Abstract
Human T-cell leukemia virus I (HTLV-I) is a deltaretrovirus that is the causative agent of adult T-cell leukemia and the neurological disorder HTLV-I-associated myelopathy/tropical spastic paraparesis. Currently, no effective antiretroviral treatment options are available to restrict the development of diseases associated with the virus. In this work, we investigated the activity of pokeweed antiviral protein (PAP) on HTLV-I, when expressed from a proviral clone in 293T cells or in an HTLV-I immortalized cell line. PAP is a plant-derived N-glycosidase that exhibits antiviral activity against a number of viruses; however, its mode of action has not been clearly defined. Here, we describe the mechanism by which PAP inhibited production of HTLV-I. We show that PAP depurinated nucleotides within the gag open reading frame and suppressed the synthesis of viral proteins in part by decreasing the translational efficiency of HTLV-I gag/pol mRNA. Observed reduction in levels of viral mRNAs were not due to enhanced degradation; rather, decreased amounts of viral transactivator protein, Tax, led to feed-back inhibition of transcription from the viral promoter. Therefore, PAP efficiently suppressed HTLV-I gene expression at both translational and transcriptional levels, resulting in substantially diminished virus production. Significantly, no changes in viability or rates of cellular transcription or translation were observed in cells expressing PAP, indicating that this protein was not toxic. Antiviral activity, together with the absence of cytotoxicity, supports further investigation of this enzyme as a novel therapeutic agent against the progression of HTLV-I infection.
Introduction
Human T-cell leukemia virus I (HTLV-I)2 is a human deltaretrovirus that causes adult T-cell leukemia/lymphoma (1) and tropical spastic paraparesis, also called HTLV-I-associated myelopathy. The latter is a chronic and progressive disease of the nervous system, characterized by muscle weakness and sensory disturbance (2, 3). HTLV-I-infected individuals have a 2–3% estimated lifetime risk of developing adult T-cell leukemia, with a period of latency from 20 to 30 years (4, 5). The majority of those infected with HTLV-I are therefore asymptomatic (6), because the virus maintains the expression of its genes at very low or undetectable levels. As a result, HTLV-I is not efficiently targeted by the immune system (7), and leukemia progresses after clonal expansion of T-cells infected with the virus (8, 9). Due to the long period of latency prior to onset of leukemia, the disease appears mainly in individuals who have been infected with HTLV-I early in life (10, 11). The virus is transmitted through body fluids, including breast milk (12, 13); therefore, mother-to-child transmission poses substantial risk for development of leukemia. Various forms of cytotoxic chemotherapy are currently used to treat adult T-cell leukemia; however, prognosis is poor, because the disease is aggressive, with a mean survival time of only 6 months. Few published accounts address the potential of antiretroviral therapy for limiting viral gene expression and/or inhibiting replication to reduce the viral load in individuals early after infection, which in turn would reduce the chances for progression of leukemia (14, 15).
In this report, we investigated the antiviral activity of a plant-derived protein against HTLV-I. Pokeweed antiviral protein (PAP) is a ribosome-inactivating protein synthesized by Phytolacca americana, the pokeweed plant (16). Like all ribosome-inactivating proteins, PAP is a glycosidase that removes a specific adenine base from the large ribosomal RNA (17). This depurination has traditionally been cited as the reason for antiviral activity, because resulting inhibition of host cell translation would limit virus proliferation (18). However, mutants of the protein have been expressed that maintain antiviral activity in the absence of rRNA depurination, thereby separating the effect on viruses from host cell translation (19, 20). Pokeweed antiviral protein has been shown to depurinate some viral RNAs, indicating that antiviral activity may be due to broader substrate specificity that is not limited to rRNA depurination (21). Incubation of purified PAP with lymphocytes prior to their infection with HIV-I reduced the number of virus particles released into the medium (22). In addition, incubation of HIV-I genomic RNA with purified PAP resulted in release of purines, suggesting that PAP depurinated the RNA in this assay (23); however, the effect of PAP on retroviral gene expression in cells has not been described. Accordingly, our goal was to examine whether PAP is able to target and inhibit HTLV-I and to determine the mechanism of such activity.
We show here that PAP efficiently inhibited HTLV-I gene expression at both translational and transcriptional levels in the absence of host cell toxicity. PAP depurinated nucleotides within the gag open reading frame of genomic RNA and reduced its translational efficiency in vitro. Decreased amounts of the viral transactivator protein, Tax, led to feed-back inhibition of HTLV-I gene expression, diminishing gag/pol, env, and tax/rex mRNA levels. Due to this combined effect, PAP significantly reduced virus production.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Human embryonic kidney 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. Jurkat cells and HTLV-I-infected human T-cell line (MT-2), obtained from the AIDS Research and Reference Reagent Program, were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics. All cells were grown in a humidified incubator with 5% CO2 at 37 °C. Purified PAP was isolated by ion exchange chromatography from pokeweed leaves (16, 24) and tested for contaminating nuclease activity as described previously (19).
Plasmids and Transfections
The pACH (wild-type) and pACH-EN (Envelope-null) proviral clones of HTLV-I have been described elsewhere (25, 26). The plasmids pcF-PAP and pcF-PAPx were generated by PCR amplification of the coding region for the mature form of wild-type PAP and its active site mutant, PAPx (E176V) (27), using the primers 3×FLAG-mPAPf (5′-GGGGGAAGCTTGTGAATACAATCATCTACAATG-3′) and 3×FLAG-mPAPr (5′-GGGGGGGATCCTCAAGTTGTCTGACAGCTCCCACCAAC-3′) and pcPAP and pcPAPx plasmids as templates (28). The PCR products were digested with BamHI and HindIII and cloned into p3×FLAG-CMV 7.1 vector (Invitrogen). The HTLV-I reporter construct, pGL3-LTR-Luc, containing the HTLV-I long terminal repeat upstream of the luciferase gene, was generously provided by Dr. K. Shimotohno (Kyoto University, Japan).
Transfections were performed as described previously (28). Briefly, 293T cells were seeded at a density of 2 × 105 cells/10-cm plate 24 h prior to transfection. Cells were refed 3 h prior to transfection, following which a total of 25 μg of plasmid DNA was transfected into cells by calcium phosphate co-precipitation. The medium was changed 16 h following transfection, and cells were harvested 24 h later. Jurkat and MT-2 cells were transfected using Lipofectamine 2000 (Invitrogen), following the manufacturer's instructions. Briefly, cells were refed with fresh RPMI 1640 supplemented with 10% fetal bovine serum without antibiotics and plated at a density of 5 × 105 cells/well in 6-well plates prior to transfection. Plasmid DNA and Lipofectamine 2000 reagent were separately mixed with RPMI 1640 without serum or antibiotics, incubated at room temperature for 5 min, and then combined and incubated for an additional 25 min. The mixture was added dropwise to the cells, and the cells were harvested 24 h later. 293T, Jurkat, and MT-2 cells were also co-transfected with pCMV-β-Gal plasmid (1 μg), and transfection efficiency was estimated by staining cells for the presence of β-galactosidase. Briefly, cells were fixed in a solution of 2% glutaraldehyde and 1% formamide for 5 min at room temperature. After fixation, cells were washed in phosphate-buffered saline and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) for 2 h at 37 °C. Cells were viewed at 4× magnification with light microscopy, and the proportion of blue cells to the total number of cells in a field of view was calculated. To test for potential PAP effect on a transiently transfected gene, 293T cells were co-transfected with pcGFP (1 μg), encoding the open reading frame for green fluorescent protein, and increasing amounts of pcF-PAP.
Protein Analyses
p19 matrix levels in the supernatant of cells and Gag protein levels in cell lysates were determined using the RETRO-TEK HTLV-I p19 antigen ELISA kit (Zeptometrix) exactly according to the manufacturer's instructions. Immunoblot analysis of cellular proteins was performed as described previously (28). PAP was detected with an anti-FLAG monoclonal antibody (1:2,500; Sigma), and HTLV-I Tax protein was detected with a polyclonal Tax antibody (1:5,000; AIDS Research and Reference Reagent Program) from 60 μg of cell lysate/sample. The immunoblots were reprobed with a monoclonal antibody specific for β-actin (1:5,000; Sigma). The amount of GFP expressed in cells co-transfected with pcGFP and increasing amounts of pcF-PAP was estimated by probing immunoblots with a polyclonal antibody specific for GFP (1:5000; Cell Signaling).
To determine the direct effect of PAP on viral RNA translation, transcript of the gag open reading frame was incubated with purified PAP and translated in a cell-free system. A portion of the HTLV-I proviral clone corresponding to the gag open reading frame (nucleotides 804–2099 in pACH) was subcloned into HindIII and BamHI sites of pcDNA3. Run-off transcripts were generated from the linearized plasmid using T7 RNA polymerase. Following in vitro transcription, RNA (2 μg) was incubated with purified PAP (25 and 50 ng) in 1× ribosome-inactivating protein buffer (60 mm KCl, 10 mm Tris-HCl, pH 7.4, 10 mm MgCl2) for 30 min at room temperature, extracted with phenol/chloroform/isoamyl alcohol, and ethanol-precipitated. Negative control samples were treated with buffer alone. The treated RNA samples were supplied as template to a rabbit reticulocyte in vitro translation system (Promega; 50-μl reactions incubated at 30 °C for 1.5 h), and the amount of synthesized p19 matrix protein was measured by ELISA.
To measure the amount of luciferase gene expression from cells co-transfected with pGL3-LTR-Luc plus pACH-EN and either pcF-PAP, pcF-PAPx, or pcDNA3, luciferase activity was measured 24 h after transfection using a luciferase assay system (Promega) exactly according to the manufacturer's instructions.
Toxicity Assay and Metabolic Labeling
The effect of PAP on cell viability was analyzed using an MTT conversion assay (Roche Applied Science) as recommended by the manufacturer. Briefly, 293T cells were transfected in 10-cm plates using Lipofectamine. Following transfection (40 h), cells were resuspended, and 103 cells were seeded into a 96-well plate in replicates of five. MTT reagent (20 μl) was added to each well, and the plate was placed in a humidified incubator at 37 °C for 4 h. Solubilizing buffer (100 μl) was then added to each well, and the plate was incubated overnight, followed by measurement of absorbance at A595 nm. To test for the potential toxic effect of PAP beyond 40 h post-transfection, cells were transfected with 2 μg of pcF-PAP and analyzed by the MTT assay over a 4-day period.
The effect of PAP on cellular translation and transcription was analyzed 40 h following transfection of 293T cells as described elsewhere (29) with modifications. For analysis of cellular translation, medium was removed from the cells and replaced with an equal volume of labeling medium (Dulbecco's modified Eagle's medium lacking cysteine and methionine, supplemented with 10% fetal bovine serum, 0.2 mm cysteine, and 4 mm l-glutamine). Thirty minutes later, the cells were labeled by replacing the medium with fresh labeling medium, supplemented with 30 μCi/ml [35S]methionine, and harvested at the indicated time points. Cycloheximide was added to one set of cells (25 μg/ml) at time 0 to serve as experimental control for inhibition of total cellular translation. The cells were then lysed in lysis buffer (150 mm NaCl, 10 mm Hepes KOH pH 7.4, 1% Triton X-100, 0.1% SDS, and protease inhibitor mixture (Roche Applied Science), and total protein was precipitated with 25% trichloroacetic acid containing bovine serum albumin (0.1 mg/ml) and filtered through glass fiber filters. Filters were washed four times with 5% trichloroacetic acid. Retained radioactively labeled protein was quantified by scintillation counting of the filters. Analysis of cellular transcription was performed by metabolic labeling of cells with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 30 μCi/ml [3H]uridine for the indicated time periods. One set of cells was treated with actinomycin D (5 μg/ml) at time 0 and served as experimental control for inhibition of total cellular transcription. Total cellular RNA was isolated and filtered through glass fiber filters. The filters were washed four times with 5% trichloroacetic acid containing 20 mm sodium pyrophosphate and once with 100% ethanol, and then dried and retained radiolabel was quantified by scintillation counting.
RNA Analyses
The degree of 28 S ribosomal RNA depurination by PAP was determined by primer extension analysis as described previously (28). Briefly, total cellular RNA was extracted from cells using TriReagent (Bioshop), following the manufacturer's instructions. RNA (3 μg) was hybridized to 5 × 105 cpm of each end-labeled “depurination” and “28 S rRNA control” primer and extended by reverse transcription. The depurination primer (5′-AGTCATAATCCCACAGATGGT-3′) annealed 65 nucleotides downstream of nucleotide A4324, whereas the 28 S rRNA control primer (5′-TTCACTCGCCGTTACTGAGG-3′) annealed 100 nucleotides downstream of the 28 S rRNA 5′-end. Extension of these primers resulted in 65-nucleotide (depurination) and 100-nucleotide (28 S rRNA) cDNA products. To confirm the location of 28 S rRNA depurination, a dideoxynucleotide sequencing ladder of the 28 S rDNA was generated using the depurination primer. The amount of depurinated rRNA in each sample was estimated by densitometry of band intensities of the A4324 cDNA products relative to the 28 S ribosomal cDNA products.
To determine whether PAP targeted HTLV-I genomic RNA for depurination, 293T cells were co-transfected with pACH-EN (5 μg) and pcF-PAP (0, 0.25, or 0.5 μg), and total RNA was isolated 40 h after transfection. In addition, total RNA (10 μg) isolated from cells transfected only with pACH-EN (5 μg) was incubated with purified PAP (10, 25, or 50 ng) in 1× ribosome-inactivating protein buffer for 30 min at room temperature, extracted with phenol/chloroform/isoamyl alcohol, and ethanol-precipitated. Negative control samples were treated with buffer alone. The treated RNA samples and total RNA from cells expressing PAP were tested for depurination by primer extension using a primer specific to the 5′ portion of the gag open reading frame of genomic viral RNA, HTLV- I (R1125) (5′-GATTGTTGGCTTGGACACGGAGG-3′), which annealed 705 nucleotides downstream of the gag initiation codon. Briefly, 2.5 × 105 cpm of end-labeled primer was added to 3 μg of RNA, denatured at 95 °C for 3 min, and then chilled on ice. Samples were brought to 10 μl in 1× buffer (75 mm KCl, 50 mm Tris-HCl, pH 8.3, 10 mm dithiothreitol, 3 mm MgCl2) and incubated at room temperature for 20 min. Following this annealing step, 10 μl of extension buffer (1 mm dNTPs, 20 units of RNase inhibitor, 100 units of Superscript II in 1× buffer) was added to each sample and incubated at 48 °C for 90 min. The reaction was terminated by the addition of 12.5 μl of formamide buffer. The extension products were resolved on a 7 m urea, 6% acrylamide gel and detected and quantified using a PhosphorImager. To identify the depurinated nucleotides, dideoxynucleotide sequencing of pACH-EN was performed with the same reverse primer used for the primer extension assay. To test whether PAP targeted a cellular mRNA for depurination, 293T cells were transfected with 2 μg of pcF-PAP or pcDNA3, and total mRNA was isolated from cells 40 h following transfection. Primers specific to GAPDH mRNA, positioned to anneal approximately every 100 nucleotides along the length of this mRNA, were incubated with 10 μg of total mRNA and extended by reverse transcriptase as described above. The cDNA products were resolved by electrophoresis as described above, and primer extension patterns were examined for the presence of depurination. Dideoxynucleotide sequencing of GAPDH cDNA (accession number M33197) was performed with the same reverse primers used for the primer extension assay.
The levels of HTLV-I mRNAs in 293T cells transfected with increasing amounts of pcF-PAP plasmid were measured by an RNase protection assay from samples of total cellular RNA 40 h following transfection. The stability of HTLV-I mRNAs was also analyzed by adding actinomycin D (5 μg/ml) to cells 40 h following transfection, harvesting them at the indicated time points, and measuring viral mRNA levels with the same assay. An antisense probe was synthesized by PCR amplification of nucleotides 4900–5300 of the pACH clone using ACH4893F (5′-AACCAACTGCCACAAAACCCGATG-3′) forward and ACH5285R2 (5′-TAATACGACTCAC TATAGGCAGGGTTTAGAGTGGTATGAGGAG-3′) reverse primers. The reverse primer contained the T7 RNA polymerase promoter sequence, which was used to transcribe the 400-nucleotide negative strand RNA probe in the presence of 50 μCi of [α-33P]CTP. Total RNA from each sample (25 μg) was dissolved in 21 μl of hybridization buffer (0.4 m NaCl, 40 mm PIPES, pH 6.4, 1 mm EDTA, 80% formamide) containing 2 × 105 cpm of antisense probe. The mixture was denatured by incubation at 70 °C for 20 min, followed by hybridization at 50 °C overnight. RNase digestion mixture (200 μl; 300 mm NaCl, 10 mm EDTA, 10 mm Tris-HCl, pH 7.5, 0.72 unit/μl RNase T1, and 0.0036 unit/μl RNase A) was added to each sample and incubated at room temperature for 20 min, followed by treatment with 17 μl of (1:4) proteinase K (10 mg/ml), SDS (10%) solution. Samples were incubated for 30 min at 37 °C, phenol/chloroform-extracted, and precipitated in ethanol using 20 μg of tRNA as carrier. Pellets were dissolved in 15 μl of formamide loading buffer and separated on a 7 m urea, 6% acrylamide gel. The different sizes of protected antisense probe were visualized and quantified with a PhosphorImager. A probe was also generated for detection of GAPDH mRNA by PCR amplification of GAPDH cDNA using pTRI-GAPDH-Human (Ambion) as template and pcGAPDH-T7f (5′-TAATACGACTCACTATAGGCTCGAGATGATGTTCTGGGCAGCC-3′) and pcGAPDHr (5′-GCGCGGGCCCAGTCCATGCCATCACTGCCACCCAGAAG-3′) primers. This template was used to transcribe a labeled negative strand probe of 80 nucleotides using T7 RNA polymerase in the presence of 50 μCi of [α-33P]CTP. GAPDH mRNA was also detected by Northern blot. Total cellular RNA (10 μg), the same samples used for RNase protection, was separated through 1% agarose, transferred to nylon membrane, and probed for GAPDH mRNA with the same negative strand transcript used for RNase protection.
Immunocytochemistry
To determine the cellular localization of PAP, 293T cells were transfected with 2 μg of pcF-PAP or pcDNA3 and grown on glass coverslips for 40 h following transfection. Cells were then fixed in 4% paraformaldehyde and permeabilized in 0.1% Triton X-100 for 5 min at room temperature. Cells were blocked in 10% normal goat serum in phosphate-buffered saline for 1 h at room temperature. F-PAP expression was detected by incubating cells with a FLAG-specific monoclonal antibody (1:200; Sigma) overnight at 4 °C and a fluorescein isothiocyanate-conjugated secondary antibody (1:500; Jackson Immunochemicals) for 45 min at room temperature. Cells were also stained with propidium iodide (300 nm in phosphate-buffered saline) for 5 min at room temperature to visualize nuclei. Cells were then mounted onto microscope slides with gold anti-fade reagent (Invitrogen) and examined at ×60 magnification with a confocal microscope (Olympus Fluoview) for fluorescein isothiocyanate (488 nm) and propidium iodide (543 nm) excitation signals.
RESULTS
PAP Reduces the Levels of HTLV-I Proteins
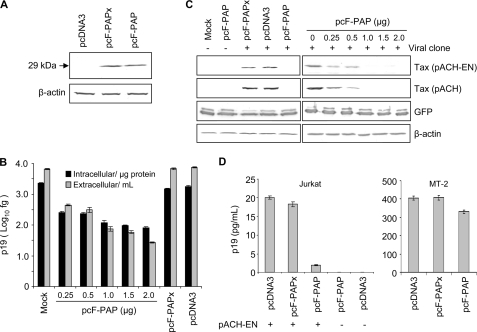
To test the antiviral activity of PAP against HTLV-I, 293T cells were co-transfected with HTLV-I-infectious (pACH) or non-infectious (pACH-EN) proviral clones with increasing amounts of pcF-PAP plasmid encoding FLAG-tagged PAP. The pACH-EN clone is an Envelope-null mutant of pACH that contains a premature stop codon near the beginning of the env open reading frame, rendering the virus particles produced from transfected cells non-infectious (25). As a negative control for PAP activity, we co-transfected cells with pcF-PAPx plasmid encoding FLAG-tagged PAPx, the active site mutant of PAP (E176V), which is unable to depurinate RNA molecules (27). Cells were also transfected with the empty vector, pcDNA3, as a control for potential promoter effect. Immunoblot analysis indicated that both F-PAP and F-PAPx were expressed at their mature size of 29 kDa and to similar levels in 293T cells (Fig. 1A). The effect of PAP on HTLV-I viral particle production was analyzed by measuring the amount of p19 matrix protein released from cells co-transfected with pACH-EN and pcF-PAP, pcF-PAPx, or pcDNA3 plasmids. Transfection of increasing amounts of pcF-PAP plasmid caused a dose-dependent decline in viral production up to 100-fold upon transfection with 2 μg of this plasmid, relative to transfection with pcDNA3 vector control (Fig. 1B). The amount of Gag protein in cell lysates was also quantified by p19 ELISA for the same cell treatments and indicated a substantial decrease in the amount of this protein in cells expressing PAP. This decline agreed with immunoblot analysis of the levels of intracellular Tax protein (Fig. 1C). As expected, expression of F-PAPx did not markedly lower the level of these viral proteins. Levels of β-actin were constant with increasing PAP concentration, indicating that PAP did not decrease the amount of this endogenous protein in cells. To test whether production of a transiently expressed protein would be altered by PAP, a construct encoding GFP was co-transfected with increasing amounts of pcF-PAP. The level of GFP did not substantially decline with increasing PAP concentration, suggesting that PAP did not alter the level of this protein. However, PAP significantly decreased the levels of HTLV-I p19 and Tax proteins in 293T cells.
FIGURE 1.
PAP decreases HTLV-I protein levels. 293T cells were co-transfected with pACH-EN or pACH (5 μg; plus and minus signs) and pcF-PAP (0.25, 0.5, 1, 1.5, or 2 μg), pcF-PAPx (2 μg), pcDNA3 (2 μg), or pcGFP (1 μg). A, cell lysates (60 μg) were separated through 12% SDS-PAGE, transferred to nitrocellulose, and probed for F-PAP and F-PAPx using a FLAG-specific monoclonal antibody (1:2,500). The molecular mass of F-PAP and F-PAPx is indicated (29 kDa). Immunoblots were also probed with a monoclonal antibody against β-actin (1:5,000). B, measurement of p19 levels in cell medium and Gag protein levels in cell lysates by p19 ELISA 40 h following transfection. The bars represent means ± S.E. for three independent experiments. C, immunoblot analysis of cell lysates (60 μg) using anti-Tax polyclonal antibody (1:5,000), GFP-specific polyclonal antibody (1:5,000), and β-actin-specific monoclonal antibody (1:5,000). GFP and β-actin were probed from lysates of cells transfected with pACH-EN. D, Jurkat cells were transfected with 5 μg of PACH-EN and 2 μg of pcF-PAP, pcF-PAPx, or pcDNA3. As controls, Jurkat cells were also transfected with either 2 μg of pcF-PAP or pcDNA3 in the absence of pACH-EN. MT-2 cells were transfected with 2 μg of pcF-PAP, pcF-PAPx, or pcDNA3. p19 ELISA was performed on cell medium 24 h following transfection. Bars, means ± S.E. for three independent experiments.
The effect of PAP on p19 levels was also examined in medium from Jurkat cells transfected with the Envelope-null proviral clone (pACH-EN), because these cells more closely approximate the T-lymphocytes normally infected by HTLV-I. Production of virus from Jurkat cells expressing PAP was substantially reduced, ∼10-fold, compared with cells expressing the active site mutant PAPx (Fig. 1D). To confirm the effect of PAP on HTLV-I protein levels in their natural context, pcF-PAP, pcF-PAPx, or pcDNA3 plasmids were transfected into MT-2 cells, T-cells chronically infected with HTLV-I. Consistent with our results from 293T and Jurkat cells, PAP reduced the amount of virus produced from MT-2 cells (Fig. 1D). The decline in viral production in MT-2 cells was ∼25%, which may reflect the lower efficiency of lymphocyte transfection compared with 293T cells. Typically, the transfection efficiency of MT-2 cells was ∼6–7% of that achieved for 293T cells, based on the percentage of cells stained positive for β-galactosidase. This, in combination with the fact that MT-2 cells are chronically infected with HTLV-I, as opposed to being transfected with the proviral clone together with pcF-PAP, may have contributed to the lower effect of PAP relative to that seen in 293T and Jurkat cells.
PAP Is Not Toxic to Cells
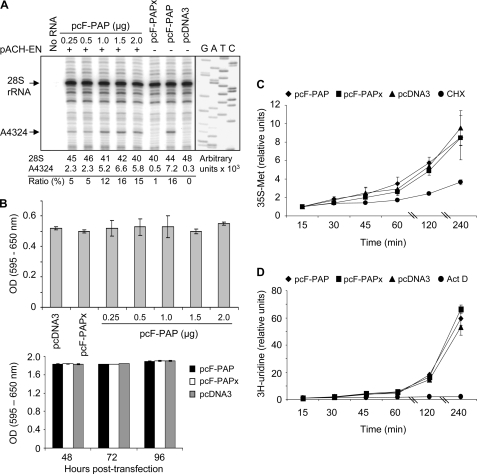
Since PAP is a ribosome-inactivating protein, we examined the extent to which it depurinated the 28 S rRNA to determine if reduction in HTLV-I proteins was primarily related to PAP effect on the translation machinery. Total cellular RNA was isolated from cells transiently transfected with pcF-PAPx, pcDNA3, or increasing amounts of pcF-PAP plasmids, followed by primer extension analysis of the 28 S rRNA. Sequencing of 28 S cDNA with the same primer was used to provide a sequence ladder that allowed for identification of depurination at the expected nucleotide A4324. PAP depurinated ∼16% of the total cellular 28 S rRNA, based on densitometry of band intensities of A4324 cDNA products relative to 28 S ribosomal cDNAs for each sample (Fig. 2A). These observations suggest that the substantial reduction in viral proteins was not due entirely to depurination of rRNA and subsequent general inhibition of translation. However, given depurination of the 28 S rRNA in 293T cells and the resultant potential for toxicity, we tested for the effect of PAP on cellular viability with an MTT conversion assay. Transient transfection of pcF-PAP plasmid (up to 2 μg) did not cause toxicity in 293T cells when measured 2 days following transfection (Fig. 2B). To test further the possibility that 2 days would not be sufficient to observe the effects of rRNA depurination, we extended the assay and performed an MTT time course for 4 days (Fig. 2B). The viability of cells expressing PAP did not decline relative to cells expressing PAPx or vector control, suggesting that the amount of PAP required to suppress viral protein levels and to depurinate the rRNA by 16% did not affect the viability of cells.
FIGURE 2.
Depurination of rRNA by PAP does not cause toxicity or inhibition of cellular transcription or translation. 293T cells were co-transfected with pACH-EN (5 μg; plus and minus signs) and pcF-PAP (0.25, 0.5, 1, 1.5, or 2 μg), pcF-PAPx (2 μg), or pcDNA3 (2 μg). A, cells were harvested 40 h following transfection, and primer extension analysis was performed on total cellular RNA (3 μg) to detect the level of rRNA depurination at A4324 by PAP relative to the total amount of 28 S rRNA. Dideoxynucleotide sequencing of 28 S rDNA with the same primer used for reverse extension confirmed the location of depurination. The percentage of depurination was estimated by densitometry of band intensities of the A4324 cDNA product relative to the 28 S ribosomal cDNA product. B, viability of cells expressing PAP was tested by an MTT conversion assay 40 h following transfection. MTT assays were also performed on cells transfected with pcF-PAP, pcF-PAPx, or pcDNA3 (2 μg) over a 4-day period (48–96 h). Values are means ± S.E. for three independent experiments. C, metabolic labeling of 293T cells treated with cycloheximide (25 μg/ml) or transfected with 2 μg of pcF-PAP, pcF-PAPx, or pcDNA3. Cells were incubated with [35S]methionine for the indicated time periods, followed by trichloroacetic acid precipitation and quantification of the incorporated radioactivity. D, metabolic labeling of 293T cells treated with actinomycin D (5 μg/ml) or transfected with 2 μg of pcF-PAP, pcF-PAPx, or pcDNA3. Cells were incubated with [3H]uridine for the indicated time periods, followed by trichloroacetic acid precipitation and quantification of the incorporated radioactivity. Values in C and D represent the amount of radioactivity incorporated at each time point relative to the first time point for each set of samples. Values are means ± S.E. for three independent experiments.
To ensure that PAP expression did not markedly affect overall cellular translation or transcription through direct or indirect mechanisms, we metabolically labeled 293T cells transfected with pcF-PAP, pcF-PAPx, or pcDNA3 plasmids and harvested the cells at various time points. Cells treated with cycloheximide or actinomycin D were also labeled and served as positive controls for inhibition of cellular translation and transcription, respectively. PAP expression did not suppress the rates of translation or transcription; therefore, based on these general cellular processes, we concluded that PAP was not toxic to these cells (Fig. 2, C and D).
PAP Reduces the Levels and Translatability of HTLV-I mRNAs
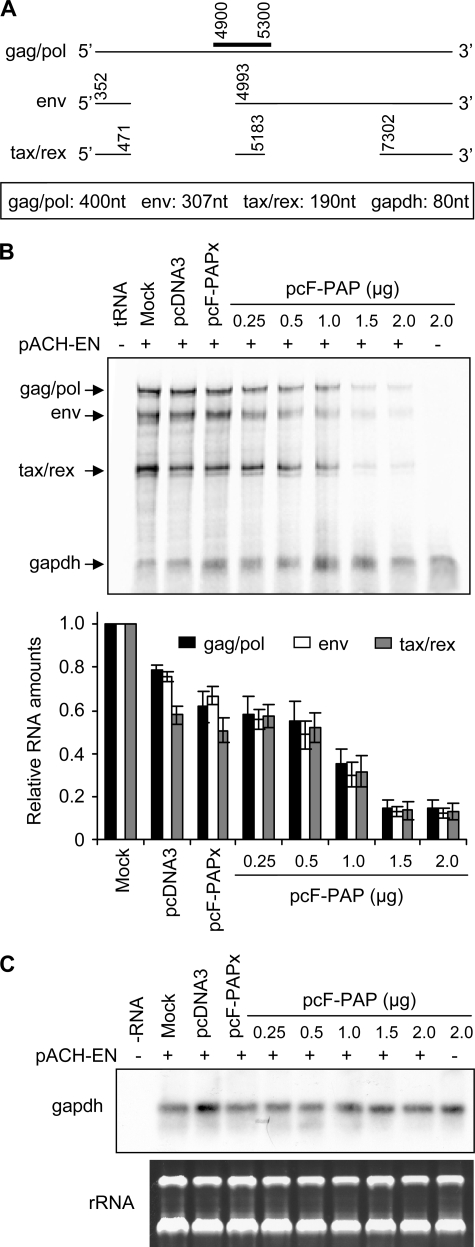
To examine whether reduced HTLV-I protein accumulation in the presence of PAP was due to decreased amounts of viral mRNAs, we performed an RNase protection assay to measure the level of HTLV-I gag/pol, env, and tax/rex mRNAs in 293T cells expressing PAP, PAPx, or pcDNA3 vector control. Fig. 3A illustrates a partial transcription map of HTLV-I, showing the annealing site of the complementary riboprobe and its expected sizes following the assay. Transient transfection of 2 μg of pcF-PAP reduced the levels of all three HTLV-I mRNAs by ∼4-fold compared with PAPx-expressing cells (Fig. 3B). All three mRNAs were reduced to a similar extent, indicating that the mRNA splicing pattern was not affected. As expected, there was no marked difference in mRNA levels between cells transfected with pcF-PAPx and pcDNA3 vector control. The level of GAPDH mRNA was also not visibly reduced by PAP expression. To confirm that PAP did not substantially affect GAPDH mRNA levels, total RNA from samples analyzed by RNase protection was also probed for this message by Northern blot (Fig. 3C). Results indicated that PAP did not appear to decrease GAPDH mRNA levels.
FIGURE 3.
PAP reduces total HTLV-I mRNA levels. A, schematic representation of the partial HTLV-I transcription map and the major mRNA splice junctions. The annealing site for the HTLV-I specific negative strand riboprobe is indicated above the gag/pol mRNA. The expected product sizes from the RNase protection assay are indicated below the diagram. As a control for total RNA amount, a riboprobe specific for GAPDH was annealed to samples and resulted in an 80-nucleotide fragment. B, 293T cells were co-transfected with pACH-EN (5 μg; plus and minus signs) and pcF-PAP (0.25, 0.5, 1, 1.5, or 2 μg), pcF-PAPx (2 μg), or pcDNA3 (2 μg). The cells were harvested 40 h later, and an RNase protection assay was performed on total cellular RNA (25 μg) or tRNA (20 μg), following hybridization with HTLV-I and GAPDH-specific riboprobes. The intensity of each band was quantified using a PhosphorImager, and the ratio of HTLV-I mRNAs to GAPDH mRNA for each sample relative to mock-transfected cells was plotted. Bars, means ± S.E. for three independent experiments. C, the same total cellular RNA samples (10 μg) described in B were probed for the presence of GAPDH mRNA by Northern blot. These RNAs were also visualized by staining with ethidium bromide.
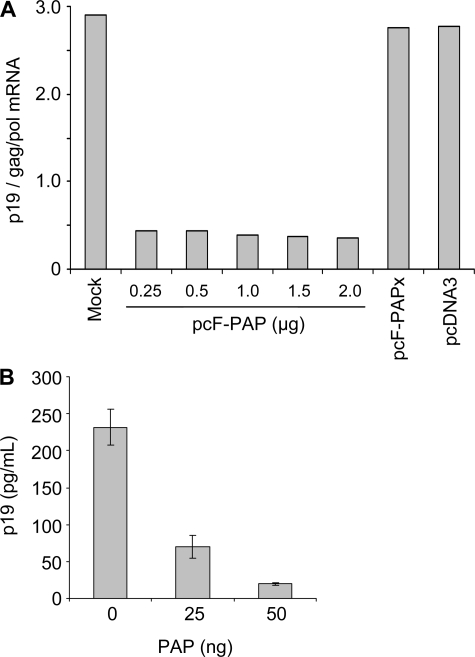
Decreased viral protein expression may have also resulted from reduced fitness of the viral mRNA for translation. To assess this possibility, we plotted the ratio of Gag protein in cell lysates estimated by p19 ELISA (Fig. 1B) to the relative abundance of gag/pol mRNA (Fig. 3B) with increasing amounts of pcF-PAP transfected into 293T cells. We chose this mRNA because p19 ELISA allows a more accurate estimation of protein amounts than immunoblot analysis of the other HTLV-I proteins. As expected, we observed a decrease in the amount of Gag protein relative to gag/pol mRNA in cells expressing PAP (Fig. 4A). Therefore, reduced translational efficiency of gag/pol mRNA may have contributed to the observed decline in Gag protein products. However, it should be noted that the amount of gag/pol mRNA present in the cytoplasm for translation may have also been decreased by PAP expression. Specifically, the viral protein Rex is a post-transcriptional regulator of HTLV-I gene expression involved in transport of viral mRNA from the nucleus to the cytoplasm (30, 31). Given that PAP reduced tax/rex mRNA levels, it is likely that Rex synthesis would be limited. Therefore, the observed decline in Gag levels may be due to decreased mRNA fitness, combined with decreased levels of cytosolic viral mRNAs.
FIGURE 4.
PAP inhibits translation of gag mRNA. A, analysis of gag/pol mRNA translational efficiency upon expression of PAP. The ratio of Gag protein values from cell lysates (from Fig. 1B) to the relative abundance of HTLV-I gag/pol mRNA within the respective samples (from Fig. 3B) was plotted. Data shown represent the ratio of means of three independent experiments. B, translation of Gag protein in reticulocyte lysate from gag transcript treated with PAP. Transcript of the gag open reading frame (2 μg) was incubated with purified PAP (25 or 50 ng) and extracted from PAP following incubation. Negative control samples were incubated in buffer alone (0 ng). The treated RNA was used as template for in vitro translation in rabbit reticulocyte lysate, and the levels of Gag protein synthesized were measured by p19 ELISA. Bars, means ± S.E. for three independent experiments.
To examine the direct effect of PAP on viral mRNA translational efficiency, a transcript of the gag open reading frame was incubated with PAP, extracted to exclude PAP following incubation, and the RNA was used as template in a cell-free translation system. Translation of viral mRNA was substantially inhibited by pretreatment with PAP, as shown by decreased levels of Gag protein measured by p19 ELISA (Fig. 4B). These data suggest that PAP reduced the fitness of this viral transcript for translation, resulting in decreased viral protein synthesis.
PAP Depurinates HTLV-I Genomic RNA
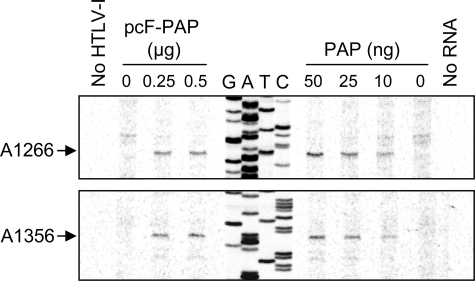
To determine whether PAP was directly targeting the viral RNA, we probed the 5′ region of the gag open reading frame within the genomic viral RNA, isolated from cells expressing PAP, for missing purines. We chose this portion of mRNA because it is unique to the gag/pol mRNA; therefore, the primer would anneal to only a single mRNA. In addition, we observed decreased expression of p19 matrix protein, which is a cleavage product of the precursor Gag protein encoded by this open reading frame. PAP depurinated two nucleotides within an ∼200-nucleotide region of the RNA, namely A1266 and A1356 (Fig. 5). Depurination sites were identified by sequencing of the proviral DNA with the same primer used for reverse extension of the gag open reading frame, and these bases are numbered relative to the first nucleotide of the 5′ U3 region of the proviral DNA (accession number L03561). Total RNA from cells transfected with pACH-EN was also incubated with purified PAP and examined for depurination using the same primer specific to the gag open reading frame. Again, the same two nucleotides were depurinated within this region of the viral RNA, illustrating that PAP targeted particular nucleotides for removal of purines. We suggest that depurination decreased fitness of the viral RNA and contributed to reduced viral protein synthesis.
FIGURE 5.
PAP depurinates the gag open reading frame of genomic HTLV-I RNA. Total RNA was isolated from 293T cells 40 h following co-transfection with pACH-EN (5 μg) and pcF-PAP (0, 0.25, or 0.5 μg). Total RNA was also isolated from 293T cells transfected only with pACH-EN (5 μg), followed by incubation with purified PAP (10, 25, or 50 ng) or buffer alone (0). Samples were analyzed by primer extension using a primer specific to the gag open reading frame of HTLV-I genomic RNA. No HTLV-I, extension from RNAs isolated from cells transfected without pACH-EN; No RNA, extension without RNA template. cDNA extension products were separated through a 7 m urea, 6% acrylamide gel and visualized with a PhosphorImager. The same primer was used to determine the position of depurination by dideoxynucleotide sequencing of pACH-EN. Depurinated nucleotides are numbered according to the first nucleotide of the 5′ U3 region of the proviral DNA (accession number L03561).
To address the possibility that PAP may be depurinating cellular messages in addition to viral RNAs, even in the absence of cell toxicity, we examined a cellular mRNA, specifically GAPDH mRNA, for depurination by primer extension. Total mRNAs were extracted from cells expressing F-PAP or vector control, and primers specific for GAPDH mRNA, spaced ∼100 nucleotides along the length of this message, were extended by a reverse transcriptase. Examination of the extension patterns revealed no depurination within the GAPDH mRNA (supplemental Fig. 1). Bands present in both PAP-expressing and vector control RNA samples indicated nonspecific strong stops for the reverse transcriptase, and only bands unique to RNA from PAP-expressing cells were scored as depurinations. The lack of detectable depurination in this cellular mRNA supports our notion that the antiviral activity of PAP is due to its depurination of viral RNAs rather than a general decline in cell health resulting from depurination of cellular mRNAs. These results are consistent with our previous work in which we generated a mutant of Brome mosaic virus 3 that could not be depurinated because every targeted purine was substituted for a pyrimidine. The amount of protein translated from this modified RNA3 was unaffected by PAP, demonstrating that depurination of viral RNA was directly responsible for the antiviral activity of PAP rather than indirect effects on cells (32).
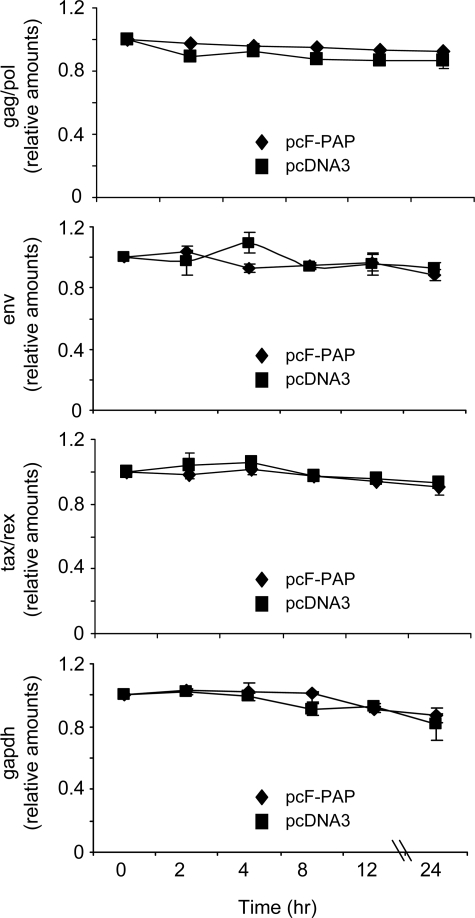
PAP Does Not Alter HTLV-I mRNA Stability
Inefficiently translated RNAs are often targeted for accelerated degradation by mRNA surveillance pathways, such as nonsense-mediated decay and nonstop decay (33, 34). We hypothesized that decreased levels of HTLV-I mRNAs were due to depurination, which would trigger their enhanced decay. To examine this possibility, we measured the levels of viral mRNAs and GAPDH mRNA over time, following the inhibition of general transcription. Comparison of the RNA levels between PAP-expressing and control cells showed that PAP did not reduce the stability of gag/pol, env, tax/rex, or GAPDH mRNAs (Fig. 6). We acknowledge that stopping cellular transcription as a consequence of this time course could have substantial effects on the health of cells; however, we conclude that increased degradation of viral RNAs was not responsible for the decline in levels observed in PAP-expressing cells.
FIGURE 6.
PAP does not decrease HTLV-I mRNA stability. 293T cells were co-transfected with 5 μg of pACH-EN and 2 μg of pcF-PAP or pcDNA3. The stability of HTLV-I gag/pol, env, tax/rex, and GAPDH mRNAs was analyzed 40 h following transfection by treatment of cells with actinomycin D (5 μg/ml). The cells were harvested at the indicated time points, followed by isolation of total RNA and RNase protection assay. RNA abundance values are ratios of each mRNA (viral and GAPDH) band intensity relative to the initial time point (t = 0). Values represent means ± S.E. for three independent experiments.
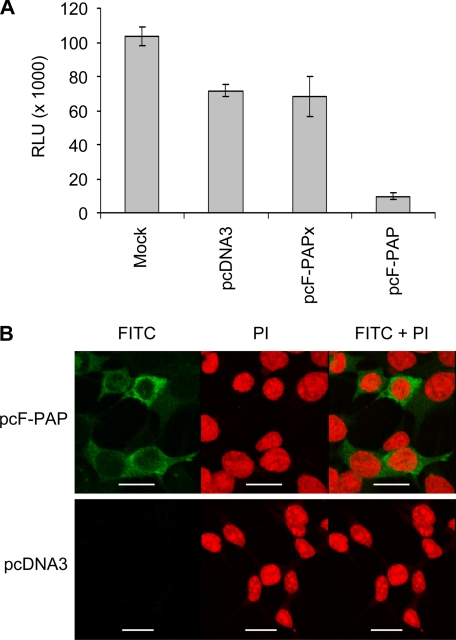
PAP Reduces Transactivation of HTLV-I 5′ Long Terminal Repeat (LTR)
Reduction in the levels of HTLV-I mRNAs may have been due to decreased synthesis rather than enhanced degradation. HTLV-I Tax protein is the transactivator of viral gene expression from the promoter found in the U3 region of the 5′-LTR (35). Because PAP diminished the level of Tax to undetectable amounts based on immunoblot analysis, we investigated whether the decline in viral mRNAs was caused by reduced transactivation of the LTR by Tax. We tested the degree of LTR transactivation using an HTLV-I reporter construct, pGL3-LTR-Luc, co-transfected with pcF-PAP, pcF-PAPx, or pcDNA3 along with pACH-EN into 293T cells, followed by a luciferase assay on the cell lysates. Cells expressing PAP showed an ∼7-fold decline in the level of Tax-mediated luciferase expression relative to PAPx and pcDNA3 vector control cells (Fig. 7A), indicating that Tax-mediated transcription from the HTLV-I 5′-LTR was inhibited by PAP.
FIGURE 7.
PAP suppresses Tax-mediated transcription from the HTLV-I 5′-LTR. A, 293T cells were co-transfected with 1 μg of pGL3-LTR-Luc, 5 μg of pACH-EN, and 2 μg of pcF-PAP, pcF-PAPx, or pcDNA3. A luciferase assay was performed 24 h following transfection to determine the degree of Tax-mediated transcription from the 5′-LTR. Values represent means ± S.E. for three independent experiments. B, 293T cells were transfected with 2 μg of pcF-PAP or pcDNA3. Cells were fixed 40 h following transfection and stained for the presence of F-PAP using a FLAG-specific monoclonal antibody (1:200) and a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (1:500). Cells were also stained with propidium iodide (PI) to visualize nuclei and were examined by confocal microscopy. Scale bar, 20 μm.
To determine the cellular localization of PAP, 293T cells transfected with pcF-PAP or pcDNA3 were fixed and incubated with primary antibody specific to FLAG and fluorescein isothiocyanate-conjugated secondary antibody. Examination by confocal microscopy of cells transfected with pcF-PAP revealed green fluorescence within the cytosol and not within nuclei (Fig. 7B). Cells transfected with pcDNA3 served as a negative control for autofluorescence. Counterstaining of cells with propidium iodide indicated nuclei. The cytosolic localization of PAP supports our contention that PAP indirectly reduces transcription of the viral genome, probably through its effect on Tax levels, rather than directly influencing transcription within the nucleus.
DISCUSSION
PAP exhibits antiviral activity against a number of different viruses; however, the mechanistic details of this activity have not been described (19, 22, 36). As a ribosome-inactivating protein, PAP also depurinates a conserved adenine within the sarcin/ricin loop of the large rRNA, which has been reported to inhibit the binding of elongation factor 2 and slow the rate of elongation during protein translation (37, 38, 39). This inhibition of translation has traditionally been cited to explain antiviral activity, because a positive correlation has been shown between ribosome depurination and decline of virus infection (18, 40). However, very few ribosome-inactivating proteins are antiviral, suggesting that merely inhibiting ribosomes does not impart antiviral qualities. Moreover, our results show that PAP inhibited HTLV-I gene expression in the absence of cytotoxicity. In addition to their potential to inhibit translation, many ribosome-inactivating proteins induce a ribotoxic stress response that results in cell death (41, 42). This response is characterized by damage to the rRNA, which triggers activation of a stress-induced kinase that may culminate in programmed cell death (43). In contrast, we have demonstrated previously that PAP expression in 293T cells resulted in cell survival rather than apoptosis (28). In the current study, the amount of PAP expression that caused 100-fold decline in HTLV-I protein levels was not sufficient to be toxic to cells, although 16% of the 28 S rRNA was depurinated. This level of rRNA depurination did not affect the rate of cellular translation, indicating that reduction in the level of viral proteins was not due to general defects in the cellular translation machinery as a result of rRNA depurination. Together with our findings indicating minimal PAP effect on cellular transcription rates, we conclude that antiviral activity was not a consequence of cytotoxicity.
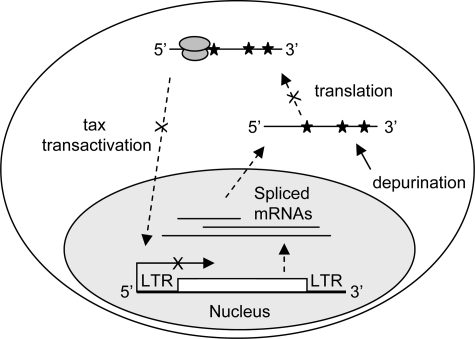
In addition to demonstrating lack of harm to host cells, examining the potential of PAP as a therapeutic option to limit HTLV-I progression would require understanding the steps in the virus life cycle affected by depurination. Here, we describe the mechanism through which PAP inhibited virus production (Fig. 8). Specifically, transient expression of PAP in 293T cells caused a substantial decline in the level of intracellular Gag and virus-associated p19 matrix protein and intracellular Tax protein. We show that the 5′ region of the gag/pol mRNA was depurinated by PAP and that this mRNA was less efficiently translated in a cell-free system, suggesting that depurination contributed to diminished levels of viral proteins. We have shown previously that depurination of Brome mosaic virus RNA3 caused ribosomes to stall at the missing base during elongation (32). Expression of PAP also reduced HTLV-I gag/pol, env, and tax/rex mRNA levels in a dose-dependent manner. Decreased tax/rex mRNA levels may have also resulted in diminished Rex synthesis, which in turn would have reduced the amount of cytoplasmic gag/pol mRNA for translation. Therefore, post-transcriptional effects of PAP contributed to the decline in viral protein synthesis. Diminished levels of viral mRNAs were not due to enhanced degradation but rather were probably caused by limited synthesis, based on the results of our reporter assay. HTLV-I Tax protein is encoded from the HTLV-I doubly spliced tax/rex mRNA and is the transactivator of the viral promoter located within the 5′-LTR (35). Since Tax is required for enhanced transcription from the viral promoter, we expected that undetectable Tax would result in decreased levels of all HTLV-I-encoded mRNAs. Therefore, feedback inhibition of viral gene expression manifested as an efficient decline in the level of viral mRNAs and proteins, hence the significant reduction in virus production.
FIGURE 8.
Model for effect of PAP on HTLV-I. Shown is a schematic representation of steps in the HTLV-I life cycle that are affected by PAP (indicated by ×). Stars represent depurination of the viral mRNA.
Apart from pokeweed antiviral protein, other enzymes exist that modify the genomes of some viruses. For example, APOBEC3G is a family member of cytidine deaminases that acts as a host defense factor against some retroviruses (reviewed in Refs. 44 and 45). These enzymes convert cytosines of negative-strand viral cDNA to uracils during reverse transcription, resulting in guanosine to adenosine hypermutations in the coding strand (46–48). The uracil-containing cDNA may be targeted for degradation, or fixation of hypermutation in proviral sequences may result in synthesis of inactive or truncated viral proteins. HTLV-I appears to be relatively resistant to the antiviral effects of APOBEC3G, and mutations of guanosines to adenosines are rarely observed (49). Interestingly, a peptide motif in the C terminus of HTLV-I Nucleocapsid protein has been shown to inhibit ABOBEC3G packaging into nascent virions, thereby limiting its antiviral effect (50). Therefore, HTLV-I appears to evade this cellular factor that causes mutation of its genome. We hypothesize that evolution of virus resistance to pokeweed antiviral protein may be difficult, given that the timing of the PAP effect described here does not rely on the protein being packaged into viral particles and that no mechanisms for repair of depurinated RNA have been described. The subsequent decline in protein synthesis from the damaged viral RNA and its effect on transcriptional and post-transcriptional events suggest that PAP may have an application as part of an antiviral strategy in the future.
Supplementary Material
Acknowledgments
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: MT-2 cells (catalog number 237) from Dr. Douglas Richman and antiserum to HTLV-I Tax (catalog number 712) from Dr. Kuan-Teh Jeang. We also thank Dr. K. Shimotohno (Kyoto University, Japan) for providing the HTLV-I reporter construct, pGL3-LTR-Luc.
This work was supported, in whole or in part, by National Institutes of Health Public Health Service Grants CA94056, CA63417, CA10073, CA10521 and CA109678 (to L. R.). This work was also supported by the Natural Sciences and Engineering Research Council of Canada and a Premier's Research Excellence Award (to K. A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- HTLV-I
- human T-cell leukemia virus I
- PAP
- pokeweed antiviral protein
- ELISA
- enzyme-linked immunosorbent assay
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PIPES
- 1,4-piperazinediethanesulfonic acid
- LTR
- long terminal repeat
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein.
REFERENCES
- 1.Uchiyama T., Yodoi J., Sagawa K., Takatsuki K., Uchino H. (1977) Blood 50, 481–492 [PubMed] [Google Scholar]
- 2.Gessain A., Barin F., Vernant J. C., Gout O., Maurs L., Calender A., de Thé G. (1985) Lancet 2, 407–410 [DOI] [PubMed] [Google Scholar]
- 3.Osame M., Matsumoto M., Usuku K., Izumo S., Ijichi N., Amitani H., Tara M., Igata A. (1987) Ann. Neurol. 21, 117–122 [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi K., Takatsuki K. (1993) Clin. Haematol. 6, 899–915 [DOI] [PubMed] [Google Scholar]
- 5.Tokudome S., Tokunaga O., Shimamoto Y., Miyamoto Y., Sumida I., Kikuchi M., Takeshita M., Ikeda T., Fujiwara K., Yoshihara M. (1989) Cancer Res. 49, 226–228 [PubMed] [Google Scholar]
- 6.Tajima K. (1990) Int. J. Cancer 45, 237–243 [DOI] [PubMed] [Google Scholar]
- 7.Yoshida M. (2001) Annu. Rev. Immunol. 19, 475–496 [DOI] [PubMed] [Google Scholar]
- 8.Taylor G. P., Matsuoka M. (2005) Oncogene 24, 6047–6057 [DOI] [PubMed] [Google Scholar]
- 9.Franchini G., Nicot C., Johnson J. M. (2003) Adv. Cancer Res. 89, 69–132 [DOI] [PubMed] [Google Scholar]
- 10.Murphy E. L., Hanchard B., Figueroa J. P., Gibbs W. N., Lofters W. S., Campbell M., Goedert J. J., Blattner W. A. (1989) Int. J. Cancer 43, 250–253 [DOI] [PubMed] [Google Scholar]
- 11.Takatsuki K., Yamaguchi K., Kawano F., Hattori T., Nishimura H., Tsuda H., Sanada I., Nakada K., Itai Y. (1985) Cancer Res. 45, 4644s–4645s [PubMed] [Google Scholar]
- 12.Höllsberg P., Hafler D. A. (1993) N. Engl. J. Med. 328, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 13.Moriuchi M., Inoue H., Moriuchi H. (2001) J. Virol. 75, 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill P. S., Harrington W., Jr., Kaplan M. H., Ribeiro R. C., Bennett J. M., Liebman H. A., Bernstein-Singer M., Espina B. M., Cabral L., Allen S. (1995) N. Engl. J. Med. 332, 1744–1748 [DOI] [PubMed] [Google Scholar]
- 15.Machuca A., Soriano V. (2000) J. Acquir. Immune Defic. Syndr. 24, 189–193 [DOI] [PubMed] [Google Scholar]
- 16.Irvin J. D. (1975) Arch Biochem. Biophys. 169, 522–528 [DOI] [PubMed] [Google Scholar]
- 17.Endo Y., Tsurugi K., Lambert J. M. (1988) Biochem. Biophys. Res. Commun. 150, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 18.Foà-Tomasi L., Campadelli-Fiume G., Barbieri L., Stirpe F. (1982) Arch. Virol. 71, 323–332 [DOI] [PubMed] [Google Scholar]
- 19.Picard D., Kao C. C., Hudak K. A. (2005) J. Biol. Chem. 280, 20069–20075 [DOI] [PubMed] [Google Scholar]
- 20.Tumer N. E., Hwang D. J., Bonness M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3866–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudak K. A., Wang P., Tumer N. E. (2000) RNA 6, 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarling J. M., Moran P. A., Haffar O., Sias J., Richman D. D., Spina C. A., Myers D. E., Kuebelbeck V., Ledbetter J. A., Uckun F. M. (1990) Nature 347, 92–95 [DOI] [PubMed] [Google Scholar]
- 23.Rajamohan F., Venkatachalam T. K., Irvin J. D., Uckun F. M. (1999) Biochem. Biophys. Res. Commun. 260, 453–458 [DOI] [PubMed] [Google Scholar]
- 24.Wang P., Tumer N. E. (1999) Nucleic Acids Res. 27, 1900–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukahara T., Wielgosz M. M., Ratner L. (2001) J. Virol. 75, 9553–9559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimata J. T., Wong F. H., Wang J. J., Ratner L. (1994) Virology 204, 656–664 [DOI] [PubMed] [Google Scholar]
- 27.Hur Y., Hwang D. J., Zoubenko O., Coetzer C., Uckun F. M., Tumer N. E. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8448–8452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan Tung K. W., Mansouri S., Hudak K. A. (2008) Int. J. Biochem. Cell Biol. 40, 2452–2461 [DOI] [PubMed] [Google Scholar]
- 29.Butsch M., Boris-Lawrie K. (2000) J. Virol. 74, 11531–11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hidaka M., Inoue J., Yoshida M., Seiki M. (1988) EMBO J. 7, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gröne M., Koch C., Grassmann R. (1996) Virology 218, 316–325 [DOI] [PubMed] [Google Scholar]
- 32.Gandhi R., Manzoor M., Hudak K. A. (2008) J. Biol. Chem. 283, 32218–32228 [DOI] [PubMed] [Google Scholar]
- 33.Conti E., Izaurralde E. (2005) Curr. Opin. Cell Biol. 17, 316–325 [DOI] [PubMed] [Google Scholar]
- 34.Wagner E., Lykke-Andersen J. (2002) J. Cell Sci. 115, 3033–3038 [DOI] [PubMed] [Google Scholar]
- 35.Sodroski J., Rosen C., Goh W. C., Haseltine W. (1985) Science 228, 1430–1434 [DOI] [PubMed] [Google Scholar]
- 36.Tomlinson J. A., Walker V. M., Flewett T. H., Barclay G. R. (1974) J. Gen. Virol. 22, 225–232 [DOI] [PubMed] [Google Scholar]
- 37.Stirpe F., Bailey S., Miller S. P., Bodley J. W. (1988) Nucleic Acids Res. 16, 1349–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gessner S. L., Irvin J. D. (1980) J. Biol. Chem. 255, 3251–3253 [PubMed] [Google Scholar]
- 39.Nilsson L., Nygård O. (1986) Eur. J. Biochem. 161, 111–117 [DOI] [PubMed] [Google Scholar]
- 40.Taylor S., Massiah A., Lomonossoff G., Roberts L. M., Lord J. M., Hartley M. (1994) Plant J. 5, 827–835 [DOI] [PubMed] [Google Scholar]
- 41.Narayanan S., Surolia A., Karande A. A. (2004) Biochem. J. 377, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouyang D. Y., Wang Y. Y., Zheng Y. T. (2005) Cell Mol. Immunol. 2, 419–425 [PubMed] [Google Scholar]
- 43.Iordanov M. S., Pribnow D., Magun J. L., Dinh T. H., Pearson J. A., Chen S. L., Magun B. E. (1997) Mol. Cell. Biol. 17, 3373–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bieniasz P. D. (2004) Nat. Immunol. 5, 1109–1115 [DOI] [PubMed] [Google Scholar]
- 45.Cullen B. R. (2006) J. Virol. 80, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. (2003) Nature 424, 99–103 [DOI] [PubMed] [Google Scholar]
- 47.Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. (2003) Nature 424, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris R. S., Sheehy A. M., Craig H. M., Malim M. H., Neuberger M. S. (2003) Nat. Immunol. 4, 641–643 [DOI] [PubMed] [Google Scholar]
- 49.Ohsugi T., Koito A. (2007) J. Virol. Methods 139, 93–96 [DOI] [PubMed] [Google Scholar]
- 50.Derse D., Hill S. A., Princler G., Lloyd P., Heidecker G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2915–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.