Abstract
The members of the AGC kinase family frequently exhibit three conserved phosphorylation sites: the activation loop, the hydrophobic motif (HM), and the zipper (Z)/turn-motif (TM) phosphorylation site. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates the activation loop of numerous AGC kinases, including the protein kinase C-related protein kinases (PRKs). Here we studied the docking interaction between PDK1 and PRK2 and analyzed the mechanisms that regulate this interaction. In vivo labeling of recombinant PRK2 by 32Pi revealed phosphorylation at two sites, the activation loop and the Z/TM in the C-terminal extension. We provide evidence that phosphorylation of the Z/TM site of PRK2 inhibits its interaction with PDK1. Our studies further provide a mechanistic model to explain different steps in the docking interaction and regulation. Interestingly, we found that the mechanism that negatively regulates the docking interaction of PRK2 to the upstream kinase PDK1 is directly linked to the activation mechanism of PRK2 itself. Finally, our results indicate that the mechanisms underlying the regulation of the interaction between PRK2 and PDK1 are specific for PRK2 and do not apply for other AGC kinases.
The regulation of protein function by phosphorylation and dephosphorylation is a key mechanism of intracellular signaling pathways in eukaryotic organisms. Protein phosphorylation is catalyzed by protein kinases, which are themselves often regulated by phosphorylation (1). The specificity of protein kinases is essential for their cellular functions. In some groups of protein kinases, the specificity is achieved by means of “docking interactions.” Protein kinase docking interactions involve a recognition site on the kinase or a flanking domain that is different from the active site. The most notable example, MAP kinases, uses a docking interaction to specifically recognize substrates, upstream kinases, and phosphatases. Despite the large amount of data on protein kinase docking interactions, e.g. in the MAP kinase field, there is very little information on how these essential interactions are regulated (2–4).
3-Phosphoinositide-dependent protein kinase 1 (PDK1)3 belongs to the AGC family of protein kinases and is the activation loop kinase for several other AGC kinases (5). A key feature of the AGC kinase family members except PDK1 is the presence of a C-terminal extension (CT) to the catalytic core that contains a conserved hydrophobic motif (HM) harboring a phosphorylation site. In many AGC kinases, the HM mediates a docking interaction with PDK1. For example, p90 ribosomal S6 kinase (RSK), p70 S6 kinase (S6K) and serum- and glucocorticoid-induced protein kinase (SGK) interact with PDK1 upon phosphorylation of the HM site (6–9). The phosphorylated HM binds to a HM-binding pocket in the catalytic core of PDK1 that was originally termed the PIF-binding pocket (6, 10).
Besides its role in the docking of substrates to PDK1, the HM/PIF-binding pocket was also identified as a ubiquitous and key regulatory site in likely all AGC kinases (7, 11). Thus, in AGC kinases studied up to now, the HM/PIF-binding pocket serves as an intramolecular docking site for the phosphorylated HM. In summary, the HM has a dual function in AGC kinase activation, (i) mediating the intermolecular interaction with PDK1 and (ii) acting as an intramolecular allosteric activator that stabilizes the active conformation of the kinase domain via binding to the HM/PIF-binding pocket.
The CT of AGC kinases additionally contains a second regulatory phosphorylation site traditionally termed the “turn motif” (TM), and more recently the zipper (Z) site. The Z/TM phosphate interacts with a binding site within the kinase domain, acting like a zipper which serves to support the intramolecular binding of the phosphorylated HM to the HM/PIF-binding pocket (12). Hence, AGC kinases are synergistically activated by phosphorylation at the activation loop, the HM, and the Z/TM sites.
Protein kinase C-related protein kinases (PRKs) (13) (also named PAK for protease-activated kinase (14–16) and PKN for protein kinase N (17)) represent a subfamily of AGC kinases. So far, three PRK isoforms were identified, PRK1, PRK2, and PKN3, which are effectors of the small GTP-binding protein Rho. PRKs, as well as the Rho-associated kinases (ROCKs), are considered to be the protein kinases that mediate the phosphorylation events downstream of Rho activation and both can be inhibited by the highly specific protein kinase inhibitor Y27632 (18). The most notable role described for PRK2 is the control of entry into mitosis and exit from cytokinesis (19). In addition, PRK2 phosphorylates the hepatitis C virus (HCV) RNA polymerase (20). In support of a function in HCV RNA replication, PRK2 inhibitors like Y27632 suppress HCV replication (21).
The N-terminal region of PRK2 possesses three Rho effector (HR1) domains (13), a pseudosubstrate region that is thought to have an autoinhibitory function (22) and a C2-like domain, which is a potential binding site for lipid activators. The C-terminal region of PRK2 harbors the HM that mediates the docking interaction with the HM/PIF-binding pocket in its upstream kinase PDK1 (10, 23). Interestingly, PRKs and also atypical protein kinase Cs (PKCs, PKCζ, and PKCι/λ), contain an acidic residue instead of a phosphorylatable amino acid at the site equivalent to the HM phosphorylation site in other AGC kinases. Therefore, the molecular events that regulate the interaction of PRK2 and PKCζ with PDK1 must be different from the mechanism characterized for S6K, SGK, and RSK.
In the present work we extended and refined the model of docking interaction between PRK2 and PDK1 and characterized C-terminal regions of PRK2 that participate in the regulation of this interaction. The work sheds light on the common as well as specific mechanisms that operate in the regulation of PDK1 docking interaction with its different substrates.
EXPERIMENTAL PROCEDURES
General Materials and Methods
Complete protease inhibitor mixture tablets were from Roche. Protein concentration was estimated using a Coomassie reagent from Perbio. Glutathione-Sepharose was from Amersham Biosciences. A phosphospecific antibody which recognizes the phosphorylated activation loop of several AGC kinases was from Upstate Biotechnology. A phosphospecific antibody which recognizes the phosphorylated Z/TM of PKCβ (phosphor T641) was from Abcam. Anti-GST (B-14) was from Santa Cruz Biotechnology. Fluorescently labeled (IRDye680, IRDye800) secondary antibodies were from LiCor. Chemiluminescent substrate used in immunoblot (Roti-lumin) was from Roth. Immunoblot stripping buffer (Restore) was from Pierce. Human embryonic kidney (HEK) 293 cells (ATCC collection) were cultured on 10-cm dishes in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Invitrogen). Materials for mammalian tissue culture were from Greiner. Molecular biology techniques were performed using standard protocols. Site-directed mutagenesis was performed using a QuikChange kit (Stratagene) following the instructions provided by the manufacturer. DNA constructs used for transient transfection were purified from bacteria using a Qiagen plasmid Mega kit according to the manufacturer's protocol. DNA sequences were verified by automatic DNA sequencing (Applied Biosystems 3100 Genetic Analyzer). Okadaic acid was from Calbiochem, Orthovanadate from Aldrich and LY294002 from Sigma.
In Vivo 32P Labeling of HEK293 Cells Transfected with FLAG-PRK2 and FLAG-PKCζ
HEK293 cells were transfected with pCMV5 plasmids coding for FLAG-PRK2 or FLAG-PKCζ using a modified calcium phosphate procedure (23). 36 h after transfection, the cells were washed with phosphate-free Dulbecco's modified minimal essential medium and incubated for 12 h with [32P]Pi (1 mCi/ml). The cells were lysed with 1.0 ml of ice-cold lysis buffer (50 mm Tris/HCl, pH 7.5, 1 mm EDTA, 1 mm EGTA, 1% (v/v) Triton X-100, 1 mm sodium orthovanadate, 10 mm sodium β-glycerophosphate, 50 mm sodium fluoride, 5 mm sodium pyrophosphate, 0.27 m sucrose, 0.1% (v/v) 2-mercaptoethanol, and protease inhibitor mixture (one tablet per 50 ml)). The lysates were centrifuged at 4 °C for 10 min at 13,000 × g, and the supernatants were incubated for 1 h on a shaking platform with 30 μl of anti-FLAG-Sepharose. The resin was washed four times with 1.0 ml of lysis buffer containing 0.5 m NaCl, and twice with buffer A (50 mm Tris/HCl, pH 7.5, 0.1 mm EGTA, 0.1% (v/v) 2-mercaptoethanol). The matrix-bound proteins were alkylated with 4-vinylpyridine, and analyzed by SDS-PAGE. After autoradiography, the 32P-labeled bands corresponding to FLAG-tagged proteins were eluted from the gel, digested with trypsin, and analyzed by chromatography on a Vydac C18 (150 mm × 2.1 mm) column. Radioactive polypeptides were identified by a combination of mass spectrometry and Edman sequencing as described previously (24).
Peptides
Polypeptides T308tide: KTFCGTPEYLAPEVRR, Crosstide: GRPRTSSFAEG, and PIFtide: REPRILSEEEQEMFRDFDYIADWC, were synthesized by us and the purity verified by HPLC. The identity of polypeptides was confirmed by N-terminal sequencing and mass spectrometry. PIFtide IL/AA: REPRAASEEEQEMFRDFDYIADWC was synthesized by JPT Peptide technologies.
Expression and Purification of Protein Kinases
For the expression and purification of protein kinases fused to GST, pEBG2T-derived plasmids were transfected by a polyethyleneimine (PEI) method (25) (10 μg plasmid/10-cm dish) into HEK293 cells. The cells were lysed after 36 h in lysis buffer. Lysates were frozen in liquid nitrogen and kept at −80 °C until required. Purification involved incubation of the cleared lysate with glutathione-Sepharose, four washes with 0.5 m NaCl in lysis buffer, followed by 10 washes with buffer A, and elution with the same buffer containing 20 mm glutathione. GST fusion proteins were aliquoted, snap-frozen in liquid nitrogen and kept at −80 °C until use. Purity at this stage was between 35 and 85% for different constructs, as estimated by SDS-PAGE and staining with Coomassie Brilliant Blue R250.
Protein Kinase Activity Tests
PRK2 activity assay was performed in a 20-μl mix containing 50 mm Tris-HCl, pH 7.5, 0.05 mg/ml bovine serum albumin, 0.1% (v/v) 2-mercaptoethanol, 10 mm MgCl2, 100 μm [γ-32P]ATP (5–50 cpm/pmol), 0.003% Brij, 1–100 ng of PRK2, and Crosstide (100 μm) as the substrate. The results were similar with proteins expressed and purified in parallel in at least two separate occasions.
PDK1 activity assays were performed essentially as previously described (26), in the presence of 150–300 ng PDK1 and T308tide (100 μm) as the substrate. PDK1 activity was also measured on the GST-CT-PRK2 pull-down. In this case, the assay was performed in a final volume of 20 μl containing excess PIFtide (50 μm), to release PDK1 from GST-CT-PRK2.
Linearity of the PRK2 activity tests was verified routinely by performing the assay on serial dilutions of the enzymes. Independently of the mutant presented in this study, PRK2 had higher specific activity when measured at more diluted concentrations. The lack of linearity could be explained by a mechanism of inhibition by oligomerization. Experiments were repeated at least twice, although most of the experiments were repeated multiple times, with similar results.
Protein-Protein Interaction Assay
The protein-protein interaction experiments were performed by co-transfection of HEK293 cells as previously performed (27). Cells were cotransfected with 10 μg of the wild-type PDK1 plasmid and 10 μg of PKCζ or the wild-type or mutant PRK2. 36-h post-transfection, the cells were lysed in 0.6 ml of lysis buffer. The lysates were cleared by centrifugation at 13,000 × g for 10 min at 4 °C, and 0.5 ml of supernatant was incubated for 2 h at 4 °C with 30 μl of glutathione-Sepharose. The beads were washed twice in lysis buffer containing 0.5 m NaCl, followed by two further washes in buffer A. The beads were resuspended in 30 μl of buffer containing 100 mm Tris/HCl, pH 6.8, 4% (w/v) SDS, 20% (v/v) glycerol, and 200 mm dithiothreitol and subjected to SDS-polyacrylamide gel electrophoresis. The gels were either stained with Coomassie Brilliant Blue or analyzed by immunoblotting with either anti-FLAG or anti-Myc antibodies (described below). In all figures we show duplicates of independent transfection and independent pull-down experiments performed in parallel. Experiments were performed at least twice, although most experiments were repeated multiple times, with similar results. The bands corresponding to the interaction between PDK1 and PRK2 were quantified using the program MultiGauge V3.0 (Fujifilm). For quantification, the amount of bound PDK1 was normalized over the amount of immobilized PRK2.
BiaCore Studies
Binding of GST-PDK1 to PIFtide or PIFtide IL/AA was analyzed by surface plasmon resonance on a BiaCore 3000 system using a streptavidin-coated Sensor chip (SA) and biotin-polypeptides (∼20 RUs), as previously described (10).
RESULTS
Phosphorylation of PRK2 and PKCζ at the Activation Loop and Z/TM Site in Vivo
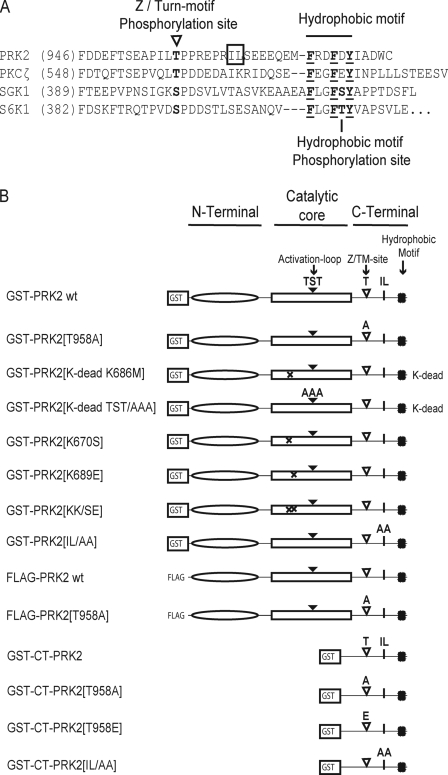
FLAG-tagged PRK2 and PKCζ were overexpressed in HEK293 cells, in vivo-labeled with 32Pi, and the phosphorylated sites were identified via Edman degradation sequencing (see “Experimental Procedures”). The results of this analysis showed that in PRK2 and PKCζ two sites were phosphorylated in vivo: (i) the activation loop site (Thr-816 in PRK2) and (ii) the Z/TM site (Thr-958 in PRK2; depicted in Fig. 1A). No other phosphorylated sites were identified.
FIGURE 1.
A, alignment of the C-terminal amino acid sequence of PRK2 with the equivalent region of PKCζ, S6K1, and SGK1. B, PRK2 constructs employed in this study. A, alignment shows the C-terminal sequence of PRK2 in comparison to PKCζ, SGK1, and S6K1. The sequences corresponding to the hydrophobic motif, the Z/TM phosphorylation and the hydrophobic motif phosphorylation sites (in S6K and SGK) are indicated. The hydrophobic residues Ile-965/Leu-966 (IL) of the C-terminal fragment of PRK2 are boxed. B, schematic representation of PRK2 and PRK2-derived proteins used in this study. GST-PRK2[TST/AAA] corresponds to GST-PRK2[T814A; S815A; T816A], and GST-PRK2[IL/AA] to GST-PRK2[I965A/L966A]; GST-CT-PRK2 expresses the C-terminal 67 residues from PRK2.
Role of the PRK2 Z/TM Phosphorylation Site in PRK2 Docking Interaction with PDK1
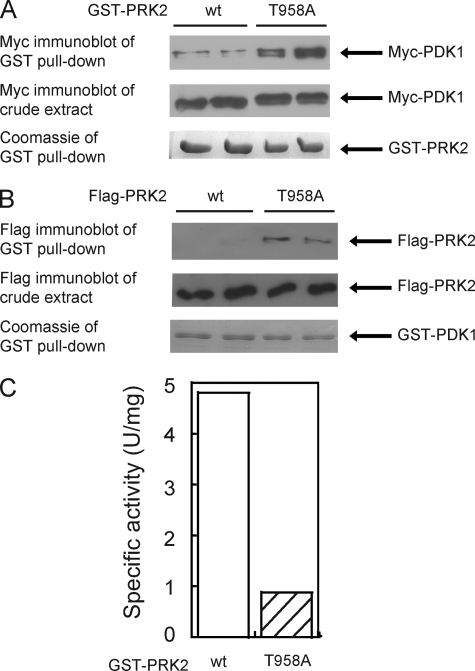
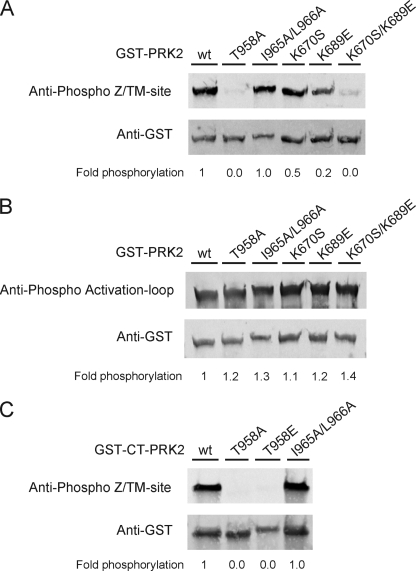
Despite the low degree of sequence identity in the C-terminal region of different AGC kinases (see Fig. 1A), the Z/TM phosphorylation site is functionally conserved in many AGC kinases (12). To analyze whether the Z/TM phosphorylation may regulate the binding of PRK2 and PKCζ to PDK1, we performed interaction studies. To this end, HEK293 cells were co-transfected with plasmids encoding Myc-PDK1 and either wild-type or Z/TM site (T958A) mutated GST-PRK2 (see Fig. 1B for all wild-type and mutant forms of PRK2 used in this study). After cell lysis, GST-PRK2 was immobilized on glutathione-Sepharose and binding of Myc-PDK1 was analyzed by immunoblot using an anti-Myc antibody. As shown in Fig. 2A, GST-PRK2[T958A] bound ∼6-fold more PDK1 than wild-type GST-PRK2. Similar results were obtained when GST-PDK1 was co-expressed with FLAG-PRK2 or FLAG-PRK2[T958A] (Fig. 2B). In vitro characterization of PRK2[T958A] indicated that the specific activity was ∼five times reduced compared with wt PRK2 (Fig. 2C). Immunoblot analysis confirmed that GST-PRK2[T958A] was not phosphorylated at the Z/TM site (Fig. 3A) while the activation loop phosphorylation was similar to that observed for GST-PRK2 wt (Fig. 3B). Hence, the mutation of the Z/TM phosphorylation site in PRK2, and not a secondary effect on the activation loop phosphorylation, increased PRK2 interaction with PDK1.
FIGURE 2.
Mutation of the Z/TM phosphorylation site Thr-958 of PRK2 increases its interaction with PDK1. A and B, HEK293 cells were transiently co-transfected with plasmid DNA encoding PDK1 and wt PRK2 or PRK2 mutated in its Z/TM phosphorylation site (T958A), respectively. GST-fused PRK2 constructs were co-transfected with Myc-PDK1 (A) and FLAG-tagged PRK2 constructs with GST-PDK1 (B). After 36 h, the cells were lysed, and the GST fusion proteins were purified with glutathione-Sepharose. Bound proteins were analyzed by SDS-PAGE followed by Coomassie staining or immunoblotting to detect co-purified proteins using an anti-Myc antibody (A) or an anti-FLAG antibody (B). To verify the similar expression of Myc-PDK1 (A) and of wild type and mutant FLAG-PRK2 (B), equal amounts of the total cell lysates were subjected to SDS-PAGE and immunoblotted using anti-Myc antibody (A) or anti-FLAG antibody (A). Duplicates of the transfection of each condition are shown. The extent of PDK1 binding to GST-PRK2 was quantified using the program MultiGauge V3.0 (Fujifilm) and normalized over the amount of immobilized PRK2. A value of 1 was assigned to wild-type PRK2. C, specific activity of wt and mutated (T958A) GST-PRK2 purified from HEK293 cells (100 ng) was measured in vitro using the polypeptide Crosstide as a substrate (see “Experimental Procedures”).
FIGURE 3.
Z/TM site and activation loop phosphorylation of PRK2 constructs employed in this study. HEK293 cells were transfected with DNA constructs encoding full-length PRK2 or the C-terminal region (CT) of PRK2 as GST fusion proteins. The cells were lysed 48-h post-transfection, and the GST fusion proteins were purified with glutathione-Sepharose. Bound proteins were eluted with SDS-sample buffer (A and B) or glutathione (C). Aliquots of the proteins were subjected to SDS-PAGE followed by immunoblotting. Phosphorylation of the Z/TM-site (A and C) and the activation loop site (B) was detected with phosphospecific antibodies for the respective site. Total protein amounts were detected using an anti-GST antibody. Secondary antibodies were fluorescently labeled and immunoblots were developed with a FLA-9000 Starion (Fujifilm). The extent of phosphorylation was quantified using the program MultiGauge V3.0 (Fujifilm) and normalized over the amount of loaded protein. A value of 1 was assigned to the phosphorylation of wt GST-PRK2 (A and B) or wt GST-CT-PRK2 (C).
In addition, we analyzed the role of Z/TM phosphorylation of PKCζ in the binding to PDK1. In contrast to PRK2, mutation of the corresponding phosphorylation site in PKCζ (T560A) had no effect on its interaction with PDK1 in co-transfection studies using either GST-PKCζ and Myc-PDK1 or FLAG-PKCζ and GST-PDK1, respectively (supplemental Fig. S1). Additionally, mutation of the Z/TM phosphorylation site in S6K and SGK did not affect their interaction with PDK1. Taken together, these results suggested that phosphorylation of the Z/TM site in PRK2 inhibited its interaction with PDK1. Interestingly, this regulatory mechanism appeared to be rather specific for PRK2.
Effect of Mutation of the PRK2 Z/TM Phosphorylation Site on the Interaction of GST-CT-PRK2 with PDK1
The increased binding of PRK2 to PDK1 caused by mutation of the Z/TM phosphorylation site could be explained by (i) an increased binding affinity of the C-terminal residues of PRK2 to PDK1 in the absence of Z/TM phosphorylation, (ii) a decreased intramolecular binding of the PRK2 C-terminal fragment to its catalytic domain in the absence of Z/TM phosphorylation or (iii) a combination of both. To distinguish between these possibilities, we co-transfected HEK293 cells with a plasmid encoding Myc-PDK1 together with plasmids coding for the C-terminal residues of PRK2 as GST-fusion proteins (GST-CT-PRK2). Wild-type GST-CT-PRK2 efficiently bound to Myc-PDK1 (Fig. 4A). However, in contrast to the effect on full-length PRK2, mutation of the Z/TM phosphorylation site to Ala on the CT construct of PRK2 (GST-CT-PRK2[T958A]) had no effect on its interaction with PDK1. This suggested that the affinity of the CT of PRK2 toward PDK1 was not increased upon mutation of this site. The interaction between the PRK2 C terminus and PDK1 was also not affected when the Z/TM phosphorylation site was mutated to glutamate (GST-CT-PRK2[T958E]; Fig. 4A), which mimics phosphorylation. As a control, we verified that GST-CT-PRK2, but not GST-CT-PRK2[T958A] or GST- CT-PRK2[T958E] was phosphorylated at the Z/TM phosphorylation site in vivo (Fig. 3C). Furthermore, transfection and purification of GST-CT-PRK2 and GST-CT-PRK2[T958A] followed by a PDK1 activity assay revealed that Ala mutation of the Z/TM phosphorylation site did not affect the ability of the overexpressed C-terminal PRK2 fragment to pull-down endogenous PDK1 (supplemental Fig. S2). Taken together, the lack of phosphorylation of the PRK2 C terminus did not result in a more pronounced binding to PDK1 as was observed for the full-length PRK2. These data strongly indicated that Z/TM phosphorylation did not modify the binding affinity of CT-PRK2 toward PDK1.
FIGURE 4.
Characteristics of the binding between the C-terminal fragment of PRK2 and PDK1. A and B, HEK293 cells were transfected with DNA constructs expressing Myc-PDK1 together with constructs expressing GST or GST-PRK2 fusion proteins, and the interaction was analyzed as described in the legend to Fig. 2. Duplicates of each condition are shown. The extent of PDK1 binding was quantified using the program MultiGauge V3.0 (Fujifilm) and normalized over the amount of immobilized PRK2. A value of 1 was assigned to wild-type PRK2. A, the GST-fused C-terminal region of PRK2 (amino acids 918–984; GST-CT-PRK2) was analyzed for binding to Myc-PDK1. Binding of GST-CT-PRK2 (wt) to PDK1 was not affected by mutation of the Z/TM phosphorylation site to Ala (T958A; 0.9-fold binding compared with wt) or Glu (T958E; 0.7-fold binding). In contrast, substitution of Ile-965 and Leu-966 to Ala (I965A/L966A) abolished this interaction (0.0-fold binding). B, full-length GST-PRK2 was mutated in its P-Z/TM binding site. Binding of PRK2 to PDK1 was increased by substitution of Lys-670 for Ser (K670S; 1.8-fold binding) or Lys-689 for Glu (K689E; 10.2-fold), similar to the mutation of the Z/TM phosphorylation site to Ala (T958A; 6.2-fold). The double mutation (K670S/K689E) did not further increase this interaction (6.5-fold binding). C, PRK2 C terminus-derived polypeptides (PIFtide, REPRILSEEEQEMFRDFDYIADWC) and PIFtide IL/AA were used to analyze the ability of the PRK2 C-terminal region to activate PDK1 in vitro. The activity of 0.2 μm PDK1 was measured in triplicates using the peptide T308tide as a substrate. Both, PIFtide wt and PIFtide mutated in Ile-965 and Leu-966 (IL/AA) activated PDK1 to a similar level. However, the AC50 of PIFtide IL/AA was 0.82 ± 0.25 units/mg, considerably higher than AC50 PIFtide wt (0.20 ± 0.04 units/mg). D, ability of biotinylated PIFtide or PIFtide IL/AA to bind to PDK1 was analyzed using surface plasmon resonance technology using SA Chips on a BiaCore system. The binding of PDK1 to biotin-peptide was performed at the indicated concentrations of PDK1. The inset shows the data at equilibrium and the estimated dissociation constant of PDK1 for each polypeptide. PDK1 is injected to the system during the “association” part of the curve. When PDK1 is not longer injected through the system, PDK1 dissociates from the chip (dissociation part of the curve). The higher slope of the dissociation from PIFtide IL/AA indicates a higher dissociation rate.
Effect of Mutation of the PRK2 P-Z/TM Binding Site on PRK2 Docking Interaction with PDK1
We next examined whether the increased interaction of GST-PRK2[T958A] with PDK1 could be due to a decreased intramolecular binding of the CT-PRK2 to its Z/TM phosphate-binding site (P-Z/TM binding site) in the catalytic domain. We previously described lysines 670 and 689 of PRK2 as key residues of its P-Z/TM binding site (12). Thus, we prepared constructs of PRK2 mutated in one or both of these residues by substituting Lys-670 for Ser and Lys-689 for Glu. Lysine 689 was predicted to be exposed to the solvent in the absence of Z/TM phosphorylation. Hence, mutation to glutamate (replacing a positive charge for a negative one) should result in the complete loss of function of the P-Z/TM binding site. The single mutation of Lys-670 (PRK2[K670S]) increased the binding to PDK1 compared with wt PRK2 (Fig. 4B). More prominently, PRK2[K689E] bound ∼10-fold higher amounts of PDK1 than PRK2 wt (Fig. 4B). The interaction with PDK1 did not further increase upon mutation of both Lys residues (PRK2[K670S/K689E]). Thus, destruction of the P-Z/TM binding site (Fig. 4) and mutation of the Z/TM phosphorylation site (Fig. 2) of PRK2 led to a similar increase in PDK1 binding. Immunoblot analysis showed that mutation of the PRK2 P-Z/TM binding site reduced its phosphorylation at the Z/TM site (Fig. 3A). In contrast, the activation loop phosphorylation of these mutants was comparable to wt PRK2 (Fig. 3B). Thus, mutation of the P-Z/TM binding-site prompted the undocking of the CT-PRK2 from its binding site and the dephosphorylation of the Z/TM site. Taken together, the data demonstrated that the mutation of the P-Z/TM binding site coupled to the dephosphorylation of the Z/TM site on PRK2 enhanced the docking to PDK1. Mechanistically, the data further indicated that the dephosphorylation of the Z/TM site increased docking to PDK1 by decreasing the affinity of the CT-PRK2 to the PRK2 P-Z/TM binding site.
Role of the PRK2 C-terminal Ile-965/Leu-966 Hydrophobic Patch in the High Affinity Interaction of GST-CT-PRK2 with PDK1
PIFtide, a 24-amino acid polypeptide derived from the CT of PRK2, interacts with higher affinity with PDK1 than the corresponding CT polypeptides from other AGC kinases (8). Besides the conserved hydrophobic residues of the HM (Phe-974, Phe-977, and Tyr-979) required for PDK1 binding (28), PRK2 possesses a second CT hydrophobic patch (Ile-965 and Leu-966) that is poorly conserved in other AGC kinases (Fig. 1A). We investigated whether Ile-965 and Leu-966 also played a role in the interaction with PDK1. In our protein-protein interaction assay, we observed that mutation of Ile-965 and Leu-966 to Ala (CT-PRK2[I965A/L966A]) abolished the interaction between GST-CT-PRK2 and PDK1 (Fig. 4A). This suggested that the high affinity interaction between the C terminus of PRK2 and PDK1 was not only mediated by the HM, but also required the non-conserved hydrophobic patch.
To further verify the role of the PRK2 residues Ile-965 and Leu-966 in the interaction with PDK1, we tested the effect of non-mutated and mutated (IL/AA) PIFtide polypeptides on the activity of PDK1. PIFtide activated PDK1 to ∼400%. The IL/AA mutant also activated PDK1 to the same level, but had ∼6-fold higher AC50 (Fig. 3C), further suggesting a decreased affinity. In agreement with this, GST-CT-PRK2 IL/AA pulled-down lower levels of endogenous PDK1 (supplemental Fig. S2).
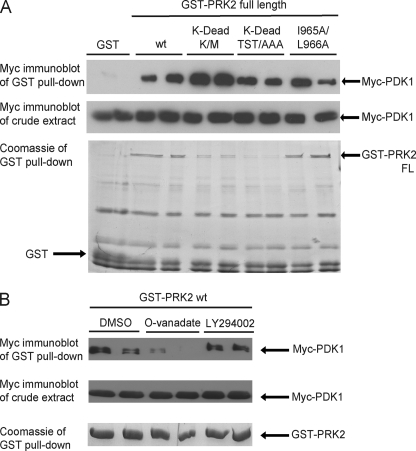
In addition, we tested the binding of the two polypeptides to PDK1 using surface plasmon resonance technology. To this end, the biotinylated polypeptides were bound to the chip and the interaction was evaluated upon injection of purified GST-PDK1 protein (Fig. 4D). In agreement with the above results, the dissociation constant of the interaction was ∼43 nm for PIFtide wt and ∼119 nm for PIFtide IL/AA. Comparison of the dissociation slope clearly showed that PDK1 dissociated at a faster rate from PIFtide IL/AA than from wt PIFtide, with more than 3-fold higher dissociation rate (Fig. 4D). In summary, the results showed that the hydrophobic patch (Ile-965/Leu-966) in the CT of PRK2 is required for the high affinity interaction with PDK1. Interestingly, both GST-PRK2 and GST-PRK2[I965A/L966A] bound similar levels of Myc-PDK1 (Fig. 5A). Thus, even if the mutation affected the interaction of the CT-PRK2 to PDK1, the mutation of the hydrophobic Ile/Leu patch did not show an effect on the interaction between full-length PRK2 and PDK1.
FIGURE 5.
Mutations that abolish the activity of PRK2 increase its interaction with PDK1. HEK293 cells were transfected with DNA constructs expressing Myc-PDK1 together with constructs expressing GST or GST-PRK2 fusion proteins. The interaction was analyzed as described in the legend to Fig. 2. Duplicates of each condition are shown. The extent of PDK1 binding was quantified using the program MultiGauge V3.0 (Fujifilm) and normalized over the amount of immobilized PRK2. A value of 1 was assigned to wild-type PRK2. A, mutation of Lys-686 in the ATP-binding site to Met (PRK2[K-dead K/M]) or substitution of Thr-814, Ser-815, and Thr-816 in the activation loop for Ala (PRK2[K-dead TST/AAA]) generated inactive, so-called kinase-dead (K-dead), PRK2 proteins. These proteins were expressed in HEK293 cells to a lower level than wild-type PRK2 (wt). However, the interaction between PDK1 and PRK2[K-dead TST/AAA] or PRK2[K-dead K/M] was even increased compared with PRK2 wt (3.7-fold for both K-dead PRK2 constructs). Mutation of Ile-965 and Leu-966 to Ala (I965A/L966A) in full-length PRK2 had no effect on the interaction with PDK1 (1.1-fold binding). B, GST-PRK2 wt was co-expressed with Myc-PDK1, and the cells were treated with 1 mm orthovanadate (O-vanadate), 50 μm LY294002, or DMSO as a control 2 h before cell lysis. The treatment with LY294002 had no effect on the interaction between PRK2 and PDK1 while the addition of orthovanadate led to a decreased binding of PRK2 to PDK1.
Increased Interaction of Kinase-dead Mutants of PRK2 with PDK1
To test if inactive forms of PRK2 have a different avidity toward PDK1, we generated two mutants of PRK2 that are virtually inactive and therefore called kinase-dead (K-dead): (i) Lys-686 in the ATP-binding site was substituted for Met (PRK2[K-dead K/M]); (ii) Thr-814, Ser-815, and Thr-816 within the activation loop were mutated to Ala (PRK2[K-dead TST/AAA]). The expression of the two K-dead mutants was lower, approximately five times less than the wt protein (Fig. 5A). Despite this, PRK2[K-dead TST/AAA] and PRK2[K-dead K/M] pulled-down ∼4-fold higher amounts of Myc-PDK1 (Fig. 5A). These results demonstrated that the inactive mutants of PRK2 bound with higher avidity to PDK1, providing evidence that PDK1 interacted more efficiently with an inactive PRK2 substrate.
Effect of Orthovanadate on the Interaction between PRK2 and PDK1
Finally, we investigated whether the interaction between PRK2 and PDK1 could be regulated by intracellular signaling events. For that purpose, we analyzed the PRK2-PDK1 interaction in the presence of compounds that affect intracellular signaling. The Tyr phosphatase inhibitor orthovanadate significantly decreased the interaction between PRK2 and PDK1 whereas this interaction was not affected by the PI3-kinase inhibitor LY294002 (Fig. 5B). Since we are not aware of a Tyr phosphorylation site on PRK2, these results suggested that the effect may be indirect. Thus, tyrosine kinases, independent from the PI3-kinase pathway, could be involved in the regulation of the PRK2-PDK1 interaction. However, the mechanisms involved remain to be studied.
DISCUSSION
Docking interactions are at the essence of the signaling specificity of a group of protein kinases, including MAP kinases and PDK1. To achieve temporal specificity, the docking interactions must be regulated. However, very little is known about the regulatory mechanisms involved. Here we describe a novel way of regulating the interaction between PDK1 and one of its substrates, PRK2. Interestingly, we discovered that the mechanism that decreases the interaction with the upstream kinase is directly linked to the mechanism of activation of the substrate kinase.
We initially observed that mutation of the Z/TM phosphorylation site to Ala increased the binding of PRK2 to PDK1 (Fig. 2). This could have been due to (i) an increased binding avidity of the C-terminal residues of PRK2 to PDK1, (ii) a decreased intramolecular binding of the PRK2 C terminus to its catalytic domain or (iii) a combination of both. The binding of the isolated PRK2 C terminus to PDK1 was neither increased by mutation of the Z/TM phosphorylation site to Ala nor decreased by mimicking phosphorylation at this site using glutamic acid (Fig. 4A). These results were in line with previous surface plasmon resonance studies, demonstrating that PDK1 interacted with equal affinity with both, an S6K polypeptide phosphorylated at the Z/TM site and the equivalent non-phosphorylated counterpart (12). The data thus excluded (i) and (iii) above as possible models to explain the Z/TM-mediated regulation of the interaction between PDK1 and PRK2. In contrast, mutation of the P-Z/TM binding site in PRK2 caused a similar increase on PDK1-binding (Fig. 4B) as observed for mutation of the Z/TM phosphorylation site. These data supported the hypothesis that the mutation of the Z/TM phosphorylation site to Ala increased the avidity of PRK2 for PDK1 by decreasing its affinity to the P-Z/TM binding site on PRK2. Thus, our results suggested that phosphorylation of the Z/TM site of PRK2 decreased the binding affinity of PRK2 for PDK1 by increasing the intramolecular binding of the CT of PRK2 to the catalytic domain of PRK2. Taken together, we conclude that Z/TM phosphorylation inhibits the interaction with its upstream kinase, PDK1.
Although PKCζ features an acidic amino acid instead of a phosphorylatable residue in its HM, similarly to PRK2, its interaction with PDK1 was not affected by mutation of the Z/TM phosphorylation site (supplemental Fig. S1). Similar results were obtained for SGK and S6K. Furthermore, in AGC kinases such as SGK, S6K, and PKB, Z/TM phosphorylation seems to be constitutive rather than regulated. Hence, the regulation of the PRK2 docking interaction with PDK1 by Z/TM phosphorylation appears to be a particular characteristic of PRK2. Moreover, our results suggest that the Z/TM kinase and phosphatase may be important factors in PRK2 regulation. So far, the kinase that phosphorylates PRK2 at the Z/TM phosphorylation site has not been identified. Recently, it was shown that the mammalian target of rapamycin complex 2 (mTORC2) is required for the Z/TM phosphorylation of several members of the PKC family as well as PKB (29, 30). These studies demonstrate that mTORC2-dependent phosphorylation of the Z/TM site is crucial for folding and stability of PKCs and PKB. Whether mTOR could also be involved in the phosphorylation of the PRK2 Z/TM site and whether this phosphorylation is important for PRK2 stability remains to be investigated.
Besides the regulation by phosphorylation, other mechanisms exist to modulate the activity of PRKs, namely interaction with the small GTPase Rho and lipid binding (31). Recently, Lim et al. (32–34) described that the PRK C terminus is required for the activation by Rho and that it is additionally involved in the regulation of PRKs by lipids. However, it is not yet clear how this is related to the regulatory mechanisms we have observed in this work.
Here we demonstrated that mutation of the Z/TM phosphorylation site on full-length PRK2 greatly decreased its specific activity in vitro (Fig. 2C) This result suggested that the Z/TM phosphorylation of PRK2 was required to achieve maximal specific activity. Together with our other findings, we conclude that the Z/TM phosphorylation site has a dual role, in regulating kinase activity and in regulating the docking interaction with PDK1.
For certain AGC kinases, Z/TM phosphorylation may have an opposite effect on the interaction with PDK1. Thus, recent peptide array data indicated that peptides derived from the CT of PKA interacted with PDK1 only when they were phosphorylated at the turn-motif site (35). In this respect, it should be mentioned that the TM site of PKA is thought to have a different structure and function than that of the Z/TM site of other AGC kinases (12). The mechanism by which these phosphopeptides, synthesized on cellulose paper, showed increased affinity for PDK1 was not investigated.
The HM is required for the specific docking interaction of PDK1 with a series of its substrates. Since PDK1 phosphorylates its substrates at the activation loop, an additional interaction site must exist along this position. However, synthetic polypeptides derived from the activation loop of PKB and S6K are extremely poor substrates for PDK1 in vitro (10), suggesting that the activation loop may not contribute significantly to this interaction. Besides the HM and the activation loop, we here identified another region in the C-terminal sequence of PRK2, the Ile-965-Leu-966 hydrophobic patch, that is required for the high affinity interaction of CT-PRK2 with PDK1 (Fig. 4, C and D). This hydrophobic patch is not conserved among AGC kinase substrates of PDK1 and may be responsible for the higher binding affinity of PDK1 to the C-terminal region of PRK2 (PIFtide) compared with the C-terminal regions of other AGC kinases (8). In agreement with this, screening data recently identified Leu-966 as a PIFtide determinant for interaction with PDK1 (35). It is not clear why the mutation of the hydrophobic patch did not affect the interaction of full-length PRK2 with PDK1. One possibility is that the hydrophobic patch is equally involved in the intermolecular docking interaction with PDK1 and the intramolecular interaction with the PRK2 catalytic core. The recent peptide array study also suggested that another motif, termed Ade (adenosine binding) motif, may be important for interaction of C-terminal regions of PRK2 with PDK1 (35). Altogether, the data suggest that the main determinants for interaction of PRK2 with PDK1 are located within the CT extension to the catalytic core.
Interestingly, our present work also shows that inactive PRK2 interacts better than active PRK2 with PDK1 (Fig. 5A). Inactive structures of AGC kinases PKB, SGK, and MSK have been published (11, 36–38). In all these inactive kinase structures, the α-B and α-C helices, which form part of the HM/PIF-binding pocket, are disordered. The higher degree of interaction of inactive PRK2 with PDK1 could be explained if the inactive structure of PRK2 was also having a disrupted HM/PIF-binding pocket, and therefore, the C terminus of PRK2 would be more readily available for interaction with PDK1.
Importantly, our recent studies on PDK1 itself show that the α-C helix is extremely mobile in solution (39), incompatible with a stable α-helix observed in active crystal structures of protein kinases. In addition, deuterium exchange experiments in solution show that the binding of the C-terminal region of PRK2 (PIFtide) or small molecule compounds to the HM/PIF-binding pocket of PDK1 stabilizes the α-C helix, and prompts allosteric effects on the ATP binding site, thereby activating PDK1 (39). Thus, there is strong support for the mechanistic model by which the docking interaction of PRK2 to the HM/PIF-binding pocket on PDK1 would prompt an allosteric activation of PDK1 and in this manner support the phosphorylation of the activation loop of PRK2.
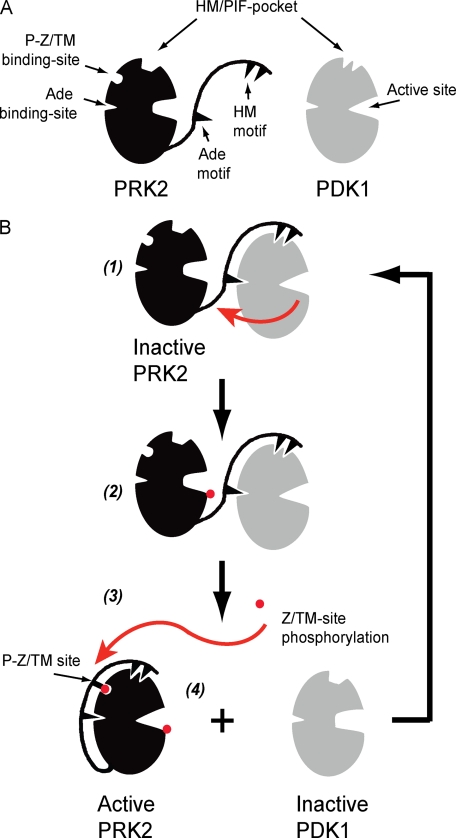
In Fig. 6B, we present a simple model describing the mechanism by which PRK2 may interact with PDK1. An inactive PRK2 molecule has a free CT and interacts with the Ade and PIF-pocket binding sites on PDK1 (1). This interaction brings PRK2 into proximity of its kinase, and it also increases the intrinsic catalytic activity of PDK1 to phosphorylate the activation loop of PRK2 (2). In a subsequent step, the PRK2 Z/TM site is phosphorylated (3) and this triggers the intramolecular interaction of the Z/TM phosphate with its P-Z/TM binding-site. At this stage, by similarity with other active AGC kinases, the model suggests that the Ade and HM motifs would also interact intramolecularly with their corresponding binding sites on the catalytic core of PRK2. However, this part of the model still requires experimental support. Altogether, these intramolecular interactions will restrict the interaction of the CT of PRK2 with PDK1, thereby promoting PRK2 dissociation from PDK1 (4). Ultimately, the activation loop and Z/TM site phosphorylations, together with the phosphate mimicking aspartic acid in the HM, act locally and allosterically to stabilize the HM/PIF-binding pocket and the active site of PRK2 in the active conformation. In this closed active conformation, PRK2 molecules have low affinity for PDK1, since the CT-interacting region is not free to interact with PDK1. Finally, the intramolecular binding of the PRK2 CT protects the phosphorylated Z/TM site from dephosphorylation, thereby also stabilizing the active conformation of PRK2.
FIGURE 6.
Model describing the regulation of the interaction between PRK2 and PDK1. A, schematic representation of PRK2 and PDK1 showing the catalytic core possessing the HM/PIF-pocket, the phospho-Z/turn-motif binding site (P-Z/TM binding-site), the Ade binding site and the active site. The Ade and HM motifs are indicated within the C-terminal extension to the PRK2 catalytic core. The catalytic core of PRK2 and PDK1 are shown in inactive and active conformations, respectively. B, CT of inactive PRK2 is free to interact with PDK1; the interaction at the HM/PIF pocket of PDK1 brings PRK2 in close proximity to PDK1 and also prompts allosteric conformational changes in PDK1 that affect the ATP binding site and activates PDK1 to phosphorylate the substrate, PRK2. The interaction between PRK2 Ade motif and PDK1 Ade binding site is hypothetical (1 and 2). Phosphorylation of the Z/TM phosphorylation site (P-Z/TM site) triggers the interaction between this phosphate and the P-Z/TM binding site, acting like a zipper enabling the interaction of the HM and Ade motif to the PRK2 catalytic core (3 and 4). The increased interaction of the PRK2 CT with its own catalytic core prompts the dissociation from PDK1. In the absence of a bound HM, the PDK1 structure is stabilized in an inactive conformation, being again available for interaction with its substrates.
Finally, it is worth noting that the mechanisms that operate in the regulation of PRK2 docking interaction with PDK1 appeared to be very different from those of other AGC kinases. Altogether, the studies on AGC kinases provide evidence for distinct intermolecular and intramolecular mechanisms of regulation that have evolved based on one key regulatory site on the kinase domain, the PIF-binding pocket, and a flexible C-terminal extension to the catalytic core.
Supplementary Material
Acknowledgments
We thank Dario Alessi for help in the in vivo labeling experiment and Maria Deak for mutagenesis of GST-PIF. We are also grateful to Katrien Busschots and Jörg Schulze for careful reading of the manuscript and comments on the work.
This work was supported by the Europrofession Foundation (Saarbrücken, Germany), BMBF Go-Bio, and DFG (BI 1044/5-1).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PDK1
- 3-phosphoinositide-dependent protein kinase-1
- PRK
- protein kinase C-related protein kinase (also termed PAK or PKN)
- PKC
- protein kinase C
- PKCζ
- atypical protein kinase C zeta
- PKB
- protein kinase B (also termed Akt)
- RSK
- p90 ribosomal S6 kinase, S6K, p70 ribosomal S6K
- SGK
- serum and glucocorticoid-induced protein kinase
- MSK
- mitogen and stress-activated protein kinase
- ROCK
- Rho-associated protein kinase
- AGC kinases
- group of protein kinases comprising PKA, PKG, and PKC
- GST
- glutathione S-transferase
- HM
- hydrophobic motif
- Z/TM phosphorylation site
- zipper/turn-motif phosphorylation site
- P-Z/TM binding site
- Z/TM phosphate binding site
- PIF
- PDK1-interacting fragment
- PIFtide
- polypeptide comprising the last 24 amino acids from PIF
- CT
- C-terminal
- PI 3-kinase
- phosphoinositide 3-kinase
- wt
- wild type
- GST
- glutathione S-transferase.
REFERENCES
- 1.Pawson T., Scott J. D. (2005) Trends Biochem. Sci. 30, 286–290 [DOI] [PubMed] [Google Scholar]
- 2.Mayor F., Jr., Jurado-Pueyo M., Campos P. M., Murga C. (2007) Cell Cycle 6, 528–533 [DOI] [PubMed] [Google Scholar]
- 3.Biondi R. M., Nebreda A. R. (2003) Biochem. J. 372, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reményi A., Good M. C., Lim W. A. (2006) Curr. Opin. Struct. Biol. 16, 676–685 [DOI] [PubMed] [Google Scholar]
- 5.Vanhaesebroeck B., Alessi D. R. (2000) Biochem. J. 346, 561–576 [PMC free article] [PubMed] [Google Scholar]
- 6.Biondi R. M., Komander D., Thomas C. C., Lizcano J. M., Deak M., Alessi D. R., van Aalten D. M. (2002) EMBO J. 21, 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frödin M., Antal T. L., Dümmler B. A., Jensen C. J., Deak M., Gammeltoft S., Biondi R. M. (2002) EMBO J. 21, 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biondi R. M., Kieloch A., Currie R. A., Deak M., Alessi D. R. (2001) EMBO J. 20, 4380–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frödin M., Jensen C. J., Merienne K., Gammeltoft S. (2000) EMBO J. 19, 2924–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biondi R. M., Cheung P. C., Casamayor A., Deak M., Currie R. A., Alessi D. R. (2000) EMBO J. 19, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J., Cron P., Thompson V., Good V. M., Hess D., Hemmings B. A., Barford D. (2002) Mol. Cell 9, 1227–1240 [DOI] [PubMed] [Google Scholar]
- 12.Hauge C., Antal T. L., Hirschberg D., Doehn U., Thorup K., Idrissova L., Hansen K., Jensen O. N., Jørgensen T. J., Biondi R. M., Frödin M. (2007) EMBO J. 26, 2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer R. H., Ridden J., Parker P. J. (1995) Eur. J. Biochem. 227, 344–351 [DOI] [PubMed] [Google Scholar]
- 14.Gabrielli B., Wettenhall R. E., Kemp B. E., Quinn M., Bizonova L. (1984) FEBS Lett. 175, 219–226 [DOI] [PubMed] [Google Scholar]
- 15.Morrice N. A., Gabrielli B., Kemp B. E., Wettenhall R. E. (1994) J. Biol. Chem. 269, 20040–20046 [PubMed] [Google Scholar]
- 16.Yu W., Liu J., Morrice N. A., Wettenhall R. E. (1997) J. Biol. Chem. 272, 10030–10034 [DOI] [PubMed] [Google Scholar]
- 17.Mukai H., Ono Y. (1994) Biochem. Biophys. Res. Commun. 199, 897–904 [DOI] [PubMed] [Google Scholar]
- 18.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt A., Durgan J., Magalhaes A., Hall A. (2007) EMBO J. 26, 1624–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S. J., Kim J. H., Kim Y. G., Lim H. S., Oh J. W. (2004) J. Biol. Chem. 279, 50031–50041 [DOI] [PubMed] [Google Scholar]
- 21.Kim S. J., Kim J. H., Sun J. M., Kim M. G., Oh J. W. (2009) J. Viral Hepat. 16, 697–704 [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa M., Shibata H., Toshimori M., Mukai H., Ono Y. (1996) Biochem. Biophys. Res. Commun. 220, 963–968 [DOI] [PubMed] [Google Scholar]
- 23.Balendran A., Biondi R. M., Cheung P. C., Casamayor A., Deak M., Alessi D. R. (2000) J. Biol. Chem. 275, 20806–20813 [DOI] [PubMed] [Google Scholar]
- 24.Campbell D. G., Morrice N. A. (2002) J. Biomol. Techniques 13, 119–130 [PMC free article] [PubMed] [Google Scholar]
- 25.Durocher Y., Perret S., Kamen A. (2002) Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engel M., Hindie V., Lopez-Garcia L. A., Stroba A., Schaeffer F., Adrian I., Imig J., Idrissova L., Nastainczyk W., Zeuzem S., Alzari P. M., Hartmann R. W., Piiper A., Biondi R. M. (2006) EMBO J. 25, 5469–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balendran A., Hare G. R., Kieloch A., Williams M. R., Alessi D. R. (2000) FEBS Lett. 484, 217–223 [DOI] [PubMed] [Google Scholar]
- 28.Balendran A., Casamayor A., Deak M., Paterson A., Gaffney P., Currie R., Downes C. P., Alessi D. R. (1999) Curr. Biol. 9, 393–404 [DOI] [PubMed] [Google Scholar]
- 29.Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukai H. (2003) J. Biochem. 133, 17–27 [DOI] [PubMed] [Google Scholar]
- 32.Lim W. G., Chen X., Liu J. P., Tan B. J., Zhou S., Smith A., Lees N., Hou L., Gu F., Yu X. Y., Du Y., Smith D., Verma C., Liu K., Duan W. (2008) Arch. Biochem. Biophys. 479, 170–178 [DOI] [PubMed] [Google Scholar]
- 33.Lim W. G., Tan B. J., Zhu Y., Zhou S., Armstrong J. S., Li Q. T., Dong Q., Chan E., Smith D., Verma C., Tan S. L., Duan W. (2006) Cell Signal. 18, 1473–1481 [DOI] [PubMed] [Google Scholar]
- 34.Lim W. G., Zhu Y., Wang C. H., Tan B. J., Armstrong J. S., Dokland T., Yang H., Zhu Y. Z., Teo T. S., Duan W. (2005) Cell Signal. 17, 1084–1097 [DOI] [PubMed] [Google Scholar]
- 35.Romano R. A., Kannan N., Kornev A. P., Allison C. J., Taylor S. S. (2009) Protein Sci. 18, 1486–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X., Begley M., Morgenstern K. A., Gu Y., Rose P., Zhao H., Zhu X. (2003) Structure 11, 21–30 [DOI] [PubMed] [Google Scholar]
- 37.Zhao B., Lehr R., Smallwood A. M., Ho T. F., Maley K., Randall T., Head M. S., Koretke K. K., Schnackenberg C. G. (2007) Protein Sci. 16, 2761–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith K. J., Carter P. S., Bridges A., Horrocks P., Lewis C., Pettman G., Clarke A., Brown M., Hughes J., Wilkinson M., Bax B., Reith A. (2004) Structure 12, 1067–1077 [DOI] [PubMed] [Google Scholar]
- 39.Hindie V., Stroba A., Zhang H., Lopez-Garcia L. A., Idrissova L., Zeuzem S., Hirschberg D., Schaeffer F., Jorgensen T. J. D., Engel M., Alzari P. M., Biondi R. M. (2009) Nat. Chem. Biol. 5, 758–764 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.