Abstract
Here we report the identification of a previously uncharacterized human protein as the human monolysocardiolipin acyltransferase-1 (MLCL AT-1). Pig liver mitochondria were treated with n-butyl alcohol followed by Q-Sepharose chromatography, preparative gel electrophoresis, cytidine diphosphate-1,2-diacyl-sn-glycerol-Sepharose chromatography, and finally monolysocardiolipin-adriamycin-agarose affinity chromatography. Elution with either monolysocardiolipin or linoleoyl coenzyme A revealed a major band at 74 kDa with high specific activity (2,300 pmol/min/mg) for the acylation of monolysocardiolipin to cardiolipin using [1-14C]linoleoyl coenzyme A as substrate. Matrix-assisted laser desorption ionization time-of-flight-mass spectrometry analysis followed by search of the Mascot protein data base revealed peptide matches consistent with a 59-kDa protein identified as unknown human protein (GenBankTM protein accession number AAX93141; nucleotide accession number AC011742.3). The purified human recombinant MLCL AT-1 protein utilized linoleoyl coenzyme A > oleoyl coenzyme A > palmitoyl coenzyme A for the specific acylation of monolysocardiolipin to cardiolipin. Expression of MLCL AT-1 in HeLa cells increased mitochondrial monolysocardiolipin acyltransferase activity and [1-14C]linoleic acid incorporated into cardiolipin, whereas RNA interference knockdown of MLCL AT-1 in HeLa cells resulted in reduction in enzyme activity and [1-14C]linoleic acid incorporated into cardiolipin. In contrast, expression of MLCL AT-1 in HeLa cells did not alter [1-14C]oleic or [1-14C]palmitate incorporation into cardiolipin indicating in vivo specificity for the remodeling of cardiolipin with linoleate. Finally, expression of MLCL AT-1 in Barth syndrome lymphoblasts, which exhibit cardiolipin levels 20% that of normal lymphoblasts, increased mitochondrial monolysocardiolipin acyltransferase activity, [1-14C]linoleic acid incorporation into cardiolipin, cardiolipin mass, and succinate dehydrogenase (mitochondrial complex II) activity compared with mock-transfected Barth syndrome lymphoblasts. The results identify MLCL AT-1 as a human mitochondrial monolysocardiolipin acyltransferase involved in the remodeling of cardiolipin.
Cardiolipin (CL)2 is a major phospholipid found in mammalian mitochondria with a multitude of biological functions (reviewed in Refs. 1–7). For example, CL is responsible for modulation of the activity of several mitochondrial enzymes involved in the generation of ATP (reviewed in Refs. 8, 9). In fact, it has been suggested that CL is the “glue” that holds the mitochondrial respiratory complex together (10). The role of CL in genetic diseases such as Barth syndrome, a rare X-linked genetic disorder, is beginning to emerge. Barth syndrome is the only known genetic disease in which the specific biochemical defect is a reduction in CL and accumulation of monolysocardiolipin (MLCL) caused by mutations in the TAZ gene (reviewed in Refs. 2, 7, 11, 12). In addition, the role that CL plays in apoptosis is now well documented (reviewed in Ref. 13). Thus, maintenance of the appropriate content and fatty acyl composition of CL in mitochondria is essential for proper cellular function.
The molecular composition of CL appears to be important for the biological function of CL. In general, there is a selection of a particular kind of fatty acid as well as restriction of the number of fatty acid species (14). The major tetra-acyl molecular species found in rat liver (∼57% of total) and bovine heart (∼48% of total) are 18:2 in each of the four fatty acyl positions of the cardiolipin molecule. Remodeling of CL is essential to obtain this enrichment of CL with linoleate because CL synthase has no molecular species substrate specificity for cytidine-5′-diphosphate-1,2-diacyl-sn-glycerol (15). In addition, the species pattern of CL precursors is similar enough to imply that the enzymes of the CL synthetic pathway are not molecular species-selective (16). Alterations in the molecular composition of CL are associated with various disease states, including diabetes and Barth syndrome (17, 18).
Remodeling of CL occurs via at least three enzymes. Mitochondrial CL was shown to be remodeled by a deacylation-reacylation cycle in which newly synthesized CL was rapidly deacylated to MLCL and then reacylated back to CL with linoleoyl-CoA (19). A mitochondrial MLCL acyltransferase (MLCL AT) activity was characterized and purified from pig liver mitochondria (20, 21). An acyl-CoA-dependent reacylation of MLCL to CL was shown to occur in rat liver microsomes (22). This enzyme was identified as acyllysocardiolipin acyltransferase-1 (ALCAT1) (23). Recently it was shown that ALCAT1 expression in endothelial and hematopoietic lineages resulted in elevated hematopoietic and endothelial genes and increased blast colonies and their progenies (24, 25). The opposite effect was observed with ALCAT1 small interfering RNA indicating that ALCAT1 may play a role in the early specification of hematopoietic and endothelial cells (24, 25). In addition to these mitochondrial and microsomal acyltransferase activities, mitochondrial CL may be remodeled by a mitochondrial CL transacylase reaction first described in rat liver (26). The Barth syndrome gene product tafazzin (TAZ) is a CL transacylase (27). Although TAZ specifically remodels mitochondrial CL with linoleic acid, TAZ alone cannot determine the fatty acid profile of mitochondrial CL (3). In this study, we identify a human protein, MLCL AT-1, with a linoleoyl coenzyme A-specific mitochondrial MLCL AT activity.
EXPERIMENTAL PROCEDURES
Materials
[1-14C]Linoleic acid, [1-14C]linoleoyl coenzyme A, [1-14C]oleoyl coenzyme A, [1-14C]oleic acid, [1-14C]palmitoyl coenzyme A, and [1-14C]palmitic acid were obtained from either DuPont or Amersham Biosciences or American Radiolabeled Chemicals Inc., St. Louis, MO. Dulbecco's modified Eagle's medium and fetal bovine serum were products of Invitrogen. Lipid standards were obtained from Serdary Research Laboratories, Englewood Cliffs, NJ. MLCL was obtained from Avanti Polar Lipids, Alabaster, NY. Thin layer chromatographic plates (silica gel G, 0.25-mm thickness) were obtained from Fisher. Ecolite scintillant was obtained from ICN Biochemicals, Montreal, Quebec, Canada. HeLa cells were obtained from American Type Culture Collection. Epstein-Barr virus-transformed BTHS lymphoblasts (patient 596) were a generous gift from Dr. Richard Kelley, The John Hopkins University. Epstein-Barr virus-transformed age-matched control lymphoblasts were obtained from Coriell Institute for Medical Research, Camden, NJ. Western blotting analysis system was used for protein expression studies and was obtained from Amersham Biosciences. Kodak X-Omat film was obtained from Eastman Kodak Co. Qiagen OneStep RT-PCR kit was used for PCR studies. All other chemicals were certified ACS grade or better and obtained from Sigma or Fisher.
Extraction of MLCL-AT Activity from Pig Liver Mitochondria
Three kg of pig liver from the local meat packers were homogenized in buffer A (10 mm Tris, 0.25 m sucrose, 2 mm EDTA, and 10 mm 2-mercaptoethanol) using a Polytron at medium speed for 5 min. The extract was then centrifuged at 2000 × g for 20 min to remove cell membranes and nuclei. The supernatant was again centrifuged at 8,500 rpm for 55 min to isolate the crude mitochondria. One liter of ice-cold buffer A was added and kept at 4 °C for 3 days with intermittent stirring at low speed with a Polytron homogenizer. Thereafter, the extract was centrifuged at 12,000 × g to produce a clarified mitochondrial extract (675 ml) that was concentrated to 170 ml by Hollow Fiber (Amicon) concentration and then freeze-dried. The freeze-dried extract was treated with 500 ml of butanol with stirring as described (28). The mitochondrial extract was then subjected to anion exchange chromatography with a Q-Sepharose column that was washed thoroughly with buffer A containing 0.2 m NaCl. MLCL AT enzyme activity was eluted from the column with 0.3 mm acetyl coenzyme A in buffer A containing 0.2 m NaCl. This acetyl coenzyme A fraction was dialyzed, freeze-dried, and then applied to a preparatory gel electrophoresis column containing 5% acrylamide. The proteins were separated on the gel with 40 mA constant current with buffer A as the elution buffer. The fractions containing MLCL AT activity were pooled and then applied to a cytidine diphosphate-1,2-diacyl-sn-glycerol-Sepharose gel and washed with buffer A containing 0.2 m NaCl. The fraction containing MLCL AT activity was eluted with 5 mm CTP and 0.2 m NaCl in buffer A. The eluted fraction was dialyzed in buffer A and then added to an MLCL-adriamycin affinity column (see below).
MLCL-Adriamycin Affinity Chromatography
The MLCL-adriamycin affinity resin was prepared by mixing 30 ml of REACTI-gel (Pierce) with 1.2 mg of adriamycin in 45 ml of 0.1 m borate buffer at 4 °C. Before addition of the partially pure MLCL AT, the gel was mixed with 1 mg of MLCL and washed with 0.1 m borate, pH 8.0. The eluted fraction from above was applied to the MLCL-adriamycin-agarose and washed thoroughly with 0.1 m borate, pH 8.0, until there was no more protein emerging from the column. Finally the MLCL AT activity was eluted from the affinity column with borate buffer containing 1 mm linoleoyl coenzyme A or 1 mm MLCL. A polyvinyl difluoride membrane blot was submitted for MALDI-TOF-mass spectrometry analysis (University of Minnesota). The peptide sequences obtained were analyzed by searching the Mascot protein data base for identification of potential homologies with other known proteins. An NCBI BLAST search for known proteins that align with the peptide sequences was performed using the Protein Data Bank.
Preparation and Purification of Recombinant Unknown Human Protein MLCL AT-1, Plasmids for Transfection and RNAi
The full-length primers for the human 59-kDa unknown protein (GenBankTM accession number AC011742.3) MLCL AT-1 containing a His6 tag in the reverse primer without stop codon was prepared from Invitrogen (custom primer design) (Table 1). The His tag was required for binding of the protein to the nickel-nitrilotriacetic acid affinity resin (see below). The primers were amplified using 1 μg of HeLa cell RNA. The cDNAs containing the full-length sequences were inserted into pcDNA 3.1 using the TOPO cloning reaction with pEXP5-CT/TOPO vector (Invitrogen). Escherichia coli One Shot® bacteria (Invitrogen) were transformed with the construct chemically with S.O.C. medium (Invitrogen) and inoculated onto ampicillin containing agar for growth overnight. In the morning, the colonies were inoculated into 5 ml of ampicillin containing LB medium and cultured at 37 °C in an orbital shaker at 220 rpm overnight. The plasmid was purified from the E. coli using the MidiPrep kit (Invitrogen). The sequences of the plasmids were verified by PCR using the specific primers and also by a DNA sequencer (Manitoba Institute of Cell Biology). The recombinant protein was expressed using the cell-free E. coli expression system (Invitrogen). The recombinant protein was purified with a nickel-nitrilotriacetic acid affinity resin (Fisher) when eluted with 200 mm imidazole. When high purity proteins were required, the resin was first of all pre-eluted with high salt (1 m NaCl) and low imidazole concentration (20 mm).
TABLE 1.
Primers used for in vitro protein expression and transfection
The cDNA containing the full-length sequence of the human unknown protein, MLCL AT-1, was inserted into pcDNA3.1/V5-His TOPO TA expression kit (Invitrogen) and grown in E. coli, and the plasmid was purified and sequence-verified as described above. The plasmid was used for transfection of HeLa cells or BTHS lymphoblasts as described below. RNAi to human MLCL AT-1 was prepared from Invitrogen using the BLOCK-iT RNAi Designer program (Table 1). The RNAi was used for transfection of HeLa cells or lymphoblasts as described below.
Culture, Radiolabeling, and Harvesting of HeLa Cells and Barth Syndrome Lymphoblasts
HeLa cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. To evaluate the effect of the human unknown protein on CL acylation with different species of fatty acid, 13 μg of MLCL AT-1 protein plasmid in 23 μl of Lipofectamine (Invitrogen) were added to HeLa cells at 50% confluence. After 24 h, 1 μCi of fatty acid ([1-14C]linoleic acid, [1-14C]oleic acid, or [1-14C]palmitic acid (bound to bovine serum albumin in a 1:1 molar ratio) was added and incubation continued for another 24 h. Cells were then harvested, and radioactivity was incorporated into CL determined as described previously (28). In other experiments, 100 nm of RNAi in 23 μl of Lipofectamine (Invitrogen) were added to HeLa cells at 50% confluence. After 24 h, 1 μCi of [1-14C]linoleic acid was added and incubation continued for another 24 h, and radioactivity was incorporated into CL determined as described above. In some experiments, cells were washed twice with ice-cold saline and harvested with 2 ml of lysis buffer (10 mm Tris-HCl, pH 7.4, 0.25 m sucrose) and then homogenized with 30 strokes of a Dounce A homogenizer. The homogenate was centrifuged at 1,000 × g for 5 min and the supernatant centrifuged at 10,000 × g for 15 min. The pellet was resuspended in 0.5 ml of homogenization buffer and used for assay of mitochondrial enzyme activities as described below.
BTHS lymphoblasts and age-matched control lymphoblasts were grown in suspension in RPMI 1640 medium containing 10% fetal bovine serum until reaching a concentration of 106 cells/ml. Lymphoblasts were pelleted and placed in Opti-MEM (Invitrogen) (5 × 106 cells/ml) and incubated with 40 μg of MLCL AT-1 plasmid, and electroporation was performed at 950 microfarads, 250 V, for 23 ms in 800 μl of Opti-MEM using a BTX Electroporation System Electrocell Manipulator 600. Cells were then incubated with 1 μCi [1-14C]linoleic acid for 4 h, and radioactivity was incorporated into CL determined as described above. In other experiments mitochondria from the above cells were prepared as above, and MLCL AT or succinate dehydrogenase activities were determined as described below. In other experiments, 100 nm small interfering RNA was transfected into lymphocytes (5 × 106 cells/ml) by electroporation as described above. Following electroporation the lymphocytes were incubated in 10 ml of Opti-MEM for 19 h. Subsequently the cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum and 1% each of antimycotic and antibiotic. Incubation at 37 °C and 5% CO2 was continued for an additional 24 h following which the cells were harvested and the mitochondria isolated as described previously. In other experiments, BTHS lymphoblasts were incubated as above, and CL was isolated and a phosphorous mass of CL determined as described previously (29).
Determination of Enzyme Activities
MLCL AT activity was determined as described previously (20, 21). Essentially human recombinant MLCL AT-1 protein (20 ng) or mitochondrial protein (20 μg) was incubated in 50 mm Tris-HCl buffer, pH 8.0, and incubated with 0.3 mm MLCL and [1-14C]linoleoyl coenzyme A (120,000 dpm/nmol) at 37 °C for 1 h or longer for smaller protein quantities. For some reactions [1-14C]linoleoyl coenzyme A was replaced with [1-14C]oleoyl coenzyme A or [1-14C]palmitoyl coenzyme A. The reaction was stopped by the addition of chloroform/methanol (2:1). The organic fraction was isolated by centrifuging the mixture after the addition of 0.9% KCl. After an additional washing of the organic fraction with theoretical upper phase, the chloroform layer was dried with nitrogen, resuspended in 25 μl of chloroform/methanol (2:1), and applied to a Whatman silica gel-coated glass thin layer plate with CL standard. [14C]CL was isolated by two-dimensional chromatography using the following solvent mixtures: first dimension (chloroform/methanol/water, 65:25:4, by volume) and second dimension (chloroform/acetone/methanol/acetic acid/water, 50:20:10:10:5, by volume). CL was visualized with iodine vapor and silica gel corresponding to the CL spot removed and placed into scintillation vials containing 5 ml of Ecolite scintillant, and radioactivity was determined in an LS 6500 liquid scintillation counter (Beckman). For kinetic analysis in some experiments MLCL AT activity of the recombinant human MLCL AT-1 protein was determined in the presence of a fixed amount of [1-14C]acyl coenzyme A with varying concentrations of MLCL and/or a fixed amount of MLCL with varying concentrations of [1-14C] acyl coenzyme A, and the reciprocal velocity versus substrate concentration was plotted. Mitochondrial succinate dehydrogenase activity was determined as described (30).
Electrophoresis and Western Blot Analysis
Proteins from the MLCL-adriamycin affinity column eluted with either linoleoyl coenzyme A or MLCL as described above were separated on the Bio-Rad mini gel electrophoresis system using 10% acrylamide containing 0.1% SDS. The disruption buffer (1×, Sigma) included SDS, 2-mercaptoethanol, and bromphenol blue dye. The electrophoresis was performed using synthetic pre-stained molecular markers from Bio-Rad. After the electrophoresis, the proteins were transferred onto polyvinylidene difluoride membranes, using Tris-glycine buffer, pH 8.3, with 20% methanol, at 15 V for 1.5 h. The proteins were probed overnight with polyclonal pig liver anti-MLCL AT antibody (21). The second antibody was anti-rabbit IgG. The protein was visualized on X-Omat film by chemiluminescence (Amersham Biosciences). In other experiments, 0.5 or 1 μg of recombinant protein from the in vitro protein translation or 12 μg of HeLa cell mitochondrial protein or 1.2 μg of lymphoblast mitochondrial protein was separated on a 10% SDS-polyacrylamide gel, blotted onto a polyvinylidene difluoride membrane, and probed with anti-MLCL AT-1 antibody as above. The MLCL-AT-1 was visualized by chemiluminescence, and the protein gel was stained with Coomassie Blue.
Other Determinations
Protein was determined as described (31). Student's t test was used for determination of statistical significance. The level of significance was defined as p < 0.05.
RESULTS
Identification of Human Mitochondrial MLCL AT-1
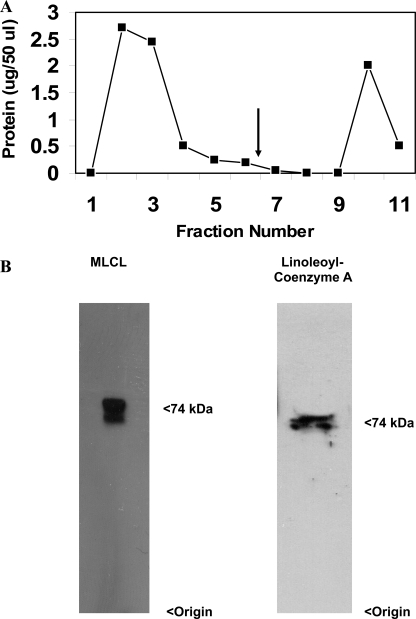
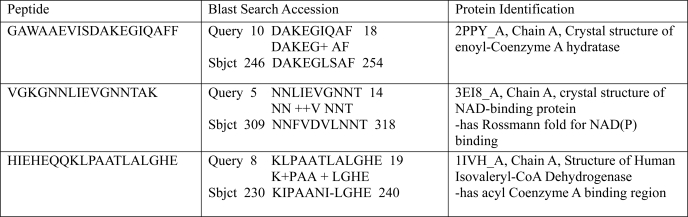
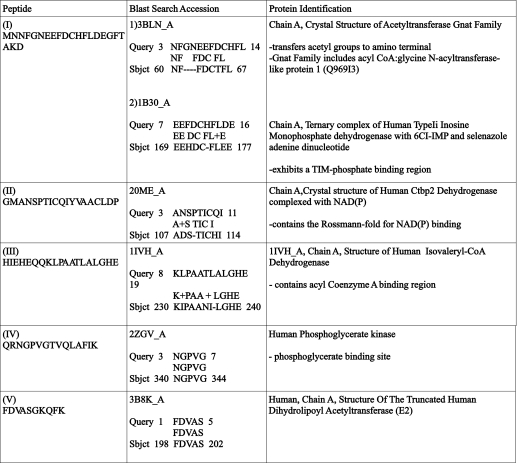
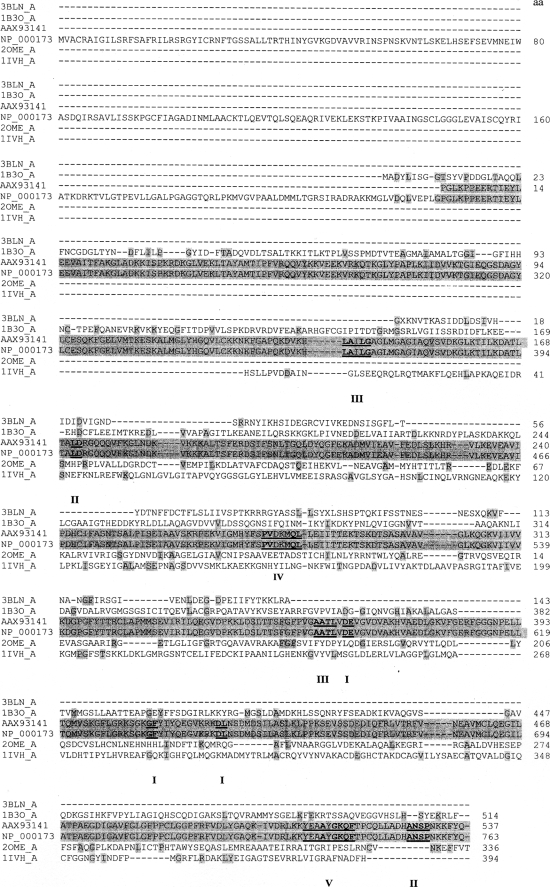
To identify the human mitochondrial MLCL AT, a pig liver mitochondrial extract was prepared as described under “Experimental Procedures.” Following elution from the cytidine diphosphate-1,2-diacyl-sn-glycerol-Sepharose gel the MLCL AT activity containing sample was dialyzed and mixed with MLCL-adriamycin-agarose and then applied to a column and extensively washed with 0.2 m NaCl in 0.1 m borate buffer, pH 9.0. MLCL AT enzyme activity was eluted with 1.0 mm MLCL (Fig. 1A). The specific activity of fraction 10 was 2,306 pmol/min/mg protein. Western blot analysis of fraction 10 using a polyclonal antibody to the pig liver MLCL AT demonstrated the presence of a protein at 74 kDa (Fig. 1B). In addition, the identical amount of MLCL AT activity could be eluted in fraction 10 with 1.0 mm linoleoyl coenzyme A. Western blot analysis of fraction 10 eluted by linoleoyl-CoA demonstrated the presence of a protein at 74 kDa (Fig. 1B). MALDI-TOF-mass spectrometry analysis of the 74-kDa protein revealed peptide matches to enoyl-coenzyme A hydratase, NAD-binding protein containing the Rossmann-fold for NAD(P) binding, and coenzyme A dehydrogenase (Table 2). These motifs are consistent with the 74-kDa pig liver mitochondrial trifunctional protein α (GenBankTM accession number NP_999127). Further peptide analysis indicated the presence of motifs for lipid, coenzyme A, and acyl coenzyme A binding and acyltransferase activity (Table 3). Alignment of human mitochondrial trifunctional protein α (GenBankTM accession number NP_000173) revealed a match with a 59-kDa human unknown protein (GenBankTM protein accession number AAX93141; nucleotide accession number AC011742.3) (Fig. 2). AAX93141 is identical to the C-terminal 59-kDa end of mitochondrial trifunctional protein. Proteins (GenBankTM accession numbers 3BLN_A; 1B3O_A; 2OME_A; and 1IVH_A) identified from the blast search were then aligned using the Cobalt multialignment program in comparison with the NP_000173 and AAX93141 (Fig. 2). Identical amino acids are highlighted, and the location of the peptides and amino acid identities are represented in boldface with the region of agreement underlined (Fig. 2).
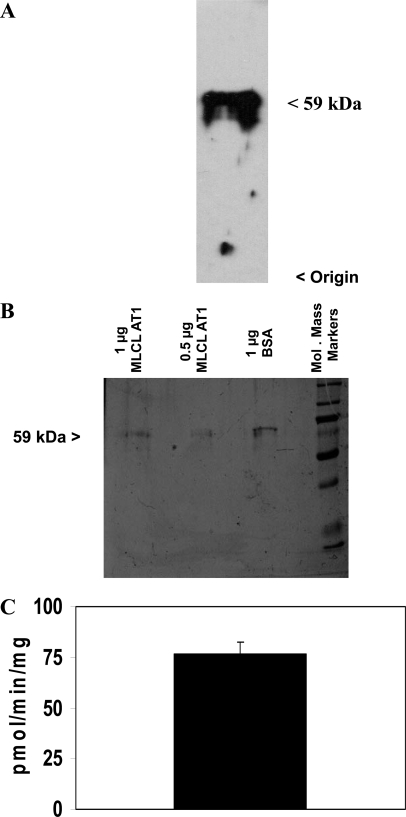
FIGURE 1.
MLCL-adriamycin-agarose affinity chromatography profile and Western blot analysis. A, 4 ml of partially purified MLCL-AT was loaded onto 1 ml of adriamycin-agarose affinity gel, which was pre-equilibrated with 0.75 mg of MLCL. The gel was washed with 12 column volumes of 0.1 m borate buffer, pH 8.0, until no more protein was eluted. Finally, MLCL-AT was eluted (arrow) by the addition of 1 mm MLCL or 1 mm linoleoyl coenzyme A. B, Western blot of fraction 10 eluted with MLCL using the polyclonal antibody to MLCL AT and Western blot of fraction 10 eluted with linoleoyl coenzyme A. Origin and molecular mass are indicated on the right. Representative blots are depicted.
TABLE 2.
Trifunctional protein motifs obtained from MALDI-TOF-mass spectrometry and BLAST search protein matches
Peptide sequences obtained from mass spectrometry on purified 74-kDa mitochondrial pig liver linoleoyl-coenzyme A monolysocardiolipin acyltransferase were analyzed by an NCBI BLAST search (Protein Data Bank) and revealed matches with alignment for the following motifs: enoyl-coenzyme A hydratase, NADB-Rossmann-fold for NAD(P) binding, and isovaleryl-CoA dehydrogenase. NADB, NAD binding.
TABLE 3.
Peptide sequences obtained from MALDI-TOF-mass spectrometry and BLAST search protein matches
Purified pig liver MLCL AT was analyzed by MALDI-TOF-mass spectrometry, and the resultant peptides were subjected to a BLAST search (Protein Data Bank), and protein matches were identified.
FIGURE 2.
Sequence alignment of AAX93141 with proteins identified through peptide matches. Proteins (3BLN_A, 1B3O_A, 2OME_A, and 1IVH_A) obtained from the BLAST search of the peptides and human trifunctional protein (NP_000173) were analyzed by the Cobalt multialignment program. The amino acid residues of the known proteins are highlighted (gray) relative to their alignment with the human MLCL AT-1 (AAX93141) and NP_000173. Matching amino acid sequences from peptides I to V from Table 3 are indicated in boldface. Protein accession number is indicated on the left.
Characterization of the MLCL AT Activity of AAX93141
Because the MLCL AT activity of the mitochondrial 74-kDa protein had been characterized previously (21), purified human recombinant 59-kDa mitochondrial trifunctional protein (protein accession number AAX93141) was prepared as outlined under “Experimental Procedures” Western blot analysis of the protein exhibited cross-reaction with the 74-kDa pig liver MLCL AT polyclonal antibody at 59 kDa (Fig. 3A). The recombinant protein was then further purified following elution from a nickel resin. SDS-PAGE analysis of the purified recombinant protein revealed the 59-kDa protein as a single band (Fig. 3B). Thus, only the 59-kDa protein was present following elution from the nickel resin. Two μg of the purified protein was examined for the ability to acylate MLCL to CL in the presence of [1-14C]linoleoyl coenzyme A. The eluate from the nickel resin prepared from the recombinant protein could acylate MLCL to CL in the presence of [1-14C]linoleoyl coenzyme A (Fig. 3C). No MLCL AT activity was observed in preparations that did not contain expression vector in the in vitro translation system. Thus, the 59-kDa human protein exhibits MLCL AT activity and was termed MLCL AT-1.
FIGURE 3.
Western blot analysis, SDS-PAGE analysis, and MLCL AT activity of the human recombinant MLCL AT-1 protein. A, purified human recombinant MLCL AT-1 was prepared and Western blot analysis performed as described under “Experimental Procedures.” A representative blot is depicted. Origin and molecular mass are indicated on the right. B, SDS-PAGE of 1 μg of bovine serum albumin (BSA) or 0.5 or 1.0 μg of purified human recombinant MLCL AT-1 following imidazole elution from the nickel column. Molecular mass markers are indicated on the right. A representative blot is depicted. C, MLCL AT activity of purified human recombinant MLCL AT-1 eluted from the nickel column. Data represent the mean ± S.D. of three preparations.
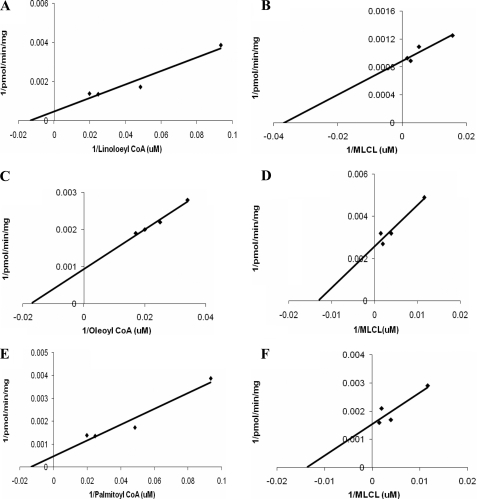
Enzyme Kinetics of MLCL AT-1
The MLCL AT enzyme activity of MLCL AT-1 was determined with varying concentrations of acyl coenzyme A in the presence of a constant amount of MLCL. The following Km values for the acyl-CoAs examined were as follows: 8 μm for linoleoyl coenzyme A, 57 μm for oleoyl coenzyme A, and 72 μm for palmitoyl coenzyme A (Fig. 4, A, C, and E). The MLCL AT enzyme activity of MLCL AT-1 was then determined with varying concentrations of MLCL in the presence of a constant amount of acyl coenzyme A. The following Km values for MLCL were determined to be 27 μm for linoleoyl coenzyme A, 71 μm for oleoyl coenzyme A, and 77 μm for palmitoyl coenzyme A (Fig. 4, B, D, and F). MLCL AT activity of MLCL AT-1 was then determined in the presence of various lysophospholipid substrates. In the presence of linoleoyl coenzyme A, MLCL was acylated to CL. In contrast the recombinant protein did not acylate any other lysophospholipids to CL, including lysophosphatidylcholine, lysophosphatidylethanolamine, lysophosphatidic acid, and lysophosphatidylglycerol (data not shown). Thus, the human recombinant MLCL AT-1 exhibited absolute specificity for MLCL and high specificity for linoleoyl coenzyme A as substrates.
FIGURE 4.
MLCL AT activity of human recombinant MLCL AT-1 in the presence of various concentrations of MLCL or acyl-CoAs. MLCL AT enzyme activity of MLCL AT-1 was determined with varying concentrations of linoleoyl coenzyme A (A) or oleoyl coenzyme A (C) or palmitoyl coenzyme A (E) in the presence of a constant amount of MLCL or determined with varying concentrations of MLCL in the presence of a constant amount of linoleoyl coenzyme A (B) or oleoyl coenzyme A (D) or palmitoyl coenzyme A (F) as described under “Experimental Procedures.” Reciprocal plots are depicted. Data represent the mean of two experiments. The results between samples differed by less than 10%.
Expression or Knockdown of MLCL AT-1 Alters in Vitro MLCL AT Activity and [1-14C]Linoleate Incorporation into CL in HeLa Cells
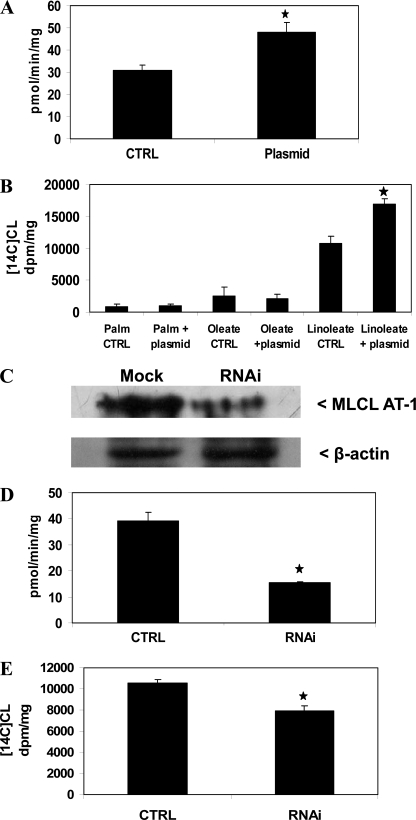
To characterize the role of MLCL AT-1 in MLCL acylation to CL, HeLa cells were mock-transfected or transfected with human MLCL AT-1, and mitochondrial fractions were prepared, and MLCL was AT activity determined. Mitochondrial MLCL AT activity increased 54% (p < 0.05) in cells transfected with MLCL AT-1 compared with mock-transfected controls (Fig. 5A). HeLa cells were then transfected with MLCL AT-1 and incubated with 0.1 μm 1-14C-labeled fatty acids. Expression of MLCL AT-1 in HeLa cells resulted in a 60% (p < 0.05) increase in incorporation of [1-14C]linoleate into CL but not [1-14C]oleate or [1-14C]palmitate incorporation into CL compared with mock-transfected controls (Fig. 5B). Thus, MLCL AT-1 exhibits MLCL AT enzyme activity and in vivo specificity for the acylation of MLCL with [1-14C]linoleate in HeLa cells.
FIGURE 5.
MLCT AT activity and 1-14C-labeled fatty acid incorporation into CL in HeLa cells expressing human recombinant MLCL AT-1 and MLCT AT-1 protein, activity, and 1-14C-labeled fatty acid incorporation into CL in HeLa cells transfected with MLCL AT-1 RNAi. A, HeLa cells were mock-transfected (CTRL) or transfected with human recombinant MLCL AT-1 (Plasmid) mitochondrial fractions prepared, and MLCL AT activity was determined as described under “Experimental Procedures.” B, HeLa cells were transfected as in A and incubated with 0.1 μm [1-14C]linoleate or [1-14C]oleate or [1-14C]palmitate (Palm), and radioactivity incorporated into CL was determined. CTRL, control. C, HeLa cells were mock-transfected (Mock) or transfected with MLCL AT-1 RNAi (RNAi), and Western blot analysis was performed as described under “Experimental Procedures.” A representative blot is shown for MLCL AT-1 and β-actin. D, HeLa cells were mock-transfected (CTRL) with or transfected with MLCL AT-1 RNAi (RNAi), and mitochondrial fractions were prepared, and MLCL AT activity was determined as described under “Experimental Procedures.” E, HeLa cells were transfected as in D and then incubated with 0.1 μm [1-14C]linoleate, and radioactivity incorporated into CL was determined. Data represent the mean ± S.D. of three experiments. *, p < 0.05.
HeLa cells were then transfected with MLCL AT-1 RNAi; mitochondrial fractions were prepared, and MLCL AT-1 protein expression and MLCL AT activity were determined. Mitochondrial MLCL AT-1 protein expression was lower in cells transfected with MLCL AT-1 RNAi compared with mock-transfected control (Fig. 5C). Mitochondrial MLCL AT was decreased 61% (p < 0.05) when cells were transfected with MLCL AT-1 RNAi compared with mock-transfected control (Fig. 5D). HeLa cells were then transfected with MLCL AT-1 RNAi and incubated with 0.1 μm [1-14C]linoleate. Expression of MLCL AT-1 RNAi in HeLa cells resulted in a 25% (p < 0.05) decrease in incorporation of [1-14C]linoleate into CL compared with mock-transfected controls (Fig. 5E). These results further confirm that MLCL AT-1 exhibits MLCL AT enzyme activity and is responsible for in vivo resynthesis of CL with [1-14C]linoleate in HeLa cells.
Expression of MLCL AT-1 in BTHS Lymphoblasts Elevates CL Formation
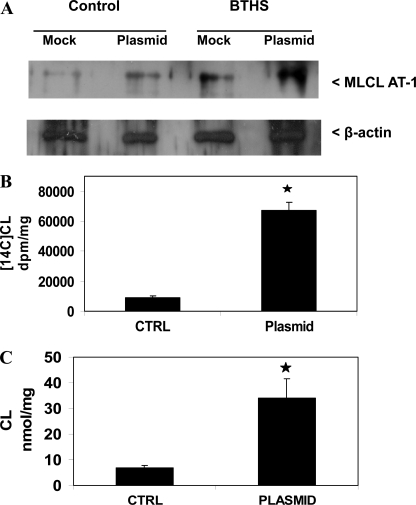
BTHS cells exhibit a reduction in CL because of a mutation in the CL remodeling gene TAZ (reviewed in Refs. 2, 11). We examined if expression of MLCL AT-1 could elevate CL formation in BTHS cells. BTHS lymphoblasts were mock-transfected or transfected with human MLCL AT-1 and MLCL AT protein, and [1-14C]linoleate incorporation into CL and CL mass was determined. Transfection with MLCL AT-1 resulted in an increase in MLCL AT-1 protein expression in both age-matched control and BTHS lymphoblasts compared with mock-transfected cells (Fig. 6A). Transfection of BTHS lymphoblasts with MLCL AT-1 resulted in a 6-fold (p < 0.05) increase in [1-14C]linoleate incorporation into CL compared with mock-transfected BTHS lymphoblasts (Fig. 6B). Transfection of BTHS lymphoblasts with MLCL AT-1 resulted in a 4.8-fold (p < 0.05) increase in CL mass compared with mock-transfected BTHS lymphoblasts (Fig. 6C). Thus, expression of MLCL AT-1 elevated the MLCL AT protein, [1-14C]linoleate incorporation into CL, and CL mass in BTHS lymphoblasts.
FIGURE 6.
MLCT AT-1 protein expression and [1-14C]linoleate incorporation into CL and CL mass in Barth syndrome lymphoblasts expressing human recombinant MLCL AT-1. A, age-matched control and BTHS lymphoblasts were mock-transfected (Mock) or transfected with human recombinant MLCL AT-1 plasmid (Plasmid), and Western blot analysis of MLCL AT-1 protein was determined as described under “Experimental Procedures.” A representative blot is shown for MLCL AT-1 and β-actin. BTHS lymphoblasts were mock-transfected (CTRL) or transfected with human recombinant MLCL AT-1 (Plasmid), and [1-14C]linoleate incorporation into CL (B) and CL mass (C) was determined. Data represent the mean ± S.D. of three experiments. *, p < 0.05.
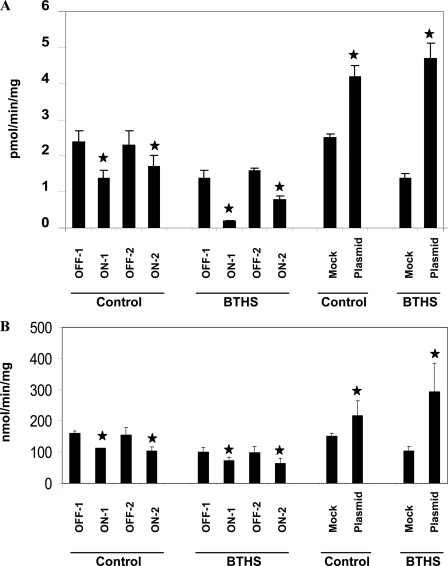
We next examined if alterations in MLCL AT-1 enzyme activity were associated with corresponding alterations in mitochondrial function in both age-matched control and BTHS lymphoblasts. Assay of mitochondrial succinate dehydrogenase, a part of the mitochondrial complex II, was used as a measure of mitochondrial function. Transfection of MLCL AT-1 RNAi decreased MLCL AT-1 activity 49% (p < 0.05) or 26% (p < 0.05) in age-matched control lymphoblasts and 85% (p < 0.05) or 42% (p < 0.05) in BTHS lymphoblasts that were transfected with on-target-1 or on-target-2 sequences, respectively, compared with age-matched control or BTHS lymphoblasts transfected with either of the off-target sequences for the MLCL AT-1 RNAi (Fig. 7A). Transfection of control lymphoblasts with MLCL AT-1 resulted in a 68% (p < 0.05) elevation in MLCL AT-1 activity compared with mock-transfected cells (Fig. 7A). In contrast, transfection of BTHS lymphoblasts with MLCL AT-1 resulted in a 3.4-fold (p < 0.05) increase in MLCL AT-1 activity compared with mock-transfected cells (Fig. 7A).
FIGURE 7.
MLCL AT-1 and succinate dehydrogenase activities in Barth syndrome lymphoblasts expressing human recombinant MLCL AT-1 or MLCL AT-1 RNAi. Control and BTHS lymphoblasts were transfected with OFF-1 or OFF-2 or ON-1 or ON-2 MLCL AT-1 RNAi plasmids or were mock-transfected (Mock) or transfected with human recombinant MLCL AT-1 plasmid (Plasmid) and MLCL AT (A) or succinate dehydrogenase (B) activities determined as described under “Experimental Procedures.” Data represent the mean ± S.D. of three experiments. *, p < 0.05.
Transfection of MLCL AT-1 RNAi decreased mitochondrial succinate dehydrogenase activity ∼30–35% (p < 0.05) in both age-matched control and BTHS lymphoblasts in cells that were transfected with either one of the two on-target sequences for the MLCL AT-1 RNAi compared with transfection with either of the off-target sequences for the MLCL AT-1 RNAi (Fig. 7B). Transfection of control lymphoblasts with MLCL AT-1 resulted in a 44% (p < 0.05) elevation in succinate dehydrogenase activity compared with mock-transfected cells (Fig. 7B). In contrast, transfection of BTHS lymphoblasts with MLCL AT-1 resulted in a 2.9-fold (p < 0.05) increase in succinate dehydrogenase activity compared with mock-transfected cells (Fig. 7B). Thus, expression or knockdown of MLCL AT-1 activity results in a corresponding alteration in mitochondrial succinate dehydrogenase activity in age-matched control and BTHS lymphoblasts.
DISCUSSION
In this study we provide clear evidence that a 59-kDa human protein, now termed MLCL AT-1, is a human mitochondrial MLCL AT. The 74-kDa pig liver protein resolved from the MLCL-adriamycin affinity column with either MLCL or linoleoyl coenzyme A upon MALDI-TOF-mass spectrometry analysis revealed peptide matches consistent with mitochondrial trifunctional protein and the unknown human protein (protein accession number AAX93141; nucleotide accession number AC011742.3). AAX93141 is identical to the C-terminal 59-kDa end of mitochondrial trifunctional protein. The purified human recombinant MLCL AT-1 exhibited MLCL AT activity using MLCL and [1-14C]fatty acyl coenzyme A as substrates. The purified human recombinant MLCL AT-1 had in vitro specificity for linoleoyl coenzyme A > oleoyl coenzyme A > palmitoyl coenzyme A and had absolute specificity for acylation of MLCL. Expression of human MLCL AT-1 in HeLa cells increased mitochondrial MLCL AT activity and [1-14C]linoleic acid incorporated into CL, whereas RNAi knockdown of human MLCL AT-1 in HeLa cells resulted in reduction in mitochondrial MLCL AT-1 protein, MLCL AT activity, and [1-14C]linoleic acid incorporated into CL. In contrast, expression of human MLCL AT-1 in HeLa cells did not alter [1-14C]oleic or [1-14C]palmitate incorporation into CL indicating in vivo specificity for CL remodeling with [1-14C]linoleate. Expression of human recombinant MLCL AT-1 in BTHS lymphoblasts increased mitochondrial MLCL AT activity, [1-14C]linoleate incorporation into CL and CL mass. Finally, expression or knockdown of MLCL AT-1 in age-matched control or BTHS lymphoblasts resulted in corresponding alterations in MLCL AT and mitochondrial succinate dehydrogenase activities. The results clearly identify the 59-kDa unknown human protein (accession number AAX93141), now termed MLCL AT-1, as a mitochondrial MLCL AT specific for the remodeling of CL.
There are at least three ways in which CL may be remodeled with linoleate (reviewed in Ref. 11). An endoplasmic reticulum (ER) lysocardiolipin acyltransferase (ALCAT1) was identified that transferred fatty acid from fatty acyl coenzyme A to MLCL to produce CL (23). The enzyme activity utilized both MLCL and dilysocardiolipin substrates and exhibited specificity for unsaturated long chain fatty acyl-CoAs. In addition, this enzyme catalyzed the acylation of a host of monoionic lysophospholipids (32, 33). This enzyme is likely identical to the acyltransferase first described in rat liver ER (22). Given that CL is localized to mitochondria the function of ALCAT1 is currently an area of active investigation. Regions of contact zones exist between ER and mitochondria (reviewed in Ref. 34). It is possible that ALCAT1 may serve as a link between CL remodeling in the ER and mitochondria at these sites. Indeed, ALCAT1 activities and mRNA expression have been shown to be altered in models of apoptosis and diabetes, conditions in which CL metabolism may be altered (35–37). In addition to the ER ALCAT1, our laboratory previously identified and purified from pig liver mitochondria a 74-kDa protein that could catalyze the acylation of MLCL with fatty acyl-CoAs (20, 21). In addition to the above two acyltransferase activities, evidence clearly supports that CL may be remodeled by a CL transacylase now identified as the Barth syndrome gene (TAZ) product tafazzin (2, 11, 12). A recent review from the laboratory that identified TAZ as the CL transacylase suggested that although evidence indicates that tafazzin clearly and specifically remodels mitochondrial CL with linoleate, the idea that tafazzin determines the fatty acid profile of CL is conceptually problematic and contradicts experimental evidence (3). For example, in hearts of AMP-activated protein kinase null mice, CDS-2, a rate-limiting enzyme of de novo CL biosynthesis, and ALCAT1 mRNA expression were reduced compared with controls, and this accompanied reduced levels of CL and linoleate in phospholipids within cardiac mitochondria (35). TAZ mRNA expression was unaltered in the hearts of these mice. In a recent study from our laboratory, tafazzin enzymatic activity was low (<1.0 pmol/min/mg protein) and its mRNA expression was unaltered in the surviving population of H9c2 cells subjected to 2-deoxyglucose-mediated apoptosis (37). In these cells, ER ALCAT1 activity was reduced, whereas mitochondrial MLCL AT activity was elevated and accompanied the increase in mitochondrial phospholipase A2 activity likely to maintain the CL fatty acyl composition that was unaltered. In addition, the presence of two patients with BTHS with exon 5 mutation in TAZ but with normal CL levels3 highlight the fact that TAZ alone may not be responsible for all mitochondrial CL remodeling. Because expression of MLCL AT-1 in BTHS lymphoblasts elevated mitochondrial MLCL AT protein activity and [1-14C]linoleic acid incorporation into CL and CL mass, it is likely that both TAZ and MLCL AT-1 are responsible for the in vivo mitochondrial remodeling of CL with linoleate. Moreover, because BTHS lymphoblasts exhibit ∼20% of CL levels compared with normal lymphoblasts (38), it is possible that MLCL AT-1 may be responsible for up to 20% of mitochondrial CL remodeling. However, it should be noted that de novo biosynthesis of CL could contribute significantly to this CL pool. Interestingly, basal protein expression of MLCL AT-1 appeared higher in BTHS lymphoblasts compared with aged-matched control lymphoblasts (Fig. 6A). It is possible that MLCL AT-1 expression is elevated as a compensatory response to the absence of a functional TAZ in BTHS cells. However, basal MLCL AT activity appeared lower in BTHS lymphoblasts compared with aged-matched control lymphoblasts (Fig. 7A). These data suggest that there may be other factors involved in the regulation of MLCL AT enzyme activity in these cells.
Expression or knockdown of MLCL AT-1 in age-matched control or BTHS lymphoblasts produced corresponding alterations in mitochondrial succinate dehydrogenase (a part of the mitochondrial complex II) activity. In addition, succinate dehydrogenase activity appeared lower in mock-transfected BTHS lymphoblasts compared with mock-transfected age-matched control lymphoblasts (Fig. 7B). A previous study had shown that mitochondrial respiratory chain supercomplexes are destabilized in BTHS patients (39). CL is required for assembly of these mitochondrial respiratory chain complexes (2, 7, 11, 12). Thus the lower succinate dehydrogenase activity observed in BTHS lymphoblasts in comparison with control cells was not surprising because BTHS lymphoblasts exhibit lower CL levels. Because expression of MLCL AT-1 in BTHS lymphoblasts greatly elevated both CL levels and succinate dehydrogenase activity, elevation of MLCL AT-1 protein expression might serve as a potential therapeutic target in BTHS patients.
Surprisingly, MLCL AT-1 does not contain conserved motifs observed in classical 1-acylglycerophosphate acyltransferases (40). Rather it exhibits structural identity to trifunctional protein (41, 42). The alignment with inosine monophosphate dehydrogenase, which contains a phosphate binding region, indicates a potential binding domain for the MLCL (43). The alignment with the isovaleryl-coenzyme A dehydrogenase with the acyl-coenzyme A binding region supports a potential binding region for linoleoyl coenzyme A (44). The glycine N-acyltransferase-like protein 1, a Gnat-related family member, transfers acyl groups to a variety of amino acid acceptors (45). This is consistent with the reaction mechanism proposed for the purified pig liver enzyme, i.e. formation of an enzyme-linoleate intermediate (21). The alignment with Ctbp2 dehydrogenase verifies the presence of the Rossmann-fold, observed in trifunctional protein (46). Finally, MLCL AT-1 does not accommodate acylation of typical lysophospholipid substrates because it has absolute substrate specificity for MLCL.
Interestingly, the Km value for linoleoyl coenzyme A of MLCL AT-1 for the acylation of MLCL to CL (8 μm) was within the range previously reported for the Km value for hydroxydecanoyl coenzyme A of the long chain l-3-hydroxyacyl-coenzyme A dehydrogenase activity of purified pig heart mitochondrial trifunctional protein (41). However, because a large section of the N-terminal of trifunctional protein is absent (>200 amino acids) in MLCL AT-1, i.e. regions that are responsible for the enoyl-coenzyme A dehydratase reaction (42), it is unlikely that MLCL AT-1 plays a direct role in mitochondrial β-oxidation. The fact that the Km value for linoleoyl coenzyme A for the 59-kDa MLCL AT-1 was 12.5-fold lower than the Km value previously reported for the 74-kDa trifunctional protein (21) suggests that the 59-kDa MLCL AT-1 protein is likely the in vivo human linoleoyl-coenzyme A monolysocardiolipin acyltransferase.
In summary, we have identified a 59-kDa human protein, MLCL AT-1 as a mitochondrial MLCL AT specific for the resynthesis of CL from MLCL and acyl coenzyme A. The existence of MLCL AT-1 in mitochondria may explain the discrepancies previously observed for tafazzin serving as the sole CL remodeling enzyme in human mitochondria. In addition, MLCL AT-1 might serve as a potential therapeutic target in BTHS patients.
This work was supported by grants from the Canadian Institutes of Health Research, the Manitoba Health Research Council, and the Barth Syndrome Foundation of Canada.
M. Schlame, personnel communication.
- CL
- cardiolipin
- MLCL
- monolysocardiolipin
- MLCL AT
- monolysocardiolipin acyltransferase
- TAZ
- tafazzin
- MALDI-TOF
- matrix activated laser desorption ionization-time of flight
- BTHS
- Barth syndrome
- ALCAT1
- acyllysocardiolipin acyltransferase-1
- MLCL AT-1
- monolysocardiolipin acyltransferase-1
- ER
- endoplasmic reticulum
- RNAi
- RNA interference.
REFERENCES
- 1.Hatch G. M. (2004) Biochem. Cell Biol. 82, 99–112 [DOI] [PubMed] [Google Scholar]
- 2.Houtkooper R. H., Vaz F. M. (2008) Cell. Mol. Life Sci. 65, 2493–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlame M. (2008) J. Lipid. Res. 49, 1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hostetler K. Y. (1982) Phospholipids ( Hawthorne J. N., Ansell G. B. eds) pp. 215–261, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 5.Schlame M., Rua D., Greenberg M. L. (2000) Prog. Lipid Res. 39, 257–288 [DOI] [PubMed] [Google Scholar]
- 6.Dowhan W. (1997) Annu. Rev. Biochem. 66, 199–232 [DOI] [PubMed] [Google Scholar]
- 7.Chicco A. J., Sparagna G. C. (2007) Am. J. Physiol. Cell Physiol. 292, C33–C44 [DOI] [PubMed] [Google Scholar]
- 8.Hoch F. L. (1992) Biochim. Biophys. Acta 1113, 71–133 [DOI] [PubMed] [Google Scholar]
- 9.Hatch G. M. (1998) Int. J. Mol. Med. 1, 33–41 [DOI] [PubMed] [Google Scholar]
- 10.Zhang M., Mileykovskaya E., Dowhan W. (2002) J. Biol. Chem. 277, 43553–43556 [DOI] [PubMed] [Google Scholar]
- 11.Hauff K. D., Hatch G. M. (2006) Prog. Lipid Res. 45, 91–101 [DOI] [PubMed] [Google Scholar]
- 12.Schlame M., Ren M. (2006) FEBS Lett. 580, 5450–5455 [DOI] [PubMed] [Google Scholar]
- 13.Ott M., Zhivotovsky B., Orrenius S. (2007) Cell Death Differ. 14, 1243–1247 [DOI] [PubMed] [Google Scholar]
- 14.Schlame M., Ren M., Xu Y., Greenberg M. L., Haller I. (2005) Chem. Phys. Lipids 138, 38–49 [DOI] [PubMed] [Google Scholar]
- 15.Hostetler K. Y., Galesloot J. M., Boer P., Van Den Bosch H. (1975) Biochim. Biophys. Acta 380, 382–389 [DOI] [PubMed] [Google Scholar]
- 16.Rüstow B., Schlame M., Rabe H., Reichmann G., Kunze D. (1989) Biochim. Biophys. Acta 1002, 261–263 [DOI] [PubMed] [Google Scholar]
- 17.Han X., Yang J., Yang K., Zhao Z., Abendschein D. R., Gross R. W. (2007) Biochemistry 46, 6417–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valianpour F., Wanders R. J., Barth P. G., Overmars H., van Gennip A. H. (2002) Clin. Chem. 48, 1390–1397 [PubMed] [Google Scholar]
- 19.Schlame M., Rüstow B. (1990) Biochem. J. 272, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma B. J., Taylor W. A., Dolinsky V. W., Hatch G. M. (1999) J. Lipid Res. 40, 1837–1845 [PubMed] [Google Scholar]
- 21.Taylor W. A., Hatch G. M. (2003) J. Biol. Chem. 278, 12716–12721 [DOI] [PubMed] [Google Scholar]
- 22.Eichberg J. (1974) J. Biol. Chem. 249, 3423–3429 [PubMed] [Google Scholar]
- 23.Cao J., Liu Y., Lockwood J., Burn P., Shi Y. (2004) J. Biol. Chem. 279, 31727–31734 [DOI] [PubMed] [Google Scholar]
- 24.Wang C., Faloon P. W., Tan Z., Lv Y., Zhang P., Ge Y., Deng H., Xiong J. W. (2007) Blood 110, 3601–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong J. W., Yu Q., Zhang J., Mably J. D. (2008) Circ. Res. 102, 1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y., Kelley R. I., Blanck T. J., Schlame M. (2003) J. Biol. Chem. 278, 51380–51385 [DOI] [PubMed] [Google Scholar]
- 27.Xu Y., Malhotra A., Ren M., Schlame M. (2006) J. Biol. Chem. 281, 39217–39224 [DOI] [PubMed] [Google Scholar]
- 28.Morton R. K. (1955) Methods Enzymol. 1, 26–51 [Google Scholar]
- 29.Hatch G. M., McClarty G. (1996) J. Biol. Chem. 271, 25810–25816 [DOI] [PubMed] [Google Scholar]
- 30.Singer T. P. (1974) Methods Biochem. Anal. 22, 123–175 [DOI] [PubMed] [Google Scholar]
- 31.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 32.Cao J., Shen W., Chang Z., Shi Y. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E647–E653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y., Chen Y. Q., Li S., Konrad R. J., Cao G. J. (2009) J. Lipid Res. 50, 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rusiñol A. E., Cui Z., Chen M. H., Vance J. E. (1994) J. Biol. Chem. 269, 27494–27502 [PubMed] [Google Scholar]
- 35.Athéa Y., Viollet B., Mateo P., Rousseau D., Novotova M., Garnier A., Vaulont S., Wilding J. R., Grynberg A., Veksler V., Hoerter J., Ventura-Clapier R. (2007) Diabetes 56, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Q., Liu J., Lu B., Feingold K. R., Shi Y., Lee R. M., Hatch G. M. (2007) Biochem. J. 401, 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danos M., Taylor W. A., Hatch G. M. (2008) Biochem. Cell Biol. 86, 11–20 [DOI] [PubMed] [Google Scholar]
- 38.Hauff K., Linda D., Hatch G. M. (2009) Biochem. J. 417, 573–582 [DOI] [PubMed] [Google Scholar]
- 39.McKenzie M., Lazarou M., Thorburn D. R., Ryan M. T. (2006) J. Mol. Biol. 361, 462–469 [DOI] [PubMed] [Google Scholar]
- 40.Beigneux A. P., Vergnes L., Qiao X., Quatela S., Davis R., Watkins S. M., Coleman R. A., Walzem R. L., Philips M., Reue K., Young S. G. (2006) J. Lipid Res. 47, 734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao K. W., Schulz H. (1996) J. Biol. Chem. 271, 17816–17820 [DOI] [PubMed] [Google Scholar]
- 42.Yang S. Y., He X. Y., Schulz H. (1995) Biochemistry 34, 6441–6447 [DOI] [PubMed] [Google Scholar]
- 43.Colby T. D., Vanderveen K., Strickler M. D., Markham G. D., Goldstein B. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volchenboum S. L., Vockley J. (2000) J. Biol. Chem. 275, 7958–7963 [DOI] [PubMed] [Google Scholar]
- 45.Kelley M., Vessey D. A. (1992) Biochem. J. 288, 315–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strausberg R. L., Feingold E. A., Grouse L. H., Derge J. G., Klausner R. D., Collins F. S., Wagner L., Shenmen C. M., Schuler G. D., Altschul S. F., Zeeberg B., Buetow K. H., Schaefer C. F., Bhat N. K., Hopkins R. F., Jordan H., Moore T., Max S. I., Wang J., Hsieh F., Diatchenko L., Marusina K., Farmer A. A., Rubin G. M., Hong L., Stapleton M., Soares M. B., Bonaldo M. F., Casavant T. L., Scheetz T. E., Brownstein M. J., Usdin T. B., Toshiyuki S., Carninci P., Prange C., Raha S. S., Loquellano N. A., Peters G. J., Abramson R. D., Mullahy S. J., Bosak S. A., McEwan P. J., McKernan K. J., Malek J. A., Gunaratne P. H., Richards S., Worley K. C., Hale S., Garcia A. M., Gay L. J., Hulyk S. W., Villalon D. K., Muzny D. M., Sodergren E. J., Lu X., Gibbs R. A., Fahey J., Helton E., Ketteman M., Madan A., Rodrigues S., Sanchez A., Whiting M., Madan A., Young A. C., Shevchenko Y., Bouffard G. G., Blakesley R. W., Touchman J. W., Green E. D., Dickson M. C., Rodriguez A. C., Grimwood J., Schmutz J., Myers R. M., Butterfield Y. S., Krzywinski M. I., Skalska U., Smailus D. E., Schnerch A., Schein J. E., Jones S. J., Marra M. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]