Abstract
The liver X receptors (LXRs) have been known as sterol sensors that impact cholesterol and lipid homeostasis, as well as inflammation. Although the hepatic functions of LXRs are well documented, whether and how LXRs play a pathophysiological role in the lung remain largely unknown. Here we show that LXRα and LXRβ are expressed in both type I and type II mouse lung epithelial cells, as well as in human lung cancer cells. To study the role of LXRα in vivo including the pulmonary function of this LXR isoform, we created LXRα knock-in (LXR-KI) mice in which a constitutively activated LXRα (VP-LXRα) was inserted into the mouse LXRα locus. We show that activation of LXR in LXR-KI mice or LXR agonist-treated wild type mice induced pulmonary expression of genes encoding multiple antioxidant enzymes. Consistent with the induction of antioxidant enzymes, LXR-KI mice and LXR ligand-treated wild type mice showed a substantial resistance to lipopolysaccharide-induced lung injury and decreased production of reactive oxygen species. In summary, we have uncovered a novel role of LXR in regulating antioxidant enzymes in the lung and the implication of this regulation in pulmonary tissue protection.
Reactive oxygen species (ROS),5 such as hydroxyl radicals, superoxide (O2˙̄), and H2O2, are highly reactive molecules produced during normal cellular processes involving oxygen, as well as during pathological responses by leukocyte enzymes. A variety of pro-inflammatory compounds, such as lipopolysaccharide (LPS), cytokines, chemokines, and lipid mediators, are capable of activating leukocytes to generate ROS. ROS causes cellular damages by reacting with macromolecules, resulting in derangements, such as mutations in DNA, alteration in protein function, and membrane damage caused by lipid peroxidation (for reviews, see Refs. 1–3).
The lung is an organ susceptible to oxidative stresses that are derived from oxygen or inflammatory responses (1, 4). The imbalance of oxidants and antioxidants plays an important role in the development of various pulmonary diseases, such as acute respiratory distress syndrome and chronic obstructive pulmonary disease (5, 6). Oxidative stress also affects inflammatory responses and alters the balance of cytokines (7). To neutralize free radicals and counteract the detrimental effect of ROS, cells express a wide array of endogenous antioxidant enzymes. These include “direct antioxidants,” such as superoxide dismutases (SODs), catalase, and glutathione peroxidase, as well as “indirect” antioxidant enzymes, such as glutathione S-transferases (GSTs), metallothioneins, and NADPH:quinone oxidoreductase (8, 9). Proper regulation of these antioxidant enzymes is essential for mammals to maintain balances between oxidants and antioxidants.
Among antioxidant enzymes, GSTs are a family of Phase II enzymes that catalyze the conjugation of the tripeptide GSH to a variety of hydrophobic, electrophilic, and cytotoxic substrates. The majority of GST substrates are either xenobiotics or products of oxidative stress that are toxic and/or carcinogenic to cells. The formation of a thioether bond between electrophiles and GSH almost always yields a conjugate that is less reactive than the parent compounds, and therefore the GST-mediated conjugation generally results in xeno- and endobiotic detoxification and cancer prevention (10).
LXRs, including the α and β isoforms, belong to the orphan nuclear receptor family of transcription factors. LXRα shows high expression in selected tissues, including the liver, lung, adipose, intestine, and kidney. In contrast, LXRβ is ubiquitously expressed (for a review, see Ref. 11). LXRs regulate gene expression by forming heterodimers with the retinoid X receptor and binding of LXR-retinoid X receptor heterodimers to LXR-responsive elements found in the target gene promoters. LXR-responsive elements are typically composed of two direct hexameric repeats separated by four nucleotides (DR4) (12, 13). Other types of LXR-responsive elements, such as IR-0 and ER-8, have also been reported (14, 15). It is widely accepted that LXRs play an important role in cholesterol metabolism and triglyceride synthesis in various tissues (16–21). LXRs have also been shown to inhibit inflammatory gene expression and prevent either bacterial or LPS-triggered inflammatory responses in macrophages (22). In addition, LXR signaling can impact antimicrobial responses by regulating macrophage gene expression and apoptosis (23). Although the hepatointestinal functions of LXRs have been well documented, whether and how LXRs play a role in the pathophysiology of the lung remain largely unknown.
In this study, we have uncovered a novel role for LXRs in preventing lung injury. The pulmonary protective function of LXRs is likely due to the positive regulation of antioxidant enzymes by these receptors.
EXPERIMENTAL PROCEDURES
Creation of LXRα-KI Mice
The LXRα knock-in targeting construct was generated by placing VP-LXRα cDNA (14) in-frame and immediately after the endogenous ATG start codon in the mouse LXRα locus. VP-LXRα cDNA was constructed by fusing the VP16 activation domain of the herpes simplex virus in-frame to the amino terminus of mouse LXRα cDNA. The SV40 poly(A) sequence was added downstream to terminate the transcription of LXRα, and the PGK-Neo selection marker was engineered thereafter. The selections of short arm and long arm sequences predict that after homologous recombination, part of exon 2, exons 3–7, and the introns in between will be replaced by the VP-LXRα-SV40-PGK-Neo cassette. The targeting construct was linearized by NotI digestion and electroporated into the Strain 129S1/X1 mouse embryonic stem cell line R1 (24) under the conditions previously described (25). After G418 (200 μg/ml) selection, embryonic stem cell clones were picked, expanded, and screened by Southern blot analysis. Positive clones were microinjected into C57BL/6J blastocysts. Chimeric male progeny were crossed with C57BL/6J females. Germ line transmission of the knock-in allele was detected in agouti progeny by Southern blot analysis. A three-primer PCR was designed for subsequent mouse genotyping, with a forward primer and a reverse primer from LXRα, yielding a 466-bp product from the wild type allele. The knock-in reverse primer is from the VP16 coding sequence and amplified a 362-bp product from the knock-in allele. The mice used in this study were maintained in the C57BL/6J-Strain 129S1/X1 mixed background. Wild type mice of the same genetic background were used where applicable.
Microarray Analysis
Lung total RNA from three LXR-KI male mice was pooled, paired with pooled samples from three WT males, and subjected to Affymetrix microarray analysis at the University of Pittsburgh Department of Pathology Gene Array Laboratory. We used the Affymetrix mouse 430A Chip (Santa Clara, CA) that contains 45,000 probe sets to analyze the expression level of over 39,000 transcripts and variants from over 34,000 well characterized mouse genes. After hybridization, the chips were scanned in an Agilent ChipScanner to detect hybridization signals. Hybridization data were exported and analyzed using GeneSpring 4.2 software (Silicon Genetics, Redwood, CA). The microarray data has been deposited to the NCBI/Gene Expression Omnibus website.
LPS-induced Lung Injury and LXR Ligand Treatment
For the LPS treatment, 12-week-old mice were anesthetized by intraperitoneal injection with ketamine (150 mg/kg) and xylazine (10 mg/kg) and subjected to intranasal instillation of LPS (10 μg/each mouse) or vehicle (saline) in a volume of 50 μl as previously described (26). LPS from Escherichia coli 0111:B4 was purchased from Sigma. The mice were sacrificed 24 h after the intranasal instillation. For LXR ligand treatment, daily gavage of TO1317 (10 mg/kg) or GW3965 (20 mg/kg) in a total volume of 100 μl was given starting 7 days prior to the LPS treatment.
Bronchoalveolar Lavage (BAL) Fluid Collection, Cell Counting, and Thiobarbituric Acid-reactive Species (TBARS) Assay
Twenty-four hours after LPS instillation, the mice were sacrificed, and BAL fluids were collected by washing three times with 0.8 ml of sterile saline. The recovered BAL fluids were centrifuged at 300 rpm for 10 min. The total BAL cells were counted using a hemocytometer. The polymorphonuclear neutrophils were selectively identified and counted after staining with the Wright solution. The cell-free supernatant fluid was collected and subjected to the measurement of protein concentration using a BCA protein assay kit (Pierce) and TBARS assay. The formation of TBARS during an acid-heating reaction was measured as previously described (27). In brief, 100 μl of BAL fluid was mixed with 0.5 ml of trichloroacetic acid (10%) and 0.5 ml of thiobarbituric acid (0.67%) and then heated in a boiling water bath for 30 min. TBARS was determined spectrophotometrically by the absorbance at 535 nm using 1,1,3,3-tetramethoxypropane as the external standard.
Measurement of Myeloperoxidase (MPO) Activity
To measure MPO activity in the lung, lung tissues were homogenized and sonicated in 50 mm KPO4 buffer containing 0.5% hexadecyltrimethylammonium bromide and 5 mm EDTA. After centrifugation at 12,000 × g for 10 min at 4 °C, the supernatants were collected and incubated in 50 mm sodium phosphate buffer (pH 6.0) containing the substrate H2O2 (0.0006%). In the presence of O-dianisidine dihydrochloride (167 μg/ml), the MPO activity was determined spectrophotometrically by measuring the change in absorbance at 460 nm over 3 min using a 96-well plate reader from Molecular Devices (Sunnyvale, CA). The results are presented as OD change/min/mg of protein (28).
SOD, Catalase, and GST Enzymatic Assays
SOD, catalase, and GST activities were measured as described previously (29). In brief, lung and tissues were homogenized in 20 mm potassium phosphate (pH 7.0) and 2 mm EDTA using a Brinkmann/Kinematica Polytron PT3000 homogenizer equipped with a PT/DA 3007/2 probe (Brinkmann Instruments, Westbury, NY). Ten percent (w/v) homogenates were clarified by centrifugation at 12,000 × g for 30 min at 4 °C, and the supernatants were immediately used for the measurement of GST activity using an assay kit from Cayman Chemical (Ann Arbor, MI). SOD activity was measured according to the method described by Paoletti and Mocali (30). Catalase activity was measured by the rate of decrease in hydrogen peroxide absorbance at 240 nm as described previously (31). One unit of catalase activity was defined as the rate constant of the first order reaction. The catalase activity was expressed as units/mg of protein.
Phospholipid Assay
One ml of cell-free BAL fluid was mixed with 3.75 ml of chloroform/methanol (1:2; v/v), followed by the addition of 1.25 ml of chloroform and 1.25 ml of double distilled H2O. The mixture was centrifuged at 1000 rpm for 10 min. The lower lipid phase was collected and dried under nitrogen gas (32). For phospholipid assay, the lipid extract was added with 0.65 ml of perchloric acid and was placed in a heat block at 180 °C for 30 min. After adding 3.3 ml of H2O, 0.5 ml of 2.5% molybdate, and 0.5 ml of 10% ascorbic acid, the sample was placed in a boiling water bath for 5 min. The absorbance of cooled samples was read at 800 nm. The solution of KH2PO4 (100 μg P/ml) served as the external standard (33).
Histology, Immunohistochemistry, and Immunofluorescence
Mouse lung tissues were inflation-fixed with 10% formaldehyde in phosphate-buffered saline at 20 cm of H2O pressure. Paraffin-embedded tissues were sectioned at 5-μm thickness, and the sections were stained by hematoxylin and eosin for general histological evaluation. For immunohistochemistry, frozen sections were fixed with acetone, and endogenous peroxidase activity was quenched with 0.6% hydrogen peroxide in methanol. The slides were blocked with 5% horse serum in phosphate-buffered saline for 45 min at room temperature. The slides were then incubated with a 1:100 dilution of mouse monoclonal anti-LXRα antibody (PP-PPZ0412–00) from Perseus Proteomics (Tokyo, Japan) overnight at 4 °C. The slides were rinsed and then incubated with a 1:200 dilution of biotinylated anti-mouse secondary antibody. The slides were rinsed again, sequentially incubated with streptavidin peroxidase followed by aminoethyl carbazole substrate solution, then rinsed, and counterstained with hematoxylin. For immunofluorescence, frozen sections were fixed with acetone, and nonspecific binding sites were blocked with 10% donkey serum for 30–60 min and rinsed for three times. The slides were incubated with a mouse anti-LXRα antibody (1:100 dilution) and a rabbit anti-surfactant protein B (SPB) antibody (1:200 dilution; Chemicon, Temecula, CA) overnight at 4 °C and subsequently with fluorescein isothiocyanate-conjugated donkey anti-mouse IgG (1:200 dilution) and Cy3-conjugated donkey anti-rabbit IgG (1:100 dilution) for 1 h in a darkroom at room temperature. The slides were rinsed three times and visualized using an Olympus FV1000 confocal microscope.
Quantitative Real Time Reverse Transcription-PCR and Northern Blot Analysis
Total RNA was extracted with the TRIzol reagent from Invitrogen. Real time PCR using SYBR Green-based assays was performed with the ABI 7300 real time PCR system as we previously described (34). All real time PCR results were normalized against the housekeeping gene cyclophilin. PCR primer sequences are listed in supplemental Table S1. When the expression of LXRs was compared between tissue types and/or cell lines, LXRα and β copy numbers were calculated according to the standard curve of five serial dilutions of double-stranded plasmid DNA ranging from 103 to 107 molecules. In these cases, LXR copy numbers were expressed as copy numbers/μg of RNA. Northern blot analysis using [32P]dCTP-labeled full-length mouse LXRα cDNA probe was performed as we previously described (28).
Cell Culture and LPS Treatment
A549 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum at 37 °C and saturated humidity (5% CO2, 95% air) in a CO2 incubator. The cells were pretreated with GW3965 (10 μm) or vehicle (Me2SO) for 24 h, followed by a 24-h treatment of LPS (100 nm). Total RNA was extracted and subjected to real time PCR analysis.
Statistical Analysis
The results are expressed as the means ± S.D. One-way analysis of variance and Tukey's test were used for statistical analysis using GraphPad Prism version 4.0. p values of less than 0.05 were considered statistically significant.
RESULTS
Both LXRα and LXRβ Are Abundantly Expressed in Mouse Lung and A549 Human Lung Cancer Cells
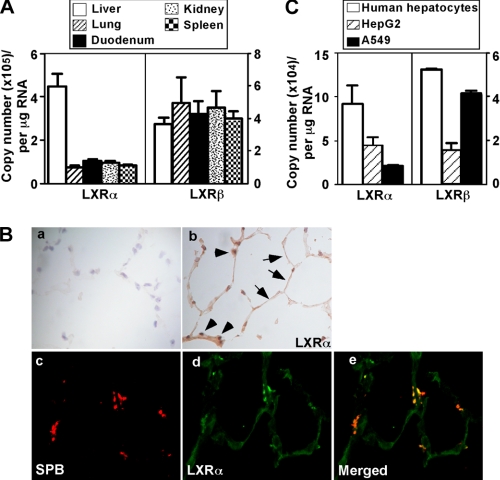
LXRα is known for its high expression in the liver, intestine, and kidney (11). We showed that LXRα was also expressed in mouse lung and human lung cancer cells. The mRNA abundance of LXRα in the mouse lung was ∼20% that of the liver based on the copy numbers determined by real time PCR (Fig. 1A). LXRβ is known to be ubiquitously expressed, and its mRNA abundance in the lung was similar to that in the liver (Fig. 1A). Immunohistochemical analysis showed that LXRα (Fig. 1B, panel b) was expressed in both type I and type II epithelial cells based on the morphologies of both cell types. It appeared that LXRα is expressed in both the nuclei and cytoplasm. The expression of LXRα in type II cells was further confirmed by immunofluorescence and confocal analysis on the expression and localization of LXRα and SPB, a specific marker of type II epithelial cells (Fig. 1B, panels c–e). It has been reported that in type II epithelial cells, SP-B precursors were detected in the endoplasmic reticulum, the Golgi complex, and the multivesicular bodies, whereas the mature SP-B was found in multivesicular and lamellar bodies, where it is stored and secreted into the alveolar space (35). Our antibody detects both premature and mature SPB and showed an uneven distribution of SPB.
FIGURE 1.
Both LXRα and LXRβ are abundantly expressed in mouse lung and A549 human lung cancer cells. A, the mRNA expression of LXRα and LXRβ in mouse lung as compared with liver, duodenum, kidney, and spleen. The mRNA expression was measured by real time PCR analysis. The copy numbers were calculated according to a standard curve of five serial dilutions of double-stranded plasmid DNA ranging from 103 to 107 molecules (n = 3 for each group). B, panels a and b, immunostaining of lung sections using normal IgG (panel a) and anti-LXRα (panel b). Arrows and arrowheads indicate type I and type II lung epithelial cells, respectively. Panels c–e, immunofluorescence to localize the expression of SPB (panel c), LXRα (panel d), and their merged image (panel e). C, the mRNA expression of LXRα and LXRβ in the lung cancer A549 cells as compared with human hepatocytes and HepG2 cells.
Both LXRα and LXRβ were also expressed in A549 cells, a human lung cancer cell line derived from type II cells (36). The mRNA abundance of LXRα in A549 was ∼30 and 50% of that of primary human hepatocytes and hepatoma HepG2 cells, respectively (Fig. 1C). The expression of LXRβ in A549 cells was similar to that of the primary hepatocytes (Fig. 1C).
Creation of LXRα Knock-in Mice That Express Constitutively Activated LXRα
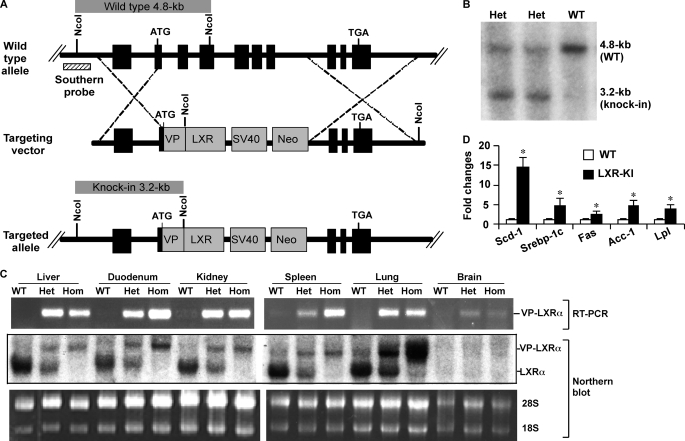
To examine the effect of LXRα activation in vivo, including the pathophysiological relevance of this activation in the lung, we created LXRα knock-in mice that express a constitutively activated LXRα (VP-LXRα). The strategy we used to create VP-LXRα knock-in (LXR-KI) mice is outlined in Fig. 2A. The VP-LXRα cDNA was constructed by fusing the VP16 activation domain of the herpes simplex virus to the amino terminus of mouse LXRα sequence (14). VP-LXRα shares the same DNA binding specificity as its wild type (WT) counterpart and activates LXR-responsive gene expression in the absence of an exogenously added ligand in cell cultures and in transgenic mice (14). In the targeting construct, VP-LXRα cDNA was placed in-frame and immediately after the endogenous ATG start codon of the mouse LXRα locus. This construct was designed so that after DNA homologous recombination, the sequence spanning part of exon 2 (starting from start codon ATG), exons 3–7, and the introns in between would be replaced by VP-LXRα. As such, VP-LXRα will be expressed under the control of the endogenous LXRα promoter, whereas the WT LXRα will be disrupted in the homozygous LXR-KI mice. The ES clones were screened by Southern blot analysis (data not shown). After ES cell blastocyst injection, germ line transmission of the knock-in allele was confirmed by Southern blot analysis. As predicted in Fig. 2A and shown in Fig. 2B, NcoI digestion of mouse tail genomic DNA produced a 4.8-kb fragment from the WT allele and a 3.2-kb fragment from the knock-in allele at the short arm region.
FIGURE 2.
Creation of LXR-KI mice that express the constitutively activated LXRα (VP-LXRα). A, depicted is the gene targeting strategy that was used to replace the endogenous LXRα gene with VP-LXRα. Part of exon 2, exons 3–7, and introns in between of the WT allele were replaced by the VP-LXRα-SV40-Neo cassette. The Southern blot probe and prediction of NcoI restriction fragment sizes are labeled. B, confirmation of homologous recombination in the heterozygous LXR-KI mice by Southern blot analysis. The mouse tail DNA was digested with NcoI. C, expression of VP-LXRα allele and/or the endogenous LXRα allele was determined by semi-quantitative reverse transcription-PCR using a pair of VP-LXRα allele-specific primers (top panel) and Northern blot analysis using a LXRα cDNA probe that detects both VP-LXRα and WT LXRα transcripts (bottom panel). Ethidium bromide staining of the agarose gel is to show the sample loading. Het, heterozygous; Hom, homozygous. D, LXR target gene expression in the liver was determined by real time PCR analysis. Acc-1, acetyl CoA carboxylase 1; Fas, fatty acid synthase; Scd-1, stearoyl CoA desaturase-1; Srebp-1c, sterol regulatory element-binding protein 1c; Lpl, lipoprotein lipase (n = 5 for each group). *, p < 0.05, compared with WT.
LXR-KI mice were viable and fertile. The phenotype of LXR-KI mice was indistinguishable from that of WT mice. Semi-quantitative reverse transcription-PCR showed that the knock-in allele was expressed in a panel of tissues of heterozygous and homozygous LXR-KI mice (Fig. 2C, top panel), and the results were confirmed by Northern analysis (Fig. 2C, bottom panel). As shown in Fig. 2C, the knock-in allele was expressed in both heterozygous and homozygous LXR-KI mice, whereas the endogenous LXRα transcript was detected only in the WT and heterozygous mice. We noticed that the expression level of the knock-in allele was not always consistent with the WT allele. For example, VP-LXRα expression in knock-in mouse liver was lower than the endogenous LXRα in the liver of WT mice (Fig. 2C). We reason this was due to the knock-in of VP-LXRα cDNA, so the efficiency of RNA splicing might be different between the knock-in and the endogenous alleles. The hepatic expression of LXR target genes, such as stearoyl CoA desaturase 1, sterol regulatory element-binding protein 1c, fatty acid synthase, acetyl CoA carboxylase 1, and lipoprotein lipase, was increased in the LXR-KI mice as expected (Fig. 2D). LXR-KI mice also showed hepatic steatosis (data not shown), suggesting that the VP-LXRα knock-in allele was fully functional in vivo.
Microarray Analysis Revealed the Regulation of Antioxidant Genes by LXR in the Lung
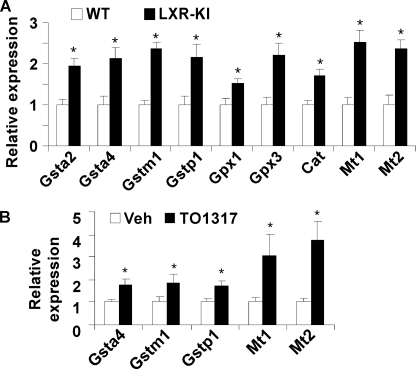
To better understand the biological consequences of LXR activation in the lung, we performed microarray analysis on lung tissues from LXR-KI mice and their wild type littermates. Total RNA from lung tissues of three LXR-KI male mice was pooled. Gene expression was compared with pooled samples from three WT male mice. In the microarray analysis, we found that the expression of several antioxidant genes was induced in the lung of LXR-KI mice. supplemental Table S2 represents a partial list of genes whose expression was induced in the lung of LXR-KI mice. These include the induction of Gsta2, Gsta4, Gstm1, Gstp1, Gpx1, Gpx3, catalase, Mt1, and Mt2. The activation of antioxidant gene expression was confirmed by real time PCR analysis (Fig. 3A). The same pattern of antioxidant gene regulation was observed in the lung of WT mice treated with the LXR agonist TO1317 (Fig. 3B).
FIGURE 3.
Activation of LXR regulated the expression of antioxidant genes in the lung. Gene expression at mRNA level was measured by real time PCR analysis. A, the expression of antioxidant genes was increased in the lung of LXR-KI mice compared with their WT littermates. B, the expression of antioxidant genes in the lung of WT mice treated with vehicle or TO1317 (20 mg/kg, daily, intraperitoneally) for 3 days (n = 3 for each group). Gpx, glutathione peroxidase; Cat, catalase; Mt, metallothioneins. *, p < 0.05, compared with WT (A) or vehicle (B) control.
Activation of LXR Conferred Resistance to LPS-induced Lung Injury
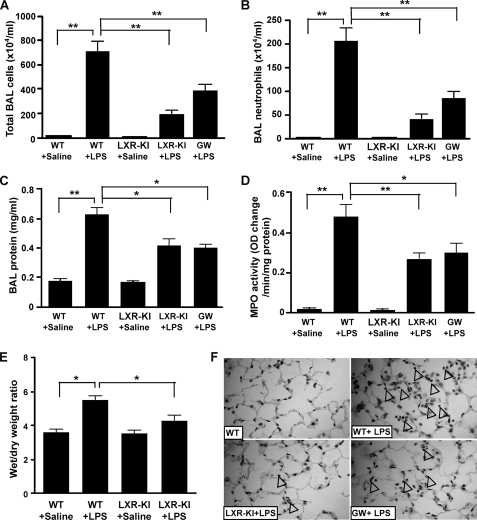
The induction of pulmonary antioxidant genes prompted us to determine whether activation of LXR in the lung confers resistance to injury caused by oxidative toxicants, such as LPS. As shown in Fig. 4A, intranasal instillation of LPS into the lung of WT mice resulted in a dramatic increase in total cell numbers in the BAL fluid as expected (26). In a sharp contrast, LPS-induced BAL cell number increase in LXR-KI mice was reduced to ∼25% of the LPS-treated WT mice (Fig. 4A). A decreased BAL cell number was also observed in WT mice treated with the LXR agonist GW3965 (Fig. 4A). A similar pattern of the inhibitory effect of the LXR-KI allele and GW3965 was observed when neutrophil infiltration (Fig. 4B), BAL protein concentration (Fig. 4C), and MPO activity (Fig. 4D) were measured in BAL fluid as the surrogate markers of lung injury. The increased BAL protein concentration indicates pulmonary microvascular leakage (37), whereas the MPO activity reflects the infiltration of lung parenchymal phagocytes (28). Consistent with the decreased BAL protein concentration, the lung wet to dry weight ratio, an indicator of edema caused by capillary leakage, was significantly lower in LPS-treated LXR-KI mice compared with their WT counterparts (Fig. 4E). At the histological level, hematoxylin and eosin staining of lung sections showed that the neutrophil infiltration readily observed in WT mice was markedly attenuated in LXR-KI and GW3965-treated WT mice (Fig. 4F).
FIGURE 4.
Activation of LXR conferred resistance to LPS-induced lung injury. WT and LXR-KI mice received intranasal instillation of saline or LPS. The fifth group of WT mice was treated with GW3965 (GW, 20 mg/kg, daily, intraperitoneally) for 7 days before being treated with LPS. Twenty-four hours after LPS instillation, the mice were sacrificed, and the lungs were lavaged to collect the BAL fluids. A, the total BAL cell numbers were counted using a hemocytometer. B, polymorphonuclear neutrophils were stained with Wright solution, and their cell numbers were counted. C, the protein concentrations of the cell-free BAL supernatants were measured. D, lung homogenates were measured for the MPO activity. E, the wet to dry weight ratio of the lung tissues. F, hematoxylin and eosin staining of lung sections derived from mice treated with saline or LPS. Arrowheads indicate neutrophil infiltration (n = 6 for each group). *, p < 0.05; **, p < 0.01, comparisons are labeled.
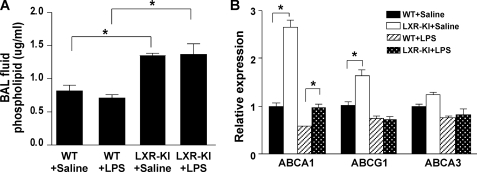
Activation of LXR Increased the Phospholipid Content in the BAL Fluid
Lung surfactant, mostly composed of phospholipids, reduces surface tension by forming a lipid monolayer at the interface of liquid and air. As such, phospholipids play an important role in protecting the lung from oxidative damage and infection. Indeed, administration of surfactant has been shown to be effective in relieving acute lung injury or acute respiratory distress syndrome (38, 39). In LXR-KI mice, the level of phospholipids in BAL fluid was significantly higher than that in WT mice regardless of the LPS treatment (Fig. 5A). ABCA1, ABCA3, and ABCG1 are the major phospholipid transporters in the lung. LXR-KI mice exhibited increased mRNA expression of ABCA1 and ABCG1 (Fig. 5B), consistent with the identities of these two transporters as LXR target genes (21, 40). Treatment with LPS tended to reduce the expression of phospholipid transporters regardless of genotypes, but the expression of ABCA1 in LXR-KI mice remained significantly higher than WT mice in the presence of LPS.
FIGURE 5.
Activation of LXR increased the phospholipid content in the BAL fluid. A, total lipids in BAL fluid were extracted by methanol and chloroform and subjected to the measurement of phospholipids as described under “Experimental Procedures.” B, the pulmonary mRNA expression of phospholipid transporters was measured by real time PCR analysis (n = 6 for each group). *, p < 0.05, comparisons are labeled.
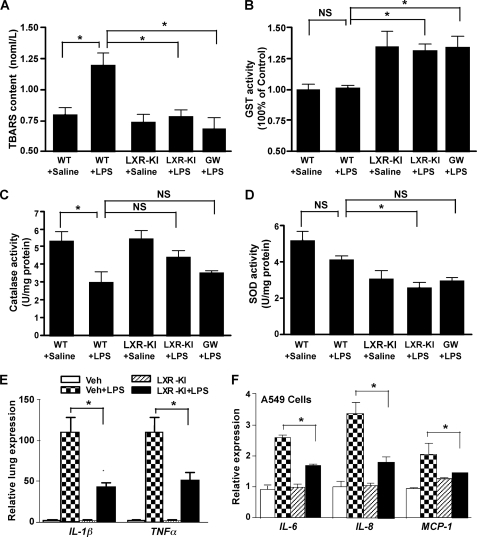
Activation of LXR Decreased LPS-induced Oxidative Stress and Inhibited Pulmonary Inflammatory Response in Vivo and in A549 Lung Cancer Cells
Oxidative stress, such as that induced by LPS, plays an important role in the pathogenesis of acute lung injury (1, 41, 42). Therefore we went on to determine whether activation of LXR relieves LPS-induced oxidative stress. The formation of TBARS was used as an index for ROS production (27). Treatment of WT mice with LPS significantly increased the TBARS content as expected (Fig. 6A). In contrast, the increased TBARS production was prevented in LPS-treated LXR-KI mice and GW3965-treated WT mice (Fig. 6A). The basal level of TBARS in LXR-KI mice was not affected (Fig. 6A). To determine the mechanism by which LXR inhibited oxidative stress, we measured the activities of antioxidant enzymes that include GST, catalase, and SOD in the mouse lung homogenates. Activation of LXR increased GST activity regardless of the LPS treatment (Fig. 6B), which was consistent with the induction of GST mRNA expression in LXR-KI mice and LXR agonist-treated WT mice (Fig. 3 and supplemental Table S1). Treatment of WT mice with LPS significantly decreased catalase activity (Fig. 6C), and LPS remained effective to reduce catalase activity in LXR-KI mice and GW3965-treated WT mice. The SOD activity was modestly decreased in LPS-treated WT mice, but the difference did not reach statistical significance (Fig. 6D). Interestingly, activation of LXR decreased both the basal and LPS-responsive SOD activities (Fig. 6D).
FIGURE 6.
Activation of LXR decreased LPS-induced oxidative stress and inhibited pulmonary inflammatory response in vivo and in A549 lung cancer cells. A, the TBARS content was measured in the BAL fluids of mice treated with saline or LPS. GW, GW3965. B–D, the lung homogenates were measured for the activities of SOD (B), catalase (C), and GST (D) as described under “Experimental Procedures” (n = 6 for each group). E, the mRNA expression of tumor necrosis factor α and IL-1β in LPS-treated WT and LXR-KI mice as measured by real time PCR analysis. F, the mRNA expression of IL-6, IL-8, and monocyte chemotactic protein-1 in A549 cells treated with LPS (100 nm) for 24 h in the absence or presence of GW3965 (10 μm). *, p < 0.05; NS, statistically not significant (p > 0.05). Veh, vehicle.
LXRs are known for their anti-inflammatory function in the macrophages. As expected, the pulmonary inflammatory responses were inhibited in LPS-treated LXR-KI mice, as evidenced by the reduced mRNA expression of IL-1β and tumor necrosis factor α in the lung (Fig. 6E). In cultured A549 cells, treatment with GW3965 decreased the mRNA expression of IL-6, IL-8, and monocyte chemotactic protein-1 (Fig. 6F), which are cytokines important for neutrophil recruitment (43). Because A549 cells are derived from type II lung epithelial cells, our results suggest that LXRs also have an anti-inflammatory effect in nonmacrophages.
DISCUSSION
Oxidative stress plays an important role in the pathogenesis of various pulmonary diseases. Much work has been done to investigate the protective role of antioxidant enzymes, including GST, glutathione peroxidase, metallothionein, SOD, and catalase, in the lung. Our results show that LXR, a nuclear receptor previously known for its role in cholesterol and lipid homeostasis, can affect the oxidative stress response by regulating the expression of antioxidant genes in the lung. Genetic (VP-LXR) or pharmacological (LXR agonist) activation of LXR in mice alleviated LPS-induced lung injury, which was associated with the activation of several antioxidant genes, including Gsta2, Gsta4, Gstm1, Gstp1, Gpx1, Gpx3, Mt1, and Mt2. The mechanism by which LXRs regulate these antioxidant genes remains to be defined. To our knowledge, no work has been published in this area. Our preliminary data suggested that GST M might be under the direct transcriptional regulation of LXRs, but future studies are necessary to further define the mechanism by which LXRs regulate this GST isoform. Because LXRs are known to inhibit inflammatory responses (21), we cannot exclude the possibility that LXR-mediated regulation of cytokine expression was indirectly involved in the regulation of antioxidant genes. Mouse genetic background is known to influence the innate immune response (44). LXR-KI mice used in this study were maintained in the C57BL/6J-Strain 129S1/X1 mixed background. It remains to be determined whether the genetic background of LXR-KI mice will influence their phenotypic exhibition.
The inhibition of the expression of inflammatory cytokines in LXR-KI mice and LXR agonist-treated WT mice may have also contributed to the protective effect. Although LXR agonists have been suggested to protect lung from LPS-induced injury by inhibiting LPS-induced cytokine production (42, 43), the use of our newly created LXR-KI mice has provided the first genetic evidence that activation of LXRα was sufficient to confer resistance to LPS-induced lung injury. The inhibition of inflammatory response in GW3965-treated A549 cells is intriguing. LXRs are known for their anti-inflammatory function in macrophages, and at least some of the inflammatory inhibitory effects of LXRs are believed to be mediated through LXR antagonism of NF-κB, a positive regulator of inflammatory genes (45, 46). Interestingly, it was reported that the anti-inflammatory activity of LXR agonist in the lung was not mediated by the NF-κB/AP-1 pathway (47). A549 cells are derived from type II lung epithelial cells. Our results suggest that LXRs also have the anti-inflammatory effect in nonmacrophages. It remains to be determined whether the anti-inflammatory activity of LXR in A549 cells is achieved by antagonizing the NF-κB/AP-1 pathway. Given the fact that the LXR-KI is a whole body knock-in mouse model, we cannot exclude the possibility that the effect of LXR-KI allele on tissues outside the lung, such as the liver, may have also contributed to the protective effect in the lung.
In addition to LXRs, several other nuclear receptors have also been implicated in oxidative stress responses. Treatment of alveolar macrophages with PPARγ agonists resulted in the suppression of LPS-induced cytokine production, inducible nitric-oxide synthase expression, and oxidative burst (48, 49). PPARγ activation has also been shown to decrease alveolar inflammation in vivo in a murine model of lung injury induced by fluorescein isothiocyanate (50). In H4IIE rat hepatoma cells, PPARγ-retinoid X receptor heterodimers induced Gsta2 gene expression by transactivating the PPAR response element in the Gsta2 gene promoter, as well as by inducing the expression of Nrf2 and CCAAT-enhancer-binding protein β, two other positive regulators of GSTs (51). Interestingly, LXRα, but not LXRβ, was functionally grouped with PPARγ based on their similarities in the tissue distribution pattern (52). Both LXRα and PPARγ play important roles in lipid metabolism and atherosclerosis (53). Our results suggest that these two receptors also share a similar function in preventing lung injury.
There are two LXR isoforms, LXRα and LXRβ. They share DNA-binding sites and many target genes. Most of the known LXR agonists can activate both LXR isoforms. However, accumulating evidence suggests that differences exist between these two isoforms (54). Because of the current lack of isoform-specific agonists, the use of isoform-specific LXR knock-out or transgenic mice represents an important strategy to understand the isoform-specific function of LXRs. In our “gain-of-function” LXR-KI mice, only LXRα is constitutively activated, which allows us to conclude that activation of LXRα alone is sufficient to prevent LPS-induced lung injury. The future creation of VP-LXRβ knock-in mice will allow a direct comparison of the function of LXRα and LXRβ in tissues including the lung. As a gain-of-function model, LXR-KI mice are superior to the fatty acid-binding protein-VP-LXRα transgenic mice, in which VP-LXRα was targeted to the liver and intestine under the control of the fatty acid-binding protein gene promoter (14). However, dictated by the fatty acid-binding protein promoter, the transgene was not targeted to tissues outside the hepato-intestinal axis that are also known to express LXRα. In contrast, LXR-KI mice normalize the tissue distribution patterns of VP-LXRα to those of the endogenous LXRα. The current study has demonstrated the utility of LXR-KI mice in studying the pulmonary function of LXRα. It is conceivable that this novel mouse model can also be used to examine the role of LXRα in many other LXRα-expressing tissues.
In summary, the current study has established a novel role of LXR in oxidative stress response and in preventing lung injury. It is hoped that drug activation of LXR may represent a novel therapeutic strategy for ROS detoxification and for the prevention and treatment of lung diseases, such as acute respiratory distress syndrome and chronic obstructive pulmonary disease.
Supplementary Material
Acknowledgments
We thank Carolyn Ferguson for technical assistance with the embryonic stem cell work, Dr. Robert Gibbs for assistance with the use of the confocal microscope, and Dr. Steve Strom for primary human hepatocytes. Normal human hepatocytes were obtained through the Liver Tissue Procurement and Distribution System, Pittsburgh, Pennsylvania, which was funded by National Institutes of Health Contract N01-DK-7-0004/HHSN267200700004C.
This work was supported, in whole or in part, by National Institutes of Health Grants ES014626 and DK076962 (to W. X.) and National Institutes of Health Shared Instrument Grant S10RR022515 (to Robert Gibbs).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- ROS
- reactive oxygen species
- BAL
- bronchoalveolar lavage
- GST
- glutathione S-transferase
- GW3965
- 3-[3-[N-(2-chloro-3-trifluoromethylbenzyl)-(2,2-diphenylethyl)amino]propyloxy]phenylacetic acid
- LPS
- lipopolysaccharide
- LXR
- liver X receptor
- TO1317 (T0901317)
- N-methyl-n-[4-(2,2,2-trifluoro-1-hydroxy-1-trifluoromethylethyl)-phenyl]-benzenesulfonamide
- KI
- knock-in
- SOD
- superoxide dismutase
- TBARS
- thiobarbituric acid-reactive species
- MPO
- myeloperoxidase
- SPB
- surfactant protein B
- WT
- wild type
- IL
- interleukin
- PPAR
- peroxisome proliferator-activated receptor.
REFERENCES
- 1.Chow C. W., Herrera Abreu M. T., Suzuki T., Downey G. P. (2003) Am. J. Respir. Cell Mol. Biol. 29, 427–431 [DOI] [PubMed] [Google Scholar]
- 2.Rahman I., Biswas S. K., Jimenez L. A., Torres M., Forman H. J. (2005) Antioxid. Redox Signal. 7, 42–59 [DOI] [PubMed] [Google Scholar]
- 3.Toyokuni S., Okamoto K., Yodoi J., Hiai H. (1995) FEBS Lett. 358, 1–3 [DOI] [PubMed] [Google Scholar]
- 4.Nakamura T., Nakamura H., Hoshino T., Ueda S., Wada H., Yodoi J. (2005) Antioxid. Redox Signal. 7, 60–71 [DOI] [PubMed] [Google Scholar]
- 5.Christofidou-Solomidou M., Muzykantov V. R. (2006) Treat Respir. Med. 5, 47–78 [DOI] [PubMed] [Google Scholar]
- 6.Rahman I. (2008) Ther. Adv. Respir. Dis. 2, 351–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mastruzzo C., Crimi N., Vancheri C. (2002) Monaldi. Arch. Chest Dis. 57, 173–176 [PubMed] [Google Scholar]
- 8.Cho H. Y., Reddy S. P., Kleeberger S. R. (2006) Antioxid. Redox Signal. 8, 76–87 [DOI] [PubMed] [Google Scholar]
- 9.Wesselkamper S. C., McDowell S. A., Medvedovic M., Dalton T. P., Deshmukh H. S., Sartor M. A., Case L. M., Henning L. N., Borchers M. T., Tomlinson C. R., Prows D. R., Leikauf G. D. (2006) Am. J. Respir. Cell Mol. Biol. 34, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes J. D., Pulford D. J. (1995) Crit. Rev. Biochem. Mol. Biol. 30, 445–600 [DOI] [PubMed] [Google Scholar]
- 11.Repa J. J., Mangelsdorf D. J. (2002) Nat. Med. 8, 1243–1248 [DOI] [PubMed] [Google Scholar]
- 12.Willy P. J., Umesono K., Ong E. S., Evans R. M., Heyman R. A., Mangelsdorf D. J. (1995) Genes Dev. 9, 1033–1045 [DOI] [PubMed] [Google Scholar]
- 13.Gong H., Xie W. (2004) Expert Opin. Ther. Targets 8, 49–54 [DOI] [PubMed] [Google Scholar]
- 14.Uppal H., Saini S. P., Moschetta A., Mu Y., Zhou J., Gong H., Zhai Y., Ren S., Michalopoulos G. K., Mangelsdorf D. J., Xie W. (2007) Hepatology 45, 422–432 [DOI] [PubMed] [Google Scholar]
- 15.Zhou J., Febbraio M., Wada T., Zhai Y., Kuruba R., He J., Lee J. H., Khadem S., Ren S., Li S., Silverstein R. L., Xie W. (2008) Gastroenterology 134, 556–567 [DOI] [PubMed] [Google Scholar]
- 16.Cummins C. L., Volle D. H., Zhang Y., McDonald J. G., Sion B., Lefrançois-Martinez A. M., Caira F., Veyssière G., Mangelsdorf D. J., Lobaccaro J. M. (2006) J. Clin. Invest. 116, 1902–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalaany N. Y., Gauthier K. C., Zavacki A. M., Mammen P. P., Kitazume T., Peterson J. A., Horton J. D., Garry D. J., Bianco A. C., Mangelsdorf D. J. (2005) Cell Metab. 1, 231–244 [DOI] [PubMed] [Google Scholar]
- 18.Laffitte B. A., Chao L. C., Li J., Walczak R., Hummasti S., Joseph S. B., Castrillo A., Wilpitz D. C., Mangelsdorf D. J., Collins J. L., Saez E., Tontonoz P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5419–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. (2000) Genes Dev. 14, 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tangirala R. K., Bischoff E. D., Joseph S. B., Wagner B. L., Walczak R., Laffitte B. A., Daige C. L., Thomas D., Heyman R. A., Mangelsdorf D. J., Wang X., Lusis A. J., Tontonoz P., Schulman I. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11896–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkateswaran A., Laffitte B. A., Joseph S. B., Mak P. A., Wilpitz D. C., Edwards P. A., Tontonoz P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12097–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph S. B., Castrillo A., Laffitte B. A., Mangelsdorf D. J., Tontonoz P. (2003) Nat. Med. 9, 213–219 [DOI] [PubMed] [Google Scholar]
- 23.Joseph S. B., Bradley M. N., Castrillo A., Bruhn K. W., Mak P. A., Pei L., Hogenesch J., O'connell R. M., Cheng G., Saez E., Miller J. F., Tontonoz P. (2004) Cell 119, 299–309 [DOI] [PubMed] [Google Scholar]
- 24.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Homanics G. E., Ferguson C., Quinlan J. J., Daggett J., Snyder K., Lagenaur C., Mi Z. P., Wang X. H., Grayson D. R., Firestone L. L. (1997) Mol. Pharmacol. 51, 588–596 [DOI] [PubMed] [Google Scholar]
- 26.Szarka R. J., Wang N., Gordon L., Nation P. N., Smith R. H. (1997) J. Immunol. Methods 202, 49–57 [DOI] [PubMed] [Google Scholar]
- 27.Ritter C., Andrades M. E., Reinke A., Menna-Barreto S., Moreira J. C., Dal-Pizzol F. (2004) Crit. Care Med. 32, 342–349 [DOI] [PubMed] [Google Scholar]
- 28.Bradley P. P., Priebat D. A., Christensen R. D., Rothstein G. (1982) J. Invest. Dermatol. 78, 206–209 [DOI] [PubMed] [Google Scholar]
- 29.Gong H., Singh S. V., Singh S. P., Mu Y., Lee J. H., Saini S. P., Toma D., Ren S., Kagan V. E., Day B. W., Zimniak P., Xie W. (2006) Mol. Endocrinol. 20, 279–290 [DOI] [PubMed] [Google Scholar]
- 30.Paoletti F., Mocali A. (1990) Methods Enzymol. 186, 209–220 [DOI] [PubMed] [Google Scholar]
- 31.Aebi H. (1984) Methods Enzymol. 105, 121–126 [DOI] [PubMed] [Google Scholar]
- 32.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 33.Rouser G., Fkeischer S., Yamamoto A. (1970) Lipids 5, 494–496 [DOI] [PubMed] [Google Scholar]
- 34.Zhou J., Zhai Y., Mu Y., Gong H., Uppal H., Toma D., Ren S., Evans R. M., Xie W. (2006) J. Biol. Chem. 281, 15013–15020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voorhout W. F., Veenendaal T., Haagsman H. P., Weaver T. E., Whitsett J. A., van Golde L. M., Geuze H. J. (1992) Am. J. Physiol. 263, L479–L486 [DOI] [PubMed] [Google Scholar]
- 36.Jia L., Xu M., Zhen W., Shen X., Zhu Y., Wang W., Wang X. (2008) Am. J. Physiol. Cell Physiol. 294, C47–C55 [DOI] [PubMed] [Google Scholar]
- 37.Jenkins J. K., Carey P. D., Byrne K., Sugerman H. J., Fowler A. A., 3rd (1991) Am. Rev. Respir. Dis. 143, 155–161 [DOI] [PubMed] [Google Scholar]
- 38.Been J. V., Zimmermann L. J. (2007) Eur. J. Pediatr. 166, 889–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willson D. F., Chess P. R., Notter R. H. (2008) Pediatr. Clin. North Am. 55, 545–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy M. A., Venkateswaran A., Tarr P. T., Xenarios I., Kudoh J., Shimizu N., Edwards P. A. (2001) J. Biol. Chem. 276, 39438–39447 [DOI] [PubMed] [Google Scholar]
- 41.Guo R. F., Ward P. A. (2007) Antioxid. Redox Signal. 9, 1991–2002 [DOI] [PubMed] [Google Scholar]
- 42.Lang J. D., McArdle P. J., O'Reilly P. J., Matalon S. (2002) Chest 122, 314S–320S [DOI] [PubMed] [Google Scholar]
- 43.Matsukawa A., Hogaboam C. M., Lukacs N. W., Lincoln P. M., Strieter R. M., Kunkel S. L. (1999) J. Immunol. 163, 6148–6154 [PubMed] [Google Scholar]
- 44.Wells C. A., Ravasi T., Faulkner G. J., Carninci P., Okazaki Y., Hayashizaki Y., Sweet M., Wainwright B. J., Hume D. A. (2003) BMC Immunol. 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castrillo A., Joseph S. B., Marathe C., Mangelsdorf D. J., Tontonoz P. (2003) J. Biol. Chem. 278, 10443–10449 [DOI] [PubMed] [Google Scholar]
- 46.Castrillo A., Joseph S. B., Vaidya S. A., Haberland M., Fogelman A. M., Cheng G., Tontonoz P. (2003) Mol. Cell 12, 805–816 [DOI] [PubMed] [Google Scholar]
- 47.Birrell M. A., Catley M. C., Hardaker E., Wong S., Willson T. M., McCluskie K., Leonard T., Farrow S. N., Collins J. L., Haj-Yahia S., Belvisi M. G. (2007) J. Biol. Chem. 282, 31882–31890 [DOI] [PubMed] [Google Scholar]
- 48.Reddy R. C., Keshamouni V. G., Jaigirdar S. H., Zeng X., Leff T., Thannickal V. J., Standiford T. J. (2004) Am. J. Physiol. Lung Cell Mol. Physiol. 286, L613–L619 [DOI] [PubMed] [Google Scholar]
- 49.Asada K., Sasaki S., Suda T., Chida K., Nakamura H. (2004) Am. J. Respir. Crit. Care Med. 169, 195–200 [DOI] [PubMed] [Google Scholar]
- 50.Christensen P. J., Goodman R. E., Pastoriza L., Moore B., Toews G. B. (1999) Am. J. Pathol. 155, 1773–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park E. Y., Cho I. J., Kim S. G. (2004) Cancer Res. 64, 3701–3713 [DOI] [PubMed] [Google Scholar]
- 52.Bookout A. L., Jeong Y., Downes M., Yu R. T., Evans R. M., Mangelsdorf D. J. (2006) Cell 126, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akiyama T. E., Sakai S., Lambert G., Nicol C. J., Matsusue K., Pimprale S., Lee Y. H., Ricote M., Glass C. K., Brewer H. B., Jr., Gonzalez F. J. (2002) Mol. Cell. Biol. 22, 2607–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albers M., Blume B., Schlueter T., Wright M. B., Kober I., Kremoser C., Deuschle U., Koegl M. (2006) J. Biol. Chem. 281, 4920–4930 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.