 30.Huang G. S., Oas T. G. (1995) Biochemistry 34, 3884–3892 [DOI] [PubMed] [Google Scholar]
- 31.Gottesman S., Roche E., Zhou Y., Sauer R. T. (1998) Genes Dev. 12, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varshavsky A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenniston J. A., Baker T. A., Fernandez J. M., Sauer R. T. (2003) Cell 114, 511–520 [DOI] [PubMed] [Google Scholar]
- 34.Lee C., Schwartz M. P., Prakash S., Iwakura M., Matouschek A. (2001) Mol. Cell 7, 627–637 [DOI] [PubMed] [Google Scholar]
- 35.Choudhury D., Thompson A., Stojanoff V., Langermann S., Pinkner J., Hultgren S. J., Knight S. D. (1999) Science 285, 1061–1066 [DOI] [PubMed] [Google Scholar]
- 36.Sauer F. G., Fütterer K., Pinkner J. S., Dodson K. W., Hultgren S. J., Waksman G. (1999) Science 285, 1058–1061 [DOI] [PubMed] [Google Scholar]
- 37.Sauer F. G., Pinkner J. S., Waksman G., Hultgren S. J. (2002) Cell 111, 543–551 [DOI] [PubMed] [Google Scholar]
- 38.Zavialov A. V., Berglund J., Pudney A. F., Fooks L. J., Ibrahim T. M., MacIntyre S., Knight S. D. (2003) Cell 113, 587–596 [DOI] [PubMed] [Google Scholar]
- 39.Fersht A. (2000) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (Julet M. R., Hadler G. L. eds) 3rd Ed., pp. 508–509 and 532–534, W. H. Freeman & Co., New York [Google Scholar]
- 40.Puorger C., Eidam O., Capitani G., Erilov D., Grütter M. G., Glockshuber R. (2008) Structure 16, 631–642 [DOI] [PubMed] [Google Scholar]
- 41.Maglica Z., Striebel F., Weber-Ban E. (2008) J. Mol. Biol. 384, 503–511 [DOI] [PubMed] [Google Scholar]
- 42.Guo F., Esser L., Singh S. K., Maurizi M. R., Xia D. (2002) J. Biol. Chem. 277, 46753–46762 [DOI] [PubMed] [Google Scholar]
- 43.Zeth K., Ravelli R. B., Paal K., Cusack S., Bukau B., Dougan D. A. (2002) Nat. Struct. Biol. 9, 906–911 [DOI] [PubMed] [Google Scholar]
- 44.Burton R. E., Siddiqui S. M., Kim Y. I., Baker T. A., Sauer R. T. (2001) EMBO J. 20, 3092–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenniston J. A., Burton R. E., Siddiqui S. M., Baker T. A., Sauer R. T. (2004) J. Struct. Biol. 146, 130–140 [DOI] [PubMed] [Google Scholar]
- 46.Best R. B., Brockwell D. J., Toca-Herrera J. L., Blake A. W., Smith D. A., Radford S. E., Clarke J. (2003) Anal. Chim. Acta 479, 87–105 [Google Scholar]
- 47.Otzen D. E., Rheinnecker M., Fersht A. R. (1995) Biochemistry 34, 13051–13058 [DOI] [PubMed] [Google Scholar]
- 48.Serrano L., Kellis J. T., Jr., Cann P., Matouschek A., Fersht A. R. (1992) J. Mol. Biol. 224, 783–804 [DOI] [PubMed] [Google Scholar]
- 49.Xu J., Baase W. A., Baldwin E., Matthews B. W. (1998) Protein Sci. 7, 158–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolin J. T., Filman D. J., Matthews D. A., Hamlin R. C., Kraut J. (1982) J. Biol. Chem. 257, 13650–13662 [PubMed] [Google Scholar]
- 51.Bohon J., Jennings L. D., Phillips C. M., Licht S., Chance M. R. (2008) Structure 16, 1157–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin A., Baker T. A., Sauer R. T. (2008) Nat. Struct. Mol. Biol. 15, 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park E., Rho Y. M., Koh O. J., Ahn S. W., Seong I. S., Song J. J., Bang O., Seol J. H., Wang J., Eom S. H., Chung C. H. (2005) J. Biol. Chem. 280, 22892–22898 [DOI] [PubMed] [Google Scholar]
- 54.Siddiqui S. M., Sauer R. T., Baker T. A. (2004) Genes Dev. 18, 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E. U., Dougan D. A., Tsai F. T., Mogk A., Bukau B. (2004) Cell 119, 653–665 [DOI] [PubMed] [Google Scholar]
- 56.Ninnis R. L., Spall S. K., Talbo G. H., Truscott K. N., Dougan D. A. (2009) EMBO J. 28, 1732–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.