Abstract
Dominant-negative interference by glycine substitution mutations in the COL7A1 gene causes dominant dystrophic epidermolysis bullosa (DDEB), a skin fragility disorder with mechanically induced blistering. Although qualitative and quantitative alterations of the COL7A1 gene product, collagen VII, underlie DDEB, the lack of direct correlation between mutations and the clinical phenotype has rendered DDEB less amenable to therapeutic targeting. To delineate the molecular mechanisms of DDEB, we used recombinant expression of wild-type (WT) and mutant collagen VII, which contained a naturally occurring COL7A1 mutation, G1776R, G2006D, or G2015E, for characterization of the triple helical molecules. The mutants were co-expressed with WT in equal amounts and could form heterotrimeric hybrid triple helices, as demonstrated by affinity purification and mass spectrometry. The thermal stability of the mutant molecules was strongly decreased, as evident in their sensitivity to trypsin digestion. The helix-to-coil transition, Tm, of the mutant molecules was 31–34 °C, and of WT collagen VII 41 °C. Co-expression of WT with G1776R- or G2006D-collagen VII resulted in partial intracellular retention of the collagen, and mutant collagen VII had reduced ability to support cell adhesion. Intriguingly, controlled overexpression of WT collagen VII gradually improved the thermal stability of the collective of collagen VII molecules. Co-expression in a ratio of 90% WT:10% mutant increased the Tm to 41 °C for G1776R-collagen VII and to 39 °C for G2006D- and G2015E-collagen VII. Therefore, increasing the expression of WT collagen VII in the skin of patients with DDEB can be considered a valid therapeutic approach.
Mutations in the collagen VII gene, COL7A1, cause dystrophic epidermolysis bullosa (DEB),3 a heritable skin fragility disorder characterized by mechanically induced blistering of the skin and mucosa, and excessive scarring (1). DEB is classified into clinical subtypes with dominant or recessive inheritance (2), and so far more than 400 different COL7A1 mutations are known, which underlie a broad spectrum of clinical presentations.
Collagen VII is the major molecular constituent of anchoring fibrils in the skin. These centro-symmetrically banded fibrils extend from the epidermal basement membrane into the underlying dermal stroma and connect the epidermis to the dermis. Collagen VII is synthesized as three identical pro-α1(VII) polypeptide chains, which are hydroxylated and glycosylated in a coordinated manner and then fold into triple-helical procollagen VII in the endoplasmic reticulum (ER). The procollagen, which contains a central collagenous triple-helix flanked by two non-collagenous domains, NC-1 and NC-2, is secreted into the extracellular space, where the C-terminal NC-2 propeptide is proteolytically removed by bone morphogenetic protein-1 (3). Subsequently, mature collagen VII undergoes a multistep fibril polymerization process to form the anchoring fibrils (4).
The pathology in DDEB has been thought to result from negative interference of mutant pro-α1(VII) chains that are incorporated into the triple-helical monomers and affect folding and registration of normal polypeptides. Typically, substitution of a glycine within the collagenous domain by a larger amino acid residue causes imperfections and delays in triple-helix folding and increased post-translational modifications (5). These can have different consequences: 1) newly synthesized mutant pro-α(VII) chains or procollagen VII molecules do not pass the ER quality control and are retained in the ER or designated for ubiquitin-proteasome degradation (6), resulting in reduced amounts of collagen VII in the skin; 2) assembly into loosely folded collagen VII monomers, which are secreted, incorporated into anchoring fibrils, and perturb the fibril architecture and render them sensitive to tissue proteases; 3) a combination of the above. All variants lead to paucity of anchoring fibrils at the dermal-epidermal junction, impaired resistance of the skin to shearing forces, and to skin blistering as a clinical symptom.
Accessibility makes the skin an ideal organ for testing of molecular therapies. Development of causal treatments for DEB is urged by the severe impact of permanent skin fragility on the life of affected individuals. Therapeutic considerations for DDEB have included an array of approaches including oligonucleotides and oligoribonucleotides (7, 8). Intriguingly, findings in a mouse model for epidermolysis bullosa simplex (EBS), a skin fragility disorder associated with dominant keratin mutations, delivered first evidence that increasing the ratio of wild-type (WT) to mutated polypeptides may improve the phenotype (9). Furthermore, our recent investigation of the collagen VII hypomorphic mouse suggested that relatively small biological changes, e.g. moderately raised levels of collagen VII, can have substantial clinical effects (10). These observations encouraged us to test the possibility that controlled overexpression of normal collagen VII may have therapeutic potential for DDEB.
Here we used protein biochemical, mass spectrometry and cell biological in vitro analysis to show that mutant α1(VII) chains can fold with WT α1(VII) chains into hybrid triple helices and exert dominant-negative interference on the protein function. The resulting destabilization and partial intracellular accumulation of the mutant molecules can be diminished by controlled overexpression of WT collagen VII.
EXPERIMENTAL PROCEDURES
Clinical and Biological Phenotypes of COL7A1 Mutations
The naturally occurring COL7A1 mutations investigated here have been shown to be associated with DEB in different families with pedigrees consistent with dominant inheritance (11, 12). The clinical phenotype of the affected individuals encompassed skin blisters, scarring, milia, and/or nail dystrophy at trauma-exposed body sites. The biological and ultrastructural phenotype included dermal-epidermal tissue separation below the lamina densa of the basement membrane, paucity of anchoring fibrils, reduced amounts of collagen VII in the skin, and variable degrees of intracellular accumulation of collagen VII in keratinocytes in vitro (11, 12).
Generation and Expression of Mutant Collagen VII
Full-length collagen VII cDNA with a FLAG tag inserted after amino acid 23 (13) was subjected to site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) with primers generating a G1776R mutant (5′-GACCGGGGTCCCCCTAGGCTGGATGGCCGGAG-3), a G2006D mutant (5′-GCGGGCTGAAGGACGATCGTGGAGACCCTGG-3), and a G2015E mutant (5′-GGCCCTCAGGAGCCACCTGGTCTAGCGCTTGGGGAGAGGG-3). The WT and mutant collagen VII cDNA were expressed under control of the CMV promoter (pcDNA3.1Zeo(−), Invitrogen) or under control of the endogenous collagen VII promoter (genomic region −495 to +1, Ref. 14), which was introduced in the pcDNA3.1Zeo(−) directly upstream of the multiple cloning site. The endogenous promoter was used for expression of normal levels of collagen VII in immortalized collagen VII-deficient keratinocytes, whereas the CMV promoter was used for expression in HEK-293T. To be able to distinguish between WT and mutant molecules, overlap extension PCR (Expand High Fidelity PCR System, Invitrogen) was used to replace the FLAG tag in WT collagen VII cDNA with an 8× His tag. In addition, to improve detection with mass spectrometry, leucine residues at position 102 and 105 in mutant collagen VII were exchanged to valine by site-directed mutagenesis using the primers 5′-CAGAGTTCGGCGTGGATGCAGTTGGCTCTGGG-3′ and 5′-CCCAGAGCCAACTGCATCCACGCCGAACTCTG-3′. Thus, we generated His-tagged WT collagen VII α-chains and FLAG-tagged mutant chains containing one glycine substitution within the triple helical domain and two leucine-to-valine substitutions close to the N terminus within the NC-1 domain.
Cell Culture and Transient Transfection
HEK-293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum, 2 mm l-glutamine, 1 mm sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (all Invitrogen). 60% confluent HEK-293T were transfected with 10 μg of cDNA per 10-cm dish using GeneJammer (Stratagene). Immortalized collagen VII-deficient RDEB keratinocytes (15) were cultured in keratinocyte-SFM supplemented with recombinant epidermal growth factor and bovine pituitary extract (Invitrogen). For transfection, 90% confluent cultures were incubated with 10 μg of DNA per 10-cm dish in Lipofectamine 2000 (Invitrogen). Co-expression of WT and mutant collagen VII was achieved by mixing the respective plasmid cDNAs prior to incubation with the transfection reagent. 48 h prior to harvest, cells were washed with PBS, and medium was replaced with serum-free DMEM (HEK-293T) or keratinocyte-SFM supplemented with 50 μg/ml ascorbate.
Protein Chemical Characterization of Mutant Collagen VII
For characterization of recombinant collagen VII (rColVII) by limited proteolysis, culture medium of transiently transfected cells were collected on ice and clarified by centrifugation (200 × g, 5 min, 4 °C). Proteins were precipitated after slow addition of 250 mg/ml ammonium sulfate overnight at 4 °C and collected by centrifugation at 16,000 × g for 1 h at 4 °C. The pellet was resuspended in PBS and dialyzed against PBS overnight. After adjustment of the pH to 2.0 with 0.2 m HCl, samples were digested with 1 μg/μl pepsin (Sigma-Aldrich) in 0.1 m acetic acid for 2 h at 4 °C. The reaction was stopped by adjusting the pH to 8.0 with 1 m NaOH. Samples were then diluted 4-fold with PBS and exposed to increasing temperatures for 1 min, followed by digestion with 1.6 μg/μl trypsin (Serva) for 2 min at 20 °C. After stopping the reaction by addition of 2 μg/μl soybean trypsin inhibitor (Sigma), samples were incubated in blue sample buffer containing 0.8 m urea, 0.0001% bromphenol blue, 2% SDS, 5% glycerine, 0.01 m dithiothreitol, 0.1 m Tris-HCl, pH 6.8, for 5 min at 95 °C.
For quantification of collagen VII in the cell cultures, the medium was precipitated with 3 volumes of ethanol at −20 °C overnight. The pellets were collected by centrifugation (16,000 × g, 30 min, 4 °C), dissolved in blue sample buffer and incubated at 95 °C for 5 min. The cell layer was washed with ice-cold PBS and lysed with 1% Nonidet P-40 in a buffer containing 0.1 m NaCl, protease inhibitor set III (Calbiochem), 25 mm Tris-HCl at pH 7.4 for 30 min on ice. The lysates were cleared by centrifugation (16,000 × g, 10 min, 4 °C). After denaturation in blue sample buffer for 5 min at 95 °C, the proteins were separated on 7% SDS-PAGE for immunoblotting with the antibody NC2–10 (16). Semiquantitative densitometry was performed with Gel-pro Express 4.0 software (Media Cybernetics Inc.).
Immunofluorescence Staining
Transiently transfected cells on cover slips were fixed with 2% paraformaldehyde in PBS for 10 min and permeabilized with 0.4% Triton X-100 for 7 min at room temperature. After blocking for 20 min in 2% bovine serum albumin, cells were incubated with the primary antibody diluted in 0.2% bovine serum albumin in PBS overnight. Collagen VII was detected either with the NC2–10 antibody (16) or with an antibody recognizing epitopes along the entire collagen VII molecule (Calbiochem). After incubation with the respective secondary antibody, the cells were extensively washed and embedded in fluorescence mounting medium (Dako) and examined with a confocal laser-scanning microscope. Confocal images were obtained using a Zeiss LSM 510 instrument equipped with a C-Apochromat 63×/1.2W corr objective (Carl Zeiss). Images of 1-μm optical slice thickness and a region of interest size of 544 × 544 pixels (58.6 μm × 58.6 μm) were recorded.
Quantification of Mutant and WT Collagen VII
For quantification of WT and mutant collagen VII after transient transfections with different mixtures of plasmids, the recombinant tagged collagens were separated using M2-agarose minicolumns (Sigma-Aldrich), which bind the FLAG tag. Medium diluted 1:10 in PBS containing 0.5% Triton X-100 was incubated overnight at 4 °C on the column. Unbound His-tagged WT collagen VII in the flow-through was collected by centrifugation at 200 × g for 20 s at 4 °C. FLAG-tagged mutant collagen VII was eluted from the column with blue sample buffer at 95 °C for 2 min.
Analysis of Collagen VII Molecule Composition by Mass Spectrometry
HEK-293T cells were transiently transfected with WT collagen VII and G2006D-L102/105V-collagen VII using equal amounts of plasmid DNA. FLAG-Tag containing collagen VII hybrids were purified from the medium using M2-agarose (Sigma-Aldrich). For this, medium was clarified from cell debris by centrifugation (200 × g, 5 min, 4 °C) and directly applied to a M2-agarose column (3.5 ml M2-agarose/22 ml medium) by gravity flow. After extensive washing with TBS (50 mm Tris/HCl, pH 7.4; 150 mm NaCl), bound protein was eluted using 100 μg/ml FLAG-peptide (Sigma-Aldrich). Input, flow-through, and eluate of the column were precipitated with 100% ethanol at −20 °C overnight. After centrifugation (16,000 × g, 30 min, 4 °C), the pellet was dissolved in blue sample buffer, denatured at 95 °C for 5 min and separated by SDS-PAGE. Coomassie Blue-stained bands of ∼290 kDa were excised from the gel and subjected to in-gel tryptic digestion for mass spectrometry. For this, gel bands were destained with 30% acetonitrile, shrunk with 100% acetonitrile, and dried in a Vacuum Concentrator (Concentrator 5301, Eppendorf, Hamburg, Germany). Digests with trypsin (Promega) were performed overnight at 37 °C in 0.05 m NH4HCO3 (pH 8). About 0.1 μg of protease was used for one gel band. Peptides were extracted from the gel slices with 5% formic acid. LC-MS/MS analyses were performed on an Agilent 6460 triple quadrupole mass spectrometer (Agilent Technologies) coupled to a 1200 Agilent nanoflow system via a HPLC-Chip cube ESI interface. Peptides were separated on a HPLC-Chip with an analytical column of 75-μm i.d. and 40-mm length and a 40-nl trap column, both packed with Zorbax 300SB C-18 (5-μm particle size). Peptides were eluted with a linear acetonitrile gradient with 1%/min at a flow rate of 300 nl/min (starting with 3% acetonitrile). MRM (multiple reaction monitoring) analyses were performed for the wild-type tryptic peptide TEFGLDALGSGGDVIR und for the tryptic peptide TEFGVDAVGSGGDVIR (L102/105V) using dynamic MRM mode. Collision voltages were optimized using Agilent Optimizer software. Two transitions were used for each peptide (TEFGLDALGSGGDVIR : 803.9 (2+) to 760.4 (y8) and 803.9 (2+) to 1059.5 (y11); TEFGVDAVGSGGDVIR: 789.9 (2+) to 760.4 (y8) and 789.9 (2+) to 1045.5 (y11), both with 21 collision voltage). Two blank runs (injection 0.1% formic acid) were performed before each sample to rule out false positive detection due to memory effects.
Cell Adhesion Assay
To exclude tag-induced differences of cell behavior, in these assays both WT and mutant collagen VII carried a FLAG tag. Coverslips were coated with 20 μg/ml purified rColVII in 20 mm carbonate buffer, pH 9.3, or with 20 μg/ml bovine serum albumin as a negative control, overnight at 4 °C and blocked with 0.5 μg/ml bovine serum albumin for 1 h at 37 °C. Then 1 × 105 human fibroblasts in serum-free DMEM or 1.5 × 105 human keratinocytes in KGM without supplements were added and incubated for 2 h at 37 °C (17). After washing with PBS, the cells were fixed with 2% paraformaldehyde for 15 min at room temperature and stained with DAPI and rhodamine-labeled phalloidin (Chemicon) for 30 min. The number of attached cells was examined with an Axiovision fluorescence microscope system (Carl Zeiss), by counting DAPI-stained nuclei in four visual fields in each probe. Spreading of human keratinocytes was evaluated by cell shape.
RESULTS
Recombinant Expression of WT and Mutant Collagen VII
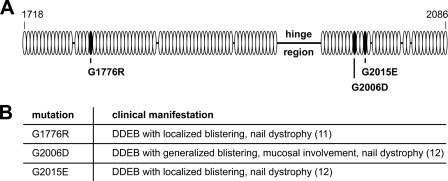
To investigate disease mechanisms of DDEB at the cellular and molecular level, WT and mutant collagen VII cDNA were expressed in HEK-293T cells or in immortalized collagen VII-deficient RDEB keratinocytes, which were homozygous for the COL7A1 mutation c.425A>G, which leads to a premature termination codon. For modeling of dominant-negative effects, we chose three naturally occurring glycine substitution mutations associated with DDEB and intracellular accumulation of collagen VII in keratinocytes in vitro. The mutation G1776R substituted a glycine within the P2 fragment of collagen VII by an arginine. The mutations G2006D and G2015E substituted glycine residues within the P1 fragment of collagen VII, in close vicinity of the hinge region, by an aspartic acid and a glutamic acid, respectively (Fig. 1). To mimic the heterozygous situation in the skin in DDEB, WT and mutated collagen VII were co-expressed using equal amounts of the respective plasmid cDNAs.
FIGURE 1.
Position of glycine substitution mutations G1776R, G2006D, and G2015E and their clinical phenotype. A, schematic representation of the mutations included in this study and their location within the triple helical domain of collagen VII. Shown is the stretch of amino acids 1718–2086 around the hinge region in the central triple helix. Open ovals, Gly-X-Y repeats; black ovals, Gly-X-Y with a glycine substitution mutation; black bar, interruptions of the Gly-X-Y repeat pattern. B, phenotype in patients carrying one COL7A1 allele with the respective mutation.
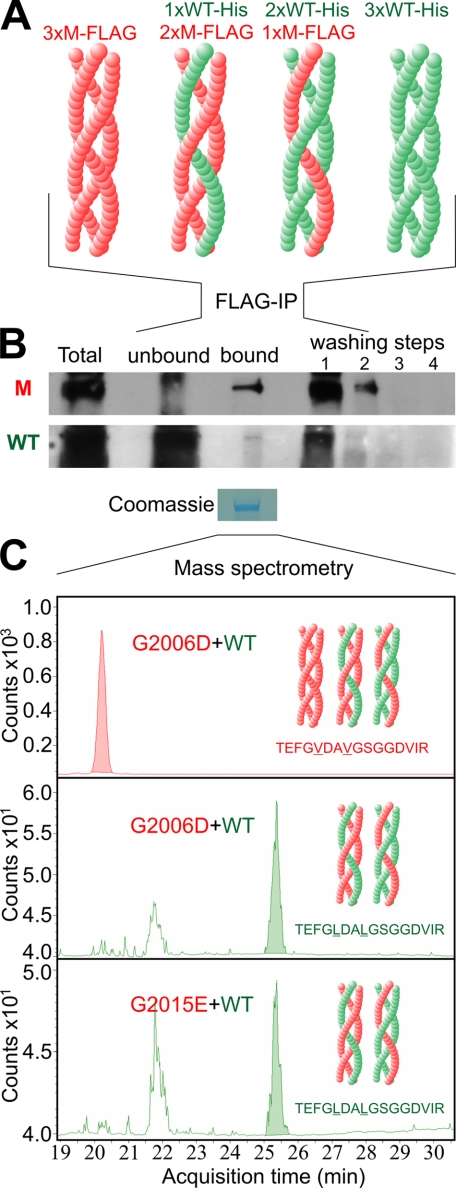
Co-expression of WT and Mutant Collagen VII Leads to Formation of Hybrid Triple Helices
Co-expression of WT and mutant collagen VII can give rise to triple helices composed of only WT α1 chains, two WT and one mutant α1 chain, one WT and 2 mutant α1 chains and only mutant α1 chains (Fig. 2A). However, it has remained unclear whether WT:mutant hybrid triple helices are actually built and secreted from the cell. To address this question, we used mass spectrometry to identify WT and mutated chains in affinity-purified recombinant collagen VII molecules. For this purpose, His-tagged WT collagen VII and mutant FLAG-tagged G1776R-L102/105V, G2006D-L102/105V, and G2015E-L102/105V collagen VII were co-expressed in HEK-293T cells, and FLAG tag-containing proteins in the culture medium were purified using an anti-FLAG column. Immunoblotting demonstrated FLAG-tagged collagen VII in the fraction eluted from the column with a FLAG peptide (Fig. 2B, upper panel). Importantly, this fraction also contained His-tagged collagen VII chains (Fig. 2B, lower panel). The unbound flow-through fraction contained predominantly His-tagged collagen VII. In the first washing step, both His- and FLAG-tagged molecules were found, suggesting that different hybrid molecules bind to the anti-FLAG column with different affinity. No collagen VII was present in the last washing steps before elution, ruling out unspecific carryover of His-tagged molecules. Control experiments using medium containing only His-tagged WT collagen VII or FLAG-tagged mutant collagen VII, mixed 1:1 prior to affinity absorption, show no His-containing collagen VII in the elution fraction (supplemental Fig. S1).
FIGURE 2.
Demonstration of WT:mutant hybrid triple helices using affinity purification and mass spectrometry. To demonstrate the formation of WT:mutant hybrid triple helices, His-tagged WT collagen VII and FLAG-tagged G1776R-L102/105V-, G2006D-L102/105V-, and G2015E-L102/105V collagen VII were co-expressed in HEK293T cells using equal amounts of the respective cDNAs. After 48 h, conditioned medium was collected and FLAG tag containing collagen VII triple helices were isolated by passing the medium over an anti-FLAG M2-agarose column and, after extensive washing, eluting the bound fraction with FLAG peptide. Presence of His-tagged WT-collagen VII and FLAG-tagged mutant collagen VII in the input, flow-through, eluate, and washing buffer was analyzed by immunoblotting using a FLAG or His tag-specific antibody. The eluate of the M2-agarose was also subjected to mass spectrometry with multiple reaction monitoring. A, schematic representation of the theoretically possible triple helices after co-expression of WT and mutant collagen VII. Red, FLAG-tagged mutant collagen VII (M-FLAG); green, His-tagged WT collagen VII (WT-His). B, immunoblot of FLAG-tagged G2006D-L102/105V collagen VII (upper panel) and His-tagged WT (lower panel) collagen VII after separation by M2-agarose column. Both FLAG-tagged mutant and His-tagged WT collagen VII are detectable in the fraction bound to the anti-FLAG M2-agarose, indicating the presence of hybrid triple helices. M, FLAG-tagged G2006D-L102/105V-collagen VII detected by anti-FLAG antibodies; WT, His-tagged WT-collagen VII detected by anti-His antibodies; Total, medium before incubation with M2-agarose; Unbound, medium after incubation with M2-agarose; Bound, fraction after elution of the column with FLAG peptide; washing steps 1–4, collagen VII in the subsequent washing steps before elution. C, collagen VII eluted from the M2-agarose was isolated by SDS-PAGE, and the Coomassie Blue-stained band was subjected to in-gel tryptic digest. Mass spectrometry with multiple reaction monitoring detected the presence of the peptide sequences TEFGVDAVGSGGDVIR and TEFGLDALGSGGDVIR representing mutant and WT collagen VII, respectively.
To corroborate the above findings, collagen VII in the elution fraction was separated by SDS-PAGE and subjected to mass spectrometry with multiple reaction monitoring. The analysis was focused on the peptide sequences TEFGVDAVGSGGDVIR and TEFGLDALGS-GGDVIR for mutant and WT collagen VII, respectively. This was necessary, since the high charge of the His and FLAG tags impaired their detection by mass spectrometry. Mass spectrometry revealed both peptides in the eluted fraction of the anti-FLAG-column in case of G2006D-collagen VII and G2015E-collagen VII (Fig. 2C), indicating the presence of hybrid collagen VII molecules containing both WT and mutant collagen α1-chains. However, this could not be shown for G1776R collagen VII (data not shown).
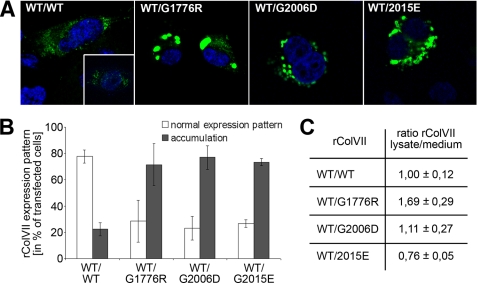
Intracellular Accumulation of Mutant Collagen VII
Immortalized RDEB keratinocytes, which are completely devoid of collagen VII (18), were transfected with the above constructs under control of the native collagen VII promoter, and the cells were analyzed by immunofluorescence staining. Newly synthesized WT collagen VII exhibited a diffuse staining pattern in the rough endoplasmic reticulum (Fig. 3A, first panel), similar to the pattern in normal human keratinocytes expressing collagen VII (Fig. 3A, first panel, inset). In contrast, cells co-expressing WT and G1776R-collagen VII showed significant intracellular accumulation in intensely stained vacuoles (Fig. 3A, second panel). Co-expression of WT with G2006D- or G2015E-collagen VII also showed strong intracellular staining (Fig. 3A, third and fourth panel), but the vacuoles were notably smaller than in cells expressing WT and G1776R-collagen VII. Determination of the number of individual cells showing intracellular accumulation or a normal collagen VII expression pattern revealed a high percentage of cells with intracellular accumulation in cultures with co-expression of WT and the mutants (Fig. 3B). Semi-quantitative estimation of the ratio of intracellular versus secreted collagen VII was obtained by immunoblotting of cell lysates and culture medium with antibodies to collagen VII. In cultures expressing only the WT construct, the ratio of collagen VII in the cell layer/medium was approximately equal and, for the sake of comparison, this ratio was set as 1 (Fig. 3C). Consequently, the value for G1776R-collagen VII-expressing cells was 1.69 ± 0.29; for G2006D-collagen VII 1.11 ± 0.27; and that for G2015E-collagen VII-expressing cells 0.76 ± 0.05, indicating that the first mutation caused significant retention of collagen VII in the cells, whereas the second and the third caused an abnormal expression pattern but less or no intracellular accumulation, as estimated in the whole culture.
FIGURE 3.
Intracellular accumulation of collagen VII containing mutations G1776R, G2006D, or G2015E. A, transfection of immortalized collagen VII-deficient keratinocytes with WT collagen VII alone shows an expression pattern (left panel) comparable to primary human keratinocytes (inset). Untransfected collagen VII-deficient keratinocytes remained completely negative (not shown). Co-transfection with cDNA for WT and mutant collagen VII in equal ratios leads to intracellular accumulation of collagen VII in case of glycine substitution G1776R (second panel from the left), glycine substitution G2006D (second panel from the right), and glycine substitution G2015E (right panel). Collagen VII is visualized by immunostaining with the NC2–10 antibody (green), nuclei are counterstained with DAPI (blue). B, percentage of transfected cells showing a normal expression pattern (white column) or intracellular accumulation (black column). C, quantification of rColVII in lysates and medium of transfected cells. Values are given as a ratio of rColVII in the cell lysate to rColVII in the medium. For comparison, the ratio observed after transfection with WT collagen VII was defined as 1.0. The data represent averages of at least two independent experiments.
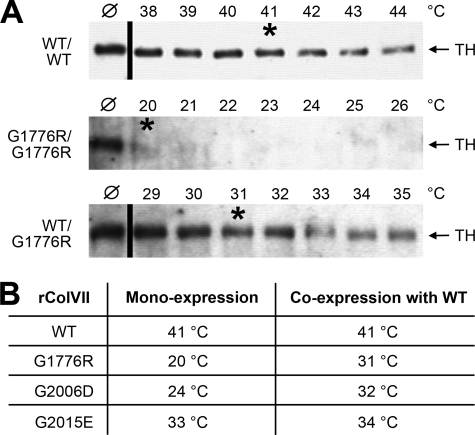
Triple Helix Stability of Collagen VII with a Glycine Substitution Is Diminished
Because intracellular accumulation of proteins often indicates faulty folding, the thermal stability of the triple helix of recombinant collagen VII was determined by measuring its resistance to limited trypsin digestion (18). Fully folded collagen triple helices are resistant to trypsin, and the proteinase sensitivity can be used as an indicator of protein folding, or unfolding, at increasing temperatures. WT and mutant collagen VII were first digested with low concentrations of pepsin to remove the non-collagenous domains, and the triple helix was then subjected to increasing temperatures between 20 and 44 °C at 1 °C steps, followed by limited digestion with trypsin. Samples were analyzed by immunoblotting, and densitometry was used to determine the temperature at which 50% of the triple-helical collagen VII was degraded by trypsin, indicating that 50% of the molecules were not folded into a tight triple helix. This temperature was defined as “melting temperature, Tm” (12).
As shown in Fig. 4A, recombinant WT collagen VII gradually looses its resistance to tryptic digestion between 38 and 44 °C, with a Tm of 41 °C (Fig. 4A, upper panel). This is in accordance with the Tm determined for authentic collagen VII produced by human keratinocytes (15). In mono-expression, G1776R-collagen VII is sensitive to tryptic digestion even after incubation at 20 °C, the lowest temperature examined (Fig. 4B, middle panel), demonstrating that this amino acid substitution profoundly influences folding of collagen VII. However, if co-expressed with WT collagen VII using equal amounts of the respective cDNAs, the thermal stability was clearly higher, with 50% of the collagen molecules sensitive to proteolysis at 31 °C (Fig. 4C, lower panel), indicating that the presence of WT polypeptide chains in the cell increases the stability in hybrid molecules. The mutations G2006D and G2015E also lead to strongly reduced resistance to proteolysis. The Tm of G2006D-collagen VII was 24 °C and that of G2015E-collagen VII 33 °C. When co-expressed with equal amounts of WT collagen VII cDNA, the Tm of G2006D-collagen VII was 32 °C and of G2015E-collagen VII 34 °C (Fig. 4B).
FIGURE 4.
Thermal stability of WT and mutant collagen VII. The thermal stability of the collagen triple helix of WT and mutant collagen VII was determined by limited proteolytic digestion. Collagen VII precipitated from the medium of transiently transfected HEK-293T cells was first subjected to pepsin digestion to remove the non-collagenous domains. After exposure to temperatures from 20 to 44 °C, the triple helices were subjected to limited trypsin digestion. The amount of rColVII resistant to tryptic digestion was determined by immunoblotting with the NC2–10 antibody. The temperature, at which 50% of the rColVII molecules were sensitive to proteolytic degradation was defined as “melting temperature, Tm”. A, sensitivity of WT rColVII to tryptic digestion (upper panel). The Tm of 41 °C (labeled with asterisk) is in agreement with previous data obtained by digesting native collagen VII produced by normal human keratinocytes (15). Mono-expression of G1776R-collagen VII resulted in a Tm below 20 °C (middle panel, Tm labeled with asterisk), whereas hybrid rColVII obtained by co-transfection of WT and G1776R-collagen VII cDNA shows a Tm of 31 °C (lower panel, Tm labeled with asterisk). ℘, collagen VII digested with trypsin at 20 °C; TH, migration position of the triple helix. B, overview of the melting temperatures of WT and mutant rColVII with the mutations G1776R, G2006D, and G2015E in mono- and co-expression with WT collagen VII, respectively.
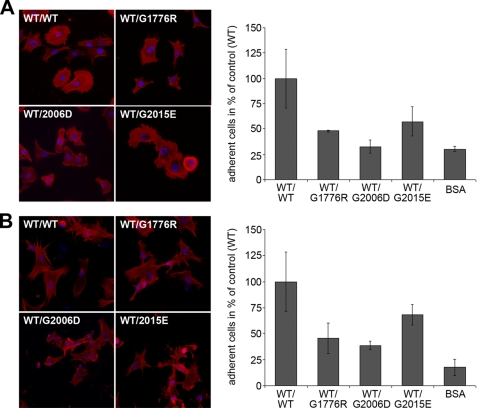
Reduced Cell Adhesion to Mutant Collagen VII
As a component of the dermal-epidermal junction and the extracellular matrix in the papillary dermis, collagen VII is in contact with both keratinocytes and fibroblasts in normal and wounded skin. It has been reported to promote fibroblast adhesion by two cell binding sites, one in the NC-1 and one in the triple helical domain (19, 17). Therefore, we compared the ability of WT collagen VII and the mutants to promote adhesion and spreading of normal human keratinocytes and fibroblasts. Purified recombinant WT collagen VII coated onto coverslips strongly promoted attachment and spreading of both cell types (Fig. 5, A and B). However, the hybrid proteins of WT and G1776R-, G2006D-, or G2015E-collagen VII were clearly less effective in enhancing fibroblast and keratinocyte adhesion. In addition, spreading of the cells on the mutated collagens was altered (Fig. 5, A and B). These observations demonstrate not only defective structural functions of G1776R-collagen VII, G2006D-collagen VII, and G2015E-collagen VII, but also uncover functional deficiency in mediating cell-matrix interactions.
FIGURE 5.
Adhesion and spreading of normal human keratinocytes and fibroblasts on WT and mutant collagen VII. For these experiments, FLAG-tagged WT and hybrid collagen VII composed of WT and mutant rColVII was produced by co-transfecting HEK293 cells with the corresponding cDNA. After FLAG tag affinity purification, the collagens were coated onto coverslips in a concentration of 20 μg/ml; bovine serum albumin coating was used as negative control. Normal human keratinocytes or fibroblasts were incubated on the coverslips for 2 h, washed, fixed, and stained with DAPI and rhodamin-coupled phalloidin. A, spreading (left panel) and adhesion (right panel) of normal human keratinocytes on WT collagen VII and on the mutants G1776R, G2006D, or G2015E. B, spreading (left panel) and adhesion (right panel) of normal human fibroblasts on WT collagen VII and the mutants G1776R, G2006D, or G2015E. All three collagen VII mutants caused decreased adhesion and abnormal spreading of both cell types. The number of attached cells was evaluated by counting the DAPI-stained nuclei (in blue). The spreading was visualized by staining the actin cytoskeleton (in red). Shown are representative data of three independent experiments.
Rescue of Dominant-negative Effects of Glycine Substitutions by Overexpression of WT Collagen VII
The dominant-negative interference of glycine substitution mutations, i.e. imperfect folding and at least partial intracellular accumulation of collagen VII, is the main cause of disease pathology in DDEB. As collagen VII forms trimers of three pro-α1(VII) chains, co-expression of mutant and WT collagen VII leads to formation of hybrid trimers consisting of either three, two, one or no WT chains or, vice versa, none, one, two, or three mutated chains. The exact mechanisms of how individual mutated chains perturb triple-helix folding of collagen VII are not fully understood, but our prediction was that higher content of mutated α1 chains is likely to increase the degree of perturbation and that overexpression of WT α1 chains will increase the probability of collagen VII trimers consisting of more WT chains.
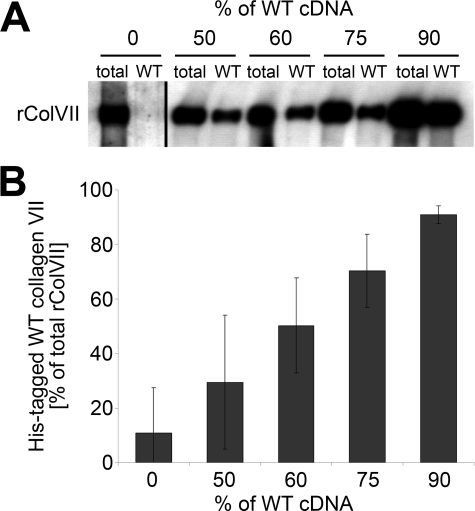
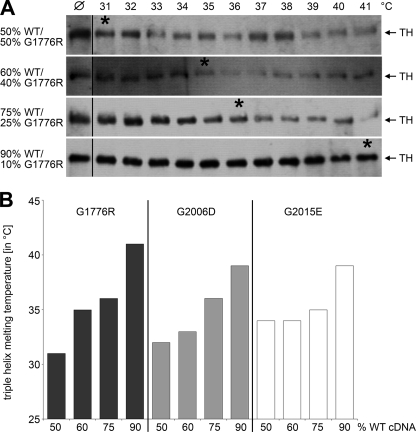
To test this hypothesis, we co-expressed cDNA for WT collagen VII and for each of the mutants in different ratios in HEK-293T cells. The ratios ranged from 50% WT:50% mutant cDNA to 90% WT:10% mutant cDNA. Secreted collagen VII molecules were precipitated from the culture medium, and the amount of WT collagen VII in the probe was determined after depletion of the mutant molecule (FLAG-tagged mutant collagen VII bound almost quantitatively to an anti-FLAG column) and quantification of His-tagged WT chains. The relative amounts of WT and mutant collagen VII cDNAs used for transfection corresponded well with the amounts of tagged WT and mutant collagen VII protein (Fig. 6). Subsequently, the thermal stability of the hybrid collagen VII molecules was analyzed using limited proteolysis as a probe. The resistance of the collagen VII molecules to tryptic digestion increased proportionally with the amount of WT protein present (Fig. 7). For hybrids of WT and G1776R-collagen VII, the temperature at which 50% of the molecules were sensitive to digestion increased from 31 °C after transfection with 50% WT and 50% mutant cDNA to 35 °C using 60% WT and 40% mutant cDNA. Further increase of the WT cDNA to 75% increased the thermal stability to a Tm of 36 °C and, intriguingly, co-expression of 90% WT and 10% G1776R mutant cDNA lead to a resistance to proteolysis similar to recombinant WT collagen VII with a Tm of 41 °C (Fig. 7A). Similarly, increasing the amount of WT collagen VII rescued the effect of the glycine substitution mutations G2006D and G2015E, although the maximal thermal stability observed was significantly lower than in WT collagen VII, with a melting temperature of 39 °C for both G2006D and G2015E (Fig. 7B). These observations indicated that controlled overexpression of WT collagen VII ameliorated the negative effects of glycine substitution mutations and enhanced secretion of a more stable collective of collagen VII molecules by HEK-293T cells.
FIGURE 6.
Quantification of mutant and WT collagen VII after co-transfection. HEK-293T cells were transiently transfected with a mixture of plasmid cDNA for His-tagged WT and FLAG-tagged mutant collagen VII. The amount of WT collagen VII in the culture medium was determined by quantitatively binding the FLAG-tagged mutant collagen VII to anti-FLAG-agarose, followed by quantification of the remaining His-tagged WT collagen VII in the unbound fraction using the NC2–10 antibody. A, immunoblot of collagen VII after incubation of hybrid molecules of His-tagged WT rColVII and FLAG-tagged mutant rColVII with an anti-FLAG column. Increasing the ratio of WT/mutant collagen VII cDNA in the transfection mix correlated with the amounts of collagen VII in the medium unable to bind to the anti-FLAG-agarose. Total, unbound fraction after incubation of medium with empty agarose; WT, unbound fraction after incubation of the medium with anti-FLAG-agarose. B, densitometric quantification of three independent immunoblots assaying the amount of His-tagged WT collagen VII after transient co-transfection of HEK-293T cells with mixtures of His-tagged WT and FLAG-tagged mutant collagen VII. Shown is the amount of His-tagged WT collagen VII in relation to the total rColVII.
FIGURE 7.
Controlled overexpression of WT collagen VII improves triple helix stability of collagen VII containing glycine substitutions. Mutant collagen VII with glycine substitutions G1776R, G2006D, or G2015E was co-expressed with increasing amounts of WT collagen VII. For determination of the thermal stability, the hybrid molecules in the medium were subjected to limited proteolytic digestion and immunoblotting with collagen VII antibodies. A, when WT collagen VII and the mutant G1776R were co-expressed in equal amounts, 50% of the hybrid molecules were sensitive to proteolysis at 31 °C (marked by asterisk). Co-expression of 60% WT collagen VII and 40% of the mutant increased the Tm to 35 °C (asterisk). Co-expression of 75% WT collagen VII and 25% mutant lead to a Tm of 36 °C (asterisk), and co-expression of 90% WT collagen VII and 10% mutant to a Tm of 41 °C (asterisk). ℘, collagen VII digested with trypsin at 20 °C; TH, triple helix. Asterisks mark the melting temperature. B, correlation of thermal stability of hybrid collagen VII composed of increasing amounts of WT and decreasing amounts of mutant collagen VII. The thermal stability of all three mutants was clearly augmented by co-expression with increasing amounts of WT cDNA.
DISCUSSION
Mutations in collagen genes underlie a wide spectrum of inherited human disorders (20, 21, 5), ranging from the Alport syndrome of the kidney, to arterial aneurysms, hemorrhagic stroke, muscular dystrophies, skeletal disorders, to Knobloch syndrome of the eye, and epidermolysis bullosa of the skin. Many of these disorders are caused by dominantly inherited glycine substitution mutations in the triple-helical domains of the collagens, which perturb collagen folding and suprastructural assembly.
To elucidate the mechanisms of dominant-negative interference by substitution of glycine residues by other amino acids, collagen I mutations in osteogenesis imperfecta (OI) have been investigated as prototypes (22). An intriguing and elusive question has been why glycine substitutions in different locations within the triple helix cause different disruptions of the collagen molecule. Only recent studies using synthetic peptides, recombinant collagen fragments (23, 24), and authentic collagen isolated from OI fibroblasts (25) have revealed the important role of the sequence microenvironment, i.e. the local helix stability around the mutated glycine and of salt bridges which function to stabilize subregions of the collagen triple-helix (26). Changed protein folding and/or disruption of the salt bridges can render the triple-helix unstable and sensitive to degradation. Similar mechanisms of dominant-negative interference underlie other diseases caused by mutations in other collagen genes (27). We have previously shown that in DDEB some, but not all, glycine substitution mutations in collagen VII lead to intracellular accumulation of collagen VII and to loss of anchoring fibrils in the skin (12). It was predicted that the mutated molecules were degraded intracellularly, at least in part, and thus caused reduction of collagen VII and paucity of anchoring fibrils in the skin.
In the present study we used recombinant expression of normal and mutated collagen VII, combined with limited pepsin/trypsin digestion, to elucidate the thermal stability and sensitivity to proteolysis of collagen molecules containing normal and mutated α1(VII) chains. As examples, three glycine substitution mutations associated with mild-to-moderate DDEB in humans were selected, G1776R, G2006D, and G2015E.
Because one of the unanswered questions about collagens is whether WT and mutant chains really fold into heterotrimeric hybrid molecules, or into separate homotrimers, i.e. a mixture of mutant and WT triple-helical molecules, we undertook affinity purification of FLAG-tagged mutant molecules and used mass spectrometry to demonstrate the presence of WT α1(VII) chains in the affinity-purified fraction. Importantly, this showed that mutant and WT chains fold into a heterotrimeric triple helix, thus proving that dominant-negative interference by glycine substitutions destabilizes mutant collagen VII. To gain more mechanistic information, it would be intriguing to isolate the different homo- and heterotrimers containing one, two, or three mutant α1(VII) chains, and characterize them using protein and physicochemical methods, as has been recently done with mutant collagen I isolated from OI fibroblasts (25). However, because of the low yields of collagen VII isolated from cell culture, such an endeavor is extremely tedious, prone to artifical effects due to protein degradation and, therefore, not practicable.
Like collagen I mutations in OI, it was evident that the collagen VII mutations had different molecular consequences. Co-expression (50% WT, 50% mutant cDNA) of G1776R-collagen with WT lead to reduced thermal stability of collagen VII, with a Tm of 31 °C, and subsequent accumulation of the hybrid molecules in the endoplasmic reticulum. The effects of co-expression of WT and G2006D-collagen VII were milder, with a Tm of 32 °C and less intracellular retention. Interestingly, co-expression of WT and G2015E-collagen VII, in which the glycine substitution is only three triplets away from G2006D, resulted in reduced thermal stability and a Tm 34 °C, but no accumulation in the endoplasmic reticulum. These observations are in keeping with genotype-phenotype correlation studies (28, 29, 17), which have not yielded a direct correlation between the thermal stability of collagen VII mutants and the DEB phenotype, although the relation between conformational heterogeneity of collagen VII and disease severity has not been mapped in a systematic manner.
The lack of direct correlation between the glycine substitution mutation and disease severity in DDEB renders this disorder less amenable to therapeutic targeting with e.g. oligonucleotide-based strategies. Nevertheless, DDEB has a severe impact on the lives of affected individuals and, therefore, biologically valid treatments are urgently needed. Since the relationship of structural disruptions caused by glycine substitution mutations in collagen genes and the phenotypes is not simple and straightforward (25), alternative therapeutic strategies should be considered. Here, we tested a rationale to ameliorate the biological phenotype of DDEB in vitro by controlled overexpression of the WT allele. The goal was to shift the ratio of normal and mutated polypeptide, which is presumed to be 50:50 in DDEB, toward normal. The prediction was that if a cell expressed more WT than mutant allele, the collective of collagen VII trimers would contain more normal α1(VII)-chains and homotrimers, fold better and form more stable and functional anchoring fibrils.
This strategy worked remarkably well in cells synthesizing both normal and mutated α1(VII) polypeptides. For all three mutations, a gradual improvement of thermal stability of collagen VII molecules was observed with increasing ratio of WT:mutant expression. For example, in case of G1776R, changing the ratio of WT:mutated collagen VII cDNA from 50:50 to 90:10 raised the Tm from 31 to 41 °C, the value for WT collagen. Similar effects were observed with the mutants G2006D and G2015E, albeit to a somewhat lesser extent. However, in both cases, a Tm of 39 °C was achieved, indicating significantly higher thermal stability of secreted collagen VII and, by extension, improved functionality.
Our findings present a proof-of-principle and lay a molecular basis for putative novel therapeutic strategies for DDEB. After titrating the amount of WT expression needed to overcome the individual threshold of intracellular accumulation and degradation of a mutant collagen VII in DDEB keratinocytes in vitro, the goal is to deliver controlled amounts of WT collagen VII into patient skin to increase the amount of functional anchoring fibrils. This could be achieved by transfecting keratinocytes and/or fibroblasts with WT collagen VII cDNA using retro- or lentiviral vectors (30, 31), Epstein-Barr virus-based systems with stable episomal maintenance (32) or phi C31 integrase-based approaches (33). Alternatively, since polymerization of anchoring fibrils takes place in the extracellular space, a different approach would be to provide additional extracellular WT collagen VII molecules for the fibril assembly. This may be feasible by intradermal injection of allogeneic normal fibroblasts, which have been shown to secrete collagen VII, deposit it at the epidermal basement membrane zone and ameliorate the DEB phenotype in a mouse model (10, 34).
Taken together, using collagen VII as an example we provide a molecular strategy for counteracting dominant-negative interference of glycine substitution mutations by shifting the balance between WT and mutant α-chains forming the collagen triple helix. This approach is also applicable to therapeutic considerations for other collagen diseases affecting other organs, which are caused by similar mutational mechanisms.
Supplementary Material
Acknowledgments
We thank Margit Schubert, Ulrike Lanner, and Björn Wienke for expert technical assistance.
This work was supported in part by grants from the Federal Ministry for Education and Research, BMBF (EB-Network, Project 9), Debra International, and the German Research Foundation (Bioss and FRIAS Lifenet programs).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- DEB
- dystrophic epidermolysis bullosa
- DDEB
- dominant dystrophic epidermolysis bullosa
- EBS
- epidermolysis bullosa simplex
- ER
- endoplasmic reticulum
- NC-1/NC-2
- non-collagenous-1/2 domains of collagen VII
- OI
- osteogenesis imperfecta
- rColVII
- recombinant collagen VII
- RDEB
- recessive dystrophic epidermolysis bullosa
- TH
- triple helix
- Tm
- melting temperature
- WT
- wild-type
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Kern J. S., Has C. (2008) Expert. Rev. Dermatol. 3, 721–733 [Google Scholar]
- 2.Fine J. D., Eady R. A., Bauer E. A., Bauer J. W., Bruckner-Tuderman L., Heagerty A., Hintner H., Hovnanian A., Jonkman M. F., Leigh I., McGrath J. A., Mellerio J. E., Murrell D. F., Shimizu H., Uitto J., Vahlquist A., Woodley D., Zambruno G. (2008) J. Am. Acad. Dermatol. 58, 931–950 [DOI] [PubMed] [Google Scholar]
- 3.Rattenholl A., Pappano W. N., Koch M., Keene D. R., Kadler K. E., Sasaki T., Timpl R., Burgeson R. E., Greenspan D. S., Bruckner-Tuderman L. (2002) J. Biol. Chem. 277, 26372–26378 [DOI] [PubMed] [Google Scholar]
- 4.Burgeson R. E. (1993) J. Invest. Dermatol. 101, 252–255 [DOI] [PubMed] [Google Scholar]
- 5.Kadler K. E., Baldock C., Bella J., Boot-Handford R. P. (2007) J. Cell Sci. 120, 1955–1958 [DOI] [PubMed] [Google Scholar]
- 6.Schröder M. (2008) Cell Mol. Life Sci. 65, 862–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto M., Sawamura D., Nishie W., Sakai K., McMillan J. R., Akiyama M., Shimizu H. (2006) J. Invest. Dermatol. 126, 2614–2620 [DOI] [PubMed] [Google Scholar]
- 8.Hengge U. R. (2008) J. Invest. Dermatol. 128, 499–500 [DOI] [PubMed] [Google Scholar]
- 9.Cao T., Longley M. A., Wang X. J., Roop D. R. (2001) J. Cell Biol. 152, 651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritsch A., Loeckermann S., Kern J. S., Braun A., Bösl M. R., Bley T. A., Schumann H., von, Elverfeldt D., Paul D., Erlacher M., Berens von, Rautenfeld D., Hausser I., Fässler R., Bruckner-Tuderman L. (2008) J. Clin. Invest. 118, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dharma B., Moss C., McGrath J. A., Mellerio J. E., Ilchyshyn A. (2001) Clin. Exp. Dermatol. 26, 93–96 [PubMed] [Google Scholar]
- 12.Hammami-Hauasli N., Schumann H., Raghunath M., Kilgus O., Lüthi U., Luger T., Bruckner-Tuderman L. (1998) J. Biol. Chem. 273, 19228–19234 [DOI] [PubMed] [Google Scholar]
- 13.Villone D., Fritsch A., Koch M., Bruckner-Tuderman L., Hansen U., Bruckner P. (2008) J. Biol. Chem. 283, 24506–24513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vindevoghel L., Chung K. Y., Davis A., Kouba D., Kivirikko S., Alder H., Uitto J., Mauviel A. (1997) J. Biol. Chem. 272, 10196–10204 [DOI] [PubMed] [Google Scholar]
- 15.Mecklenbeck S., Compton S. H., Mejía J. E., Cervini R., Hovnanian A., Bruckner-Tuderman L., Barrandon Y. (2002) Hum. Gene Ther. 13, 1655–1662 [DOI] [PubMed] [Google Scholar]
- 16.Bruckner-Tuderman L., Nilssen O., Zimmermann D. R., Dours-Zimmermann M. T., Kalinke D. U., Gedde-Dahl T., Jr., Winberg J. O. (1995) J. Cell Biol. 131, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodley D. T., Hou Y., Martin S., Li W., Chen M. (2008) J. Biol. Chem. 283, 17838–17845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruckner P., Prockop D. J. (1981) Anal. Biochem. 110, 360–368 [DOI] [PubMed] [Google Scholar]
- 19.Chen M., Marinkovich M. P., Veis A., Cai X., Rao C. N., O'Toole E. A., Woodley D. T. (1997) J. Biol. Chem. 272, 14516–14522 [DOI] [PubMed] [Google Scholar]
- 20.Gould D. B., Phalan F. C., Breedveld G. J., van Mil S. E., Smith R. S., Schimenti J. C., Aguglia U., van der Knaap M. S., Heutink P., John S. W. (2005) Science 308, 1167–1171 [DOI] [PubMed] [Google Scholar]
- 21.Myllyharju J., Kivirikko K. I. (2004) Trends Genet. 20, 33–43 [DOI] [PubMed] [Google Scholar]
- 22.Marini S., Fasciglione G. F., de Sanctis G., D'Alessio S., Politi V., Coletta M. (2000) J. Biol. Chem. 275, 18657–18663 [DOI] [PubMed] [Google Scholar]
- 23.Hyde T. J., Bryan M. A., Brodsky B., Baum J. (2006) J. Biol. Chem. 281, 36937–36943 [DOI] [PubMed] [Google Scholar]
- 24.Persikov A. V., Pillitteri R. J., Amin P., Schwarze U., Byers P. H., Brodsky B. (2004) Hum. Mutat. 24, 330–337 [DOI] [PubMed] [Google Scholar]
- 25.Makareeva E., Mertz E. L., Kuznetsova N. V., Sutter M. B., DeRidder A. M., Cabral W. A., Barnes A. M., McBride D. J., Marini J. C., Leikin S. (2008) J. Biol. Chem. 283, 4787–4798 [DOI] [PubMed] [Google Scholar]
- 26.Xu K., Nowak I., Kirchner M., Xu Y. (2008) J. Biol. Chem. 283, 34337–34344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pace R. A., Peat R. A., Baker N. L., Zamurs L., Mörgelin M., Irving M., Adams N. E., Bateman J. F., Mowat D., Smith N. J., Lamont P. J., Moore S. A., Mathews K. D., North K. N., Lamandé S. R. (2008) Ann. Neurol. 64, 294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kern J. S., Kohlhase J., Bruckner-Tuderman L., Has C. (2006) J. Invest. Dermatol. 126, 1006–1012 [DOI] [PubMed] [Google Scholar]
- 29.Mecklenbeck S., Hammami-Hauasli N., Höpfner B., Schumann H., Kramer A., Küster W., Bruckner-Tuderman L. (1999) J. Invest Dermatol. 112, 398–400 [DOI] [PubMed] [Google Scholar]
- 30.Gache Y., Baldeschi C., Del Rio M., Gagnoux-Palacios L., Larcher F., Lacour J. P., Meneguzzi G. (2004) Hum. Gene Ther. 15, 921–933 [DOI] [PubMed] [Google Scholar]
- 31.Mavilio F., Pellegrini G., Ferrari S., Di Nunzio F., Di Iorio E., Recchia A., Maruggi G., Ferrari G., Provasi E., Bonini C., Capurro S., Conti A., Magnoni C., Giannetti A., De Luca M. (2006) Nat. Med. 12, 1397–1402 [DOI] [PubMed] [Google Scholar]
- 32.Conese M., Auriche C., Ascenzioni F. (2004) Gene Ther. 11, 1735–1741 [DOI] [PubMed] [Google Scholar]
- 33.Ortiz-Urda S., Thyagarajan B., Keene D. R., Lin Q., Fang M., Calos M. P., Khavari P. A. (2002) Nat. Med. 8, 1166–1170 [DOI] [PubMed] [Google Scholar]
- 34.Kern J. S., Loeckermann S., Fritsch A., Hausser I., Roth W., Magin T. M., Mack C., Muller M. L., Paul O., Ruther P., Bruckner-Tuderman L. (2009) Mol. Ther. 17, 1605–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.