Abstract
The Wnts include a large family of secreted proteins that serve as important signals during embryonic development and adult homeostasis. In the most well understood Wnt signaling pathway, Wnt binding to Frizzled and low density lipoprotein receptor-related protein induces β-catenin protein stabilization and entry into the nucleus, resulting in changes in target gene transcription. Emerging evidence suggests that Wnt5a can inhibit Wnt/β-catenin signaling through interaction with the receptor Ror2. The Ror2 protein belongs to the receptor tyrosine kinase superfamily and contains several recognizable structural motifs. However, limited information is available regarding which specific domains are required for the inhibitory signaling activity of Wnt5a. Through mutation and deletion analysis, we have analyzed which specific domains and residues, including those necessary for tyrosine kinase activity, mediate the Wnt5a signal. To determine whether Ror2 can inhibit canonical Wnt signaling in vivo, we examined the effect of Ror2 loss on the expression of the Wnt reporter Axin2LacZ, finding increased reporter activity in Ror2 null mice, demonstrating that Ror2 can also inhibit Wnt/β-catenin signaling in the context of intact tissues.
Wnts include a large family of secreted hydrophobic proteins that regulate essential developmental processes, including embryonic development, cell growth, migration, and differentiation (1). When Wnt signaling is perturbed, cancer or degenerative diseases result (2–6). Thus, understanding the ways in which Wnt signaling is regulated is of great importance.
The large number of different Wnt family members evokes such questions as why so many Wnt proteins are necessary and whether they all signal through the same mechanism. Some clues come from studies in cell culture and intact organisms, which have shown that various intracellular signaling pathways become activated either directly or indirectly in response to different Wnt proteins (7–9). Ongoing questions in the field, thus, are how do distinct Wnt family members elicit differential cellular outcomes and which Wnts, if not all, activate each distinct pathway?
Whereas most Wnt signaling has been attributed to the activation of the Frizzled and low density lipoprotein receptor-related protein 5 and 6 (Lrp5/6) coreceptor proteins leading to β-catenin stabilization (a pathway called Wnt/β-catenin or canonical Wnt signaling), emerging evidence suggests that some Wnts can bind to alternative receptors to activate noncanonical signal transduction cascades (10, 11). For example, Wnt5a has been shown to interact with one such alternative Wnt receptor, Ror2, leading to the inhibition of Wnt/β-catenin signaling (12–14). Although it has been shown that this inhibition can occur at the level of β-catenin stabilization or T cell factor/lymphoid enhancer factor gene transcription, much remains to be understood regarding the cytosolic pathway that is activated in response to Wnt5a binding. Thus, research into the mechanism by which Ror2 functions will further our understanding of this novel regulatory pathway.
The Ror2 receptor belongs to the receptor tyrosine kinase (RTK)2 superfamily (15). This large protein family plays an important role in regulating diverse cellular processes ranging from the cell cycle, cell migration, as well as cell proliferation and differentiation (16). The Ror2 protein and its homolog Ror1 play essential roles during development (17). Mutations of the Ror2 receptor, resulting in protein misfolding or premature truncation, have been associated with human diseases such as dominant Brachydactyly B and recessive Robinow syndrome (18). These autosomal syndromes are associated with a variety of phenotypes that vary in their degree of severity depending on which specific mutation has occurred (Fig. 1A). Defects associated with these diseases include brachydactyly (a shortening of the digits), skeletal dysplasia, congenital heart disease, and craniofacial abnormalities (19, 20). Ror2 null mice exhibit skeletal abnormalities similar to the human syndromes. In addition, these mice display heart and lung defects and die perinatally, suggesting that Ror2 may play a broad role in the embryonic development of various tissues (20–22).
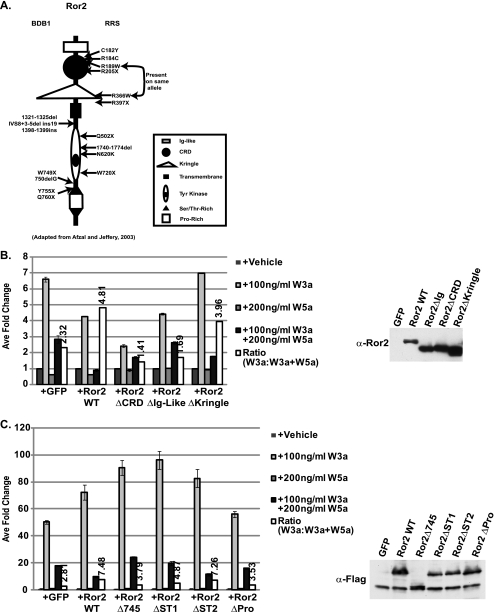
FIGURE 1.
Deletion of mRor2 extracellular and intracellular domains results in decreased Wnt5a-mediated inhibition of Wnt/β-catenin signaling. A, diagram of the major domains of the mRor2 receptor. B, extracellular domain deletion mutants of mRor2 (ΔCRD and ΔIg-like) show reduced ability to enhance Wnt5a-mediated inhibition. ∼40 h post-transfection with STF reporter and Ror2 variants, HEK293 cells were treated with vehicle, Wnt3a protein alone, Wnt5a protein alone, or Wnt3a protein plus Wnt5a protein for an additional 20 h. Following reporter assay, cell lysates were examined for receptor expression via Western blot analysis with α-Ror2 antibody. GFP, green fluorescent protein; WT, wild type. C, intracellular domain deletion mutants of mRor2 show reduced ability to enhance Wnt5a-mediated inhibition. Cells were transfected with Ror2 deletion variants described and treated as indicated above. Following reporter assay, cell lysates were examined for receptor expression via Western blot with α-FLAG antibody.
Despite the increased interest in the Ror2 receptor, relatively few efforts have been made to determine which specific residues are required for Wnt5a-mediated Ror2 inhibitory signaling (23), hereafter referred to as Wnt5a/Ror2 signaling. To that end, we performed mutation/deletion analysis of the receptor and have determined which specific intracellular domains and residues are necessary for Ror2-mediated Wnt5a signaling.
Although there is strong in vitro evidence suggesting that Ror2 can inhibit Wnt/β-catenin signaling, this has not been tested in vivo. Thus, to determine whether Ror2 can inhibit Wnt/β-catenin signaling in the context of a whole organism, we analyzed Ror2 protein expression, using a monoclonal antibody we generated, and the effect of Ror2 loss on Wnt/β-catenin signaling in Wnt reporter mice. We find that the absence of Ror2 leads to enhanced Wnt/β-catenin signaling, specifically in cells that have lost Ror2 expression. Our data, taken together, suggest that Ror2 can inhibit canonical Wnt signaling in vivo and point to Ror2 as a potential therapeutic target for human disease.
EXPERIMENTAL PROCEDURES
Mono- and Polyclonal Anti-Ror2 Antibody Generation
To generate anti-Ror2 antibodies, bases 2535–2835 of the mouse Ror2 receptor were fused in-frame to GST and maltose-binding protein (MBP) to create Ror2-GST and Ror2-MBP fusion proteins that were subsequently purified from Escherichia coli. For rabbit anti-mRor2 polyclonal antibody, purified Ror2-MBP fusion protein was injected into rabbits following standard procedures (Josman Laboratories). The final bleed was subsequently affinity-purified against the purified Ror2-GST fusion protein. For rabbit anti-phospho-Tyr-645 Ror2 antibodies, a peptide (Arg-Glu-Val-Tyr-Ser-Ala-Asp-pTyr-Tyr-Lys-Leu-Met-Gly-Asn-Ser) conjugated to keyhole limpet hemocyanin (Anaspec, San Jose, CA) was injected into rabbits following standard procedures (Josman Laboratories). The final bleed was subsequently affinity-purified against a biotinylated peptide (Anaspec, San Jose, CA).
Anti-Ror2 monoclonal antibodies were generated by injection of Ror2-MBP into mice following standard protocols. Following analysis of test bleeds, splenic cells were fused to form hybridomas. Following screening via plate ELISA, two independent cell clones were scaled up in serum-free media, and antibodies were isolated from conditioned media using standard methods. Following concentration, antibodies were dialyzed into PBS and diluted with 0.1% azide, 50% glycerol. These antibodies are specific to the mouse and human forms of Ror2 but do not detect the mRor1 homolog as would be predicted based on sequence divergence in this region (supplemental Fig. S1). Whereas both antibodies are suitable for Western blotting and other applications, the rabbit polyclonal antibody exclusively detects mRor2 in frozen tissue sections whereas the mouse monoclonal antibody exclusively detects mRor2 protein in paraffin-embedded tissues.
cDNA Constructs and Antibodies
SuperTopFlash was obtained from the Dr. Randall Moon Laboratory (24). pEF1α-myc/his A vector was obtained from Invitrogen, and pEF1a-LacZ (Invitrogen) was used for β-galactosidase activity normalization. pcDNA3-mRor2 and mRor2ΔCRD(Δ508–906) constructs were obtained from the Minami laboratory (51) and were subsequently subcloned into pEF1α vector. pEF-mRor2ΔIg(Δ166–438), pEF-mRor2ΔKringle(Δ943–1185), pEF- mRor2N620K, pEF-mRor2ΔST1(Δ2232–2346), pEF-mRor2ΔST2(Δ2580–2646), pEF-mRor2ΔPro(2349–2577), pEF- mRor2Y641F, pEF-mRor2Y645F, pEF-mRor2Y646F, and pEF-mRor2Y641,Y645F,Y646F constructs were created via sewing PCRs using overlapping primer sets. All Ror2 variants were FLAG-tagged at the C terminus and all DNA segments generated by PCR were sequenced to rule out spurious mutations.
Cell Culture, Mammalian Cell Transfection, and Luciferase Assays
HEK293 cells were cultured in Dulbecco's modified Eagle's medium, 10% fetal bovine serum, and antibiotics. Transient transfections were performed with Lipofectamine2000 (Invitrogen) on HEK293 cells plated the night before into 6-well plates. Empty vector, GFP, pEF1-mRor2, or derivatives were transfected (1 μg/well) together with the SuperTopFlash (STF) luciferase reporter (2 μg/well) and β-galactosidase (0.33 μg) (pEF-1α-LacZ, Invitrogen) plasmids. 24 h post-transfection, cells were re-plated into a 96-well plate, allowed to recover for 6–8 h, and then treated in triplicate with Wnt proteins as described in text. Luciferase assays were performed using Dual-Light reporter gene assay system (Applied Biosystems). Relative luciferase units were measured and normalized against β-galactosidase activity 48 h post-transfection. Error bars represent standard deviation, and each assay was performed at least two times.
Cell Extraction and Western Blotting
Cells were lysed in TNT buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Triton X-100) supplemented with protease and phosphatase inhibitors on ice for 10 min. After clearing lysates with high speed spin, protein supernatants were diluted into SDS sample buffer. Samples were resolved on SDS-10% polyacrylamide gels and transferred to nitrocellulose. The membranes were incubated for 1 h in blocking solution (1% bovine serum albumin, 3% nonfat dry milk in Tris-buffered saline containing 0.2% Tween 20) and then overnight at 4 °C with antibody in blocking solution.
For IP-kinase assay, cells were treated with vehicle or Wnt5a protein for 10 min and then lysed in cold TNT buffer supplemented with protease and phosphatase inhibitors. Lysates were immunoprecipitated overnight with anti-FLAG antibodies (M2, Sigma) and protein G beads at 4 °C. IPs were washed three times with kinase buffer (60 mm Tris, pH 7.3, 12 mm MgCl2) and then incubated with buffer containing 40 μm ATP with GST protein or GST protein fused to bases 1884–2006 of mRor2 for 30 min at 30 °C with rocking. Beads were spun down, and supernatants containing the GST peptides were brought up in sample buffer and analyzed via Western blot using anti-phosphotyrosine or anti-GST antibodies. For sandwich ELISA, following kinase assay, peptide supernatants were bound to a plate pre-bound with anti-phosphotyrosine antibodies. Following a 1-h room temperature incubation, wells were washed and incubated with rabbit anti-GST antibody followed by washing and incubation with alkaline phosphatase-conjugated anti-rabbit antibody. After washing, wells were incubated for 30 min at room temperature with alkaline phosphatase (p-nitrophenyl phosphate) liquid substrate, and the absorbance at 405 nm was then measured.
Immunofluorescence Staining
For immunofluorescence microscopy, 48 h post-transfection cells grown on glass coverslips were fixed with 4% paraformaldehyde at 4 °C for 10 min and permeabilized in methanol at −20 °C for 20 s. The coverslips were then exposed to primary antibodies, followed by Cy3-conjugated secondary antibodies. Slides were mounted with Vectashield mounting media with 4′,6-diamidino-2-phenylindole (Vector Laboratories), and fluorescence was examined with a Zeiss Axioplan 2 microscope.
Tissue Processing
Mouse embryos were isolated from pregnant females following timed matings and fixed in paraformaldehyde. Following PBS washes, specimens were either incubated with 30% sucrose and embedded into optimal cutting temperature for cryo-sectioning or dehydrated with increasing concentrations of ethanol, incubated in histoclear, and paraffin-embedded. Paraffin-embedded embryos were cut into 7-μm sections and stained using anti-Ror2 monoclonal antibodies at a 1:100 dilution using the Vectashield ABC kit with diaminobenzidine substrate (Vector Laboratories) or Envision+ diaminobenzidine kit (DAKO) according to the manufacturer's instructions, counterstained with hematoxylin, and mounted in nonaqueous mounting media. For fluorescent imaging of cryosections, embedded embryos were cut into 10–20-μm sections. Tissues were blocked for 1 h with PBS/Tween with 2.5% bovine serum albumin and then incubated with 1:100 anti-Ror2 polyclonal antibodies in blocking buffer for 1 h at room temperature or overnight at 4 °C. Tissues were washed with PBS/Tween, and subsequently incubated with fluorescent secondary antibodies for 1 h at room temperature. After washing, slides were mounted with ProLong Gold with 4′,6-diamidino-2-phenylindole (Invitrogen) and visualized on a Zeiss upright microscope. For LacZ staining, cryosections were incubated overnight at room temperature with LacZ staining solution and then postfixed with paraformaldehyde. Following washes, slides were counterstained with nuclear fast red and mounted in aqueous mounting media.
RESULTS
Ror2 CRD and Ig-like Domains Are Required for Wnt5a-mediated Inhibition of Wnt/β-Catenin Signaling
The Ror2 protein is a transmembrane receptor tyrosine kinase that contains several recognizable structural motifs (Fig. 1A). To measure Ror2 signaling activity, we used SuperTopFlash (STF) (24), a T cell factor/lymphoid enhancer factor luciferase reporter that serves as a transcriptional readout for Wnt/β-catenin signaling, following exposure of cells to various concentrations of purified Wnt3a or Wnt5a protein. As we have shown previously, Wnt3a activates the STF reporter in HEK293 cells, whereas Wnt5a protein treatment does not stimulate reporter activity. When cells are treated with both Wnt proteins simultaneously, Wnt5a inhibits the ability of Wnt3a to activate the reporter ∼2.5-fold, which is due to endogenous expression of Ror2 in HEK293 cells (Fig. 1B) (14).
We next took advantage of the fact that Wnt5a-mediated inhibition of STF is enhanced in the presence of overexpressed wild type mRor2 to assay the activity of mutated forms of the mRor2 molecule. To determine which extracellular domains are important for Wnt5a signaling, we created mRor2 variants wherein the cysteine-rich domain, Ig-like, and Kringle domains were deleted (Ror2ΔCRD, Ror2ΔIg, and Ror2ΔKringle, respectively) and then assayed their activity in the STF assay. Mouse monoclonal and rabbit polyclonal antibodies directed against the last 100 amino acids of the mRor2 protein were generated to monitor Ror2 expression in vitro and in vivo. These antibodies are specific to the mouse and human forms of Ror2 but do not detect the mRor1 homolog as would be predicted based on sequence divergence in this region (supplemental Fig. S1A). In addition, although these antibodies were generated against the same peptide, they detect differing epitopes (supplemental Fig. S1B). Western blot analysis of the luciferase assay lysates revealed that all Ror2 variants were expressed at equivalent levels (Fig. 1B).
Deletion of the CRD results in a mutant form (mRor2ΔCRD) that does not enhance the Wnt5a-mediated inhibition of the STF reporter (Fig. 1B). The inhibition observed is similar to that seen when cells are transfected with empty vector, providing evidence that the CRD domain is required for mRor2 to transduce the Wnt5a signal. We next assayed the Ror2ΔIg and Ror2ΔKringle constructs in the Wnt3a inhibition assay. Ig-like domains are general protein-protein binding domains found in many proteins and almost all RTKs. Kringle domains, however, contain a characteristic pattern of cysteine residues and are found in a smaller subset of proteins such as clotting factors. The Rors are the only RTK family members that possess Kringle domains, which suggests that the mRor2 Kringle domain might be an important motif necessary for conferring ligand specificity (15). Interestingly, deletion of these domains showed that the Ig-like domain is required for Wnt5a signaling, but the Kringle domain is not.
Several Ror2 Intracellular Domains Are Required for Wnt5a Signaling
Brachydactyly B and recessive Robinow syndromes are caused by a variety of genetic mutations (18). Several of these mutations (missense, nonsense, and frameshift) result in the premature truncation of the Ror2 protein (Fig. 1A). Frameshift mutations at amino acids 441, 463, or 467 are associated with one or both diseases. In addition, frameshift or missense mutations resulting in premature stop of the coding sequence cluster to the area around amino acids 749–755. To test the effect of truncation of Ror2 at similar positions in our Wnt5A/Ror2 signaling assay, we generated FLAG-tagged Ror2 variants that terminated at amino acid position 469 (mRor2Δ469) or 745 (mRor2Δ745). mRor2Δ469 lacks almost the entire intracellular domain of Ror2, whereas mRor2Δ745 is truncated immediately following the tyrosine kinase domain. Following transient transfection into HEK293 cells with the STF reporter and subsequent Wnt treatment, both intracellular deletion variants displayed significantly reduced signaling potential as compared with wild type mRor2 (supplemental Fig. S2) even though the transfected receptors were expressed in equivalent amounts. This indicates that the intracellular domain of Ror2 is required for functional Wnt5a/Ror2 signaling.
To determine the importance of specific subdomains within the intracellular portion of the receptor, we individually deleted the mRor2 serine/threonine (Ser/Thr)-rich domains 1 and 2 and the proline (Pro)-rich domain. As shown in Fig. 1C, loss of Ser/Thr-rich domain 1 (ST1) and the Pro-rich domain (Pro) reduced Ror2 signaling activity, similarly to the mRor2Δ745 mutant. Interestingly, deletion of Ser/Thr-rich domain 2 (ST2) had minimal effect on receptor activity suggesting that only the Ser/Thr-rich domain 1 and the proline-rich domain carry out crucial signaling and/or structural functions.
Tyrosine Kinase Activity Is Necessary for mRor2 Signaling
By sequence similarity to other closely related RTKs, the mRor2 receptor should be a fully functional tyrosine kinase. The asparagine residue at amino acid 620, invariant in all receptor tyrosine kinases, is likewise conserved in the Ror receptor family. In humans, mutation of this residue to lysine (N620K) results in recessive Robinow syndrome (18). Structural analyses have indicated that this residue binds to one of the magnesium ions within the catalytic cleft of tyrosine kinases with homology to Ror2 (15, 25). Although others have shown that Wnt5a is able to induce Ror2 signaling through tyrosine kinase activation (26, 27), it has not been shown that tyrosine kinase activity is required for the inhibitory effects of Wnt5a on Wnt/β-catenin signaling, leading us to investigate this using several approaches. We found that overexpression of this predicted kinase-inactive variant of Ror2, Ror2N620K, did not enhance Wnt5a inhibition of Wnt/β-catenin signaling above base line (Fig. 2A). The fact that mRor2N620K was unable to signal in a wild type fashion potentially links impaired Wnt/Ror2 signaling to recessive Robinow syndrome.
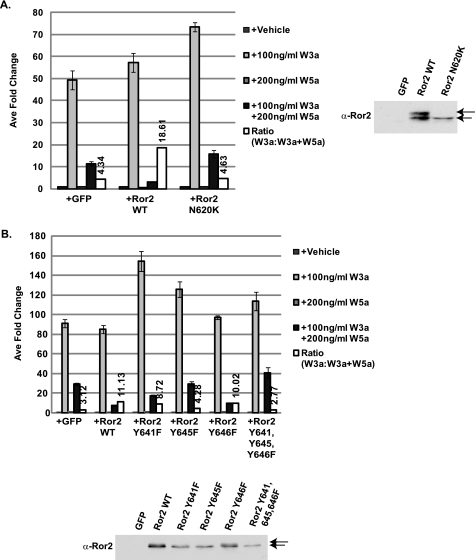
FIGURE 2.
Point mutation of the tyrosine kinase domain results in impaired signaling ability. A, N620K point mutation, which is associated with recessive Robinow syndrome, results in impaired Wnt5a-mediated inhibitory activity. ∼40 h post-transfection with STF reporter and Ror2 variants, HEK293 cells were treated with vehicle, Wnt3a protein alone, Wnt5a protein alone, or Wnt3a protein plus Wnt5a protein for an additional 20 h. Following reporter assay, cell lysates were examined for receptor expression via Western blot analysis with α-Ror2 antibody. GFP, green fluorescent protein; WT, wild type. B, point mutation of tyrosine residues within the catalytic loop of the tyrosine kinase domain results in impaired inhibitory activity. Cells were transfected with Ror2 deletion variants described in text and treated as indicated above. Following reporter assay, cell lysates were examined for receptor expression via Western blot analysis with α-Ror2 antibody.
To further interrogate the role of Ror2 tyrosine kinase activity in Wnt5a signaling, we point-mutated conserved tyrosine residues within the tyrosine kinase regulatory loop of Ror2. Mutation of tyrosine residues 641, 645, and 646 singly, and in combination, to phenylalanine resulted in varying degrees of receptor dysfunction, with Y645F displaying the greatest loss of activity. Mutation of equivalent residues within the insulin receptor regulatory loop results in an inactive kinase (28, 29). These independent mRor2 mutations in combination with the mRor2N620K data indicate the requirement of an active tyrosine kinase domain for functional Wnt5a signaling.
As shown in Fig. 2, A and B, all of the mutated receptors were expressed in equivalent amounts. Interestingly, whereas wild type Ror2 runs as a doublet following Western blot analysis with our anti-Ror2 antibodies, Ror2N620K appears as a single band. This is also what is observed when Tyr-641 and Tyr-645 are mutated to phenylalanine, the point mutations within the regulatory loop that result in the most severe loss of function. Additional mutational analysis of the Ror2 receptor revealed that only mutation of these specific residues resulted in the loss of the slower migrating band (supplemental Fig. S2). These data indicate that the wild type receptor is post-translationally modified in a manner different from the tyrosine kinase domain mutants. Because Ror2 can autophosphorylate (30), these observations likely indicate that loss of kinase activity resulted in the lack of autophosphorylation at other distant sites on the receptor and suggest that Wnt5a protein directly activates mRor2 tyrosine kinase activity.
One reason why the mutant Ror2 receptors (mRor2N620K, mRor2Y645F, etc) used in our analyses above may be nonfunctional in our Wnt5a-mediated inhibition assays is that the mutated receptors may not be properly localized to the cell surface. As assessed by fluorescence microscopy imaging and staining with either anti-FLAG or rabbit anti-Ror2 antibodies, the receptor molecules were detected on the plasma membrane (supplemental Fig. S3). Thus, not only were all of the mutant proteins expressed, they were also accessible to ligand, suggesting that the inability of the various mutants to signal is due to in part to receptor inactivation and not simply defective trafficking as has been described previously for mutations associated with recessive Robinow syndrome (31, 32).
Wnt5a Directly Modulates Ror2 Tyrosine Kinase Activity
As shown above, receptor mutagenesis suggested that mRor2 tyrosine kinase activity is necessary for receptor function. Following their activation, most tyrosine kinase receptors become autophosphorylated via cross-phosphorylation (33). Thus, to test whether Wnt5a protein directly activates Ror2 tyrosine activity, we performed IP kinase assays. Overexpressed mRor2 was immunoprecipitated from lysates made from cells treated with vehicle or Wnt5a for 10 min. The immunoprecipitated protein was then incubated with kinase buffer and either GST protein (as a control) or a protein in which GST is fused to the mRor2 tyrosine kinase domain to determine whether full-length Ror2 protein could phosphorylate its own tyrosine kinase domain following ligand-induced activation. After the kinase assay was performed, the GST peptides were analyzed for phosphotyrosine incorporation via Western blot analysis with anti-phosphotyrosine antibodies. Fig. 3A shows that immunoprecipitated mRor2 protein phosphorylated only the GST-Ror2 fusion peptide and that Wnt5a treatment of cells resulted in increased phosphotyrosine incorporation into the GST-Ror2 peptide as compared with vehicle treatment.
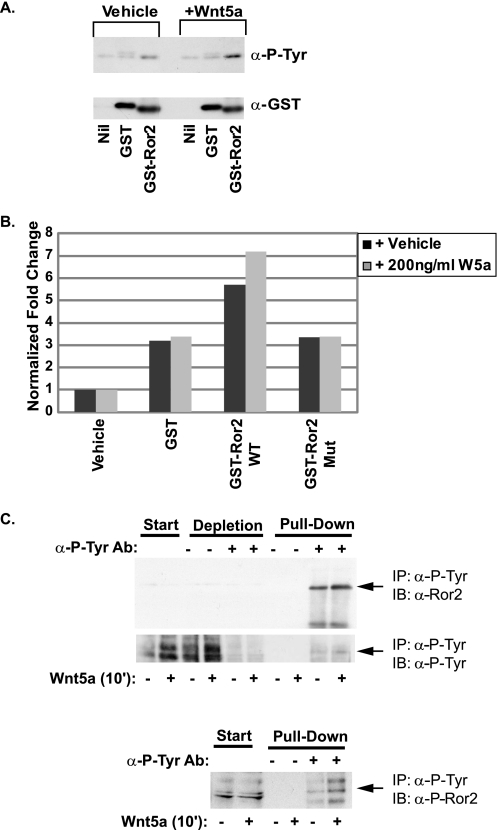
FIGURE 3.
Wnt5a protein treatment enhances Ror2 tyrosine kinase activity. A, IP-kinase assay shows that immunoprecipitated overexpressed mRor2 can phosphorylate a GST-Ror2 tyrosine kinase domain fusion protein as assessed by Western blot analysis of peptides using anti-phosphotyrosine antibodies. This activity is enhanced following treatment with Wnt5a protein. The membrane was stripped and reprobed with anti-total GST antibodies to show equivalent protein loading. B, quantitative IP-kinase assay shows that immunoprecipitated mRor2 can phosphorylate a GST-Ror2 tyrosine kinase domain fusion peptide but not a tyrosine residue mutated GST-Ror2 tyrosine kinase domain fusion peptide (GST-Ror2 Mut), as assessed by sandwich ELISA. WT, wild type. See under “Experimental Procedures” for details. C, Wnt5a treatment of HEK293 cells results in more endogenous Ror2 protein being immunoprecipitated (IP) from cell lysates using anti-phosphotyrosine antibodies. In addition, more phospho-Ror2 protein is immunoprecipitated by anti-phosphotyrosine antibodies as assessed by anti-phospho-Ror2 antibodies. Ab, antibody; IB, immunoblot.
As an independent quantitative assay for tyrosine kinase activity, the peptide supernatants were analyzed via sandwich ELISA following the kinase assay (Fig. 3B). Immunoprecipitated mRor2 protein was again able to increase phosphotyrosine incorporation into the GST-Ror2 tyrosine kinase domain fusion protein, an activity that was increased following Wnt5a stimulation. By contrast, GST protein alone or a point-mutated GST-Ror2 fusion protein (GST-Ror2 Y641A,Y645A,Y646A) showed no increase in phosphotyrosine incorporation under both conditions.
Finally, to determine whether Wnt5a activates the tyrosine kinase activity Ror2 in a cell-based assay, HEK293 cells, which express endogenous Ror2 protein, were treated with vehicle or Wnt5a for 10 min. Protein lysates were immunoprecipitated with anti-phosphotyrosine antibodies and then immunoblotted for Ror2. Following Wnt5a stimulation, more Ror2 protein was immunoprecipitated by phosphotyrosine antibodies (Fig. 3C). In addition, more phospho-Ror2 Tyr-645 was immunoprecipitated by phosphotyrosine antibodies as assessed by Western blot analysis using a phospho-Ror2 antibody we generated against the Tyr(P)-645 within the Ror2 tyrosine kinase regulatory loop (supplemental Fig. S1). Thus, in both cell-free and cell-based assays, Wnt5a enhances mRor2 tyrosine kinase activity.
mRor2 Inhibits Wnt/β-Catenin Signaling in Vivo
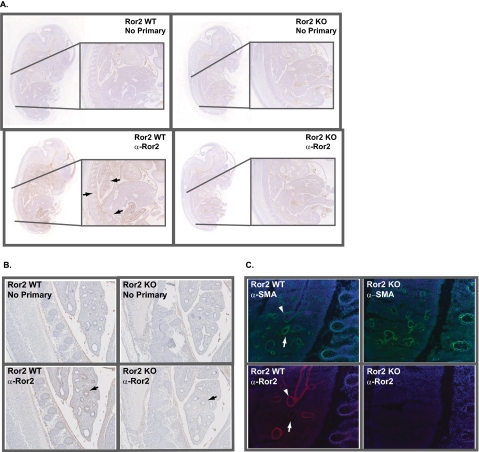
To address whether Ror2 functions in Wnt5a signaling in vivo, we first examined the location of the Ror2 protein in wild type and, as a control, in Ror2 mutant mice (22). Using the anti-Ror2 monoclonal antibody that we generated, mRor2 staining was performed on whole mount sections of paraffin-embedded E13.5 embryos. Although only background staining was observed in the absence of primary antibody or in Ror2 null embryos, sagittal sections of wild type mice showed specific staining in the developing rib and vertebral anlagen and also in the telencephalic neuroepithelium of the forebrain and snout region (Fig. 4A). Specific staining was also detectable in the developing lung, kidneys, and heart as has been described previously (34–36). This pattern of expression is similar to that of Wnt5a, which has been described to be prominently expressed in the perichondrium throughout the developing embryonic skeleton, including the vertebral column and the trachea (37).
FIGURE 4.
Embryonic mRor2 expression analysis. A, monoclonal anti-Ror2 antibody was used to stain whole mount E13.5 Ror2+/− and Ror2−/− embryos. In paraffin-embedded sections, Ror2 expression is observed in the developing rib and vertebral anlagen and also in the telencephalic neuroepithelium of the forebrain and snout regions. Staining is also observed in the developing lung and heart. B, higher magnification reveals a mesenchymal cell Ror2 staining pattern in the developing lung. C, α-SMA immunofluorescence staining of frozen sections of E14.5 Ror2+/− and Ror2−/− embryos reveals that Ror2 colocalizes with a subset of α-SMA-positive cells. WT, wild type.
Ror2 null embryos die perinatally partially due to defects in alveolar expansion of the lung (22). To examine mRor2 expression in tissues where Ror2 is known to play an important role during embryonic development, sagittal sections of E14.5 paraffin-embedded embryos were stained with anti-Ror2 monoclonal antibodies. Ror2 expression was highest in the cells that surround airway epithelial cells, in a pattern consistent with cells of mesenchymal origin (Fig. 4B).
Using our Ror2 polyclonal antibody, fluorescence imaging of frozen sections taken from E14.5 embryos revealed that Ror2-positive cells are contained in a subset of cells that stain positive for α-smooth muscle actin (α-SMA), a marker for mesenchymal cell differentiation previously shown to be present in lung airway and vasculature, confirming the mesenchymal nature of the Ror2-positive cells. Again, the staining pattern observed for Ror2 seemed to complement the previously observed expression of Wnt5a in the embryonic lung, where low level, diffuse expression of Wnt5a protein is localized to both the epithelial and mesenchymal compartments of the E12 lung (38).
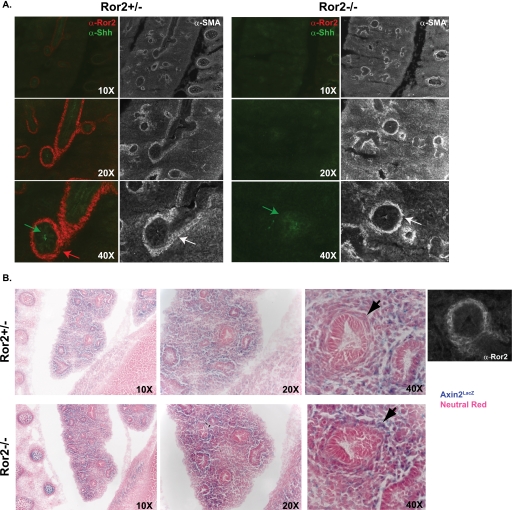
It has been shown previously that Wnt5a signaling regulates chick pulmonary development in part due to its effect on Sonic hedgehog (Shh) protein levels (39). To test whether Ror2 loss in the mouse also affects Shh protein levels in the lung, we stained 16.5 embryonic sections for Shh. In wild type E16.5 embryos, Shh protein is localized primarily to cells of the epithelial airway (Fig. 5A). No major differences in the Shh protein staining pattern or intensity were observed in E16.5 Ror2 null embryos.
FIGURE 5.
Lack of Ror2 expression results in increased Wnt/β-catenin signaling. A, Shh expression appears unaffected by Ror2 loss. E16.5 Ror2+/− and Ror2−/− embryos were stained with α-Ror2 and α-Shh antibodies; serial sections were stained with α-SMA antibodies to indicate mesenchymal staining pattern. B, loss of Ror2 results in increased Wnt/β-catenin signaling. Axin2+/LacZ Ror2+/− mice were crossed to Ror2+/− mice to produce Axin+/LacZ Ror2+/− or Axin2+/LacZ Ror2−/− embryos. Immunofluorescence staining of frozen sections of E16.5 embryos shows that cells specifically lacking Ror2 display increased LacZ expression indicating increased Wnt/β-catenin signaling.
Finally, to determine what effect mRor2 loss has on Wnt/β-catenin signaling in vivo, we crossed mRor2 mutant animals to a Wnt/β-catenin signaling reporter mouse strain: AxinlacZ/+ (40). Axin2 is a Wnt target gene in many mouse tissues (41–43). In AxinlacZ/+ mice, one copy of the Axin2 locus is replaced by the lacZ gene, expression of which serves as a read-out for Wnt/β-catenin signaling. Analysis of mRor2 null, LacZ-positive embryo (AxinlacZ/+ mRor2−/−) mice revealed that cells that no longer express Ror2 display more β-galactosidase activity than cells examined from littermate control AxinlacZ/+ Ror2+/− embryos. This increase in LacZ expression was specific to regions where cells lost Ror2 expression; no general increase in LacZ activity was observed in surrounding cells that do not normally express mRor2. These data confirm that Ror2 protein expression can affect Wnt/β-catenin signal transduction in vivo.
DISCUSSION
Even though the Ror2 receptor has been shown to bind directly to Wnt proteins by various groups, the call for “deorphanization” of this important receptor has only recently been sounded (44). Although the role of mRor2 in noncanonical Wnt signaling has been intensely investigated in recent years (12, 23, 26, 45–47), few studies have addressed the ability of mRor2 to transduce a signal mediated by Wnt5a, ultimately resulting in Wnt/β-catenin signaling inhibition. Using various approaches, we show in this study that Ror2 acts as a classical ligand-dependent tyrosine kinase receptor, the ligand being the Wnt5a protein. Through mutation and deletion analyses of the receptor, we have determined that two extracellular domains, the Ig-like and CRD, and three domains within the cytoplasmic domain, the tyrosine kinase, Ser/Thr-rich 1, and pro-rich domains, are required for Ror2 to transduce the inhibitory signal of Wnt5a. In addition, the embryonic Ror2 expression pattern was examined to help discern important anatomical sites of Ror2 function.
In some model systems, the cytoplasmic domain has been shown to be dispensable for Ror2 function, and it has been suggested that Ror2 may simply capture Wnts and present them to other cell surface receptors. In Xenopus, mutant variants of XRor2 that lack the cytoplasmic domain are able to inhibit convergence and extension movements similar to wild type XRor2 (48). In addition, deletion of the Caenorhabditis elegans CAM-1 cytoplasmic domain results in a protein that is able to negatively regulate Egl-20 (Wnt ortholog) signaling during Q cell migration in a manner indistinguishable from wild type (49, 50). With respect to the data in C. elegans, however, Green et al. (44) have shown that CAM-1/Ror exerts it function in cells adjacent to Wnt target cells, working as a “sink” to modulate Wnt signaling nonautonomously. Importantly, we show here that the cytoplasmic domain is required for Ror2 to mediate the inhibitory signal of Wnt5a in the mammalian cell lines, suggesting a different, cell-autonomous role for Wnt5A/Ror2 in mammals.
Our data demonstrate that the Ror2 tyrosine kinase domain is essential for Ror2 function. Variants of the Ror2 receptor that lack the entire cytoplasmic domain show impaired ability to transduce the inhibitory signal of Wnt5a. In addition, point mutation of a single residue (Asn-620) that, by extrapolation, should be crucial for tyrosine kinase activity results in a protein that can no longer enhance the inhibitory activity of Wnt5a. This is the first time it has been shown that a mutant allele of Ror2 associated with human disease, recessive Robinow syndrome, is also deficient in Wnt5a signal transduction. These data might help explain some of the phenotypes associated with recessive Robinow syndrome as a disease of hyperactive Wnt signaling.
The embryonic expression pattern of mRor2 was analyzed via immunostaining. Ror2 was found to be expressed in many different tissues, with the highest expression found in the developing lung where Ror2 is known to play a crucial role during development (20–22). In the embryonic lung, Ror2 was found to colocalize with α-SMA, reflecting its expression in cells of mesenchymal origin. The observation that Ror2 loss did not affect Shh protein levels in the developing lung in the stages that we examined might be explained by the fact that Ror2 acts cell-autonomously in the mesenchyme, and thus does not perturb Shh signaling in the lung epithelium.
Finally, we observed the effect Ror2 loss has on Wnt/β-catenin signaling in vivo. We found that in cells specifically expressing Ror2, Wnt/β-catenin signaling is diminished. Thus, through in vitro and in vivo analyses, we have confirmed that Ror2 acts as a bona fide inhibitor of Wnt/β-catenin signaling.
Taken together, our data show that Ror2 signals cell-autonomously and that Wnt5a induces Ror2 tyrosine kinase activity. These results highlight the fact that Ror2 is an important kinase that warrants further study in the context of human disease. As we have shown that Ror2 can inhibit canonical Wnt signaling in vivo, activation of this inhibitory signaling pathway in the context of cancers in which Wnt/β-catenin signaling is hyperactivated could serve as an important therapeutic tool.
Supplementary Material
Acknowledgments
We thank our colleagues, in particular Renee van Amerongen, for helpful discussions and comments on the manuscript.
This work was supported by the Howard Hughes Medical Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- RTK
- receptor tyrosine kinase
- STF
- SuperTopFlash
- GST
- glutathione S-transferase
- MBP
- maltose-binding protein
- ELISA
- enzyme-linked immunosorbent assay
- PBS
- phosphate-buffered saline
- IP
- immunoprecipitate
- E
- embryonic day
- α-SMA
- α-smooth muscle actin.
REFERENCES
- 1.Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 2.Malanchi I., Huelsken J. (2009) Curr. Opin. Oncol. 21, 41–46 [DOI] [PubMed] [Google Scholar]
- 3.Chien A. J., Conrad W. H., Moon R. T. (2009) J. Invest. Dermatol. 129, 1614–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 5.Fox S., Dharmarajan A. (2006) Front. Biosci. 11, 2106–2112 [DOI] [PubMed] [Google Scholar]
- 6.Polakis P. (2000) Genes Dev. 14, 1837–1851 [PubMed] [Google Scholar]
- 7.Veeman M. T., Axelrod J. D., Moon R. T. (2003) Dev. Cell 5, 367–377 [DOI] [PubMed] [Google Scholar]
- 8.Semenov M. V., Habas R., Macdonald B. T., He X. (2007) Cell 131, 1378. [DOI] [PubMed] [Google Scholar]
- 9.Hendrickx M., Leyns L. (2008) Dev. Growth Differ. 50, 229–243 [DOI] [PubMed] [Google Scholar]
- 10.Cadigan K. M., Liu Y. I. (2006) J. Cell Sci. 119, 395–402 [DOI] [PubMed] [Google Scholar]
- 11.van Amerongen R., Mikels A., Nusse R. (2008) Sci. Signal. 1, re9. [DOI] [PubMed] [Google Scholar]
- 12.Schambony A., Wedlich D. (2007) Dev. Cell 12, 779–792 [DOI] [PubMed] [Google Scholar]
- 13.Ishitani T., Ninomiya-Tsuji J., Matsumoto K. (2003) Mol. Cell. Biol. 23, 1379–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikels A. J., Nusse R. (2006) PLoS Biol. 4, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrester W. C. (2002) Cell. Mol. Life Sci. 59, 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlessinger J. (2000) Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 17.Nomi M., Oishi I., Kani S., Suzuki H., Matsuda T., Yoda A., Kitamura M., Itoh K., Takeuchi S., Takeda K., Akira S., Ikeya M., Takada S., Minami Y. (2001) Mol. Cell. Biol. 21, 8329–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afzal A. R., Jeffery S. (2003) Hum. Mutat. 22, 1–11 [DOI] [PubMed] [Google Scholar]
- 19.Afzal A. R., Rajab A., Fenske C. D., Oldridge M., Elanko N., Ternes-Pereira E., Tüysüz B., Murday V. A., Patton M. A., Wilkie A. O., Jeffery S. (2000) Nat. Genet. 25, 419–422 [DOI] [PubMed] [Google Scholar]
- 20.Oldridge M., Fortuna A. M., Maringa M., Propping P., Mansour S., Pollitt C., DeChiara T. M., Kimble R. B., Valenzuela D. M., Yancopoulos G. D., Wilkie A. O. (2000) Nat. Genet. 24, 275–278 [DOI] [PubMed] [Google Scholar]
- 21.DeChiara T. M., Kimble R. B., Poueymirou W. T., Rojas J., Masiakowski P., Valenzuela D. M., Yancopoulos G. D. (2000) Nat. Genet. 24, 271–274 [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi S., Takeda K., Oishi I., Nomi M., Ikeya M., Itoh K., Tamura S., Ueda T., Hatta T., Otani H., Terashima T., Takada S., Yamamura H., Akira S., Minami Y. (2000) Genes Cells 5, 71–78 [DOI] [PubMed] [Google Scholar]
- 23.Winkel A., Stricker S., Tylzanowski P., Seiffart V., Mundlos S., Gross G., Hoffmann A. (2008) Cell. Signal. 20, 2134–2144 [DOI] [PubMed] [Google Scholar]
- 24.Kaykas A., Yang-Snyder J., Héroux M., Shah K. V., Bouvier M., Moon R. T. (2004) Nat. Cell Biol. 6, 52–58 [DOI] [PubMed] [Google Scholar]
- 25.Vihinen M., Vetrie D., Maniar H. S., Ochs H. D., Zhu Q., Vorechovský I., Webster A. D., Notarangelo L. D., Nilsson L., Sowadski J. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12803–12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akbarzadeh S., Wheldon L. M., Sweet S. M., Talma S., Mardakheh F. K., Heath J. K. (2008) PLoS ONE 3, e1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Rubin B., Bodine P. V., Billiard J. (2008) J. Cell. Biochem. 105, 497–502 [DOI] [PubMed] [Google Scholar]
- 28.Tornqvist H. E., Gunsalus J. R., Nemenoff R. A., Frackelton A. R., Pierce M. W., Avruch J. (1988) J. Biol. Chem. 263, 350–359 [PubMed] [Google Scholar]
- 29.Wilden P. A., Backer J. M., Kahn C. R., Cahill D. A., Schroeder G. J., White M. F. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 3358–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kani S., Oishi I., Yamamoto H., Yoda A., Suzuki H., Nomachi A., Iozumi K., Nishita M., Kikuchi A., Takumi T., Minami Y. (2004) J. Biol. Chem. 279, 50102–50109 [DOI] [PubMed] [Google Scholar]
- 31.Schwarzer W., Witte F., Rajab A., Mundlos S., Stricker S. (2009) Hum. Mol. Genet., [DOI] [PubMed] [Google Scholar]
- 32.Ali B. R., Jeffery S., Patel N., Tinworth L. E., Meguid N., Patton M. A., Afzal A. R. (2007) Hum. Genet. 122, 389–395 [DOI] [PubMed] [Google Scholar]
- 33.Wilden P. A., Kahn C. R., Siddle K., White M. F. (1992) J. Biol. Chem. 267, 16660–16668 [PubMed] [Google Scholar]
- 34.Al-Shawi R., Ashton S. V., Underwood C., Simons J. P. (2001) Dev. Genes Evol. 211, 161–171 [DOI] [PubMed] [Google Scholar]
- 35.Matsuda T., Nomi M., Ikeya M., Kani S., Oishi I., Terashima T., Takada S., Minami Y. (2001) Mech. Dev. 105, 153–156 [DOI] [PubMed] [Google Scholar]
- 36.Oishi I., Takeuchi S., Hashimoto R., Nagabukuro A., Ueda T., Liu Z. J., Hatta T., Akira S., Matsuda Y., Yamamura H., Otani H., Minami Y. (1999) Genes Cells 4, 41–56 [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi T. P., Bradley A., McMahon A. P., Jones S. (1999) Development 126, 1211–1223 [DOI] [PubMed] [Google Scholar]
- 38.Li C., Xiao J., Hormi K., Borok Z., Minoo P. (2002) Dev. Biol. 248, 68–81 [DOI] [PubMed] [Google Scholar]
- 39.Loscertales M., Mikels A. J., Hu J. K., Donahoe P. K., Roberts D. J. (2008) Development 135, 1365–1376 [DOI] [PubMed] [Google Scholar]
- 40.Lustig B., Jerchow B., Sachs M., Weiler S., Pietsch T., Karsten U., van de Wetering M., Clevers H., Schlag P. M., Birchmeier W., Behrens J. (2002) Mol. Cell. Biol. 22, 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., Costantini F. (2002) Mol. Cell. Biol. 22, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H. M., Jerchow B., Sheu T. J., Liu B., Costantini F., Puzas J. E., Birchmeier W., Hsu W. (2005) Development 132, 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aulehla A., Wehrle C., Brand-Saberi B., Kemler R., Gossler A., Kanzler B., Herrmann B. G. (2003) Dev. Cell 4, 395–406 [DOI] [PubMed] [Google Scholar]
- 44.Green J. L., Inoue T., Sternberg P. W. (2008) Cell 134, 646–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto S., Nishimura O., Misaki K., Nishita M., Minami Y., Yonemura S., Tarui H., Sasaki H. (2008) Dev. Cell 15, 23–36 [DOI] [PubMed] [Google Scholar]
- 46.Saiga H., Nishimura J., Kuwata H., Okuyama M., Matsumoto S., Sato S., Matsumoto M., Akira S., Yoshikai Y., Honda K., Yamamoto M., Takeda K. (2008) J. Immunol. 181, 8521–8527 [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto H., Yoo S. K., Nishita M., Kikuchi A., Minami Y. (2007) Genes Cells 12, 1215–1223 [DOI] [PubMed] [Google Scholar]
- 48.Hikasa H., Shibata M., Hiratani I., Taira M. (2002) Development 129, 5227–5239 [DOI] [PubMed] [Google Scholar]
- 49.Forrester W. C., Kim C., Garriga G. (2004) Genetics 168, 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim C., Forrester W. C. (2003) Dev. Biol. 264, 376–390 [DOI] [PubMed] [Google Scholar]
- 51.Oishi I., Suzuki H., Onishi N., Takada R., Kani S., Ohkawara B., Koshida I., Suzuki K., Yamada G., Schwabe G. C., Mundlos S., Shibuya H., Takada S., Minami Y. (2003) Genes Cells 8, 645–654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.