Abstract
Aldosterone and endothelin-1 (ET-1) act on collecting duct cells of the kidney and are important regulators of renal sodium transport and cardiovascular physiology. We recently identified the ET-1 gene (edn1) as a novel aldosterone-induced transcript. However, aldosterone action on edn1 has not been characterized at the present time. In this report, we show that aldosterone stimulated edn1 mRNA in acutely isolated rat inner medullary collecting duct cells ex vivo and ET-1 peptide in rat inner medulla in vivo. Aldosterone induction of edn1 mRNA occurred in cortical, outer medullary, and inner medullary collecting duct cells in vitro. Inspection of the edn1 promoter revealed two putative hormone response elements. Levels of heterogeneous nuclear RNA synthesis demonstrated that edn1 mRNA stimulation occurred at the level of transcription. RNA knockdowns corroborated pharmacological studies and demonstrated both mineralocorticoid receptor and glucocorticoid receptor participated in this response. Aldosterone resulted in dose-dependent nuclear translocation and binding of mineralocorticoid receptor and glucocorticoid receptor to the edn1 hormone response elements. Hormone receptors mediated the association of chromatin remodeling complexes, histone modification, and RNA polymerase II at the edn1 promoter. Direct interaction between aldosterone and ET-1 has important implications for renal and cardiovascular function.
The steroid hormone aldosterone is critical for sodium homeostasis and blood pressure control. Aldosterone works by modulating the fine regulation of sodium reabsorption in the distal nephron and collecting duct of the kidney. Classic aldosterone action is mediated through the mineralocorticoid receptor (MR),3 a member of the nuclear receptor family of proteins that function as ligand-dependent transcription factors (1). MR acts on cells of the distal nephron and collecting duct to stimulate transcription of genes involved in transepithelial sodium transport, including the epithelial sodium channel α subunit (scnn1a, αENaC), the sodium, potassium-ATPase α1 subunit (atp1a1), and the serum- and glucocorticoid-regulated kinase-1 (sgk1) (2). The increase in expression of genes involved in sodium transport results in net sodium reabsorption followed by in increase in extracellular fluid volume and a consequent increase in blood pressure. Indeed, MR antagonists such as spironolactone and eplerenone are used clinically as diuretic and anti-hypertensive agents (3).
The mechanism of MR action is consistent with a classic steroid receptor mechanism (4). Prior to activation MR resides in the cytosol. Ligand binding induces a conformational change that releases chaperone proteins and reveals a nuclear localization signal. Nuclear MR binds directly to DNA at hormone response elements (HREs) in target genes to modulate their transcription. A typical HRE for MR consists of two receptor binding half-sites with the consensus sequence 5′-TGTTCT-3′, arranged as an inverted palindrome (1, 5). These HREs facilitate binding of steroid receptors in a dimeric conformation. Once bound to a gene promoter MR serves as a molecular platform for the recruitment of transcription factors such as the steroid receptor coactivator-1 (SRC-1) and RNA polymerase II (6–8).
MR is highly homologous to the glucocorticoid receptor (GR) (1) and can bind glucocorticoids with equal affinity to aldosterone. However, inappropriate glucocorticoid activation of MR can lead to severe hypertension (9). Aldosterone-responsive cells are protected from glucocorticoids by the activity of 11β-hydroxysteroid dehydrogenase type 2, an enzyme that converts glucocorticoids into 11-ketosteroids that have very little affinity for MR or other steroid receptors (10, 11). Alternatively, GR can bind mineralocorticoids (Kd = 14–60 nm) and may contribute to aldosterone action (1). MR and GR share 94% homology in their DNA binding domains and have conserved amino acids at each residue shown to make direct contacts with DNA (1, 12). Indeed, MR and GR are known to bind to the same HRE in several genes (13–18).
Previously, we identified endothelin-1 (edn1) as a transcript and protein that increases in response to aldosterone in inner medullary collecting duct (mIMCD-3) cells (19). Similarly, this interaction has been documented in whole kidney extracts from rat (20). The gene product of edn1 is a 212-amino acid prepropeptide that is enzymatically processed to form the biologically active 21-amino acid peptide, ET-1. ET-1 plays a complex role in cardiovascular and renal physiology. Several reports have demonstrated aldosterone induction of edn1 in vascular smooth muscle and cardiac tissue (21, 22). In these cell types, systemic ET-1 is largely vasoconstrictive and profibrotic. Indeed, both hormones have been implicated in cardiac and renal fibrosis, glomerular damage, and proteinuric disease (10, 23, 24). In the kidney, ET-1 has effects on renal hemodynamics (25, 26), sodium and water homeostasis (27–31), and acid-base balance (32). These same processes are also influenced by aldosterone. However, renal ET-1 is a well documented natriuretic peptide that directly blocks sodium transport in the tubule collecting duct (28, 33–37). The physiological importance of collecting duct ET-1 is emphasized by the fact that collecting duct cell-specific edn1 knock-out mice exhibit salt-sensitive hypertension (31). Thus, in the renal collecting duct, the actions of aldosterone and ET-1 on sodium transport directly oppose each other.
The goal of the present report was to characterize aldosterone regulation of edn1 in the renal collecting duct. Indeed, studies presented here demonstrate aldosterone-stimulated edn1 in cortical, outer medullary, and inner medullary collecting duct cells in vitro and edn1 and ET-1 peptide in the rat kidney in vivo. Putative HREs in the edn1 promoter were identified. These elements were evaluated for the recruitment of hormone receptors and the assembly of an aldosterone-dependent transcription complex.
MATERIALS AND METHODS
Chemicals
Aldosterone (Fluka), spironolactone and RU486 (Sigma) were prepared in 100% ethanol and stored at −20 °C. Collagenase type I (MP Biomedicals), hyaluronidase type IV (Sigma), and DNase I (Sigma) were prepared fresh on the day of the experiment.
Animals
Male Sprague-Dawley rats (300–350 g) were obtained from Harlan and housed at the University of Florida Animal Care Services rodent facilities. Standard rat chow and tap water were provided ad libitum. All procedures adhered to the Animal Care Services guidelines and were approved by the University of Florida Institutional Animal Care and Use Committee.
Acute Isolation of Rat IMCD Cells
Rats were euthanized by sodium pentobarbital (50 mg kg−1 body weight) and cervical dislocation. Kidneys were immediately removed, and the inner medulla was carefully dissected for acute IMCD isolation according to Stricklett et al. (39). In brief, minced tissue was incubated at 37 °C in a digestion solution containing collagenase type I (3 mg/ml), hyaluronidase type IV (2 mg/ml), and DNase I (0.1 mg/ml). Digested IMCD cells were collected by centrifugation through a sucrose buffer. Isolated IMCDs were resuspended in Dulbecco's modified Eagle's medium/F-12 and equilibrated in a 5% CO2 incubator at 37 °C for 20 min. Inner medullas from the left and right kidney of each rat were processed in tandem, but separately, to allow for a paired analysis between vehicle (0.04% ethanol) and 1 μm aldosterone treatments. Thus, each rat served as its own internal control. After 1 h cells were immediately pelleted by gentle centrifugation at 4 °C and resuspended in TRIzol® Reagent (Invitrogen) for RNA isolation as described below.
Aldosterone Administration in Rat and ET-peptide Measurement
Rats were given an intraperitoneal injection (1 ml kg−1 body weight) of aldosterone (1 mg kg−1) or vehicle (2% ethanol in saline). After 2 h rats were anesthetized with inhaled isoflurane, and kidneys were flushed by an aortic perfusion of ice-cold phosphate-buffered saline with the vena cava vented. Kidneys were removed and dissected into cortex, outer medulla, and inner medulla. Tissues were immediately snap frozen in liquid nitrogen and stored at −80 °C until use. ET-1 was extracted from renal tissues using a protocol originally described by Yorikane et al. (40). Immunoreactive ET-1 peptide was detected by chemiluminescent enzyme-linked immunosorbent assay (R&D Systems) and normalized to total protein content as determined by standard protein assays (Bio-Rad).
Cell Culture and Aldosterone Treatment
All cells were maintained in Dulbecco's modified Eagle's medium/F-12 plus 10% fetal bovine serum and 50 μg/ml gentamicin. The mpkCCDc14 cells were a kind gift of Dr. Alain Vandewalle (41), OMCD1 cells were a kind gift of Dr. Thomas DuBose (42), IMCD-K2 cells were a gift of Dr. Bruce Stanton (43), and mIMCD-3 cells were purchased from ATCC. For all hormone experiments cells were grown 24 h past confluency and changed to Dulbecco's modified Eagle's medium/F-12 plus 10% charcoal-dextran-stripped fetal bovine serum (Invitrogen). After 24 h, cells were treated with vehicle (ethanol) or aldosterone (0.01–1 μm) for 1 h.
Steady-state mRNA Determination
Hormone studies were conducted as described above on growth-arrested confluent monolayers grown in 6-well Costar Transwell plates (Corning). The final concentration of ethanol in all treatments was 0.1%. Total RNA (2 μg) was isolated from cells using TRIzol® Reagent (Invitrogen), treated with DNase I (Ambion) to eliminate genomic DNA, and reverse transcribed using oligo(dT), random hexamers, and SuperscriptTM III (Invitrogen). No reverse transcriptase served as a negative control. Resulting cDNAs (32 ng) were used as templates in duplicate QPCR reactions (Applied Biosystems). Cycle threshold (Ct) values were normalized against β-actin (actb), and relative quantification was performed using the ΔΔCt method (38). Primer/probe sets for edn1, sgk1, and actb were purchased from Applied Biosystems: rat edn1 (Rn00561129_m1), rat sgk1 (Rn00570285_m1), rat actb (4352931E), mouse edn1 (Mm00438656_m1), mouse sgk1 (Mm00441380_m1), and mouse actb (Mm00607939_s1).
Heterogeneous Nuclear RNA Assay
As described by Lipson and Baserga (44), levels of unspliced edn1 hnRNA were detected using primers designed to amplify a region between exon 4 and intron 4 (forward: 5′-gaagtgtatctatcagcagctgg-3′; reverse: 5′-agaccatgacgactctattactgg-3′). QPCR reactions were set up with 32 ng of cDNA, 200 nm of each primer, and SYBR Green mastermix (Bio-Rad). Expression of hnRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase (gapdh) mRNA (forward primer: 5′-gaagcccatcaccatcttcc-3′; reverse primer: 5′-tgatgatccttttggctcc-3′). Edn1 hnRNA and gapdh primers were validated for QPCR over 4 orders of magnitude of input cDNA for similar PCR efficiency (data not shown). Melting curves (55–95 °C) and agarose gel electrophoresis were used to verify product size.
Nuclear Translocation Western Blots
Hormone experiments were conducted in mIMCD-3 cells as described above except that cells were grown in 150-mm dishes (Corning). Cells were treated with vehicle (0.15% ethanol), aldosterone, antagonist, or aldosterone plus antagonist. Antagonists, spironolactone and RU486, were supplied at the final concentration of 10 μm in each experiment. In some cases, cells were pretreated with antagonists 1 h prior to aldosterone treatment. Cytoplasmic and nuclear extracts were obtained using the NE-PER® Reagents (Pierce Biotechnology). Protein concentrations were determined using the Bradford assay, and 90 μg was separated on a 7.5% SDS-PAGE Ready gel (Bio-Rad). Proteins were transferred to polyvinylidene difluoride overnight and visualized by Ponceau S. Membranes were blocked with 2% Rodeo blocker plus 0.05% saddle soap (U. S. Biochemical Corp.) in Tris-buffered saline. The monoclonal MR antibody was a kind gift of Drs. Elise and Celso Gomez-Sanchez (45). Detailed information on all primary and secondary antibodies used is listed in supplemental Table 1. Blots were washed with blocking solution and developed with RodeoTM ECL Western blot Detection Reagents (U. S. Biochemical Corp.). Densitometric analysis was performed with Quantity One® software (Bio-Rad).
For each nuclear translocation experiment blots were stripped using 2% SDS plus 10% β-mercaptoethanol for 30 min at 70 °C. Blots were redeveloped to ensure completely antibody stripping, washed, and reprobed using another primary antibody.
Hormone Receptor Knockdown
MR-siRNA (J-061269-09 NR3C2), GR-siRNA (J-045970-10 NR3C1), and control non-targeting siRNA (#2 D-001210-02-05) were purchased from Dharmacon (Lafayette, CO). Cells were seeded at a density of 75,000 cells per cm2 on 6-well Transwell plates (Corning) and transfected for 24 h with 2 μm siRNA in 6 μl of DharmaFect 4. At the time of transfection cells were switched to phenol-red free Dulbecco's modified Eagle's medium/F-12 plus 10% charcoal-dextran-stripped fetal bovine serum. After 24 h the cells were treated with 1 μm aldosterone or vehicle for 1 h. RNA was extracted and processed as described above for QPCR.
Chromatin Immunoprecipitation Assays
ChIP assays were performed as previously described by Leach et al. (46). Briefly, cells were fixed with 1% formaldehyde and quenched with glycine. Nuclei were isolated, and DNA was sonicated to ∼500 bp. Fragment length was verified by gel electrophoresis. Specific antibodies (supplemental Table 1) were used to immunoprecipitate DNA-protein complexes on bovine serum albumin-blocked protein-A-Sepharose beads. Cross-links were reversed, and DNA fractions were analyzed for bound edn1 by PCR. Primers: forward 5′-tctgatcggcgatactaggg-3′ and reverse 5′-cgctcttgaatcccagctac-3′, amplify a 235-bp region containing putative HREs (Fig. 5A). Standard PCR products were visualized with SYBR Green on a 5% Tris-borate-EDTA gel. Alternatively, bound edn1 was quantified by QPCR using SYBR Green mastermix (Bio-Rad). Values were normalized to total input DNA and are expressed as -fold change relative to control. Primers were validated for quantification by analyzing PCR efficiency over a serial dilution of input DNA. Melting curves confirmed specific PCR products and melting temperatures (data not shown).
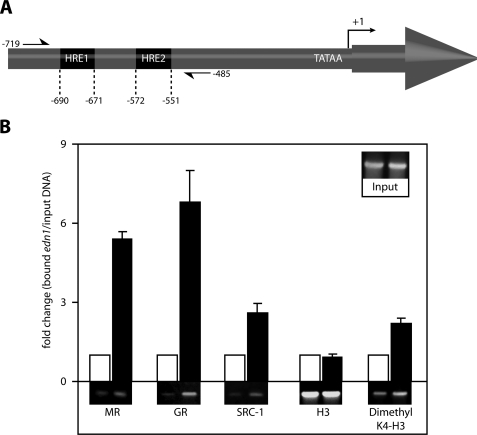
FIGURE 5.
Aldosterone recruits steroid receptors to a region of the murine edn1 promoter that contains two putative HREs. A, diagram of the edn1 promoter indicating the position of the identified HREs relative to the TATAA-box and transcriptional start site (+1) (not to scale). Primers used in ChIP assays flank both HREs (−719 to −485) and are indicated with half arrows. B, ChIP assays were used to detect protein binding to the edn1 promoter in vehicle (open bars) or 1 μm aldosterone (closed bars) treated mIMCD-3 cells. Antibodies used for immunoprecipitation are indicated below. Bound edn1 DNA was quantified by SYBR Green QPCR. Values were normalized to total input DNA (INPUT) and expressed as mean -fold change relative to vehicle ± S.E. Additionally, standard PCR products were run on a 5% TBE gel and imaged with SYBR green dye. Representative gels are displayed below their corresponding QPCR values (n ≥ 3).
Coimmunoprecipitation
Cytosolic and nuclear extracts (175 μg) obtained from vehicle- and aldosterone-treated mIMCD-3 cells were diluted to a final volume of 250 μl in phosphate-buffered saline with fresh protease inhibitors (Roche Applied Science). Diluted samples were pre-cleared with 30 μl of bovine serum albumin-blocked protein-A Sepharose beads and 0.4 μg of normal mouse IgG (Santa Cruz Biotechnology) for 30 min at room temperature with end-over-end rotation. Following gentle centrifugation supernatants (240 μl) were collected and subjected to immunoprecipitation with anti-MR or anti-GR (see supplemental Table 1) and 40 μl of blocked protein-A-Sepharose beads for 1 h. Beads were pelleted and washed three times with ice-cold phosphate-buffered saline plus protease inhibitors. Wash supernatants were removed with flat-head gel-loading tips (USA Scientific) after each wash. Washed samples were resuspended in 40 μl of 2× lithium dodecylsulfate plus 10% β-mercaptoethanol, boiled for 5 min, and subjected to Western blotting as described above.
DNA-affinity Purification Analysis
Cytosolic and nuclear extracts obtained from mIMCD-3 cells as described above were subjected to DNA-affinity purification analysis (DAPA) as described by Deng et al. (47). Double-stranded DNA probes were biotinylated on 5′-ends (Sigma Genosys) and were homologous to HRE 1 (half-sites underlined): 5′-agacttggtggaaggggtggtggtggaaaagt-3′ or HRE 2: 5′-ggatgtacctgacaaaaccacattgttgttgttatc-3′ in the edn1 promoter (Fig. 6, A and B). Probes were immobilized on 50 μl of streptavidin-coated agarose beads and incubated with 175 μg of cellular extract in the presence of freshly prepared protease inhibitors (Roche Applied Science) for 1 h at room temperature with end-over-end rotation. Beads were pelleted. Supernatants were removed and used for input controls by Western blotting for actin. Pelleted beads were washed four times with ice-cold phosphate-buffered saline plus protease inhibitors. After the final wash, all liquid was aspirated from the beads with flat-headed gel loading tips, and 50 μl of 2× lithium dodecylsulfate (Invitrogen) plus β-mercaptoethanol. Samples were boiled for 5 min, chilled on ice, and loaded onto a 7.5% Tris-HCl SDS-PAGE Ready Gel (Bio-Rad) for electrophoresis. Purified proteins were identified by Western analysis as described above except that blots were washed with Tris-buffered saline plus 0.05% saddle soap without blocking reagent. Equal loading was controlled for by Bradford assay, input control Western blots against actin, and reprobing DAPA blots with actin.
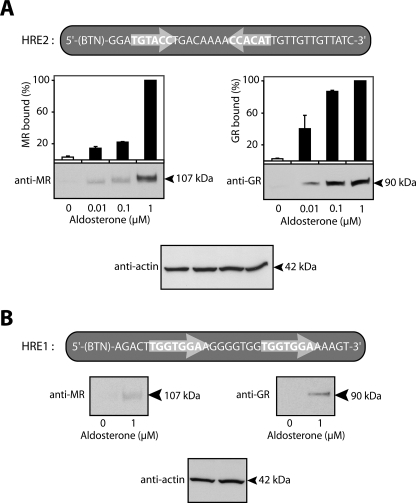
FIGURE 6.
High resolution mapping of steroid receptor binding to HRE1 and HRE2 in the edn1 promoter. DAPA experiments were performed on nuclear extracts from vehicle- and aldosterone-treated mIMCD-3 cells to map receptor binding to either HRE with higher resolution. Sequences of the HRE1 and HRE2 DAPA probes are indicated above their corresponding immunoblots. Half-site sequences and orientation are indicated with arrows. A, representative immunoblots following DAPA experiments using HRE2 as bait are shown for MR (left panel) and GR (right panel). Immunoblots were quantified by densitometry. Values were normalized by setting total intensity units calculated for MR (or GR) in the presence of 1 μm aldosterone to 100%. Equal loading was verified by immunoblot against actin (bottom panel) (n ≥ 3). B, representative immunoblots are shown for MR (left panel), GR (right panel), and actin (lower panel) from DAPA experiments using HRE1 as bait. Densitometry was not performed because MR and GR were not consistently detected at concentrations of aldosterone lower than 1 μm (n ≥ 3).
Statistics
Data are presented as the means ± S.E. Unless otherwise stated all experiments were performed in duplicate at least three independent times. Statistical significance was calculated using the two-tailed Student's t test, and p < 0.05 was considered significant.
RESULTS
Aldosterone Stimulates ET-1 in Rat Inner Medulla
Renal collecting duct cells are a target cell type for aldosterone action in the body. Indeed, our original report showed stimulation of edn1 mRNA by aldosterone occurred in mIMCD-3 cells (19). To determine if collecting duct cells were also the target cell type in the animal, aldosterone studies were conducted on acutely isolated rat IMCD cells ex vivo. Following a brief equilibration, IMCD cells isolated from a single rat were separated for a paired analysis between vehicle- and 1 μm aldosterone treatments. After 1 h, aldosterone led to a 41 ± 6% increase in edn1 mRNA expression compared with control (Fig. 1A). The observed stimulation in edn1 exceeded the 28 ± 5% increase in the mRNA of the well established aldosterone response gene sgk1 (48, 49).
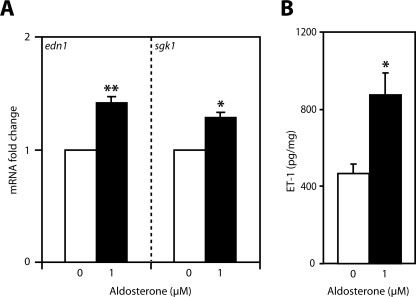
FIGURE 1.
Aldosterone stimulates inner medullary ET-1 expression in rat. A, IMCD cells were acutely isolated from rat and treated with vehicle (open bars) or 1 μm aldosterone (closed bars) for 1 h ex vivo. Levels of edn1 and sgk1 mRNA were determined by QPCR, normalized to β-actin, and expressed as -fold change relative to vehicle ± S.E. (n = 4, *, p < 0.05, **, p < 0.005). B, inner medullary ET-1 peptide levels were measured in kidneys isolated from rats injected with aldosterone (1 mg kg−1 body weight, intraperitoneally) or vehicle for 2 h. Inner medullary ET-1 peptide levels were determined by enzyme-linked immunosorbent assay and normalized to total protein content. Values are expressed as ET-1 (picograms/mg of protein) ± S.E. (n ≥ 8 per group; *, p < 0.05).
Animal studies were extended to investigate the level of ET-1 peptide in rat kidney. Consistent with published reports, basal levels of ET-1 were 50 times greater in the inner medulla compared with cortex or outer medulla (supplemental Table 2) (50). Aldosterone injection (1 mg kg−1 body weight, intraperitoneally) resulted in an approximate 2-fold increase in inner medullary ET-1 levels compared with control (Fig. 1B). The observation that aldosterone stimulated ET-1 in inner medulla in vivo, combined with the known role of ET-1 in modulating renal sodium transport (27, 28), supports the hypothesis that ET-1 is a regulator of aldosterone action in the mammalian kidney.
Aldosterone Stimulates Dose-dependent Transcription of Edn1 in Collecting Duct Cells
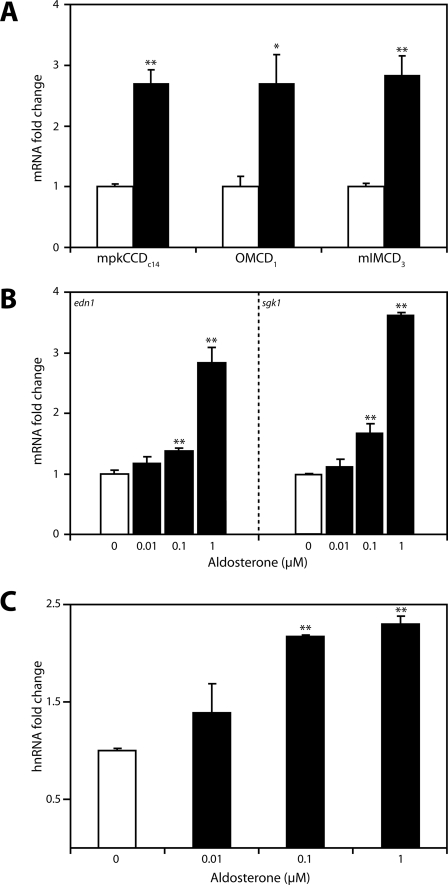
To determine if the stimulation of edn1 by aldosterone was specific to IMCD cells or was a more generalized collecting duct cell response, the effect of aldosterone was evaluated in three renal cell lines thought to be representative of cortical, outer medullary, and inner medullary collecting duct cells. These cell lines were mpkCCDc14 (41), OMCD1 (42), and mIMCD-3 (19, 51), respectively. Aldosterone (1 μm) led to an approximate 3-fold increase in edn1 mRNA at 1 h in each cell line (Fig. 2A). Aldosterone also stimulated a 2.5 ± 0.4-fold increase in edn1 mRNA in mIMCD-K2 cells, an independently derived IMCD cell model (43) (data not shown). Taken together, this evidence indicates that aldosterone induction of edn1 mRNA occurs in multiple collecting duct cells in vitro and is likely to occur along the length of the collecting duct in vivo.
FIGURE 2.
Aldosterone stimulates edn1 mRNA and hnRNA in collecting duct cells. A, growth-arrested confluent monolayers of mpkCCDc14, OMCD1, or mIMCD-3 cells were treated with vehicle (open bars) or 1 μm aldosterone (closed bars) for 1 h. Steady-state edn1 mRNA was measured by QPCR, normalized to β-actin, and expressed as mRNA -fold change relative to vehicle ± S.E. B, aldosterone dose-response studies were conducted on mIMCD-3 cells treated with vehicle or aldosterone (0.01, 0.1, or 1 μm) for 1 h. Steady-state mRNA of edn1 or sgk1 was quantified as above. C, levels of edn1 hnRNA were also determined from dose-response studies conducted on mIMCD-3 cells. hnRNA was measured by SYBR Green QPCR, normalized to gapdh, and expressed as hnRNA -fold change relative to vehicle ± S.E. (n ≥ 3, *, p < 0.05, **, p < 0.005).
To more fully characterize aldosterone induction of edn1, aldosterone dose-response studies were conducted in mIMCD-3 cells. This cell line was selected because the inner medulla is an important site for ET-1 action in vivo and because this cell line has been validated as a model for aldosterone action (52–56). Edn1 mRNA exhibited a dose-dependent increase in the presence of aldosterone that was significant at concentrations as low as 0.1 μm (Fig. 2B). Importantly, mRNA induction of edn1 paralleled that of sgk1 (Fig. 2B), which demonstrated that hormone concentrations were appropriate to reproduce an aldosterone response in vitro. In addition, aldosterone treatment resulted in an increase in functional ET-1 peptide release from mIMCD-3 cells (supplemental Fig. 1), which further validates this cell line as a model of aldosterone action on ET-1.
In general, aldosterone action is mediated at the level of transcription (57). To test the hypothesis that the increase in edn1 mRNA in response to aldosterone also occurred at the level of transcription, the concentration of edn1 hnRNA was determined. Levels of hnRNA can generally be used to measure transcriptional activity of a specific gene, because pre-splicing hnRNA molecules are not subject to factors affecting overall mRNA stability (58, 59). Consistent with levels of steady-state mRNA (Fig. 2B), aldosterone stimulated a dose-dependent increase in edn1 hnRNA (Fig. 2C). Given the transcriptional mechanism of aldosterone, the increase in edn1 hnRNA supports the hypothesis that the induction of edn1 occurs by transcription.
Aldosterone Action on Edn1 Involves Both MR and GR
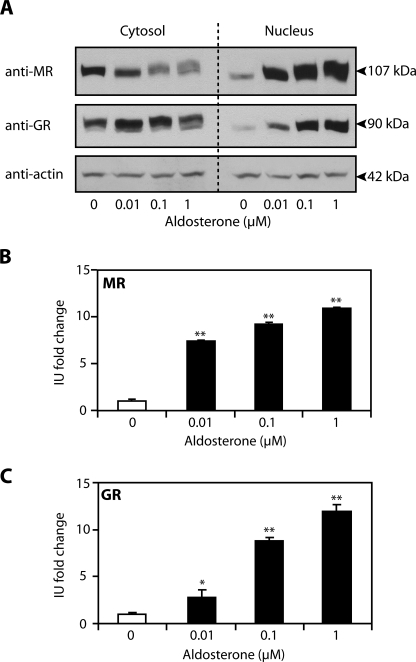
Activation of MR is central to the mechanism by which aldosterone modulates transcription of target genes. However, aldosterone-regulated gene transcription may also be mediated through GR. Therefore, several approaches were used to investigate the contribution of each receptor in mediating aldosterone action. First, a Western blotting approach was adopted to follow nuclear translocation of MR (Fig. 3A, top panel) or GR (Fig. 3A, middle panel) in response to aldosterone treatment. Aldosterone (1 μm) resulted in comparable 10.9 ± 0.2 and 11.9 ± 0.8-fold increases in the abundance of nuclear MR and GR, respectively (Fig. 3, A–C). Furthermore, nuclear translocation of MR and GR was dose-dependent and occurred at concentrations of aldosterone as low as 0.01 μm. Of note, this concentration of hormone failed to stimulate a detectable increase in edn1 or sgk1 mRNA (Fig. 3 versus Fig. 2B). These observations provide strong evidence that aldosterone action is mediated through both MR and GR in mIMCD-3 cells.
FIGURE 3.
Aldosterone action in mIMCD-3 cells is mediated through MR and GR. A, receptor nuclear translocation was followed by Western blot for MR (top panel) and GR (middle panel) on cytosolic and nuclear extracts obtained from mIMCD-3 cells treated with vehicle or aldosterone (0.01, 0.1, or 1 μm) for 1 h. Blots were stripped and reprobed for actin (lower panel) to verify equal loading. B and C, densitometry was used to quantify the relative abundance of MR (B) and GR (C) in the nucleus. Values are expressed as the -fold change in intensity units (IU) relative to vehicle control ± S.E. (n ≥ 3; *, p < 0.05; **, p < 0.005).
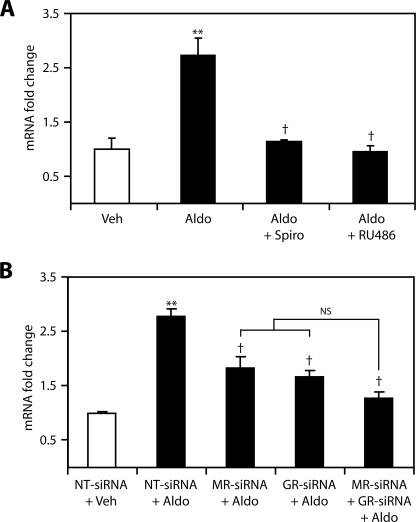
To determine if each receptor was directly involved in edn1 transcription receptor blockade experiments were performed on mIMCD-3 cells. Consistent with previously reported data (19), inhibition of MR or GR with spironolactone or RU486, respectively, completely blocked aldosterone induction of edn1 mRNA (Fig. 4A). To corroborate these findings, siRNA gene silencing was used to specifically knock down MR or GR expression. Independent transfections of MR-siRNA or GR-siRNA resulted in nearly 90% knockdown of their relevant receptor mRNAs in mIMCD-3 cells (80). In the presence of aldosterone, transfection of MR-siRNA or GR-siRNA inhibited the induction of edn1 mRNA by 35 ± 8% and 40 ± 4%, respectively (Fig. 4B). Cotransfection of MR-siRNA and GR-siRNA together resulted in a 54 ± 4% reduction in edn1 mRNA. However, the effect of cotransfection was not significantly additive compared with either siRNA transfected alone (Fig. 4B). The additive trend by siRNA knockdown most likely reflects the different mechanism of receptor inhibition given that pharmacological blockade of either receptor alone completely prevented the induction of edn1. Nevertheless, the observation that targeted inhibition of GR blocked aldosterone-induced edn1 demonstrates that MR was not able to compensate for the loss of GR. Thus, these data indicate that both MR and GR are functionally required for the aldosterone-mediated induction of edn1.
FIGURE 4.
Blockade of MR or GR inhibits aldosterone induction of edn1 mRNA. A, the effect of pharmacological inhibition of MR or GR on aldosterone-stimulated edn1 mRNA was evaluated in mIMCD-3 cells. Cells were treated with vehicle (veh, open bars), 1 μm aldosterone (aldo, closed bars), or 1 μm aldosterone in the presence of either 10 μm spironolactone (spiro) or 10 μm RU486 for 1 h. Edn1 mRNA levels were determined by QPCR as above and are expressed as the mean -fold change relative to vehicle ± S.E. (n ≥ 3; **, p < 0.005 relative to vehicle; †, p < 0.005 relative to aldosterone). B, the effect of siRNA silencing of MR or GR on aldosterone-stimulated edn1 mRNA was evaluated. Cells were transfected with control non-target (NT)-siRNA, MR-siRNA, or GR-siRNA 24 h prior to being treated with vehicle or 1 μm aldosterone for 1 h. Changes in mRNA were measured using QPCR as above. Values are expressed as mean -fold change relative to vehicle-treated cells transfected with NT-siRNA ± S.E. (n = 3; **, p < 0.005 relative to NT-siRNA plus vehicle; †, p < 0.05 relative to NT-siRNA plus aldosterone, NS = not significant).
Aldosterone Modulates Hormone Receptor Binding to the Edn1 Promoter
The observation that both MR and GR were involved in aldosterone-mediated edn1 transcription suggested that the edn1 promoter contained functional HREs. Inspection of 1990 bp of the proximal murine edn1 promoter and 5′-untranslated region revealed two putative HREs designated HRE1 and HRE2 (Fig. 5A). In contrast to a classic element that contains receptor binding half-sites separated by three nucleotides, the identified HREs each have half-sites separated by eight nucleotides. Furthermore, each of the identified HREs are different from one another in that the downstream HRE1 consists of two directly repeated half-sites, whereas the upstream HRE2 has half-sites arranged as an imperfect inverted palindrome (Fig. 6, A and B).
ChIP assays were performed on vehicle or 1 μm aldosterone-treated mIMCD-3 cells in order to determine if MR or GR interacted directly with the edn1 promoter. PCR primers were designed to flank both HREs (Fig. 5A). After 1 h, aldosterone treatment resulted in a 5.4 ± 0.3-fold increase in MR and a 6.8 ± 1.2-fold increase in GR bound to the edn1 promoter (Fig. 5B). Furthermore, the aldosterone-dependent association of MR and GR was accompanied by a 2.6 ± 0.4-fold increase in the transcriptional coactivator SRC-1 associated with the edn1 promoter. Regional histone H3 lysine 4 residues were dimethylated after treatment with aldosterone. This particular histone modification is widely associated with transcriptionally active promoters (60). Taken together, these data are consistent with the concept that the edn1 promoter is more active in the presence of aldosterone.
The association of MR and GR in the same region of the edn1 promoter suggested that the receptors bound directly to one or both of the identified HREs. Accordingly, higher resolution DAPA experiments were employed to map the aldosterone-dependent recruitment of MR and GR to either HRE1 or HRE2. DAPA allows one to use a small region of double-stranded DNA, in this case HRE1 or HRE2 of the edn1 promoter, as bait in an affinity purification protocol to pull down the protein complex bound to the DNA (47). Aldosterone induced dose-dependent association of MR and GR with HRE2 (Fig. 6A). Both receptors were also recruited to HRE1 in the presence of 1 μm aldosterone (Fig. 6B). However, the relative abundance of either receptor recruited to HRE1 was ∼10% of the total abundance of MR or GR bound to HRE2. Furthermore, receptors could not be consistently detected at lower concentrations of hormone suggesting that the upstream HRE2 is the primary response element governing edn1 induction by aldosterone.
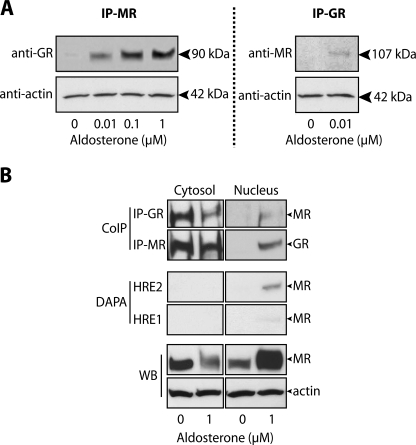
The observation that both MR and GR interacted with the same HRE suggested that both receptors might also be in the same transcription complex in the nucleus. Indeed, coimmunoprecipitation experiments performed on mIMCD-3 cells revealed that MR and GR were present in the same protein complex in the nucleus of aldosterone but not vehicle-treated cells (Fig. 7A). The interaction of MR and GR in the nucleus was dose-dependent and occurred at concentrations of aldosterone as low as 0.01 μm. Coimmunoprecipitation experiments were also conducted on cytosolic extracts from vehicle- and 1 μm aldosterone-treated mIMCD-3 cells (Fig. 7B, top panels). Both MR and GR were detected in the same protein complex in the presence or absence of aldosterone in the cytosol. Conversely, neither MR nor GR were precipitated from cytosolic extracts by DAPA using either HRE as bait (Fig. 7B, middle panels). Together these data indicate that MR and GR are in the same protein complex in the cytosol prior to hormone activation. However, the association of either MR or GR with DNA is exclusive to aldosterone-activated receptors localized in the nucleus.
FIGURE 7.
Aldosterone-dependent association of MR and GR by coimmunoprecipitation. A, nuclear extracts from mIMCD-3 cells treated with vehicle or aldosterone were subjected to coimmunoprecipitation (IP) with anti-MR and subsequently immunoblotted for GR. As a control, nuclear extracts from vehicle- and 1 μm aldosterone-treated cells were subjected to the reverse immunoprecipitation by anti-GR followed by immunoblot against anti-MR (n ≥ 3). B, coimmunoprecipitation (CoIP), and DAPA experiments were conducted on cytosolic extracts (left panels) from mIMCD-3 cells treated with vehicle or 1 μm aldosterone. Parallel experiments conducted on nuclear extracts are shown as a reference (right panels). Coimmunoprecipitation experiments were conducted as above using either anti-GR or anti-MR as bait. Similarly, cellular extracts were subjected to DAPA using either HRE2 or HRE1 as bait. Western blots against MR and actin are shown as controls.
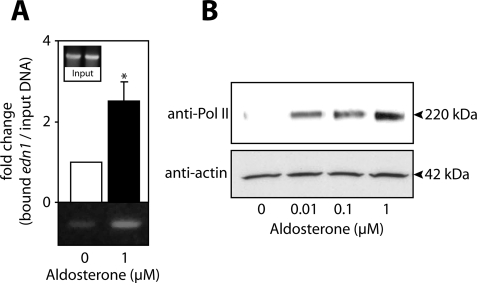
To test whether aldosterone-activated MR and GR could recruit RNA polymerase II to the edn1 promoter, ChIP and DAPA experiments were performed. ChIP analysis revealed that RNA polymerase II was present on the edn1 promoter in aldosterone-treated cells at levels 2.5 ± 0.5-fold higher than control (Fig. 8A). Likewise, DAPA experiments showed dose-dependent recruitment of RNA polymerase II to HRE2 (Fig. 8B). In summary, aldosterone stimulates edn1 in collecting duct cells by a mechanism involving the assembly of a transcription complex at HRE2 in the edn1 promoter that contains MR, GR, SRC-1, and RNA polymerase II.
FIGURE 8.
Aldosterone-dependent recruitment of RNA polymerase II to the edn1 promoter. A, ChIP assays were used to detect RNA polymerase II (Pol II) binding to the edn1 promoter in vehicle (open bars)- or 1 μm aldosterone (closed bars)-treated mIMCD-3 cells. Bound edn1 DNA was quantified as above by SYBR Green QPCR or imaged by running standard PCR products on a 5% TBE gel. QPCR values are normalized to total input DNA and expressed as mean -fold change relative to vehicle ± S.E. (n ≥ 3). B, dose-dependent recruitment of RNA polymerase II to HRE2 was evaluated by DAPA on vehicle- and aldosterone-treated mIMCD-3 cells. Representative immunoblots for Pol II and actin loading controls are shown (n = 3).
DISCUSSION
In this report the regulation of edn1 by aldosterone was characterized in renal collecting duct cells. Aldosterone stimulated edn1 in rat IMCD cells ex vivo, as well as four independent collecting duct cell models in vitro. We report the first direct evidence of aldosterone induction of ET-1 peptide in rat inner medulla in vivo. Coimmunoprecipitation experiments showed that MR and GR were present in the same protein complex in the cytosol prior to hormone activation. Nuclear translocation, pharmacological inhibition, siRNA silencing, ChIP, and DAPA experiments all demonstrated that both MR and GR were involved in mediating aldosterone action on the edn1 gene. Receptors bound directly to the edn1 promoter to facilitate the assembly of a transcription complex that included the transcription coactivator SRC-1 and RNA polymerase II.
Supporting the hypothesis that ET-1 modulates aldosterone action in the kidney was the observation that aldosterone induction of edn1 mRNA occurred in collecting duct cells, the target cell type for aldosterone action. Furthermore, induction of ET-1 peptide was detected in the renal inner medulla of rats given a 2-h injection of aldosterone. Inner medullary ET-1 is well characterized natriuretic peptide that stimulates compounds such as nitric oxide and cGMP (55, 61). Similarly, ET-1 potently inhibits sodium transport through the epithelial sodium channel in collecting duct cells (34, 36). Consequently, aldosterone induction of edn1 may represent an important negative feedback loop on aldosterone-stimulated sodium reabsorption in the collecting duct. Indeed, the renal ET-1 pathway is differentially regulated in animals with mineralocorticoid-induced hypertension (62–66). However, direct support for this concept comes from studies conducted on collecting duct cell-specific edn1 knock-out mice. These mice exhibit severe salt-sensitive hypertension that is effectively remediated by either epithelial sodium channel or MR antagonists (31, 67).
Given the known functions of aldosterone and ET-1 in collecting duct cells, studies were conducted to more fully characterize aldosterone regulation of edn1 in mIMCD-3 cells. Aldosterone treatment resulted in dose-dependent increases in edn1 and sgk1 mRNA and edn1 hnRNA. This increase in hnRNA is consistent with the classic transcriptional mechanism of aldosterone.
Two HREs were mapped in the edn1 promoter that each contained receptor binding half-sites separated by eight nucleotides in different orientations. Variations in spacer regions have been reported for several aldosterone response genes (14, 15) and may influence cooperative binding of multiple hormone receptors (15). Indeed, both MR and GR interacted at the same HRE in the edn1 promoter. Half-site orientation is also known to affect receptor binding as well as transcriptional activation (5). Although GR can bind to directly repeated half-sites with low affinity (68), structural studies revealed that GR preferentially binds to palindromic DNA sequences as a dimer in a “head-to-head” conformation (12). Consistent with these reports, both MR and GR demonstrated a stronger affinity for HRE2 in comparison to HRE1. Moreover, only HRE2 could recruit RNA polymerase II. Similarly, the aldosterone response gene scnn1a also contains two HREs in different orientations. Only the inverted HRE was capable of stimulating transcription (69).
Interestingly, inhibition of hormone receptors with spironolactone and RU486 resulted in hormone receptor nuclear translocation and edn1 promoter binding (data not shown). However, these antagonists are known to inhibit hormone receptors by altering receptor conformation and disrupting coactivator recruitment (70). Indeed, proper transcriptional coactivation by MR and GR requires the recruitment of SRC-1 (6, 71). ChIP analysis revealed that SRC-1 was recruited to the edn1 promoter in the presence of aldosterone.
Multiple molecular studies in this report show that MR and GR were actively recruited to the edn1 gene to mediate aldosterone action. Both hormone receptors have documented roles in regulating aldosterone response genes, including scnn1a (69), sgk1 (18), and atp1a1 (16, 72). Both receptors have also been reported to mediate aldosterone stimulated sodium transport in the collecting duct (14, 73). However, the role of GR in aldosterone action is actively debated due to the concept that GR is not active in aldosterone responsive cells that express 11β-hydroxysteroid dehydrogenase type 2 (10, 24, 74–76). However, it is important to note that 11β-hydroxysteroid dehydrogenase type 2 metabolites also lack an affinity for GR. Thus, GR would be readily available for activation by another high affinity ligand such as aldosterone. Our studies show that MR and GR are present in the same protein complex. Several methods, including fluorescence resonance energy transfer (77), have demonstrated heterodimerization between MR and GR (74, 77–79). Moreover, these heterodimers exhibited distinct transcriptional properties (78). Indeed, aldosterone action mediated by two hormone receptors with different transcriptional properties would certainly provide a collecting duct cell with a higher degree of adaptability in the regulation of sodium transport.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of anti-MR antibody from Drs. Elise and Celso Gomez-Sanchez. We also thank Dr. Donald E. Kohan for his advice and suggestions and Dr. Marc Bailly for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1-DK049750 (to C. S. W.) from USPHS. This work was also supported by the Research Service of the Department of Veteran Affairs, the Department of Medicine of the University of Florida, and American Heart Association Predoctoral (AHA071572B) and Postdoctoral (AHA0825467E) Fellowships (to L. R. S. and M. L. G., respectively).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Fig. 1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Fig. 1.
- MR
- mineralocorticoid receptor
- GR
- glucocorticoid receptor
- ChIP
- chromatin immunoprecipitation
- HRE
- hormone response element
- SRC-1
- steroid receptor coactivator-1
- ET-1
- endothelin-1
- IMCD
- inner medullary collecting duct
- QPCR
- quantitative PCR
- hnRNA
- heterogeneous nuclear RNA
- siRNA
- small interference RNA
- DAPA
- DNA-affinity purification analysis.
REFERENCES
- 1.Arriza J. L., Weinberger C., Cerelli G., Glaser T. M., Handelin B. L., Housman D. E., Evans R. M. (1987) Science 237, 268–275 [DOI] [PubMed] [Google Scholar]
- 2.Viengchareun S., Le Menuet D., Martinerie L., Munier M., Pascual-Le Tallec L., Lombès M. (2007) Nucl. Recept. Signal 5, e012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McManus F., McInnes G. T., Connell J. M. (2008) Nat. Clin. Pract. Endocrinol. Metab. 4, 44–52 [DOI] [PubMed] [Google Scholar]
- 4.Beato M., Klug J. (2000) Hum. Reprod. Update 6, 225–236 [DOI] [PubMed] [Google Scholar]
- 5.Funder J. W. (1993) Science 259, 1132–1133 [DOI] [PubMed] [Google Scholar]
- 6.Li Y., Suino K., Daugherty J., Xu H. E. (2005) Mol. Cell 19, 367–380 [DOI] [PubMed] [Google Scholar]
- 7.Hellal-Levy C., Fagart J., Souque A., Wurtz J. M., Moras D., Rafestin-Oblin M. E. (2000) Mol. Endocrinol. 14, 1210–1221 [DOI] [PubMed] [Google Scholar]
- 8.Pascual-Le Tallec L., Lombès M. (2005) Mol. Endocrinol. 19, 2211–2221 [DOI] [PubMed] [Google Scholar]
- 9.Ulick S., Levine L. S., Gunczler P., Zanconato G., Ramirez L. C., Rauh W., Rösler A., Bradlow H. L., New M. I. (1979) J. Clin. Endocrinol. Metab. 49, 757–764 [DOI] [PubMed] [Google Scholar]
- 10.Odermatt A., Atanasov A. G. (2009) Steroids 74, 163–171 [DOI] [PubMed] [Google Scholar]
- 11.Funder J. W., Pearce P. T., Smith R., Smith A. I. (1988) Science 242, 583–585 [DOI] [PubMed] [Google Scholar]
- 12.Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. (1991) Nature 352, 497–505 [DOI] [PubMed] [Google Scholar]
- 13.Itani O. A., Liu K. Z., Cornish K. L., Campbell J. R., Thomas C. P. (2002) Am. J. Physiol. Endocrinol. Metab. 283, E971–E979 [DOI] [PubMed] [Google Scholar]
- 14.Mick V. E., Itani O. A., Loftus R. W., Husted R. F., Schmidt T. J., Thomas C. P. (2001) Mol. Endocrinol. 15, 575–588 [DOI] [PubMed] [Google Scholar]
- 15.Ou X. M., Storring J. M., Kushwaha N., Albert P. R. (2001) J. Biol. Chem. 276, 14299–14307 [DOI] [PubMed] [Google Scholar]
- 16.Kolla V., Robertson N. M., Litwack G. (1999) Biochem. Biophys. Res. Commun. 266, 5–14 [DOI] [PubMed] [Google Scholar]
- 17.Derfoul A., Robertson N. M., Lingrel J. B., Hall D. J., Litwack G. (1998) J. Biol. Chem. 273, 20702–20711 [DOI] [PubMed] [Google Scholar]
- 18.Webster M. K., Goya L., Ge Y., Maiyar A. C., Firestone G. L. (1993) Mol. Cell. Biol. 13, 2031–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gumz M. L., Popp M. P., Wingo C. S., Cain B. D. (2003) Am. J. Physiol. Renal Physiol. 285, F664–F673 [DOI] [PubMed] [Google Scholar]
- 20.Wong S., Brennan F. E., Young M. J., Fuller P. J., Cole T. J. (2007) Endocrinology 148, 1511–1517 [DOI] [PubMed] [Google Scholar]
- 21.Wolf S. C., Schultze M., Risler T., Rieg T., Lang F., Schulze-Osthoff K., Brehm B. R. (2006) Biochem. Pharmacol. 71, 1175–1183 [DOI] [PubMed] [Google Scholar]
- 22.Doi T., Sakoda T., Akagami T., Naka T., Mori Y., Tsujino T., Masuyama T., Ohyanagi M. (2008) Am. J. Physiol. Heart Circ. Physiol. 295, H1279–H1287 [DOI] [PubMed] [Google Scholar]
- 23.Barton M., Yanagisawa M. (2008) Can. J. Physiol. Pharmacol. 86, 485–498 [DOI] [PubMed] [Google Scholar]
- 24.Funder J. W., Mihailidou A. S. (2009) Mol. Cell. Endocrinol. 301, 2–6 [DOI] [PubMed] [Google Scholar]
- 25.Baylis C. (1999) Clin. Exp. Pharmacol. Physiol. 26, 253–257 [DOI] [PubMed] [Google Scholar]
- 26.Inscho E. W., Imig J. D., Cook A. K., Pollock D. M. (2005) Br. J. Pharmacol. 146, 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohan D. E. (2009) Kidney Int. 76, 481–486 [DOI] [PubMed] [Google Scholar]
- 28.Kohan D. E. (2006) Curr. Opin. Nephrol. Hypertens. 15, 34–40 [DOI] [PubMed] [Google Scholar]
- 29.Ge Y., Ahn D., Stricklett P. K., Hughes A. K., Yanagisawa M., Verbalis J. G., Kohan D. E. (2005) Am. J. Physiol. Renal Physiol. 288, F912–F920 [DOI] [PubMed] [Google Scholar]
- 30.Ge Y., Stricklett P. K., Hughes A. K., Yanagisawa M., Kohan D. E. (2005) Am. J. Physiol. Renal Physiol. 289, F692–F698 [DOI] [PubMed] [Google Scholar]
- 31.Ahn D., Ge Y., Stricklett P. K., Gill P., Taylor D., Hughes A. K., Yanagisawa M., Miller L., Nelson R. D., Kohan D. E. (2004) J. Clin. Invest. 114, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khanna A., Simoni J., Wesson D. E. (2005) J. Am. Soc. Nephrol. 16, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 33.Bugaj V., Pochynyuk O., Mironova E., Vandewalle A., Medina J. L., Stockand J. D. (2008) Am. J. Physiol. Renal Physiol. 295, F1063–F1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrera M., Garvin J. L. (2005) Am. J. Physiol. Renal Physiol. 288, F58–F64 [DOI] [PubMed] [Google Scholar]
- 35.Gilmore E. S., Stutts M. J., Milgram S. L. (2001) J. Biol. Chem. 276, 42610–42617 [DOI] [PubMed] [Google Scholar]
- 36.Hoffman A., Abassi Z. A., Brodsky S., Ramadan R., Winaver J. (2000) Hypertension 35, 732–739 [DOI] [PubMed] [Google Scholar]
- 37.Gallego M. S., Ling B. N. (1996) Am. J. Physiol. Renal Physiol. 271, F451–F460 [DOI] [PubMed] [Google Scholar]
- 38.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 39.Stricklett P. K., Hughes A. K., Kohan D. E. (2006) Am. J. Physiol. Renal Physiol. 290, F1315–F1319 [DOI] [PubMed] [Google Scholar]
- 40.Yorikane R., Sakai S., Miyauchi T., Sakurai T., Sugishita Y., Goto K. (1993) FEBS Lett. 332, 31–34 [DOI] [PubMed] [Google Scholar]
- 41.Bens M., Vallet V., Cluzeaud F., Pascual-LeTallec L., Kahn A., Rafestin-Oblin M. E., Rossier B. C., Vandewalle A. (1999) J. Am. Soc. Nephrol. 10, 923–934 [DOI] [PubMed] [Google Scholar]
- 42.Guntupalli J., Onuigbo M., Wall S., Alpern R. J., DuBose T. D., Jr. (1997) Am. J. Physiol. Cell Physiol. 273, C558–C571 [DOI] [PubMed] [Google Scholar]
- 43.Kizer N. L., Lewis B., Stanton B. A. (1995) Am. J. Physiol. Renal Physiol. 268, F347–F355 [DOI] [PubMed] [Google Scholar]
- 44.Lipson K. E., Baserga R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 9774–9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez-Sanchez C. E., de Rodriguez A. F., Romero D. G., Estess J., Warden M. P., Gomez-Sanchez M. T., Gomez-Sanchez E. P. (2006) Endocrinology 147, 1343–1348 [DOI] [PubMed] [Google Scholar]
- 46.Leach K. M., Vieira K. F., Kang S. H., Aslanian A., Teichmann M., Roeder R. G., Bungert J. (2003) Nucleic Acids Res. 31, 1292–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng W. G., Zhu Y., Montero A., Wu K. K. (2003) Anal. Biochem. 323, 12–18 [DOI] [PubMed] [Google Scholar]
- 48.Vallon V., Huang D. Y., Grahammer F., Wyatt A. W., Osswald H., Wulff P., Kuhl D., Lang F. (2005) Am. J. Physiol. Regul. Integr. Comp. Physiol. 289, R395–R401 [DOI] [PubMed] [Google Scholar]
- 49.Flores S. Y., Loffing-Cueni D., Kamynina E., Daidié D., Gerbex C., Chabanel S., Dudler J., Loffing J., Staub O. (2005) J. Am. Soc. Nephrol. 16, 2279–2287 [DOI] [PubMed] [Google Scholar]
- 50.Kitamura K., Tanaka T., Kato J., Eto T., Tanaka K. (1989) Biochem. Biophys. Res. Commun. 161, 348–352 [DOI] [PubMed] [Google Scholar]
- 51.Rauchman M. I., Nigam S. K., Delpire E., Gullans S. R. (1993) Am. J. Physiol. Renal Physiol. 265, F416–F424 [DOI] [PubMed] [Google Scholar]
- 52.Kohan D. E., Hughes A. K., Perkins S. L. (1992) J. Biol. Chem. 267, 12336–12340 [PubMed] [Google Scholar]
- 53.Boesen E. I., Pollock D. M. (2007) Am. J. Physiol. Renal Physiol. 292, F185–F191 [DOI] [PubMed] [Google Scholar]
- 54.Vassileva I., Mountain C., Pollock D. M. (2003) Hypertension 41, 1359–1363 [DOI] [PubMed] [Google Scholar]
- 55.Pollock D. M. (2000) Curr. Opin. Nephrol. Hypertens. 9, 157–164 [DOI] [PubMed] [Google Scholar]
- 56.Kitamura K., Tanaka T., Kato J., Ogawa T., Eto T., Tanaka K. (1989) Biochem. Biophys. Res. Commun. 162, 38–44 [DOI] [PubMed] [Google Scholar]
- 57.Bhargava A., Wang J., Pearce D. (2004) Mol. Cell. Endocrinol. 217, 189–196 [DOI] [PubMed] [Google Scholar]
- 58.Palii S. S., Thiaville M. M., Pan Y. X., Zhong C., Kilberg M. S. (2006) Biochem. J. 395, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiaville M. M., Dudenhausen E. E., Zhong C., Pan Y. X., Kilberg M. S. (2008) Biochem. J. 410, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang G., Lin J. C., Wei V., Yoo C., Cheng J. C., Nguyen C. T., Weisenberger D. J., Egger G., Takai D., Gonzales F. A., Jones P. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edwards R. M., Pullen M., Nambi P. (1992) Am. J. Physiol. Renal Physiol. 263, F1020–F1025 [DOI] [PubMed] [Google Scholar]
- 62.Montezano A. C., Callera G. E., Mota A. L., Fortes Z. B., Nigro D., Carvalho M. H., Zorn T. M., Tostes R. C. (2005) Peptides 26, 1454–1462 [DOI] [PubMed] [Google Scholar]
- 63.Pollock D. M., Allcock G. H., Krishnan A., Dayton B. D., Pollock J. S. (2000) Am. J. Physiol. Renal Physiol. 278, F279–F286 [DOI] [PubMed] [Google Scholar]
- 64.Matsumura Y., Kuro T., Kobayashi Y., Konishi F., Takaoka M., Wessale J. L., Opgenorth T. J., Gariepy C. E., Yanagisawa M. (2000) J. Cardiovasc. Pharmacol. 36, S86–S89 [DOI] [PubMed] [Google Scholar]
- 65.Hsieh T. J., Lin S. R., Lee Y. J., Shin S. J., Lai Y. H., Hsu C. H., Tsai J. H. (2000) Am. J. Physiol. Renal Physiol. 279, F112–F121 [DOI] [PubMed] [Google Scholar]
- 66.Matsumura Y., Kuro T., Konishi F., Takaoka M., Gariepy C. E., Yanagisawa M. (2000) Br. J. Pharmacol. 129, 1060–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ge Y., Huang Y., Kohan D. E. (2008) Can. J. Physiol. Pharmacol. 86, 329–336 [DOI] [PubMed] [Google Scholar]
- 68.Aumais J. P., Lee H. S., DeGannes C., Horsford J., White J. H. (1996) J. Biol. Chem. 271, 12568–12577 [DOI] [PubMed] [Google Scholar]
- 69.Sayegh R., Auerbach S. D., Li X., Loftus R. W., Husted R. F., Stokes J. B., Thomas C. P. (1999) J. Biol. Chem. 274, 12431–12437 [DOI] [PubMed] [Google Scholar]
- 70.Rogerson F. M., Fuller P. J. (2003) Mol. Cell. Endocrinol. 200, 45–55 [DOI] [PubMed] [Google Scholar]
- 71.He Y., Szapary D., Simons S. S., Jr. (2002) J. Biol. Chem. 277, 49256–49266 [DOI] [PubMed] [Google Scholar]
- 72.Whorwood C. B., Ricketts M. L., Stewart P. M. (1994) Endocrinology 135, 901–910 [DOI] [PubMed] [Google Scholar]
- 73.Bens M., Vallet V., Cluzeaud F., Pascual-Letallec L., Kahn A., Rafestin-Oblin M. E., Rossier B. C., Vandewalle A. (1999) J. Am. Soc. Nephrol. 10, 923–934 [DOI] [PubMed] [Google Scholar]
- 74.Nishi M., Kawata M. (2007) Neuroendocrinology 85, 186–192 [DOI] [PubMed] [Google Scholar]
- 75.Gaeggeler H. P., Gonzalez-Rodriguez E., Jaeger N. F., Loffing-Cueni D., Norregaard R., Loffing J., Horisberger J. D., Rossier B. C. (2005) J. Am. Soc. Nephrol. 16, 878–891 [DOI] [PubMed] [Google Scholar]
- 76.Morris D. J., Latif S. A., Brem A. S. (2009) Steroids 74, 1–6 [DOI] [PubMed] [Google Scholar]
- 77.Nishi M., Tanaka M., Matsuda K., Sunaguchi M., Kawata M. (2004) J. Neurosci. 24, 4918–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu W., Wang J., Sauter N. K., Pearce D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 12480–12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savory J. G., Préfontaine G. G., Lamprecht C., Liao M., Walther R. F., Lefebvre Y. A., Haché R. J. (2001) Mol. Cell. Biol. 21, 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gumz M. L., Stow L. R., Lynch I. J., Greenlee M. M., Rudin A., Cain B. D., Weaver D. R., Wingo C. S. (2009) J. Clin. Invest. 119, 2423–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.