Abstract
CcpN, a transcriptional repressor from Bacillus subtilis that is responsible for the carbon catabolite repression of three genes, has been characterized in detail in the past 4 years. However, nothing is known about the actual repression mechanism as yet. Here, we present a detailed study on how CcpN exerts its repression effect at its three known target promoters of the genes sr1, pckA, and gapB. Using gel shift assays under non-repressive and repressive conditions, we showed that CcpN and RNA polymerase can bind simultaneously and that CcpN does not prevent RNA polymerase (RNAP) binding to the promoter. Furthermore, we investigated the effect of CcpN on open complex formation and demonstrate that CcpN also does not act at this step of transcription initiation at the sr1 and pckA and presumably at the gapB promoter. Investigation of abortive transcript synthesis revealed that CcpN acts differently at the three promoters: At the sr1 and pckA promoter, promoter clearance is impeded by CcpN, whereas synthesis of abortive transcripts is repressed at the gapB promoter. Eventually, we demonstrated with Far Western blots and co-elution experiments that CcpN is able to interact with the RNAP α-subunit, which completes the picture of the requirements for the repressive action of CcpN. On the basis of the presented results, we propose a new working model for CcpN action.
CcpN, a transcriptional repressor from Bacillus subtilis, mediates CcpA-independent carbon catabolite repression of at least three genes: sr1, encoding a small RNA and pckA and gapB (1, 2), encoding two gluconeogenic enzymes (3, 4). Since its discovery in 2005, CcpN has been thoroughly investigated. Binding properties and binding motifs were examined, revealing that CcpN possesses two asymmetric binding sites that are bound cooperatively and positioned differently at the three regulated promoters (5): At the sr1 promoter, binding sites are located upstream of the −35 region and between the −35 and the −10 region, while binding sites cover the −35 as well as the −10 region at the pckA promoter. One operator at the gapB promoter overlaps the −10 region, the second one is located around +20. ATP and low pH have been identified as signals required for CcpN-mediated repression (6) and the detailed biophysical properties of CcpN-DNA interaction have been reported (7). In addition, it has been shown that CcpN controls central carbon fluxes in the metabolism of B. subtilis and that the growth defect of CcpN knock-out mutants is caused by ATP dissipation via extensive futile cycling (8). It has been demonstrated that a CcpN knock-out is able to increase the industrial production of riboflavin in B. subtilis by a deregulation of the gapB gene (9). However, nothing is known about the actual repression mechanism of CcpN as yet.
Initiation of transcription is a stepwise process (10), beginning with binding of RNA polymerase (RNAP)2 to the promoter and formation of a loose closed complex, which is then rearranged into a tighter closed complex. This is followed by the melting of DNA around the transcriptional start site, called the open complex. RNAP can subsequently form the initiation complex and begin to transcribe the DNA, often producing short abortive transcripts resulting from failed attempts to leave the promoter. Eventually, RNAP escapes the promoter and forms the elongation complex. Transcriptional repressors can act at any of these steps, beginning with steric hindrance of RNAP binding, like the Fur protein of Escherichia coli (11) over the inhibition of open complex formation, like B. subtilis Spo0A at the abrB promoter (12) to the prevention of promoter clearance, as observed with the phage Φ29 protein p4 at the viral A2c promoter (13). Different mechanisms of transcriptional repression have already been reviewed in detail (14).
Whereas steric hindrance of RNA polymerase binding does not involve direct repressor-RNAP contacts, repression of other steps in the transcription initiation process often does. In most of those cases, contacts between a transcriptional repressor and the C-terminal domain of the α-subunit of RNAP are described, as for the p4 protein at the A2c promoter or for the repressor Spx from B. subtilis (13, 15). However, interactions with other subunits of RNAP have also been proposed, for example for the Rsd protein of E. coli or the main carbon catabolite mediator of B. subtilis, CcpA (16, 17). A special case of repressors that interact with RNA polymerase subunits are anti-σ-factors. These proteins can sequester free σ-factor and are thus able to influence the expression of whole regulons (18, 19).
In this work, we present a detailed analysis of the action of CcpN at all steps of transcription initiation and show that it prevents promoter clearance at the sr1 and pckA promoter, while displaying a rare effect at the gapB promoter: It allows the formation of the open complex, but prevents the synthesis of abortive transcripts. Furthermore, we demonstrate that CcpN is able to interact with the α-subunit of RNAP and probably regulates the sr1 and pckA promoters this way. Eventually, we present a new working model for CcpN-mediated transcriptional repression in regard to the specific operator positions and promoter sequences.
EXPERIMENTAL PROCEDURES
Strains and Media Used in This Study
B. subtilis strain NIG2001 was used for expression of His-tagged B. subtilis RNA polymerase (20) and strain DB104 (21) was used for the preparation of B. subtilis protein crude extracts. E. coli strain TG1 (pREP4, pQGDR) was used for overexpression and purification of CcpN-His5 and strain BL21 (DE3) (pETSigA) was used for overexpression and purification of His-tagged B. subtilis SigA (4, 22). All strains were grown in TY medium (16 g Bacto tryptone, 10 g of yeast extract, 5 g of NaCl in 1 liter) with the respective antibiotics.
Protein Purification
CcpN overexpression and purification with a Ni2+-nitrilotriacetic acid (NTA)-agarose column and by anion exchange chromatography was performed as published before (6). Expression and purification of His-tagged B. subtilis RNA polymerase and His-tagged SigA with a Ni2+-NTA-agarose column was carried out according to the protocols established by Fujita and Sadaie (20, 22).
Gel Shift Assays
Binding reactions were performed in a final volume of 10 μl in either 0.5× TBE and 10 mm MgCl2 for the formation of closed complexes or in in vitro transcription buffer (40 mm Tris acetate, pH 7.3, 10 mm magnesium acetate, 100 mm potassium acetate, and 20% glycerol) for the formation of open complexes, 0.05 g/liter herring sperm DNA as nonspecific competitor, and 1 nm end-labeled DNA fragment. Where indicated, 3 μm CcpN-His5, 3 μm RNAP-His6, 3 mm ATP, HCl to a final pH of 6.5 or 0.1 g/liter of heparin were added. After incubation at 37 °C for 15 min, the reaction mixtures were denatured and separated on 5% native polyacrylamide gels run at room temperature for 1 h at 230 V. Gels were dried and subjected to PhosphorImaging (Fujix BAS 1000).
Open Complex Formation Assays
Binding reactions were performed in a final volume of 10 μl in 50 mm sodium-cacodylate buffer (pH 7.3) using 1 nm of an end-labeled DNA fragment. Where indicated, 100 nm CcpN-His5, 100 nm of native RNAP, 3 mm ATP, and/or HCl to a final pH of 6.5 were added. After 15 min of incubation at 37 °C, 1 μl of DEPC (final concentration of 10%) was added, and the reaction was incubated at 37 °C for another 10 min. The reaction was stopped by the addition of 50 μl of stop solution (1.5 m NaAc, 0.1 g/liter tRNA) and precipitated with ethanol, followed by dissolving of the pellet in 10% piperidine and cleavage at the modified sites for 30 min at 90 °C. Subsequently, the cleavage reaction was precipitated with ethanol again, and the pellet dissolved in formamide loading dye to a final activity of 2000 cpm/μl. Afterwards, 3 μl were denatured and separated on a 6% denaturing polyacrylamide gel. Gels were dried and subjected to PhosphorImaging.
In Vitro Transcription
In vitro transcription reactions at the pckA and gapB promoters were performed in a final volume of 20 μl in in vitro transcription buffer in the presence of 3 mm ATP, 0.1 mm CTP, and GTP, 0.01 mm UTP, and 0.011 μm [α-32P]UTP. For the sr1 promoter, 0.1 mm UTP and 0.011 μm [α-32P]ATP were used to allow detection of abortive transcripts. Where indicated, HCl to a final pH of 6.5 and CcpN-His5 were added, followed by 100 nm double-stranded DNA template and 100 nm RNAP-His6. The reaction was gently mixed and incubated for 30 min at 37 °C. Half of the reaction was ethanol-precipitated with potassium acetate to keep unincorporated [α-32P]NTPs in solution and then dissolved in 10 μl of distilled water. One volume of formamide loading dye was added to each half of the reaction, followed by denaturation for 5 min at 90 °C, quick cooling on ice, and analysis on a 6% denaturing polyacrylamide gel to detect full-length transcripts or on a 23% denaturing polyacrylamide gel to detect abortive transcripts. Electrophoresis was performed at 300 V/25 mA for 50 min. Gels were dried and subjected to PhosphorImaging.
Western and Far Western Blotting
For Western blotting, samples were separated on a 15.5% SDS-polyacrylamide gel and subsequently blotted using polyvinylidene difluoride membrane (Carl Roth). A polyclonal antiserum from rabbit against CcpN-His5 as primary antibody and horseradish peroxidase-coupled anti-rabbit secondary antibody (Santa Cruz Biotechnology, Inc.) was used, both with a dilution of 1:2000. Blots were developed by diaminobenzidine reaction, digitized with a ScanPrisa 640U (Acer) scanner, and analyzed with TINA-PC BAS 2.08e software. For Far Western blotting, two identical sets of protein samples were separated by SDS-PAGE and subsequently blotted. SDS was then removed from the blot membranes by washing with PBST. After blocking, the part of the membranes containing the first set of samples was incubated with blocking buffer again for 1 h at room temperature, while the membrane containing the other set was incubated with 200 nm CcpN-His5 in blocking buffer. Both membranes were then washed with PBST and incubated with primary and secondary antibody as described in the Western blotting procedure.
Co-elution
B. subtilis DB104 was grown to an A560 of 4 in 150 ml of TY medium, cells were harvested, resuspended in 15 ml of PBS and sonicated three times for 10 min. 180 μl of phenylmethylsulfonyl fluoride (17 g/liter in isopropanol) were added prior to sonication. After centrifugation at 4 °C, the supernatant was obtained and incubated with 300 nm of purified RpoA-His6, SigA-His6 or without any protein for 1 h at room temperature. The samples were then purified using a Ni2+-NTA-agarose column, and the eluates analyzed on a 15.5% PAA-SDS-gel.
RESULTS
CcpN Does Not Inhibit Formation of the Closed Complex
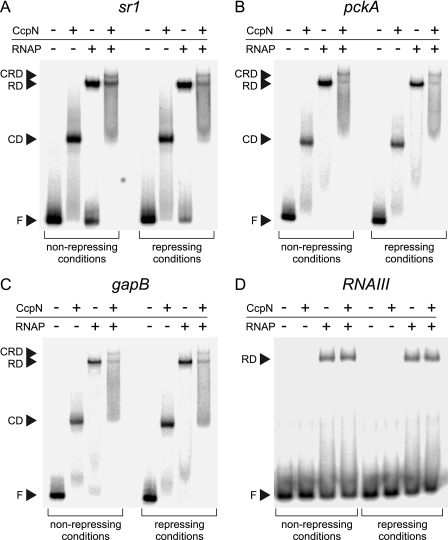
Transcriptional repressors can act during a variety of different steps in transcription initiation. To investigate whether CcpN exerts its repression effect by preventing RNA polymerase binding to the promoter, we performed gel shift assays using 89-bp end-labeled double-stranded DNA fragments carrying the sr1, pckA, gapB, or RNAIII (as a negative control that is unable to bind CcpN) promoters (Fig. 1). Purified CcpN-His5 and purified His-tagged B. subtilis RNA polymerase alone and together were incubated with the labeled DNA fragment and complex formation was analyzed on native polyacrylamide gels. The presence of CcpN or RNA polymerase alone resulted in a single band corresponding to the respective protein-DNA complex at all three promoters. When both proteins were present, an additional band was visible at all three promoters, emerging from a complex of DNA, CcpN, and RNA polymerase. As expected, the control promoter of RNAIII showed only a single band corresponding to an RNAP-DNA complex, but no CcpN-DNA complex. All experiments were performed under non-repressive (0 mm ATP, pH 7.3) and under repressive conditions (3 mm ATP, pH 6.5) to assay whether CcpN is able to prevent RNA polymerase binding to the promoter sequence. For analysis under repressive conditions, both ATP and low pH were also present in the gel and in the running buffer to ensure that the conditions did not change during electrophoresis. At all three promoters, the band representing the CcpN-RNAP-DNA complex did not change in intensity when comparing non-repressive with repressive conditions, indicating that CcpN is not able to prevent the formation of the closed complex.
FIGURE 1.
Electrophoretic mobility shift assays (EMSAs) with His-tagged CcpN and RNA polymerase at the sr1 (A), pckA (B), gapB (C), and RNAIII (D) promoters. The presence or absence of 3 μm CcpN-His5 and 3 μm RNAP-His6 is indicated above each lane. Experiments were performed under non-repressing (pH 7.0, 0 mm ATP) or repressing (pH 6.5, 3 mm ATP) conditions. Autoradiograms of the gels are shown. F, free DNA; CD, CcpN-DNA complex; RD, RNAP-DNA complex; CRD, CcpN-RNAP-DNA complex.
CcpN Does Not Inhibit Open Complex Formation
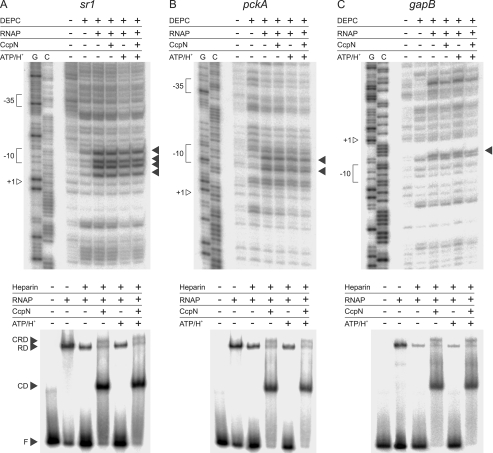
The next step in transcription initiation is the formation of the open complex, involving melting of the DNA at the promoter region. To detect formation of an open complex, a double-stranded DNA fragment was probed for the presence of single-stranded regions under different conditions using DEPC (Fig. 2), which is known to react preferentially with single-stranded regions in B-form DNA (23). Usually KMnO4 is used for detection of single-stranded regions, but did not work under our buffer conditions. Therefore, DEPC was used, although it has the disadvantage of producing weaker signals at stacked adenosine residues. As shown in Fig. 2, signals emerged at all three promoters upon addition of RNA polymerase that were not present in the negative control, where only DEPC was added. These signals persisted in the presence of CcpN (non-repressive conditions) as well as in the presence of CcpN, ATP and low pH (repressive conditions) at all investigated promoters. Thus, one can conclude that CcpN is not able to prevent formation of the open complex at any of the three promoters. To corroborate these findings, another assay for open complex formation using heparin as a probe has been performed, mainly confirming the results of the DEPC probing. Interestingly, the amount of RNAP-DNA complex decreases significantly upon heparin addition at the gapB promoter, indicating that RNAP does not seem to be able to be efficiently converted into a stable open complex. However, the fraction of stable complex that is formed cannot be destabilized by CcpN even under repressive conditions.
FIGURE 2.
Open complex formation assay at the sr1 (A), pckA (B), and gapB (C) promoters. Probing with DEPC is shown at the top while the corresponding heparin-probing is shown below. For DEPC-probing, DEPC (10%), RNAP (100 nm), CcpN-His5 (100 nm), were added where indicated. Bands showing the presence of single-stranded DNA regions, and therewith open complexes are indicated by arrows. G, G→A sequencing reaction, C, C+T sequencing reaction. Positions of +1, the −10 and −35 box are indicated. Please note that the noncoding strand was used for sr1 and pckA, while the coding strand was used for gapB. For heparin-probing, Heparin (0.1 g/liter) CcpN-His5 (3 μm), RNAP-His6 (3 μm), ATP (3 mm), or HCl (to a final pH of 6.5) were added where indicated. F, free DNA; CD, CcpN-DNA complex; RD, RNAP-DNA complex; CRD, CcpN-RNAP-DNA complex. Autoradiograms of the gels are shown.
CcpN Acts Differently at the Three Promoters
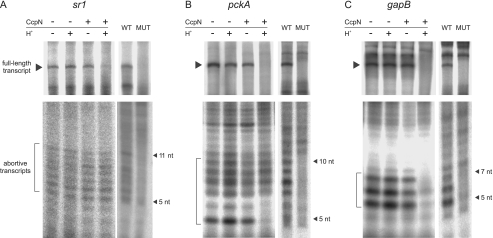
Because formation of the open complex is not impeded by CcpN at any promoter, it can either prevent the synthesis of abortive transcripts or promoter clearance. To investigate this issue, in vitro transcription reactions under non-repressive and repressive conditions were performed and analyzed on two different denaturing polyacrylamide gels: 6% gels were used to detect full-length transcripts while 23% gels were used to detect abortive transcripts (Fig. 3). Because there is no uridine within the first 11 bases of the sr1 transcript, [α-32P]ATP instead of UTP was used for labeling. This resulted in very faint bands for both the full-length and the abortive transcripts, because all in vitro transcription reactions were performed in the presence of 3 mm ATP necessary to observe the repressive effect of CcpN. To ensure that the observed abortive transcripts are produced by the analyzed promoters rather than non-promoter sites on the template, templates with mutated promoters were investigated (Fig. 3). Indeed, certain transcripts within the expected size of 3–11 nt are no longer produced from the mutated fragments, indicating that they emerge from the investigated promoters. At all three promoters, formation of full-length or abortive transcripts was not influenced in the presence of CcpN or low pH alone. Fig. 3A shows that abortive transcripts are produced at the sr1 promoter in all four lanes, even under repressive conditions, while synthesis of the full-length transcript is significantly repressed in the presence of CcpN, ATP, and low pH. At the pckA promoter, most of the abortive transcripts are still produced during CcpN-mediated repression, however, the smallest two transcripts are lost. Nevertheless, Fig. 3B clearly shows that abortive transcription in general is not affected by CcpN. A completely different picture can be found at the gapB promoter (Fig. 3C). Here, bands corresponding to abortive transcripts are hardly or not at all detectable under repressive conditions. Thus, one can conclude that CcpN acts at the sr1 and pckA promoters by preventing RNA polymerase from leaving the promoter and proceeding with transcription, while still allowing the production of short abortive transcripts. At the gapB promoter, on the contrary, CcpN appears to impede transcription initiation itself, resulting in the inability to produce abortive transcripts. However, the conversion from closed to open complex at this promoter is inefficient under our experimental condition. Thus, it is possible that CcpN acts as an inhibitor of open complex formation under conditions where the closed to open complex transition is more efficient.
FIGURE 3.
In vitro transcription and detection of abortive transcripts at the sr1 (A), pckA (B), and gapB (C) promoters. Transcription was performed in in vitro transcription buffer (see “Experimental Procedures”) using 100 nm DNA template and 100 nm His-tagged B. subtilis RNA polymerase. 300 nm CcpN-His5 was added, or pH was lowered where indicated. Half of each reaction was separated on either a 6% denaturing polyacrylamide gel to detect the full-length transcripts, indicated by an arrow or on a 23% denaturing polyacrylamide gel to detect abortive transcripts, indicated by a bracket. Control experiments to the right of each panel show which of the abortive transcripts are produced by the investigated promoters. WT, wild-type promoter, MUT, mutated promoter, where the −10 regions have been replaced by the sequence GCCGAT (sr1) or GCCGCT (pckA and gapB). The estimated size of the abortive transcripts on each gel is indicated by arrows. Autoradiograms of the gels are shown.
CcpN Is Able to Interact with RNA Polymerase
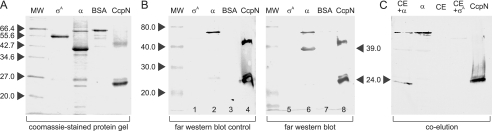
Because CcpN is able to prevent RNA polymerase from leaving the promoter, we wanted to find out whether this is due to a direct interaction. To this end, we purified the RNA polymerase α-subunit (RpoA) as well as the B. subtilis major σ-factor SigA. Fig. 4A shows the two proteins, along with BSA and purified CcpN. While SigA is apparently pure, the α-subunit contains some impurities, although in a much lower concentration than the protein itself. Two gels with identical protein samples have subsequently been subjected to far Western blotting to analyze possible interactions between CcpN and these proteins (Fig. 4B). The left panel shows the control blot that was only incubated with primary (anti-CcpN) and secondary antibody. As expected, CcpN itself produced a very strong signal, indicating that the antibodies work as intended. However, there are also two signals in the lane with the RpoA preparation: A very intensive signal corresponding to the largest impurity, indicating extensive antibody cross-reaction and a weak signal at the 27 kDa impurity. The RpoA band itself did not produce a signal, demonstrating that the anti-CcpN-antibody did not bind to it unspecifically. Furthermore, there were no antibody cross reactions with either SigA or bovine serum albumin. The right panel shows the experiment itself, where the blot has been incubated with CcpN before the application of the first antibody. Strikingly, a band emerges that corresponds exactly to the 39-kDa band comprised of RpoA, indicating that CcpN is able to specifically interact with the RNA polymerase α-subunit. Neither SigA nor bovine serum albumin showed any interaction with CcpN at all.
FIGURE 4.
CcpN-RpoA interaction studies. The corresponding molecular weights of the marker bands are indicated beside the marker lanes. A, 15.5% SDS-polyacrylamide gel of different purified proteins. MW, molecular weight marker; σ, purified SigA-His6; α, purified RpoA-His6; BSA, bovine serum albumin; CcpN, purified CcpN-His5. 1 μg of each protein was loaded into each lane. B, Far Western blot of the protein gel shown in A. Equal amounts of protein were loaded into lanes 1–4 and 5–8, respectively. Proteins were renatured after blotting by washing with SDS-free phosphate-buffered saline. Lanes 1–4 are control lanes and were just blocked, incubated with rabbit-anti-CcpN antibody and subsequently with horseradish peroxidase coupled anti-rabbit antibody. Lanes 5–8 are the sample lanes and were treated like lanes 1–5, but were incubated with 200 nm CcpN-His5 after blocking and before incubation with anti-CcpN antibody. The blots were developed using horseradish peroxidase catalyzed conversion of diaminobenzidine. PC-BAS 2.08e software was used for quantification. C, co-elution of RpoA-His6 and CcpN. The lanes were loaded as follows: CE+α: RpoA-His6 preincubated with B. subtilis DB104 protein crude extract (see “Experimental Procedures”) and subsequently purified using a Ni2+-NTA-agarose column; α, RpoA-His6 without preincubation with B. subtilis DB104 protein crude extract; CE, B. subtilis DB104 protein crude extract, purified; CE+σ, SigA-His6 preincubated with B. subtilis DB104 protein crude extract and subsequently purified; CcpN, purified CcpN-His5. Equal amounts of eluate were loaded into each lane.
To corroborate these findings, we investigated whether CcpN can be co-eluted with an α-subunit preparation. To this end, a crude extract of B. subtilis DB104 was incubated with RpoA-His6 and subsequently purified using a Ni2+-NTA-agarose column. As controls, RpoA-His6 alone, the crude extract alone as well a crude extract preincubated with SigA-His6 was purified in the same manner. Fig. 4C shows the results of these experiments. It can be clearly seen that only in the case where the crude extract was preincubated with RpoA, a new band emerges that corresponds to native CcpN. As expected, this band runs marginally faster than the purified CcpN because of the lack of the His tag used for CcpN purification. Taking these results and the far Western blot together, one can conclude that CcpN is able to specifically interact with the α-subunit of RNA polymerase.
DISCUSSION
Repression Mechanism of CcpN
Here, we report the elucidation of the repression mechanism employed by the transcriptional repressor CcpN from B. subtilis. Gel shift assays demonstrated that CcpN does not prevent RNA polymerase binding and that both proteins can bind simultaneously to the promoter. Interestingly, CcpN and RNA polymerase, although able to bind simultaneously, appear to compete for binding to the used DNA fragments. Fig. 1 clearly shows that the bands for all three complexes are significantly weaker when both proteins are present than the complexes where only one of the proteins is present. Because CcpN and RNA polymerase concentrations have been chosen to reflect their actual concentrations in vivo 24),3 it is conceivable that there is also a competition between these two proteins for promoter binding within the cell. This finding would also explain the observations made by Servant et al. (3) who reported a significant derepression of the pckA and gapB promoters in a ccpN knock-out mutant, even under gluconeogenic conditions where CcpN is not active, a feature that was also reported for other transcriptional repressors, although not to such a huge extent (25).
Repressors that bind simultaneously with RNA polymerase, either at overlapping or at different sites, often repress transcription by preventing melting of DNA at the transcriptional start site, i.e. formation of the open complex. Such transcription factors are for example E. coli MerR at the merT promoter (26, 27), which binds together with RNAP at opposite sites of the DNA helix, or the KorB protein of broad host range plasmid RK2 (28), whose binding sites do not overlap those of RNAP. CcpN features both versions of operator sites; some overlap with RNAP binding sites whereas some do not (5). However, open complex formation assays clearly ruled out the possibility that CcpN acts by preventing DNA melting at the sr1 and pckA and probably at the gapB promoter.
The inhibition of the synthesis of abortive transcripts, as observed by us at the gapB promoter, is a case rarely reported in literature. The H-NS protein at the rrnB P1 promoter or the FIS protein at the gyrB promoter are two examples for this kind of repression (29, 30). For H-NS, a binding pattern similar to CcpN has been reported, where the operator overlaps the RNAP binding site. H-NS is then able to alter the DNA structure at this position, allowing the formation of open complexes, but preventing subsequent transcription. A similar mode of action is conceivable for CcpN at the gapB promoter, although an inhibition of open complex formation by CcpN under conditions where open complexes are formed efficiently cannot be ruled out. DNase I footprints have revealed the appearance of several hypersensitive sites upon CcpN binding at this promoter, which is usually a good indication for structural alterations of the DNA (3, 4). At the sr1 and pckA promoter, however, abortive transcripts are readily formed, but escape of RNA polymerase from the promoter is inhibited. Prevention of promoter clearance is usually mediated by one of two different ways: A repressor can bind downstream of RNAP and simply create a roadblock before a stable elongation complex can be formed. This has for example been shown for CcpA-mediated regulation of the treP gene in B. subtilis, and even as a proof of principle with an artificial construct using the Lac repressor (31, 32). Regarding the operator positions at the sr1 and pckA promoters, this mechanism appears to be highly unlikely, which favors the second alternative possibility: An interaction between the repressor molecule and parts of the RNA polymerase. It is known that the polymerase can be stalled at promoters with close-to-consensus sequences, resulting from a extremely tight binding that subsequently makes promoter clearance very difficult (33). Transcriptional repressors, which usually bind their operator sequences with high affinity, can mimic the aforementioned effect by binding RNAP and keeping it in place. Examples for this mechanism include the phage Φ29 protein p4 at the phage A2c promoter (34) and the Gal repressor (35).
CcpN Interacts with the RNAP α-Subunit
With respect to our finding that CcpN is able to specifically interact with the RNA polymerase α-subunit, we conclude that CcpN acts as a repressor at the sr1 and pckA promoters by keeping RNAP in place through the aforementioned interaction. There are various reports about the α-subunit, and especially the C-terminal domain, being an interaction interface for transcriptional repressors, as mentioned above. However, interactions with the α-subunit have also been reported for activators, like CcpA at the ackA promoter (17, 36) or SoxS during oxidative stress conditions (38). Considering the binding site position of CcpN at the sr1 and pckA promoters, an interaction with the α-subunit appears very conceivable. It has been shown that up elements in B. subtilis have a slightly broader tolerance regarding location than in E. coli (39, 40), reaching approximately from −40 to −66, which would position the α-C-terminal domain to be able to interact with CcpN at these promoters.
At the gapB promoter, however, an interaction with the α-subunit can be excluded, because both operator sites are to far downstream to allow any contact between the two proteins. Two possibilities are conceivable for how CcpN exerts its action here: Either CcpN alters the DNA structure as mentioned above, or it interacts with an RNAP subunit other than the α-subunit or the σ-factor, because the first one cannot be contacted and no interaction has been detected with the latter. This ultimately leads to the inability to produce transcripts, probably by inhibiting transcription initiation, although an inhibition of open complex formation under different conditions is also conceivable. Reports of transcription factors that interact with e.g. the β-subunit are quite uncommon. One of these examples is the AsiA protein from bacteriophage T4 (41), another being the Rsd protein of E. coli (16), both of which have been shown to be able to interact with the core RNA polymerase. If CcpN actually interacts with parts of the RNAP other than the α-subunit, this needs to be experimentally determined. However, the relatively small size of CcpN, leaving little space for extensive interaction surfaces and the fact that DNA structure is altered upon CcpN binding, seem to favor the possibility of repression by DNA structure rearrangements.
The example of CcpN shows that one single repressor can exert repression in very different ways, depending on how its operators are positioned relative to the RNA polymerase binding sites. Varying binding site distribution is quite common, for example as found in the case of CytR from E. coli (37) and many more. Interestingly, cases where variations in operator positioning result in different repression mechanisms have not been frequently reported in literature. However, this is mostly because the actual repression mechanism for these proteins has not been elucidated. A well documented example where operator site positions have an impact on the repression mechanism is cre element positioning, allowing CcpA to exert a broad range of repression or even activation mechanisms on its targets (17).
Taking all results together, a quite clear picture of the repression mechanism of CcpN can be established where CcpN and the α-subunits are in a spatial position that allows interaction and subsequent promoter arrest at the sr1 and pckA promoters, but not at the gapB promoter. Here, repression by modification of the DNA structure appears to be a probable alternative.
Supplementary Material
Acknowledgments
We thank M. Salas for the gift of native purified B. subtilis RNA polymerase as well as for the strain for the overproduction of His-tagged α-subunit. Furthermore, we would like to thank M. Fujita who kindly sent us the strain for overproduction of His-tagged σ-factor.
This work was supported by Grant BR1552/6-3 from the Deutsche Forschungsgemeinschaft (to S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
A. Licht, unpublished observation.
- RNAP
- RNA polymerase
- NTA
- nitrilotriacetic acid
- DEPC
- diethylpyrocarbonate.
REFERENCES
- 1.Fillinger S., Boschi-Muller S., Azza S., Dervyn E., Branlant G., Aymerich S. (2000) J. Biol. Chem. 275, 14031–14037 [DOI] [PubMed] [Google Scholar]
- 2.Yoshida K., Kobayashi K., Miwa Y., Kang C. M., Matsunaga M., Yamaguchi H., Tojo S., Yamamoto M., Nishi R., Ogasawara N., Nakayama T., Fujita Y. (2001) Nucleic Acids Res. 29, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Servant P., Le Coq D., Aymerich S. (2005) Mol. Microbiol. 55, 1435–1451 [DOI] [PubMed] [Google Scholar]
- 4.Licht A., Preis S., Brantl S. (2005) Mol. Microbiol. 58, 189–206 [DOI] [PubMed] [Google Scholar]
- 5.Licht A., Brantl S. (2006) J. Mol. Biol. 364, 434–448 [DOI] [PubMed] [Google Scholar]
- 6.Licht A., Golbik R., Brantl S. (2008) J. Mol. Biol. 380, 17–30 [DOI] [PubMed] [Google Scholar]
- 7.Zorrilla S., Ortega A., Chaix D., Alfonso C., Rivas G., Aymerich S., Lillo M. P., Declerck N., Royer C. A. (2008) Biophys. J. 95, 4403–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tännler S., Fischer E., Le Coq D., Doan T., Jamet E., Sauer U., Aymerich S. (2008) J. Bacteriol. 190, 6178–6187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tännler S., Zamboni N., Kiraly C., Aymerich S., Sauer U. (2008) Metab. Eng. 10, 216–226 [DOI] [PubMed] [Google Scholar]
- 10.Record M. T., Jr., Reznikoff W. S., Craig M. L., McQuade K. L., Schlax P. J. (1996) Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, American Society for Microbiology, Washington DC [Google Scholar]
- 11.Escolar L., Pérez-Martín J., de Lorenzo V. (1998) J. Bacteriol. 180, 2579–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene E. A., Spiegelman G. B. (1996) J. Biol. Chem. 271, 11455–11461 [DOI] [PubMed] [Google Scholar]
- 13.Monsalve M., Mencía M., Salas M., Rojo F. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8913–8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojo F. (2001) Curr. Opin. Microbiol. 4, 145–151 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Nakano S., Choi S. Y., Zuber P. (2006) J. Bacteriol. 188, 4300–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilag L. L., Westblade L. F., Deshayes C., Kolb A., Busby S. J., Robinson C. V. (2004) Structure 12, 269–275 [DOI] [PubMed] [Google Scholar]
- 17.Kim J. H., Yang Y. K., Chambliss G. H. (2005) Mol. Microbiol. 56, 155–162 [DOI] [PubMed] [Google Scholar]
- 18.Hughes K. T., Methee K. (1998) Annu. Rev. Microbiol. 52, 231–286 [DOI] [PubMed] [Google Scholar]
- 19.Campbell E. A., Westblade L. F., Darst S. A. (2008) Curr. Opin. Microbiol. 11, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita M., Sadaie Y. (1998) Gene 221, 185–190 [DOI] [PubMed] [Google Scholar]
- 21.Kawamura F., Doi R. H. (1984) J. Bacteriol. 160, 442–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita M., Sadaie Y. (1998) J. Biochem. 124, 89–97 [DOI] [PubMed] [Google Scholar]
- 23.Scholten P. M., Nordheim A. (1986) Nucleic Acids Res. 14, 3981–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner R. (2000) Transcriptional Regulation in Prokaryotes, Oxford University Press, New York [Google Scholar]
- 25.Barragán M. J., Blázquez B., Zamarro M. T., Mancheño J. M., García J. L., Díaz E., Carmona M. (2005) J. Biol. Chem. 280, 10683–10694 [DOI] [PubMed] [Google Scholar]
- 26.Heltzel A., Lee I. W., Totis P. A., Summers A. O. (1990) Biochemistry 29, 9572–9584 [DOI] [PubMed] [Google Scholar]
- 27.Summers A. O. (1992) J. Bacteriol. 174, 3097–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams D. R., Motallebi-Veshareh M., Thomas C. M. (1993) Nucleic Acids Res. 21, 1141–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schröder O., Wagner R. (2000) J. Mol. Biol. 298, 737–748 [DOI] [PubMed] [Google Scholar]
- 30.Schneider R., Travers A., Kutateladze T., Muskhelishvili G. (1999) Mol. Microbiol. 34, 953–964 [DOI] [PubMed] [Google Scholar]
- 31.Ujiie H., Matsutani T., Tomatsu H., Fujihara A., Ushida C., Miwa Y., Fujita Y., Himeno H., Muto A. (2009) J. Biochem. 145, 59–66 [DOI] [PubMed] [Google Scholar]
- 32.Lopez P. J., Guillerez J., Sousa R., Dreyfus M. (1998) J. Mol. Biol. 276, 861–875 [DOI] [PubMed] [Google Scholar]
- 33.Ellinger T., Behnke D., Knaus R., Bujard H., Gralla J. D. (1994) J. Mol. Biol. 239, 466–475 [DOI] [PubMed] [Google Scholar]
- 34.Monsalve M., Mencia M., Rojo F., Salas M. (1996) EMBO J. 15, 383–391 [PMC free article] [PubMed] [Google Scholar]
- 35.Choy H. E., Park S. W., Aki T., Parrack P., Fujita N., Ishihama A., Adhya S. (1995) EMBO J. 14, 4523–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turinsky A. J., Grundy F. J., Kim J. H., Chambliss G. H., Henkin T. M. (1998) J. Bacteriol. 180, 5961–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collado-Vides J., Magasanik B., Gralla J. D. (1991) Microbiol. Rev. 55, 371–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah I. M., Wolf R. E., Jr. (2004) J. Mol. Biol. 343, 513–532 [DOI] [PubMed] [Google Scholar]
- 39.Gourse R. L., Ross W., Gaal T. (2000) Mol. Microbiol. 37, 687–695 [DOI] [PubMed] [Google Scholar]
- 40.Meijer W. J., Salas M. (2004) Nucleic Acids Res. 32, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Severinova E., Severinov K., Darst S. A. (1998) J. Mol. Biol. 279, 9–18 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.