Abstract
The class B scavenger receptor CD36 has numerous ligands that include modified forms of low density lipoprotein, fibrillar amyloid, apoptotic cells, and Plasmodium falciparum-infected red blood cells, linking this molecule to atherosclerosis, Alzheimer disease, malaria, and other diseases. We studied the signaling events that follow receptor engagement and lead to CD36 and ligand internalization. We show that oxidized low density lipoprotein or antibody-induced clustering of CD36 triggers macropinocytosis and internalization of the receptor-ligand complex. Remarkably, however, CD36 internalization is independent of macropinocytosis and occurs by a novel endocytic mechanism that depends on actin, but not dynamin. This actin-driven endocytosis requires the activation Src family kinases, JNK, and Rho family GTPases, but, unlike macropinocytosis, it is not affected by inhibitors of phosphatidylinositol 3-kinase or Na/H exchange. Manipulation of this unique mode of internalization may prove helpful in the prevention and management of the wide range of diseases in which CD36 is implicated.
Uptake and storage of cholesterol by macrophages are key contributors to the formation of atherosclerotic plaque. Endothelial cells, seemingly activated by the deposition of modified forms of low-density lipoprotein (LDL),3 release chemokines that recruit macrophages into the vascular intima. Infiltrated macrophages can then readily oxidize and take up the modified LDL. Accumulation of lipids derived from oxidized LDL (oxLDL) transforms macrophages into foam cells, which release excess cytokines, triggering an inflammatory cascade. In addition, foam cells express proteases and other factors that contribute to plaque rupture and subsequent thrombosis.
OxLDL particles are recognized by a variety of receptors, including the class A scavenger receptor SR-A and the class B scavenger receptor CD36. CD36 is thought to be responsible for ∼50% of oxLDL uptake by murine and human macrophages (1–3). In addition, this protein mediates cholesterol uptake from high density lipoprotein (4) and is also a receptor for internalization of oxidized high density lipoprotein (5).
CD36 encodes a protein with two transmembrane domains located near the N and C termini, leaving only short cytoplasmic tails at each end. Despite having small intracellular domains, engagement of CD36 by its cognate ligands triggers signaling reactions that lead to the internalization of the resulting complex. However, the precise pathways that are activated and the specific mode of internalization remain unclear.
Jones and Willingham (6) demonstrated that, in macrophages, modified LDL stimulates ruffling activity and the formation of phase-bright macropinosomes. By transmission electron microscopy they found that gold-conjugated modified LDL associated with ruffles and was present within macropinosomes. These observations underlie the widely accepted view that uptake of modified LDL occurs by macropinocytosis. However, Zeng et al. (7) showed that internalized DiI-oxLDL and CD36 were found in moderately sized cytoplasmic structures that co-localized with a glycosylphosphatidylinositol-anchored protein, suggesting uptake via lipid raft endocytosis. Moreover, Sun et al. (8) reported that uptake of oxLDL by CD36 was independent of actin but dependent on dynamin. The results of these two studies are not easily reconciled with mediation by macropinocytosis, an actin-dependent process that generates large vacuoles, and they suggest instead that CD36 is internalized by a more conventional endocytic pathway.
It is not clear whether the apparent discrepancy stems from the engagement of different receptors in the different biological systems used in these studies. Jones and Willingham used macrophages, whereas Zeng et al. and Sun et al. studied, respectively, Chinese hamster ovary and COS cells heterologously transfected with CD36. The types and abundance of receptors capable of binding modified LDL in all likelihood differ greatly in these systems, and heterologous (over)expression in immortalized cells is liable to produce results of questionable biological relevance.
In view of this uncertainty and considering the important and versatile roles of CD36, we set out to reexamine the mode of internalization of this receptor. We used both primary and cultured macrophages and selectively targeted CD36 using specific antibodies. The responses triggered by selective CD36 cross-linking were also compared with those elicited by oxLDL. We show that clustering CD36 initiates a signaling cascade that results in the activation of both macropinocytosis and internalization of CD36. Remarkably, however, CD36 internalization is largely independent of macropinocytosis and occurs by a novel dynamin-independent, actin-driven process that requires activation of Src family and c-Jun N-terminal kinases.
EXPERIMENTAL PROCEDURES
Antibodies
Mouse anti-human CD36 IgG (clone 131.2) was prepared by Narendra N. Tandon. The following antibodies were purchased from the sources indicated: mouse anti-mouse CD36 IgA and mouse anti-GAPDH (Chemicon); anti-mouse IgA Texas Red (Bio/FX); anti-CD36 IgM (BD Pharmingen); anti-mouse Alexa Fluor 488 (Molecular Probes/Invitrogen); anti-mouse IgG Cy3, anti-mouse F(ab′)2 Cy3, anti-rabbit horseradish peroxidase, and anti-mouse horseradish peroxidase (Jackson Immunological Laboratories); Alexa Fluor 488-conjugated anti-phosphotyrosine primary antibody (PY-20 clone) (BioLegend); anti-mouse IgA-fluorescein isothiocyanate (Sigma-Aldrich); mouse anti-phosphotyrosine (4G10) (Upstate); rabbit anti-phospho-JNK (Cell Signaling); and mouse anti-vinculin (Chemicon).
Tissue Culture and Macrophage Preparation
Human peripheral macrophages were prepared according to McGilvray et al. (9). Wild-type and CD36-knock-out mouse peritoneal macrophages were a gift from Kevin Kain (University Health Network, Toronto, Canada) and prepared according to Patel et al. (10). U937 cells (ATCC) were differentiated with 1 μm phorbol 12-myristate 13-acetate (Bioshop Canada) for 2 days. RAW 264.7 cells (ATCC) were grown in Dulbecco's modified Eagle's medium (Wisent) with 10% fetal bovine serum. During live experiments, cells were maintained in HEPES-buffered RPMI 1640 medium (H-RPMI; Wisent).
CD36 Cross-linking and Internalization
Total CD36 immunostaining was accomplished by fixing RAW 264.7, differentiated U937, or human peripheral macrophages with 4% PFA (Electron Microscopy Sciences), permeabilizing with 0.1% Triton X-100, blocking with 5% serum, and labeling with anti-CD36 IgA or anti-human CD36 IgG, followed by anti-mouse IgA Texas Red or anti-mouse IgG Cy3 secondary antibody, respectively.
CD36 receptors were internalized by cross-linking with a primary antibody followed by a secondary antibody and warming, as follows. RAW 264.7 cells were chilled for 3 min in ice-cold H-RPMI followed by a 5-min incubation in ice-cold 5% donkey serum (Sigma-Aldrich), a 10-min incubation in ice-cold anti-CD36 IgA, and a 10-min incubation in ice-cold anti-mouse IgA Texas Red or donkey anti-mouse F(ab′)2 Cy3. Cells were washed twice with ice-cold H-RPMI before and after antibody labeling steps. Differentiated U937 cells and human peripheral macrophages were similarly labeled using either mouse IgG clone 131.2 or mouse anti-CD36 IgM, followed by anti-mouse IgG Cy3. In cases where only surface labeling was to be examined, cells were washed in ice-cold H-RPMI and fixed in 4% PFA. For internalization protocols, cells were warmed at 37 °C in H-RPMI for 30 min in a non-CO2 incubator.
Acid Wash
In most cases, antibodies bound to the noninternalized CD36 were stripped off with an acid wash protocol: Cells were treated for 2 min with ice-cold acid wash buffer (0.5 m glacial acetic acid, 150 mm sodium chloride, pH 2.5) followed by recovery in ice-cold H-RPMI for 2 min. Both steps were repeated once.
OxLDL Treatment
Wild-type, CD36-knock-out mouse peritoneal macrophages and RAW 264.7 cells were serum-starved in H-RPMI for 1 h and treated with DiI-labeled oxidized LDL (1 μg/ml; Intracel). After 30 min, cells were acid-washed twice. Peritoneal cells were fixed in 4% PFA, and internalized oxidized LDL was imaged. RAW 264.7 cells were fixed in methanol at −20 °C for 20 min, blocked with 5% serum, and stained with anti-CD36 IgA followed by anti-mouse IgG Alexa Fluor 488.
Transfections
For transfection experiments, RAW 264.7 cells were transfected with 2 μg of DNA (wild-type Dynamin1, K44A dominant negative Dynamin1, PAK1-PBD-YFP, AKT1-PH-GFP, Rac1 DN, Cdc42 DN, RhoA DN, or Arf6 DN) using 6 μl of FuGENE HD (Roche Applied Science) according to the manufacturer's instructions. To examine PAK1 recruitment during CD36 cross-linking, cells were transfected with PAK1-PBD-YFP, labeled for CD36, warmed in H-RPMI for 3 min, fixed, and imaged on a spinning disc microscope. In all cases, cells were examined 16 h after transfection.
Pharmacological Analysis of CD36 Uptake
For chemical inhibition, RAW 264.7 cells were pretreated with inhibitors as follows: latrunculin B (10 μm, 30 min; Calbiochem), cytochalasin D (5 μm, 30 min; Calbiochem), Clostridium difficile neurotoxin B (0.5 μg/ml, 3 h; Tech Lab), JNK inhibitor SP600125 (20 μm, 1 h; Calbiochem), wortmannin (250 nm, 30 min; Calbiochem), PP1 (10 μm, 30 min; Alexis Biochemicals); or LY294002 (100 μm, 30 min; Biomol) before labeling with antibody (see “CD36 Cross-linking”). Cells were warmed for 30 min and fixed (without acid wash), and CD36 internalization was imaged by spinning disc microscopy. For data quantification, the same experiment included an acid wash protocol prior to fixation.
To determine the dependence of oxLDL-induced CD36 uptake on actin, Src family kinases, and phospho-JNK, RAW 264.7 cells were serum-starved for 4 h and incubated in the presence of unlabeled oxLDL (50 μg/ml; Intracel) and the appropriate inhibitor for 30 min before fixation. Total CD36 was detected by permeabilizing cells and labeling with anti-CD36 IgA and anti-mouse secondary antibody.
Immunoblotting
RAW 264.7 cells were grown to 90% confluence in 6-well dishes, serum-starved in H-RPMI for 4 h, then treated with or without PP1. These cells were further treated as follows: no treatment or labeled with CD36 primary with or without secondary antibody in the cold, with the exception that the secondary antibody used was anti-mouse CD36 F(ab′)2 labeled in ice-cold H-RPMI for 10 min. Cells were then induced to internalize CD36 by incubating in prewarmed H-RPMI at 37 °C for 3 min, with or without PP1, lysed in Laemmli sample buffer containing 10 mm N-ethylmaleimide (Bioshop Canada), 1 mm sodium orthovanadate (Sigma-Aldrich), 100 nm okadaic acid (Bioshop Canada), 1 mm sodium fluoride (Sigma-Aldrich), and mammalian protease inhibitors (Sigma-Aldrich). Equal amounts of lysates were resolved on a 10% SDS-polyacrylamide gel, transferred to nitrocellulose, blocked overnight with 3% bovine serum albumin (EMD Biosciences) in 1× TBST (0.05% Tween 20), and probed with mouse anti-phosphotyrosine antibody (4G10) and goat anti-mouse horseradish peroxidase secondary antibody. Blots were stripped and reprobed with mouse anti-GAPDH primary antibody.
Macropinocytosis Assays
RAW 264.7 cells were serum-starved in H-RPMI without CO2 for 1 h and then treated with inhibitors as follows; no treatment, wortmannin (250 nm, 30 min), LY294002 (100 μm, 30 min), amiloride (0.5 mm, 10 min; Sigma-Aldrich), or HOE-694 (10 μm, 10 min) in H-RPMI at 37 °C. To induce macropinocytosis, cells were then incubated in H-RPMI containing 40 ng/ml mouse M-CSF (Sigma-Aldrich), 150 μg/ml sulforhodamine B (Molecular Probes), and the appropriate inhibitor for 30 min at 37 °C without CO2. The negative control contained sulforhodamine but no M-CSF.
All experiments were performed three times. To measure sulforhodamine uptake, cells were fixed in 4% PFA, and multiple focal planes were immediately imaged on a DRM/IRE2 Leica fluorescent microscope, using OpenLab software 5.5.0 (Improvision). Images were analyzed in Photoshop by counting only macropinosomes larger than 0.7 μm. For each experimental condition, the total number of macropinosomes/cell was counted, averaged, and normalized to the control (no inhibitor).
To determine whether the inhibition of macropinosome formation can lead to inhibition of CD36 internalization, cells were pretreated with agents shown to inhibit macropinocytosis (0.5 mm amiloride for 10 min or 10 μm HOE-694 for 10 min). Cells were fixed in 4% PFA, permeabilized, and stained 30 min with 10 μg/ml DAPI (Molecular Probes).
Scanning Electron Microscopy of CD36 Uptake Compared with Ruffling
To induce CD36 uptake for scanning electron microscopy, RAW 264.7 cells were labeled with CD36 antibodies (see “CD36 Cross-linking”), and warmed for 3 min at 37 °C. To demonstrate ruffles induced by M-CSF, cells were treated with mouse M-CSF and sulforhodamine B for 3 min at 37 °C. Cells were fixed in 2% glutaraldehyde and 0.2 m sodium phosphate, pH 7.4. Microscopy was performed on an FEI XL30 environmental scanning electron microscope.
Image Quantification
All images except those for macropinocytosis assays were obtained on a Zeiss Axiovert 200M spinning disc confocal microscope, using Volocity v.4.3.2 software (Improvision). Macropinocytosis images were acquired on a DRM/IRE2 Leica fluorescent microscope, using OpenLab software v. 5.5.0. Scanning electron microscope images were acquired at a magnification of ×5000.
Data for the graphs in Figs. 2–7 were obtained by analyzing images taken with a spinning disc microscope. At least three assays were performed per data set. Z-stacks (30–40 layers) were flattened using Volocity software v. 4.3.2 to a single layer using the Extended Focus function while maintaining pixels at subsaturating levels, allowing an arbitrary pixel range from 0 to 64,535. Pixel densities were analyzed in ImageJ v.1.38 (National Institutes of Health) to obtain an arbitrary amount of CD36 internalization/number of cells. These data were normalized to the control (untreated cells), standard deviations were calculated, and the data were plotted in Excel.
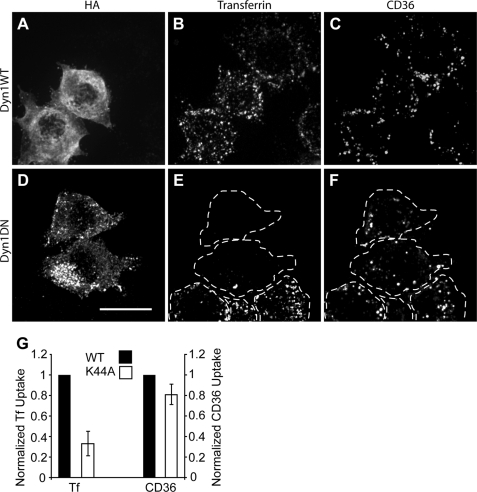
FIGURE 2.
Effect of dynamin1 K44A dominant negative mutant on CD36 internalization. RAW 264.7 cells were transfected with hemagglutinin (HA)-tagged wild-type (WT) dynamin1 (Dyn1; A–C) or dynamin1 K44A (D–F), cross-linked in the cold with anti-CD36 IgA antibody plus labeled secondary antibody, incubated with transferrin-Alexa Fluor 647, fixed, and stained for hemagglutinin (A and D) and imaged for transferrin (B and E) and CD36 (C and F). DN, dominant negative. Dashed lines indicate the outline of the cells. Scale bar, 10 μm. G, for quantification, the same assay was done with the addition of two acid washes prior to fixation. Data are means ± S.E. of three experiments with at least 100 cells counted in each case. Transferrin (Tf) and internalized CD36 data are normalized to their respective wild-type controls (black bars).
FIGURE 3.
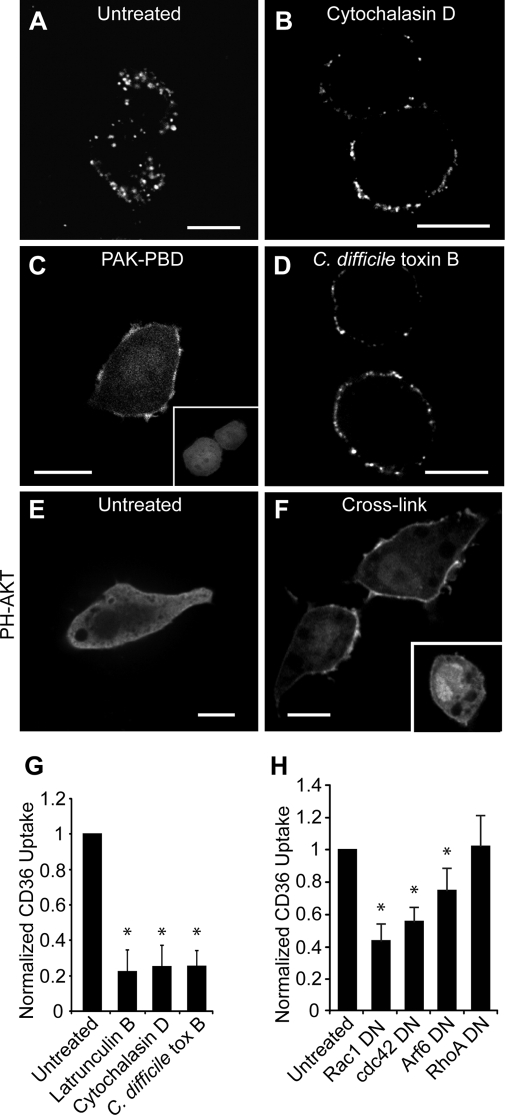
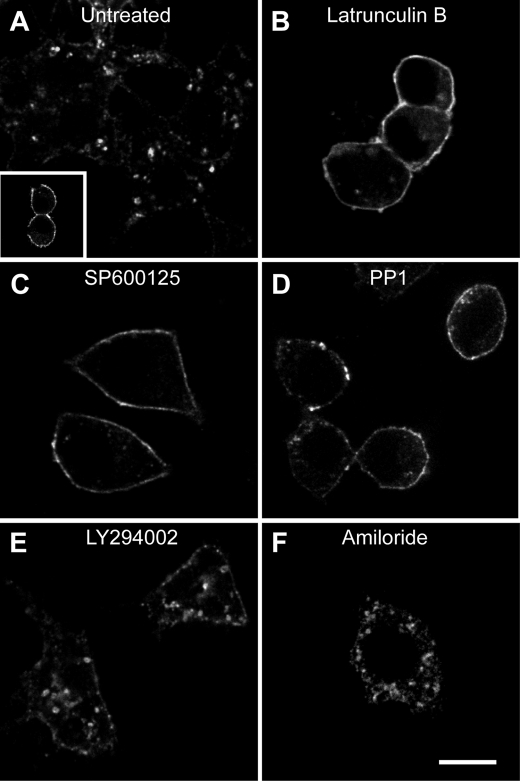
Actin inhibitors block internalization of CD36. RAW 264.7 cells were pretreated as follows: no treatment (A), cytochalasin D (B), or C. difficile toxin B (D). CD36 receptors were then cross-linked, and cells were warmed, fixed, and imaged. Acid wash was omitted to preserve surface labeling. C, Rac1 activation was demonstrated by recruitment of PAK1-PBD-YFP to the membrane. RAW 264.7 cells transfected with PAK-PBD-YFP were imaged following CD36 receptor cross-linking with (C) or without (C, inset) anti-CD36 antibodies. E and F, cross-linking activates PI3K. RAW 264.7 cells transiently expressing PH-AKT-GFP were treated without (E) or with (F) CD36 cross-linking antibody and warming. F, inset, cross-linking was done in the presence of LY294002. To quantify CD36 internalization (G), the same assay was performed with the addition of two acid washes. Cells were warmed to 37 °C, and internalized CD36 was imaged live on a spinning disc microscope. H, to demonstrate dependence of CD36 internalization on Rho or Arf family small GTPases, RAW 264.7 cells were transfected with dominant negative (DN) DNA constructs of Rac1, Cdc42 Arf6 or RhoA, and internalized CD36 receptors were quantified as above. Scale bars, 10 μm. *, p < 0.05 compared with untreated control.
FIGURE 4.
Macropinocytosis is induced by cross-linking CD36, but CD36 internalization is not sensitive to inhibitors of macropinocytosis. A, untreated RAW 264.7 cells take up little sulforhodamine B by macropinocytosis, but treatment with M-CSF (B) or cross-linking with antibody (C) stimulates macropinocytosis. Sulforhodamine is indicated in red. Scale bar, 10 μm. D, inhibitors of macropinocytosis block sulforhodamine uptake. RAW 264.7 cells were pretreated with no inhibitor, LY294002, amiloride, or the amiloride analog HOE-694 followed by cross-linking with antibody or mouse M-CSF followed by warming with sulforhodamine. Data are means ± S.E. of three experiments with at least 500 cells counted in each case. E, internalization of CD36 after cross-linking is not blocked by inhibitors of macropinocytosis. RAW 264.7 cells were left untreated or were pretreated with wortmannin, LY294002, amiloride, or HOE-694 prior to cross-linking CD36 with antibody in the cold, and warming in the presence of appropriate inhibitors. Finally cells were treated with two acid washes and fixed. Data are means ± S.E. of three experiments with at least 100 cells counted in each case. Data are normalized to the untreated control.
FIGURE 5.
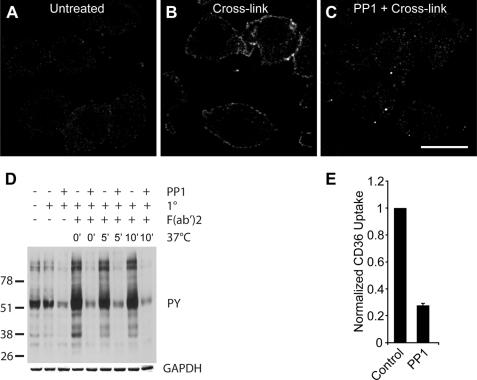
Cross-linking of CD36 receptors causes tyrosine phosphorylation. A–C, RAW 264.7 cells untreated (A), cross-linked with anti-CD36 (B), or cross-linked after pretreatment with the Src family tyrosine kinase inhibitor PP1 (C) were fixed and stained for phosphotyrosine. Scale bar, 10 μm. D, immunoblot of tyrosine phosphorylation subsequent to CD36 cross-linking and internalization. RAW 264.7 cells were pretreated without or with PP1 followed by cross-linking of CD36 with primary IgA antibody in the cold, secondary F(ab′)2 antibody and warming for 0 min, 5 min, or 10 min. Immunoblot was probed for phosphotyrosine (4G10), stripped, and reprobed for GAPDH. E, dependence of CD36 internalization on tyrosine phosphorylation. RAW cells were pretreated without (control) or with PP1 prior to cross-linking of CD36 receptors and warming in the presence of PP1. Cells were acid-washed twice before fixation and imaging on a spinning disc confocal microscope. Data are normalized to the control.
FIGURE 6.
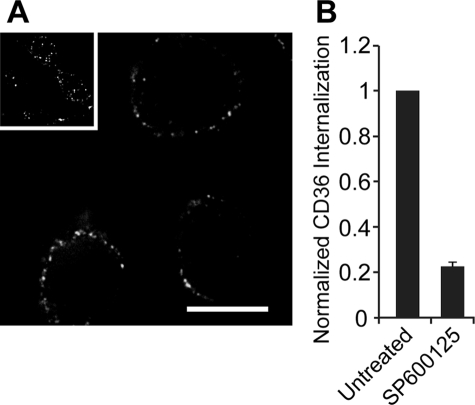
JNK inhibitor SP600125 blocks internalization of cross-linked CD36. A, RAW cells were treated without (inset) or with (main panel) SP600125 prior to cross-linking of CD36 and warmed with or without SP600125 present, and then fixed and imaged on a spinning disc confocal microscope. Scale bar, 10 μm. B, quantification of A. As in A, but with the addition of two acid wash steps before fixation. Images were acquired and quantified. Data are normalized to the untreated control and represent means ± S.E. of three experiments with at least 100 cells counted in each case.
FIGURE 7.
Inhibitors that block internalization of cross-linked CD36 also block oxidized LDL-dependent internalization of CD36. A, untreated RAW cells have internalized CD36 following incubation with nonlabeled oxLDL. Inset, no oxLDL added. B–F, RAW cells were pretreated with latrunculin B (B), SP600125 (C), PP1 (D), LY294002 (E), or amiloride (F) before incubation with nonlabeled oxLDL and stained for total CD36. Scale bar, 10 μm.
RESULTS
Anti-CD36 Antibodies and OxLDL Trigger Internalization of CD36
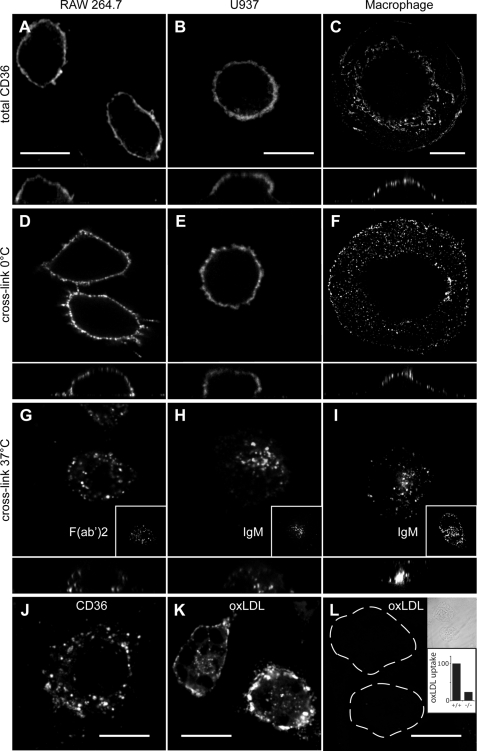
We first examined the steady-state distribution of unstimulated CD36 in fixed and permeabilized macrophages. For comparative purposes, we examined the murine RAW macrophage-like cell line, the human U937 monocyte cell line differentiated to macrophages by phorbol 12-myristate 13-acetate, and primary human macrophages. As shown in Fig. 1, A–C, confocal microscopy revealed that virtually all of the endogenous CD36 resides at the cell surface of resting macrophages. The primary macrophages adopt a “fried egg” appearance on the coverslip so that confocal images of surface CD36 frequently appeared punctate, as seen in Fig. 1C. However, this was due to tangential sectioning through the plasma membrane of these flat cells, and reconstructions of the vertical profile (Z plane; shown beneath each image) clearly demonstrated that the staining was at the cell surface. Predominant cell membrane localization had previously been observed when CD36 was expressed in Chinese hamster ovary cells (7).
FIGURE 1.
Distribution of endogenous CD36 following antibody and oxLDL cross-linking. A–C, distribution of CD36 on permeabilized resting RAW 264.7 cells (A), U937 cells (B), or human primary macrophages (C). D–F, cross-linking antibodies at 0 °C did not alter distribution of CD36 on RAW cells (D), U937 cells (E), or primary human macrophages (F). G–I, warming of cross-linked cells to 37 °C resulted in internalization of CD36. G, cross-linking in murine RAW cells involved an IgA primary antibody or a F(ab′)2 fragment (inset). H and I, cross-linking of human U937 cells or primary macrophages involved IgG (clone 131.2) or IgM antibodies (insets). J, oxLDL also triggered CD36 internalization in RAW cells. K and L, CD36 is the major receptor for oxLDL. Wild-type mouse peritoneal macrophages (K) or CD36−/− knock-out mouse peritoneal macrophages (L) were incubated with DiI-labeled oxidized LDL, and noninternalized oxLDL was removed by acid wash. Because the cells in L bind little oxLDL and are not readily visible by fluorescence microscopy, they are outlined in the main panels and shown by differential interference contrast image in the top inset. The lower inset shows quantitation of oxLDL uptake in murine wild-type (+/+) and CD36-deficient (−/−) macrophages, quantified from experiments like K and L. Data are means of three experiments, each counting at least 50 cells of each type. To facilitate comparison between experiments, data were normalized to wild type. In images A–I, the x-z plane is shown below. Scale bars, 10 μm.
To determine whether antibody-induced cross-linking could trigger signaling through CD36 and drive its internalization, live cells were first incubated in the cold in the presence of anti-CD36 antibodies, followed by a labeled secondary antibody. When maintained at 0 °C, CD36 remained at the cell surface but gained a slightly more punctate appearance in all three cell types (Fig. 1, D–F). When cells that had been cross-linked in the cold were warmed to 37 °C, a striking redistribution of CD36 labeling was observed. After 30 min, CD36 was found in a punctate pattern within intracellular compartments (Fig. 1, G–I). To validate the intracellular localization of the punctate fluorescence and to quantify the efficiency of the internalization protocol, cells were treated as above and then fixed without permeabilization and immunostained with primary and a different secondary antibody to detect residual exofacial CD36. Little CD36 remained at the surface under the conditions used (data not shown). Two lines of evidence indicate that the internalization was not caused by engagement of Fcγ receptors, which are abundant on the surface of macrophages. First, extensive internalization was observed when CD36 was cross-linked in RAW cells using a primary IgA antibody followed by fluorescently labeled anti-mouse F(ab′)2 secondary antibodies (Fig. 1G, inset). Second, potent internalization was obtained by exposing U937 (Fig. 1H, inset) and primary human macrophages (Fig. 1I, inset) to anti-human CD36 IgM antibodies, which do not interact with Fc receptors.
CD36 is thought to be responsible for ∼50% of oxLDL uptake by murine and human macrophages (1–3). To determine whether oxLDL would trigger internalization of CD36, RAW cells were incubated in the presence of oxLDL for 30 min and then fixed, permeabilized, and immunostained for CD36. As shown in Fig. 1J, CD36 was internalized into punctate organelles in the cytoplasm following exposure to oxLDL, resembling the pattern observed when antibodies triggered internalization. Similar results were reported earlier in J774 cells transfected with GFP-tagged CD36 (7). To confirm that CD36 is at least partly responsible for oxLDL uptake in these cells, primary macrophages isolated from wild-type or CD36-deficient mice were incubated with DiI-labeled oxLDL. As shown in Fig. 1, K and L, wild-type macrophages bound and internalized a much greater amount of oxLDL than did CD36−/− macrophages. Quantification of three independent experiments revealed that CD36−/− macrophages only internalized 22.6 ± 0.9% as much oxLDL as control macrophages. Together, these results suggest that under the experimental conditions used, CD36 is responsible for the majority of oxLDL uptake and that the internalization of CD36 triggered by oxLDL can be mimicked by cross-linking the receptors with anti-CD36 antibodies.
Internalization of CD36 Is Independent of Dynamin
The recent study by Sun et al. (8) suggested that in COS7 cells, uptake of oxLDL by CD36 depends on dynamin. We therefore examined whether the uptake of endogenous CD36 in macrophages also relies on dynamin. To test this, RAW cells were transiently transfected with either wild-type or dominant negative forms of dynamin1. As before, cells were incubated in the cold in the presence of anti-CD36 and a fluorescently labeled secondary antibody and then warmed to 37 °C for 30 min, acid-washed, and fixed for microscopy. To ensure that dynamin inhibition had in fact occurred, we also labeled the cells with a brief pulse of fluorescent transferrin. Uptake of transferrin is known to occur by clathrin- and dynamin-dependent endocytosis (11). As shown in Fig. 2, A–C, the cells transfected with wild-type dynamin, like their untransfected neighbors, internalized both transferrin and CD36. Of note, there was little overlap in the distribution of these two markers, raising the possibility that they were internalized by different means. Indeed, cells transfected with dominant negative dynamin1 (Fig. 2, D–F) were unable to take up transferrin but internalized CD36 to levels comparable with their untransfected neighbors and wild-type dynamin control cells. Note that the acid washing technique removes bound surface transferrin that is not internalized. The results of three independent experiments, quantified and presented in Fig. 2G, demonstrate that internalization of endogenous CD36 is largely independent of dynamin in macrophages.
Internalization of CD36 Depends on Actin
Because macropinocytosis had been invoked as a mechanism for oxLDL uptake (6), we tested whether antibody-dependent internalization of CD36 required filamentous actin. Cells were pretreated for 30 min with either latrunculin B or cytochalasin D before cross-linking CD36 with primary and secondary antibodies. Fig. 3B shows that cytochalasin D blocked internalization (quantified in Fig. 3G) and caused CD36 to aggregate into large clusters on the cell surface (compare Fig. 3, A and B). Similar results were obtained with latrunculin B (Fig. 3G).
Actin polymerization often follows activation of Rho family GTPases, and Rac has been implicated in forming large membrane ruffles of the type involved in macropinocytosis. To determine whether Rac or Cdc42 activation is occurring during CD36 cross-linking, RAW cells were transfected with YFP fused to the Rac binding domain of the Rac effector PAK1 (PAK1-PBD-YFP). This fusion protein binds to Rac and Cdc42 only when in the active, GTP-bound form but not when bound to GDP. In resting cells, PAK1-PBD-YFP is distributed diffusely throughout the cytoplasm (Fig. 3C, inset) but is recruited to the plasma membrane upon cross-linking of CD36 (main panel in Fig. 3C). This implies that Rac and/or Cdc42 is activated upon CD36 engagement and raises the possibility that the GTPases may contribute to the internalization of the receptors. To test this possibility, cells were preincubated with C. difficile toxin B, which glucosylates and inhibits Rho family proteins. Following treatment with this toxin, CD36 internalization was impaired (Fig. 3, D and G).
To define the contribution of Rho family GTPases further, we transfected RAW cells with constructs that express dominant negative forms of Rac1, Cdc42, or RhoA and measured internalization in transfected cells. As shown in Fig. 3H, Rac1 and Cdc42 dominant negative proteins significantly inhibited CD36 uptake by 56 and 45%, respectively (p < 0.01 in both cases). Because CD36 is also known to associate with integrins, we considered whether its internalization may occur by the Arf6 pathway implicated in integrin recycling (12). Dominant negative Arf6 only reduced CD36 internalization by 25%, although this decrease was also significant (p < 0.05) (Fig. 3H).
The preceding findings, which implicate Rac/Cdc42 and actin polymerization, are consistent with the notion that CD36 is internalized by macropinocytosis, as suggested by Jones and Willingham (6). Because generation of phosphatidylinositol 3,4,5-trisphosphate (PIP3) is an obligatory feature of macropinocytosis, we determined whether this phosphoinositide also accumulates upon CD36 clustering. The distribution of PIP3 was analyzed by transfecting RAW cells with a reporter construct consisting of the Pleckstrin homology domain of AKT linked to GFP. In unstimulated cells, this probe is found predominantly in the cytoplasm, indicating that PIP3 levels are low (Fig. 3E). However, following cross-linking of CD36, AKT-GFP rapidly translocated to the plasma membrane, reflecting increased levels of PIP3 and/or phosphatidylinositol 3,4-bisphosphate, which is a product of dephosphorylation of PIP3 and is also recognized by the probe (Fig. 3F). To ensure that the relocalization of the probe was due to activation of phosphatidylinositol 3-kinase (PI3K), cells were pretreated with the specific inhibitor, LY294002. This treatment blocked the accumulation of AKT-GFP at the membrane (Fig. 3F, inset), confirming that CD36 clustering leads to activation of PI3K.
CD36 Cross-linking Triggers Macropinocytosis, but CD36 Internalization Is Independent of Macropinocytosis
Activation of PI3K and Rac, followed by actin polymerization, are hallmarks of macropinocytosis (13). We therefore tested whether CD36 internalization occurred via the formation of macropinosomes. To assay macropinocytosis we incubated RAW cells in the presence of sulforhodamine as a fluid-phase marker. The macrophages were treated with either M-CSF, a well established inducer of macropinocytosis, or with anti-CD36 primary plus secondary antibodies to induce cross-linking, as above. Because sulforhodamine can also be taken up by endocytosis, only structures larger than 0.7 μm in diameter were scored as macropinosomes. As shown in Fig. 4A, few macropinosomes were seen in untreated cells. In contrast, M-CSF induced the appearance of macropinosomes in a considerable fraction of the cells (Fig. 4B), consistent with earlier findings in primary macrophages (14). Importantly, CD36 cross-linking also induced the formation of macropinosomes in a significant fraction of the cells (Fig. 4C).
To ensure that the large structures containing the fluid-phase marker are indeed macropinosomes, we performed experiments in the presence of LY294002. This inhibitor precluded accumulation of sulforhodamine in M-CSF-stimulated cells (Fig. 4D), confirming that PI3K activity is essential for macropinocytosis (15). The accumulation of the fluid-phase marker was also obliterated by LY294002 (Fig. 4D), consistent with mediation by macropinocytosis. Macropinosome formation is also sensitive to amiloride. Although this drug has multiple cellular targets, its effect on macropinocytosis is likely mediated by inhibition of Na+/H+ exchange because it is replicated by HOE-694. At low micromolar concentrations this agent is a rather selective inhibitor of NHE1, the ubiquitous isoform of the Na+/H+ exchanger (16). We initially confirmed that both amiloride and HOE-694 inhibited the formation of macropinosomes elicited by M-CSF (Fig. 4D) and proceeded to study the effect of these drugs on CD36-induced sulforhodamine uptake. As illustrated in Fig. 4D, both NHE1 antagonists effectively eliminated the formation of large fluid-phase vacuoles, confirming that the structures formed upon engagement of CD36 are bona fide macropinosomes.
The effectiveness of LY294002, amiloride, and HOE-694 as inhibitors of macropinocytosis enabled us to test whether this process is responsible for the internalization of CD36. Remarkably, under conditions that virtually eliminated macropinosome formation, the intake of CD36 was inhibited only to a marginal, statistically insignificant degree (Fig. 4E). These results imply that, although CD36 effectively induces macropinocytosis, this process accounts for only a small fraction of the internalization of the receptor. The remainder occurs by a pathway that, unlike macropinocytosis, is independent of PIP3 and insensitive to inhibition of NHE1.
Tyrosine Kinase Activation Is Required for CD36 Uptake
The preceding results indicate that in macrophages CD36 is internalized, for the most part, by a unique endocytic process that is distinct from macropinocytosis, yet dependent on actin remodeling. The data also imply that PI3K is not an essential component of the pathway leading to actin-dependent endocytosis. We therefore proceeded to investigate the signals required for this specialized endocytic process. In several cell types, CD36 is associated with and can activate Src family kinases. To examine the possible role of these kinases, we assessed tyrosine phosphorylation by both immunofluorescence and immunoblotting. Otherwise untreated RAW cells labeled with the anti-phosphotyrosine-specific antibody PY20 showed little immunoreactivity (Fig. 5A), In contrast, cells where CD36 had been cross-linked displayed distinct immunostaining of the plasma membrane (Fig. 5B). This signal was not detected in cells pretreated with PP1, suggesting that it arose from activation of Src family kinases (Fig. 5C). Immunoblotting of cell lysates (Fig. 5D) confirmed these observations. A marked increase in the tyrosine phosphorylation of several proteins was apparent following CD36 cross-linking. The phosphorylation, which persisted for at least 10 min, was apparent even at 0 °C, indicating the ability of Src kinases to function at reduced temperatures. Accordingly, the phosphorylation detected by immunoblotting was also inhibited by PP1 (Fig. 5D).
To determine whether activation of Src kinases is required for the internalization of CD36, we tested the effect of PP1 on receptor endocytosis. As shown in Fig. 5E, PP1 markedly reduced the internalization of CD36 following antibody-mediated cross-linking, indicating that activation of Src family kinases is essential for the process.
Jun Kinase Activity Is Required for CD36 Internalization
Recent studies have shown that JNK2 is activated in macrophages following exposure to modified LDL, a substrate of CD36, and that this activity is required for foam cell formation (17). To determine whether the activation of JNK2 is essential for CD36 uptake, we treated cells with a JNK inhibitor, SP600125. Fig. 6A illustrates that pretreatment with SP600125 blocked internalization of CD36. Quantification of these results is presented in Fig. 6B. Therefore, activation of JNK, likely downstream of Src kinases, is required for the actin-dependent endocytosis of CD36.
OxLDL-dependent Internalization of CD36 Occurs by a Novel, Actin-dependent Process
As found by others and shown also in Fig. 1J, oxLDL can drive the internalization of CD36, resembling the effects of direct cross-linking of CD36 with specific antibodies. It is therefore conceivable that oxLDL internalization utilizes the novel actin-dependent internalization pathway described above. This notion was tested experimentally using kinase and actin polymerization inhibitors. As in the case of antibody-mediated cross-linking, the endocytosis of CD36 triggered by oxLDL was precluded by latrunculin (Fig. 7, A and B). Similarly Src kinase and JNK inhibition prevented the uptake of CD36 induced by oxLDL (Fig. 7, C and D). In sharp contrast, neither amiloride nor LY294002 significantly affected CD36 internalization. These findings imply that, although both modified LDL (6) and antibody-mediated CD36 clustering (Fig. 4) can trigger macropinocytosis, this process does not contribute importantly to the overall internalization of the receptors.
DISCUSSION
This study was intended to resolve an ongoing controversy regarding the mode of oxLDL uptake by macrophages, a process of enormous clinical importance. Some studies suggested that oxLDL was internalized by macropinocytosis (6), whereas others proposed that it occurred by a pathway akin to conventional endocytosis, dependent on dynamin but not requiring actin. Remarkably, our studies using both oxLDL and anti-CD36 antibodies demonstrated that neither of the preexisting hypotheses was entirely correct. Rather, CD36 engagement triggers internalization of the receptor by an unprecedented mechanism that entails activation of Rac and/or Cdc42, and possibly Arf6, and the subsequent remodeling of actin to generate small endocytic vesicles that are distinct from macropinosomes. The internalization process differs from macropinocytosis not only in the size of the vacuoles generated, but also in that neither PI3K nor Na+/H+ exchange is required for CD36 uptake.
Our pharmacological evidence indicates that Src family kinases are essential for CD36 endocytosis. These findings are compatible with the reported association between the receptors and Src-related kinases (18). The pathway linking kinase activity with actin remodeling, however, is less clear. In microglia, which engage β-amyloid via CD36, Src family kinases have been proposed to signal through focal adhesion complex-associated proteins. In these cells, clustering CD36 induces the activation of Pyk, paxillin, and p130Cas, leading to actin remodeling and stimulated cell migration. Whether the same pathway is responsible for CD36 internalization remains to be established. It is noteworthy, however, that CD36 associates constitutively with integrins in a variety of cell types, including phagocytes (19, 20).
The possible sources of apparent disagreement between our results and those published earlier merit discussion. Sun et al. (8) and Zeng et al. (7) used overexpression of CD36 in nonmyeloid cell lines to characterize oxLDL uptake. We hypothesize that differences in the choice of experimental system account for the discrepancies between their studies and ours. CD36 appears to exist in stable complexes with kinases, integrins, and/or tetraspanins (18–21). Alteration of the stoichiometry of such complexes by heterologous expression is likely to result in atypical forms of CD36 that may become internalized by other, default pathways. To that extent, we feel that analysis of cells that express CD36 endogenously, such as macrophages, yields more reliable results.
Like us, Jones and Willingham (6) utilized macrophages to analyze the uptake of modified LDL. They reported that exposure of the cells to acetylated or oxidized, but not native, LDL prompted the formation of ruffles and large vacuoles. Because the modified LDL molecules were trapped within such vacuoles, macropinocytosis was suggested as the mechanism underlying internalization. Our studies confirmed the occurrence of macropinocytosis but additionally demonstrated that the fraction of CD36 internalized by this process is comparatively small. Instead, the majority of the CD36 was detected in small vesicles, formed by a process that was insensitive to the conventional inhibitors of macropinocytosis. Such smaller vesicles containing modified LDL were also detected by Jones and Willingham. Thus, our findings and theirs are not incompatible, and only our conclusions regarding the relative contribution of macropinosomes and endosomes differ somewhat.
The concentration and solubility of modified LDL may dictate the primary route of internalization. Large aggregates that deposit onto the extracellular matrix are likely to be degraded by a process akin to frustrated phagocytosis prior to internalization. At low concentrations, soluble or loosely adherent modified LDL will be taken up by the actin-dependent endocytosis process described in this paper. Under these conditions, the effectiveness of macropinocytosis is likely to be limited. However, as the concentration of the protein increases, considerable amounts of modified LDL may be trapped within the lumen of forming macropinosomes. Indeed, even native LDL uptake can lead to foam cell formation when macropinocytosis is activated by phorbol esters (22) or M-CSF (23), independently of scavenger receptor activation. Perhaps the development of foam cells in vivo involves a combination of steps where comparatively low levels of modified LDL trigger internalization primarily by a nonmacropinosome mediated process while also simultaneously stimulating macropinocytosis. This would in turn result in the uptake of the more abundant soluble LDL, thereby exacerbating the problem and ultimately leading to foam cell development. If this model holds true, the distinction between oxLDL uptake and bulk LDL uptake may provide an opportunity to develop selective therapeutic approaches to prevent atherogenesis by allowing the clearance of harmful, modified LDL species while preventing the accidental uptake of “innocent bystander” LDL in macropinosomes.
In summary, we found that in addition to macropinocytosis, engagement of CD36 triggers a novel type of actin-dependent endocytosis. The precise mechanism connecting CD36 to the cytoskeleton is still unclear, but we speculate that integrins may provide a physical bridge and/or a signaling intermediate. In this regard, it is noteworthy that in myeloid cells inactive (unengaged) integrins associate with the cytoskeleton and also with CD36. A similar complex may also contribute to the phagocytosis of malaria-infected red cells and to the clearance of β-amyloid by microglia.
Acknowledgments
We thank Dr. H. J. Lang (Aventis Pharma Deutschland, Frankfurt am Main, Germany) for the gift of HOE-694 (3-methylsulfonyl-4-piperido-benzoyl-guanidine methanesulfonate) and Adam Durbin (Hospital for Sick Children, Toronto, Canada) for assistance with p-JNK blots. Wild-type Dynamin1 and K44A dominant-negative Dynamin1 DNA constructs were provided by Sandra L. Schmid (The Scripps Research Institute, La Jolla, CA). PAK1-PBD-YFP was obtained from Joel A. Swanson (University of Michigan Medical School, Ann Arbor, MI). AKT1-PH-GFP was from Tobias Meyer (Stanford University, Stanford, CA). Dominant negative DNA constructs Rac1, cdc42, RhoA, and Arf6 were provided by Gary Bokoch (The Scripps Research Institute).
This work was supported by Canadian Institutes of Health Research Grant FRN68956.
- LDL
- low density lipoprotein
- oxLDL
- oxidized LDL
- DN
- dominant negative
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- M-CSF
- macrophage colony-stimulating factor
- PBD
- PAK-binding domain
- PFA
- paraformaldehyde
- PI3K
- phosphatidylinositol 3-kinase
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- YFP
- yellow fluorescent protein
- DiI
- 1,1′-didodecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- PP1
- 4-amino-5-(4- methylphenyl)-7-(t-butyl)pyrazolo[3,4-d]-pyrimidine.
REFERENCES
- 1.Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., Protter A. A. (1993) J. Biol. Chem. 268, 11811–11816 [PubMed] [Google Scholar]
- 2.Nozaki S., Kashiwagi H., Yamashita S., Nakagawa T., Kostner B., Tomiyama Y., Nakata A., Ishigami M., Miyagawa J., Kameda-Takemura K. (1995) J. Clin. Invest. 96, 1859–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunjathoor V. V., Febbraio M., Podrez E. A., Moore K. J., Andersson L., Koehn S., Rhee J. S., Silverstein R., Hoff H. F., Freeman M. W. (2002) J. Biol. Chem. 277, 49982–49988 [DOI] [PubMed] [Google Scholar]
- 4.Graf G. A., Connell P. M., van der Westhuyzen D. R., Smart E. J. (1999) J. Biol. Chem. 274, 12043–12048 [DOI] [PubMed] [Google Scholar]
- 5.Thorne R. F., Mhaidat N. M., Ralston K. J., Burns G. F. (2007) FEBS Lett. 581, 1227–1232 [DOI] [PubMed] [Google Scholar]
- 6.Jones N. L., Willingham M. C. (1999) Anat. Rec. 255, 57–68 [DOI] [PubMed] [Google Scholar]
- 7.Zeng Y., Tao N., Chung K. N., Heuser J. E., Lublin D. M. (2003) J. Biol. Chem. 278, 45931–45936 [DOI] [PubMed] [Google Scholar]
- 8.Sun B., Boyanovsky B. B., Connelly M. A., Shridas P., van der Westhuyzen D. R., Webb N. R. (2007) J. Lipid Res. 48, 2560–2570 [DOI] [PubMed] [Google Scholar]
- 9.McGilvray I. D., Serghides L., Kapus A., Rotstein O. D., Kain K. C. (2000) Blood 96, 3231–3240 [PubMed] [Google Scholar]
- 10.Patel S. N., Serghides L., Smith T. G., Febbraio M., Silverstein R. L., Kurtz T. W., Pravenec M., Kain K. C. (2004) J. Infect. Dis. 189, 204–213 [DOI] [PubMed] [Google Scholar]
- 11.Herskovits J. S., Burgess C. C., Obar R. A., Vallee R. B. (1993) J. Cell Biol. 122, 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellinen T., Ivaska J. (2006) J. Cell Sci. 119, 3723–3731 [DOI] [PubMed] [Google Scholar]
- 13.Swanson J. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Racoosin E. L., Swanson J. A. (1992) J. Cell Sci. 102, 867–880 [DOI] [PubMed] [Google Scholar]
- 15.Araki N., Johnson M. T., Swanson J. A. (1996) J. Cell Biol. 135, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Counillon L., Franchi A., Pouysségur J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahaman S. O., Lennon D. J., Febbraio M., Podrez E. A., Hazen S. L., Silverstein R. L. (2006) Cell Metab. 4, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang M. M., Bolen J. B., Barnwell J. W., Shattil S. J., Brugge J. S. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7844–7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bamberger M. E., Harris M. E., McDonald D. R., Husemann J., Landreth G. E. (2003) J. Neurosci. 23, 2665–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koenigsknecht J., Landreth G. (2004) J. Neurosci. 24, 9838–9846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao W. M., Vasile E., Lane W. S., Lawler J. (2001) Blood 97, 1689–1696 [DOI] [PubMed] [Google Scholar]
- 22.Kruth H. S., Jones N. L., Huang W., Zhao B., Ishii I., Chang J., Combs C. A., Malide D., Zhang W. Y. (2005) J. Biol. Chem. 280, 2352–2360 [DOI] [PubMed] [Google Scholar]
- 23.Zhao B., Li Y., Buono C., Waldo S. W., Jones N. L., Mori M., Kruth H. S. (2006) J. Biol. Chem. 281, 15757–15762 [DOI] [PubMed] [Google Scholar]