Abstract
Unlike other antiapoptotic members of the Bcl-2 family, Bfl-1 does not contain a well defined C-terminal transmembrane domain, and whether the C-terminal tail of Bfl-1 functions as a membrane anchor is not yet clearly established. The molecular modeling study of the full-length Bfl-1 performed within this work suggests that Bfl-1 may co-exist in two distinct conformational states: one in which its C-terminal helix α9 is inserted in the hydrophobic groove formed by the BH1–3 domains of Bfl-1 and one with its C terminus. Parallel analysis of the subcellular localization of Bfl-1 indicates that even if Bfl-1 may co-exist in two distinct conformational states, most of the endogenous protein is tightly associated with the mitochondria by its C terminus in both healthy and apoptotic peripheral blood lymphocytes as well as in malignant B cell lines. However, the helix α9 of Bfl-1, and therefore the binding of Bfl-1 to mitochondria, is not absolutely required for the antiapoptotic activity of Bfl-1. A particular feature of Bfl-1 is the amphipathic character of its C-terminal helix α9. Our data clearly indicate that this property of helix α9 is required for the anchorage of Bfl-1 to the mitochondria but also regulates the antiapoptotic function Bfl-1.
Apoptosis is a highly regulated process that plays a key role in maintaining cellular homeostasis, and a delicate balance between proapoptotic and antiapoptotic regulators of apoptosis pathways ensures the proper survival of cells in a variety of tissues. Imbalance between proapoptotic and antiapoptotic proteins occurs in diseases such as cancer, where an overexpression of antiapoptotic proteins endows cells with a selective survival advantage that promotes malignancy. Bcl-2 family members are essential regulators of the intrinsic apoptotic pathway, which act at the level of mitochondria as initiators of cell death (1). This family comprises nearly 20 proteins divided into three main groups. Antiapoptotic members such as Bcl-2, Bcl-xL, Bcl-w, Bfl-1, and Mcl-1 promote cell survival, whereas proapoptotic members such as Bax and Bak function as death effectors. The life and death balance is displaced in favor of cell death by proapoptotic BH3-only proteins such as Bim, Bad, Bid, Puma, and Noxa, which interact with antiapoptotic proteins and inactivate their function (2) or directly interact with and activate the Bax-like proteins (3).
Distinct subcellular localizations of antiapoptotic members have been reported correlating with the accessibility of their C-terminal tail. The C-terminal tail of the antiapoptotic proteins Bcl-2, Bcl-xL, and Bcl-w possess a hydrophobic region known to be a membrane anchor domain. Thus, Bcl-2 localizes to mitochondria as well as to the endoplasmic reticulum and nuclear membranes (4, 5, 6), and deletion of its C-terminal amino acids abrogates its targeting to the outer mitochondrial membrane (7). In contrast, in healthy cells, Bcl-xL and Bcl-w localize mainly in the cytosol because their C-terminal tails are sequestered. Bcl-xL exists as a homodimer through the exchange of the C-terminal tail bound in the hydrophobic groove of the reciprocal dimer partner (8), whereas the C-terminal tail of Bcl-w occupies its own hydrophobic groove in the monomer form (9, 10). It has been proposed that, following apoptotic stimuli, interaction of the BH3 domain from BH3-only proteins with the hydrophobic groove of Bcl-w or Bcl-xL liberates their C-terminal tail and then the two proteins translocate to the mitochondria (8, 11).
Unlike Bcl-2, Bcl-xL, and Bcl-w, Bfl-1 and its murine homolog, A1, do not contain a well defined C-terminal transmembrane domain (12, 13). C-terminal ends of these two proteins are similar and contain several hydrophilic residues that interrupt their putative transmembrane hydrophobic domain. Whether the C-terminal tail of Bfl-1 functions as a membrane anchor remains to be clarified. Immunofluorescence analyses in an earlier study have shown that overexpressed human Bfl-1 is predominantly localized in the endoplasmic/nuclear envelope regions (14). Then, recent independent studies, with Bfl-1-overexpressing cells, suggested that Bfl-1 localizes to the mitochondria (15, 16, 17) and that the C-terminal end of Bfl-1 is important for anchoring Bfl-1 to the mitochondria due to GFP-Bfl-1 being associated to the mitochondria, whereas GFP-Bfl-1, devoid of its C-terminal tail, also localizes in the cytosol (16, 18). However, localization of endogenous Bfl-1 has never been investigated. In this study, we present a molecular modeling study of full-length Bfl-1 (FL-Bfl-1), based on the crystal structure of a truncated form of Bfl-1 (residues 1–149) in complex with the BIM-BH3 peptide (Protein Data Bank code 2VM6).4 Our model suggests that Bfl-1 may co-exist in two distinct conformational states, the first one with its C-terminal helix α9 (residues 155–175) inserted in the hydrophobic groove formed by the BH1–3 domain of Bfl-1, and the second one with its C-terminal tail. Interestingly, helical wheel projection of the C-terminal helix of Bfl-1 highlights its amphipathic character, a feature of transmembrane helices or membrane anchors. These observations incited the reinvestigation of the subcellular localization of Bfl-1 in both malignant B cell lines and peripheral blood lymphocytes (PBLs).5 We demonstrate here that endogenous Bfl-1 is preferentially anchored to the mitochondria in malignant B cell lines but also in healthy PBLs. Moreover, we show that both the anchorage of Bfl-1 to the mitochondria and the anti-apoptotic function of the protein are dependent on the amphipathic nature of the C-terminal helix.
EXPERIMENTAL PROCEDURES
Molecular Modeling of Bfl-1
Modeling of the three-dimensional structure was carried out with the program MODELLER 9 version 3 (19). The full-length sequence of human Bfl-1 in the UniProt Database (accession number Q16548) was used as query sequence. The crystal structure of the human Bcl-2-related protein A1 in complex with the BIM-BH3 peptide (Protein Data Bank code 2VM6)4 was used as a three-dimensional template for homology modeling. The accuracy of the model was improved by several iterations with MODELLER alternately with manual corrections of the three-dimensional model using the graphics program TURBO-FRODO (20). The stereochemical quality of the final model was assessed using the validation program PROCHECK (21).
Material and Cell Culture
All media and cell culture reagents were purchased from Invitrogen. IM9, BP3, and PBLs were cultured in RPMI complete medium supplemented with 10% fetal bovine serum, 2 mm glutamine, 10 mm Hepes, and 40 μg/ml gentamycin. GPE, HeLa 293T, and NIH-3T3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm glutamine, 10 mm Hepes, and 40 μg/ml gentamycin. PBLs were collected from healthy donors (Etablissement Français du Sang). Separation of PBLs was obtained by standard Ficoll/Percoll density gradient centrifugation. After two washes in phosphate-buffered saline, PBLs were activated with interleukin-2 (50 units/ml) and phytohemagglutinin (5 μg/ml) for 4 days. Dead cells were removed by centrifugation on a layer of Ficoll and washed twice in phosphate-buffered saline. Viable cells were then cultured at 1 × 106/ml.
Plasmid Constructions
Human FLAG-FL-Bfl-1 cloned in pEGZ vector was a gift from Ingolf Berberich (University of Würzburg, Würzburg, Germany) and human FLAG-Δα9-Bfl-1-(1–151) was amplified by PCR from a FLAG-FL-Bfl-1 template and inserted into EcoR1-BamH1 sites of the pEGZ vector. Mutant forms of FL-Bfl-1 (mut-FL-Bfl-1 and mutant HR7-FL-Bfl-1) were amplified by PCR from FL-Bfl-1 template with primers containing mutated sites and inserted into EcoR1-BamH1 sites of the pEGZ vector. Mutant forms of FL-Bfl-1 were inserted into EcoR1-BamH1 sites of the pMIG vector (kind gift of Dr. Janet Maryanski). pEYFP-N1 Bax was a gift from Junying Yuan (Harvard Medical School, Boston, MA). The molecular identity of all constructs was confirmed by sequencing (Genome express).
Generation of Stably Transfected Cell Lines
GPE packaging cells were transfected with an empty pMIG vector, a pMIG vector expressing FL-Bfl-1 or mutated FL-Bfl-1 constructs, or pEGZ vector expressing Δα9-Bfl-1. 48 h after transfection, cells were harvested and FACS-sorted according to GFP expression using the FACSVantage SE Option Diva (digital system, Becton Dickinson). GPE viral supernatant was added to NIH-3T3 cells. 2 days after infection, NIH-3T3 cells were FACS-sorted according to GFP expression. Stability of infection was checked with GFP expression.
Apoptosis Assays
NIH-3T3 cells were cultured overnight in 24-well plates with 1 × 105 cells/well in complete medium. Then, cells were washed twice with phosphate-buffered saline and cultured in a serum-lacking medium for the indicated time. Cell death was evaluated by propidium iodide staining and analyzed by FACS with the CellQuest software (Becton Dickinson).
Rescue assays
HeLa cells were plated in 24 well plate with 1,25.105 cells/well in complete medium and transfected with pECFP-N1-Bax alone or with different Bfl-1 constructs expressed in pEGZ vector. Transfections were performed using Jet-PEI reagent (Polyplus transfection) according to manufacturer instructions. 48 h following transfection, cell death was evaluated by propidium iodide staining and analyzed by FACS with the CellQuest Software (Becton Dickinson).
Subcellular Fractionation
5 × 107 cells were harvested, washed in phosphate-buffered saline, pH 7.2, and centrifuged. Cells were permeabilized for 5 min on ice at a density of 5 × 107 cells/ml in cytosolic extraction buffer (250 mm sucrose, 70 mm KCl, 137 mm NaCl, 4.3 mm Na2HPO4, pH 7.2, protease inhibitor (Sigma) and 25 or 200 μg/ml digitonin for PBLs or other cells, respectively). Plasma membrane permeabilization was confirmed by staining with 0.2% Trypan Blue solution. Cells were centrifuged at 10,000 × g for 10 min at 4 °C, the supernatants (cytosolic fractions) were saved, and the pellets were solubilized in the same volume of mitochondrial lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 2 mm EDTA, 2 mm EGTA, 0.2% Triton X-100, 0.3% Nonidet P-40, and protease inhibitors (Sigma)) and incubated for 20 min on ice. The mitochondria-enriched fractions were then centrifuged at 10,000 × g for 10 min at 4 °C, and the supernatant from high speed centrifugation was used as the mitochondrial fraction.
Western Blot Analysis
Protein extracts (25 or 50 μg) were analyzed by Western blot as described previously (22) using the following antibodies: anti-Bfl-1 serum (gift from J. Borst, The Netherlands Cancer Institute, The Netherlands), anti-Bcl-2 (Bcl-2 oncoprotein, Dako), anti-Bcl-xL (BD Pharmingen), anti-Bax (R&D Systems), and anti-FLAG M2 monoclonal antibody F3165 (Sigma). Anti-actin antibody Ac15 (Sigma) or rabbit polyclonal anti-human Mn-SOD antibody (Stressgen) were used as a loading or a mitochondrial fraction controls.
In Vitro Mitochondrion Assays
[35S]Met proteins FL-Bfl-1 and Δα9-Bfl-1 were synthesized from cDNAs of FLAG-Bfl-1-pEGZ vector using the TnT-coupled transcription/translation system (Promega). Plasmid (2 μg) was transcribed and then translated using T7 polymerase and Flexi® rabbit reticulocyte lysate by incubation at 30 °C for 2 h. Then, 4 fmol of the [35S]Met protein was incubated in the presence of 100 μg of rat liver mitochondria or bovine serum albumin for 60 min at 30 °C and centrifuged for 15 min at 8,000 × g at 4 °C. The association with the mitochondria was analyzed by an alkaline treatment of the mitochondria (incubation of mitochondrial pellet in 100 mm Na2CO3, 300 mm sucrose, pH 11.3, for 30 min on ice), which removes proteins that are associated with mitochondrial membranes but not inserted into the membrane. The supernatant and mitochondrial pellets were analyzed by SDS-PAGE followed by exposition in a PhosphorImager.
Protein Purification
GST fusion proteins containing Δα9-Bfl-1, FL-Bfl-1, or mut-FL-Bfl-1 were expressed from pGEX 4T-1 plasmid in XL1-Blue cells (Stratagene). The cells were grown in 1 liter of TY media (1.6% (w/v) tryptone, 1% (w/v) yeast extract, and 85 mm NaCl) with 50 μg/ml ampicillin at 37 °C to an A600 nm of 0.8 followed by the addition of isopropyl-β-d-thiogalactopyranoside (0.4 mm) and incubated at 25 °C for 7 h. The cells were recovered in 10 ml lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Tween 20, 0.1% 2-β-mercaptoethanol supplemented with protease inhibitor mixture (Roche Diagnostics), 0.5 mg/ml lysozyme and benzonase (250 units/μl)) 30 min at 30 °C, followed by three freezing cycles. The cellular debris were removed by centrifugation at 6,000 × g for 20 min, and the resulting supernatants were incubated with 2 ml of glutathione-Sepharose (GE Healthcare) at 4 °C for 3 h. The resin was washed three times with buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% Tween 20, and 0.1% 2-β-mercaptoethanol) followed by elution of GST fusion proteins in 5 mm of reduced glutathione dissolved in 50 mm Tris-HCl, pH 8.0.
Fluorescence Polarization Assays
Fluorescence polarization assays were done using recombinant GST-Δα9-Bfl-1 and fluorescein isothiocyanate-conjugated Bim BH3 (fluorescein isothiocyanate-DMRPEIWIAQELRRIGDEFNAYYAR). Various concentrations of recombinant proteins were incubated with 15 nm fluorescein isothiocyanate-conjugated Bim or Bak-BH3 peptide in the dark. Fluorescence polarization was measured using a FusionTM Packard equipment. IC50 determinations were performed using GraphPad Prism software (GraphPad, Inc., San Diego, CA).
RESULTS
Molecular Modeling of FL-Bfl-1
Modeling of the three-dimensional structure of FL-Bfl-1 was done based on the crystal structure of the human Bcl-2-related protein A1 in complex with the BIM-BH3 peptide (Protein Data Bank code 2VM6).4 The latter only comprises residues 1–149. The remaining residues (150–175) were deduced and modeled using the coordinates of Cα atoms of the BIM-BH3 peptide bound to the protein. Finally, disordered residues 25–30 (not present in the experimental structure) have been modeled as well.
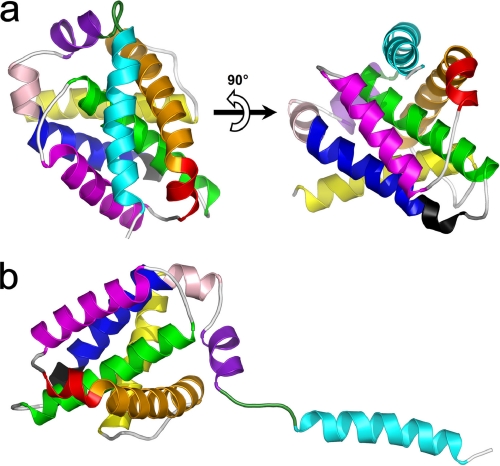
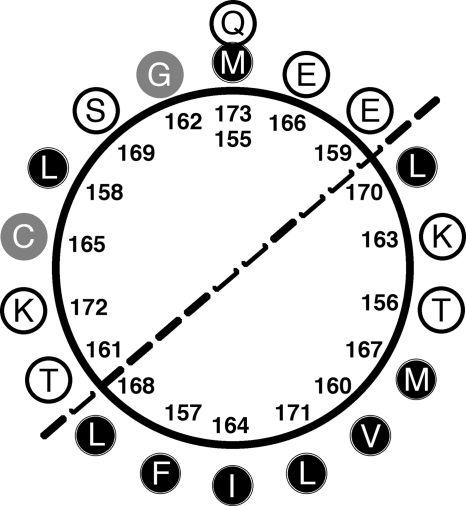
The obtained three-dimensional model includes all 175 amino acid residues forming human FL-Bfl-1. The overall quality of the model as assessed by a Ramachandran plot was high, with 96.2% of the residues in most favored regions and no residues in a disallowed region. For main chain bond lengths, bond angles, and planar groups 98.7, 95, and 100% were within the allowed limits, respectively. The model of human FL-Bfl-1 displays a compact globular structure with approximate dimensions of 30 × 30 × 30 Å3. The overall structure is mainly made up of 9 α-helices and a small 310-helix (Fig. 1a). Like other members of the Bcl-2 family, α5, a central hydrophobic α-helix, was surrounded by a set of amphipathic helices packed against the latter. Interestingly, the C-terminal helix α9 was located at the surface of the protein, diagonally to the α5-helix, and was directly connected to α8 by a loop region defined by residues 149–156 (Fig. 1a). When comparing our model to the structure of Bcl-xL/Bak (23) or Bcl-xL/Bim (24) complexes, it was observed that α9 was located at exactly the same position as the helical fragments of Bak or Bim. This position corresponds to the surface of the hydrophobic BH3 binding groove previously described for Bcl-xL and also present in our model. Helices α2, α3, and α4 formed the two inner sides of the cavity, whereas α5 (and to a lower extent α2) defined its base (Fig. 1a). However, due to the presence of α9, this cavity was not accessible. The present model displayed a small number of interactions between α9 and the BH3-binding cleft. Only one hydrogen bond (Gln-73–O to Lys-172–NZ) was predicted. In addition, several hydrophobic interactions were possible, implicating residues Phe-157, Leu-158, Gly-162, Ile-164, Cys-165, Glu-166, Leu-168, Ser-169, Leu-171, and Lys-172 from helix α9 and 13 residues from the hydrophobic cleft, namely Ser-43, Val-44, Glu-47, Val-48, Asn-51, Leu-52, Val-74, Lys-77, Glu-78, Arg-88, Thr-91, Lys-147, and Phe-148. These interactions were mainly located in the C-terminal half of the helix. We therefore hypothesized that at least the first half of helix α9 has a relatively low affinity for the hydrophobic cleft and that its presence at this location may be fluctuating. Moreover, the length of the loop, which links α8 to α9, is sufficient (six residues) for allowing α9 to shift in and out of the cleft (Fig. 1, a and b). Inspection of the helical wheel projection of α9 clearly shows its amphipathic character (Fig. 2). One side of the helix pointing toward α2 and α3 (left face on Fig. 2) contains mainly hydrophilic amino acids against a majority of hydrophobic residues (pointing toward α4 and α5, right face on Fig. 2) on the other side. Hydrophobic residues are Phe-157, Val-160, Ile-164, Met-167, Leu-168, Leu-170, and Leu-171. α-Helices of amphipathic character are known to potentially interact with biological membranes and to constitute transmembrane helices or membrane anchors. The Bfl-1 model presented here seems ideal for interacting with the biological membrane because α9 is located in the C terminus of Bfl-1.
FIGURE 1.
Three-dimensional model of full-length human Bfl-1. a, ribbon representations of Bfl-1 in its compact form with helix α9 placed in the hydrophobic BH3-binding cleft. b, Bfl-1 in its extended form with helix α9 protruding from the globular core. α-helices are colored sequentially from N- to C-terminal ends as follows: yellow, α1 (aa 2–21); orange, α2 (aa 32–51); red, α3 (aa 53–58); purple, α4 (aa 64–79); green, α5 (aa 86–106); blue, α6 (aa 116–130); pink, α7 (aa 132–136); magenta, α8 (aa 139–148); and cyan, α9 (aa 155–173). The 310-helix (aa 113–115) is shown in black. The loop connecting α8 and α9 is shown in dark green (aa 149–154). Orientation of the model in b is deduced from that of a by a 90° clockwise rotation in the plane.
FIGURE 2.
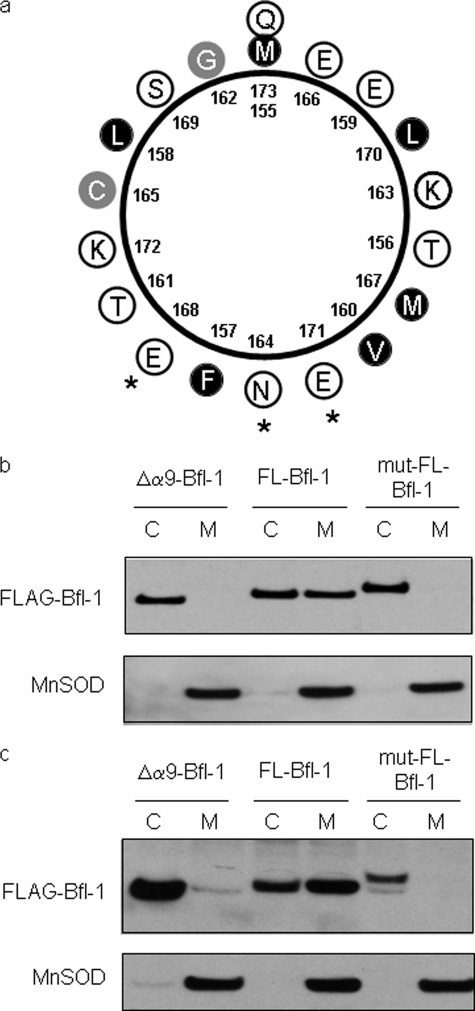
Helical wheel projection showing the amphipathic character of helix α9 (amino acids 155–173). Hydrophobic residues are shown by black circles, hydrophilic residues are shown by white circles, and other residues are presented by gray circles.
Endogenous Bfl-1 Is Localized at the Mitochondria
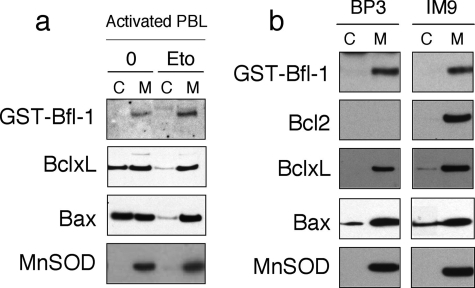
Because our model suggests that Bfl-1 may co-exist in two different conformations and that helix α9 may regulate the interaction of Bfl-1 with biological membranes, we investigated subcellular localization of endogenous Bfl-1 in healthy and etoposide-treated PBLs as well as in two malignant B cell lines. Localization of Bfl-1 was assessed by Western blot analysis following a cell fractionation assay. Endogenous Bfl-1 was found in the mitochondrial pellet in both healthy and apoptotic PBLs and in the two B cell lymphoma cell lines BP3 and IM9 (Fig. 3). In healthy PBLs, Bfl-1 localization was therefore different from that of Bcl-xL, which was present in both cytosolic and mitochondrial fractions. As described previously, we observed that most of the Bcl-xL protein re-localized to the mitochondria following apoptotic stimuli. Similar to Bcl-xL, Bax was observed in both cytosolic and mitochondrial fractions in healthy PBLs and was mostly found at the mitochondria in etoposide-treated PBLs (Fig. 3a). Of note that in malignant B cell lines Bcl-xL and Bax were mainly localized at the mitochondria as well as Bfl-1 and Bcl-2 (Fig. 3b).
FIGURE 3.
Subcellular localization of endogenous Bfl-1 in human PBLs and tumoral cell lines. a, activated PBLs were treated with 0 μm (0) or 2 μm etoposide (Eto) for 12 h to induce apoptosis. Cytosolic (C) and mitochondrial (M) protein extracts were prepared and protein was assessed by Western blot using anti-Bfl-1, anti-Bcl-xL, and anti-Bax antibodies. Mn-SOD is a marker of mitochondrial fractions. Data are representative of three independent experiments. b, BP3 and IM9 cells were harvested, and cytosolic and mitochondrial protein extracts were prepared, and proteins were assessed by Western blot as in a. Data are representative of three independent experiments.
Helix α9 Mediates Bfl-1 Targeting to the Mitochondria
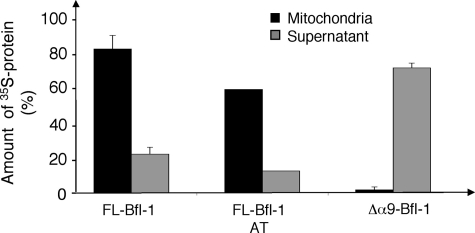
We hereafter investigated the role of helix α9 in the association of Bfl-1 with mitochondria. In a cell-free system, we analyzed the binding of in vitro-translated FL-Bfl-1 and Δα9-Bfl-1 to purified rat liver mitochondria. We observed that 80% of the FL-Bfl-1 associated with mitochondria. This association was not altered by an alkaline treatment of mitochondria pellets, indicating that FL-Bfl-1 is strongly inserted into mitochondria. Deletion of helix α9 abolished the association of Bfl-1 with mitochondria, indicating that this helix mediates Bfl-1 targeting to the mitochondria (Fig. 4).
FIGURE 4.
Importance of helix α9 in the mitochondrial localization of Bfl-1. FL-Bfl-1 or Δα9-Bfl-1 was translated in vitro in the presence of radioactive methionine and then incubated with purified liver rat mitochondrial. After separation by centrifugation, supernatant and mitochondrial pellets were analyzed by SDS-PAGE and quantified using a PhosphorImager. Localization of FL-Bfl-1 was assessed with or without alkali treatment (AT) before separation. Results are expressed as mean ± S.E. of three independent experiments.
The Amphipathic Character of the C-terminal Helix α9 Is Required for Mitochondrial Localization of Bfl-1
One special feature of the C-terminal helix α9 is its amphipathic nature (Fig. 2), a feature already stressed by Ko et al. (25). Amphipathic helices are known to potentially interact with biological membranes and to constitute transmembrane helices or membrane anchors (26). We therefore tested whether the amphiphathic character of the C-terminal helix α9 was required for Bfl-1 anchorage to the mitochondria. Bfl-1 localization was tested, by subcellular fractionation and Western blot analysis, in 293T cells expressing either FL-Bfl-1, Δα9-Bfl-1 proteins, or a mut-FL-Bfl-1 variant that contains the triple mutation I164N/L168E/L171E. This latter triple mutant annihilates the amphipathic nature of the C-terminal helix α9 (Fig. 5a). We observed that when overexpressed in 293T cells, FL-Bfl-1 was localized in both cytosolic and mitochondrial fractions. In agreement with our previous data obtained in the cell-free system, Δα9-Bfl-1 is predominantly localized in the cytosolic fraction (Fig. 5b). Interestingly, the triple mutation I164N/L168E/L171E abrogated mitochondrial localization of Bfl-1. Altogether, these data indicate that when overexpressed in 293T cells, and in contrast to endogenous protein (Fig. 3), FL-Bfl-1 is localized both in the cytosol and at the mitochondria. Furthermore, our data also indicate that the C-terminal helix α9 and especially its amphipathic character is required for Bfl-1 anchorage to the mitochondria.
FIGURE 5.
Role of the amphipathic helix α9 for Bfl-1 subcellular localization. a, helical wheel projection showing the mutated-Bfl-1 α9-helix. An asterisk indicates mutated residues. Hydrophobic residues are shown by black circles, hydrophilic residues are shown by white circles, and other residues are presented by gray circles. b, cytosolic (C) and mitochondrial (M) protein extracts were prepared from transiently transduced 293T cells with FLAG-Δα9-Bfl-1pEGZ, FLAG-FL-Bfl-1pEGZ, or FLAG-mut-FL-Bfl-1pEGZ. Bfl-1 proteins were assessed by Western blot using anti-FLAG antibody. Mn-SOD is a marker of mitochondrial fractions. c, cytosolic and mitochondrial protein extracts were prepared from stably transduced NIH-3T3 cells with FLAG-Δα9-Bfl-1pEGZ, FLAG-FL-Bfl-1pMIG, or FLAG-mut-FL-Bfl-1pMIG. Bfl-1 proteins were assessed by Western blot using anti-FLAG antibody.
The Amphipathic Character of the C-terminal Helix α9 Is Required for Prosurvival Activity of Bfl-1
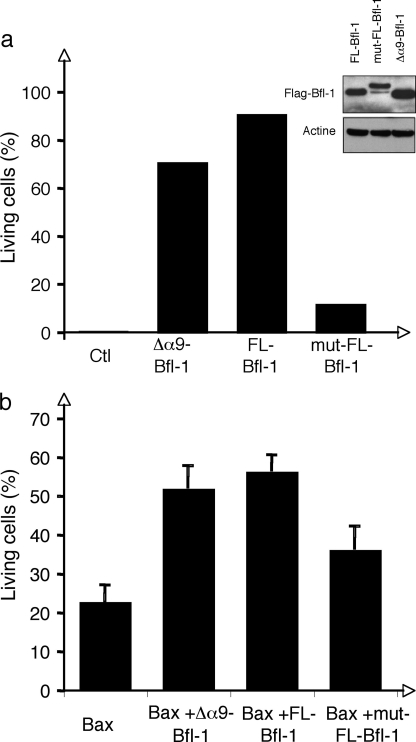
Because deletion of the helix α9 or loss of its amphipathic character affected Bfl-1 mitochondrial localization, we tested, in two experimental models, whether such deletion or mutations have an effect on the prosurvival activity of Bfl-1. First, we analyzed survival of NIH-3T3 cells stably expressing either FLAG-tagged FL-, Δα9-, or mut-FL-Bfl-1 proteins in response to serum withdrawal. We found that both FL- and Δα9-Bfl-1 proteins efficiently protected cells from apoptosis 72 h after serum deprivation (Fig. 6a). In contrast, our data clearly indicated that the triple I164N/L168E/L171E mutation in helix α9 dramatically reduced prosurvival activity of the Bfl-1 protein. Hereafter, the capacity of FL-, Δα9-, or mut-FL-Bfl-1 proteins to rescue Bax-mediated apoptosis in Hela cells was tested. We observed that both FL- and Δα9-Bfl-1 proteins efficiently rescued Hela cells from Bax-mediated apoptosis enabling survival of 50–60% of cells at 48 h. However, and in agreement with results obtained in NIH-3T3 cells, the mut-FL-Bfl-1 variant showed diminished antiapoptotic activity (Fig. 6b). These data support the idea that helix α9 and therefore the binding of Bfl-1 to mitochondria, is not absolutely required for the antiapoptotic activity of Bfl-1. Nonetheless, these results clearly indicate that the amphipathic character of the C-terminal helix α9 regulates the prosurvival function of Bfl-1.
FIGURE 6.
Role of the amphipathic helix α9 for Bfl-1 anti-apoptotic activity. a, NIH3T3 cells were stably transduced with empty vector (Ctl), FLAG-Δα9-Bfl-1pEGZ, FLAG-FL-Bfl-1pMIG, or FLAG-mut-FL-Bfl-1pMIG. Apoptosis was induced by serum deprivation. Cell death was measured by propidium iodide staining and FACS analysis 72 h following serum withdrawal. Data are representative of three independent experiments. Expression of FL-, mut-, and Δα9-Bfl-1 proteins was controlled by Western blot using anti-FLAG antibody and is shown in the inset. b, HeLa cells were transiently transfected with Bax-pEGFP-N1 vector (Bax), Bax-pEGFP-N1 + FLAG-Δα9-Bfl-1pEGZ (Δα9-Bfl-1), Bax-pEGFP-N1 + FLAG-FL-Bfl-1pEGZ (FL-Bfl-1) or Bax-pEGFP-N1 + FLAG-mut-FL-Bfl-1pEGZ (mut-FL-Bfl-1). Cell death was measured by propidium iodide staining at 48 h. Data are presented as percent of living cells among GFP-positive cells. Data are presented as a mean of five (Bax, FL-Bfl-1, and mut-FL-Bfl-1) or two (Δα9-Bfl-1) independent experiments.
As suggested by our molecular modeling (Fig. 1), when the FL-Bfl-1 protein is not attached to the mitochondria, its C-terminal helix α9 may be located to the surface of the hydrophobic groove, thus limiting the access for binding of pro-apoptotic BH3-only proteins and therefore its antiapoptotic activity. To validate this hypothesis, we evaluated binding affinity of the Bim- or Bak-BH3 peptide toward recombinant FL-Bfl-1, mut-FL-Bfl-1, or Δα9-Bfl-1 proteins using a fluorescence polarization assay. As expected, Bim- and Bak-BH3 bound to recombinant Δα9-Bfl-1 protein with an EC50 of 66 and 167 nm, respectively (Table 1). On the contrary, we could not detect any binding of these peptides either with FL-Bfl-1 or mut-FL-Bfl-1. These results suggest that when FL-Bfl-1 protein (either wild-type or mutant) is not anchored to the mitochondrial membrane, it adopts a conformation in which helix α9 is located within the hydrophobic pocket and thereby limits the access BH3 domains of proapoptotic proteins.
TABLE 1.
Fluorescence Polarization Assay analysis of Bfl-1 proteins binding to BH3 peptides
Various concentrations of GST-Δα9-Bfl-1, GST-FL-Bfl-1, and GST-mut-FL-Bfl-1 proteins were incubated with 15 nm fluorescein isothiocyanate-conjugated peptides. Fluorescence polarization was measured after 10 min. The concentrations to achieve 50% maximal binding (EC50) were calculated from the data and are presented in Table 1. These data are representative of three independent experiments.
| Protein | Bim-BH3 | Bak-BH3 |
|---|---|---|
| Δα9-Bfl-1 | 66 nm | 167 nm |
| FL-Bfl-1 | <1 μm | <1 μm |
| mut-FL-Bfl-1 | <1 μm | <1 μm |
Altogether, results obtained by fluorescence polarization assays of offer a rationale for the observation that both FL- and Δα9-Bfl-1 are efficient in protecting NIH-3T3 cells from apoptosis. The hydrophobic groove is indeed fully accessible in Δα9-Bfl-1, but it is also accessible in the FL-Bfl-1 that is anchored to mitochondria membrane through its C-terminal domain. On the contrary, mut-FL-Bfl-1 does not interact with the mitochondria and may adopt a conformation with its helix α9 localized within the hydrophobic groove and therefore prevent its prosurvival property.
DISCUSSION
Structural studies of Bcl-2 family members showing a remarkably similar fold despite an overall divergence in amino acid sequence and function (proapoptotic versus antiapoptotic), have provided important insights into their molecular reaction mechanisms. Molecular modeling of FL-Bfl-1 presented herein indicates that the C-terminal helix α9 may interact with the hydrophobic groove. Similar observations were previously made for the human prosurvival protein Bcl-w (9, 10) and for the proapoptotic protein Bax (27). In accordance with the Bcl-w structure, it has been reported that both endogenous and overexpressed Bcl-w were found in the cytosolic fraction of healthy cell lysates and become inserted into the mitochondrial fraction in response to various apoptotic stimuli (10, 11), following displacement of the C-terminal region by BH3-only proteins. For Bax, it has been demonstrated that its C terminus is not implicated in its mitochondrial localization but plays a role in the dimerization process (28).
In the case of Bfl-1, our model indicates that helix α9 may adopt two distinct conformational states. One is with helix α9 located in the hydrophobic binding groove giving rise to a compact molecule and where it may play a regulatory role by limiting the access to the hydrophobic BH3-binding cleft. In a second model, the molecule is more extended, with helix α9 protruding from the globular core of the protein and therefore potentially involved in interactions with membranes. Interestingly, helical wheel projection of helix α9 brings to the fore its amphipathic character, a characteristic of transmembrane helices or membrane anchors. We demonstrated here that endogenous Bfl-1, in healthy and etoposide-treated PBLs but also in malignant B cell lines, is localized to the mitochondria. Reinforcing this observation, we showed that in cell-free assays, the majority of FL-Bfl-1 is associated with mitochondria and that this association resists an alkaline treatment, thereby indicating a strong insertion of Bfl-1 into the mitochondria. In contrast to FL-Bfl-1, Δα9-Bfl-1 does not associate with mitochondria, demonstrating that helix α9 is absolutely required for Bfl-1 targeting to the mitochondria. Furthermore, as opposed to what has been described for Bax (28) and Bcl-xL (8), the C-terminal tails are not required for dimerization of Bfl-1 (data not shown).
A particular feature of Bfl-1 is the amphipathic character of helix α9. Our results clearly indicate that this property is required for the localization of Bfl-1 to the mitochondria but also for the prosurvival activity of Bfl-1. This amphipathic character was suggested earlier by Ko et al. (25). These authors showed that a peptide corresponding to the C-terminal residues of Bfl-1 (residues 147–175) localizes to the mitochondria and induces mitochondrial permeability transition and cell death, supporting previous reports suggesting that proteolysis or N-terminal deletion of Bfl-1 converts this molecule from an antiapoptotic into a proapoptotic protein (16, 18, 29). Ko et al. (25) suggested that the amphipathic nature of the peptide is essential for its proapoptotic function but not for its localization to the mitochondria. They rather proposed that lysine residues 147 and 151, located within the loop connecting helices α8 and α9, and lysine 172, located in helix α9 (Fig. 1), are critical for targeting of the Bfl-1 C-terminal to the mitochondria. As reported by Ko et al. (25), we confirmed that the triple mutation (E159Q/K163L/E166Q) within helix α9 does not affect mitochondrial localization of Bfl-1. However, we expected this result, as our analysis indicated that these mutations did not affect the amphipathic character of the C-terminal tail of Bfl-1 (supplemental Fig. 1, a and b). Finally, when introduced in the full-length form of the protein, these mutations did not either affect the antiapoptotic function of Bfl-1 (supplemental Fig. 1c).
These data suggest that even though Bfl-1 may co-exist in two distinct conformational states, most of the endogenous Bfl-1 seems to be tightly associated with the mitochondria via its C-terminal end and that the amphipathic nature of helix α9 is critical for that localization. Therefore, Bfl-1 behaves differently from Bcl-w (11) but also behaves differently from Bcl-xL (8) and Mcl-1 (30) that partially exist in the cytosol and also attached to the mitochondria. Following apoptotic stimuli, cytosolic Bcl-w and Bcl-xL are translocated to the mitochondria, whereas Mcl-1 is rapidly degraded by the proteasome.
An interesting observation is that whereas both Δα9-Bfl-1 and mut-FL-Bfl-1 localize in the cytosol, they display different activities. In agreement with previous data, we observe that Δα9-Bfl-1 remains functional (16), but in contrast that mut-FL-Bfl-1 loses its antiapoptotic property. These findings suggest that mut-FL-Bfl-1 putatively adopts a conformation in which helix α9 is located in the hydrophobic binding groove and thereby limits the access. Indeed, when looking at the three-dimensional model of mut-FL-Bfl-1 (results not shown) it appears that the triple mutation I164N/L168E/L171E induces the loss of a few hydrophobic interactions between helix α9 and the groove. However, this loss is largely compensated by the formation of four additional hydrogen bonds located in the C-terminal half of helix α9 and implicating the following residues: Asn-164/Arg-88, Asn-164/Glu-78, Glu-168/Glu-78, and Glu-171/Lys-77. Moreover, due to subtle rearrangements of side chains neighboring the three mutated residues, new hydrophobic contacts could be formed in the N-terminal half of α9. All of these newly established interactions are in favor of helix α9 being more stabilized in the hydrophobic binding groove rather than in an open conformation of mut-FL-Bfl-1, which seems more unlikely. This model is reinforced by fluorescence polarization assays showing that BH3 peptides cannot bind to mut-FL-Bfl-1 or FL-Bfl-1 proteins, suggesting that the hydrophobic groove is not fully accessible in these constructs.
Supplementary Material
Acknowledgments
We thank Y. Leverrier for critical reading of the manuscript. We also thank C. Bella and O. de Bouteiller for expertise with cell sorting and D. Nègre and B. Boson for production of lentivirus particles.
This work was supported by institutional grants from INSERM and Université Claude Bernard Lyon I. This work was also supported by the Institut National du Cancer, the Association pour la Recherche sur le Cancer and the Ligue contre le Cancer (Comité de Savoie, du Rhône et de la Drôme), and the Arthritis Foundation Courtin.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
M. D. Herman, T. Nyman, M. Welin, L. Lehtiö, S. Flodin, L. Trésaugues, T. Kotenyova, A. Flores, and P. Nordlund, unpublished data.
- PBL
- peripheral blood lymphocytes
- FACS
- fluorescence-activated cell sorter
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- FL
- full-length
- aa
- amino acid(s).
REFERENCES
- 1.Cory S., Adams J. M. (2002) Nat. Rev. Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 2.Willis S. N., Adams J. M. (2005) Curr. Opin. Cell Biol. 17, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 4.Kaufmann T., Schlipf S., Sanz J., Neubert K., Stein R., Borner C. (2003) J. Cell. Biol. 160, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lithgow T., van Driel R., Bertram J. F., Strasser A. (1994) Cell Growth & Differ. 5, 411–417 [PubMed] [Google Scholar]
- 6.Akao Y., Otsuki Y., Kataoka S., Ito Y., Tsujimoto Y. (1994) Cancer Res. 54, 2468–2471 [PubMed] [Google Scholar]
- 7.Nguyen M., Millar D. G., Yong V. W., Korsmeyer S. J., Shore G. C. (1993) J. Biol. Chem. 268, 25265–25268 [PubMed] [Google Scholar]
- 8.Jeong S. Y., Gaume B., Lee Y. J., Hsu Y. T., Ryu S. W., Yoon S. H., Youle R. J. (2004) EMBO J. 23, 2146–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denisov A. Y., Madiraju M. S., Chen G., Khadir A., Beauparlant P., Attardo G., Shore G. C., Gehring K. (2003) J. Biol. Chem. 278, 21124–21128 [DOI] [PubMed] [Google Scholar]
- 10.Hinds M. G., Lackmann M., Skea G. L., Harrison P. J., Huang D. C., Day C. L. (2003) EMBO J. 22, 1497–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson-Annan J., O'Reilly L. A., Crawford S. A., Hausmann G., Beaumont J. G., Parma L. P., Chen L., Lackmann M., Lithgow T., Hinds M. G., Day C. L., Adams J. M., Huang D. C. (2003) J. Cell. Biol. 162, 877–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Sa-Eipper C., Chinnadurai G. (1998) Oncogene 16, 3105–3114 [DOI] [PubMed] [Google Scholar]
- 13.Herold M. J., Zeitz J., Pelzer C., Kraus C., Peters A., Wohlleben G., Berberich I. (2006) J. Biol. Chem. 281, 13663–13671 [DOI] [PubMed] [Google Scholar]
- 14.D'Sa-Eipper C., Subramanian T., Chinnadurai G. (1996) Cancer Res. 56, 3879–3882 [PubMed] [Google Scholar]
- 15.Werner A. B., de Vries E., Tait S. W., Bontjer I., Borst J. (2002) J. Biol. Chem. 277, 22781–22788 [DOI] [PubMed] [Google Scholar]
- 16.Kucharczak J. F., Simmons M. J., Duckett C. S., Gélinas C. (2005) Cell Death Differ. 12, 1225–1239 [DOI] [PubMed] [Google Scholar]
- 17.Duriez P. J., Wong F., Dorovini-Zis K., Shahidi R., Karsan A. (2000) J. Biol. Chem. 275, 18099–18107 [DOI] [PubMed] [Google Scholar]
- 18.Ko J. K., Choi K. H., Kim H. J., Choi H. Y., Yeo D. J., Park S. O., Yang W. S., Kim Y. N., Kim C. W. (2003) FEBS Lett. 551, 29–36 [DOI] [PubMed] [Google Scholar]
- 19.Fiser A., Sali A. (2003) Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 20.Roussel A., Cambillau C. (1993) TRUBO-FRODO, Biographics, Architecture et Fonction des Macromolécules Biologiques, Marseille, France [Google Scholar]
- 21.Laskowski R. A., MacArthur M. W., Moss D. S. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 22.Brien G., Trescol-Biemont M. C., Bonnefoy-Berard N. (2007) Oncogene 26, 5828–5832 [DOI] [PubMed] [Google Scholar]
- 23.Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. (1997) Science 275, 983–986 [DOI] [PubMed] [Google Scholar]
- 24.Liu X., Dai S., Zhu Y., Marrack P., Kappler J. W. (2003) Immunity 19, 341–352 [DOI] [PubMed] [Google Scholar]
- 25.Ko J. K., Choi K. H., Pan Z., Lin P., Weisleder N., Kim C. W., Ma J. (2007) J. Cell Sci. 120, 2912–2923 [DOI] [PubMed] [Google Scholar]
- 26.Penin F., Brass V., Appel N., Ramboarina S., Montserret R., Ficheux D., Blum H. E., Bartenschlager R., Moradpour D. (2004) J. Biol. Chem. 279, 40835–40843 [DOI] [PubMed] [Google Scholar]
- 27.Suzuki M., Youle R. J., Tjandra N. (2000) Cell 103, 645–654 [DOI] [PubMed] [Google Scholar]
- 28.Er E., Lalier L., Cartron P. F., Oliver L., Vallette F. M. (2007) J. Biol. Chem. 282, 24938–24947 [DOI] [PubMed] [Google Scholar]
- 29.Yang W. S., Ko J. K., Park S. O., Choi H. Y., Kim Y. N., Kim C. W. (2005) J. Cell. Biochem. 94, 1234–1247 [DOI] [PubMed] [Google Scholar]
- 30.Nijhawan D., Fang M., Traer E., Zhong Q., Gao W., Du F., Wang X. (2003) Genes Dev. 17, 1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.