Abstract
Ligand-dependent corepressor LCoR interacts with the progesterone receptor (PR) and estrogen receptor ERα in the presence of hormone. LCoR contains tandem N-terminal PXDLS motifs that recruit C-terminal-binding protein (CtBP) corepressors as well as a C-terminal helix-turn-helix (HTH) domain. Here, we analyzed the function of these domains in coregulation of PR- and ERα-regulated gene expression. LCoR and CtBP1 colocalize in nuclear bodies that also contain CtBP-interacting protein CtIP and polycomb group repressor complex marker BMI1. Coexpression of CtBP1 in MCF7 or T47D breast cancer cells augmented corepression by LCoR, whereas coexpression of CtIP did not, consistent with direct interaction of LCoR with CtBP1, but not CtIP. The N-terminal region containing the PXDLS motifs is necessary and sufficient for CTBP1 recruitment and essential for full corepression. However, LCoR function was also strongly dependent on the helix-turn-helix domain, as its deletion completely abolished corepression. LCoR, CtBP, and CtIP were recruited to endogenous PR- and ERα-stimulated genes in a hormone-dependent manner. Similarly, LCoR was recruited to estrogen-repressed genes, whereas hormone treatment reduced CtBP1 binding. Small interfering RNA-mediated knockdown of LCoR or CtBP1 augmented expression of progesterone- and estrogen-stimulated reporter genes as well as endogenous progesterone-stimulated target genes. In contrast, their ablation had gene-specific effects on ERα-regulated transcription that generally led to reduced gene expression. Taken together, these results show that multiple domains contribute to LCoR function. They also reveal a role for LCoR and CtBP1 as attenuators of progesterone-regulated transcription but suggest that LCoR and CtBP1 can act to enhance transcription of some genes.
The progesterone receptor (PR)6 and estrogen receptor α (ERα) are members of the nuclear receptor superfamily of hormone-regulated transcription factors whose functions have been implicated in a broad range of physiological responses (1–7). During activation of gene transcription, agonist-bound nuclear receptors recruit so-called coactivators such as p160 proteins (2, 8–10). Coactivators act in part by functioning as histone acetyltransferases (HATs) or by recruiting HAT activity to target promoters. Notably, screens designed to identify coactivators revealed that agonist-bound nuclear receptors also recruit proteins with corepressor properties, such as nuclear receptor-interacting protein 1 (RIP140/NRIP1) (11–14) or LCoR (11, 15). We identified LCoR as a protein that interacted with the estrogen-bound ligand binding domain of ERα. Transiently expressed LCoR repressed hormone-dependent transactivation by a range of nuclear receptors. This included particularly efficacious repression of progesterone-stimulated transcription. LCoR transcripts are detectable as early as the two-cell stage of embryonic development, and the protein is robustly expressed in numerous fetal and adult tissues, including progesterone and estrogen target tissues such as breast, endometrium, and placenta (15).
LCoR acts as a scaffold for both histone deacetylases (HDACs) and C-terminal binding protein (CtBP) corepressors and functions by both HDAC-dependent and -independent mechanisms in a receptor-specific manner. For example, whereas the HDAC inhibitor trichostatin A abolished LCoR-dependent corepression of estrogen-stimulated reporter gene expression, PR-stimulated expression was largely resistant to trichostatin A (15). LCoR contains two adjacent sequences, (P/V)XLDLX(K/R) or (P/V)XLDLXXK, near its N terminus that correspond to extended PXDLS consensus motifs recognized by CtBPs (13). CtBP1 was identified as a protein that bound the C-terminal region of human adenoviral E1A (16) and is the founding member of a well conserved family of proteins (17–19). Highly homologous CtBP2 was identified from EST databases (20). Remarkably, CtBPs bind NAD(H) and are related to 2-hydroxy acid dehydrogenases, and CtBP1 has weak dehydrogenase activity. In addition, dinucleotide binding stimulates protein oligomerization activity and corepression.
CtBPs interact directly with several transcriptional coregulatory proteins, many of which share the PXDLS motifs described above (13, 17, 21). For example, a screen for CtBP cofactors identified CtBP interacting protein (CtIP) (22), which also binds BRCA1 and retinoblastoma gene product (Rb) tumor suppressor proteins (23, 24). Like LCoR, CtBPs can function by HDAC-dependent or -independent mechanisms depending on the promoter tested (17). They are components of several multisubunit assemblies, including polycomb repressor PRC1 complexes (25–27). Targeted ablation of CtBP1 or -2 expression in mice revealed that the two proteins play important and overlapping roles in mouse development (19, 28).
In addition to its N-terminal CtBP interaction motifs and a central HDAC binding domain (29), LCoR contains a C-terminal helix-turn-helix (HTH) domain that is homologous to motifs encoded by the Eip93F (CG18389 gene product from transcript CG18389-Rb) and MBLK-1 genes of Drosophila and Honeybee (Apis mellifera) (30), respectively. The LCoR HTH domain also bears 35% homology to pipsqueak motifs (PSQ). PSQ motifs are repeated four times in the DNA binding region of the Drosophila pipsqueak transcription factor, which plays a role in gene silencing (31). Multiple repeats of the domain are required for PSQ DNA binding (31), and mutation of one of the two HTH motifs in the MBLK-1 gene strongly reduced site-specific DNA binding (30). The PSQ domain is homologous to unique motifs found in a number of prokaryotic and eukaryotic proteins that interact with DNA, such as recombinases (31, 32), raising the possibility that LCoR itself may interact with DNA. Other studies have shown that HTH domains can function in protein-protein interactions, where the HTH motif, combined with other domains, can induce formation of multisubunit complexes (33). Such proteins with multidomains can act as scaffolds between the basal transcription machinery and transcription factors (33).
The accompanying paper showed that LCoR recruits HDAC6 through a central domain (29). In this study we have analyzed the roles of domains controlling recruitment of CtBPs and the C-terminal HTH motif in corepression by LCoR. We were primarily interested in determining the roles of these domains in corepression of the PR, as our previous work showed that the efficacious corepression of PR-driven gene reporter gene expression by LCoR appeared to be largely insensitive to HDAC inhibition (15). We find that both the PXDLS motifs and the HTH domain are required for corepression of both the PR and ERα, as disruption of either region markedly attenuated LCoR function. LCoR colocalizes with CtBP1 and CtIP in nuclear foci, and CtBP1 is corecruited with LCoR to PR and ERα target genes in a hormone-dependent manner. Ablation of LCoR and/or CtBP1 enhanced progesterone-stimulated gene expression in T47D breast cancer cells. In contrast, loss of LCoR or CtBP1 had gene-specific effects on ERα-regulated genes and generally led to reduced target gene expression.
EXPERIMENTAL PROCEDURES
Antibodies
A rabbit polyclonal antipeptide antibody was raised against LCoR amino acids 20–36 (QDPSQPNSTKNQSLPKA) fused to keyhole limpet hemocyanin and purified over a peptide affinity column (Bethyl Laboratories, Montgomery, TX). Rabbit polyclonal α-CtBP (sc-11390), goat polyclonal α-CtBP1 (sc-5963), goat polyclonal α-CtIP (sc-5970), goat polyclonal α-Bmi1 (sc-8906), rabbit polyclonal α-Bmi1 (sc-10745), mouse monoclonal glyceraldehyde-3-phosphate dehydrogenase (sc-69778), rabbit polyclonal ERα (sc-543), rabbit polyclonal OCTA-Probe (sc-807), goat anti-mouse IgG (sc-2005), goat anti-rabbit IgG (sc-2004), normal mouse IgG (sc-2025), normal rabbit IgG (sc-2027), and protein A-agarose (sc-2001) and protein G Plus-agarose (sc-2002) were from Santa Cruz Biotechnology (Santa Cruz, CA). Cy3-donkey polyclonal α-goat (705-165-147) and Cy2-goat polyclonal α-rabbit (711-225-152), Cy3-donkey polyclonal α-rabbit (711-165-152), and Cy2-donkey polyclonal α-mouse (715-225-150) were purchased from Jackson ImmunoResearch (West Grove, PA). Mouse monoclonal α-FLAG M2 (F3165) and α-FLAG M2 horseradish peroxidase (HRP)-conjugate (A-8592), monoclonal α-rabbit HRP conjugate (A2074), and rabbit polyclonal α-goat HRP conjugate (A5420) were from Sigma. Rabbit polyclonal CtBP1 (07-306) was purchased from Millipore (Temecula, CA). Rabbit polyclonal LCoR (18-003-44018) was purchased from GenWay Biotech (San Diego, CA). Rabbit polyclonal PR (ab68195) and rabbit polyclonal CtIP (ab70163) were purchased from Abcam (Cambridge, MA).
Recombinant Plasmids
PSG5/LCoR, FLAG-LCoR/pcDNA3.1 and LCoR derivatives mutagenized in the CtBP binding motifs, PLDLTVR (LCoR amino acids 64–70; m1) and VLDLSTK (LCoR amino acids 82–88; m2), and the double mutant (m1m2) have been described (3). cDNAs mutated in the CtBP binding motifs were subcloned downstream of FLAG in pCDNA3.1. FLAG-LCoRΔHTH/pcDNA3.1 was made using the QuikChange mutagenesis kit (Stratagene 200518, La Jolla, CA) as per the manufacturer's instructions. Primers were designed to delete amino acids 350–395 from LCoR. The new construct was sequenced to confirm proper deletion LCoR.
Cell Culture and Transfections
All cells were cultured under the recommended conditions. For immunocytochemistry, MCF7 cells grown on collagen IV-treated microscope slides in 6-well plates in DMEM supplemented with 10% FBS. Cells were prepared for immunocytochemistry as described below. For immunoprecipitation of tagged proteins, MCF7 cells in 100-mm dishes were transfected with 10 μl of Lipofectamine containing 10 μg of pSG5 vectors containing FLAG-LCoR, FLAG-m1, FLAG-m2, or FLAG-m1 + m2. For analysis of the effects of CtBP1 and CtIP on LCoR corepression, MCF7 cells (60–70% confluent) were grown in DMEM without phenol red supplemented with 10% FBS on 6-well plates. Cells were transfected in medium without serum (Opti-MEM (Invitrogen) with Lipofectamine 2000 (Invitrogen). The amounts of expression vectors were as follows: 100 ng of ERα or human PR expression vectors (as indicated), 100 ng of LCoR/pcDNA3.1, 250 ng of ERE3-TATA/pXP2 or pGRE5/pXP2 reporter plasmid, 250 ng of internal control vector pCMV-βgal. Quantities of expression vectors used are indicated in the legends to Figs. 3 and 5. The medium was replaced 18 h after transfection by a medium containing charcoal-stripped serum and ligand (10 nm) for 30 h as indicated. For the Luciferase reporter assay, cells were harvested in 250 μl of reporter lysis buffer (Promega).
FIGURE 3.
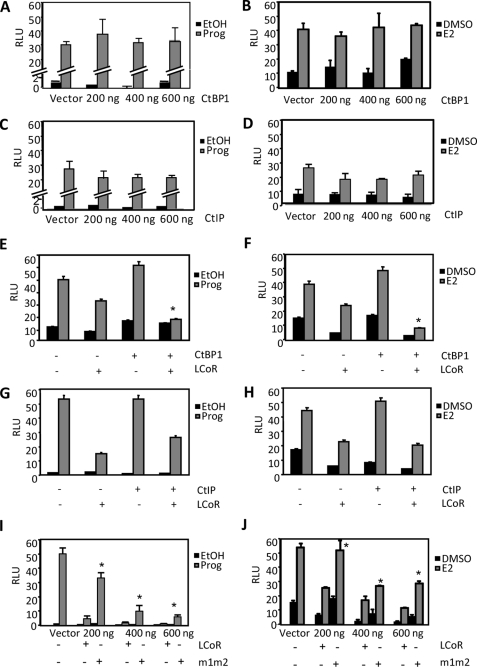
Roles of CtBP1 and CtIP in LCoR-dependent corepression in MCF7 cells. Cells were transiently transfected with expression vectors for either PR (100 ng) or ERα (100 ng) and their corresponding reporter plasmids (250 ng) for 18 h. Media was then changed, and cells were treated with vehicle or hormone for 30 h. A and B, shown are dose-response curves analyzing the effects of CtBP1 on reporter gene expression in cells treated with P4 (10 nm; A) or E2 (10 nm; B). Increasing amounts of CtBP1 were transfected (200, 400, and 600 ng). C and D, shown are dose-response curves analyzing the effects of CtIP on reporter gene expression in cells treated with P4 (C) or E2 (D). Increasing amounts of CtIP1 were transfected (200, 400, and 600 ng). E and F, shown is analysis of the effects of coexpression of LCoR and CtBP1 on hormone-dependent gene expression. Cells were transiently transfected with either vector alone, LCoR alone (100 ng), CtBP1 alone (200 ng), or with both LCoR and CtBP1 and treated with P4 (E) or E2 (F). *, p < 0.05 for results of LCoR and CtBP1 coexpression versus LCoR expression alone. G and H, shown is analysis of the effects of coexpression of LCoR and CtIP on hormone-dependent gene expression. Cells were transiently transfected with either vector alone, LCoR alone, CtIP1 alone, or with both LCoR and CtIP1 and treated with P4 (G) or E2 (H). I and J, shown are dose-response curves of either LCoR or m1m2 in cells treated with P4 (I) or E2 (J). Increasing amounts of wild-type or mutant LCoR were transfected (200, 400, and 600 ng). *, p < 0.05 for results of corresponding wild-type LCoR versus mutant form m1m2. RLU, relative luciferase units.
FIGURE 5.
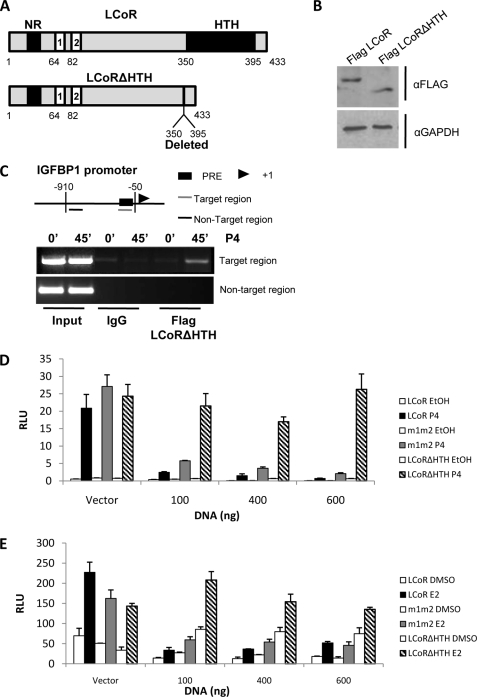
The HTH domain of LCoR is essential for corepression. A, shown are schematic representations of full-length LCoR (upper panel) and a deletion mutant lacking the HTH domain (lower panel; LCoRΔHTH) in which amino acids 350–395 were deleted, leaving the C-terminal portion of the protein (395–433 amino acids) intact. B, LCoR and LCoRΔHTH are expressed equally in T47D cells. Shown is a Western blot of T47D cell extracts expressing FLAG-LCoR or FLAG-LCoRΔHTH blotted for FLAG (first row) or loading control glyceraldehyde-3-phosphate dehydrogenase (GAPDH, second row). C, LCoRΔHTH was recruited to the progesterone target gene encoding IGFBP1. ChIP assays in T47D cells treated with P4 (10 nm) for 45 min and immunoprecipitated with FLAG are shown. The upper panel shows a schematic view of IGFBP1 promoter. PRE, progesterone response element. D and E, deletion of HTH domain of LCoR abolishes corepressor function is shown. Dose-response curves were performed analyzing corepression in the presence of increasing amounts of LCoR or LCoRΔHTH expression vectors (0, 100, 400, and 600 ng, as indicated). T47D cells were treated with P4 (10 nm; panel D) and MCF7 cells were treated with E2 (10 nm; panel E). Cells were transiently transfected with expression vectors of either PR (100 ng) or ERα (100 ng) and their corresponding reporter plasmid (250 ng) for 18 h. Media was then changed, and cells were treated for 30 h. RLU, relative luciferase units.
Immunocytochemistry and Immunoprecipitations
Cells were cultivated on collagen IV-treated microscope slides in 6-well plates, fixed with 2% paraformaldehyde for 15 min at room temperature, washed (3×) with phosphate-buffered saline and permeabilized with 0.2% Triton X-100, 5% BSA, 10% horse serum in phosphate-buffered saline. MCF7 cells were then incubated with α-LCoR (1:500) and goat polyclonal antibodies against CtBP1, CtIP, or Bmi1 (1:50) in buffer B (0.2% Triton X-100, 5% BSA in phosphate-buffered saline) for 1 h at room temperature. Cells were washed (3×) with phosphate-buffered saline and incubated with goat anti-rabbit-Cy2 and donkey anti-goat Cy3 (1:300) in buffer B for 1 h at room temperature. Slides were mounted with Immuno-Fluore Mounting Medium (ICN, Aurora, Ohio) and visualized using a Zeiss LSM 510 confocal microscope.
For immunoprecipitation of endogenous CtBP, CtIP, or Bmi1, MCF7 cells in 150-mm dishes were lysed for 3 min at 4 °C in 1 ml of LB (150 mm NaCl, 10 mm Tris-HCl, pH 7.4, 0.2 mm sodium orthovanadate, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 0.5% IGEPAL CA-630, protease inhibitor mixture). Cell debris was pelleted by centrifugation (14,000 rpm, 5 min), and proteins were immunoprecipitated with 4 μg of αCtBP or αCtIP or polyclonal rabbit αBMI1 or control rabbit or goat IgG at 4 °C overnight followed by 2 h of incubation at 4 °C with protein A-agarose (for αCtBP, αBmi1, control rabbit IgG) or protein A+G-agarose (for αCtIP or control goat IgG). Beads were washed (3×) with LB. Bound immunocomplexes were boiled in Laemmli buffer, separated by 10% SDS/PAGE, blotted on polyvinylidene difluoride membranes with α-LCoR (1/1000), α-CtBP1, α-CtBP2, α-CtIP, or α-BMI1 (1:100), and detected by enhanced chemiluminescence (PerkinElmer Life Sciences). For immunoprecipitation of tagged proteins, transfected MCF7 cells were lysed for 30 min at 4 °C in 1 ml of LB, 48 h after transfection. Supernatants were cleared and incubated overnight with 4 μg of αCtBP or α-FLAG M2 antibody followed by a 2-h incubation with protein A-agarose or protein A+G-agarose beads, respectively. Beads were washed (3×) with LB and Western-blotted as above. Dilutions of specific antibodies used for Western blotting were α-CtBP1 (1:100) and α-FLAG M2-peroxidase (1:100).
Western Blotting
A Western blot was performed as previously described (34) using MCF7 cells extracts. Cells were grown in 10-cm dishes (70% confluent) and transiently transfected with 500 ng of FLAG-tagged full-length of mutated LCoR. Cells were harvested 30 h later.
Chromatin Immunoprecipitation (ChIP) Assays and reChIP Assays
ChIP and reChIP assays were performed as previously described (35) in MCF7 and T47D cells. Cells were grown in 10-cm dishes (70% confluent) and transiently transfected with 500 ng of FLAG-tagged LCoR (or FLAG-tagged LCoRΔHTH as indicated). After transfection, cells were starved for 2 days in DMEM-phenol free and FBS-free media and treated with 2.5 μm α-amanitin (Sigma, A2263) for 2 h before hormone treatment to properly synchronize cells. Cells were collected, and cofactor recruitment was evaluated on promoter regions containing either estrogen or progesterone response element (as indicated) of target genes. For ChIP primers sequences, please refer to supplemental Table 1.
siRNA Knockdowns
siRNAs were purchased from Thermo Scientific Dharmacon (Lafayette, CO). The following ON-TARGETplus SMART pool siRNAs were used: LCoR (L-026303-00), CtBP1 (L-008609-00), CyPB (D-001820-10), and non-targeting (D-001818-10). siRNAs were resuspended per the manufacturer's instructions. Transfections were done in 6-well plates as described previously. Lipofectamine 2000 (10 μl) was used as the transfection reagent. DMEM phenol-free with 10% stripped FBS was added 12 h after transfection. For Western blot analysis, cells were collected 48 h after transfection. Luciferase reporter assays after siRNA knockdowns were performed as follows. 100 ng of ERα expression vector and 250 ng of ERE3-TATA-pXP2 vector were transfected with the corresponding siRNA. DMEM phenol-free with 10% stripped FBS was added 12 h after transfection. Ligand was added 36 h after transfection, and cells were collected 24 h later. Luciferase activity was measured as previously described.
RNA Isolation, cDNA Synthesis, and Quantitative Real-time (qRT)- PCR
Cells were grown in 100-mm dishes. Media were replaced with charcoal-stripped medium-containing ligand. Total RNA was extracted with TRIZOL reagent. cDNA synthesis was performed with iScript cDNA synthesis kit (Bio-Rad) according to the instructions of the manufacturer. The MiniOpticon real-time PCR system with the iQ SYBR Green Supermix (Bio-Rad) was used for qRT-PCR expression analysis of target genes. The program used was as follows: 1) incubation at 94 °C for 60 s, 2) incubation at 95 °C for 20 s, 3) incubation at 60 °C for 30 s (decreasing temperature by 1° per cycle), 4) incubation at 72 °C for 30 s, 5) plate reading, 6) repetition from step 2 five more times, 7) incubation at 95 °C for 20 s, 8) incubation at 57.5 °C for 30 s, 9) incubation at 72 °C for 30 s, 10) plate reading, 11) repetition from step 7 thirty-five more times, 12) performance of melting curve and end. Results were normalized to β-actin mRNA expression. For qRT-PCR primers sequences, please refer to supplemental Table 2.
RESULTS
Association of LCoR with CtBP1 and CtIP
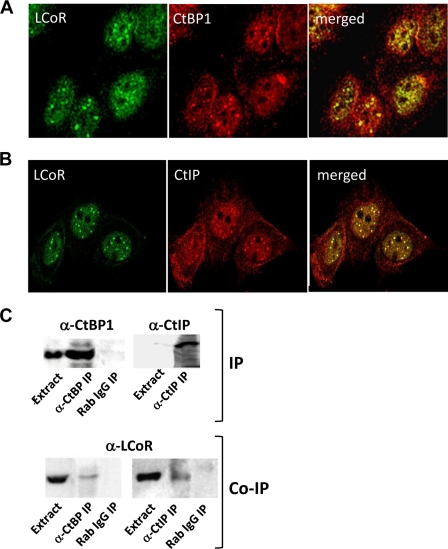
Colocalization of LCoR with CtBP1 in MCF7 cells was confirmed by immunocytochemical analyses (Fig. 1A). Both proteins were broadly distributed in the nucleus and were also concentrated in discrete nuclear bodies. A similar colocalization of CtBP2 and LCoR was also observed (data not shown). Given the extensive overlap of CtBPs and LCoR, we determined whether LCoR colocalized with CtIP, which was identified as a CtBP-interacting protein containing an extended PXDLS motif (22). Similar to results obtained with CtBP, CtIP and LCoR showed strongly overlapping patterns of expression concentrated in discrete nuclear bodies (Fig. 1B). In addition, endogenous LCoR coimmunoprecipitates with antibodies directed against either CtBPs or CtIP (Fig. 1C).
FIGURE 1.
The association of endogenous LCoR with endogenous CtBP1 and CtIP (A and B) is shown. Confocal microscopic analysis of the subcellular colocalization of LCoR with CtBP1 (A) and CtIP (B) by immunocytochemistry (see “Experimental Procedures” for details) is shown. C, shown is the analysis of the association of LCoR with CtBP1 and CtIP by coimmunoprecipitation (Co-IP). Extracts of MCF7 cells were immunoprecipitated (IP) with antibodies against CtBP1 or CtIP as indicated and probed by Western blotting for enrichment of target proteins in immunoprecipitates. Immunoprecipitates were also probed for the coimmunoprecipitation of LCoR.
Other studies have shown that CtIP interacts directly with the retinoblastoma gene product (22) and link CtBP1 and Rb to polycomb group repressor complexes (26, 36). PRC1 complexes form large foci containing numerous factors, including BMI1 polycomb ring finger oncogene (BMI1), visible as discrete nuclear structures (37). Indeed, we found that BMI1 and LCoR coimmunoprecipitated and colocalized in nuclear bodies (supplemental Fig. 1). Taken together, these studies show that LCoR extensively colocalizes with CtBP1 and CtIP in the nucleus, including in PRC1 complexes.
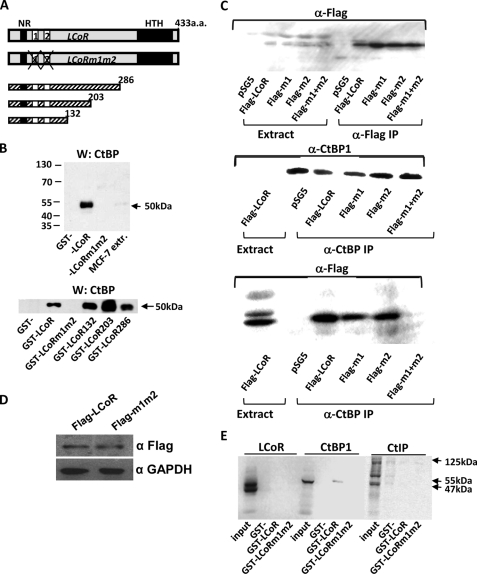
The specificity of the interactions of LCoR with CtBP1 were further analyzed in vitro using glutathione S-transferase (GST) fused to a series of C-terminal deletion mutants of LCoR or LCoR mutant m1m2, which lacks the tandem PXDLS motifs (Fig. 2A). CtBP1 bound to full-length LCoR (Fig. 2B, top) and all C-terminal deletion mutants (Fig. 2B, bottom) but not to LCoR m1m2 (Fig. 2A). Moreover, tagged LCoR mutated in either one of the two PXDLS motifs coimmunoprecipitated with endogenous CtBPs from extracts of MCF7 cells (m1, amino acids 64–70 deleted; m2, amino acids 82–88 deleted; Fig. 2C, bottom panel). In contrast, no coimmunoprecipitation was observed in cells expressing the m1m2 derivative lacking both sites (Fig. 2C, bottom), although the wild-type and m1m2 proteins were expressed at similar levels (Fig. 2D). Taken together, these results show that the integrity of at least one of the two PXDLS motifs of LCoR is required for binding to CtBP1.
FIGURE 2.
Direct association of LCoR with CtBP1 but not CtIP. A, shown are a schematic representation of the primary structure of LCoR along with LCoR mutated in the PXDLS motifs that bind CtBPs (m1m2) and C-terminal deletion mutants of LCoR. aa, amino acids. B, shown is GST pulldown analysis of the interaction of CtBP1 present in MCF7 cell extracts (MCF7 extr.) with GST fusions of wild-type LCoR or mutants described in A. CtBP1 bound to fusion proteins was detected by Western (W) blotting. C, mutation of both CtBP binding sites of LCoR disrupts its interaction with CtBPs in MCF7 cell extracts. MCF7 cells were transfected with FLAG-tagged wild-type LCoR or tagged LCoR mutated in one (m1 or m2) or both (m1m2) CtBP binding sites, as indicated. Top panel, extracts and immunoprecipitations (IP) with the anti-FLAG antibody of transfected MCF7 cells show that tagged proteins are expressed at similar levels in all cases. Middle panel, control immunoprecipitation with anti-CtBP1 antibody and Western blots shows that CtBP1 is expressed at similar levels in all cases. Bottom panel, shown is coimmunoprecipitation of tagged LCoR derivatives from extracts of transfected MCF7 cells. D, shown are Western blots of MCF7 extracts expressing FLAG-LCoR or FLAG-m1m2 blotted for FLAG (first row) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; second row), used as the loading control. E, GST pulldown assays show no direct binding between LCoR and CtIP. Binding of CtBP1 to GST-LCoR fusions was used as a positive control.
We also analyzed the association of LCoR with CtIP by GST pull down assay. The coimmunoprecipitation of CtIP and LCoR is remarkable given that CtIP and LCoR interact with CtBPs through common motifs. However, no evidence was found for LCoR binding directly to CtIP in vitro (Fig. 2E), suggesting that their association in cells is indirect.
CtBP1 but Not CtIP Augments LCoR-dependent Corepression
The function of CtBP1 and CtIP as corepressors of progesterone- or estrogen-regulated gene expression in the absence or presence of LCoR was analyzed in MCF7 (Fig. 3) and T47D (supplemental Fig. 2) breast cancer cells. Neither protein repressed (hormone-dependent) gene expression in MCF7 cells in the absence of LCoR when coexpressed in dose-response experiments with either the PR or ERα (Fig. 3, A–D). On the other hand, cotransfection of a limiting amount (200 ng) of a CtBP1 expression vector (Fig. 3, E and F) significantly augmented LCoR-dependent corepression, whereas coexpression of CtIP had no effect on LCoR function (Fig. 3, G and H). This is consistent with the direct interaction of CtBP1, but not CtIP, with LCoR, observed above. In agreement with a role of CtBP recruitment in LCoR function, corepression of progesterone- or estrogen-induced gene expression by the m1m2 mutant of LCoR lacking the CtBP binding motifs was markedly attenuated (Fig. 3, I and J). Similar results were obtained in T47D cells (supplemental Fig. 2).
CtBP1 Is Corecruited with LCoR to PR- and ERα-stimulated Target Genes in the Presence of Hormone
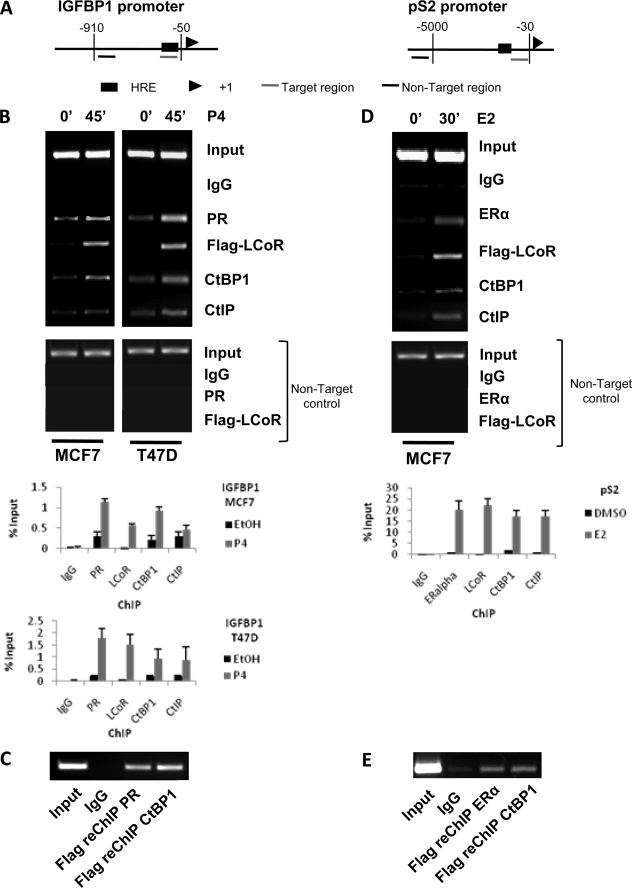
The binding of LCoR and CtBP1 to hormone-responsive promoters was also analyzed by ChIP assay. As we do not have an antibody that immunoprecipitates endogenous LCoR efficiently, LCoR was expressed as a FLAG-tagged protein in these experiments. Treatment of T47D or MCF7 cells with hormone induced PR binding to the progesterone response element of the insulin-like growth factor binding protein 1 (IGFBP1) promoter (Fig. 4, A–C, left-hand side). Similarly, LCoR and CtBP1 as well as CtIP were recruited to the promoter under the same conditions in both cell lines. Non-target controls showed that the binding of cofactors and nuclear receptors was specific for their corresponding hormone response elements (Fig. 4, A and B). In re-ChIP experiments, proteins from progesterone-treated T47D cells were immunoprecipitated with an anti-FLAG antibody to concentrate LCoR and were re-immunoprecipitated with antibodies against either the PR or CtBP1 (Fig. 4C), which confirmed that LCoR is recruited to the same promoters in vivo as the PR and CtBP1. Similar results were obtained when recruitment of ERα, LCoR, CtBP1, and CtIP to the estrogen-responsive trefoil factor 1 (pS2) promoter was analyzed in MCF7 cells (Fig. 4, A, right-hand side, D, and E). Note that essentially identical results were obtained in multiple biological replicates with both cell lines.
FIGURE 4.
ChIP and reChIP assays of protein association with progesterone and estrogen target genes. MCF7 and T47D cells were transiently transfected with FLAG-tagged LCoR, synchronized for 2 h with α-amanitin (2.5 μm), and treated with P4 (10 nm) for 45 min or E2 (10 nm) for 30 min, as indicated. Cell extracts were collected and immunoprecipitated with IgG or antibodies against PR, ERα, FLAG, CtBP1, or CtIP, as indicated. A, shown is a schematic representation of the progesterone-sensitive IGFBP1 promoter (left-hand side) and estrogen-stimulated pS2 promoter (right-hand side). The hormone response element (HRE), transcription start site, and both PCR-amplified sequences (target and non-target control regions) are depicted. B, shown is a ChIP assay of factor binding to the IGFBP1 promoter in MCF7 cells (left-hand side) and T47D cells (right-hand side). Results of semiquantitative and quantitative PCR analyses are presented. Note that no signal was detected by quantitative PCR in the non-target controls. C, shown are reChIP assay in the extracts of T47D cells treated with P4 and immunoprecipitated with FLAG. A second round of immunoprecipitations with IgG, PR, or CtBP1 was performed, as indicated. D, ChIP assays of factor binding to the pS2 promoter in MCF7 cells treated with E2 are shown. E, a reChIP assay in extracts of MCF7 cells treated with E2 and immunoprecipitated with FLAG is shown. A second round of immunoprecipitations with IgG, ERα, or CtBP1 was performed, as indicated.
The recruitment of CtIP is intriguing, given its indirect association with LCoR and the lack of effect of CtIP on hormone-dependent transactivation either in the presence or absence of LCoR (Fig. 3). The data raise the possibility that at least a portion of CtIP may be recruited to hormone-responsive promoters through its colocalization with LCoR in PRC1 complexes. Consistent with this hypothesis, we found that PRC1 marker BMI1 was also recruited to the pS2 promoter in the presence of estradiol (supplemental Fig. 1).
Deletion of the HTH Domain of LCoR Abolishes Corepressor Function
Function of the C-terminal HTH motif of LCoR in corepression of progesterone- and estrogen-regulated gene expression was analyzed by deletion of the domain (LCoRΔHTH; Fig. 5A) and comparison of the function of the resulting mutant to full-length LCoR. Western blotting and ChIP assays showed that the ΔHTH mutant was expressed at similar levels in T47D cells as full-length LCoR and that it was recruited to the IGFBP1 promoter in the presence of progesterone (Fig. 5, B and C). Remarkably, however, the LCoRΔHTH protein was essentially devoid of corepressor activity on either progesterone- or estrogen-responsive promoters in transient expression experiments (Fig. 5, D and E). In similar studies (not shown) corepressor activity of the LCoRΔHTH mutant was even more attenuated than that of LCoR derivative m1m2 lacking PXDLS motifs, identifying the HTH region as being critical to LCoR corepressor function.
Function of LCoR and CtBP1 as Attenuators of Progesterone-regulated Gene Expression
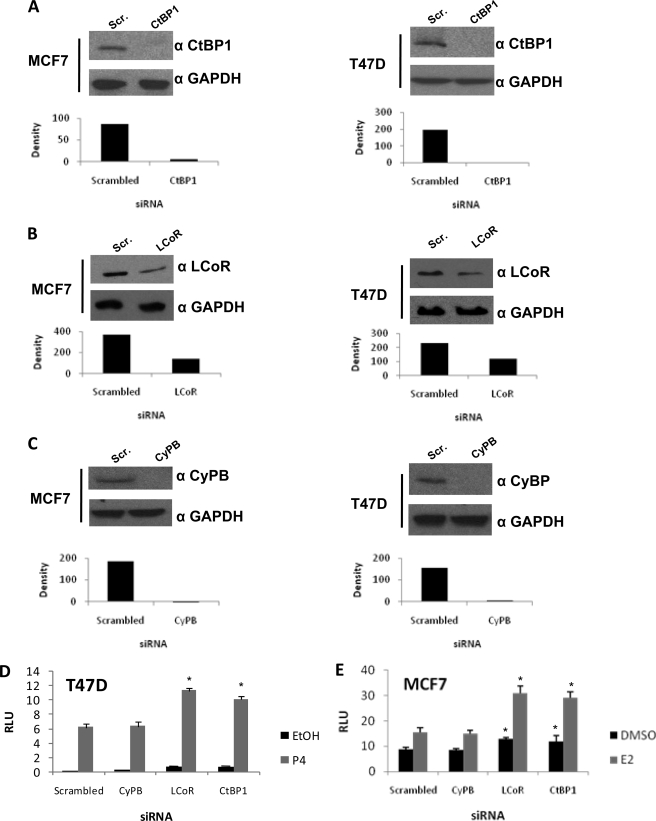
To further address the function of LCoR and CtBP1 in regulating hormone-dependent gene transcription, we knocked down expression of both proteins using siRNAs in T47D and MCF7 cells. Knockdown of cyclophilin B was used as the control for off-target effects of siRNA in these studies (Fig. 6, A–C). Loss of either LCoR or CtBP1 augmented progesterone-induced reporter gene expression in T47D cells (Fig. 6D) and estrogen-stimulated reporter expression in MCF7 cells (Fig. 6E), consistent with a role in corepression of hormone-inducible gene transcription.
FIGURE 6.
siRNA knockdown of LCoR and CtBP1 expression in MCF7 and T47D cells. A, B, and C, Western blots of MCF7 and T47D extracts are shown. Cells were transfected for 48 h with pools of scrambled siRNAs (Scr.) or siRNAs targeting LCoR (A) or CtBP1 (B) as well as siRNAs targeting CyPB (C) to control for off-target effects. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used as a control. Densities of bands on Western blots, scanned with Bio-Rad Gel Doc XR system and analyzed with Quantity One, are presented below the images of the blots. D and E, luciferase reporter assays in siRNA-transfected cells. A PR expression vector was transfected along with scrambled, LCoR, CtBP1, or CyPB siRNAs (D), or an ERα expression vector was transfected along with scrambled, LCoR, CtBP1, or CyPB siRNAs (E). After 24 h of treatment with P4 (10 nm; D) or E2 (10 nm; E), cells were harvested, and luciferase activity was measured. Data are shown as relative luciferase units (RLU). Data are the averages of three or more independent experiments, and error bars represent the S.E.; *, p < 0.05 for results of specific knockdown (LCoR, CtBP1, or CyPB) versus results with scrambled siRNA.
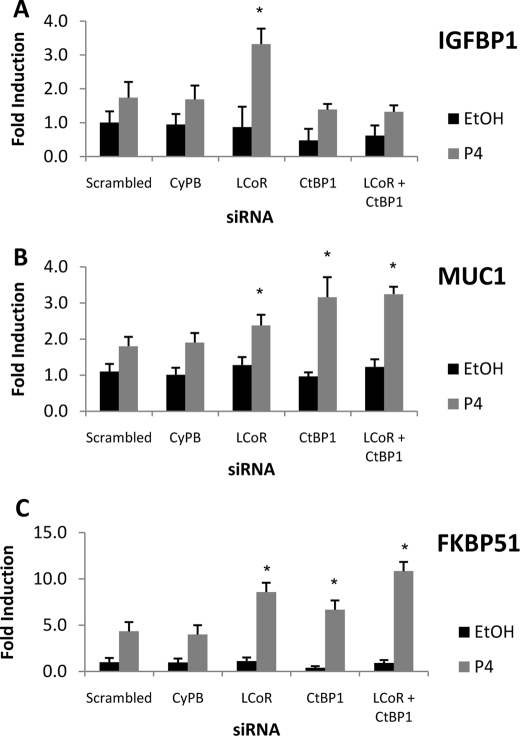
We extended this analysis to the regulation in T47D cells of progesterone target genes encoding IGFBP1, mucin 1 (MUC1) and FK506-binding protein 5 (FKBP51) (4, 38, 39) (Fig. 7). Knockdown of LCoR markedly enhanced progesterone-stimulated expression of the IGFBP1 gene (Fig. 7A). Unexpectedly, ablation of CtBP1 expression alone or in combination with LCoR slightly attenuated basal and hormone-induced expression of the gene. In contrast, loss of LCoR or CtBP1 individually or in combination substantially augmented progesterone-stimulated expression of the mucin 1 (MUC1) and FK506-binding protein 5 (FKBP51) genes (Fig. 7, B and C). Taken together, the data in Figs. 6 and 7 provide evidence for roles of LCoR and CtBP1 as hormone-recruited attenuators of progesterone-regulated gene transcription. However, they also suggest that CtBP1 function may enhance the expression of some genes, similar to what was observed below in an analysis of estrogen-regulated gene expression.
FIGURE 7.
Effects of LCoR and CtBP1 ablation in T47D cells on regulation of endogenous PR target genes. Cells were transfected with the corresponding siRNAs (scrambled, CyPB, LCoR, or CtBP1 or LCoR and CtBP1) for 36 h, then treated with vehicle (EtOH) or P4 (10 nm) for 24 h. qRT-PCR was performed to analyze regulated expression of IGFBP1 (A), mucin 1 (MUC1; B), or FK506-binding protein 5 (FKBP51; C), and β-actin expression was used as an internal control. Results are shown as -fold induction. Data are the averages of three or more independent experiments. Error bars represent the S.E.; *, p < 0.05 for results of specific knockdown (LCoR, CtBP1, or LCoR and CtBP1) versus scrambled results with scrambled siRNA.
Ablation of LCoR or CtBP1 Diminishes Expression of Estrogen Target Genes in a Gene-specific Manner
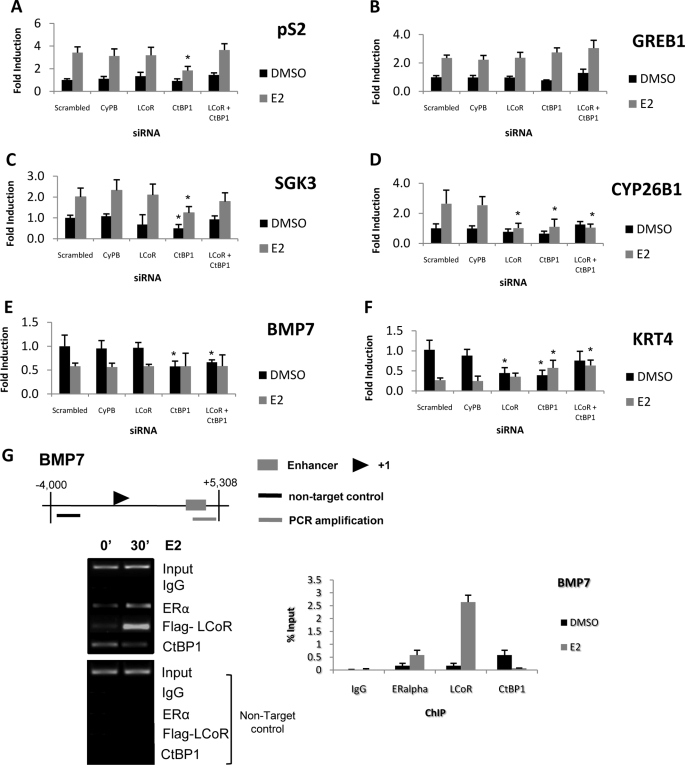
The effects of LCoR and/or CtBP1 knockdown on expression of a series of ERα target genes (40) were examined in MCF7 cells. Unlike the general stimulatory effect on progesterone-induced gene expression, knockdown of LCoR elicited distinct and gene-specific effects on estrogen target gene regulation. Notably, loss of CtBP1 but not LCoR attenuated estrogen-induced pS2 transcription (Fig. 8A). This effect of CtBP1 ablation is in agreement with previous studies showing that CtBP1 or CtBP2 knockdown attenuated estrogen-stimulated pS2 expression in MCF7 cells (41) and is consistent with the reduced expression of the IGFBP1 gene seen above in CtBP1-deficient cells. Although loss of CtBP1 had no effect on regulation of the GREB1 gene (the gene regulated by estrogen in breast cancer; Fig. 8B), its knockdown attenuated both basal and estrogen-regulated expression of the serum/glucocorticoids-regulated kinase 3 (SGK3) and cytochrome P450, family 26, subfamily b, polypeptide 1 (CYP26B1) genes (Fig. 8, C and D). Knockdown of LCoR had no effect on basal or estrogen-induced expression of GREB1 or SGK3, whereas estrogen-induced expression of CYP26B1 was eliminated (Fig. 8D), mimicking the effect of CtBP1 ablation. These results are remarkable given the augmented progesterone-stimulated gene transcription seen in LCoR-deficient cells.
FIGURE 8.
Effects of LCoR and/or CtBP1 ablation in MCF7 cells on regulation of endogenous estrogen target genes. Cells were transfected with corresponding siRNAs (scrambled, CyPB, LCoR, or CtBP1 or both LCoR and CtBP1) for 36 h, then treated with vehicle (DMSO) or E2 (10 nm) for 24 h. qRT-PCR was performed to analyze regulated expression of pS2 (A), GREB1 (B), SGK3 (C), CYP26B1 (D), BMP7 (E), or keratin 4 KRT4; F), and β-actin was used as an internal control. Results are shown as -fold induction. Data are the averages of three or more independent experiments. Error bars represent the S.E.; *, p < 0.05 for results of specific knockdown (LCoR, CtBP1, or both LCoR and CtBP1) versus scrambled results. G, shown are ChIP assays analyzing factor binding to the BMP7 promoter (upper panel) in MCF7 cells treated with E2 (10 nm). Cell extracts were collected and immunoprecipitated with IgG or antibodies against ERα, FLAG, or CtBP1, as indicated. Results of semiquantitative and quantitative PCR analyses are presented. Note that no signal was detected by quantitative PCR in the non-target controls.
Given the unexpected effects of knockdowns on estrogen-induced expression, we also analyzed the potential roles of LCoR and CtBP1 on genes whose transcription is repressed by estrogen (40). Loss of CtBP1, but not LCoR, attenuated basal expression of the bone morphogenetic protein 7 (BMP7) gene (Fig. 8E), whereas loss of either protein attenuated basal expression of the keratin 4 (KRT4) gene (Fig. 8F). Effects of ablation on estrogen-repressed transcription were variable, with no change in estrogen-repressed BMP7 transcription and a slight increase in expression of the keratin 4 gene. In general, these effects are not consistent with CtBP1 or LCoR functioning as corepressors on these genes.
We further analyzed the molecular basis of these results by performing ChIP assays on the BMP7 promoter, where the ERα binding region has been identified (42). Remarkably, we found that whereas LCoR was strongly recruited to the promoter after 30 min of estrogen treatment, CtBP1 partially dissociated from the promoter, indicating that the two factors function independently (Fig. 8G). ReChIP analysis found evidence for corecruitment of LCoR with ERα, but not CtBP1, on the BMP7 promoter after 30 min of estrogen treatment (not shown). These results are consistent with an effect of CtBP1 ablation on basal but not estrogen-regulated expression of the BMP7 gene.
DISCUSSION
We have analyzed the roles of LCoR and its cofactor CtBP1 in coregulation of progesterone and estrogen receptor-regulated gene expression. Our previous findings showed that LCoR was a particularly efficacious inhibitor of progesterone-regulated reporter gene expression and that corepression was largely resistant to HDAC inhibitor trichostatin A (15). This study showed that CtBP1 did not repress hormone-regulated gene expression on its own in transient expression studies but did contribute to corepression by LCoR. GST pulldown experiments and coimmunoprecipitations with a series of LCoR mutants suggested that the N-terminal region of LCoR containing the tandem PXDLS motifs was necessary and sufficient for interactions of CtBP1 with LCoR. Other experiments showed that deletion of the C-terminal HTH domain severely abrogated corepression, although this domain had no apparent role in CtBP1 recruitment. The HTH domain is distinct from the central region of LCoR identified in the accompanying manuscript (29) as being required for interaction with HDAC6. Together, these findings emphasize that LCoR is a multidomain protein that exerts its coregulator function through diverse mechanisms. siRNA-mediated knockdown of LCoR expression established its role in attenuation of progesterone-regulated gene transcription, as hormone-stimulated expression was enhanced on all three PR target genes tested in cells lacking LCoR.
The function of LCoR in corepression of PR-stimulated gene expression may be of considerable physiological importance. Progesterone signaling is essential for normal development and homeostasis of a number of physiological processes including female sexual behavior, ovulation, protection against seizures, maintenance of quiescence of the uterus during pregnancy, induction of germ cell maturation, and oocyte maturation (1, 5, 6). Progestins and anti-progestins are used clinically in contraception, hormone replacement therapy, induction of labor, treatment of endometriosis, and endometrial cancer (43). Moreover, expression of the PR along with ERα and human epidermal growth factor receptor 2 (HER2) are used as predictive markers for breast cancer therapy (44).
There is considerable overlap in the expression patterns of LCoR and the PR. LCoR is widely expressed throughout development and in the adult (15). In tissue blots we observed the highest expression of LCoR in the placenta along with robust expression in several fetal tissues. Notably, the placenta is a site of progesterone biosynthesis, and in situ hybridization analysis of near-term placenta (15) revealed that LCoR mRNAs were predominantly expressed in the syncytiotrophoblast layer of terminally differentiated cells, a site of PR expression and signaling (45, 46). The syncytiotrophoblast layer acts as a barrier between maternal circulation and the fetus, and its function is critical for controlling maternal signals that modulate fetal metabolism and development (47).
Compared with the substantial increases observed in progesterone-stimulated gene expression, the effects of LCoR or CtBP1 ablation on estrogen target genes were distinct and gene-specific. We observed no effect of LCoR knockdown on expression of three of four estrogen-stimulated genes studied. This may reflect a redundancy in corepressor function on the genes tested. For example, knockdown in MCF7 cells of NRIP1, another corepressor recruited in the presence of hormone, had gene-specific effects on estrogen-dependent transactivation (48). LCoR ablation did augment both basal and hormone-stimulated expression of an estrogen-sensitive reporter gene, pointing to a potential role as an attenuator of ERα signaling. Remarkably, however, its knockdown blocked estrogen-stimulated expression of CYP26B1. This may reflect its function as a cofactor of CtBP1 on the CYP26B1 promoter, as ablation of CtBP1 also abolished estrogen-induced transcription. Notably, we found in the accompanying manuscript (29) that knockdown of HDAC6 had no effect on CYP26B1 regulation. A diminution in estrogen-stimulated gene expression in the absence of CtBP1 was also observed on two of three other estrogen-stimulated genes tested. Although these results were unexpected, they are consistent with other findings that knockdown of CtBP1 led to reduced expression of the ATP binding cassette, subfamily B (MDR/TAP) member 1 (MDR1) gene (49).
We also found that LCoR and CtBP1 are corecruited to the progesterone-stimulated IGFBP1 gene and the ERα target gene pS2. This behavior is in apparent contrast to the results of Stossi et al. (41), who found estrogen-induced dissociation of CtBP1 from the pS2 promoter. They found that CtBP1 controlled estrogen-mediated gene repression and that CtBP1 was recruited to repressed genes in the presence of estradiol. On the other hand, our observations are entirely consistent with other findings (50) in which peak recruitment of CtBP1 to the pS2 promoter was observed after 30 min of estradiol treatment.
CtBP1 and LCoR were extensively colocalized in the nucleus, including in pronounced foci. This pattern was similar to the colocalization of LCoR with CtIP and PRC1 marker BMI1 and strongly suggests that a substantial portion of CtBP1-bound LCoR is associated with PRC1 complexes. We found no evidence for direct interaction of LCoR with CtIP (Fig. 2) or BMI17 by GST pulldown assay. However, their indirect association is supported by coimmunoprecipitation of LCoR with both proteins and their hormone-dependent recruitment to estrogen or progesterone-regulated genes. Polycomb group proteins and complexes, including PRC1, form nuclear foci visualized as PcG (polycomb group proteins) bodies (51). Immunocytochemical studies have estimated these bodies to measure between 0.2 and 1.5 μm and to vary greatly in composition, suggesting that they represent foci with numerous independently functioning transcriptional regulatory complexes (25, 52).
More than 30 transcription factors have been shown to interact with CtBP1 (53). Recent studies characterized a CtBP-corepressor complex that contained a great number of proteins with opposing enzymatic activities. This included histone modifying enzymes (53) and other coregulatory proteins that can either activate or repress transcription depending on the context. Additionally, LCoR was identified as one of the components of a CtBP-corepressor complex purified from the nuclear extracts of HeLa cells (54). Taken together, these studies suggest that LCoR is associated with several distinct multisubunit transcriptional regulatory complexes; hence, implying the importance of LCoR in transcriptional control.
Supplementary Material
Acknowledgments
We are grateful to Jacynthe Laliberté for technical assistance with confocal microscopy. A special thank you goes to Dr. Myles Brown (Harvard Medical School) for providing the locations of EREs and ER-regulated enhancers.
This work was supported by Canadian Institutes of Health Research Grant MT-11704 (to J. H. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental and Tables 1 and 2.
A. Palijan, I. Fernandes, M. Verway, M. Kourelis, Y. Bastien, L. E. Tavera-Mendoza, A. Sacheli, V. Bourdeau, S. Mader, and J. H. White, unpublished data.
- PR
- progesterone receptor
- ChIP
- chromatin immunoprecipitation
- CtBP
- C-terminal binding protein
- CtIP
- CtBP-interacting protein
- CyPB
- cyclophilin B
- E2
- estradiol
- ERα
- estrogen receptor α
- ERE
- estrogen response element
- HDAC
- histone deacetylase
- HTH
- helix-turn-helix
- LCoR
- ligand-dependent corepressor
- MBLK-1
- honeybee MBlk-1-related factor
- P4
- progesterone
- PRC1
- polycomb group repressor complexes
- pS2
- trefoil factor 1
- PSQ
- pipsqueak motif
- siRNA
- small interfering RNA
- qRT
- quantitative real-time
- SGK3
- serum/glucocorticoids-regulated kinase 3
- BMP7
- bone morphogenetic protein 7
- DMEM
- Dulbecco's modified Eagle's medium
- BSA
- bovine serum albumin
- GST
- glutathione S-transferase
- IGFBP1
- insulin-like growth factor binding protein 1
- CYP26B1
- cytochrome P450, family 26, subfamily b, polypeptide 1.
REFERENCES
- 1.Edwards D. P. (2005) Annu. Rev. Physiol. 67, 335–376 [DOI] [PubMed] [Google Scholar]
- 2.Hall J. M., McDonnell D. P. (2005) Mol. Interv. 5, 343–357 [DOI] [PubMed] [Google Scholar]
- 3.Wong C. W., McNally C., Nickbarg E., Komm B. S., Cheskis B. J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99, 14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Kim J. J., Buzzio O. L., Li S., Lu Z. (2005) Biol. Reprod. 73, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boonyaratanakornkit V., McGowan E., Sherman L., Mancini M. A., Cheskis B. J., Edwards D. P. (2007) Mol. Endocrinol. 21, 359–375 [DOI] [PubMed] [Google Scholar]
- 6.Aoyagi S., Archer T. K. (2007) Mol. Endocrinol. 21, 843–856 [DOI] [PubMed] [Google Scholar]
- 7.Gronemeyer H. (1991) Annu. Rev. Genet. 25, 89–123 [DOI] [PubMed] [Google Scholar]
- 8.McKenna N. J., O'Malley B. W. (2002) Cell 108, 465–474 [DOI] [PubMed] [Google Scholar]
- 9.Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 10.Zhao L. J., Subramanian T., Zhou Y., Chinnadurai G. (2006) J. Biol. Chem. 281, 4183–4189 [DOI] [PubMed] [Google Scholar]
- 11.White J. H., Fernandes I., Mader S., Yang X. J. (2004) Vitam. Horm. 68, 123–143 [DOI] [PubMed] [Google Scholar]
- 12.Gurevich I., Flores A. M., Aneskievich B. J. (2007) Toxicol. Appl. Pharmacol. 223, 288–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vo N., Fjeld C., Goodman R. H. (2001) Mol. Cell. Biol. 21, 6181–6188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White K. A., Yore M. M., Deng D., Spinella M. J. (2005) J. Biol. Chem. 280, 7829–7835 [DOI] [PubMed] [Google Scholar]
- 15.Fernandes I., Bastien Y., Wai T., Nygard K., Lin R., Cormier O., Lee H. S., Eng F., Bertos N. R., Pelletier N., Mader S., Han V. K., Yang X. J., White J. H. (2003) Mol. Cell 11, 139–150 [DOI] [PubMed] [Google Scholar]
- 16.Boyd J. M., Subramanian T., Schaeper U., La Regina M., Bayley S., Chinnadurai G. (1993) EMBO J. 12, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinnadurai G. (2002) Mol. Cell 9, 213–224 [DOI] [PubMed] [Google Scholar]
- 18.Chinnadurai G. (2007) Int. J. Biochem. Cell Biol. 39, 1593–1607 [DOI] [PubMed] [Google Scholar]
- 19.Mani-Telang P., Arnosti D. N. (2007) Dev. Genes Evol. 217, 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsanis N., Fisher E. M. (1998) Genomics 47, 294–299 [DOI] [PubMed] [Google Scholar]
- 21.Chinnadurai G. (2009) Cancer Res. 69, 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinnadurai G. (2006) Biochim. Biophys. Acta 1765, 67–73 [DOI] [PubMed] [Google Scholar]
- 23.Meloni A. R., Smith E. J., Nevins J. R. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 9574–9579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X., Chen J. (2004) Mol. Cell. Biol. 24, 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otte A. P., Kwaks T. H. J. (2003) Curr. Opin. Genet. Dev. 13, 448–454 [DOI] [PubMed] [Google Scholar]
- 26.Sewalt R. G., Gunster M. J., van der Vlag J., Satijn D. P., Otte A. P. (1999) Mol. Cell. Biol. 19, 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez C., Sánchez I., Demmers J. A., Rodriguez P., Strouboulis J., Vidal M. (2007) Mol. Cell. Proteomics 6, 820–834 [DOI] [PubMed] [Google Scholar]
- 28.Hildebrand J. D., Soriano P. (2002) Mol. Cell. Biol. 22, 5296–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palijan A., Fernandes I., Bastien Y., Tang L., Verway M., Kourelis M., Tavera-Mendoza L. E., Li Z., Bourdeau V., Mader S., Yang X. J., White J. (2009) J. Biol. Chem. 284, 30264–30274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi H., Kage E., Sawata M., Kamikouchi A., Ohashi K., Ohara M., Fujiyuki T., Kunieda T., Sekimizu K., Natori S., Kubo T. (2001) Insect Mol. Biol. 10, 487–494 [DOI] [PubMed] [Google Scholar]
- 31.Lehmann M., Siegmund T., Lintermann K. G., Korge G. (1998) J. Biol. Chem. 273, 28504–28509 [DOI] [PubMed] [Google Scholar]
- 32.Siegmund T., Lehmann M. (2002) Dev. Genes Evol. 212, 152–157 [DOI] [PubMed] [Google Scholar]
- 33.Aravind L., Anantharaman V., Balaji S., Babu M. M., Iyer L. M. (2005) FEMS Microbiol. Rev. 29, 231–262 [DOI] [PubMed] [Google Scholar]
- 34.Tavera-Mendoza L. E., Quach T. D., Dabbas B., Hudon J., Liao X., Palijan A., Gleason J. L., White J. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8250–8255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavera-Mendoza L., Wang T. T., Lallemant B., Zhang R., Nagai Y., Bourdeau V., Ramirez-Calderon M., Desbarats J., Mader S., White J. H. (2006) EMBO Rep. 7, 180–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fasano C. A., Dimos J. T., Ivanova N. B., Lowry N., Lemischka I. R., Temple S. (2007) Cell Stem Cell. 1, 87–99 [DOI] [PubMed] [Google Scholar]
- 37.Ren X., Vincenz C., Kerppola T. K. (2008) Mol. Cell. Biol. 28, 2884–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horne A. W., Lalani E. N., Margara R. A., White J. O. (2006) Reproduction 131, 733–742 [DOI] [PubMed] [Google Scholar]
- 39.Hubler T. R., Denny W. B., Valentine D. L., Cheung-Flynn J., Smith D. F., Scammell J. G. (2003) Endocrinology 144, 2380–2387 [DOI] [PubMed] [Google Scholar]
- 40.Bourdeau V., Deschênes J., Métivier R., Nagai Y., Nguyen D., Bretschneider N., Gannon F., White J. H., Mader S. (2004) Mol. Endocrinol. 18, 1411–1427 [DOI] [PubMed] [Google Scholar]
- 41.Stossi F., Madak-Erdogan Z., Katzenellenbogen B. S. (2009) Mol. Cell. Biol. 29, 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusumegi T., Tanaka J., Kawano M., Yonemoto J., Tohyama C., Sone H. (2004) J. Biochem. Mol. Toxicol. 18, 1–11 [DOI] [PubMed] [Google Scholar]
- 43.Richer J. K., Jacobsen B. M., Manning N. G., Abel M. G., Wolf D. M., Horwitz K. B. (2002) J. Biol. Chem. 277, 5209–5218 [DOI] [PubMed] [Google Scholar]
- 44.Duffy M. J. (2005) Clin. Chem. 51, 494–503 [DOI] [PubMed] [Google Scholar]
- 45.Ghosh D., Dhara S., Kumar A., Sengupta J. (1999) Hum. Reprod. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 46.Patel F. A., Clifton V. L., Chwalisz K., Challis J. R. G. (1999) J. Clin. Endocrinol. Metab. 84, 291–299 [DOI] [PubMed] [Google Scholar]
- 47.Pepe G. J., Albrecht E. D. (1995) Endocr. Rev. 16, 608–648 [DOI] [PubMed] [Google Scholar]
- 48.Castet A., Herledan A., Bonnet S., Jalaguier S., Vanacker J. M., Cavaillès V. (2006) Mol. Endocrinol. 20, 1035–1047 [DOI] [PubMed] [Google Scholar]
- 49.Jin W., Scotto K. W., Hait W. N., Yang J. M. (2007) Biochem. Pharmacol. 74, 851–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perissi V., Scafoglio C., Zhang J., Ohgi K. A., Rose D. W., Glass C. K., Rosenfeld M. G. (2008) Mol. Cell 29, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hernández-Muñoz I., Taghavi P., Kuijl C., Neefjes J., van Lohuizen M. (2005) Mol. Cell. Biol. 25, 11047–11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spector D. L. (2001) J. Cell Sci. 114, 2891–2893 [DOI] [PubMed] [Google Scholar]
- 53.Kuppuswamy M., Vijayalingam S., Zhao L. J., Zhou Y., Subramanian T., Ryerse J., Chinnadurai G. (2008) Mol. Cell. Biol. 28, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi Y., Sawada J., Sui G., Affar el B., Whetstine J. R., Lan F., Ogawa H., Luke M. P., Nakatani Y., Shi Y. (2003) Nature 422, 735–738 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.