Abstract
GPIHBP1, a glycosylphosphatidylinositol-anchored endothelial cell protein of the lymphocyte antigen 6 (Ly6) family, binds lipoprotein lipase (LPL) avidly and is required for the lipolytic processing of triglyceride-rich lipoproteins. GPIHBP1 contains two key structural motifs, an acidic domain and an Ly6 motif (a three-fingered domain specified by 10 cysteines). The acidic domain is required for LPL binding, but the importance of the Ly6 domain is less clear. To explore that issue, we transfected cells with a wild-type GPIHBP1 expression vector or mutant GPIHBP1 vectors in which specific cysteines in the Ly6 domain were changed to alanine. The mutant GPIHBP1 proteins reached the cell surface, as judged by antibody binding studies and by the ability of a phosphatidylinositol-specific phospholipase C to release these proteins from the cell surface. However, cells expressing the cysteine mutants could not bind LPL. The acidic domain of the cysteine mutants appeared to remain accessible, as judged by binding studies with an antibody against the acidic domain. We also developed a cell-free assay of LPL binding. We created a rat monoclonal antibody against the carboxyl terminus of mouse GPIHBP1 and used that antibody to coat agarose beads. We then tested the ability of soluble forms of GPIHBP1 that had been immobilized on monoclonal antibody-coated beads to bind LPL. In this assay, wild-type soluble GPIHBP1 bound LPL avidly, but the cysteine mutants did not. Thus, our studies suggest that a structurally intact Ly6 domain (in addition to the acidic domain) is essential for LPL binding.
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1)2 is an endothelial cell protein that is required for the lipolytic processing of triglyceride-rich lipoproteins in the plasma (1). In the absence of GPIHBP1, lipolysis of plasma lipoproteins is virtually abolished, leading to severe hypertriglyceridemia (1). Expression of GPIHBP1 in cultured cells confers the ability to bind lipoprotein lipase (LPL) (1). That finding, along with the fact that GPIHBP1 is located in endothelial cells, led Beigneux et al. (1) to hypothesize that GPIHBP1 serves as an endothelial cell platform for lipolysis.
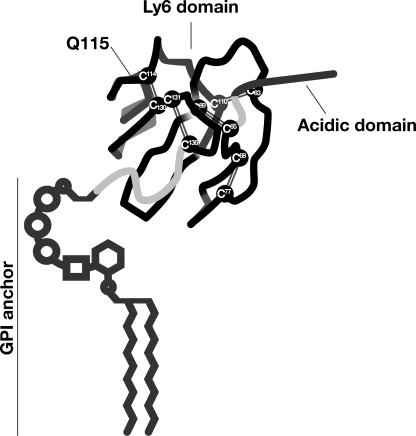
The discovery of the role of GPIHBP1 in lipolysis prompted interest in defining which portions of GPIHBP1 are important for its function (e.g. for its ability to bind LPL). Mature GPIHBP1 is a relatively short protein with only two noteworthy structural domains (Fig. 1). First, the amino terminus of the protein contains a strongly acidic domain (amino acids 24–48 in the mouse sequence) with a large number of aspartates and glutamates (2). This negatively charged domain is absolutely critical for binding LPL, a protein that contains two well characterized positively charged “heparin-binding domains” (3–5). Second, GPIHBP1 contains a cysteine-rich Ly6 domain (amino acids 63–135 in the mouse sequence). The function of the Ly6 domain in LPL binding is less clearly defined.
FIGURE 1.
Schematic of human GPIHBP1, showing the location of the acidic domain and the 10 highly conserved cysteines of the Ly6 domain. The location of Gln115 is also shown; a Q115P mutation was identified in association with chylomicronemia in a young man (15). This figure is modified, with permission, from a figure published in Ref. 22.
The Ly6 domain is an ancient motif containing either 8 or 10 cysteines that are organized in a highly characteristic spacing pattern (6, 7). Mammalian genomes contain ∼25–30 Ly6 proteins, of which the best studied are urokinase-type plasminogen activator receptor and CD59. In those proteins, as well as in other Ly6 proteins, crystallographic studies have shown that each of the cysteines is involved in a disulfide bond, producing a three-fingered structural motif (8–10).
In the case of urokinase-type plasminogen activator receptor and CD59, the Ly6 motif is crucial for ligand binding (8–13). But in the case of GPIHBP1, one could argue that this domain might be dispensable, simply because it is plausible that the acidic domain would be sufficient for binding the positively charged domains within LPL. On the other hand, other considerations suggest that the GPIHBP1 Ly6 domain might actually be important for GPIHBP1 function. First, the cysteines that define the Ly6 domain are perfectly conserved in the GPIHBP1 of every mammalian species from platypus to human, suggesting that the three-fingered structure of this domain is important (14). Second, Beigneux et al. (15) found that a missense mutation adjacent to a conserved cysteine (Q115P) impaired the ability of GPIHBP1 to bind LPL. The latter observation led us to entertain the possibility that the Ly6 domain could be functionally important in LPL binding.
To explore the functional relevance of the GPIHBP1 Ly6 domain, we decided to mutate the highly conserved cysteines in GPIHBP1 (Fig. 1), because those mutations would be predicted to disrupt the structure of the three-fingered motif. For other Ly6 proteins, mutation of conserved cysteines appears to impair protein function. Mutation of several (but not all) of the Ly6 cysteines in CD59 reduces the ability of that molecule to inhibit complement-mediated membrane attack (13). Also, missense mutations involving cysteines were found in a secreted Ly6 protein, SLURP1, in association with a recessive palmoplantar keratoderma (16). Unfortunately, the function and the binding partner for SLURP1 are unknown, so no one has been able to use biochemical assays to rigorously assess the impact of cysteine mutations on protein function. In contrast, GPIHBP1 has a well established function—binding LPL, and a cell-based system for assessing LPL binding has been developed (1, 15, 17).
In the current study, we sought to assess the impact of Ly6 cysteine mutations on the LPL binding capacity of GPIHBP1. But as we embarked on these studies, we worried that the cell-based assay system might be inadequate, for the simple reason that cysteine mutations sometimes interfere with the ability of proteins to reach the cell surface (13, 18). Accordingly, we also developed a cell-free, monoclonal antibody-based assay system for assessing the ability of GPIHBP1 to bind LPL. Both the cell-based binding system and the cell-free system were used to test the possibility that the Ly6 domain of GPIHBP1, like the acidic domain, is critical for binding LPL.
EXPERIMENTAL PROCEDURES
GPIHBP1 Constructs and Cell Transfections
Expression vectors for S-protein-tagged versions of mouse and human GPIHBP1 have been described previously (1). Constructs for soluble untagged and S-protein-tagged mouse GPIHBP1 proteins were created with the QuikChange site-directed mutagenesis kit (Stratagene) by truncating the protein (i.e. by replacing the GPI anchoring signal with a stop codon, G198X). Another soluble S-protein-tagged mouse GPIHBP1 construct was generated by introducing a premature stop codon at residue 136 (N136X). Also, site-directed mutagenesis was used to create human and mouse GPIHBP1 constructs in which specific cysteines within the Ly6 domain were changed to alanine and also an S-protein-tagged mouse GPIHBP1 construct in which the sole N-linked glycosylation site was replaced with glutamine (N76Q). We also performed studies with mutant GPIHBP1 proteins in which all of the aspartates and glutamates between residues 24 and 33 or residues 38 and 48 were replaced with alanine (17) and a mutant GPIHBP1 protein containing a Q114P mutation (15). The integrity of all constructs was verified by DNA sequencing. To express the GPIHBP1 proteins, 1–5 × 106 CHO pgsA-745 cells (19) were electroporated with 2–5 μg of plasmid DNA with the nucleofector II apparatus (Lonza) and the Cell Line T Nucleofector kit (Lonza).
Production of LPL for Binding Assays
A stable CHO cell line expressing a V5-tagged human LPL was obtained from Dr. Mark Doolittle (University of California, Los Angeles) and then adapted for growth in 95% serum-free/protein-free medium (CHO III from Gibco). Conditioned medium was concentrated 10-fold with Amicon Ultra 30,000 centrifugal filters (Millipore). The presence of LPL in the conditioned media was assessed by Western blotting with a mouse monoclonal antibody against the V5 tag (1:200; Invitrogen).
Production of Soluble Mouse GPIHBP1 for LPL Binding Studies
CHO pgsA-745 cells were electroporated with 5 μg of plasmid DNA and grown in 6-well plates in serum-free medium. Cell culture medium was collected 3 days later and concentrated 20-fold with Amicon Ultra 10,000 centrifugal filter units (Millipore). The presence of soluble GPIHBP1 in the conditioned medium was assessed by Western blotting with a rabbit polyclonal antibody against mouse GPIHBP1 (1:1000; Novus).
Development of a Rat Monoclonal Antibody against Mouse GPIHBP1 and a Mouse Monoclonal Antibody against Human GPIHBP1
Mouse and human GPIHBP1 proteins expressed in Escherichia coli were used to immunize rats or mice, respectively. Splenocytes from immunized animals were fused with myeloma cells, and stable hybridomas were selected. A mouse monoclonal antibody, clone 14, against human GPIHBP1 bound to the human GPIHBP1 expressed by CHO cells that had been transfected with a human GPIHBP1 expression vector, as judged by Western blotting. A rat monoclonal antibody against mouse GPIHBP1, clone 11A12, was evaluated for its ability to bind to both wild-type and truncated GPIHBP1 constructs (see “Results”). Hybridomas were adapted to serum/protein-free medium, and immunoglobulins in the medium were purified on a protein G-agarose column (Roche Applied Science). The purity of immunoglobulins was documented with Coomassie Blue-stained SDS-polyacrylamide gels.
Testing the Ability of Soluble GPIHBP1 to Bind LPL
We tested the ability of LPL to bind to soluble GPIHBP1 proteins that had been immobilized on monoclonal antibody 11A12-coated agarose beads. First, antibody 11A12 was conjugated to beaded agarose with the Aminolink Plus kit (Pierce). Next, 50 μl of concentrated medium containing soluble GPIHBP1 and 20 μl of concentrated medium containing V5-tagged human LPL were incubated overnight with 40 μl of antibody 11A12-coated agarose beads in 200 μl of PBS containing 0.2% Nonidet P-40. The beads were then washed three times in PBS containing 0.2% Nonidet P-40. Finally, the bound GPIHBP1 (along with any LPL that had been bound by GPIHBP1) was eluted from the beads with 200 μl of 0.1 m glycine, pH 2.5 (this step was repeated three times). Centrifugation steps were performed at 500 × g for 2 min. The amounts of GPIHBP1 and LPL in each fraction were assessed by Western blotting.
Binding of Human LPL to GPIHBP1-expressing CHO cells
5 × 106 CHO pgsA-745 cells were electroporated with 5 μg of plasmid DNA and seeded in triplicate wells of a 24-well tissue culture plate. 24 h after the electroporation, the cells were incubated for 2 h at 4 °C with V5-tagged human LPL (400 μl/well). In some wells, heparin was added to the incubation medium (final concentration, 500 units/ml). At the end of the incubation period, the cells were washed six times in ice-cold PBS containing 1 mm MgCl2 and 1 mm CaCl2, and cell extracts were collected in radioimmuno precipitation assay buffer (PBS containing 1% Triton X-100, 0.5% deoxycholate, and 0.02% SDS) along with complete mini EDTA-free protease inhibitors (Roche Applied Science). The amounts of LPL and GPIHBP1 in cell extracts were assessed by Western blotting.
Western Blot Studies
Proteins were size-fractionated on 12% Bis-Tris SDS-polyacrylamide gels and then transferred to nitrocellulose membranes for Western blotting. The antibody dilutions for Western blots were 1:1000 for a goat polyclonal antibody against the S-protein tag (Abcam); 1:1000 for a rabbit polyclonal antibody against the acidic domain of mouse GPIHBP1 (17); 1:200 for a mouse monoclonal antibody against the V5 tag (Invitrogen); 1:500 for a rabbit polyclonal antibody against β-actin (Abcam); 1:5000 for an IRdye800-conjugated donkey anti-goat IgG (Li-Cor); 1:500 for an IRdye800-conjugated donkey anti-mouse IgG (Li-Cor); and 1:2000 for an IRdye680-conjugated donkey anti-rabbit IgG (Li-Cor). Antibody binding was detected with an Odyssey infrared scanner (Li-Cor).
Assessing Amounts of GPIHBP1 Proteins at the Surface of Transfected Cells
To determine whether GPIHBP1 proteins reached the cell surface, we tested whether GPIHBP1 could be released into the medium with a phosphatidylinositol-specific phospholipase C (PIPLC) (1). CHO pgsA-745 cells (1 × 106 cells) were electroporated with 2 μg of plasmid DNA and seeded in duplicate wells of a 24-well plate. 24 h after the electroporation, the cells were incubated with PIPLC (5 units/ml) in serum-free medium for 1 h at 37 °C. At the end of the incubation, the medium was harvested, and cell extracts were collected in radioimmuno precipitation assay buffer containing complete mini EDTA-free protease inhibitors (Roche Applied Science).
In other experiments, we assessed the amount of GPIHBP1 expression on the surface of cells compared with the total amount of GPIHBP1 within cells. CHO pgsA-745 cells (2 × 106 cells) were electroporated with 4 μg of plasmid and then seeded in four wells of a 24-well plate. 24 h after the electroporation, the cells were incubated with a goat polyclonal antibody against the S-protein tag (1:400 for 2 h at 4 °C) in ice-cold PBS containing 1 mm MgCl2 and 1 mm CaCl2 (PBS/Mg/Ca). At the end of the incubation period, the cells were washed six times in ice-cold PBS/Mg/Ca, and the cell extracts were collected in radioimmuno precipitation assay buffer containing complete mini EDTA-free protease inhibitors (Roche Applied Science). To judge the amount of GPIHBP1 on the cell surface, we performed Western blots on cell extracts with an IRdye680-conjugated donkey anti-goat IgG (Li-Cor; 1:800). To assess the total amount of GPIHBP1 in cells, we simultaneously performed a Western blot with the rat monoclonal antibody 11A12 (1:100) or the mouse monoclonal antibody (clone 14) (1:100). The binding of antibodies was detected with an IRdye800-conjugated donkey anti-rat IgG (Li-Cor, 1:2000) or donkey anti-mouse IgG (Li-Cor; 1:2000). The intensity of each band was quantified with an Odyssey infrared scanner (Li-Cor). The ratio of GPIHBP1 on the cell surface (680-nm channel) compared with total GPIHBP1 (800-nm channel) was determined for each GPIHBP1 construct and expressed as percentage of the ratio observed with wild-type GPIHBP1.
Assessing the Accessibility of the Acidic Domain in Mutant GPIHBP1 Proteins
To assess the accessibility of the acidic domain in mutant GPIHBP1 proteins, GPIHBP1-expressing CHO pgsA-745 cells were incubated with a rabbit polyclonal antibody against the acidic domain of mouse GPIHBP1 (17) (1:400 for 2 h at 4 °C) in ice-cold PBS/Mg/Ca. After washing the cells, the amount of rabbit IgG bound to the cell surface was determined by Western blotting with an IRdye680-conjugated donkey anti-rabbit IgG (Li-Cor; 1:800). To assess the total amount of GPIHBP1 in cells, we used the goat polyclonal antibody against the S-protein tag (Abcam; 1:1000), followed by an IRdye800-conjugated donkey anti-goat IgG (Li-Cor; 1:5000). Western blots were performed with the Odyssey infrared scanner (Li-Cor).
Immunofluorescence Microscopy
CHO pgsA-745 cells (1 × 106 cells) were electroporated with 2 μg of plasmid and plated on coverslips in 24-well plates. The cells were fixed in 3% paraformaldehyde, blocked with blocking buffer (PBS/Mg/Ca containing 10% donkey serum), and incubated with a fluorescein isothiocyanate-conjugated goat antiserum against the S-protein tag (Abcam; 1:400). In some experiments, the cells were permeabilized with 0.2% Triton X-100. After washing, the cells were stained with 4′,6-diamidino-2-phenylindole to visualize DNA. The images were obtained with an Axiovert 200M microscope equipped with a 40× objective, an AxioCam MRm, and an ApoTome and were processed with the AxioVision 4.2 software (all from Zeiss).
RESULTS
Cell Surface Expression of Mutant GPIHBP1 Proteins Containing Cysteine-to-Alanine Substitutions within the Ly6 Domain
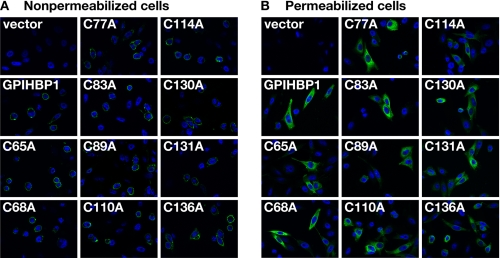
To assess the impact of specific cysteine mutations on the trafficking of GPIHBP1 to the cell surface, CHO pgsA-745 cells were transiently transfected with wild-type and mutant human GPIHBP1 constructs, and levels of GPIHBP1 expression were assessed by immunohistochemistry in nonpermeabilized cells (Fig. 2A) and cells that had been permeabilized with 0.2% Triton X-100 (Fig. 2B). Each of the GPIHBP1 mutants (C65A, C68A, C77A, C83A, C89A, C110A, C114A, C130A, C131A, and C136A) was easily detected at the surface of nonpermeabilized cells (Fig. 2A).
FIGURE 2.
Mutant GPIHBP1 proteins with cysteine-to-alanine mutations within the Ly6 domain reach the cell surface. CHO pgsA-745 cells were electroporated with an empty vector, a construct encoding an S-protein-tagged human GPIHBP1, or mutant GPIHBP1 constructs in which specific cysteines within the Ly6 domain were replaced with alanine. The presence of GPIHBP1 in nonpermeabilized (A) and permeabilized (B) cells was assessed by immunofluorescence microscopy with a goat antiserum against the S-protein tag (green). The cell nuclei were visualized with 4′,6-diamidino-2-phenylindole (blue).
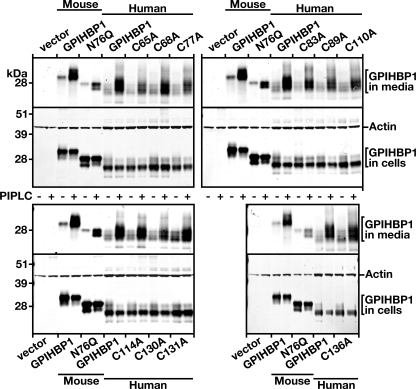
We also assessed the ability of PIPLC to release wild-type and mutant GPIHBP1 proteins from the cell surface. For these experiments, we transfected CHO pgsA-745 cells with wild-type and mutant GPIHBP1 constructs or mouse wild-type and nonglycosylated (N76Q) GPIHBP1 constructs. After 24 h, the cells were treated with PIPLC, and the amount of GPIHBP1 released into the medium was assessed by Western blotting. Nonglycosylated GPIHBP1 is retained in the endoplasmic reticulum (20); thus, it was not surprising that the amount of GPIHBP1 released into the medium by PIPLC was lower with the N76Q mutant than with wild-type GPIHBP1 (Fig. 3). Significant amounts of the cysteine mutants could be released from the surface of cells with PIPLC, although the absolute amount released tended to be slightly lower than the amount released from cells expressing wild-type GPIHBP1 (Fig. 3).
FIGURE 3.
Mutant GPIHBP1 proteins with cysteine-to-alanine mutations within the Ly6 domain can be released from the cell surface with a PIPLC. CHO pgsA-745 cells were electroporated with expression vectors encoding wild-type mouse GPIHBP1, a mutant mouse GPIHBP1 (N76Q) that eliminates the sole N-linked glycosylation site, wild-type human GPIHBP1, and mutant human GPIHBP1 proteins with specific cysteine-to-alanine mutations in the Ly6 domain. All of the GPIHBP1 constructs contained an S-protein tag. Amounts of GPIHBP1 in the medium were assessed by Western blotting, both in untreated cells and in cells that had been treated with PIPLC (5 units/ml for 1 h at 37 °C).
The amount of GPIHBP1 at the cell surface, relative to the amount in the cells, was also assessed by Western blotting. CHO pgsA-745 cells were transiently transfected with S-protein-tagged wild-type and mutant human GPIHBP1 constructs and S-protein-tagged wild-type and nonglycosylated mouse GPIHBP1 (N76Q) constructs. After incubating the cells with an antibody against the S-protein tag, cell extracts were prepared. The ratio of cell surface expression of GPIHBP1 (as judged by binding of the S-protein antibody) to the total cellular GPIHBP1 expression (as judged by antibody against GPIHBP1) was determined for each GPIHBP1 protein with Western blots (see “Experimental Procedures”). As expected, the amount of the nonglycosylated GPIHBP1 at the cell surface was low, only 31% of the amount of wild-type GPIHBP1 (Fig. 4). The levels of the cysteine mutants at the cell surface also tended to be somewhat lower, ranging from 67 to 87% of wild-type GPIHBP1 (Fig. 4).
FIGURE 4.
Assessing relative amounts of GPIHBP1 on the surface of cells with an antiserum against the S-protein tag. CHO pgsA-745 cells were electroporated with vectors encoding wild-type mouse GPIHBP1, a mutant mouse GPIHBP1 (N76Q) in which the sole N-linked glycosylation site was eliminated, wild-type human GPIHBP1, and mutant human GPIHBP1 proteins with specific cysteine-to-alanine mutations in the Ly6 domain. All of the GPIHBP1 constructs contained an S-protein tag. 24 h after the electroporation, the medium was removed, and the cells were incubated for 2 h at 4 °C with a goat antibody against the S-protein tag. After removing the antibody, the cells were washed six times with ice-cold PBS. The cell extracts were prepared, and Western blots were performed with an anti-goat secondary antibody and either a rat monoclonal against mouse GPIHBP1 (in the case of cells transfected with mouse GPIHBP1 constructs) or a mouse monoclonal against human GPIHBP1 (in the case of cells transfected with human GPIHBP1 constructs). Band intensities were quantified with an Odyssey infrared scanner (Li-Cor). The signal corresponding to the goat anti-S-protein tag IgG was normalized to the binding of the rat antibody against mouse GPIHBP1 (for experiments with mouse GPIHBP1) or the monoclonal antibody against human GPIHBP1 (for experiments with human GPIHBP1) and expressed relative to wild-type control (set at 100%).
Binding of LPL to Mutant GPIHBP1 Proteins with Cysteine-to-Alanine Substitutions within the Ly6 Domain
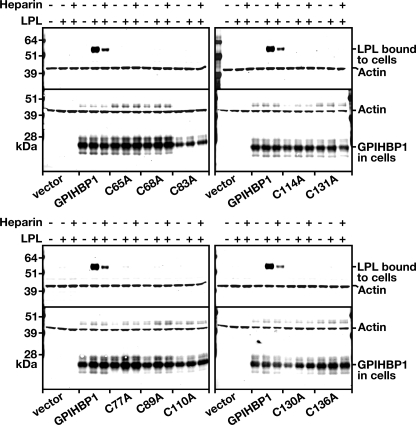
The fact that significant amounts of the cysteine mutants were expressed at the cell surface allowed us to test their ability to bind LPL. V5-tagged human LPL was incubated with CHO pgsA-745 cells expressing either wild-type or mutant GPIHBP1, and the amount of LPL bound to the cells was assessed by Western blotting (Fig. 5). Cells expressing wild-type GPIHBP1 bound LPL avidly, and this binding was largely blocked by heparin (Fig. 5). In contrast, cells expressing the cysteine mutants bound little if any LPL (Fig. 5).
FIGURE 5.
Reduced binding of human LPL to cells expressing mutant versions of GPIHBP1 with specific cysteine-to-alanine mutations in the Ly6 domain. CHO pgsA-745 cells were electroporated with an S-protein-tagged wild-type GPIHBP1 construct or mutant constructs with specific cysteine-to-alanine mutations. 24 h after the electroporation, the cells were incubated with V5-tagged human LPL in the presence or absence of heparin (500 units/ml). After washing the cells six times, the cell extracts were prepared, and the level of LPL bound to the cells was assessed by Western blotting with a V5 tag-specific antibody. Simultaneously, the level of GPIHBP1 in cell extracts was assessed by Western blotting with an antibody against the S-protein tag. Actin was used as a loading control.
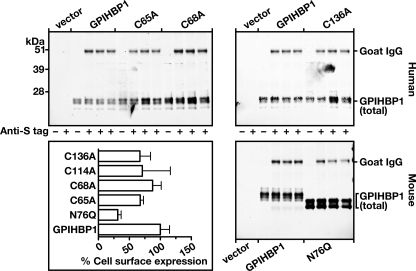
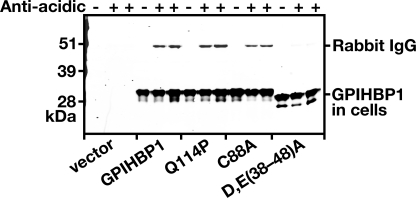
We considered the possibility that the absence of LPL binding to the cysteine mutants might be secondary to major conformational changes in GPIHBP1 that would limit accessibility to the acidic domain of GPIHBP1 (i.e. that the altered conformation of the Ly6 domain would cause the acidic domain to be buried). If this were the case, one might reasonably expect to find impaired binding of an anti-acidic domain antiserum to the cysteine mutants. We previously generated a rabbit antiserum against the acidic domain of GPIHBP1 and showed that it blocked LPL binding to GPIHBP1 (17). In the current study, we tested the ability of that antiserum to bind to wild-type and mutant GPIHBP1 molecules on the surface of live CHO cells. As expected, the rabbit antiserum did not bind to cells that had been transfected with empty vector or to cells expressing a mutant GPIHBP1 in which all of the aspartates and glutamates between residues 38 and 48 were replaced with alanine (D/E(38–48)A) (Fig. 6). However, the antiserum bound equally well to cells expressing wild-type GPIHBP1, GPIHBP1-Q114P, and GPIHBP1-C88A (Fig. 6), suggesting that the acidic domains in those mutants remain accessible to large proteins such as IgG.
FIGURE 6.
Mutations in mouse GPIHBP1 that interfere with LPL binding (Q114P and C88A) do not appear to interfere with the accessibility of antibodies against the acidic domain of GPIHBP1. CHO pgsA-745 cells were electroporated with vectors encoding wild-type mouse GPIHBP1, GPIHBP1 with a Q114P mutation, GPIHBP1 with a C88A mutation, and GPIHBP1 in which all of the aspartates and glutamates between amino acids 38 and 48 were replaced with alanines (D/E(38–48)A). All of the GPIHBP1 constructs contained an S-protein tag. The next day the medium was removed, and the cells were incubated for 2 h at 4 °C with a rabbit polyclonal antibody raised against a peptide spanning the entire acidic domain of mouse GPIHBP1 (17). After removing the antibody, the cells were washed six times with ice-cold PBS. The cell extracts were prepared for Western blots with a goat antibody against the S-protein tag (to detect total GPIHBP1 in cells) and a donkey anti-rabbit secondary antibody (to detect binding of the anti-acidic domain rabbit antibody).
A Rat Monoclonal Antibody against Mouse GPIHBP1
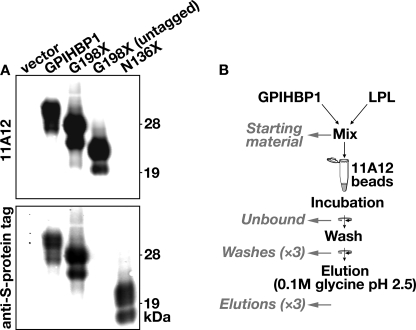
We generated a new rat monoclonal antibody, 11A12, against mouse GPIHBP1 (see “Experimental Procedures”). Antibody 11A12 bound to wild-type GPIHBP1 and to a soluble GPIHBP1 truncated before the GPI-anchoring motif (G198X); however, the antibody did not bind to a soluble GPIHBP1 protein truncated after the last cysteine of the Ly6 domain (N136X) (Fig. 7A). These data suggest that the epitope for antibody 11A12 resides within the carboxyl-terminal portion of GPIHBP1 between residues 136 and 198. Antibody 11A12 did not bind to human GPIHBP1 (not shown).
FIGURE 7.
Schematic of a new method to assess the binding of LPL to soluble mouse GPIHBP1 captured on monoclonal antibody 11A12-coated agarose beads. A, Western blots to characterize antibody 11A12, a new rat monoclonal antibody against mouse GPIHBP1. The following GPIHBP1 proteins were examined: an S-protein-tagged GPIHBP1, S-protein-tagged and untagged “soluble” GPIHBP1 (G198X, truncated immediately before the GPI anchoring site), and S-protein-tagged “truncated” GPIHBP1 (N136X, truncated immediately after the Ly6 domain). Western blots were performed with antibody 11A12 and a goat antibody against an S-protein tag (located at the amino terminus of GPIHBP1 proteins); the binding of antibody 11A12 and the goat anti-S protein tag antibody was detected with donkey anti-rat (top) and anti-goat (bottom) secondary antibodies, respectively. B, a new monoclonal antibody-based assay to assess the binding of LPL to GPIHBP1. Agarose beads were coated with monoclonal antibody 11A12 as described under “Experimental Procedures.” The beads were then incubated with S-protein-tagged soluble GPIHBP1 and V5-tagged human LPL (from concentrated cell culture medium). The beads were washed with the incubation buffer, and the GPIHBP1 was eluted from the beads with 0.1 m glycine, pH 2.5. Western blots were performed to detect soluble GPIHBP1 and LPL in the following fractions: the samples loaded onto the beads (Starting material), the Unbound fraction, three sequential washes of the beads, and three sequential elutions of the beads with 0.1 m glycine, pH 2.5.
A Monoclonal Antibody-based, Cell-free Assay to Assess LPL Binding to GPIHBP1
We expressed soluble versions of GPIHBP1 (proteins without a GPI anchor, truncated at residue 198). Soluble GPIHBP1 protein, along with LPL, were incubated with agarose beads coated with antibody 11A12 (Fig. 7B). After washing the beads three times with the incubation buffer (see “Experimental Procedures”), the soluble GPIHBP1 (along with any GPIHBP1-bound LPL) was eluted from the agarose beads with 0.1 m glycine, pH 2.5 (Fig. 7B). The amounts of GPIHBP1 and LPL in each fraction, at each step of the assay, were assessed by Western blotting (Fig. 7B).
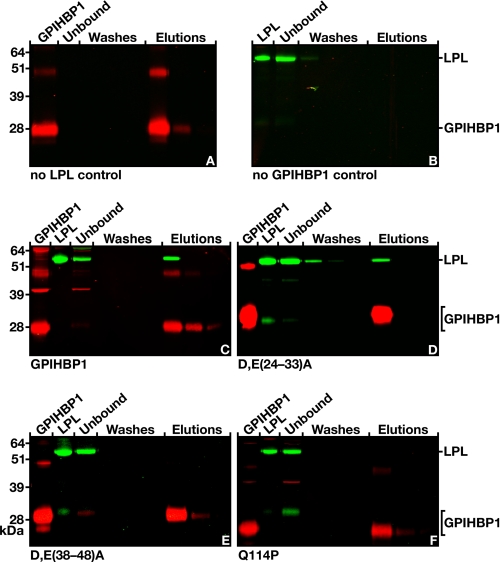
In preliminary studies, we found that the antibody 11A12-coated beads captured GPIHBP1 and that the soluble GPIHBP1 was readily eluted with 0.1 m glycine, pH 2.5 (Fig. 8A). Not surprisingly, no GPIHBP1 was captured when the beads were coated with control rat IgG (supplemental Fig. S1). The antibody 11A12-coated beads had no ability to bind to LPL in the absence of GPIHBP1 (Fig. 8B). However, LPL was captured by GPIHBP1 when both GPIHBP1 and LPL were incubated with the beads, and both GPIHBP1 and LPL were eluted from the beads with 0.1 m glycine, pH 2.5 (Fig. 8C).
FIGURE 8.
Testing the ability of human LPL to bind to soluble mouse GPIHBP1 that had been captured by antibody 11A12-coated agarose beads. A, binding of soluble GPIHBP1 alone (no LPL control) to antibody 11A12-coated beads. The soluble GPIHBP1 was eluted with 0.1 m glycine, pH 2.5. B, absence of LPL binding to antibody 11A12-coated beads (no GPIHBP1 control). C, binding of LPL to soluble wild-type GPIHBP1 that had been captured on antibody 11A12-coated beads; GPIHBP1 and LPL were both released with 0.1 m glycine, pH 2.5. D, binding of LPL to a mutant version of soluble GPIHBP1 (D/E(24–33)A) (17) that had been captured on antibody 11A12-coated beads. In D/E(24–33)A, all of the aspartates and glutamates between residues 24 and 33 were changed to alanine. E, absence of LPL binding to a mutant version of soluble GPIHBP1 (D/E(38–48)A) (17) that had been captured on antibody 11A12-coated beads. In D/E(38–48)A, all of the aspartates and glutamates between residues 38 and 48 were changed to alanine. F, absence of LPL binding to a mutant version of soluble GPIHBP1 (Q114P) (15) that had been captured on antibody 11A12-coated beads.
We next used this assay to evaluate LPL binding properties of mutant GPIHBP1 proteins. Changing all of the aspartates and glutamates between residues 38 and 48 to alanine (D/E(38–48)A) abolished the ability of GPIHBP1 to bind LPL in cell-based assays (17), whereas changing all of the aspartates and glutamates between residues 24 and 33 to alanine (D/E(24–33)A) had only small effects on LPL binding (17). The monoclonal antibody-based assay yielded results that were highly consistent with the cell-based assays reported earlier (17). When a soluble version of the D/E(24–33)A mutant and LPL were added to the beads, both proteins bound and could be eluted with 0.1 m glycine, pH 2.5 (Fig. 8D). On the other hand, when the soluble D/E(38–48)A mutant and LPL were incubated with the beads, all of the LPL appeared in the unbound fraction, and only the mutant GPIHBP1 was bound by the beads (Fig. 8E). We also tested the LPL binding properties of a soluble version of GPIHBP1 harboring a Q114P substitution. In cell-based binding assays, this mutation nearly abolishes LPL binding (15). Consistent with those results, we found that a secreted GPIHBP1 harboring a Q114P mutation could not bind LPL (i.e. GPIHBP1 was bound by the beads, but the LPL appeared in the unbound fraction) (Fig. 8F).
Using the Cell-free Assay System to Examine the Impact of Cysteine-to-Alanine Substitutions on LPL Binding
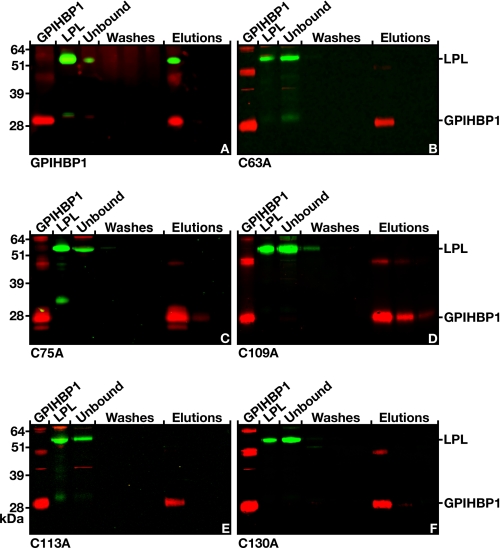
In cell-based LPL binding assays (Fig. 5), GPIHBP1 proteins harboring a cysteine-to-alanine mutation could not bind LPL. However, in view of the fact that at least some of the cysteine mutants reached the cell surface less efficiently than wild-type GPIHBP1 (Figs. 3 and 4), we considered it to be important to test the LPL binding properties of the cysteine mutants in the cell-free, monoclonal antibody-based assay (Fig. 9). In these experiments, the soluble version of wild-type GPIHBP1 bound LPL (Fig. 9A), whereas soluble versions of GPIHBP1 with C63A, C75A, C109A, C113A, and C130A mutations did not bind LPL (Fig. 9, B–F). Similarly, soluble versions of GPIHBP1 harboring C66A, C81A, C88A, C129A, or C135A mutations were unable to bind LPL (supplemental Fig. S2).
FIGURE 9.
Testing the ability of human LPL to bind to cysteine mutant forms of GPIHBP1 captured by monoclonal antibody 11A12-coated agarose beads. A, Western blot showing the binding of LPL to wild-type soluble GPIHBP1 that had been immobilized on antibody 11A12-coated beads. GPIHBP1 and LPL were both released by 0.1 m glycine, pH 2.5. B–F, Western blots showing the absence of LPL binding to mutant soluble GPIHBP1 proteins harboring specific cysteine-to-alanine mutations within the Ly6 domain of GPIHBP1. Structural studies of three Ly6 proteins has revealed that the first cysteine forms a disulfide bond with the fifth cysteine, the second with the third, the fourth with the sixth, the seventh with the eight, and the ninth with the tenth (8–10). The five cysteine mutants (C63A, C75A, C109A, C113A, and C130A) shown here were predicted to interrupt five different disulfide bonds in GPIHBP1 (10). GPIHBP1 mutants corresponding to the opposite cysteine in the disulfide pair (C66A, C81A, C88A, C129A, and C135A) were also created and tested in the monoclonal antibody-based assay; none of these mutants bound LPL (see supplemental Fig. 2).
DISCUSSION
GPIHBP1 is an Ly6 protein of endothelial cells that binds LPL and is crucial for the lipolytic processing of triglyceride-rich lipoproteins (1). Earlier studies demonstrated that the acidic domain of GPIHBP1 is absolutely required for LPL binding (17). In the current study, we investigated the hypothesis that the structural integrity of the Ly6 domain of GPIHBP1 also might be required for LPL binding. Our approach was to assess the LPL binding properties of GPIHBP1 mutants harboring cysteine-to-alanine mutations in the Ly6 domain, substitutions that would be predicted to perturb the conserved three-fingered structure of that region. We found that LPL binding was abolished with all 10 of the cysteine mutants, a finding confirmed with two independent assays. In the first assay, we found that cells expressing cysteine mutants could not bind LPL when it was added to the incubation medium. The second assay relied on the development of a new rat monoclonal antibody, 11A12, that binds to the carboxyl-terminal region of mouse GPIHBP1. Antibody 11A12-coated agarose beads readily capture soluble wild-type GPIHBP1, which in turn binds LPL. However, when any of the 10 cysteines in the soluble GPIHBP1 were changed to alanine, the immobilized GPIHBP1 was incapable of binding LPL.
We feared that mutation of cysteines within the Ly6 domain of GPIHBP1 might prevent GPIHBP1 from trafficking to the cell surface, a result that would have made us completely reliant on the monoclonal antibody-based LPL binding system. Before embarking on our experiments, our fears appeared well founded; mutation of several of the cysteines within CD59 blocked trafficking of the molecule to the cell surface (13), and mutations of cysteines in the ligand-binding repeats of the LDL receptor have similar effects (18). However, as it turned out, mutations of cysteines in GPIHBP1 had only minor effects on cell surface expression of GPIHBP1. Under the experimental conditions that we used (CHO cells and GPIHBP1 expression vectors driven by a cytomegalovirus promoter), cell surface expression of the cysteine mutants was at most ∼33% lower than the wild-type proteins. Interestingly, the cysteine mutants reached the cell surface significantly better than a nonglycosylated GPIHBP1 mutant, which is known to be retained within the endoplasmic reticulum (20). From the perspective of the current study, the fact that the cysteine mutants reached the cell surface at a relatively high efficiency was a welcome surprise because it meant that we were able to document the LPL binding properties of the cysteine mutants with two completely independent assay systems.
The parsimonious interpretation of our findings is that the interaction between GPIHBP1 and LPL requires both the acidic domain and the Ly6 domain. However, one could conceivably argue that the acidic domain is all that is required for LPL binding and that the Ly6 mutations merely change the conformation of the protein, burying the acidic domain and limiting its accessibility to LPL. However, we think that this scenario is somewhat unlikely, for several reasons. First, the Ly6 domains of two Ly6 family members, urokinase-type plasminogen activator receptor and CD59, are known to participate in ligand binding, and the strong conservation of the Ly6 domain of GPIHBP1 supports the idea that it is functionally important. Second, because all 10 cysteine-to-alanine mutations blocked LPL binding, one would have to argue that all 10 of the mutations blocked accessibility of the acidic domain in a very similar fashion, and that seems unlikely to us. Third, the binding of an anti-acidic domain antiserum to a GPIHBP1-C88A mutant was normal, making it unlikely that the acidic domain is buried and inaccessible to LPL. Each of these considerations leads us to suspect that both the acidic domain and the Ly6 domain have essential roles in LPL binding. We speculate that the highly acidic domain in GPIHBP1 (which is always present but is poorly conserved in terms of its precise amino acid sequence) may represent a rather promiscuous “negatively charged bait” for attracting positively charged proteins and that the “lock and key” specificity for LPL binding may depend on the Ly6 domain.
If we are correct in our suggestion that two different domains in GPIHBP1 participate in LPL binding, then what about LPL? Does LPL have two GPIHBP1-binding domains? The principal heparin-binding domain of LPL involves several positively charged residues (Lys430, Arg432, Lys434, Lys440, and Lys441) and is required for LPL binding to negatively charged heparan sulfate proteoglycans (5). These very same amino acid residues are required for the binding of LPL to GPIHBP1 (17). Of course, these findings beg the question of which region of LPL interacts with the Ly6 domain of GPIHBP1. By analogy with the two-domain scenario for GPIHBP1, it is tempting to speculate that two GPIHBP1-binding domains exist in LPL and that the Ly6-binding domain still needs to be identified. Ultimately, crystallographic studies of GPIHBP1-LPL interactions will likely be required for definitive answers.
Several missense mutations in SLURP1 have been identified in association with Mal de Meleda, the recessive palmoplantar keratoderma (16). Interestingly, two of these mutations, C77R and C99Y, involve the seventh and tenth cysteine, respectively, of the Ly6 domain); another involves the introduction of a proline adjacent to a conserved cysteine (L98P) (16). The latter mutation was intriguing because we have already identified, in a human patient with severe chylomicronemia, a GPIHBP1 mutation involving the introduction of a proline adjacent to a cysteine (15). That mutation, Q115P, dramatically reduced the ability of GPIHBP1 to bind LPL. Our current findings, along with the Mal de Meleda experience (16), should prompt human geneticists to look for mutations in conserved cysteines in GPIHBP1 in human patients with chylomicronemia. Indeed, while we were preparing this manuscript, Dallinga-Thie et al. (21) reported (at the 2009 Arteriosclerosis, Thrombosis, and Vascular Biology meeting) a C65Y mutation in the Ly6 domain of GPIHBP1 in a young boy with severe chylomicronemia. We suspect that human geneticists will identify additional GPIHBP1 cysteine mutations in short order.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL094732 (to A. P. B.), P01 HL090553 (to S. G. Y.), and R01 HL087228 (to S. G. Y.). This work was also supported by Scientist Development Award from the American Heart Association National Office (to A. P. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- GPIHBP1
- glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1
- GPI
- glycosylphosphatidylinositol
- LPL
- lipoprotein lipase
- PIPLC
- phosphatidylinositol-specific phospholipase C
- CHO
- Chinese hamster ovary
- Ly6
- lymphocyte antigen 6
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Beigneux A. P., Davies B. S., Gin P., Weinstein M. M., Farber E., Qiao X., Peale P., Bunting S., Walzem R. L., Wong J. S., Blaner W. S., Ding Z. M., Melford K., Wongsiriroj N., Shu X., de Sauvage F., Ryan R. O., Fong L. G., Bensadoun A., Young S. G. (2007) Cell Metab. 5, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ioka R. X., Kang M. J., Kamiyama S., Kim D. H., Magoori K., Kamataki A., Ito Y., Takei Y. A., Sasaki M., Suzuki T., Sasano H., Takahashi S., Sakai J., Fujino T., Yamamoto T. T. (2003) J. Biol. Chem. 278, 7344–7349 [DOI] [PubMed] [Google Scholar]
- 3.Hata A., Ridinger D. N., Sutherland S., Emi M., Shuhua Z., Myers R. L., Ren K., Cheng T., Inoue I., Wilson D. E., Iverius P. H., Lalouel J. M. (1993) J. Biol. Chem. 268, 8447–8457 [PubMed] [Google Scholar]
- 4.Ma Y., Henderson H. E., Liu M. S., Zhang H., Forsythe I. J., Clarke-Lewis I., Hayden M. R., Brunzell J. D. (1994) J. Lipid Res. 35, 2049–2059 [PubMed] [Google Scholar]
- 5.Sendak R. A., Bensadoun A. (1998) J. Lipid Res. 39, 1310–1315 [PubMed] [Google Scholar]
- 6.Fry B. G., Wüster W., Kini R. M., Brusic V., Khan A., Venkataraman D., Rooney A. P. (2003) J. Mol. Evol. 57, 110–129 [DOI] [PubMed] [Google Scholar]
- 7.Mallya M., Campbell R. D., Aguado B. (2006) Protein Sci. 15, 2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Llinas P., Le Du M. H., Gårdsvoll H., Dano K., Ploug M., Gilquin B., Stura E. A., Ménez A. (2005) EMBO J. 24, 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieffer B., Driscoll P. C., Campbell I. D., Willis A. C., van der Merwe P. A., Davis S. J. (1994) Biochemistry 33, 4471–4482 [PubMed] [Google Scholar]
- 10.Huang Y., Fedarovich A., Tomlinson S., Davies C. (2007) Acta Crystallogr. D. Biol. Crystallogr. 63, 714–721 [DOI] [PubMed] [Google Scholar]
- 11.Ploug M. (1998) Biochemistry 37, 16494–16505 [DOI] [PubMed] [Google Scholar]
- 12.Gårdsvoll H., Danø K., Ploug M. (1999) J. Biol. Chem. 274, 37995–38003 [DOI] [PubMed] [Google Scholar]
- 13.Petranka J., Zhao J., Norris J., Tweedy N. B., Ware R. E., Sims P. J., Rosse W. F. (1996) Blood Cells Mol. Dis. 22, 281–296 [DOI] [PubMed] [Google Scholar]
- 14.Young S. G., Davies B. S., Fong L. G., Gin P., Weinstein M. M., Bensadoun A., Beigneux A. P. (2007) Curr. Opin. Lipidol. 18, 389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beigneux A. P., Franssen R., Bensadoun A., Gin P., Melford K., Peter J., Walzem R. L., Weinstein M. M., Davies B. S., Kuivenhoven J. A., Kastelein J. J., Fong L. G., Dallinga-Thie G. M., Young S. G. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 956–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Favre B., Plantard L., Aeschbach L., Brakch N., Christen-Zaech S., de Viragh P. A., Sergeant A., Huber M., Hohl D. (2007) J. Invest. Dermatol. 127, 301–308 [DOI] [PubMed] [Google Scholar]
- 17.Gin P., Yin L., Davies B. S., Weinstein M. M., Ryan R. O., Bensadoun A., Fong L. G., Young S. G., Beigneux A. P. (2008) J. Biol. Chem. 283, 29554–29562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein J. L., Brown M. S., Anderson R. G., Russell D. W., Schneider W. J. (1985) Annu. Rev. Cell Biol. 1, 1–39 [DOI] [PubMed] [Google Scholar]
- 19.Esko J. D., Stewart T. E., Taylor W. H. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beigneux A. P., Gin P., Davies B. S., Weinstein M. M., Bensadoun A., Ryan R. O., Fong L. G., Young S. G. (2008) J. Lipid Res. 49, 1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dallinga-Thie G. M., Beigneux A. P., Franssen R., Hertecant J., Gin P., Weinstein M. M., Kastelein J. J., Bensadoun A., Fong L. G., Young S. G. (2009) Arterioscler. Thromb. Vasc. Biol. 29, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beigneux A. P., Weinstein M. M., Davies B. S., Gin P., Bensadoun A., Fong L. G., Young S. G. (2009) Curr. Opin. Lipidol. 20, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.