Abstract
In eukaryotes, each subcellular compartment harbors a specific group of proteins that must accomplish specific tasks. Nfs1 is a highly conserved mitochondrial cysteine desulfurase that participates in iron-sulfur cluster assembly as a sulfur donor. Previous genetic studies, in Saccharomyces cerevisiae, have suggested that this protein distributes between the mitochondria and the nucleus with biochemically undetectable amounts in the nucleus (termed “eclipsed distribution”). Here, we provide direct evidence for Nfs1 nuclear localization (in addition to mitochondria) using both α-complementation and subcellular fractionation. We also demonstrate that mitochondrial and nuclear Nfs1 are derived from a single translation product. Our data suggest that the Nfs1 distribution mechanism involves at least partial entry of the Nfs1 precursor into mitochondria, and then retrieval of a minor subpopulation (probably by reverse translocation) into the cytosol and then the nucleus. To further elucidate the mechanism of Nfs1 distribution we determined the N-terminal mitochondrial sequence of Nfs1 by Edman degradation. This led to the discovery of a novel mitochondrial processing enzyme, Icp55. This enzyme removes three amino acids from the N terminus of Nfs1 after cleavage by mitochondrial processing peptidase. Intriguingly, Icp55 protease (like its substrate Nfs1) appears to be dual distributed between the nucleus and mitochondria.
Eclipsed distribution is a specific case of the dual protein subcellular localization in which one compartment contains the vast majority of a specific protein while a second compartment contains only a minute (eclipsed) amount (1–3). Therefore, eclipsed distributed proteins are hard to detect, and it is hard to discriminate between the specific functions of each isoprotein. For example, aconitase, a well characterized mitochondrial protein that participates in the tricarboxylic acid cycle, has been recently shown to be present in minute amounts in the cytosol, where it is crucial for the function of the glyoxylate shunt (2, 3).
The protein NifS was first identified and characterized in Azotobacter vinelandii, as part of the nitrogen fixation (nif) operon (4). This enzyme is a pyridoxal 5′-phosphate-dependent cysteine desulfurase. In eukaryotes, an NifS-like protein termed Nfs1 has been identified, which shows a highly conserved sequence homology. The Saccharomyces cerevisiae mitochondrial Nfs1 has been shown to participate in iron-sulfur cluster assembly (5). Nakai and coworkers identified an NLS5-like sequence, RRRPR, in the mature sequence of yeast Nfs1, that is highly conserved in eukaryotic Nfs1 proteins (e.g. in yeast, mouse, and human) (6). When this NLS-like sequence was mutated to RRGSR, the mutant gene could not complement cell growth of a chromosomal NFS1-depleted strain. Nevertheless, this mutation did not affect the function of Nfs1 in the biogenesis of iron-sulfur proteins (5, 6). Nfs1 performs an additional function in the post-transcriptional modification of mitochondrial and cytosolic tRNAs (7). However, recent evidence suggests that nuclear Nfs1 does not serve as the sulfur donor for cytosolic tRNAs (8).
In S. cerevisiae and in human cells, Nfs1 cysteine desulfurases are encoded by single genes. The sorting mechanism that governs the simultaneous distribution of this protein into several subcellular compartments is unknown. In yeast, attempts to detect the endogenous Nfs1 in the nucleus by immunoblotting or immunofluorescence failed (6, 9, 10) most probably because Nfs1 is present at extremely low (eclipsed) levels in the nucleus. In human cells, the subcellular distribution of transiently expressed Nfs1 in mitochondria, cytosol, and nucleus was claimed to be achieved by alternative translation initiation from a single transcript (11). As described for yeast Nfs1, the low native amounts of extramitochondrial mammalian Nfs1 hampered its physical detection in the nucleus (5, 10, 12).
In this study, we characterized the processing pathway of yeast Nfs1 during its translocation into the mitochondrial matrix. To our surprise, we discovered that yeast Nfs1 undergoes two steps of proteolytic processing; first it is cleaved by the mitochondrial processing peptidase (MPP), which removes its mitochondrial targeting sequence (MTS) and then it is cleaved by a newly discovered peptidase, designated Icp55, which removes three amino acids from its N terminus. In addition, we used an α-complementation assay to detect the nuclear localization of Nfs1, and we provide evidence that the mitochondrial and nuclear Nfs1 isoforms are derived from a single translation product.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Growth Conditions
Strains
S. cerevisiae strains used were BY4741 (Mata; his3Δ1; leu2Δ0; met15Δ0; ura 3Δ0), W303B1 (Matα; leu2–3,112; trp1–1; can1–100; ura3–1; ade2–1; his3–11,15), Δicp55 (Δyer078c; derived from a W303B1 strain and was kindly provided by Thomas Langer (13)), and Tet-NFS1 (pNFS1:: kanR-tet07-TATA URA3::CMV-tTA; MATa; his3–1; leu2–0; met15–0) (14).
Plasmids Constructs
NFS1 gene was amplified using yeast genomic DNA as template by PCR using the indicated oligonucleotides (Table 1) and was cloned into vector p425Gal10 (15). All mutations were created using the QuikChange® II kit (Stratagene) or PCR reactions using either the indicated oligonucleotides (Table 1). Nfs1-α was created as was described elsewhere (16) pHxK-α, pKGD2-α, cytosolic ω (pωc), and mitochondrial ω (pωm) were also described elsewhere (16). Nuclear ω (pωNLS)-LacZ ω fragment was amplified from pωc template using the indicated primers (Table 1); the reverse primer harbors the SV40-T antigen NLS (17), and the PCR product was cloned into pRS426-Met25 vector.
TABLE 1.
Oligonucleotides (5′ to 3′) used in this study
| Mutant name | Forward | Reverse |
|---|---|---|
| Nfs1 | GGATCCATGTTGAAATCAACTGCTAC | AAGCTTTCAATGACCTGACCATTT |
| Nfs1-6His | AAGCTTTCAGTGATGGTGATGGTGATGCCCGGGATGACCTGACCATTTGATG | AAGCTTTCAGTGATGGTGATGGTGATGCCCGGGATGACCTGACCATTTGATG |
| Nfs1-M85L | GCTTCTGGTAGCACTGCATTGAGCCATGCTTATCAAG | CTTGATAAGCATGGCTCAATGCAGTGCTACCAGAAGC |
| Nfs1-Shift | GGATCCATGTTGAAATCAACTCGCTACAAG | AAGCTTTCAATGACCTGACCATTT |
| Nfs1-ΔMTS | CGGATCCCAGAATGTCCCCTCCTGCAGC | GCTGCAGGAGGGGACATTCTGGGATCCG |
| Nfs1-UP | GGGCTTGTTTGGTAAGCGGAGAATTCTATTCCCCTCCTGCAGCAG | CTGCTGCAGGAGGGGAATAGAATTCTCCGCTTACCAAACAAGCCC |
| Nfs1-Y34L | GTTTGGTAAGCAGGAGATTCCTTTCCCCTCCTGCAGCAGGCG | CGCCTGCTGCAGGAGGGGAAAGGAATCTCCTGCTTACCAAAC |
| Nfs1-F33S,Y35F | GTTTGGTAAGCAGGAGATCCTTTTCCCCTCCTGCAGCAGGCG | CGCCTGCTGCAGGAGGGGAAAAGGATCTCCTGCTTACCAAAC |
| Nfs1-P37A | GGTAAGCAGGAGATTCTATTCCCCTGCTGCAGCAGGCGTGAAGTTAG | CTAACTTCACGCCTGCTGCAGCAGGGGAATAGAATCTCCTGCTTACC |
| Nfs1-S35A | GTTTGGTAAGCAGGAGATTCTATGCCCCTCCTGCAGCAGGC | GCCTGCTGCAGGAGGGGCATAGAATCTCCTGCTTACCAAAC |
| Nfs1-P36D,P37D | GGTAAGCAGGAGATTCTATTCCGATGATGCAGCAGGCGTGAAGTTAG | CTAACTTCACGCCTGCTGCATCATCGGAATAGAATCTCCTGCTTACC |
| Nfs1-P36A | GGTAAGCAGGAGATTCTATTCCGCTCCTGCAGCAGGCGTGAAG | CTTCACGCCTGCTGCAGGAGCGGAATAGAATCTCCTGCTTACC |
| Nfs1-Shift | GGATCCATGTTGAAATCAACTCGCTACAAG | AAGCTTTCAATGACCTGACCATTTG |
| Nfs1-M85L | GCTGCGACCTCTCTATTTGGATGTGCAAGCTAC | GTAGCTTGCACATCCAAATAGAGAGGTCGCAGC |
| Icp55-ΔNLS | GAATTGCAGAGTTCGGTAAGATCGCGTCCCCTCAAGAGTTGAG | CTCAACTCTTGAGGGGACGCGATCTTACCGAACTCTGCAATTC |
| CCCCTCAAGAGTTGGGAATTATGAGGGGAGCTGGCCAAATATC | GATATTTGGCCAGCTCCCCTCATAATTCCCAACTCTTGAGGGG | |
| pωNLS | CCAAGCTTGGGATGATTACGGATTCACTGG | CGGGATCCCGTTAATCTTCAACCTTTCTCTTCTTCTTTGGACCACCACCTTTTTGACACCAGACCAAC |
| For two-step PCR procedure | ||
| Nfs1-mutNLS | GGATCCATGTTGAAATCAACTGCTAC | ATCCACCTCCTACATAGATGGCACCTATTC |
| CATCTATGTAGGAGGTGGATCAAGAGTTAGATTAGAACC | AAGCTTTCAATGACCTGACCATTTG |
Growth Conditions
Strains harboring the appropriate plasmids were grown overnight at 30 °C in synthetic depleted (SD) medium containing 0.67% (w/v) yeast nitrogen base 2% galactose or 2% glucose (w/v), supplemented with the appropriate amino acids (50 μg/ml). For agar plates, 2% agar was added and when indicated 0.08% X-gal was added, dissolved in 100% N,N-dimethylformamide and phosphate buffer (25 mm sodium phosphate buffer titrated to pH 7.0). Plates containing 2% yeast extract, 1% peptone, and 2% glucose (w/v) (YPD) supplemented with 120 μg/ml tetracycline where used for conditional knockout of Nfs1 for the Tet-NFS1 strain.
Metabolic Labeling
Cultures or induced cultures (in galactose) were harvested and labeled with 10 μCi/ml [35S]methionine and further incubated for 30 min at 30 °C. When required, 20 μm carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added before labeling. Labeling was stopped by addition of 10 mm sodium azide. Labeled cells were collected by centrifugation, resuspended in Tris/EDTA buffer, pH 8.0, containing 1 mm phenylmethylsulfonyl fluoride, broken with glass beads for 5 min, and centrifuged to obtain the supernatant fraction. Supernatants were denatured by boiling in 1% SDS, immunoprecipitated with anti-Nfs1 or anti-α rabbit antiserum and protein A-Sepharose (Amersham Biosciences), and then analyzed by SDS-PAGE.
Subcellular Fractionation
Induced yeast cultures were grown to an absorbance (A) of 1.5 at 600 nm. Mitochondria were isolated as described previously (18). Spheroplasts were prepared in the presence of Zymolyase-20T (MP Biomedicals, Irvine, CA). Each of our subcellular fractionation experiments was assayed for cross-contaminations by using anti-Aco1 antibody as a mitochondrial marker and anti-hexokinase 1 (anti-HxK1) antibody as a cytosolic marker.
For isolation of nucleus, 200 ml of induced yeast culture was grown to 1.5 A at 600 nm. Cells were centrifuged at 5000 × g for 6 min and washed with phosphate-citrate buffer (pH = 6.5). Cells were digested to spheroplasts by Zymolyase-20T (MP Biomedicals, Irvine, CA). Spheroplasts were then precipitated at 1200 × g for 5min and resuspended in lysis buffer (50 mm EDTA, 50 mm Tris-HCl, pH 7.6, 1% Triton X-100). Cells debris were centrifuged at 200 × g for 10 min at 4 °C, twice. Total sample was taken before nucleus precipitation. Nuclei were precipitated at 1500 × g for 20 min and then washed in the lysis buffer without Triton X-100. Nuclei were lysed with a high salts concentration buffer (20 mm Hepes, pH 7.6, 25% glycerol, 420 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride). Samples were centrifuged at 14,000 × g for 20 min to remove nucleus debris and DNA. Total and nuclear fractions were assayed with anti-histone H4 antibody, which was kindly provided by Amikam Cohen (Hebrew University of Jerusalem).
α-Complementation Assay
Yeast cells were transformed with plasmids encoding various α fusion proteins and with pωc, pωm, or pωNLS. Colonies were streaked on X-gal plates and incubated at 30 °C for 72 h.
Protein Purification for Edman Degradation
Induced yeast cultures were grown to 1.5 A at 600 nm. Cells were lysed with glass beads in Buffer A (6 m guanidine hydrochloride, 0.1 m NaH2PO4, 10 mm Tris, pH adjust to 8 with NaOH). Purification proceeded according to Qiagen protocol of nickel-nitrilotriacetic acid-Sepharose (Qiagen). Purification quality was assessed with Coomassie Blue SDS-PAGE staining. The purified protein was subjected to an Edman degradation analysis.
RESULTS
Nfs1 Is Processed by Two Peptidases, MPP and the Novel Icp55
Nfs1 is a highly conserved mitochondrial cysteine desulfurase in all organisms. In S. cerevisiae and in other eukaryotes the protein harbors a typical N-terminal MTS and is localized to the mitochondria. As outlined in the introduction, a low abundant form of Nfs1 in the nucleus may be targeted by a nuclear localization sequence (NLS) (6, 9). To characterize the Nfs1 distribution mechanism, one of our first efforts was to identify its exact MTS and determine the MPP-cleavage site at the N terminus of Nfs1. Nfs1 fused to a six-histidine tag was purified from a wild-type (WT) yeast strain and was subjected to Edman degradation analysis. The N-terminal sequence detected was PAAGVKLDD, indicating that Nfs1 is cleaved between two proline residues at positions 36 and 37 (Fig. 1A).
FIGURE 1.
Processing of Nfs1. A, the wild type Nfs1 N-terminal amino acid sequence. Arginines that can be potential MPP recognition elements are indicated as enlarged R letters. Arrows indicate MPP and Icp55 cleavage sites that were determined by Edman degradation. B, expression of Nfs1 in WT and Δicp55 strains was analyzed by Western blotting using Nfs1 antiserum. C, cleavage of Nfs1 mutant proteins by MPP (left arrows) and Icp55 (right arrows) as determined by Edman degradation. Substituted amino acids are highlighted, the lack of cleavage is indicated by an “X,” and a gray arrow indicates partial cleavage. D, metabolic labeling of Nfs1-α and Nfs1-UP-α in the absence or presence of CCCP. p, precursor; m, mature.
The detected cleavage site according to this Edman degradation analysis was unexpected, because typical MPP cleavage sites usually include arginine residues at one or more of the positions −2, −3, and/or −10 relative to the cleavage site (19, 20). In fact, cleavage between phenylalanine 33 and tyrosine 34 would place arginines at positions −2 and −3 relative to this site according to the MPP preferences, referred to above (see highlighted arginines at positions −2 and −3 relative to this cleavage site in Fig. 1A). Another prediction based on the bioinformatic MitoProtII program (21, 22), places the cleavage site for yeast Nfs1 between tyrosine 34 and serine 35 (arginines would be at positions −2 and −10 relative to this cleavage site).
Thus, we hypothesized the existence of an additional proteolytic step (besides that performed by MPP), which cleaves Nfs1 immediately downstream of the predicted MPP cleavage site. This could result in the N-terminal sequence detected by Edman degradation of Nfs1 (between proline 36 and proline 37). We screened yeast databases (Saccharomyces Genome Database and Yeast Proteome Database) for known and putative mitochondrial proteases. We were particularly interested in enzymes with aminopeptidase activity, that could theoretically remove between two and three amino acids. Among the candidate enzymes we found a group of putative Xaa-Pro aminopeptidases that presumably remove Xaa-Pro from the N terminus of substrate proteins. Only one putative Xaa-Pro aminopeptidase, Icp55 (YER078c), was predicted to be localized in the mitochondria according to the databases.
To directly examine if Nfs1 is a substrate of Icp55, Western blot analysis was performed on extracts of yeast cells harboring a deletion of the ICP55 gene (Δicp55) in comparison with WT strains. Nfs1, from the Δicp55 strain exhibited a slightly slower migrating band, indicating a higher molecular weight than the corresponding protein band from WT strains (Fig. 1B). Edman degradation analysis performed on Nfs1 purified from the knockout strain, Δicp55, detected three additional amino acids, YSP, at the N terminus of Nfs1 (Fig. 1A). Thus, Nfs1 is subjected to two steps of processing: first by MPP, which cleaves the precursor between phenylalanine 33 (Phe-33) and tyrosine 34 (Tyr-34), and second by a newly discovered processing enzyme, Icp55, which cleaves between proline 36 (Pro-36) and proline 37 (Pro-37) removing three amino acids residues (YSP).
To investigate the properties of the novel peptidase, we first examined whether Icp55 can cleave the full-length Nfs1 precursor (i.e. the unprocessed form). A mutant MPP recognition sequence was constructed by replacing the two arginine codons at positions −2 and −3, which are expected to prevent cleavage by MPP (Fig. 1C, UP, unprocessed). As anticipated, the mutant protein migrated on SDS-PAGE according to a molecular weight of the Nfs1 precursor (Fig. 1D, compare lanes 2 and 3). This indicates that, in the absence of Nfs1 processing by MPP, the Nfs1 precursor is not cleaved by Icp55, because we can only detect the full-length Nfs1 precursor. Thus, Icp55 is in fact a mitochondrial processing enzyme whose recognition sequence is exposed only after removal of the MTS by MPP.
To gain insight into the requirements for Icp55 processing we changed the amino acid sequence in the vicinity of the Icp55 cleavage site. Each mutant protein was subjected to Edman degradation following expression in both the WT strain (expressing endogenous Icp55) and in the chromosomal knockout strain, Δicp55 (Fig. 1C). Only in one case, replacement of two prolines (Pro-36 and Pro-37) by two aspartates, was the cleavage prevented by Icp55. These two prolines seem to be of importance for the Icp55 cleavage reaction, because the replacement of a single proline changed the specificity to a new cleavage site without eliminating the cleavage reaction per se (Fig. 1C). Replacement of the first proline (Pro-36) by alanine moved the cleavage site between Tyr-34 and Ser-35, whereas exchange of the second (Pro-37) by alanine moved the cleavage between residue Ser-35 and Pro-36 (Fig. 1C). Another residue that we thought may be of importance is the N-terminal tyrosine exposed by MPP cleavage. Replacement of Tyr-34 by a phenylalanine had no effect on Icp55 cleavage between the prolines (Pro-36 and Pro-37) while its exchange by leucine brought about incomplete cleavage at the same site. Possibly an aromatic residue at this position may be of significance. Replacement of Ser-35 by alanine did not prevent cleavage but also moved the cleavage site between Tyr-34 and Ala-35 (Fig. 1C). These data clearly show that changing amino acids in the vicinity of the cleavage site affect cleavage efficiency and/or the specific site cleaved. The specificity of Icp55 cleavage remains to be identified.
Detection of Nfs1 in Mitochondria and the Nucleus, but Not in the Cytosol
There are a number of studies that suggest a nuclear function for yeast Nfs1 (5–7), however, due to the minute amounts of the protein in the nuclear fraction these data are indirect. The difficulty in detection prompted us to employ our α-complementation assay, which has been previously shown to be a sensitive in vivo approach for detecting eclipsed distributed proteins (1, 16). The α-complementation is based on the ability of two artificially expressed peptide fragments of Escherichia coli β-galactosidase (designated α 77 amino acids and ω 993 amino acids) to assemble in vivo into an enzymatically active complex only when present in the same compartment (16) (Fig. 2A). We fused the α fragment of β-galactosidase to the C terminus of Nfs1 (Nfs1-α) and expressed this fusion protein in yeast, whereas the ω polypeptide was expressed either in the cytosol (ωc), mitochondria (ωm), or nucleus (ωNLS). Although ωc and ωm have been described previously (16), ωNLS is described for the first time here as a way to get an indication of nuclear localization. To create ωNLS, we attached the NLS of SV-40 T-Antigen (PKKKRKV) (17) to the C terminus of the ω fragment. We have performed subcellular fractionation and can show that the ωNLS fusion protein exhibits a significant presence in the nucleus (data not shown).
FIGURE 2.
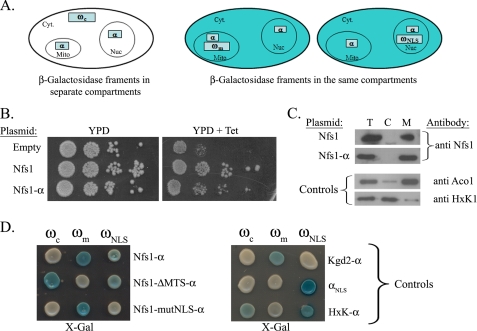
Nfs1-α is also localized to the nucleus. A, illustration of the α-complementation assay: the α-complementation assay is based on co-localization of β-galactosidase fragments, α and ω, within the same compartment (mitochondria (middle panel), or nucleus (right panel)). Enzymatic activity can be detected by production of blue colonies on X-gal plates. In the case that the two fragments are in separate compartments white colonies should appear (left panel). Cyt, cytosol; Mit, mitochondria; Nuc, nucleus. B, Tet-NFS1 strains harboring the indicated plasmids were grown on YPD plates in the presence or absence of tetracycline (the repressor of the Tet promoter) at 30 °C for 48 h. C, subcellular fractionation of yeast cells expressing Nfs1 or Nfs1-α. The total (T), cytosol (C), and mitochondria (M) fractions were analyzed by Western blotting using the indicated antibodies. Controls: HxK1 (hexokinase 1), a cytosolic marker, Aco1 (aconitase), a mitochondrial marker. D, yeast cultures co-expressing cytosolic ω (ωc), mitochondrial ω (ωm), or nuclear ω (ωNLS) together with the indicated Nfs1-α mutants were grown on galactose medium containing X-gal. Blue colonies detect α fragments that are associated with the indicated ω fragments. Nfs1-ΔMTS-α is a mutant that lacks the MTS, and Nfs1-mutNLS-α is a mutant in which four amino acids of the NLS sequence were mutated. Controls: αNLS-α fragment fused to the NLS of SV40 T-antigen, HxK1-α - (hexokinase 1) a cytosolic marker fused to the α fragment, Kgd2-α - a mitochondrial marker fused to the α fragment.
The first step in using the Nfs1-α fusion protein for analysis of Nfs1 distribution was to show that the apparent characteristics of the fusion protein are identical to those of the wild-type protein. Described below are the fusion protein's enzymatic activity in vivo (Fig. 2B), subcellular distribution (Fig. 2C), and processing in mitochondria (Fig. 3A). The Nfs1-α fusion protein can complement the Nfs1 activity in the Tet-NFS1 strain (Fig. 2B). This yeast strain contains a Tet-off promoter upstream of the endogenous NFS1 gene and can be repressed by adding tetracycline to the medium (14). The Tet-NFS1 strain expressing an empty vector does not grow on YPD plates containing tetracycline, whereas the same strain expressing the fusion protein (Nfs1-α) readily grows on the same media (Fig. 2B). Subcellular fractionation experiments reveal that the majority of the Nfs1-α molecules are found in the mitochondrial fraction (Fig. 2C), a pattern that is reminiscent of the Nfs1 wild-type protein (Fig. 2C, top). The quality of each fractionation was monitored with antibodies against mitochondrial (Aco1) and cytosolic (hexokinase 1 (HxK1)) markers (Fig. 2C). In addition, Nfs1-α is processed in the mitochondria efficiently, as will be described in the next section (Fig. 3A). Hence, we conclude that Nfs1-α exhibits the same subcellular distribution as wild-type Nfs1.
FIGURE 3.
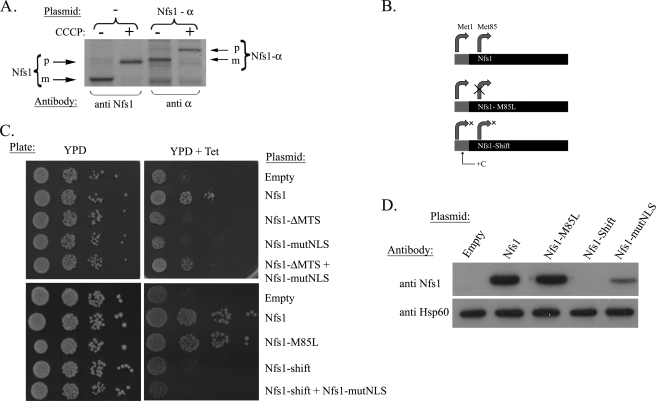
The NFS1 gene encodes a single gene product. A, metabolic labeling of wild-type strains expressing Nfs1 or Nfs1-α was performed in the absence or presence of CCCP (membrane potential uncoupler). p, precursor; m, mature. B, schematic representation of Nfs1 mutants. Gray and black boxes represent the MTS and mature sequences, respectively, curved arrows represent the potential translation initiation sites. Nfs1-M85L mutant protein contains a missense mutation in which Met-85 was changed to Leu. Nfs1-Shift has an additional nucleotide (C) inserted between the first and second AUG that causes early termination of translation starting from the first AUG. C, the Tet-NFS1 strain expressing the various Nfs1 mutant proteins were examined for growth on YPD plates in the presence or absence of tetracycline (Tet). D, Western blot analysis of the Tet-NFS1 strain expressing the mutant proteins above. The cells were grown on galactose media containing tetracycline for 8 h. Hsp60 antibody was used to control equivalent loading.
To examine the subcellular localization of the Nfs1-α fusion protein using the α-complementation assay, plasmids encoding Nfs1-α together with a cytosolic, mitochondrial, or nuclear localized ω fragments (ωm, ωc, and ωNLS, respectively) were simultaneously coexpressed in yeast WT cells. When these cells were grown on agar plates containing the X-gal indicator and galactose as the carbon source, blue colonies were obtained for cells coexpressing Nfs1-α either with ωm or with ωNLS, but not with the ωc fragment (Fig. 2D). These results indicate that a subpopulation of the Nfs1-α molecules is located in the nucleus and in fact no Nfs1-α is present in the cytosol (Fig. 2D), providing direct evidence for the dual distribution of this protein. As a negative control, we show that an exclusive mitochondrial protein fused to the α fragment of Kgd2-α (dihydrolipoyl transsuccinylase, a component of the mitochondrial α-ketoglutarate dehydrogenase complex), produces non-colored colonies when coexpressed with ωNLS or with ωc (Fig. 2D). The α-fragment itself, however, when attached to NLS as a positive control, produces blue colonies exclusively when coexpressed with the nuclear ω fragment (Fig. 2D). The cytosolic control HxK1-α fusion protein (HxK1 fused to α) produces blue colonies when coexpressed not only with ωc but also with ωNLS. This result is due to incomplete localization of ωNLS to the nucleus, and a subfraction of it remains in the cytosol. As a consequence, blue colonies appear for cells coexpressing ωNLS and any cytosolic protein fused to the α fragment. In this regard, because Nfs1-α is not a cytosolic protein (white colonies with ωc), blue colonies with ωNLS obviously detect the nuclear localization of the protein.
In support of this analysis a Nfs1-α mutant, lacking the MTS (ΔMTS-Nfs1-α), as expected, appears to be located in the nucleus but not in mitochondria. Surprisingly, ΔMTS-Nfs1-α appeared to also be in the cytosol, suggesting that the nuclear import of ΔMTS-Nfs1-α is somehow saturated, which in turn may suggest the participation of currently unknown factors in this process.
In addition, we mutated the NLS sequence of Nfs1-α similarly (not identically) to that described by Nakai and coworkers (6) from RRRPR to GGGSR (designated Nfs1-mutNLS-α), which prevents the targeting of the protein to the nucleus. As shown in Fig. 2C mutant Nfs1-mutNLS-α is exclusively located in mitochondria and is absent from the nucleus and cytosol (Fig. 2D). Thus, we conclude that Nfs1 is dual targeted between the mitochondria and nucleus, and it appears that both the NLS and the MTS are required for this dual localization.
Yeast Nfs1 Is a Dual Localized, Single Translation Product
There are various mechanisms by which the same protein can be localized to more than one subcellular compartment. In one possible mechanism two translation products are made, one harboring and one lacking a targeting signal (23, 24). An alternative dual targeting mechanism involves a single translation product which, nevertheless, distributes between two compartments. To detect the Nfs1 precursor(s) in comparison to the mature protein, we performed metabolic labeling of yeast cells in which import of proteins was blocked by the membrane potential uncoupler CCCP. If there are two Nfs1 translation products, we should expect two bands when the import to the mitochondria is blocked by CCCP, corresponding to an MTS containing precursor and a mature form, lacking the MTS (higher and lower molecular weights, respectively). However, in the presence of CCCP only the precursor (p) form of the protein appeared for both Nfs1 and Nfs1-α (Fig. 3A, compare lanes 1 to 2 and 3 to 4), which is consistent with a single Nfs1 gene product. This obviously does not rule out the possibility that a second translation product (nuclear derivative) is not detected, because it is made in minute amounts.
To address this question of a low abundance single translation product we generated a NFS1 mutant in which the second AUG, the only alternative in-frame initiation codon, was mutated to UUG (changing the methionine codon 85 to leucine, Nfs1-M85L, Fig. 3B). As shown in Fig. 3C, the Nfs1-M85L mutant protein readily grows on tetracycline-containing YPD plates like the wild-type Nfs1 (compare seventh and eighth rows), indicating that the single translation product (starting at the first AUG) is giving rise to both the mitochondrial and the nuclear protein. As a control we created a frameshift mutation (nucleotide insertion of C) between the first and the second AUGs, preventing meaningful translation from the first AUG of NFS1, but this should not effect translation from the second AUG (Met-85, Fig. 3B). This Nfs1-Shift mutant as expected could not grow in the presence of tetracycline (Fig. 3C, ninth row) due to the absence of mitochondrial Nfs1. However, if translation initiation from the second AUG (Met-85), enabling nuclear Nfs1 targeting, actually exists in vivo, one should be able to fully complement this activity with exclusive mitochondrial Nfs1. To address this question we used the exclusive mitochondrial Nfs1-mutNLS mutant as described earlier. Cells harboring plasmids encoding both the mitochondrial (Nfs1-mutNLS) and nuclear (ΔMTS-Nfs1)-targeted versions can in fact grow on tetracycline plates (Fig. 3C, right panel fifth row). However, strains co-expressing Nfs1-shift and Nfs1-mutNLS could not grow on tetracycline plates (Fig. 3C), indicating that Nfs1 is not translated from the second AUG and thus cannot complement the Nfs1 nuclear function. Consistently, no expression of Nfs1-shift was detected by Western blot (Fig. 3D, fourth lane). These results suggest that yeast NFS1 encodes a single, full-length translation product, which distributes between the mitochondria and nucleus.
Nuclear Nfs1 Is Processed by MPP
What is the mechanism by which the single Nfs1 translation product is dual localized in yeast? Two scenarios are possible: First, the precursor may be efficiently targeted to mitochondria, but a small fraction of molecules could, after being processed by MPP, move back by reverse translocation into the cytosol. In this case the nuclear protein should be processed, thus lacking the MTS. Such a mechanism resembles the previously described dual localization of fumarase and aconitase in yeast (1, 23–28). In the second scenario, some precursor Nfs1 molecules never reach the mitochondria and are directly targeted from the cytosol to the nucleus (2, 5). A somewhat similar mechanism has been described for the protein Adk1 in its distribution between the cytosol and mitochondria (29). The nuclear fraction according to this mechanism should contain the full-length Nfs1 precursor, including the MTS. To distinguish between these possibilities, we asked whether the precursor of Nfs1 can be functional in the nucleus. As described in the previous sections we have mutated the Nfs1 MPP cleavage site (Fig. 1A), which results in the expression of an unprocessed form of Nfs1 (Nfs1-UP). As shown in Fig. 1D, the processing of Nfs1-UP-α was examined by metabolic labeling in the presence (+) or absence (−) of CCCP. For Nfs1-UP we detected identical precursor size bands in the presence and absence of the uncoupler (see differences in size for Nfs1-α and no change for the Nfs1-UP-α mutant).
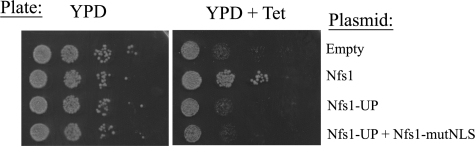
The activity of the Nfs1-UP mutant protein in vivo was analyzed in the conditional yeast Tet-NFS1 strain. This strain expressing the Nfs1-UP mutant protein did not show growth in the presence of tetracycline (Fig. 4, right panel). To examine whether the Nfs1-UP mutant is inactive in the nucleus, we used the Nfs1-mutNLS. We found that Nfs1-UP cannot complement Nfs1-mutNLS for growth of the Tet-NFS1 strain (Fig. 4, left panel last row), indicating that the Nfs1-UP mutant protein is inactive in the nucleus. Based on these findings we propose that Nfs1 needs to be processed by MPP to be functional in the nucleus. These data support a mechanism that involves reverse translocation of processed Nfs1, i.e. the retrograde movement of Nfs1 (first scenario above).
FIGURE 4.
The unprocessed Nfs1 mutant is inactive in the nucleus. The Tet-NFS1 cells expressing the indicated Nfs1 mutant proteins were grown on YPD plates in the presence or absence of tetracycline (Tet).
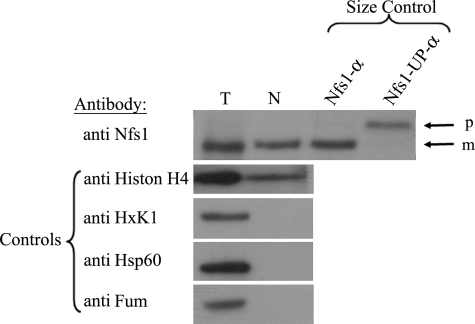
To gain additional support for the notion that the Nfs1 nuclear form is processed, a WT culture overexpressing Nfs1-α under the GAL10 promoter was subjected to a nuclear subcellular fractionation experiment. The nuclear fraction (N) was concentrated 24-fold more than the total cell extract (T), and aliquots were analyzed by Western blotting. The quality of the fractionation was monitored by antibodies against nuclear (histone acetyl H4), mitochondrial (Hsp60, Fumarase), and cytosolic (Hexokinase1, Fumarase) markers (Fig. 5). Nuclear Nfs1-α displays an apparent molecular weight that is indistinguishable from the mature mitochondrial form detected in the total cell extract and markedly differs from the precursor size (Fig. 5, compare the Nfs1-α in the first and second lanes to the third and fourth lanes). Taken together, these results strongly support a mechanism in which the nuclear Nfs1 is partially translocated into mitochondria (N terminus), and, after processing by MPP, undergoes a retrograde movement back into the cytosol and then the nucleus.
FIGURE 5.
All the Nfs1 molecules are processed in mitochondria. Subcellular fractionation of the WT strain overexpressing Nfs1-α. Aliquots of the total (T) and nuclear (N) fractions where analyzed by Western blotting. The nuclear fraction is concentrated approximately twenty-four times more than the total fraction. p, precursor; m, mature. Markers: Nfs1-α and Nfs1-UP-α. Controls: Hsp60 (mitochondria), HxK1 (cytosol), Histone H4 (nucleus), and Fum1 (mitochondria and cytosol).
Icp55-α Distributes Like Its Substrate, Nfs1, between Mitochondria and the Nucleus
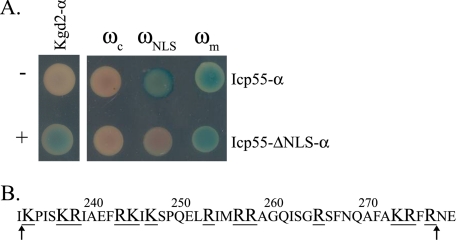
In this study we have shown that the protein Nfs1 is dual localized and that it is a substrate of a novel processing enzyme, Icp55. We asked whether Icp55 is also dual localized to the mitochondria and nucleus like its substrate, Nfs1. Again, our α-complementation assay was employed and blue colonies appeared only when Icp55-α was co-expressed with mitochondrial or nuclear localized ω fragments but not with the cytosolic ω fragment (Fig. 6A, top row). This distribution pattern resembles the pattern of Nfs1-α. The Icp55 sequence was scanned for potential nuclear localization signals. Because we did not find any consensus sequence similar to that of T-antigen NLS or the bipartite NLS of the nucleoplasmin (30, 31), we searched for regions enriched in positively charged amino residues (32). As shown in Fig. 6B, a region of positively charged amino acid residues (underlined and enlarged in Fig. 6B) was detected and subsequently deleted (42 amino acids between Lys-234 and Arg-276), creating a mutant that presumably lacks a putative NLS (Icp55-ΔNLS-α). This mutant was also subjected to the α-complementation assay and, in contrast to the wild type Icp55, it exhibited a blue colony phenotype only when expressed with the mitochondrial ω fragment and not when expressed with the nuclear ω fragment (Fig. 6B, lower row). These results indicate that the Icp55 “NLS” mutant was not targeted to the nucleus and, furthermore, supports the notion that Icp55 is also a dual distributed protein between mitochondria and nucleus. Thus, cleavage of the three residues YSP might occur in the nucleus.
FIGURE 6.
Icp55-α distributes between the mitochondria and the nucleus. A, blue colonies detect α fragments that are in the cytosol, mitochondria, or nucleus as described in the legend of Fig. 2. Controls: mitochondrial Kgd2-α co-expressed with ωm (+) or ωc (−). B, the putative Icp55 NLS region is presented with positively charged amino acids enlarged and underlined. The sequence between the two arrows was deleted from the Icp55-ΔNLS-α mutant protein.
DISCUSSION
In this study we provide insight into the distribution process of the yeast enzyme Nfs1 between mitochondria and the nucleus. Previously, Nakai and co-workers showed evidence that suggested that Nfs1 is also localized to the nucleus based on an NLS-like sequence they detected. Here, we present physical evidence for Nfs1 dual localization. First, we demonstrate that Nfs1-α is associated with ω fragment both in the nucleus and in the mitochondria, but not in the cytosol. Mutations in the NLS sequence eliminate the association of the two fragments, α and ω, in the nucleus. In addition, subcellular fractionation clearly detects overproduced Nfs1 in the nucleus. This enzyme is, in fact, dual localized in an “eclipsed” distribution pattern with the majority of the protein in mitochondria and a minute amount in the nucleus.
With respect to the mechanism of Nfs1 distribution, the results presented in this study are consistent with a single translation product of this protein giving rise to both localizations: (i) elimination of the second potential translation initiation codon (Nfs1-M85L) exhibited full functional complementation; (ii) a frameshift mutation between the first AUG (translation initiation codon) and the second AUG (Nfs1-shift) did not provide a functional nuclear Nfs1; (iii) only one protein species corresponding to the precursor was detected when processing by MPP was prevented and only the mature protein was detected (also in the nucleus) when processing was permitted; and (iv) the NFS1 gene in many eukaryotes does not contain any alternative start codons at the N terminus, which may encode alternative functional Nfs1 derivatives (5). In contrast, the human nuclear form of Nfs1 was suggested to be produced by alternative translation initiation (11). Our results suggest that a single Nfs1 translation product is generated in yeast, which is then distributed between the mitochondria and nucleus.
What is the precise mechanism of Nfs1 targeting and subcellular distribution? Our data show that nuclear Nfs1 depends on MPP processing for function and, hence, suggest that this isoform may first be translocated into mitochondrial matrix before its targeting and import into the nucleus guided by the internal NLS. There is no evidence for an export mechanism of matrix-located proteins out of the organelle. One possible mechanism, that could explain the data, involves reverse translocation and was first described for the tricarboxylic acid cycle enzyme, fumarase (18, 25, 27, 28, 33). According to this mechanism the extramitochondrial localization is achieved by retrograde movement of processed protein (MTS removed by MPP) through the TOM and TIM23 translocation channels back to the cytosol during translocation. This mechanism involves only partial import of the N terminus of the protein into the mitochondrial matrix prior to reverse translocation. Nuclear localization of Nfs1 involving reverse translocation is supported by several findings indicating that this isoform was produced as a precursor with an MTS, which then was removed as follows. (i) Unprocessed Nfs1 cannot complement nuclear activity in a strain depleted for Nfs1, whereas a version of the protein lacking the MTS can complement the Nfs1 nuclear function. This rules out the possibility that Nfs1 that is not initially targeted to the mitochondria as a precursor is functional in the nucleus. (ii) The mitochondrial and nuclear Nfs1 isoforms are generated from one single product (see above). (iii) Subcellular fractionation detects nuclear Nfs1 at a molecular weight precisely corresponding to the mature protein. Thus, removal of the MTS by MPP from the Nfs1 single translation product requires at least partial translocation of the N terminus into mitochondria. Moreover, following a block in mitochondrial protein import (by CCCP), we can only detect the precursor form of Nfs1, which indicates there is no other processing enzyme outside mitochondria, which can remove the MTS from the nuclear Nfs1. Because there is no evidence for export of proteins out of mitochondrial matrix (as mentioned earlier), the only known mechanism that can enable mitochondrially processed proteins to leave mitochondria is reverse translocation of a translocation intermediate spanning both import channels. According to this mechanism all of the Nfs1 precursor molecules are first partially targeted to the mitochondria and, after removal of the MTS by MPP, a small fraction still spanning the membranes moves back and is then targeted to the nucleus. We currently do not know the driving force for reverse translocation of Nfs1 back through the mitochondrial membranes. This could be, among other possibilities, the specific folding traits of the protein that block forward movement as in the case of fumarase (26, 27) or the specific N-terminal amino acid sequence of Nfs1 that causes inefficient binding between the motor mtHsp70 and the mature Nfs1, allowing a “slippage” of some of the protein molecules back into the cytosol.
A second topic of our study is the description of the novel mitochondrial processing enzyme, Icp55. We show that Nfs1 undergoes a two-step processing in the mitochondrial matrix; by MPP and then by a new peptidase, Icp55, which removes three amino acids, YSP from Nfs1 after MPP cleavage. Known processing peptidases in mitochondria include the following: (i) MPP, mentioned above, which is a conserved hetero-dimeric metallopeptidase that cleaves off sorting sequences in the matrix space (34); (ii) the MIP, which is a monomeric metallopeptidase that removes an octapeptide from preproteins after their processing by MPP (34); (iii) the inner membrane peptidase is a heterotrimeric complex that is located within the inner membrane (34) and cleaves several nuclearly encoded proteins in the intermembrane space after they have first been cleaved by MPP in the matrix (34); and (iv) a number of proteases, including AAA proteases, Rhomboid protease Pcp1, and the metalloprotease Atp23 that perform specific processing events (34). The physiological role of the different processing enzymes is not always understood; however, severe phenotypes upon gene inactivation suggest that they have important functions. Icp55 joins this important list of processing peptidases.
Based on domain homology, Icp55 was suspected to be an X-Pro aminopeptidase (EC 3.4.11.9), which catalyzes the release of any N-terminal amino acid that is linked with proline (NCBI, Conserved Domain Data base, CDD family no. cd01085). This description does not fit the observed Icp55 cleavage reactions for both the Nfs1 wild-type and mutant N termini as determined by sequential Edman degradation. Although proline appeared to be important for the overall processing reaction, this residue does not have to be located at the +2 position (relative to the MPP cleavage site) as would be expected for an X-Pro aminopeptidase. The prolines in Nfs1 are located further in, at positions +3 and/or +4. Furthermore, exchange of only one of these prolines did not fully abolish cleavage. In this regard, Icp55 exhibits similar characteristics to the other mitochondrial processing peptidases referred to above suggesting that this peptidase exhibits rather low sequence specificity. Moreover, our data show that Icp55 can cleave Nfs1 only after MPP processing has occurred, similarly to MIP and inner membrane peptidase. What is the physiological importance of cleavage by Icp55? Cleavage by Icp55 can change the N-terminal amino acid, which for example can affect the characteristics of the protein and/or its stability. For Nfs1 cleavage by MPP exposes Tyr at the N terminus, whereas the secondary cleavage by Icp55 exposes Pro at the N terminus. We assume that Icp55 has additional substrates in the mitochondria and the nucleus besides Nfs1 and may have a wide range of effects under specific conditions.
The α-complementation assay in combination with a preliminary mutational analysis intriguingly suggest that Icp55 may also be a dual distributed protein located in the same compartments as its Nfs1 substrate, the mitochondria, and nucleus. Whether Icp55 processes Nfs1 before it moves back into the cytosol or whether processing by Icp55 occurs in the nucleus remains to be determined. At present due to the low levels of nuclear Nfs1 it is impossible to subject nuclear Nfs1 to standard biochemical analysis. Whether these preliminary findings define a general concept of protein targeting, in that combinations of proteins that interact with one another are identically dual targeted to the same organelles, remains to be explored.
Acknowledgments
We thank Ariel Gaaton (Hebrew University of Jerusalem) for Edman degradation. Special thanks to Yudit Karp for dedicated assistance.
Footnotes
- NLS
- nuclear localization sequence
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- MPP
- mitochondrial processing peptidase
- MTS
- mitochondrial targeting sequence
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- HxK1
- hexokinase 1
- WT
- wild type.
REFERENCES
- 1.Regev-Rudzki N., Karniely S., Ben-Haim N. N., Pines O. (2005) Mol. Biol. Cell 16, 4163–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Regev-Rudzki N., Pines O. (2007) BioEssays 29, 772–782 [DOI] [PubMed] [Google Scholar]
- 3.Shlevin L., Regev-Rudzki N., Karniely S., Pines O. (2007) Traffic 8, 169–176 [DOI] [PubMed] [Google Scholar]
- 4.Zheng L., White R. H., Cash V. L., Jack R. F., Dean D. R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2754–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mühlenhoff U., Balk J., Richhardt N., Kaiser J. T., Sipos K., Kispal G., Lill R. (2004) J. Biol. Chem. 279, 36906–36915 [DOI] [PubMed] [Google Scholar]
- 6.Nakai Y., Nakai M., Hayashi H., Kagamiyama H. (2001) J. Biol. Chem. 276, 8314–8320 [DOI] [PubMed] [Google Scholar]
- 7.Nakai Y., Umeda N., Suzuki T., Nakai M., Hayashi H., Watanabe K., Kagamiyama H. (2004) J. Biol. Chem. 279, 12363–12368 [DOI] [PubMed] [Google Scholar]
- 8.Nakai Y., Nakai M., Lill R., Suzuki T., Hayashi H. (2007) Mol. Cell Biol. 27, 2841–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kispal G., Csere P., Prohl C., Lill R. (1999) EMBO J. 18, 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakai Y., Yoshihara Y., Hayashi H., Kagamiyama H. (1998) FEBS Lett. 433, 143–148 [DOI] [PubMed] [Google Scholar]
- 11.Land T., Rouault T. A. (1998) Mol. Cell 2, 807–815 [DOI] [PubMed] [Google Scholar]
- 12.Fosset C., Chauveau M. J., Guillon B., Canal F., Drapier J. C., Bouton C. (2006) J. Biol. Chem. 281, 25398–25406 [DOI] [PubMed] [Google Scholar]
- 13.Kambacheld M., Augustin S., Tatsuta T., Müller S., Langer T. (2005) J. Biol. Chem. 280, 20132–20139 [DOI] [PubMed] [Google Scholar]
- 14.Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., Trochesset M., Morse D., Krogan N. J., Hiley S. L., Li Z., Morris Q., Grigull J., Mitsakakis N., Roberts C. J., Greenblatt J. F., Boone C., Kaiser C. A., Andrews B. J., Hughes T. R. (2004) Cell 118, 31–44 [DOI] [PubMed] [Google Scholar]
- 15.Mumberg D., Müller R., Funk M. (1994) Nucleic Acids Res. 22, 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karniely S., Rayzner A., Sass E., Pines O. (2006) Exp. Cell Res. 312, 3835–3846 [DOI] [PubMed] [Google Scholar]
- 17.Lanford R. E., Kanda P., Kennedy R. C. (1986) Cell 46, 575–582 [DOI] [PubMed] [Google Scholar]
- 18.Knox C., Sass E., Neupert W., Pines O. (1998) J. Biol. Chem. 273, 25587–25593 [DOI] [PubMed] [Google Scholar]
- 19.Ito A. (1999) Biochem. Biophys. Res. Commun. 265, 611–616 [DOI] [PubMed] [Google Scholar]
- 20.Niidome T., Kitada S., Shimokata K., Ogishima T., Ito A. (1994) J. Biol. Chem. 269, 24719–24722 [PubMed] [Google Scholar]
- 21.Andreoli C., Prokisch H., Hortnagel K., Mueller J. C., Munsterkotter M., Scharfe C., Meitinger T. (2004) Nucleic Acids Res. 32, D459–D462; database issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prokisch H., Andreoli C., Ahting U., Heiss K., Ruepp A., Scharfe C., Meitinger T. (2006) Nucleic Acids Res. 34, D705–D711; database issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danpure C. J. (1995) Trends Cell Biol. 5, 230–238 [DOI] [PubMed] [Google Scholar]
- 24.Karniely S., Pines O. (2005) EMBO Rep. 6, 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regev-Rudzki N., Yogev O., Pines O. (2008) J. Cell Sci. 121, 2423–2431 [DOI] [PubMed] [Google Scholar]
- 26.Sass E., Blachinsky E., Karniely S., Pines O. (2001) J. Biol. Chem. 276, 46111–46117 [DOI] [PubMed] [Google Scholar]
- 27.Sass E., Karniely S., Pines O. (2003) J. Biol. Chem. 278, 45109–45116 [DOI] [PubMed] [Google Scholar]
- 28.Yogev O., Karniely S., Pines O. (2007) J. Biol. Chem. 282, 29222–29229 [DOI] [PubMed] [Google Scholar]
- 29.Schricker R., Angermayr M., Strobel G., Klinke S., Korber D., Bandlow W. (2002) J. Biol. Chem. 277, 28757–28764 [DOI] [PubMed] [Google Scholar]
- 30.Dingwall C., Robbins J., Dilworth S. M., Roberts B., Richardson W. D. (1988) J. Cell Biol. 107, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalderon D., Richardson W. D., Markham A. F., Smith A. E. (1984) Nature 311, 33–38 [DOI] [PubMed] [Google Scholar]
- 32.Makkerh J. P., Dingwall C., Laskey R. A. (1996) Curr. Biol. 6, 1025–1027 [DOI] [PubMed] [Google Scholar]
- 33.Karniely S., Regev-Rudzki N., Pines O. (2006) J. Mol. Biol. 358, 396–405 [DOI] [PubMed] [Google Scholar]
- 34.Koppen M., Langer T. (2007) Crit. Rev. Biochem. Mol Biol. 42, 221–242 [DOI] [PubMed] [Google Scholar]