Abstract
GATA-1 is a lineage-restricted transcription factor that plays essential roles in hematopoietic development. The Gata1 gene hematopoietic enhancer allowed Gata1 reporter expression in erythroid cells and megakaryocytes of transgenic mice. The Gata1 hematopoietic enhancer activity is strictly dependent on a GATA site located in the 5′ region of the enhancer. However, the importance of the GC-rich region adjacent to the 3′-end of this GATA site has been also suggested. In this study, we show that this GC-rich region contains five contiguous deoxyguanosine residues (G5 string) that are bound by multiple nuclear proteins. Interestingly, deletion of one deoxyguanosine residue from the G5 string (G4 mutant) specifically eliminates binding to ZBP-89, a Krüppel-like transcription factor, but not to Sp3 and other binding factors. We demonstrate that GATA-1 and ZBP-89 occupy chromatin regions of the Gata1 enhancer and physically associate in vitro through zinc finger domains. Gel mobility shift assays and DNA affinity precipitation assays suggest that binding of ZBP-89 to this region is reduced in the absence of GATA-1 binding to the G1HE. Luciferase reporter assays demonstrate that ZBP-89 activates the Gata1 enhancer depending on the G5 string sequence. Finally, transgenic mouse studies reveal that the G4 mutation significantly reduced the reporter activity of the Gata1 hematopoietic regulatory domain encompassing an 8.5-kbp region of the Gata1 gene. These data provide compelling evidence that the G5 string is necessary for Gata1 gene expression in vivo and ZBP-89 is the functional trans-acting factor for this cis-acting region.
The differentiation and lineage-specification of hematopoietic cells are coordinated by the combinatorial functions of lineage-restricted and general transcription factors. The precise expression profile of lineage-restricted transcription factors is critical for promoting the differentiation program. GATA-1 is a lineage-restricted transcription factor required for normal erythropoiesis and megakaryopoiesis. The expression of Gata1 is restricted to erythroid cells, megakaryocytes, eosinophils, mast cells, and dendritic cells within the hematopoietic system (1, 2). We previously generated transgenic mice expressing a β-galactosidase (lacZ) reporter gene driven by the Gata1 hematopoietic regulatory domain (Gata1-HRD)2 (3, 4). The Gata1-HRD (5) is an 8.5-kbp region of the murine Gata1 gene and comprises a 3.9-kbp region 5′ of the proximal first exon (IE) and the entire first intron. Expression of the Gata1-HRD-driven reporter recapitulated endogenous GATA-1 expression in erythroid cells and megakaryocytes (3, 4). Furthermore, transgenic expression of GATA-1 cDNA driven by the Gata1-HRD could abolish lethal anemia in Gata1 germline mutant mice, demonstrating that the Gata1-HRD can drive sufficient transcriptional activity in vivo (6, 7).
Four discrete cis-acting regions have been identified as playing essential roles in the activity of the Gata1-HRD. These regions are: the Gata1 gene hematopoietic enhancer (G1HE, also referred to as hypersensitive site I, mouse hypersensitive site-3.5); the two upstream promoter elements containing a double GATA site and a CACCC box, referred to as hypersensitive site II; and a region containing multiple GATA repeats located in the first intron (3, 4, 8–11). The G1HE is located 3.9-2.6 kbp 5′ to the hematopoietic cell-specific first exon (IE). This region allows high expression levels of reporter genes in erythroid cells and megakaryocytes, as demonstrated by transgenic mouse reporter analysis (4, 11, 12). G1HE activity is strictly reliant on a GATA site located in the 5′ region of the G1HE, because disruption of this GATA site completely abolished reporter expression in both lineages. Interestingly, however, we and others demonstrated that sequential deletion in the 3′ region of the G1HE reduced reporter expression in fetal liver, even in the presence of the critical GATA site (4, 11). These results suggest that an additional cis-acting element 3′ of the GATA site is required for full activity of the G1HE. However, the properties of this additional region have not been fully characterized.
A Krüppel-type zinc finger transcription factor ZBP-89 (also referred to as zfp148, BERF-1, and BFCOL1 (13–15)) was recently identified as a novel GATA-1-associated transcription factor in both erythroid cells and megakaryocytes (16). A high level of ZBP-89 chromatin occupancy at the G1HE was found in both erythroid and megakaryocytic cell lines (16, 17). Moreover, erythroid and megakaryocyte differentiation is severely attenuated in the absence of ZBP-89 in zebrafish and mouse embryos (16, 18), overlapping with the phenotype of Gata1-deficient animals (19–21). These findings confirmed that ZBP-89 plays essential roles for Gata1 gene regulation during hematopoietic development. ZBP-89 binds to a GC-rich sequence that is also bound by numerous Krüppel-like transcription factors (KLFs), such as Sp1, Sp3, and EKLF/KLF1. Sp1 and EKLF interacted physically with GATA-1 and synergistically activated reporter transcription in transient transfection assays (22, 23). Thus, a functional redundancy might exist among KLFs in the regulation of Gata1 gene expression. In addition, because ZBP-89, Sp1, and EKLF associate with GATA-1, these factors might be able to act as cofactors and direct DNA binding of these factors might not be necessary.
To test these possibilities, we examined the GC-rich region in the G1HE by transgenic mouse reporter assays and gel mobility shift assays (EMSAs). We found that five contiguous deoxyguanosine residues (G5 string) within the GC-rich sequence are essential for Gata1 gene reporter expression in vivo. Interestingly, a series of EMSAs demonstrated that deletion of one deoxyguanosine residue from the G5 string (G4 mutant) specifically abolished binding to ZBP-89, but not to Sp3 and other binding factors. Furthermore, we show that the G4 mutation significantly reduced the Gata1-HRD reporter activity in fetal liver of transgenic mice. These data indicate that the G5 string is necessary for Gata1 gene expression and binding of ZBP-89 to this region is critical for the G1HE activity in vivo.
EXPERIMENTAL PROCEDURES
Constructs
IE3.9LacZ and IE3.9intLacZ reporter genes were constructed as described previously (3). For assembling the mutated and deleted reporter plasmids, a 1.3-kb G1HE fragment was excised from IE3.9intLacZ and cloned into pBluescript II KS+ plasmid between the XhoI and EcoRI sites (pBSG1HE). Fragments 1–235 and 1–207 of the G1HE were amplified by PCR using pBSG1HE as a template and inserted into the 5′-end of IE2.6LacZ that was constructed as described previously (3). Mutations were created in pBSG1HE using a QuikChangeTM site-directed mutagenesis kit according to the manufacturer's directions. For IE3.9intG4LacZ, the mutated G1HE fragment was removed from pBSG1HE and replaced by the G1HE in IE3.9intLacZ. For 1–235mutIE2.6LacZ and 1–235IE2.6G4LacZ constructs, the G1HE-(1–235) fragments were amplified using mutated pBSG1HE as a template and cloned into the 5′-end of IE2.6LacZ. The GATA-1 plasmids for maltose-binding protein (MBP) were constructed by means of a pMAL-c2 vector as described previously (24). The prokaryotic expression plasmid pGEXZBP-89 for generating the glutathione S-transferase (GST)-ZBP-89 protein was a generous gift from Dr. Juanita L. Merchant, University of Michigan. The cDNAs for selected domains of ZBP-89 were generated by PCR and cloned into a pGEX vector with modified restriction enzyme sites. The eukaryotic expression plasmid pCDNA3.1BFCOL1 was a generous gift from Dr. Ken-ichi Isobe, University of Nagoya. The expression plasmid for the C-terminal Myc-tagged BFCOL-1 (ZBP-89-Myc) was made from pCDNA3.1BFCOL1. The cDNAs for GATA-1 and its deletion mutants were cloned into a pEF-BOS eukaryotic expression vector (25) as described previously (26). For the luciferase reporter constructs, the G1HE-(124–235) fragment was amplified by PCR and cloned into the thymidine kinase minimal promoter reporter plasmid pT81-luc (27).
Generation and Analyses of Transgenic Mice
Transgenic reporter mice were generated by microinjection into fertilized BDF-1 eggs using standard procedures (28). Transgenic founders were dissected on embryonic day 14.5 and transgene integration was verified by PCR amplification of a lacZ gene fragment (4). Reporter expression was examined by 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) staining using cryosections of fetal liver as described previously (4). At least two separate fetal liver sections stained with X-gal were prepared for each embryo. At least 200 erythroid cells and 50 megakaryocytes were counted in each fetal liver section. Fetal liver with more than 10% of the total cells counted expressing β-galactosidase was scored as positive. We did not observe a founder with an X-gal staining that was positive only for erythroid cells, but not for megakaryocytes, or vice versa. Micrograph images were taken with a BX40F4 research microscope (Olympus) with an UplanF1 objective lens (20 × 0.40 or 40 × 0.75). Data acquisition was carried out with a DP70 digital camera and DP-controller software (Olympus).
EMSA
Nuclear extract preparation was performed as previously reported (4). The plasmid pGEXZBP-89 DNA binding domain (DBD) was generated by PCR. GST fusion proteins were expressed in the Escherichia coli strain BL21(DE3)pLysS (Invitrogen) and purified as previously reported (15). Probe labeling and the DNA-binding assay were performed as previously described (26). For supershift assays, a mouse monoclonal antibody for Sp1, a rabbit polyclonal antibody for Sp3 (sc-420 and sc-644, respectively, Santa Cruz), and a rabbit polyclonal antibody for ZBP-89 (code number 100-401-685, Rockland) were used. The p21 and p21 mut sequences shown in Fig. 3, A and B, are as follows: p21 (forward), 5′-ggttggtcctgcctctgagggggcggggcctgggccgag-3′; p21 (reverse), 5′-ctcggcccaggccccgccccctcagaggcaggaccaacc-3′; p21 mut (forward), 5′-ggttggtcctgcctctgagggttcggggcctgggccgag-3′; and p21 mut (reverse), 5′-ctcggcccaggccccgaaccctcagaggcaggaccaacc-3′.
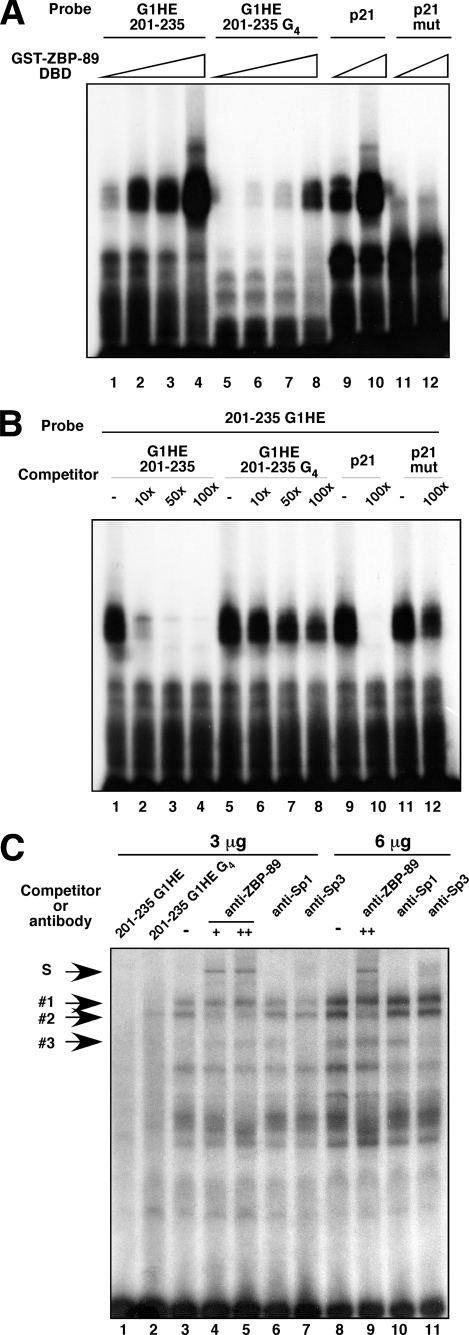
FIGURE 3.
The G5 string in G1HE-(201–235) is required for ZBP-89 binding. A, binding of the recombinant ZBP-89 DNA binding domain (amino acids 169–281) GST fusion protein (GST-ZBP-89 DBD) to the G1HE-(201–235) region. One μg of recombinant protein was incubated with probe. Note that binding of GST-ZBP-89 DBD to the probe was markedly reduced when one deoxyguanosine residue was deleted in G1HE-(215–219) (G1HE 201–235 G4). A known ZBP-89 binding sequence from the mouse p21waf1 promoter and its mutant (p21 and p21 mut, respectively) (35) were used as controls. The sequences of the p21waf1 and p21waf1 mut oligonucleotides are given under “Experimental Procedures.” B, binding of GST-ZBP-89 DBD to the G1HE-(201–235) probe either in the absence or presence of unlabeled competitors. G1HE-(201–235), G1HE-(201–235) G4, p21waf1, and p21waf1 mut competitors were added to the binding reactions in the amounts indicated. C, endogenous ZBP-89 binding to G1HE-(201–235) was detected by EMSA supershift assays in K562 nuclear extracts. K562 nuclear extracts (3 or 6 μg) were incubated with the G1HE-(201–235) probe either in the absence or presence of competitors or antibodies. A 100-fold molar excess of unlabeled G1HE-(201–235) or G1HE-(201–235) G4 was added in lanes 1 and 2, respectively. Antibodies for ZBP-89 (lanes 4, 5, and 9), Sp1 (lanes 6 and 10), and Sp3 (lanes 7 and 11) were added to determine supershifted complexes (S). DNA-protein complexes are indicated by 1, 2, and 3.
Chromatin Immunoprecipitation (ChIP) and ReChIP Assays
The ChIP assay was performed using mouse erythroleukemia (MEL) cells. A total of 106 cells per immunoprecipitation (IP) were fixed with 0.4% formaldehyde for 10 min at room temperature. Cross-links were quenched with 125 mm glycine. Cells were washed with phosphate-buffered saline (PBS), resuspended in SDS cell lysis buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% SDS, 1 mm EDTA and protease inhibitor mixture) and sonicated for four 15-s pulses using a TOMY UD-201 sonicator. The lysates were diluted 5 times with ChIP dilution buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100 and protease inhibitor mixture). The diluted samples were used for IP with a rat monoclonal antibody for GATA-1 (N6, sc-265, Santa Cruz) or a rabbit polyclonal antibody for ZBP-89 (number 100-401-685, Rockland). To prepare antibody-bead complexes, 15 μl of Dynabeads-Protein A (Invitrogen) was equilibrated with PBS for 30 min at 4 °C. The beads were resuspended with 675 μl of PBS containing 50 μg/ml bovine serum albumin, and 5 μg of anti-rat IgG rabbit antibody (Jackson ImmunoResearch) as a secondary antibody and incubated on a rotator for 2 h at 4 °C. Then the beads were washed twice with PBS and resuspended with 675 μl of PBS containing 50 μg/ml bovine serum albumin and 1 μg of anti-GATA-1 antibody as a primary antibody, incubated on a rotator for 2 h at 4 °C. The antibody-bead complexes were then washed twice with ChIP dilution buffer. For ZBP-89 ChIP, 3 μg of anti-ZBP-89 antibody was conjugated directly to Dynabeads-Protein A without a secondary antibody. The prepared antibody-bead complexes were resuspended with the diluted sample and incubated on a rotator overnight at 4 °C. The immune complexes were washed twice with ChIP dilution buffer, once with Wash buffer I (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.1% SDS and protease inhibitor mixture), once with Wash buffer II (10 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA, 0.25 m LiCl, 0.5% deoxycholate, 0.5% Nonidet P-40 and protease inhibitor mixture), and twice with TE buffer. The TE buffer was replaced with 300 μl of elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, and 1% SDS). After incubation for 2 h at 65 °C, Proteinase K was added and incubated overnight at 65 °C. The DNA fragments were extracted with phenol-chloroform, precipitated with ethanol, and suspended with 50 μl of TE buffer. Purified DNA from IP was analyzed in triplicates by a Mx3000P real time PCR system (Stratagene) with 2 μl of DNA solution and Platinum SYBR Green qPCR SuperMix (Invitrogen). The results were normalized with those from input DNA without IP. The PCR primers were as follows: 5′-tccaggaatgaagaaatggg-3′ (forward) and 5′-gtatgggggagtctcattgg-3′ (reverse) for G1HE; 5′-ggtccaggaaaaggcataag-3′ (forward) and 5′-tactgcccacctctatcagg-3′ (reverse) for a Gata1 coding region (exon 6) as a negative control. These primer pairs amplified a single product confirmed by 2% agarose gel electrophoresis and melting-curve analysis. For reChIP assays, the first immunoprecipitated complexes were eluted from Dynabeads-Protein A with reChIP elution buffer (20 mm dithiothreitol and 0.01% Triton X-100 in PBS) at 37 °C for 30 min. The eluted complex was diluted 50 times with ChIP dilution buffer and the ChIP procedure was repeated again.
GST Pulldown Assay
MBPs for GATA-1 were prepared and the binding assay with GST-ZBP-89 was performed as previously described (29). Immunoblotting was performed with anti-MBP antibody (sc-808, Santa Cruz).
Transfection of Small Interfering RNA (siRNA)
The siRNA for human GATA-1 (D-009610), ZBP-89 (D-012658), Sp3 (D-23096), and control (D-001210) were purchased from Thermo Scientific. One hundred pmol of siRNA was transfected into K562 cells (2 × 106 cells per transfection) by using the Cell Line Nucleofector Kit V (Lonza) according to the manufacturer's protocol. Cells were harvested 24 h after the transfection and nuclear extracts or total RNA were prepared for analyses.
Western Blotting
Western blotting was performed as described previously (26). GATA-1 (C20) and Lamin B (M20) antibodies were purchased from Santa Cruz Biotechnology.
Densitometry Analysis
For densitometry analysis, x-ray films were scanned by LAS-3000 image analyzer and density measurements were done by using MultiGauge version 3.0 software (FUJI FILM).
Real Time Reverse Transcription PCR Analysis
Total RNA was isolated by using ISOGEN (NIPPON GENE). Reverse transcription reactions were performed by using Superscript II (Invitrogen) according to the manufacturer's protocol. The quantitative real time PCRs were performed as described in ChIP assays. Primer sequences are as follows: GATA-1 (forward), 5′-gctcaactgtatggagggga-3′; GATA-1 (reverse), 5′-cagttgaggcagggtagagc-3′; ZBP-89 (forward), 5′-cgcatttagaagatgcgtca-3′; ZBP-89 (reverse), 5′-attttgctccagtggctgtt-3′; and glyceraldehyde-3-phosphate dehydrogenase (forward), 5′-agatcatcagcaatgcctcc-3′; glyceraldehyde-3-phosphate dehydrogenase (reverse), 5′-tgtggtcatgagtccttcca-3′.
DNA Affinity Precipitation Assay (DAPA)
DAPA was performed according to the methods of Billon et al. (30) with minor modifications. K562 cells were transfected with the expression plasmid for Myc-tagged ZBP-89 by using the Cell Line Nucleofector Kit V. Nuclear extracts were collected 24 h after the transfection. To prepare DNA probes, the G1HE-(131–230) oligonucleotides (wild type, 5′-gattcccttatctatgccttcccagctgcctccctgctggctgaactgtggccacagacttctgggccttgcaccccctccacagggatgggggagggaa-3′; GATA box mutant, 5′-gattccctggcttatgccttcccagctgcctccctgctggctgaactgtggccacagacttctgggccttgcaccccctccacagggatgggggagggaa-3′; and G5 strings mutant, 5′-gattcccttatctatgccttcccagctgcctccctgctggctgaactgtggccacagacttctgggccttgcacgcactccacagggatgcacgagggaa-3′) were biotinylated at their 5′ termini (Operon Biotechnologies) and annealed with the corresponding antisense-strand oligonucleotides. The DAPA was performed by mixing 1 μg of the biotinylated probe, 20 μg of salmon testes DNA, and 200 μg of K562 nuclear extracts in binding buffer containing 20 mm HEPES, pH 7.9, 10% glycerol, 50 mm KCl, 0.2 mm EDTA, 1.5 mm MgCl2, 10 μm ZnCl2, 1 mm dithiothreitol, and 0.25% Triton X-100. The sample was incubated on ice for 50 min and 25 μl of NeutrAvidin-agarose beads (Thermo Scientific) were added to the reactions. After a 2-h incubation at 4 °C, the agarose beads were washed 4 times in binding buffer and then once in PBS. The DNA-bound protein was eluted by adding 25 μl of 2× SDS sample buffer and heating at 95 °C for 5 min. The eluted protein was resolved on SDS-protein gel and Western blot analysis was performed using anti-GATA-1 antibody (C20) and anti-c-Myc antibody (Sigma).
Transfection Reporter Assays
Quail fibrosarcoma QT6 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. A total of 5 × 104 cells per transfection were seeded on 12-well plates 24 h before transfection. Cells were co-transfected with various combinations of a firefly luciferase reporter (either wild type or mutant), expression plasmids, and a Renilla luciferase control plasmid (pRLTK luc). Cells were harvested 48 h after transfection. Cell lysis and luciferase activity measurements were performed with a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol.
Statistical Analysis
Data were shown as average ± S.E. Statistical analysis was performed using Student's t test.
RESULTS
A G5 String in the 5′ Region of the G1HE Is Critical for Gata1 Gene Reporter Expression in Transgenic Mouse Fetal Liver
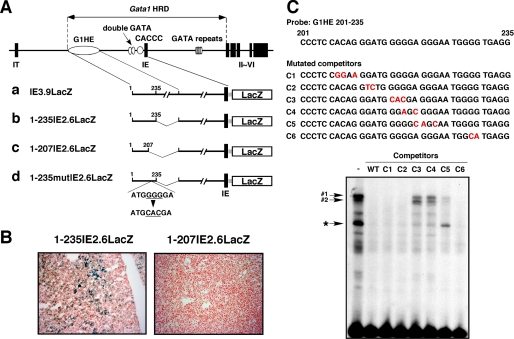
Previously, we and other groups found that 3′-deletion of the G1HE reduced Gata1 reporter expression in the fetal livers of transgenic mice, even in the presence of the critical GATA box located in the 5′ region of the G1HE (4, 11). In this study, we sought to identify the cis-acting element required for reporter expression in fetal livers of transgenic mice. We generated several reporter constructs (Fig. 1A) and examined their expressions in the fetal livers of transgenic founders on embryonic day 14.5 (E14.5). When the lacZ reporter gene was linked to the IE exon and its 3.9-kbp upstream region (IE3.9LacZ), both erythroid cells and megakaryocytes in fetal livers were stained with X-gal in 9 of 17 transgenic embryos (Table 1). Similar results were obtained when the 3′ 236–1174-bp portion of the G1HE was deleted (1–235IE2.6LacZ, see Table 1 and Fig. 1B). In contrast, no X-gal positive embryos were obtained when base pairs 208–1174 of the G1HE was deleted (1–207IE2.6LacZ, Table 1 and Fig. 1B). These results indicate that a critical cis-acting element for reporter expression in the fetal liver resides within the 208–235-bp region of the G1HE.
FIGURE 1.
Contiguous deoxyguanosine residues located at 215–219 of the G1HE are critical for Gata1 gene reporter expression in transgenic mice. A, mouse Gata1 locus with two cell-type specific first exons (IT and IE), five coding exons (II–VI), and four cis-regulatory elements (G1HE, double GATA, CACCC, and GATA repeats). a–d, structures of the reporter constructs used for the transgenic mouse reporter assays. B, representative X-gal staining of fetal liver prepared from 1–235IE2.6LacZ and 1–207IE2.6LacZ transgenic embryos. C, top, sequence of the G1HE-(201–235) probe and the competitors (C1–C6) used for EMSA. The mutated residues of the competitors are shown in red. Bottom, the G1HE-(201–235) region was used as a probe in EMSA using K562 nuclear extracts. Binding reactions were performed using 3 μg of nuclear extract and probe either in the absence or presence of unlabeled competitors. A 100-fold molar excess of wild type (WT) and mutated competitors (C1–C6) were used. DNA-protein complexes are indicated as 1, 2, and asterisk (*).
TABLE 1.
Expression of LacZ in the E14.5 fetal livers of transgenic founders
| Constructs | Number of founders | Number of LacZ-positive embryos |
|---|---|---|
| IE3.9LacZ | 17 | 9 |
| 1–235IE2.6LacZ | 15 | 7 |
| 1–207IE2.6LacZ | 16 | 0 |
| 1–235mutIE2.6LacZ | 17 | 0 |
To define the sequences obligatory for transcription factor binding within this region of the G1HE, we performed an EMSA using nuclear extracts prepared from K562 human leukemia cells that express GATA-1 abundantly. We employed oligonucleotides based on the G1HE-(201–235) bp sequence as a probe. This region contains a high GC content (69%) and is conserved in the corresponding region of the human GATA1 gene, except that 209A is replaced by G in human. Multiple retarded bands were observed using nuclear extracts from K562 cells (Fig. 1C). To determine the sequence critical for the formation of complexes, we generated six unlabeled competitors (C1–C6) based on serial mutations of the G1HE-(201–235) sequence and examined their abilities to compete with probe binding (Fig. 1C). A 100-fold molar excess of unlabeled competitor without mutation (wild type), as well as C1, C2, and C6 mutated competitors completely disrupted the formation of all complexes. Notably, competitor C6 that disrupts GGGTG (a complementary sequence of CACCC), a consensus binding sequence for KLFs, was still able to eliminate the binding of nuclear proteins to the probe. On the other hand, competitors C3 and C4 failed to disrupt the formation of the two slowly migrating complexes 1 and 2. C3 and C4 bind poorly to the other rapid migrating complexes. In addition, competitor C5 failed to eliminate the rapid migrating complex indicated by the asterisk. This competitor also binds complexes 1 and 2 poorly. C3 and C4 involved mutations in the deoxyguanosine residues in base pairs 216–219. C5 involved mutations in residues adjacent to the 3′-end of the deoxyguanosine stretch. Together, these findings indicate that the five contiguous deoxyguanosine residues (G5 string) in this region are essential for binding of nuclear proteins to the probe in K562 cells. In addition, the residues adjacent to the 3′-end of the G5 string contribute the complex binding. The results were consistently observed using nuclear extract prepared from MEL cells (data not shown).
To test whether the G5 string in the G1HE is critical for Gata1 gene expression in vivo, a transgenic mouse reporter assay was performed by employing a reporter construct of the 3′-deleted G1HE region with mutations disrupting the G5 string (1–235mutIE2.6LacZ, Fig. 1A, d). The mutated sequence in the reporter was identical to that of competitor C3 used in the EMSA. Of the 17 transgenic founders, no X-gal positive embryos were observed in fetal livers at E14.5. These outcomes assign residency of the G5 string in the 5′ section of the G1HE and deem it vital for Gata1 gene expression in vivo.
A Nuclear Protein That Specifically Binds to a G5, but Not to a G4, String Bound to the G1HE-(201–235) Probe
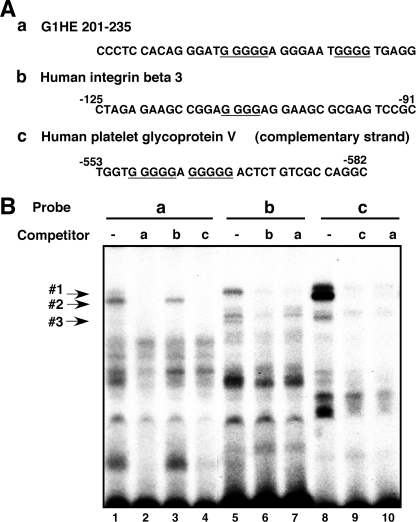
A GC-rich sequence is frequently observed in regulatory regions of megakaryocyte-specific genes, such as INTEGRIN α2 and β3, PLATELET GLYCOPROTEIN V, and CYCLIN D3 (31–34). Some of these regions contain G-strings similar to that in G1HE-(201–235). To characterize the DNA-protein complex formed on G1HE-(201–235), we performed a series of EMSA with G1HE-(201–235) and two probes containing G-strings derived from megakaryocytic genes. The sequences of the probes are shown in Fig. 2A and the G-strings are underlined. These sequences are the regions from −125 to −91 bp of the human INTEGRIN β3 gene (probe b) (32) and from −582 to −553 bp of the human PLATELET GLYCOPROTEIN V gene (probe c) (33). The transcription start site was designated as +1 for both genes. G1HE-(201–235) (probe a) contains a G5 string at 215–219 bp and a G4 string at 227–230 bp. Probe b contains a G4 string and probe c contains two G5 strings on the complementary strand. We observed multiple retarded bands for all probes using nuclear extracts from K562 cells (Fig. 2B). Despite the same amount of nuclear proteins being used, the formation of probe-protein complex 2 was much more evident with probe c containing two G5 strings than with probe a containing a single G5 string (Fig. 2B, lanes 1 and 8). In contrast, probe-protein complex 2 was absent on probe b that lacks a G5 string (Fig. 2B, lane 5) and did not appear even when the amount of nuclear protein was doubled (3 to 6 μg) (data not shown). Probe-protein complex 3 was not clearly observed on probe a, but did appear upon doubling the amount of nuclear protein (data not shown). Competitor b at a 100-fold molar excess failed to compete with probe a for protein complex 2 (lane 3). In contrast, competitor c completely abolished the formation of complexes 1, 2, and 3 on probe a (lane 4). Competitor a abolished the formation of these complexes on probes b and c (lanes 7 and 10, respectively). Collectively, these results suggest that complex 2 contains a nuclear protein that specifically binds to a G5, but not to a G4, string in these probes.
FIGURE 2.
Formation of nuclear protein complexes on G-string containing probes. A, sequences of the G1HE-(201–235) (a) and the promoter regions of the human INTEGRIN β3 (b) and human PLATELET GLYCOPROTEIN V (c) genes used as probes for EMSA. The complementary sequence of the human PLATELET GLYCOPROTEIN V gene promoter is shown for comparison of sequence similarities. G-strings are underlined. B, nuclear extracts (3 μg of protein) from K562 cells were incubated with probes either in the absence or presence of unlabeled competitors. A 100-fold molar excess of unlabeled oligonucleotides (a, b, and c) were used as competitors. DNA-protein complexes are indicated by 1, 2, and 3.
The G5 String in G1HE-(201–235) Is Required for ZBP-89 Binding
A recent report showed that the Krüppel-type zinc finger transcription factor ZBP-89 binds to the 3′ region of the G1HE in L8057 megakaryocytic cells (17). To examine whether or not ZBP-89 is the factor that specifically binds to a G5, but not to a G4, string, a purified GST fusion protein of the DBD of ZBP-89 (amino acids 169–281, GST-ZBP-89 DBD) was examined for its ability to bind to the G1HE-(201–235) probe and G1HE-(201–235) G4 mutant probe (Fig. 3, A and B). G1HE-(201–235) G4 is a mutant with one deoxyguanosine deletion from 215 to 219-bp region of the G1HE. We found that a much greater amount of GST-ZBP-89 DBD protein was needed to bind to the G1HE-(201–235) G4 probe than to the wild type probe (Fig. 3A, lanes 5–8 and 1–4, respectively). Consistent with DNA binding studies in an earlier report (35), the GST-ZBP-89 DBD bound to wild type, but not to mutant, oligonucleotides derived from the p21 promoter that is a target gene of ZBP-89 (Fig. 3A, lanes 9–12). It is noteworthy that the G5 string in the p21 promoter probe was lost in the mutation (wild type, agggggc; mutant, agggttc; the entire sequence of the probes are shown under “Experimental Procedures”). Furthermore, unlabeled G1HE-(201–235) G4 oligonucleotide, as well as p21 mut, inefficiently competed with the probe for binding to GST ZBP-89 DBD, even in the presence of a 100-fold molar excess of competitor relative to probe (Fig. 3B, lanes 6–8 and 12). In contrast, only a 10-fold molar excess of unlabeled G1HE-(201–235) competitor was able to displace most of the probe binding (Fig. 3B, lane 2). Together, these results indicate that ZBP-89 requires the G5 string at base pairs 215–219 of the G1HE for binding and that its DNA binding ability was significantly attenuated by a single deoxyguanosine deletion from G1HE-(215–219).
We then examined if complex 2 that specifically formed on G1HE-(201–235) (Fig. 2B) contains ZBP-89 in the K562 nuclear extract. We performed a series of EMSA supershift assays using K562 nuclear extract and anti-ZBP-89 antibody (Fig. 3C). As expected, anti-ZBP-89 antibody specifically shifted complex 2, but not complexes 1 and 3, to a slower migrating complex (indicated as S in Fig. 3C). Neither anti-Sp1 nor anti-Sp3 antibodies affected the formation of complex 2. Complex 3 was shifted in the presence of anti-Sp3 antibody, suggesting that complex 3 contains Sp3. The formation of complex 1 was unaffected by any of the antibodies tested in this study. Similar results were observed in nuclear extracts from MEL cells (data not shown). In summary, these results indicate that complex 2 requiring the G5 string in G1HE-(215–219) contains ZBP-89 in nuclear extracts from K562 and MEL cells.
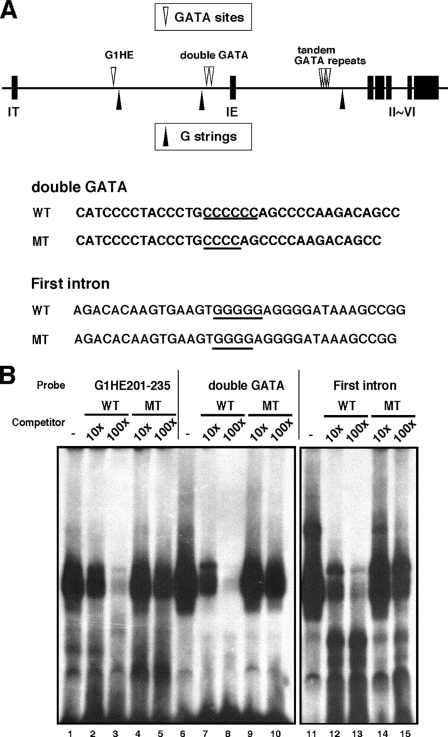
ZBP-89 Binds to the G5 String Containing Sequences in Two Additional Regulatory Elements in the Mouse Gata1 Gene
Guyot et al. (17) demonstrated that occupation of a chromatin region by ZBP-89 was also detected in the IE promoter and mHS+3.5, the intronic cis-acting region of the mouse Gata1 gene in L8057 cells. Importantly, these cis-acting elements contain critical GATA binding sites necessary for activating Gata1 reporter gene expression in vivo (Fig. 4A, lanes 8–10). These are the double GATA sites in the IE promoter region and the 13 GATA repeats found in tandem in the intronic region. We tested whether ZBP-89 binds to these sequences by a series of EMSA using GST-ZBP-89 DBD (Fig. 4B). Indeed, GST-ZBP-89 DBD formed a complex with both the double GATA and the intron probes (Fig. 4B). Moreover, similar to the results with the G1HE-(201–235) probe (Figs. 3B and 4B), mutant competitors with a deoxycytidine or deoxyguanosine deletion failed to eliminate GST-ZBP-89 DBD binding to the probes even in the presence of a 100-fold molar excess of competitor (Fig. 4B, lanes 10 and 15). These results indicate that ZBP-89 specifically requires the C6 and G5 strings in the probes originating from the double GATA and intronic regions, respectively.
FIGURE 4.
ZBP-89 binds to the G-strings in two additional regulatory elements of the mouse Gata1 gene. A, schematic of the cis-acting regions of the mouse Gata1 gene containing G-strings and GATA sites. The sequences of the probes and competitors used for EMSA are indicated. B, binding of GST-ZBP-89 DBD to the G1HE-(201–235) (lanes 1–5), double GATA (lanes 6–10), and first intron (lanes 11–15) probes was examined by EMSA. One μg of recombinant protein was incubated with probe. Note that wild type (WT) competitor (lanes 2, 3, 7, 8, 12, and 13), but not the G4 (MT) competitor (lanes 4, 5, 9, 10, 14, and 15), displaced probe binding of GST-ZBP-89 DBD.
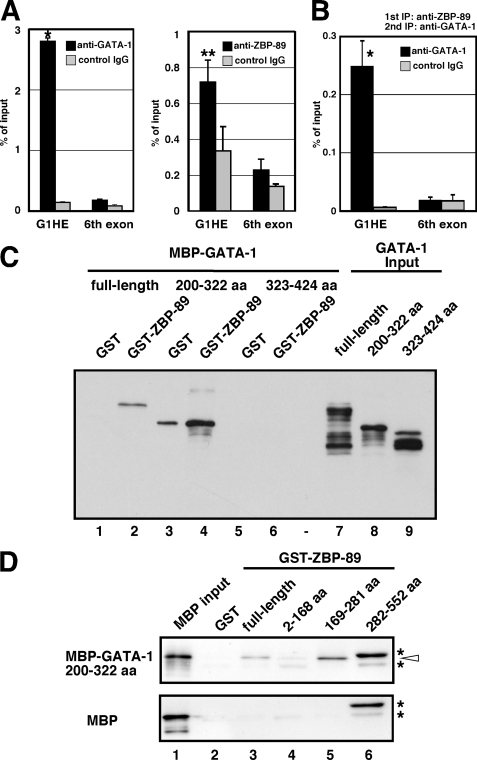
ZBP-89 and GATA-1 Bind to the G1HE Chromatin Region and Physically Associate in the Absence of DNA
ZBP-89 was identified as a component of multiprotein complexes involving GATA-1 and friend of GATA-1 (FOG-1) in erythroid and megakaryocytic cells (16). ZBP-89 and GATA-1 occupy a chromatin region that includes G1HE-(201–235), as shown by our ChIP assays using MEL cells (Fig. 5A), which is consistent with previous reports (16, 17). In contrast, control ChIP assays examining the coding sequence of Gata1 at the sixth exon show only the background level of PCR amplification. We next performed a ChIP-reChIP assay using MEL cells (Fig. 5B). We first precipitated the nuclear protein-DNA complex with anti-ZBP-89 antibody, and the second immunoprecipitation was performed using anti-GATA-1 antibody. The G1HE region was amplified with real time PCR after the second round of immunoprecipitation. In contrast, the coding region of Gata1 at the sixth exon was not amplified from the same templates. These results indicate that GATA-1 and ZBP-89 co-occupy the G1HE chromatin region. Next, we examined the interaction domains of GATA-1 and ZBP-89 by a GST pull-down assay (Fig. 5, C and D). Full-length and the zinc finger region (amino acids 200–322, lane 4), but not with the C-terminal region (amino acids 323–424, lane 6), of GATA-1 was pulled down by GST-ZBP-89 (Fig. 5C), which is akin to the co-immunoprecipitation assay reported previously (16). To identify the GATA-1-interacting domain of ZBP-89, various ZBP-89 deletion mutants were expressed as GST fusion proteins. These proteins were tested for interaction with the GATA-1 (amino acids 200–322) fused to MBP (Fig. 5D). We found that MBP-GATA-1 (amino acids 200–322) was pulled down by full-length and the zinc finger region (amino acids 169–281), but not the N-terminal (amino acids 2–168) and C-terminal (amino acids 553–794, data not shown) regions of ZBP-89 (Fig. 5D, upper panel). Protein pulled down by GST-ZBP-89 (amino acids 282–552) appeared in a different size as the input protein that also appeared in the control experiment using MBP alone (Fig. 5D, asterisks, lane 6). These results indicate that GATA-1 and ZBP-89 physically associate in the absence of DNA, and the zinc finger regions of both proteins are mapped as interaction sites by GST pull-down assays.
FIGURE 5.
ZBP-89 physically interacts with GATA-1 zinc finger regions. A, binding of GATA-1 (left panel) and ZBP-89 (right panel) to the G1HE chromatin region was examined by ChIP assays. Cross-linked chromatin from MEL cells was immunoprecipitated using anti-GATA-1 or anti-ZBP-89 antibody and was quantified by real time PCR (black bars). Rat IgG and rabbit IgG were used as controls for anti-GATA-1 and anti-ZBP-89 antibodies, respectively (gray bars). A primer set that amplifies the Gata1 sixth exon coding region was used as a negative control. Values from PCR amplicon using immunoprecipitated chromatin relative to those of input are shown. Results are shown as averages ± S.E. of the data obtained from 6 independent assays. *, p < 0.01; **, p < 0.05 compared with control IgG. B, binding of GATA-1 and ZBP-89 to the chromatin region of G1HE was examined by ChIP-reChIP assay. Cross-linked chromatin from MEL cells was immunoprecipitated with anti-ZBP-89 antibody and the second IP was performed using anti-GATA-1 antibody. Rat IgG was used as a control for anti-GATA-1 antibody (gray bars). A primer set that amplifies the Gata1 sixth exon was used as a negative control. Values from PCR amplicon using immunoprecipitated chromatin relative to those of input are shown. Results are shown as averages ± S.E. of the data obtained from three independent assays. C and D, interaction of ZBP-89 and GATA-1 tested by a GST pull-down assay. C, full-length ZBP-89 GST fusion protein was incubated with GATA-1 proteins fused to MBP. The full-length (lanes 1 and 2), the zinc finger region (amino acids 200–322; lanes 3 and 4), and the C-terminal region (amino acids 323–424; lanes 5 and 6) of the GATA-1 proteins were examined in the assay. As input, 10% of the MBP-GATA-1 proteins tested in the interaction assay were loaded onto the gel (lanes 7–9). D, mapping of the GATA-1-interacting domain of ZBP-89. The zinc finger region of GATA-1 fused to MBP (upper panel) or MBP alone (lower panel) was tested for interaction with various GST-ZBP-89 proteins. Input of MBP protein (lane 1), GST alone (lane 2), the full-length (lane 3), and 3 deletion mutants (lanes 4–6) of ZBP-89 were examined in the assay. White arrowhead corresponds to MBP-GATA-1 (amino acids 200–322). Asterisks indicate the undefined proteins that appeared only in the presence of GST-ZBP-89 (amino acids 282–552).
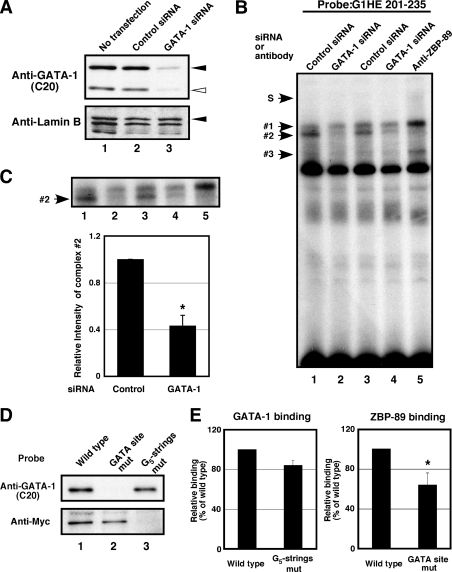
GATA-1 Affects the Binding of ZBP-89 to the G1HE Region
To examine whether GATA-1 affects ZBP-89 binding to the G5 string, GATA-1 siRNA was transfected into K562 cells and the formation of the nuclear protein complex was examined by EMSA. The expression level of GATA-1 was markedly reduced in the presence of GATA-1 siRNA compared with control siRNA (Fig. 6A, upper panel, lane 3). Consistent with the results of Fig. 3C, we observed that band 2 was supershifted by anti-ZBP-89 antibody (Fig. 6B, lane 5). Interestingly, formation of this complex was reduced to 0.43 ± 0.09 relative to that of control in GATA-1 knockdown cells by densitometry analysis (Fig. 6, B and C, lanes 1–4). Western blot analysis demonstrated that the ZBP-89 protein level was not largely affected by GATA-1 siRNA in K562 cells (supplemental Fig. S1A). We then examined whether GATA-1 binding to the G1HE region was affected by ZBP-89 (supplemental Fig. S1). To this end, we performed EMSA by using ZBP-89 knockdown K562 cells and the G1HE-(121–158) probe that contains a GATA site. Western blot analysis showed that the ZBP-89 protein was reduced in the presence of ZBP-89 siRNA (supplemental Fig. S1A). We observed two slowly migrating complexes that were eliminated by G1HE-(121–158), but not by the GATA mutant competitor (supplemental Fig. S1B, lanes 5 and 6). Formation of both complexes was not largely affected by ZBP-89 siRNA (supplemental Fig. S1, B and C). Collectively, these results indicate that binding of ZBP-89 to the G1HE region was reduced in GATA-1 knockdown cells, whereas GATA-1 binding to this region was not largely affected by ZBP-89 siRNA.
FIGURE 6.
GATA-1 affects the binding of ZBP-89 to the G1HE region. A, 100 pmol of GATA-1 or control siRNA was transfected into K562 cells. The expression of GATA-1 protein was determined by immunoblotting. Ten μg of nuclear extracts were resolved by SDS-PAGE. The anti-lamin B antibody was used for loading control. Lane 1, no transfection; lane 2, control siRNA-transfected cells; lane 3, GATA-1 siRNA-transfected cells. Black arrowheads indicate the bands of the expected size. White arrowhead corresponds to GATA-1s, an alternative translation product of GATA-1 (48). B, formation of a nuclear protein complex on G1HE-(201–235) was examined by EMSA. Nuclear extracts (3 μg) were prepared from K562 cells transfected with control (lanes 1 and 3) or GATA-1 (lanes 2 and 4) siRNA and incubated with probe. Antibodies for ZBP-89 were added to nuclear extracts from untransfected K562 cells (lane 5). DNA-protein complexes 1–3 correspond to those shown in Fig. 3C. S, a complex supershifted by anti-ZBP-89 antibody. C, densitometry measurements of the ZBP-89 containing complex (2) shown in B. Upper panel shows the magnified view of B, lanes 1–5. Measurements of number 2 density were performed by using this image and two additional data of EMSA. These samples were prepared from independent siRNA transfections (n = 4). The densities of number 2 in the GATA-1 siRNA-transfected cells relative to those in the control siRNA-transfected cells are shown as averages ± S.E. *, p < 0.05. D, DAPA on the G1HE-(131–230) region. Biotinylated G1HE-(131–230) oligonucleotide (wild type, GATA site mutant, and G5 string mutants) was incubated with nuclear extracts from K562 cells transfected with the ZBP-89-Myc expression plasmid. DNA-protein complexes were recovered with NeutrAvidin-agarose beads, denatured, and resolved by SDS-PAGE. The GATA-1 (upper panel) and ZBP-89-Myc (lower panel) in these complexes was detected by immunoblotting. E, densitometry measurements of GATA-1 (left panel) and ZBP-89-Myc (right panel) proteins shown in D. Data were obtained from four independent experiments. The amount of GATA-1 or ZBP-89-Myc protein bound to the wild type probe was set at 100. *, p < 0.05. Error bars are 1 S.E.
Because we have used the probe that contains either the GATA site or the G5 string in EMSA, we next performed DAPA by using the G1HE-(131–230) probe that contains binding sites for both proteins (Fig. 6, D and E). The nuclear protein in K562 cells bound to the biotinylated probe was precipitated by NeutrAvidin-agarose beads and the GATA-1 or ZBP-89 protein was detected by Western blot analysis. In this experiment, the ZBP-89 protein with a Myc tag at the C terminus was expressed in K562 cells and determined by anti-Myc antibody. We found that both GATA-1 and ZBP-89-Myc proteins were pulled down by the biotinylated G1HE-(131–230) probe (Fig. 6D, lane 1). The GATA-1 binding was completely abolished by disrupting the GATA site in the probe, indicating that GATA-1 specifically binds to the GATA site of this probe. It should be noted that there is a complementary G5 string at position 199–203 bp of the G1HE (C5 string). This region was not completely included in our EMSA probe spanning 201–235 bp of the G1HE. When we examined the G1HE-(131–230) probe with a mutation only in the G5 string at position 215 to 219, the binding of ZBP-89-Myc was reduced but not completely abolished in DAPA (data not shown). Mutations in both the C5 and G5 strings completely eliminated the binding of ZBP-89-Myc (Fig. 6D, lower panel, lane 3). These results suggest that ZBP-89 binds to either the G1HE-(199–203) or –(215–219) regions. Consistent with our observations in EMSA, ZBP-89-Myc binding was reduced to 0.64 ± 0.12 in the GATA site-mutated probe relative to the wild type, despite that no mutation was created in the G5 and C5 strings (Fig. 6E, right panel). In contrast, GATA-1 binding was not significantly affected by mutations in the G5 and C5 strings (Fig. 6E, left panel). Taken together with the EMSA data, these results suggest that GATA-1 binding to the GATA site may facilitate ZBP-89 binding to the G1HE region.
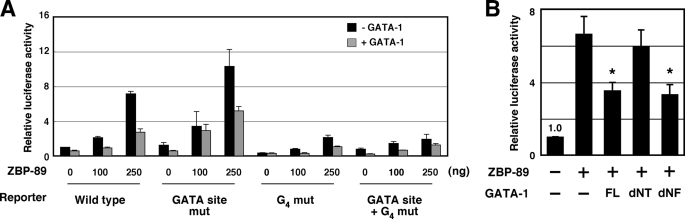
The G5 String Is Required for ZBP-89-dependent Activation of the G1HE Luciferase Reporter
To examine how ZBP-89 affects the activity of G1HE, a series of luciferase reporter assays were performed with non-hematopoietic QT-6 cells (Fig. 7). A luciferase reporter construct was prepared using the G1HE-(124–235) region linked to a minimal thymidine kinase promoter (G1HE-(124–235)-luc). The reporter was activated by co-transfection of a ZBP-89 expression plasmid in a dose-dependent manner (Fig. 7A). This activation was reduced by the G4 mutation (G4 mut), but not by the GATA box mutation (GATA box mut) in the G1HE-(124–235)-luc reporter (Fig. 7A). The reporter activity was significantly reduced when both the G5 string and the GATA site were mutated (Fig. 7A, GATA box + G4 mut). These results indicate that the G5 string is necessary for the ZBP-89-dependent activation of G1HE. Somehow unexpectedly, the reporter activation by ZBP-89 was reduced in the presence of GATA-1. In addition, GATA-1 repressed the reporter activation by ZBP-89 even in the absence of its own binding site (Fig. 7A, GATA box mut). Although we should consider that activation or repression of the reporter in the transient transfection assays might be influenced by cellular context, it is possible that GATA-1 may limit ZBP-89-mediated activation of the G1HE by interacting with ZBP-89. We then examined whether GATA-1 lacking the N-terminal region (1–83 amino acids, dNT) or the N-terminal zinc finger region (197–232 amino acids, dNF) were also able to reduce the ZBP-89 activity (Fig. 7B). The ZBP-89 activity was reduced by wild type and the dNF mutant, but not by the dNT mutant of GATA-1. These results suggest that the N-terminal region of GATA-1 is responsible for the inhibition of ZBP-89 activity.
FIGURE 7.
The G5 string is required for ZBP-89-dependent activation of the G1HE-(124–235)-luc reporter. A, QT-6 cells were co-transfected with 20 ng of wild type or mutant G1HE-(124–235)-luc reporter, 5 ng of a Renilla luciferase control reporter, and 0, 100, or 250 ng of a ZBP-89 expression plasmid with (gray bars) or without (black bars) 100 ng of a GATA-1 expression plasmid. Wild type and mutant (GATA site mut, G4 mut, and GATA site + G4 mut) G1HE-(124–235) reporters were examined. B, QT-6 cells were co-transfected with 100 ng of an expression plasmid of GATA-1 (wild type or mutant), 250 ng of a ZBP-89 expression plasmid, 20 ng of G1HE-(124–235)-luc reporter, and 5 ng of a Renilla luciferase control reporter. The reporter activity of the expression vector with no insert was set to 1.0. FL, full-length; dNT, the N-terminal deleted GATA-1 mutant protein; dNF, the N-finger deleted GATA-1 mutant protein. *, p < 0.05 versus ZBP-89 without the GATA-1 expression plasmid.
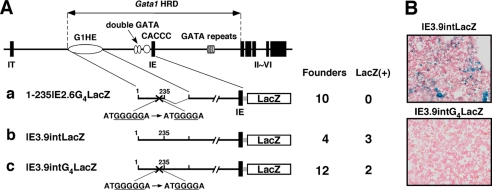
The G5 String in the 5′ Region of G1HE Is Critical for Complete Gata1-HRD Reporter Activity in Vivo
Because ZBP-89 activated the G1HE-(124–235)-luc reporter, we next examined whether ZBP-89 siRNA reduces the endogenous GATA-1 mRNA level in K562 cells comparing the effect of Sp3 siRNA. Unexpectedly, however, the GATA-1 mRNA level in K562 cells was not largely affected by ZBP-89 siRNA and was similar in the ZBP-89 and Sp3 knockdown cells (data not shown). It is possible that ZBP-89 may play critical roles during erythroid cell differentiation that is incapable of evaluating in K562 cells. Therefore, we performed the transgenic mouse reporter assays using reporter constructs with one deoxyguanosine deletion in the G1HE-(215–219) sequence (Fig. 8A). Our data of DNA binding assays demonstrated that one deoxyguanosine deletion in the G5 string abrogated ZBP-89 binding, whereas other KLF factors were capable of binding to this deletion mutant. We first tested a reporter construct of the G1HE-(1–235) region with one deoxyguanosine deletion in the G5 string linked to IE2.6LacZ (1–235IE2.6G4LacZ). As shown in Table 1, 7 of 15 transgenic founders were positive for X-gal staining for the 1–235IE2.6LacZ reporter without mutation. In contrast, no X-gal positive fetal livers were found in 10 transgenic founders for 1–235IE2.6G4LacZ at E14.5. Finally, we examined the requirement of the G5 string in a reporter encompassing the entire Gata1-HRD region (Fig. 8, A and B). In our previous transgenic mouse reporter assays performed by the same procedures, we found that 3 of 4 transgenic lines and 6 of 9 transgenic founders were positive for X-gal staining in fetal liver (3, 4). In addition, we previously demonstrated that the reporter expression driven by the Gata1-HRD region nearly recapitulates endogenous Gata1 gene expression in vivo by using GFP and luciferase reporter mice (36, 37). Furthermore, transgenic expression of GATA-1 cDNA driven by this region could rescue Gata1 germline mutant mice from lethal anemia in multiple transgenic lines (6, 7, 26). Consistent with these previous observations, we observed that 3 of 4 transgenic founders were positive for X-gal staining in fetal liver at E14.5. In contrast, only two founders were positive for X-gal staining out of 12 transgenic embryos harboring IE3.9intG4LacZ, a mutant reporter with one deoxyguanosine deletion in the Gata1-HRD region. Together, these data indicate that the G5 string in the 5′ region of the G1HE is essential for complete Gata1-HRD activity and plays critical roles for Gata1 gene expression in vivo.
FIGURE 8.
The G5 string in the G1HE is required for complete Gata1-HRD activity in fetal liver of transgenic mouse. A, structures of the reporter constructs, number of founders, and number of LacZ-positive embryos in the transgenic mouse reporter assays. The expression of the reporter gene was examined by X-gal staining of fetal liver at E14.5. B, representative X-gal staining of fetal liver prepared from IE3.9intLacZ and IE3.9intG4LacZ transgenic embryos.
DISCUSSION
There is a line of evidence suggesting that Gata1 gene expression is regulated dynamically during erythroid differentiation (12, 36). We have recently examined the differentiation stage-specific Gata1 gene expression in erythroid cells by using transgenic mouse lines expressing green fluorescent protein from mouse Gata1 locus in a bacterial artificial chromosome (G1BAC-GFP). We found that the Gata1 gene expression is up-regulated at burst-forming unit erythroid stage progenitors, peaked at colony-forming unit erythroid stage progenitors, and down-regulated during erythroblast maturation (12). This strict control of Gata1 gene expression seems essential for normal erythropoiesis, because sustained overexpression of GATA-1 at a later stage of erythropoiesis was lethal to transgenic mice due to impaired erythroid cell maturation (38, 39). The G1HE plays a central role for the dynamic regulation of Gata1 gene expression. Deletion of the G1HE or the N-terminal core region (1–235 amino acids) of the G1HE from the G1BAC-GFP resulted in a marked reduction of reporter expression in erythroid lineage of transgenic mice, especially in BFU-E stage progenitors (12). We previously identified a GATA site in G1HE (4) and this GATA site plus E-box nearby have been the only cis-acting elements identified in the enhancer (4, 11). The E-box resides 10 bp 3′ to the GATA site and the GATA-E-box has been considered as the binding site for a multiprotein complex including GATA-1, TAL-1, Ldb1, LMO2, and E2A (40). Surprisingly, however, a mutation in the E-box did not affect the Gata1 gene reporter expression in transgenic mice (4, 11). Therefore, an assessment of cis-acting element function should be carefully performed in vivo and such information would be important to understand how GATA-1 and its partners contribute to Gata1 gene regulation. This study provides compelling evidence that the G5 string in the G1HE is necessary for Gata1 gene expression in transgenic mouse fetal liver. Besides the GATA-E-box complex, the G5 string is the first element in the G1HE that has been proved to be necessary for Gata1 gene expression in vivo. We also identified ZBP-89 as a functional trans-acting factor binding to this cis-acting element.
The G5 string resides in a GC-rich region in the G1HE. In contrast to the GATA sites, numerous cell-type specific and ubiquitous KLFs can bind to GC-rich regions. In general, it is hard to determine which factor actually binds to a GC-rich region and contributes gene expression. A GC-rich region is also found in the upstream promoter region of the Gata1 gene (hypersensitive site II). This region contains a CACCC box that is critical for Gata1 gene expression (10). As we noticed that a complementary CACCC sequence resides at residues 226–231 of the G1HE, we assumed that this CACCC box might be a functional binding site for nuclear proteins. Unexpectedly, however, a series of EMSA demonstrated that the G5 string, but not the CACCC box, is the binding site for nuclear proteins in K562 cells. It has been reported that GATA-1 associates with several KLF factors such as EKLF, Sp1, Sp3, and ZBP-89 (16, 22, 23). Given that five or more G-strings are not found in the hypersensitive site II, it is conceivable that the two GC-rich regions in the hypersensitive site II and G1HE are bound by different KLF factors and thereby play distinct roles in Gata1 gene regulation.
ZBP-89 is a widely expressed transcription factor that harbors four zinc fingers. Unlike other KLFs that have three contiguous C2H2-type zinc fingers, ZBP-89 possesses a fourth C2HC-type zinc finger. In addition, all zinc fingers are located in the N-terminal region of ZBP-89, whereas the zinc finger regions of KLFs are generally found in the C-terminal region (41–43). Disparity in the DNA-binding sequences between ZBP-89 and other KLF factors might be caused by these differences. ZBP-89 contributes to the regulation of a variety of genes such as gastrin, Type I collagen, ORNITHINE DECARBOXYLASE, VIMENTIN, and p21waf1 (14, 15, 35, 44–46). Recently, occupation of the chromatin region by ZBP-89 has been shown for several erythroid and megakaryocytic genes by ChIP assays. These include the genomic locus of α-Globin (47), β-Globin HS2, and c-Mpl (16). In many cases, ZBP-89 functions as a repressor protein. In the case of the human ORNITHINE DECARBOXYLASE gene, ZBP-89 has been shown to compete with Sp1 for DNA binding and oppose transcriptional activation by Sp1 (45). ZBP-89 also acts as a transcriptional activator depending on target gene and cell type context. In the case of the p21waf1 promoter, ZBP-89 directly interacts with histone acetyltransferase p300 and activates gene transcription (44). The ZBP-89 consensus site has been reported to be GGGAGG or its converse CCTCCC that has been found in p21waf1 and other target gene promoters (35). G1HE-(201–235) contains this consensus sequence overlapping with the G5 string (TGGGGGAGGGA). The G5 string is observed in the consensus sequence of other target genes, such as mouse Type I collagen and human ORNITHINE DECARBOXYLASE genes (14, 45). Collectively, these observations strongly suggest the requirement of 5 or more G-strings for the binding of ZBP-89. We should note that there is another G5 string at position 199–203 of G1HE in a complementary strand. Our data of DAPA demonstrated that ZBP-89 binds to this sequence of naked DNA. On the other hand, the transgenic mouse reporter assays demonstrated that IE3.9intG4LacZ resulted in reduced frequency of reporter expression although the G5 string at G1HE-(199–203) was not mutated. Although contribution of the G1HE-(199–203) region to Gata1 gene regulation in vivo remains to be determined, it is possible that the two G5 strings are both required for Gata1 gene expression. Alternatively, the G5 string at G1HE-(215–219) may play dominant roles over the other G5 string in vivo.
Our data of EMSA and DAPA suggest that DNA binding of ZBP-89 is facilitated by GATA-1 binding to the G1HE. Given that ZBP-89 was found in multiprotein complexes containing GATA-1 and FOG-1 (16) and that ZBP-89 physically associates with GATA-1 in vitro, it is conceivable that ZBP-89 is recruited by GATA-1 to the G1HE region through a protein-protein interaction. It remains an open question why GATA-1 binding to the G1HE was not affected by ZBP-89 in our study. Because our DNA binding assays demonstrated that other KLF factors can bind to the G4 mutant probe, these factors might compensate for the absence of ZBP-89 binding and support the GATA-1 binding.
The NT domain of GATA-1 has been considered as a transcription activation domain, whereas the NF domain provides an interaction surface for FOG-1 and other transcription factors (1). The N-terminal truncated form of GATA-1 (GATA-1s) has been reported as a product of somatic mutations of the GATA1 gene in children with Down syndrome-related acute megakaryoblastic leukemia (48). In this study, we observed that deletion of NT, but not NF, from GATA-1 canceled the repression of ZBP-89-mediated reporter activation in transfection reporter assays. Because the NT region of GATA-1 was not required for association with ZBP-89 in our GST pull-down assays, this domain might act as a interaction surface for transcriptional repressors.
In summary, our data suggest that the G5 string located in the 5′ region of the G1HE is necessary for Gata1 gene expression. Although several KLFs are able to bind to this region, the G4 mutant assays performed both in vivo and in vitro strongly suggest that ZBP-89 plays indispensable roles in activation of the G1HE. Besides the GATA site, the G5 string is the first element in the G1HE that has been proved to be necessary for Gata1 gene expression in vivo. Given that ZBP-89 has been identified as a component of multiprotein complexes containing GATA-1 and FOG-1 (16), the GATA site and the G5 string might play a fundamental role for the formation of multiprotein complexes and Gata1 gene regulation.
Supplementary Material
Acknowledgments
We thank Juanita L. Merchant for providing pGEXZBP-89, and Ken-ichi Isobe for pCDNA3.1BFCOL1. We appreciate JIMRO Co., Ltd. for providing a place for radioisotope experiments. We also acknowledge Yukie Kawatani, Reiko Kawai, and Naomi Kaneko for technical assistance, Tania O'Connor for critical reading of the manuscript, and Mikiko Suzuki, Takashi Moriguchi, Makoto Kobayashi, and Naoko Minegishi for helpful discussions.
This work was supported by grants from the Naito Foundation (to K. O.), Japan Science and Technology Corporation-ERATO Environmental Response Project (to M. Y.), and the Ministry of Education, Science, Sports and Culture (to M. Y. and K. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HRD
- hematopoietic regulatory domain
- G1HE
- Gata1 gene hematopoietic enhancer
- KLF
- Krüppel-like transcription factors
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactoside
- DBD
- DNA binding domain
- ChIP
- chromatin immunoprecipitation
- MEL
- mouse erythroleukemia
- IP
- immunoprecipitation
- DAPA
- DNA affinity precipitation assay
- GST
- glutathione S-transferase
- FOG-1
- friend of GATA-1
- MBP
- maltose-binding protein
- EMSA
- electrophoretic mobility shift assay
- PBS
- phosphate-buffered saline
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein.
REFERENCES
- 1.Ferreira R., Ohneda K., Yamamoto M., Philipsen S. (2005) Mol. Cell Biol. 25, 1215–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutiérrez L., Nikolic T., van Dijk T. B., Hammad H., Vos N., Willart M., Grosveld F., Philipsen S., Lambrecht B. N. (2007) Blood 110, 1933–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onodera K., Takahashi S., Nishimura S., Ohta J., Motohashi H., Yomogida K., Hayashi N., Engel J. D., Yamamoto M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4487–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishimura S., Takahashi S., Kuroha T., Suwabe N., Nagasawa T., Trainor C., Yamamoto M. (2000) Mol. Cell. Biol. 20, 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motohashi H., Katsuoka F., Shavit J. A., Engel J. D., Yamamoto M. (2000) Cell 103, 865–875 [DOI] [PubMed] [Google Scholar]
- 6.Takahashi S., Shimizu R., Suwabe N., Kuroha T., Yoh K., Ohta J., Nishimura S., Lim K. C., Engel J. D., Yamamoto M. (2000) Blood 96, 910–916 [PubMed] [Google Scholar]
- 7.Ferreira R., Wai A., Shimizu R., Gillemans N., Rottier R., von Lindern M., Ohneda K., Grosveld F., Yamamoto M., Philipsen S. (2007) Blood 109, 5481–5490 [DOI] [PubMed] [Google Scholar]
- 8.Nicolis S., Bertini C., Ronchi A., Crotta S., Lanfranco L., Moroni E., Giglioni B., Ottolenghi S. (1991) Nucleic Acids Res. 19, 5285–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohneda K., Shimizu R., Nishimura S., Muraosa Y., Takahashi S., Engel J. D., Yamamoto M. (2002) Genes Cells 7, 1243–1254 [DOI] [PubMed] [Google Scholar]
- 10.Tsai S. F., Strauss E., Orkin S. H. (1991) Genes Dev. 5, 919–931 [DOI] [PubMed] [Google Scholar]
- 11.Vyas P., McDevitt M. A., Cantor A. B., Katz S. G., Fujiwara Y., Orkin S. H. (1999) Development 126, 2799–2811 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki M., Moriguchi T., Ohneda K., Yamamoto M. (2009) Mol. Cell. Biol. 29, 1163–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antona V., Cammarata G., De Gregorio L., Dragani T. A., Giallongo A., Feo S. (1998) Cytogenet. Cell Genet. 83, 90–92 [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa T., Takeuchi A., Miyaishi O., Isobe K., de Crombrugghe B. (1997) J. Biol. Chem. 272, 4915–4923 [DOI] [PubMed] [Google Scholar]
- 15.Merchant J. L., Iyer G. R., Taylor B. R., Kitchen J. R., Mortensen E. R., Wang Z., Flintoft R. J., Michel J. B., Bassel-Duby R. (1996) Mol. Cell. Biol. 16, 6644–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo A. J., Moran T. B., Schindler Y. L., Choe S. K., Langer N. B., Sullivan M. R., Fujiwara Y., Paw B. H., Cantor A. B. (2008) Mol. Cell. Biol. 28, 2675–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guyot B., Murai K., Fujiwara Y., Valverde-Garduno V., Hammett M., Wells S., Dear N., Orkin S. H., Porcher C., Vyas P. (2006) J. Biol. Chem. 281, 13733–13742 [DOI] [PubMed] [Google Scholar]
- 18.Li X., Xiong J. W., Shelley C. S., Park H., Arnaout M. A. (2006) Development 133, 3641–3650 [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara Y., Browne C. P., Cunniff K., Goff S. C., Orkin S. H. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12355–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyons S. E., Lawson N. D., Lei L., Bennett P. E., Weinstein B. M., Liu P. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi S., Onodera K., Motohashi H., Suwabe N., Hayashi N., Yanai N., Nabesima Y., Yamamoto M. (1997) J. Biol. Chem. 272, 12611–12615 [DOI] [PubMed] [Google Scholar]
- 22.Gregory R. C., Taxman D. J., Seshasayee D., Kensinger M. H., Bieker J. J., Wojchowski D. M. (1996) Blood 87, 1793–1801 [PubMed] [Google Scholar]
- 23.Merika M., Orkin S. H. (1995) Mol. Cell. Biol. 15, 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa K., Kobayashi M., Masumi A., Lyons S. E., Weinstein B. M., Liu P. P., Yamamoto M. (2003) Mol. Cell. Biol. 23, 8295–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima S., Nagata S. (1990) Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu R., Takahashi S., Ohneda K., Engel J. D., Yamamoto M. (2001) EMBO J. 20, 5250–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordeen S. K. (1988) BioTechniques 6, 454–458 [PubMed] [Google Scholar]
- 28.Hogan B., Beddington R., Constantini F., Lacy E. (1994) Manipulating the Mouse Embryo, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Shimizu R., Trainor C. D., Nishikawa K., Kobayashi M., Ohneda K., Yamamoto M. (2007) J. Biol. Chem. 282, 15862–15871 [DOI] [PubMed] [Google Scholar]
- 30.Billon N., Carlisi D., Datto M. B., van Grunsven L. A., Watt A., Wang X. F., Rudkin B. B. (1999) Oncogene 18, 2872–2882 [DOI] [PubMed] [Google Scholar]
- 31.Jacquelin B., Rozenshteyn D., Kanaji S., Koziol J. A., Nurden A. T., Kunicki T. J. (2001) J. Biol. Chem. 276, 23518–23524 [DOI] [PubMed] [Google Scholar]
- 32.Jin Y., Wilhide C. C., Dang C., Li L., Li S. X., Villa-Garcia M., Bray P. F. (1998) Blood 92, 2777–2790 [PubMed] [Google Scholar]
- 33.Lepage A., Uzan G., Touche N., Morales M., Cazenave J. P., Lanza F., de La Salle C. (1999) Blood 94, 3366–3380 [PubMed] [Google Scholar]
- 34.Wang Z., Zhang Y., Lu J., Sun S., Ravid K. (1999) Blood 93, 4208–4221 [PubMed] [Google Scholar]
- 35.Hasegawa T., Xiao H., Isobe K. (1999) Biochem. Biophys. Res. Commun. 256, 249–254 [DOI] [PubMed] [Google Scholar]
- 36.Suzuki N., Suwabe N., Ohneda O., Obara N., Imagawa S., Pan X., Motohashi H., Yamamoto M. (2003) Blood 102, 3575–3583 [DOI] [PubMed] [Google Scholar]
- 37.Suzuki M., Ohneda K., Hosoya-Ohmura S., Tsukamoto S., Ohneda O., Philipsen S., Yamamoto M. (2006) Blood 108, 726–733 [DOI] [PubMed] [Google Scholar]
- 38.Whyatt D. J., Karis A., Harkes I. C., Verkerk A., Gillemans N., Elefanty A. G., Vairo G., Ploemacher R., Grosveld F., Philipsen S. (1997) Genes Funct. 1, 11–24 [DOI] [PubMed] [Google Scholar]
- 39.Whyatt D., Lindeboom F., Karis A., Ferreira R., Milot E., Hendriks R., de Bruijn M., Langeveld A., Gribnau J., Grosveld F., Philipsen S. (2000) Nature 406, 519–524 [DOI] [PubMed] [Google Scholar]
- 40.Wadman I. A., Osada H., Grütz G. G., Agulnick A. D., Westphal H., Forster A., Rabbitts T. H. (1997) EMBO J. 16, 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaczynski J., Cook T., Urrutia R. (2003) Genome Biol. 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lomberk G., Urrutia R. (2005) Biochem. J. 392, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki T., Aizawa K., Matsumura T., Nagai R. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 1135–1141 [DOI] [PubMed] [Google Scholar]
- 44.Bai L., Merchant J. L. (2000) J. Biol. Chem. 275, 30725–30733 [DOI] [PubMed] [Google Scholar]
- 45.Law G. L., Itoh H., Law D. J., Mize G. J., Merchant J. L., Morris D. R. (1998) J. Biol. Chem. 273, 19955–19964 [DOI] [PubMed] [Google Scholar]
- 46.Wieczorek E., Lin Z., Perkins E. B., Law D. J., Merchant J. L., Zehner Z. E. (2000) J. Biol. Chem. 275, 12879–12888 [DOI] [PubMed] [Google Scholar]
- 47.Vernimmen D., De Gobbi M., Sloane-Stanley J. A., Wood W. G., Higgs D. R. (2007) EMBO J. 26, 2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wechsler J., Greene M., McDevitt M. A., Anastasi J., Karp J. E., Le Beau M. M., Crispino J. D. (2002) Nat. Genet. 32, 148–152 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.