Abstract
Single-stranded DNA-binding protein 1 (SSB1) plays an important role in the DNA damage response and maintenance of genomic stability. Here, by using protein affinity purification, we have identified Integrator3 (INT3) as a novel partner of SSB1. INT3 forms a complex with SSB1 by constitutively interacting with SSB1 regardless of DNA damage. However, following DNA damage, along with SSB1, INT3 relocates to the DNA damage sites and regulates the accumulation of TopBP1 and BRCA1 there. Moreover, INT3 controls DNA damage-induced Chk1 activation and G2/M checkpoint activation. In addition, INT3 is involved in homologous recombination repair by regulating Rad51 foci formation following DNA damage. Taken together, these results demonstrate that INT3 plays a key role in the DNA damage response.
The DNA damage response, including DNA damage checkpoint activation and DNA damage repair, ensures genomic stability under genotoxic stress. Among various types of DNA damage, DNA double-strand breaks (DSBs)3 are the most deleterious, easily causing chromosomal loss, fusion, and translocation. However, cells can sense and repair DNA DSBs by activating evolutionarily conserved pathways (1–3). Following DNA DSBs, ATM, ATR, and DNAPK, a family homologous to phosphoinositide 3-kinases (4, 5), are activated and phosphorylate histone H2AX at the DNA damage sites (6). Subsequently, phospho-H2AX (γH2AX) provides the platform for accumulation of a larger group of DNA damage response factors, such as MDC1, BRCA1, 53BP1, and TopBP1 (2, 7–9), at the DNA damage sites. Translocalization of these proteins to the DNA DSBs facilitates DNA damage checkpoint activation by activating downstream Chk1/Chk2 kinases, which arrest the cell cycle at G1, S, or G2 phase (10). In addition, it also enhances the efficiency of DNA damage repair by recruiting and stabilizing the DNA repair machinery at the DNA damage sites (11).
Among these important mediators, single-stranded DNA (ssDNA)-binding proteins play important roles during the DNA damage response. For example, following DNA damage, the MRN complex recognizes DNA DSB ends and processes the blunt ends into ssDNA overhangs (12). The replication protein A (RPA) complex, a group of ssDNA-binding proteins, immediately coats these ssDNA overhangs and loads and activates the ATR·ATRIP complex at the DNA damage sites (13). Meanwhile, the RPA complex protects ssDNA from nucleolytic resection and facilitates Rad51 filament formation along ssDNA overhangs, which is a key step for homologous recombination repair (14). Moreover, RPA70 and RPA32 subunits in the complex could recruit several DNA damage response factors to the DNA damage sites that enhance the efficacy of DNA damage repair (15).
Besides the RPA complex, several other ssDNA-binding proteins have been identified to participate in the DNA damage response recently. One of them is ssDNA-binding protein 1 (SSB1) (16). Human SSB1 is a 211-amino acid polypeptide with an N-terminal oligosaccharide/oligonucleotide-binding (OB) domain. It has been shown that SSB1 is phosphorylated by ATM and relocates to the DNA damage site following DNA DSBs. Loss of SSB1 impairs DNA damage-induced checkpoint activation and induces genomic instability. Like the RPA complex, SSB1 participates in homologous recombination by facilitating Rad51·ssDNA filament formation and stabilizing Rad51 at the DNA damage sites. Interestingly, SSB1 has a homolog SSB2 that contains an almost identical OB domain at the N terminus. However, the function of SSB2 in the DNA damage response is not clear yet.
To examine the molecular mechanism and functional pathway of SSB1 and SSB2 in the DNA damage response, we have searched for functional partners of SSB1 and SSB2 by using protein affinity purification. We have found Integrator3 (INT3) to be a common partner of both SSB1 and SSB2. Like SSB1, following DNA damage, INT3 relocates to the DNA damage sites and regulates ATR activation. Moreover, INT3 not only participates in DNA damage checkpoint activation but also regulates homologous recombination repair. Taken together, we have found a novel mediator in the DNA damage response.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
For protein affinity purification and other analyses, full-length cDNA of SSB1, SSB2, or INT3 was cloned into pS-FLAG-SBP vector. For truncation mutants, SSB1, SSB2, or INT3 cDNA fragments were PCR-amplified and cloned into Myc-tagged or HA-tagged vector modified from pcDNA3.1. For GST fusion proteins, INT3 full-length, N1 (amino acids 1–306), N2 (amino acids 1–628), C1 (amino acids 593–1042), or SSB1 full-length was cloned into the pGEX-4T vector. The siRNA sequences targeting SSB1 and SSB2 are CCAACAAGGCGGUGCAGAAdTdT and AGAGUGAACAGAAGAAUAAdTdT, respectively. The sequence of INT3 siRNA is GGACAAAGUACUCCAGCUAdTdT. siRNAs were transfected into the cells using Oligofectamine (Invitrogen) according to the manufacturer's instructions.
Rabbit anti-SSB1 and anti-SSB2 antibodies were raised against GST-SSB1 (full-length) or GST-SSB2 (full-length), respectively. Two rabbit anti-INT3 antibodies were raised GST-INT3 (amino acids 1–306) and GST-INT3 (amino acids 593–1042). Anti-Myc antibody was purchased from Covance. Anti-FLAG and anti-β-actin were purchased from Sigma. Anti-Chk1 and phospho-Chk1 (phosphoserine 345) antibodies were purchased from Cell Signaling Technology. Anti-53BP1, phospho-H2AX, BRCA1, Rad51, and TopBP1 antibodies have been described previously (17–19).
Cell Culture
Human cancer cell lines were maintained in RPMI 1640 medium with 10% fetal calf serum. MDC1-deficient mouse embryo fibroblasts (MEFs), H2AX-deficient MEFs, 53BP1-deficient MEFs, and RNF8-deficient MEFs were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum. All of the above cells were cultivated at 37 °C in 5% CO2 (v/v). For ionizing radiation (IR), cells were irradiated by using a JL Shepherd 137Cs radiation source at the indicated doses.
Cell Lysis, Immunoprecipitation, and Western Blotting
Cells were lysed with NETN300 buffer (0.5% Nonidet P-40, 50 mm Tris-HCl, pH 8.0, 2 mm EDTA, and 300 mm NaCl) unless otherwise specified. Immunoprecipitation and Western blotting were performed following standard protocol as described previously (20).
Protein Affinity Purification
Purification of SFB triple-tagged protein was described (20, 21). To search for binding partners of SSB1 and SSB2, 1 liter of HEK293T cells stably expressing SFB-SSB1 or SFB-SSB2 was harvested and washed with phosphate-buffered saline. Cells were lysed with 30 ml of ice-cold NETN300 buffer. The soluble fraction was incubated with 0.5 ml of streptavidin-conjugated beads. The beads were washed three times with NETN300 buffer. Associated proteins were eluted with 2 mm biotin in phosphate-buffered saline and incubated further with 50 μl of S beads (Novagen). The bound proteins were eluted with SDS sample buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromphenol blue, 0.125 m Tris-HCl) and analyzed with SDS-PAGE and mass spectrometry. 293T cells with empty vector were used as purification controls.
Immunofluorescence Staining
Cells grown on coverslips were fixed with 3% paraformaldehyde for 20 min and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline for 5 min at room temperature. Samples were blocked with 5% goat serum and then incubated with primary antibody for 60 min. Samples were washed three times and incubated with secondary antibody for 30 min. The coverslips were mounted onto glass slides and visualized by a fluorescence microscope. For SSB1 and INT3 staining, cells were treated with 0.5% Triton X-100 for 5 min before fixation. To examine ionizing radiation-induced foci (IRIF), cells were cultured on coverslips and treated with 10 Gy of IR followed by recovery for 4 h unless otherwise specified. To count DNA damage-induced foci, we usually choose nine fields from each slide and count 500 cells. To avoid nonspecific staining, cells with at least 3 clear foci were considered as foci-positive cells. To quantify the co-localization of INT3 foci and γH2AX foci, 100 INT3 foci-positive cells were randomly picked up. Pictures were merged by Adobe Photoshop. Every INT3 focus in every cell was examined whether or not it was positively stained with γH2AX.
In Vitro Binding Assays
Ten micrograms of anti-SSB1 antibodies was used to immobilize GST-SSB1 onto protein A beads and incubated with 1 μg of GST-INT3 in the presence of 0.5 mg/ml bovine serum albumin, 100 mm NaCl, and 20 mm sodium phosphate, pH 7.4. After a 1-h incubation at 4 °C, the beads were washed, and bound proteins were analyzed by immunoblotting.
G2/M Checkpoint Assay
U2OS cells were treated with control siRNA, SSB1 siRNA, or INT3 siRNA. siRNA-treated cells were irradiated with or without IR (2 Gy). After recovery for 1 h, cells were fixed with 70% ethanol and stained with rabbit anti-histone H3 phosphoserine 10 antibody, pH 3, followed by incubation with fluorescein isothiocyanate-conjugated goat anti-rabbit secondary antibody. The stained cells were treated with RNase, incubated with propidium iodide, and then analyzed by flow cytometry.
Homologous Recombination Assay
The assay was established and modified by Dr. Jasin's group (22). U2OS cells stabilized with a single copy of DR-GFP were transfected with control siRNA or SSB1 siRNA or INT3 siRNA. siRNA-treated cells were infected by adenovirus-encoded I-SecI (adeno-I-SecI). Cells were harvested 2 days after infection and subjected to flow cytometric analysis. The GFP-positive cell population was measured.
RESULTS
INT3 Associates with Both SSB1 and SSB2
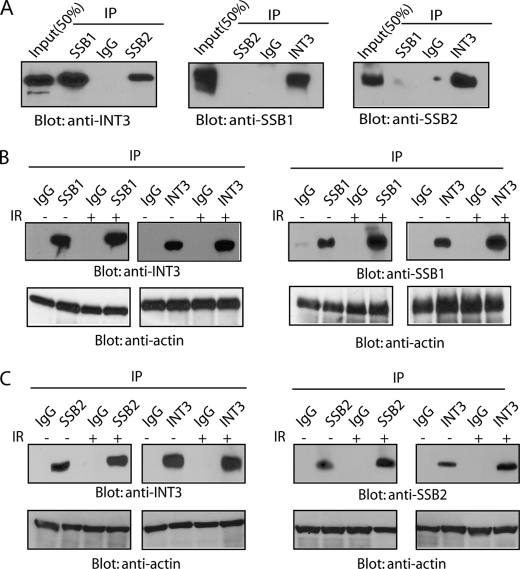
To explore the functional partner of SSB1 and SSB2, we generated HEK293T cells stably expressing streptavidin-FLAG-S protein (SFB)-tagged SSB1 or SSB2 and used tandem affinity purification to examine the associated proteins of SSB1 and SSB2. Mass spectrometry analysis revealed that INT3, a subunit of the Integrator complex, which was reported to associate with RNA polymerase II for RNA 3′ end processing (23), was present in both SSB1 and SSB2 affinity purifications (supplemental Tables 1 and 2). To validate the interactions between INT3 and SSB1 or SSB2, we first generated anti-SSB1 and anti-SSB2 antibodies. As shown in supplemental Fig. S1A, anti-SSB1 and anti-SSB2 antibodies recognize special bands around 36 and 28 kDa, respectively. We then performed co-immunoprecipitation (co-IP) and reverse co-IP of the endogenous proteins using these antibodies and found that INT3 associated with both SSB1 and SSB2 (Fig. 1A). Consistent with the purification results, in which we did not detect SSB1 from SSB2 affinity purifications and vice versa (supplemental Tables 1 and 2), endogenous SSB1 and SSB2 cannot interact with each other (Fig. 1A). These results suggest that SSB1 and SSB2 form two different complexes with INT3. Because SSB1 has been shown to participate in the DNA damage response, we wonder whether DNA damage affects the interaction between SSB1 and INT3. As shown in Fig. 1B and supplemental Fig. S1, B and C, we only observed a moderate increase of the SSB1·INT3 complex following IR treatment. Given that DNA damage does not modulate the expression of INT3, but SSB1 (Fig. 1B) (16), it is likely that the subtle increase of the SSB1·INT3 complex following DNA damage is due to DNA damage-induced SSB1 expression (supplemental Fig. S1, B and C); and the interaction between SSB1 and INT3 occurs constitutively regardless of the DNA damage. A similar constitutive interaction was also observed between SSB2 and INT3 (Fig. 1C and supplemental Fig. S1, B and C). Because INT3 has been reported as a subunit in the Integrator complex that associates with RNA polymerase II, we explored whether other subunits in the Integrator complex also interact with SSB1. FLAG-tagged INT3, INT11, and INT12 were expressed in HEK293T cells and subjected to co-IP with SSB1 antibody. As shown in supplemental Fig. S2A, only INT3, but not INT11 and INT12, associated with SSB1, indicating that SSB1 specifically interacts with INT3. To examine whether INT3 interacts with INT11 and INT12, we performed co-IP by expressing FLAG-tagged INT3, INT11, and INT12 in HEK293T cells. We were unable to detect the direct association between INT3 and INT11 or INT12 (supplemental Fig. S2B). Moreover, by performing a co-IP assay, we failed to detect any interaction between endogenous INT3 and RNA polymerase II (supplemental Fig. S2C). Taken together, these results indicated that the function of SSB1/SSB2-associated INT3 may be independent of the Integrator complex and RNA processing.
FIGURE 1.
INT3 associates with SSB1 and SSB2. A, INT3 interacts with SSB1 and SSB2. 293T cells were lysed with NETN300 buffer and then analyzed by immunoprecipitation (IP) and Western blotting with anti-SSB1, anti-SSB2, or anti-INT3 antibodies, respectively. Whole cell lysates were blotted and are shown as the input. An irrelevant IgG was used as the immunoprecipitation control. B and C, the interactions between SSB1 and INT3 (B) and SSB2 and INT3 (C) are independent of DNA DSBs. HEK293T cells were treated with 0 or 10 Gy of IR. One hour after IR, cells were lysed and analyzed with the indicated antibodies. A blot with anti-β-actin antibody was used as protein-loading control.
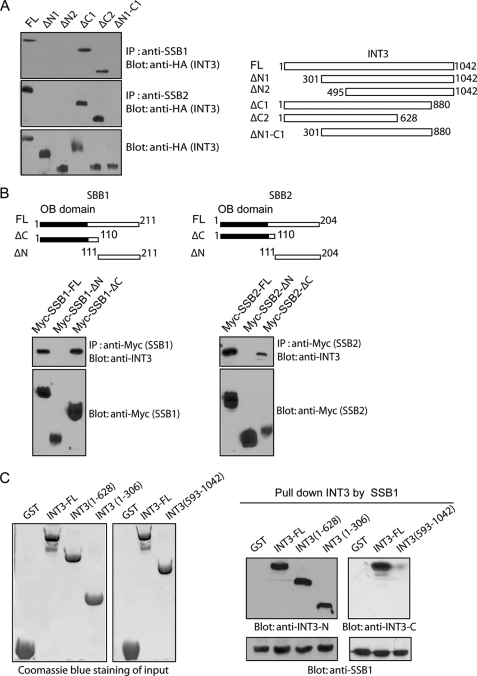
N Terminus of INT3 Directly Binds OB Domain of SSB1 or SSB2
To examine which region of INT3 is responsible for the interactions with SSB1, HA-tagged wild-type INT3 and a series of deletion mutants were expressed in HEK293T cells and subjected to co-IP with SSB1 or SSB2 antibodies. Results showed that N1 (deletion of amino acids 1–300) and N2 (deletion of amino acids 1–494) mutants of INT3 failed to associate with SSB1 or SSB2 (Fig. 2A). Interestingly, this N-terminal region of INT3 is evolutionary conserved (supplemental Fig. S3), indicating that this region could serve as a protein-protein binding motif. To map the INT3-interacting region on SSB1 and SSB2, N-terminal and C-terminal deletion mutants of SSB1 and SSB2 were generated and examined. As shown in Fig. 2B, the N-terminal OB domain deletion mutants of SSB1 and SSB2 abolish the interaction with INT3, suggesting that the conserved OB domain not only is a ssDNA-binding domain but also interacts with its partners.
FIGURE 2.
N terminus of INT3 interacts with OB domain of SSB1 and SSB2. A, the N terminus of INT3 interacts with SSB1 or SSB2. HA-tagged wild-type INT3 and deletion mutants were expressed in HEK293T cells. Cell lysates were analyzed by the indicated antibodies. The expression levels of exogenous INT3 proteins were examined by Western blotting with anti-HA antibody. IP, immunoprecipitation. FL, full-length. B, OB domain of SSB1 or SSB2 interacts with INT3. Myc-tagged wild-type SSB1, SSB2, and its deletion mutants were expressed in HEK293T cells. The interactions among SSB1, SSB2, and INT3 were examined by immunoprecipitation and Western blotting with the indicated antibodies. C, the N terminus of INT3 interacts with SSB1 directly. Anti-SSB1 antibodies were used to immobilize purified full-length SSB1 protein onto protein A beads and then incubated with purified GST, full-length INT3, the N terminus of INT3 (amino acids 1–306 or 1–628) or the C terminus of INT3 (amino acids 593–1042), respectively. The beads were washed, and bound proteins were analyzed by Western blotting using anti-INT3 N-terminal or C-terminal antibodies, respectively. The inputs of recombinant GST proteins were analyzed by Coomassie Blue staining.
To examine whether the N terminus of INT3 binds directly to SSB1, we expressed and purified recombinant full-length and the N terminus (amino acids 1–306 and 1–628) and C terminus (amino acids 593–1042) of INT3 and full-length SSB1 in bacteria. SSB1 antibodies were used to immobilize purified full-length SSB1 protein onto protein A beads and then incubated with purified wild-type or mutant INT3 proteins. The beads were washed, and bound proteins were analyzed by Western blotting using anti-INT3 N-terminal or C-terminal antibodies, respectively. As shown in Fig. 2C, full-length and the N terminus of INT3, but not the C terminus of INT3, interacted directly with SSB1 in vitro, suggesting that the N terminus of INT3, but not the C terminus INT3, interacts directly with SSB1. Taken together, these results demonstrate that INT3 is a binding partner of SSB1.
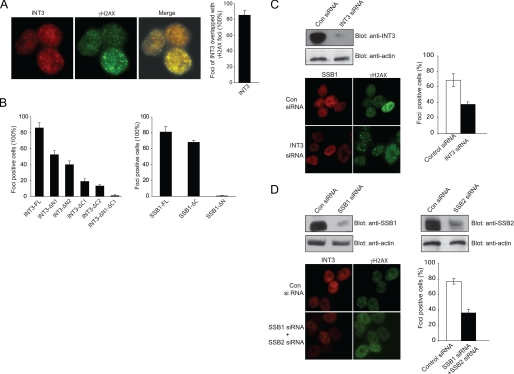
INT3 Relocates to the DNA Damage Sites Following DNA DSBs
It has been shown that SSB1 relocated to the DNA damage sites and co-localized with γH2AX following IR treatment (16). Because INT3 is a binding partner of SSB1, we examined IRIF formation of INT3. As shown in Fig. 3A, INT3, like SSB1, formed IRIF and co-localized with γH2AX, indicating that INT3 and SSB1 may function together in the DNA damage response. To examine which region targets INT3 to the DNA damage sites, we have examined the IRIF of INT3 deletion mutants. Interestingly, C-terminal deletion mutations significantly disrupted the IRIF of INT3, although N-terminal truncation mutations also impaired the foci formation of INT3. Moreover, when both N and C termini of INT3 are deleted, the foci formation of INT3 was completely abolished (Fig. 3B), suggesting that both N and C termini of INT3 are important for foci formation of INT3 after DNA damage. Similarly, we also examined the IRIF of SSB1 mutants and found that the OB domain of SSB1 is essential to target SSB1 to the DNA damage sites (Fig. 3B). Down-regulation of INT3 by siRNA modestly abrogated the IRIF of SSB1 (Fig. 3C). Meanwhile, knockdown SSB1 and SSB2 together also impaired the IRIF of INT3 (Fig. 3D). These results suggest that the interaction between INT3 and SSB1 could be important to stabilize each other at the DNA damage sites.
FIGURE 3.
INT3 relocates to DNA damage sites following DNA DSBs. A, INT3 co-localizes with γH2AX following IR treatment. U2OS cells were treated with 10 Gy of IR. Cells were stained with the indicated antibodies. Foci-positive cells were calculated by examining 500 cells in at least 9 fields. The percentage of INT3 foci that co-localize with γH2AX was quantified. The error bar was calculated from three independent experiments. B, both the N terminus and C terminus of INT3 and the OB domain of SSB1 are important for its translocation to DNA damage sites. Wild-type INT3 or its deletion mutants and wild-type SSB1 and its mutants were expressed in HeLa cells. Cells were treated with 10 Gy of IR. The percentage of foci-positive cells was counted from 500 cells of each of three independent experiments. FL, full-length. C, knockdown of INT3 modestly impairs IRIF of SSB1. U2OS cells were treated with control (Con) siRNA or INT3 siRNA for 48 h and then irradiated with 10 Gy of IR. Cells were fixed 6 h after IR and co-stained with the indicated antibodies. Foci-positive cells were counted from 500 cells of each of three independent experiments. D, knockdown of SSB1 and SSB2 together impairs IRIF of INT3. U2OS cells were treated with the indicated siRNA for 48 h and then irradiated with 10 Gy of IR. Foci-positive cells were enumerated.
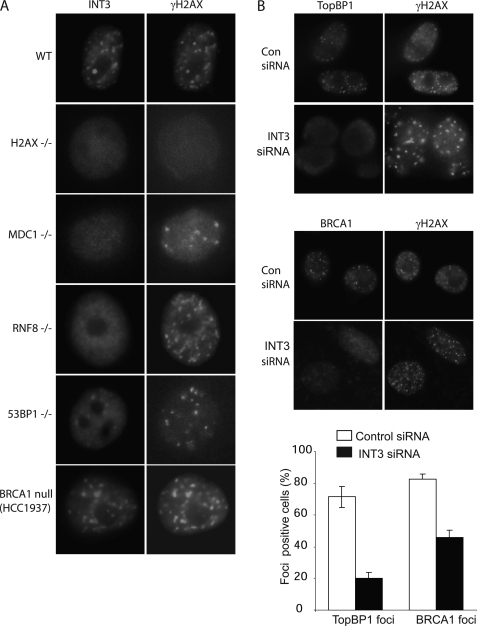
IRIF of INT3 Is Controlled by H2AX, MDC1, and RNF8; INT3 Regulates TopBP1 and BRCA1 IRIF
Following DNA damage, H2AX is rapidly phosphorylated on chromatin surrounding DNA lesions by ATM, ATR, and DNAPK, subsequently providing the platform to recruit and stabilize MDC1 and RNF8 at DSBs (2, 9). This pathway regulates several downstream proteins to the DNA damage sites, such as 53BP1 and the RAP80·BRCA1 complex (2). To examine the potential position of INT3 in the DNA damage response pathway, we have examined the IRIF of INT3 in several DNA damage response-deficient cells. As shown in Fig. 4A, the IRIF of INT3 was abolished in H2AX-, MDC1-, and RNF8-deficient cells, but was intact in the BRCA1- and 53BP1-deficient cells, suggesting that H2AX, MDC1, and RNF8 are important for recruiting or stabilizing INT3 at the DNA damage sites. To examine whether INT3 acts upstream of BRCA1 and 53BP1 following DNA damage, we examined the IRIF of BRCA1 and 53BP1 in the INT3 knockdown cells and found that BRCA1 IRIF, but not 53BP1 IRIF, was impaired by depleting INT3 (Fig. 4B and supplemental Fig. S4). Moreover, the IRIF of TopBP1 was significantly disrupted by INT3 siRNA treatment (Fig. 4B). TopBP1 is one of the factors to activate the ATR pathway following DNA damage (24, 25). Thus, our results indicate that INT3 might be important for ATR activation following DNA damage. In addition, down-regulation of INT3 by siRNA does not affect the mRNA level of BRCA1, TopBP1, and 53BP1 (supplemental Fig. S5). Taken together, these results demonstrate that the IRIF of INT3 is controlled by H2AX, MDC1, and RNF8; INT3 is important for the IRIF of TopBP1 and BRCA1.
FIGURE 4.
IRIF of INT3 is controlled by H2AX, MDC1, and RNF8; INT3 regulates TopBP1 and BRCA1 IRIF. A, IRIF of INT3 is dependent on H2AX, MDC1, and RNF8 but not 53BP1 or BRCA1. Wild-type (WT), H2AX-, MDC1-, RNF8-, and 53BP1-deficient MEFs and BRCA1-deficient HCC1937 cells were transfected with FLAG-INT3 and then treated with 10 Gy of IR. Cells were fixed 6 h after IR and immunostained with anti-FLAG (INT3) and γH2AX antibodies. B, knockdown of INT3 impairs IRIF of TopBP1 and BRCA1. U2OS cells were treated with control siRNA or INT3 siRNA. Cells were irradiated with 10 Gy of IR. Six hours after IR, cells were fixed and co-stained with the indicated antibodies.
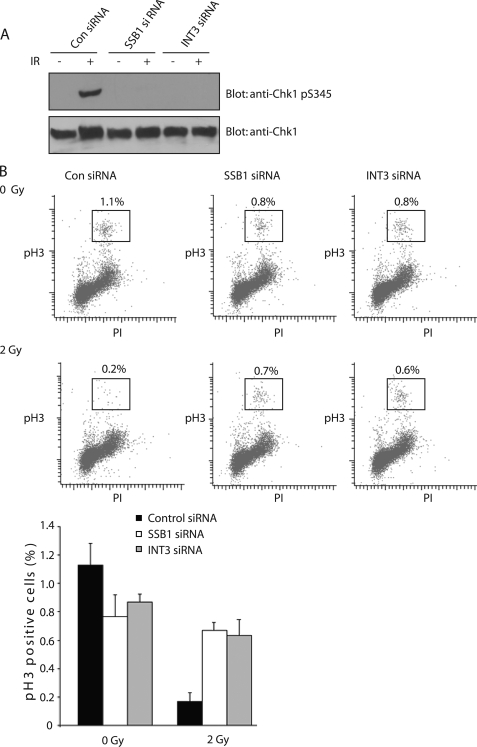
INT3 Regulates Chk1 Activation and G2/M Checkpoint Activation following DNA Damage
Because INT3 might be involved in ATR activation and SSB1 has been shown to regulate Chk1 phosphorylation (16), we have examined the role of INT3 in ATR-dependent signaling. As shown in Fig. 5A, knockdown INT3 abolished DNA damage-induced Ser345 phosphorylation of Chk1, which is a surrogate marker of Chk1 activation (26–28). Because Chk1 controls activation of several checkpoints following DNA damage, including G2/M checkpoint activation, loss of Chk1 activation abrogates the G2/M checkpoint (26–28). Thus, we examined the role of INT3 in G2/M checkpoint activation using a previously established G2/M checkpoint assay (21). Following DNA damage, a normal cell with a G2/M checkpoint is arrested at the G2/M transition to allow DNA lesions to be repaired. Mitotic cells (marked by phosphohistone H3) are significantly reduced. If the G2/M checkpoint is abolished, cells could not be arrested at the G2/M transition following DNA damage. The mitotic population will not change as dramatically as that in control cells. As shown in Fig. 5B, when cells were treated with a control siRNA, their mitotic population was dramatically reduced (from 1.1% to 0.2%) after treatment with 2 Gy of IR, suggesting that the DNA damage-induced G2/M checkpoint is activated and blocks the cell cycle at the G2/M transition. In contrast, in cells treated with either SSB1 siRNA or INT3 siRNA, the mitotic fractions are just slightly reduced after IR, suggesting that the DNA damage-induced G2/M checkpoint is disrupted in the absence of either SSB1 or INT3. These data suggest that SSB1 and INT3 are essential for normal DNA damage-induced G2/M checkpoint activation.
FIGURE 5.
INT3 is required for Chk1 activation and G2/M checkpoint. A, knockdown of INT3 disrupts IR-induced Ser345 phosphorylation of Chk1. U2OS cells were treated with control siRNA, INT3 siRNA, or SSB1 siRNA. siRNA-treated cells were irradiated with 10 Gy of IR. One hour after IR treatment, cell lysates were analyzed by immunoprecipitation and Western blotting with the indicated antibodies. The protein loading control was examined with anti-β actin antibody. B, INT3 participates in DNA damage-induced G2/M checkpoint activation. U2OS cells were treated with the indicated siRNA and then irradiated with or without 2 Gy of IR. One hour after IR treatment, cells were harvested and stained with phosphohistone H3 (phosphoserine 10) antibody and quantified by FACS analysis. Data represent averages from three independent experiments.
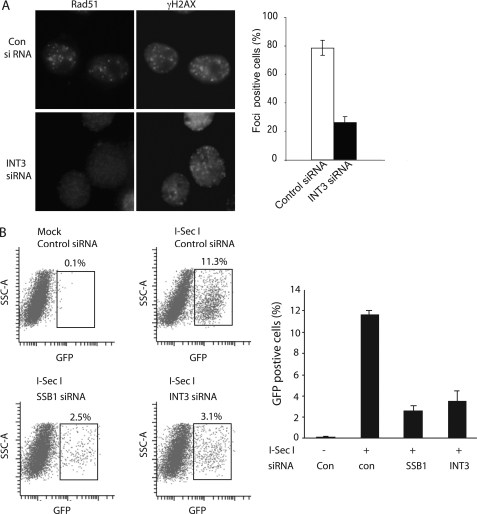
INT3 Controls Rad51 Foci Formation and Homologous Recombination
Similar to the RPA complex, SSB1 has been shown to facilitate Rad51 and ssDNA to form nucleoprotein filament and to regulate homologous recombination repair (16). Thus, we first examined whether INT3 affects Rad51 foci formation after IR. As shown in Fig. 6A, depletion of INT3 dramatically reduced the IRIF of Rad51. To examine the role of INT3 in the homologous recombination, we used an I-SceI-dependent GFP reporter assay to measure homologous recombination in SSB1- or INT3-delepted cells. This assay depends on the reconstitution of GFP as a consequence of repairing an I-SceI-induced DSB (29). I-Sce1 is a yeast DNA restriction enzyme but does not have any restriction site in whole human genomic DNA. A plasmid (DR-GFP reporter construct) with two mutant GFP genes (mGFP1 and mGFP2) was stably transfected into U2OS cells. These two mutant GFP genes contain different mutations and will not produce wild-type GFP that transmits green fluorescence. One of these two mutant GFP genes (mGFP1) harbors a unique I-Sce1 site (mGFP1). I-Sce1 will generate a DSB in mGFP1. By using another mutant GFP gene (mGFP2) as a template, this break in mGFP1 will be repaired by endogenous homologous recombination machinery, and a wild-type GFP will be generated and excite green fluorescence. Homologous recombination-positive cells can be measured by flow cytometry. After double-transfected SSB1 siRNA, INT3 siRNA, or control siRNA, U2OS cells that stably integrated a DR-GFP reporter construct were infected by adeno-I-SecI. Forty-eight hours after infection, the cells were harvested, and the number of GFP-positive cells was assessed by flow cytometry. As shown in Fig. 6B, the GFP-positive population was raised from 0.1% to 11.3% following I-SceI expression in the cells treated with control siRNA, suggesting that I-SceI-induced homologous recombination repair is activated in the cells treated with control siRNA. However, compared with control siRNA-treated cells, the GFP-positive population was dramatically reduced in the cells treated with SSB1 siRNA or INT3 siRNA, suggesting that I-SceI-induced homologous recombination repair is abrogated in the absence of SSB1 and INT3. Taken together, these results suggest that INT3, like SSB1, plays an important role in homologous recombination.
FIGURE 6.
INT3 is important for IRIF of Rad51 and homologous recombination. A, knockdown of INT3 impairs IRIF of Rad51. U2OS cells were treated with control siRNA or INT3 siRNA for 48 h and then irradiated with 10 Gy of IR. Cells were fixed 6 h after IR and co-stained with the indicated antibodies. B, knockdown of INT3 impairs homologous recombination. U2OS-DR-GFP cells were treated with indicated siRNAs. The siRNA-treated cells were then infected by adeno-I-SecI. Two days later, cells were harvested and subjected to flow cytometric analysis to analyze the homologous recombination efficiency. The percentage of GFP-positive cells is shown. Data represent averages from three independent experiments.
DISCUSSION
SSB1 plays a major role in the DNA damage response. Here, we have identified INT3 as a functional partner of SSB1 in the DNA damage response. The SSB1·INT3 complex regulates both DNA damage checkpoint activation and DNA damage repair. Like SSB1, INT3 relocates to DNA lesions following IR. Regarding the DNA damage-induced foci formation, INT3 is downstream of H2AX, MDC1, and RNF8, but upstream of TopBP1, which is important to activate ATR kinase following DNA damage. By examining Chk1 phosphorylation, we found that INT3 regulates ATR-dependent signaling, which mediates DNA damage-induced cell cycle checkpoints. The IRIF of TopBP1 is dependent on its BRCT domain (18), a phosphoprotein-binding module (17). Although we do not detect TopBP1 during the protein affinity purification, it is possible that other mediators may exist between the SSB1·INT3 complex and TopBP1 following DNA damage. Moreover, TopBP1 has been shown to associate with BRCA1 (30). It is likely that INT3 modulates a TopBP1·BRCA1 complex following DNA damage. Alternatively, INT3 may indirectly regulate BRCA1 through TopBP1-dependent ATR activation.
Both SSB1 and INT3 are important for Rad51 IRIF and Rad51-mediated homologous recombination repair. Because SSB1 recognizes ssDNA, as proposed, this complex could be important to stabilize Rad51 at the DNA lesions and facilitate Rad51 ssDNA filament formation (16). Future structural analysis will reveal how Rad51 fits in or replace SSB1·INT3 at 3′ overhangs at the lesions. Recently, similar findings reported by two other groups further confirmed the role of the SSB1 complex in the DNA damage response, especially homologous combination repair (31, 32).
Besides SSB1, we also characterized SSB2. It associates with INT3 through the N-terminal ssDNA-binding domain. SSB1 and SSB2 share very similar ssDNA-binding domains at the N terminus, but with distinct C-terminal tails (16). Like SSB1, SSB2 interacts with INT3 through its N-terminal OB domain. However, compared with SSB1, the expression of SSB2 is very low in normal culture cells (supplemental Fig. S6). After knockdown SSB2 by siRNA, we could not detect any effect on ATR-dependent signaling, G2/M checkpoint activation, and homologous recombination. Thus, it is possible that SSB2 is merely a redundant homolog of SSB1 during regular DNA damage response and only functions in some special circumstances.
Supplementary Material
Acknowledgments
We thank Dr. Eric Fearon for sharing experimental equipment, Dr. Khanna for reagents, and Dr. Lin-Yu Lu for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA132755 and CA130899 (to X. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S6, and three supplemental tables.
- DSB
- double-strand break
- adeno-I-SecI
- adenovirus-encoded I-SecI
- co-IP
- co-immunoprecipitation
- γH2AX
- phospho-H2AX
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- INT3
- Integrator3
- IR
- ionizing radiation
- IRIF
- IR-induced foci
- MEF
- mouse embryonic fibroblast
- OB domain
- oligosaccharide/oligonucleotide-binding domain
- RPA
- replication protein A
- siRNA
- small interfering RNA
- ssDNA
- single-stranded DNA
- SSB1 and SSB2
- ssDNA-binding protein 1 and 2, respectively.
REFERENCES
- 1.Rouse J., Jackson S. P. (2002) Science 297, 547–551 [DOI] [PubMed] [Google Scholar]
- 2.Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 3.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 4.Abraham R. T. (2004) DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 5.Khanna K. K., Lavin M. F., Jackson S. P., Mulhern T. D. (2001) Cell Death Differ. 8, 1052–1065 [DOI] [PubMed] [Google Scholar]
- 6.Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 7.Stucki M., Jackson S. P. (2006) DNA Repair 5, 534–543 [DOI] [PubMed] [Google Scholar]
- 8.Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) Curr. Biol. 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Capetillo O., Allis C. D., Nussenzweig A. (2004) J. Exp. Med. 199, 1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartek J., Lukas J. (2003) Cancer Cell 3, 421–429 [DOI] [PubMed] [Google Scholar]
- 11.Lisby M., Rothstein R. (2004) Curr. Opin. Cell Biol. 16, 328–334 [DOI] [PubMed] [Google Scholar]
- 12.Takeda S., Nakamura K., Taniguchi Y., Paull T. T. (2007) Mol. Cell 28, 351–352 [DOI] [PubMed] [Google Scholar]
- 13.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 14.West S. C. (2003) Nat. Rev. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 15.Fanning E., Klimovich V., Nager A. R. (2006) Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard D. J., Bolderson E., Cubeddu L., Wadsworth R. I., Savage K., Sharma G. G., Nicolette M. L., Tsvetanov S., McIlwraith M. J., Pandita R. K., Takeda S., Hay R. T., Gautier J., West S. C., Paull T. T., Pandita T. K., White M. F., Khanna K. K. (2008) Nature 453, 677–681 [DOI] [PubMed] [Google Scholar]
- 17.Yu X., Chini C. C., He M., Mer G., Chen J. (2003) Science 302, 639–642 [DOI] [PubMed] [Google Scholar]
- 18.Yamane K., Wu X., Chen J. (2002) Mol. Cell. Biol. 22, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward I. M., Minn K., van Deursen J., Chen J. (2003) Mol. Cell. Biol. 23, 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., Yu X. (2009) Curr. Biol. 19, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H., Chen J., Yu X. (2007) Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 22.Weinstock D. M., Nakanishi K., Helgadottir H. R., Jasin M. (2006) Methods Enzymol. 409, 524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baillat D., Hakimi M. A., Naar A. M., Shilatifard A., Cooch N., Shiekhattar R. (2005) Cell 123, 265–276 [DOI] [PubMed] [Google Scholar]
- 24.Kumagai A., Lee J., Yoo H. Y., Dunphy W. G. (2006) Cell 124, 943–955 [DOI] [PubMed] [Google Scholar]
- 25.Cimprich K. A., Cortez D. (2008) Nat. Rev. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. (2000) Genes Dev. 14, 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Girona A., Tanaka K., Chen X. B., Baber B. A., McGowan C. H., Russell P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11289–11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao H., Piwnica-Worms H. (2001) Mol. Cell. Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moynahan M. E., Chiu J. W., Koller B. H., Jasin M. (1999) Mol. Cell 4, 511–518 [DOI] [PubMed] [Google Scholar]
- 30.Greenberg R. A., Sobhian B., Pathania S., Cantor S. B., Nakatani Y., Livingston D. M. (2006) Genes Dev. 20, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J., Gong Z., Ghosal G., Chen J. (2009) Mol. Cell 35, 384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Bolderson E., Kumar R., Muniandy P. A., Xue Y., Richard D. J., Seidman M., Pandita T. K., Khanna K. K., Wang W. (2009) J. Biol. Chem. 284, 23525–23531 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.