Abstract
The molecular determinants necessary and sufficient for recognition of its specific DNA target are contained in the C-terminal domain (H-NSctd) of nucleoid-associated protein H-NS. H-NSctd protects from DNaseI cleavage a few short DNA segments of the H-NS-sensitive hns promoter whose sequences closely match the recently identified H-NS consensus motif (tCG(t/a)T(a/t)AATT) and, alone or fused to the protein oligomerization domain of phage λ CI repressor, inhibits transcription from the hns promoter in vitro and in vivo. The importance of H-NS oligomerization is indicated by the fact that with an extended hns promoter construct (400 bp), which allows protein oligomerization, DNA binding and transcriptional repression are highly and almost equally efficient with native H-NS and H-NSctd::λCI and much less effective with the monomeric H-NSctd. With a shorter (110 bp) construct, which does not sustain extensive protein oligomerization, transcriptional repression is less effective, but native H-NS, H-NSctd::λCI, and monomeric H-NSctd have comparable activity on this construct. The specific H-NS-DNA interaction was investigated by NMR spectroscopy using monomeric H-NSctd and short DNA duplexes encompassing the H-NS target sequence of hns (TCCTTACATT) with the best fit (8 of 10 residues) to the H-NS-binding motif. H-NSctd binds specifically and with high affinity to the chosen duplexes via an overall electropositive surface involving four residues (Thr109, Arg113, Thr114, and Ala116) belonging to the same protein loop and Glu101. The DNA target is recognized by virtue of its sequence and of a TpA step that confers a structural irregularity to the B-DNA duplex.
Nucleoid-associated DNA-binding protein H-NS plays the dual role of architectural protein of the nucleoid and regulator of expression of a large number of genes. Differential display analysis of transcriptomes of wild type and hns null mutants of Escherichia coli has revealed that the number of genes whose expression is directly or indirectly controlled by H-NS corresponds to ∼5% of the total chromosomal genes (1). Two types of repression mechanisms (promoter exclusion and RNA polymerase entrapment) have been documented (for reviews see Refs. 2 and 3). Both mechanisms depend upon the recognition of DNA bends and bending of linear tracts of DNA, two H-NS properties that in turn depend upon the DNA binding and oligomerization capacity of this protein (4–6). However, because the molecular basis of the preference that H-NS displays for some promoter regions but not for others (for reviews see Refs. 2 and 7–9) is far from clear, the mechanism by which H-NS performs its selective transcriptional repressor activity is understood only in part.
H-NS binds preferentially to DNA bends (4, 10–13) endowed with special geometric features (i.e. planar bends) (14, 15), but the quantitative aspects of this preference remained a matter of dispute, also because the very extended, ill-defined, and diffuse footprinting patterns often produced by H-NS on its target promoters (16, 17) suggested that H-NS was devoid of any DNA binding specificity (18). Thus, the selectivity by which H-NS targets the promoters of some genes and not of others was difficult to explain. However, contrary to general opinion, recent data have shown that H-NS recognizes a specific 10-bp-long consensus sequence (19). Furthermore, it was suggested that the consensus sequences represent the specific target where H-NS nucleation initially starts before subsequent protein oligomerization leads to the assembly of the transcriptional repression complex.
Indeed, following initial binding to nucleation sites strategically located in a given promoter, H-NS-H-NS interactions could promote bridging between the two sides of a DNA bend (17, 18, 20–22) and/or the lateral condensation of distant stretches of duplex DNA (5), two processes considered to be key features by which H-NS exercises its roles of transcriptional repressor and architectural organizer of the nucleoid (19).
The protein oligomerization functions of H-NS is generally attributed to the N-terminal domain (H-NSntd), whereas the C-domain2 (H-NSctd) is considered responsible for DNA binding (for reviews see Refs. 9 and 23). However, important roles in protein oligomerization and DNA binding have been attributed also to H-NSctd and to H-NSntd, respectively (24–26).
In this article we show that H-NSctd fused to the protein oligomerization domain of the bacteriophage λCI repressor, or even the isolated, monomeric H-NSctd can (auto)repress, both in vivo and in vitro, the H-NS-sensitive hns promoter with the same specificity, albeit not with the same efficiency, as native H-NS. Furthermore, we show that the isolated H-NSctd is able to selectively protect the same 10-bp consensus sequence recognized by the whole protein on the same promoter. These findings demonstrate that the H-NSctd contains all the molecular determinants necessary and sufficient for recognition and binding to a specific DNA target and that the interaction with these site(s) likely reflects a key step in the mechanism of transcriptional repression by which H-NS controls the expression of its own gene and, likely, that of entire groups of select genes.
In light of the above premises, in this article we have used NMR spectroscopy to investigate the residues of H-NSctd involved in the specific recognition of its target DNA as well as the structural features of the DNA target itself. We conclude from our results that the specific binding of H-NSctd to its consensus target entails an interaction between a tetrapeptide loop of the protein and 5 bp in the duplex where its structure deviates from the canonical B-DNA form.
EXPERIMENTAL PROCEDURES
Buffers
Buffer A contained 10 mm Tris-HCl, pH 7.7, 60 mm NH4Cl, 10 mm magnesium acetate, 5 mm β-mercaptoethanol, 0.2 mm phenylmethylsulfonyl fluoride, and 0.2 mm benzamidine; Buffer B contained 20 mm Tris-HCl, pH 7.7, 1.0 mm EDTA, pH 8.0, 5% glycerol, 100 mm NaCl, 5 mm β-mercaptoethanol, 0.2 mm phenylmethylsulfonyl fluoride, and 0.2 mm benzamidine; Buffer C contained 20 mm Tris-HCl, pH 7.7, 0.1 mm EDTA, pH 8.0, 100 mm NaCl; Buffer D contained 25 mm Tris-HCl, pH 7.7, 1 mm EDTA, pH 8.0, 25 mm NaCl, 5 mm β-mercaptoethanol 0.2 mm phenylmethylsulfonyl fluoride, and 0.2 mm benzamidine; Buffer E contained 25 mm Tris-HCl, pH 7.7, 100 mm NaCl, 1 mm EDTA, pH 8.0; Buffer F contained 40 mm HEPES-KOH, pH 8.0 buffer containing 50 mm KCl, 10 mm magnesium acetate, 5 mm dithiothreitol, 0.005% (v/v) Nonidet P-40; and Buffer G contained 10 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 100 mm NaCl, 10 mm KCl.
Production and Purification of H-NS, H-NSctd, [15N]H-NSctd, and [15N/13C]H-NSctd
The full-length protein was overproduced in E. coli UT5600 harboring pcI857 and pPLc2833, which carries hns under the control of the λPL promoter. During the exponential phase the cells, grown at 30 °C in LB containing ampicillin (60 mg/l) and kanamycin (25 mg/l), were induced to overexpress H-NS by a 30–42 °C temperature shift. After 30 min at 42 °C and 30 min at 37 °C, the cells were harvested, resuspended in 20 ml of Buffer A, and ruptured by sonication. The cell lysate was centrifuged at 12,000 rpm for 1 h at 4 °C to obtain the S30 cell fraction. The NH4Cl concentration was increased to 1 m, and the cell extract subjected to ultracentrifugation at 80 K rpm (Sorvall S80AT3–0181 rotor) for 1 h at 4 °C to yield a postribosomal (S100) fraction that was extensively dialyzed against buffer B. The extract was then loaded on a phosphocellulose column and eluted by a linear gradient of NaCl (100–700 mm) in buffer B, and the fractions containing H-NS, identified by 16% SDS-PAGE were pooled and dialyzed against buffer B, loaded on a heparin-Sepharose column, and eluted by a linear gradient of NaCl (100–700 mm) in buffer B. The pooled fractions containing H-NS were concentrated and subjected to gel filtration through a Superdex 75-HR column (Amersham Biosciences) eluting with buffer B containing 0.7 m NaCl. The H-NS-containing fractions were pooled and dialyzed against buffer C.
The C-terminal domain of H-NS (residues 89–136) was overproduced in E. coli 71-18 harboring both pEV1, carrying the coding sequence for H-NS89–136 and pcI857. Induction of H-NSctd was carried out essentially as described for the full-length protein but for the fact that for the isotopic labeling the cells were grown in minimal medium (50 mg/liter MgSO4, 4.4 g/liter KH2PO4, 12 g/liter K2HPO4, 7 mg/liter (NH4)2Fe(SO4)2·6H2O, 0.6 g/liter NH4Cl, 5 g/liter glucose, 10 mg/liter thiamine hydrochloride) supplemented with ampicillin (60 mg/liter) and kanamycin (25 mg/liter) as well as with 0.5 g/liter 15NH4Cl or with 0.5 g/liter 15NH4Cl and 3 g/liter 13C-glucose. Purification of the H-NSctd was carried out following a protocol similar to that used for the full-length protein, but for the use of buffer D and the elution from the column obtained with a 25–600 mm linear NaCl gradient and for the presence of 6 m urea in the buffer used for gel filtration. The purified proteins were resuspended in buffer E and stored at −80 °C.
DNase I Footprinting
The DNA fragment containing the hns promoter was generated by PCR using Taq platinum polymerase (Invitrogen) and two primers, one of which was end-labeled with [γ-32P]ATP (3,000 Ci/mmol) using phage T4 polynucleotide kinase. The primers (5′-GCTTCGCTCATTGTAGTAATC-3′ and 5′-AGTCCATGCTCTTATTGCGAC-3′) specifically amplify a 360-bp fragment from the template pKK400 (13). The PCR product was purified on a GenElute PCR clean-up column (Sigma). The labeled fragment (<2 nm) containing the hns promoter was incubated at 20 °C with the indicated concentrations of protein. The nuclease reaction was carried out for 30 s at 20 °C with 10 μg/ml of DNase I (Sigma) in 20 μl of Buffer F. The reaction was stopped by placing the samples on ice and adding 75 μl of 100 mm EDTA. After phenol chloroform extraction, the samples were precipitated with 3 volumes of ethanol in the presence of 20 μg/ml of glycogen and 1 μg/ml of calf thymus DNA. The samples were then resuspended in 5 μl of formamide blue (90% (v/v) formamide, 10 mm EDTA, pH 8.0, 0.025% (w/v) bromphenol blue, 0.025% (w/v) xylene cyanol) and loaded on 8% (w/v) denaturing polyacrylamide gels.
Plasmid Construction and in Vivo Studies
The plasmids used for the in vivo studies were constructed as follows. The DNA encoding the oligomerization domain of the λCI repressor was first placed into pBAD under the control of the ara promoter using the BglII and EcoRI sites. Then a fragment comprising the ara promoter and the oligomerization domain of λCI repressor was excised with HincII and PstI and placed into pACYC177 to give pACYC177-λCIctd. Finally, the PCR product of the DNA encoding the C-terminal domain of H-NS(Δ1–488) was inserted into the NcoI and BglII sites of pACYC177-λCIctd to yield the plasmid pACYC-H-NSctd:: λCIctd encoding the chimeric protein H-NS(Δ1–88)::CI(Δ1–92). To construct pACYC177-H-NSwt and pACYC177-H-NSctd, the plasmids expressing H-NSwt and the C-terminal domain of H-NS(Δ1–88) under the control of the ara promoter, the PCR products of the hns sequence encoding the entire H-NS protein or its C-terminal domain were inserted into the NcoI and PstI sites of pACYC177. The pACYC constructs were transformed into E. coli YK4124, an hns null strain (27) carrying pKK400, pKK110, or pBR328, three plasmids encoding the cat reporter gene (13). Because in our hands, the ara promoter was rather leaky, its induction proved to be unnecessary to obtain the in trans expression of H-NSwt, H-NS(Δ1–88), and H-NS(Δ1–88)::CI(Δ1–92). The cellular level of these proteins was estimated, using a protocol similar to that previously described (28), by semi-quantitative Western blotting using polyclonal antibodies raised against H-NSwt and calibration curves constructed with known amounts of the three purified proteins. The ability of H-NSwt, H-NSctd::λCIctd, or H-NSctd to repress transcription of the cat reporter gene was measured by testing the chloramphenicol acetyltransferase activity expressed by the cells (29).
Electrophoretic Gel Shift
Each reaction mixture contained ∼5 ng of the appropriate DNA fragment (end-labeled with [32P]dATP by fill-in reaction using Klenow fragment of DNA polymerase), 50 ng of poly(IdT) DNA competitor, and the indicated amounts of purified proteins in 20 μl of Buffer G containing 1 mm spermidine, 0.5 mm dithiothreitol, and 5% glycerol. After 10-min incubation at 22 °C, the samples were subjected to electrophoresis at room temperature on a 7% polyacrylamide gel in Tris acetate buffer.
In Vitro Transcription
The reaction was carried out in 30 μl of Buffer G containing 2 mm spermidine, 2 mm dithiothreitol, 0.1 mg/ml bovine serum albumin, 0.1 μg of pKK400 DNA, 0.5 mm of the four NTPs, and the indicated amounts of purified proteins. The mixtures were preincubated at 37 °C for 10 min and, after the addition of 0.2 unit of E. coli RNA polymerase, incubated at the same temperature for 30 min. The reactions were stopped with 5 mm EDTA, and the samples were precipitated with ethanol after the addition of 1 m ammonium acetate and 1 μg of tRNA. Electrophoretic analysis of the product, followed by Northern blotting and hybridization (30), showed that chloramphenicol acetyltransferase mRNA is the only product obtained. The DNA probe used to reveal the chloramphenicol acetyltransferase mRNA produced in the transcription reaction was a DNA fragment containing the entire cat gene labeled with [32P]dATP using a random primer kit (Amersham Biosciences).
NMR Methods
Typical conditions for NMR experiments consist of samples in 10–50 mm phosphate buffer at pH 6.5. The protein resonances were assigned using a double-labeled 15N-13C sample and recording 15N HSQC, 13C HSQC, 15N total correlated spectroscopy-HSQC, 15N NOESY-HSQC, HNHA, CBCA(CO)NH, CBCANH, HBHA(CO)NH spectra. NMR experiments were performed on Bruker AVANCE instruments operating at 400, 600, 750, and 900 MHz with the temperature set to 300 °K. Data were processed using NMRPipe (31) and analyzed using NMRView (32) or Sparky.
Analysis of the titration of H-NSctd with the DNA fragment was performed considering the ΔδHN and the ΔδN difference shifts of the complex and the free protein domain as well as combining both shifts according to the relationship: Δδ = [ΔδHN2 + (ΔδN/R)2]1/2, where R = 10. All of the protons of the 20-, 15-, and 10-bp target fragments were assigned from two-dimensional NOESY and total correlated spectroscopy spectra obtained in both H2O and D2O following the established procedure (33).
RESULTS
In previous studies we have used two-hybrid systems to study the in vivo oligomerization properties of H-NS as well as of its fragments and mutants and have shown that changes of some environmental parameters can affect the efficiency by which tetramers of H-NS (the biologically active form of this protein) are formed (24).
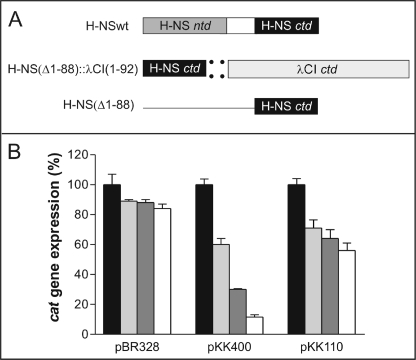
To study the H-NS-DNA interaction here we have used a similar two-hybrid approach. However, instead of using chimerae having the DNA binding capacity of a lambdoid repressor and the protein oligomerization property of H-NS, we have constructed a chimera in which the DNA binding property is provided by H-NSctd (H-NS(Δ1–88)) and the protein oligomerization function is provided by the C-domain of bacteriophage λ CI repressor (CI(Δ1–92)) (Fig. 1A). This chimera was tested in vivo for its capacity to repress transcription from the hns promoter, which is subject to transcriptional autorepression by H-NS (13, 17, 34). For this purpose two plasmid constructs, pKK400 and pKK110 (schematically represented in Ref. 13), were used. Both constructs contain the promoter-less reporter gene cat placed under the control of the core elements of the hns promoter. The construct pKK400 contains all three H-NS-binding regions found in hns, namely the promoter-proximal site and the two extended tracts of DNA flanking the intrinsic DNA bend located at approximately −150; pKK110, on the other hand, lacks the entire upstream region and contains only the promoter-proximal H-NS-binding site (13, 17). As a result, transcription from pKK400 is strongly repressed by H-NS, which, exploiting its oligomerization capacity, can build an extended multimeric nucleoprotein complex, thereby preventing the RNA polymerase from forming a functional transcription initiation complex. On the other hand, transcription from pKK110 is inhibited with lower efficiency by H-NS, which can bind to only one DNA region and inhibits transcription by competing with RNA polymerase for binding to the same DNA tract (13, 17). Cells harboring pBR328 were used as a control because in this plasmid the cat gene is expressed from its natural promoter, which is not H-NS-sensitive, unlike the hns promoter. These three plasmids (pBR328, pKK400, and pKK110) were transformed into hns null mutant cells which either: (a) do not express any H-NS; (b) express full-length H-NS (H-NSwt) in trans under the control of the ara promoter; (c) express the H-NSctd::λCIctd chimera; or (d) express a protein fragment (H-NSctd) consisting of the C-terminal domain. The level of cat expressed from the three plasmid constructs was then measured in the aforementioned genetic backgrounds. Because semi-quantitative Western blotting (see “Experimental Procedures”) indicated that the three forms of H-NS (H-NSwt, the H-NSctd::λCIctd chimera, and H-NSctd) were present in vivo in stoichiometrically comparable amounts, the level of cat measured should represent a good estimate of the in vivo repressor capacity of these proteins (see “Discussion”).
FIGURE 1.
In vivo transcriptional repression activity of H-NSwt and of its oligomerizing and nonoligomerizing C-terminal domain (H-NSctd) as a function of the nature of the promoter. A, schematic representation from top to bottom of the domain structure of H-NSwt, H-NSctd::λCIctd [H-NS(Δ1–88)::λCI(Δ1–92)] chimera, and H-NSctd [H-NS (Δ1–88)]. B, in vivo transcription of cat in cells harboring the indicated plasmids (pBR328, pKK400, and pKK110) and expressing no H-NS (black), H-NSwt (white), H-NSctd (light gray), and the H-NSctd::λCIctd chimera (dark gray). The percentage of expression of the reporter gene cat (ordinate) is calculated taking as 100% of expression in the absence of H-NS (hns null mutant).
As seen from the results obtained, cat transcription from pBR328 is essentially unaffected by H-NSwt, by the H- NSctd::λCIctd chimera, and by the isolated H-NSctd, its level of expression being in all cases ∼90% of the control lacking H-NS (Fig. 1B). This is the expected result for a promoter that is not under transcriptional control of H-NS. In contrast, cat expression from pKK400 was found to be severely repressed (∼90%) by H-NSwt, by the H-NSctd::λCIctd chimera (∼70%), and, albeit with a lower efficiency, by the isolated monomeric H-NSctd (∼40%). Furthermore, in agreement with published results (13), H-NSwt was able to repress cat expression from pKK110, albeit to a lesser extent than from pKK400 (∼40 versus ∼90%). It is remarkable, however, that with the pKK110 plasmid, cat is repressed to almost the same level by H-NSwt (∼44%), by the HNSctd::λCIctd chimera (∼37%), and by H-NSctd (∼32%) (Fig. 1B).
Taken together, these results indicate that the highest repression capacity of H-NS requires protein oligomerization, which brings together H-NS molecules bound to distant portions of the promoter, an event that is possible in the pKK400 plasmid where the hns promoter contains multiple H-NS-binding sites but not in pKK110, which contains a single H-NS-binding region (13, 17). The similar repression capacity of the oligomerization-incompetent H-NSctd and of the oligomerization-competent H-NSwt and H-NSctd::λCIctd chimera adds further support to this premise. Furthermore, the good performance of the H-NSctd::λCIctd chimera in inhibiting cat transcription from pKK400 indicates that the C-terminal domain of the λCI repressor can efficiently replace H-NSntd in promoting H-NSctd oligomerization, yielding a transcriptional repressor with properties similar to those of H-NSwt and that the N-terminal domain of this protein does not significantly contribute to the recognition and binding of the DNA target, a property that is instead due to the C-domain of the protein. The premise that all molecular determinants for the recognition of a target promoter are contained within the C-domain of H-NS is further supported by the finding that, despite its lack of binding cooperativity, the monomeric H-NSctd is capable of selective transcriptional repression vis-à-vis an H-NS-sensitive promoter.
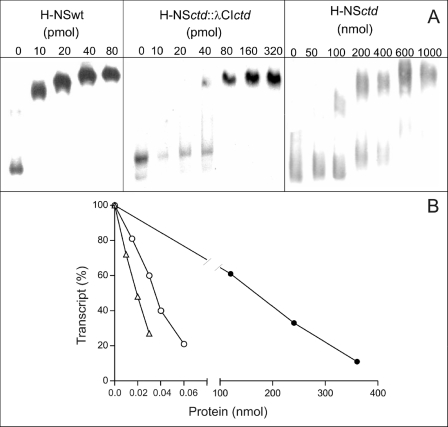
Consistent with the in vivo results are in vitro data in which purified H-NSwt, H-NSctd::λCIctd chimera, and H-NSctd were compared for their DNA binding capacity and for their effect on hns::cat transcription. As seen from the electrophoretic band shift experiment shown in Fig. 2A, increasing concentrations of all three proteins retarded the mobility of a 400-bp DNA fragment (excised from pKK400) containing the complete hns promoter region.
FIGURE 2.
In vitro DNA binding and transcriptional repression activity of H-NSwt and its oligomerizing and nonoligomerizing C-terminal domain (H-NSctd). A, electrophoretic mobility shift of a 400-bp DNA fragment containing the entire hns promoter region as a function of the indicated concentrations of H-NSwt (left top panel), H-NSctd::λCIctd chimera (center top panel), and H-NSctd (right top panel). The 400-bp DNA fragment used in the experiment has been excised from the pKK400 plasmid by BamHI/HindIII digestion. The other experimental conditions have been described (13, 17). B, cat transcription in the presence of increasing concentrations of H-NSwt (▵), H-NSctd::λCIctd chimera (○), and H-NSctd (●) indicated in the abscissa. The template used for transcription was supercoiled pKK400, and the level of transcription obtained in the absence of added protein is taken as 100%. The transcription reaction was performed as described (13, 17) for 30 min at 37 °C. The values reported in the figure represent the average of quadruplicate samples taken for each point.
In addition to demonstrating that all three proteins can bind to this DNA fragment, these results indicate that the three proteins each have different affinity for this fragment. In fact, although the level of retardation reaches saturation with approximately the same amounts (in the pmol range) of both H-NSwt and H-NSctd::λCIctd chimera, a comparable retardation level requires much larger amounts (i.e. in the nmol range) of H-NSctd. From these results, we estimate that the affinity of the H-NSctd::λCIctd chimera for the DNA is ∼2–4 times lower than that of H-NSwt, whereas that of H-NSctd is ∼4 orders of magnitude lower.
Because the gel shift experiments are carried out in the presence of an excess of nonspecific DNA, it is unlikely that the electrophoretic retardation of the 400-bp fragment by the three proteins reflects nonspecific binding. This premise is confirmed by the finding that this interaction produces a specific biological activity such as transcriptional repression. In fact, all three proteins are able to inhibit transcription of supercoiled pKK400, albeit with different efficiency (Fig. 2B); in fact, although 50% inhibition is obtained with 20 pmol of H-NSwt and 40 pmol of H-NSctd::λCIctd, ∼180 nmol of H-NSctd are necessary to obtain the same effect (Fig. 2B). Thus, the different efficiency of the three proteins in inhibiting transcription closely reflects their different affinity for the DNA seen in the band shift experiments.
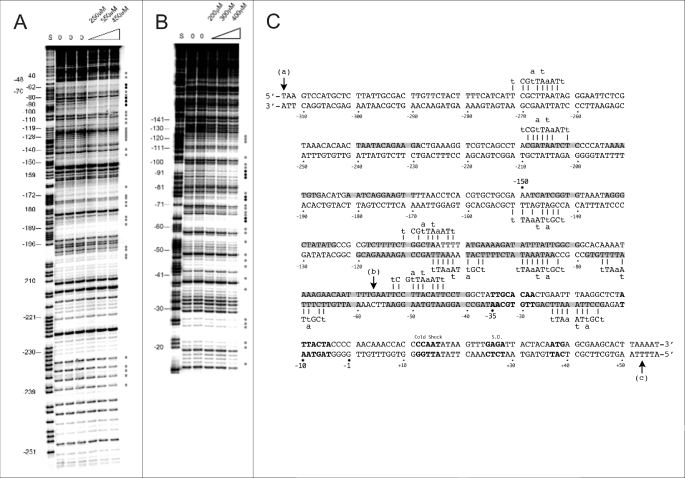
Taken together, the above results not only confirm the known DNA binding capacity of H-NSctd (35, 36) but also reveal its unexpected binding specificity. In fact, H-NSctd targets the same promoter as native H-NS and is almost as efficient as H-NS in binding to DNA and in inhibiting transcription when this otherwise monomeric domain is allowed to oligomerize by the presence of the C-domain of the phage λ repressor. Thus, the lack of oligomerization capacity, which prevents binding cooperativity, is the most likely reason for the reduced DNA affinity of H-NSctd and for its reduced efficiency in causing transcription inhibition compared with the native protein. Further evidence for the DNA binding specificity of H-NSctd comes from the results of its DNaseI footprints on the hns promoter. As seen from Fig. 3(A and B), the isolated, monomeric H-NSctd protects, on both DNA strands, a small number of short segments whose sequences display 7–8 of 10 matches (Fig. 3C) with the recently identified target sequence (i.e. tCG(t/a)T(a/t)AATT) of H-NS (19).
FIGURE 3.
Protection of the hns promoter by H-NSctd. A and B, electrophoretic separation of the fragments obtained following DNaseI footprinting (see “Experimental Procedures”) of the coding (A) and noncoding (B) strands of the hns promoter. Protected nucleotides are indicated by gray or black dots depending upon the intensity of the protection. C, sequence of the hns promoter fragment cloned into pKK400. The numbers indicate nucleotide positions with respect to the transcription start. The sites affected by H-NSctd are highlighted in gray, whereas the −35 and −10 core elements of the promoter, the cold shock recognition signal, the Shine-Dalgarno sequence (S.D.), and the initiation triplet are indicated (from top left) in bold. The H-NS-binding motifs are indicated above or below the DNA duplex, and the short vertical bars indicate the base match between motifs and promoter sequence. Vertical arrows (a) and (b) indicate the 5′ ends of the 400-bp DNA promoter fragment present in pKK400 and of the 110-bp fragment in pKK110, respectively, whereas arrow (c) indicates the 3′ terminus of both fragments.
To pursue our studies aimed at characterizing the structural bases of the recognition of the target DNA by H-NSctd, we undertook the characterization of the two ligands by NMR spectroscopy. For this purpose, we used 15N/13C labeled H-NSctd and the tCGATAAATT sequence located between −44 and −53 of the hns promoter (Fig. 3C). We selected this from among the H-NSctd-protected sequences as being the one displaying the highest homology with the consensus and prepared three synthetic double-stranded DNA fragments of 10, 15, and 20 bp containing this sequence (see “Experimental Procedures”).
Initial experiments, carried out by NMR titration, showed that H-NSctd binds selectively the 15-bp DNA fragment, whereas under the same conditions, it does not bind to a DNA fragment having identical size and base composition but a different sequence (data not shown). We estimate the Kd of this specific H-NSctd-DNA complex to be ∼4 μm, a value at least 3 orders of magnitude lower than that of the complex formed by H-NSctd with the nonspecific DNA fragment.
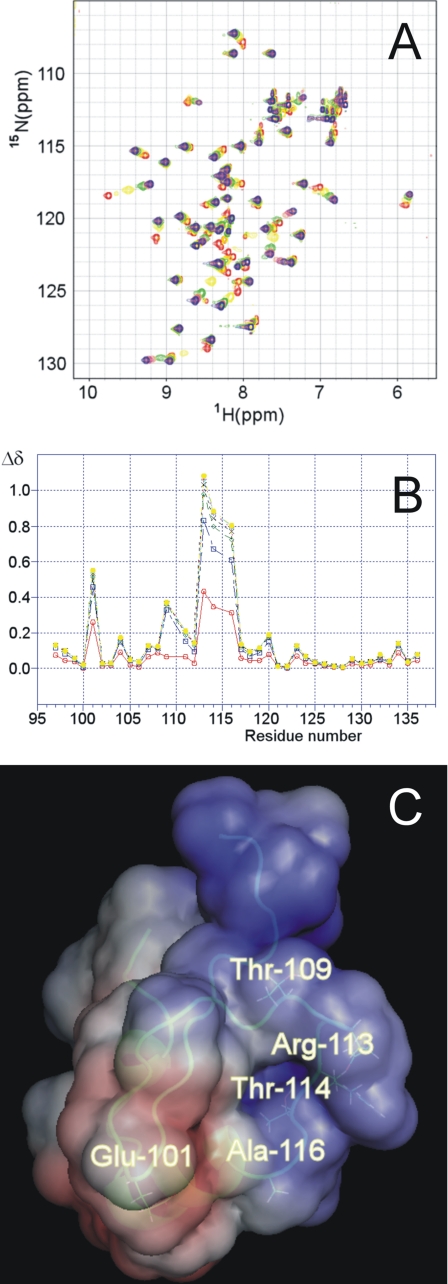
After assigning all of the resonances corresponding to free H-NSctd, the interaction of this protein with the DNA fragment was characterized by looking at the variations in the 15N-HSQC spectrum of the labeled protein upon the addition of increasing amounts of the 20-bp DNA fragment. The two-dimensional 15N-HSQC spectra collected upon the addition of increasing amounts of DNA show that signals shift in a continuous way, sometimes broadening, indicating that they are in a regime of moderate to fast exchange. A safe way to follow the changes observed during the titration consists in assigning and comparing the proton signals in the free and in the bound state. Thus, assignment of the protein signals at 1:1 DNA to protein ratio was performed by experiments similar to those used to assign the free protein. In Fig. 4A the two-dimensional 15N-HSQC spectra of this titration are superimposed and shown in different colors, and the chemical shift variations recorded for all of the residues in the presence of increasing amounts of DNA are shown in Fig. 4B. These titration experiments identify Arg113, Thr114, and Ala116 as the amino acids of H-NSctd most affected by the interaction with DNA. These three residues belong to a single tetrapeptide loop (the fourth amino acid being Pro115, which is not visible in 15NHSQC spectra). Minor effects caused by the addition of the DNA fragment are seen also with Glu101, Thr109, and Gln111. It is interesting to note that Trp108, whose intrinsic fluorescence was reported to increase upon the addition of DNA (37), is not affected. In fact, unlike the signal corresponding to the HN of its backbone, which “senses” only very slightly the presence of DNA (Fig. 4B), neither the HN nor the CH groups of the aromatic ring are affected (not shown). The localization of the residues affected by the interaction with DNA within the three-dimensional structure of H-NSctd is highlighted in Fig. 4C. As seen from this figure, with the exception of Glu101, all of the other residues are found in the same peptide loop that protrudes from the same side of the molecule characterized by a homogeneous positive electrostatic potential (Fig. 4C).
FIGURE 4.
Identification and localization of the sites of H-NSctd active in target DNA recognition. A, superimposition of the 15N-HSQC spectra of H-NSctd collected at 600 MHz in the presence of increasing amounts of target DNA 20-bp fragment. The stoichiometric ratios DNA/H-NSctd were: 0.0 (red), 0.2 (yellow), 0.4 (green), 0.6 (coral), 0.8 (magenta), 1.0 (orange), 1.2 (gold), and 1.4 (blue). B, chemical shift variation in the presence of increasing amounts of DNA for the residues belonging to H-NSctd; the combined Δδ (see “Experimental Procedures”) are reported (for the following DNA/H-NSctd stoichiometric ratios: 0.2 (red), 0.4 (blue), 0.6 (green), 0.8 (black), 1.0 (pink), 1.2 (light blue), and 1.4 (yellow). C, localization of the most affected residues within the three-dimensional structure of H-NS. The structure of H-NSctd is taken from Ref. 36. The electrostatic surface of the molecule is also shown with negative and positive potentials in red and blue, respectively.
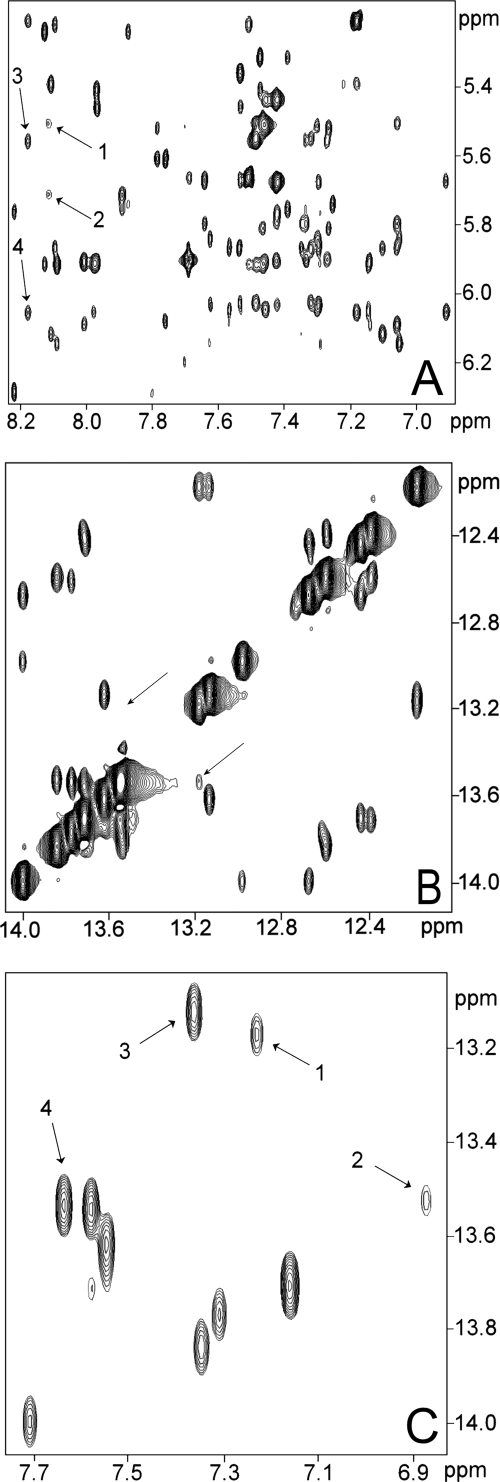
To look at the H-NSctd-DNA interaction from the point of view of the nucleic acid, we decided to assign the proton signals of this 20-bp fragment using conventional NMR techniques (see “Experimental Procedures”). Because it has been noticed that the H-NS consensus sequence contains a TpA step (19) and several observations made in the past have led to the conclusion that these steps determine a structural instability with conformational averaging occurring at the level of the adenine (Ref. 38 and references therein), the NMR spectra of the DNA target of H-NS were analyzed to investigate the possible existence of some structural feature that might represent a recognition signal for H-NS. Analysis of the nonexchangeable protons of the 20-bp DNA using a NOESY spectrum recorded in D2O with a short mixing time (50 ms), a condition under which the cross-peak intensity is directly related to the interproton distance, showed anomalously weak NOE signals at A31H8-A31H1′, A31H8-T30H1′ (Fig. 5A) and A11H8-T10H2″ (data not shown) corresponding to the TpA step. Furthermore, the NOESY spectrum collected in H2O shows a reduced or absent cross-peak between the T30H1 and T10H1 protons, which should have been visible at the positions indicated by the arrows (Fig. 5B). The normal intensity of the NOEs between these imino protons and their neighboring nucleotides indicates that the observed low NOE intensity cannot be attributed exclusively to exchange with the solvent but is caused mainly by a longer NH-NH proton distance because of a local conformational distortion.
FIGURE 5.
The 20-bp DNA target of H-NS contains a structural anomaly. A, connectivities present in the 900-MHz NOESY spectrum of a 20-bp DNA fragment containing the binding target of H-NS in the middle at short mixing time (50 ms) showing the intra and sequential connectivities between H6 or H8 protons and H1′ protons; the arrows refer to sequential connectivity of the H8 proton of A31 and H1′ of T30 (arrow 1) and to the intraconnectivity with its own H1′ proton (arrow 2). The lower than expected intensity of these cross-peaks can be appreciated from the comparison with the intensity of the cross-peaks resulting from the sequential and intraconnectivities of the H8 proton of A11, which are indicated by arrows 3 and 4, respectively. B, 900-MHz NOESY spectrum of the same 20-bp DNA fragment showing the imino to imino connectivities. The arrows indicate the expected connectivity between T10 and T30. C, 900-MHz NOESY spectrum of the same 20-bp DNA fragment showing the interstrand connectivities between adenine H2 protons and HN imino protons. The arrows indicate connectivity between H2 protons of A11 and HN imino proton of T30 (arrow 1) and to the connectivity of H2 protons of A31 and HN imino proton of T10 (arrow 2). The lower than expected intensity of these cross-peaks can be appreciated from the comparison with the intensity of the cross-peaks resulting from the connectivity of H2 of A13 and HN of T28 (arrow 3) and with the intensity of the cross-peak between H2 of A4 and HN of T37 (arrow 4). The numbering schemes and sequences of the DNA duplex are as follows: 1TGAATTCCTTACATTCCTGG20, 5′ to 3′; and 40ACTTAAGGAATGTAAGGACC21, 3′ to 5′.
The interstrand NOEs between T10H1 and A31H2 as well as between T30H1 and A11H2 are also significantly weaker than those of the other base pairs (Fig. 5C). Furthermore, in all of the DNA fragments analyzed in our study, the H2 signal of the adenine of the TpA step (corresponding to A31 in the 20-bp fragment) shows broadening and is shifted upfield, resonating at ∼6.8 ppm. Similar features have previously been reported and can be regarded as diagnostic indications for a partial conformational averaging implicating the adenine ring (38).
Taken together, our data indicate that the structure of the DNA fragment specifically targeted by H-NSctd contains a local distortion of the B-DNA structure. In fact, combined analysis of the short mixing time NOESY and primitive exclusive correlation spectroscopy spectra shows that the sugar moieties belonging to the TpA step adopt the C2′ endo conformation typical of canonical B-DNA.
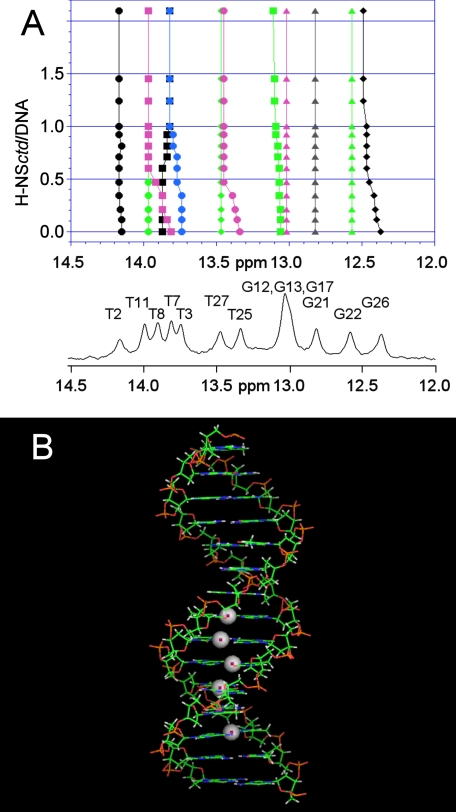
Following the above described NMR analyses carried out with the specific 20-bp DNA fragment, the H-NSctd interactions with the shorter (15 and 10 bp) DNA fragments were also analyzed. These analyses confirmed all of the data reported above, including the structural anomaly corresponding to the TpA step (independent of its position in the oligomers). Furthermore, these fragments were used in titration experiments with increasing amounts of H-NSctd, and the chemical shift variations of each HN imino proton were recorded to determine the effect of the protein on its target.
As seen from the results obtained with the 15-bp fragment and H-NSctd (Fig. 6A), the interaction does not involve random points of the DNA. In fact, substantial chemical shift variations are seen only for a select group of resonances of the spectrum corresponding to some heterocyclic bases of the double helix. Indeed, all the imino protons influenced by the interaction belong to a restricted portion of the DNA target, which likely represents the “minimum binding site” of H-NSctd. In agreement with the previous results, the 10-bp fragment consisting of the “minimum binding site” is still able to bind H-NSctd with high affinity. This target includes the site where the “structurally anomalous” TpA step is located (Fig. 6B) so that it appears clear that a structural distortion brought about by a TpA step within a particular DNA sequence is an important feature of the target recognized by H-NSctd and, most likely, by intact H-NS. Finally, because the observed shifts of the imino protons likely reflect changes in base stacking, it can be surmised that the DNA duplex adopts a new conformation upon H-NSctd binding. Furthermore, because in the H-NSctd-bound state there are minor changes of the chemical shifts of the H6/H8 protons and major changes of the H2 protons of the adenine in the TpA step, it can be surmised that the peptide loop of H-NSctd interacts with the minor groove of the DNA.
FIGURE 6.
Sites of target DNA involved in H-NSctd recognition. A, 400-MHz NMR spectrum of the imino resonances of the 15-bp DNA alone and in the presence of increasing amounts of H-NSctd. The DNA numbering schemes are as follows: 1CTTACATTCCTGGCT15, 5′ to 3′; and 30GAATGTAAGGACCGA16, 3′ to 5′. B, model of the 15-bp DNA fragment. The spheres represent the imino protons that show the largest chemical shift differences upon addition of H-NSctd.
DISCUSSION
Until recently, nucleoid-associated protein H-NS was considered to be a protein with a moderate binding preference for intrinsically curved DNA but lacking any sequence selectivity. These features were difficult to reconcile with the long recognized role of H-NS in the transcriptional regulation of a select group of bacterial genes, comprising, in particular, some of the genes associated with pathogenicity and responsiveness to environmental changes. However, comparison of short sequences targeted with high affinity by H-NS within some H-NS-sensitive promoters (i.e. proV, hns, fis) led to the identification of a 10-bp-long, somewhat degenerate consensus sequence that was suggested to represent the specific target where H-NS nucleation may start (19). This sequence is characterized by a high (78%) AT content and by the presence of a centrally positioned TpA step (19).
The present study provides the first, unambiguous indication that the H-NSctd alone is endowed with remarkable selectivity in its DNA binding properties, being able to recognize the aforementioned short DNA consensus target. When fused to a heterologous protein oligomerization domain (39), H-NSctd proved able to inhibit transcription in vitro and in vivo with almost the same efficiency as H-NSwt. Furthermore, even as a monomeric domain H-NSctd displayed transcriptional repressor activity toward the same promoters, albeit with lower efficiency than native H-NS.
Concerning the relative efficiency by which the different forms of H-NS inhibit transcription, it should be noted that although in vivo the inhibitory efficiency of H-NSctd is closer to that displayed by H-NSwt and by the H-NS-λCI chimera, in vitro it is lower by approximately 3 orders of magnitude as judged from the concentrations of the proteins required to attain the same level of inhibition in the in vitro transcription tests or to elicit the same extent of DNA band shift. Because, as mentioned above, the levels of the three proteins expressed in vivo under control of the ara promoter are virtually the same, we are inclined to think that the binding/inhibition efficiency determined in vitro closely reflects the actual activity of the proteins, whereas the inhibitory efficiency of H-NSwt (and possibly of the H-NS-λCI chimera) is underestimated, compared with that of H-NSctd, in the in vivo experiments. This underestimation likely results from the fact that a fraction of the H-NSwt is sequestered in vivo in nucleoprotein complexes with proteins other than H-NS and is therefore not available for transcriptional repression. It should be recalled, in this connection, that H-NS plays a role in the architectural organization of the nucleoid (5, 18, 40) and that its capacity to form complexes with several other proteins is well documented (see Refs. 9, 41, and 42 and references therein).
Overall, our results demonstrate that, despite its small dimension (47 residues), H-NSctd represents the key structural-functional module of H-NS. In fact, in addition to being capable of nonspecific binding to DNA (4, 35, 36) and of participating in protein oligomerization (4, 24), this module can also recognize a specific DNA sequence that turns out to have, as shown here by the NMR data, an irregular duplex structure. Conversely, the present results do not support the premise that H-NSntd might participate in conferring DNA binding specificity (26).
The small size of both the H-NSctd and its specific DNA consensus target provided a unique opportunity to use NMR spectroscopy to gain a deeper understanding of the molecular basis for the specific DNA-protein recognition, which underlies the H-NS-DNA interaction, allowing us to identify the regions of the DNA duplex and of H-NSctd that specifically interact with each other.
As to the protein, the NMR titration experiments identified the active site of H-NSctd implicated in the interaction with DNA as consisting of three main (Arg113, Thr114, and Ala116) and two minor (Thr109 and Glu101) residues. With the exception of Glu101, these residues belong to the flexible loop spanning from Thr109 to Ala116 and together define a DNA-binding surface characterized by an overall positive electrostatic potential (Fig. 4C).
The results reported here should be compared with those obtained in an earlier NMR study in which the H-NS-DNA interaction was analyzed using a larger H-NS fragment and a different DNA segment (36). The protein previously used was 30 residues longer (spanning from 59 to 136) and because of the presence of both C-domain and linker was a dimer, whereas the H-NSctd used here lacks the linker and is therefore monomeric. The DNA used by Shindo et al. (36) was a curved yet not sequence-specific 14-mer that displayed a much lower (∼3 orders of magnitude) affinity for the protein than that of the fragment used in the present study, which contains the H-NS consensus sequence motif. These differences are likely responsible for the differences between the present and the previously published data (36). In fact, although the implication in DNA binding of the Thr109–Ala116 loop can be derived also from the previous study, insofar as Thr114 was found to be one of the most affected residues, neither Arg113 nor Ala116 had been detected before among the implicated residues. Furthermore, although qualitatively similar results were obtained for Glu101 and Thr109, the previous study detected an additional set of residues that were affected by the presence of DNA (i.e. Val81, Ser83, Arg89, Gln91, Arg92, and Lys95). Given the specificity and the high affinity by which H-NSctd interacts with the consensus sequence, it is clear that these additional residues, which belong to the linker, are not involved in the recognition of the specific target but instead may be involved in a nonspecific DNA-protein interaction and/or in a DNA-promoted dimerization of the protein. On the other hand, the two residues of the loop (Arg113 and Ala116), whose involvement is clearly demonstrated by our results, could be the residues implicated in sequence recognition.
As far as the DNA target of H-NSctd is concerned, the NOESY experiments using either the 20-bp fragment (Fig. 5) or two shorter (15 and 10 bp) fragments reveal an unexpected pattern of NOE intensities, corresponding exclusively to the region of the duplex where H-NS binds, suggesting that the H-NS target might be characterized by some kind of structural anomaly because of the presence of the TpA base step. That this might indeed be the case is in line with previous studies showing that a TpA step might confer instability to the duplex (38) and with the evidence that the sequences recognized by H-NS in proV are characterized by thermal instability (43). Furthermore, experiments in which the electrophoretic retardation of nicked 300-bp DNA fragments was studied as a function of the nature of the base pair near the nick have led to the conclusion that, from the energetic point of view, both base stacking and base pairing contributions are close to zero at TpA steps (44).
Because we found that all of the resonances of the imino protons are present in the expected region (i.e. from 12.0 to 14.0 ppm), indicating that the DNA retains its double-stranded character along the fragment, our data and the above-mentioned considerations suggest that in the H-NS-binding region there is a local distortion of the B-DNA duplex in which the stacking and orientation between adjacent nucleotides is slightly altered, resulting in a higher axial and torsional flexibility. Thus, although a full clarification of the structural details of the H-NS target sequence must await the complete elucidation of the three-dimensional structure of this DNA fragment by multidimensional NMR spectroscopy, it can at least be concluded at this stage that a TpA step within a specific AT-rich DNA sequence having a reduced width of the minor groove represents a characteristic by which DNA targets belonging to different promoters are recognized by H-NS.
Acknowledgments
We are particularly indebted to Dr. Rainer Wechselberger (Bijvoet Center for Biomolecular Research, Utrecht University) for assistance with NMR spectroscopy experiments, to Dr. Fabio Polticelli (University of Rome 3) for electrostatic calculations, and to Stefano Stella (presently at University of California, Los Angeles) for technical help in performing in vivo and in vitro biological tests.
This work was supported by Programmi di Ricerca di Rilevante Interesse Nazionale Grants PRIN 2003 (to C. L. P. and M. S.) and PRIN 2005 (to C. L. P. and R. S.) from the Ministero dell'Istruzione, dell'Università e della Ricerca and by Contract RII3-026145 from the European Commission 6th Framework for Access to Research Infrastructures.
- C-domain
- C-terminal domain
- HSQC
- heteronuclear single quantum coherence
- NOESY
- nuclear Overhauser effect spectroscopy.
REFERENCES
- 1.Hommais F., Krin E., Laurent-Winter C., Soutourina O., Malpertuy A., Le Caer J. P., Danchin A., Bertin P. (2001) Mol. Microbiol. 40, 20–36 [DOI] [PubMed] [Google Scholar]
- 2.Pon C. L., Stella S., Gualerzi C. O. (2005) in DNA Conformation and Transcription ( Ohyama T. ed) pp. 52–65, Landes Bioscience, Austin, TX [Google Scholar]
- 3.Dame R. T., Wyman C., Wurm R., Wagner R., Goosen N. (2002) J. Biol. Chem. 277, 2146–2150 [DOI] [PubMed] [Google Scholar]
- 4.Spurio R., Falconi M., Brandi A., Pon C. L., Gualerzi C. O. (1997) EMBO J. 16, 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dame R. T. (2005) Mol. Microbiol. 56, 858–870 [DOI] [PubMed] [Google Scholar]
- 6.Dame R. T., Wyman C., Goosen N. (2001) Biochimie 83, 231–234 [DOI] [PubMed] [Google Scholar]
- 7.Tobe T., Yoshikawa M., Mizuno T., Sasakawa C. (1993) J. Bacteriol. 175, 6142–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ussery D. W., Hinton J. C., Jordi B. J., Granum P. E., Seirafi A., Stephen R. J., Tupper A. E., Berridge G., Sidebotham J. M., Higgins C. F. (1994) Biochimie 76, 968–980 [DOI] [PubMed] [Google Scholar]
- 9.Dorman C. J. (2004) Nat. Rev. Microbiol. 2, 391–400 [DOI] [PubMed] [Google Scholar]
- 10.Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. (1989) EMBO J. 8, 4289–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada H., Muramatsu S., Mizuno T. (1990) J. Biochem. 108, 420–425 [DOI] [PubMed] [Google Scholar]
- 12.Yamada H., Yoshida T., Tanaka K., Sasakawa C., Mizuno T. (1991) Mol. Gen. Genet. 230, 332–336 [DOI] [PubMed] [Google Scholar]
- 13.Falconi M., Higgins N. P., Spurio R., Pon C. L., Gualerzi C. O. (1993) Mol. Microbiol. 10, 273–282 [PubMed] [Google Scholar]
- 14.Zuber F., Kotlarz D., Rimsky S., Buc H. (1994) Mol. Microbiol. 12, 231–240 [DOI] [PubMed] [Google Scholar]
- 15.Rimsky S., Zuber F., Buckle M., Buc H. (2001) Mol. Microbiol. 42, 1311–1323 [DOI] [PubMed] [Google Scholar]
- 16.Rimsky S., Spassky A. (1990) Biochemistry 29, 3765–3771 [DOI] [PubMed] [Google Scholar]
- 17.Falconi M., Brandi A., La Teana A., Gualerzi C. O., Pon C. L. (1996) Mol. Microbiol. 19, 965–975 [DOI] [PubMed] [Google Scholar]
- 18.Luijsterburg M. S., Noom M. C., Wuite G. J., Dame R. T. (2006) J. Struct. Biol. 156, 262–272 [DOI] [PubMed] [Google Scholar]
- 19.Lang B., Blot N., Bouffartigues E., Buckle M., Geertz M., Gualerzi C. O., Mavathur R., Muskhelishvili G., Pon C. L., Rimsky S., Stella S., Babu M. M., Travers A. (2007) Nucleic Acids Res. 35, 6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falconi M., Colonna B., Prosseda G., Micheli G., Gualerzi C. O. (1998) EMBO J. 17, 7033–7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falconi M., Prosseda G., Giangrossi M., Beghetto E., Colonna B. (2001) Mol. Microbiol. 42, 439–452 [DOI] [PubMed] [Google Scholar]
- 22.Prosseda G., Falconi M., Giangrossi M., Gualerzi C. O., Micheli G., Colonna B. (2004) Mol. Microbiol. 51, 523–537 [DOI] [PubMed] [Google Scholar]
- 23.Rimsky S. (2004) Curr. Opin. Microbiol. 7, 109–114 [DOI] [PubMed] [Google Scholar]
- 24.Stella S., Spurio R., Falconi M., Pon C. L., Gualerzi C. O. (2005) EMBO J. 24, 2896–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badaut C., Williams R., Arluison V., Bouffartigues E., Robert B., Buc H., Rimsky S. (2002) J. Biol. Chem. 277, 41657–44166 [DOI] [PubMed] [Google Scholar]
- 26.Bloch V., Yang Y., Margeat E., Chavanieu A., Augé M. T., Robert B., Arold S., Rimsky S., Kochoyan M. (2003) Nat. Struct. Biol. 10, 212–218 [DOI] [PubMed] [Google Scholar]
- 27.Yasuzawa K., Hayashi N., Goshima N., Kohno K., Imamoto F., Kano Y. (1992) Gene 122, 9–15 [DOI] [PubMed] [Google Scholar]
- 28.Brandi A., Spurio R., Gualerzi C. O., Pon C. L. (1999) EMBO J. 18, 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldenberg D., Azar I., Oppenheim A. B., Brandi A., Pon C. L., Gualerzi C. O. (1997) Mol. Gen. Genet. 256, 282–290 [DOI] [PubMed] [Google Scholar]
- 30.La Teana A., Brandi A., Falconi M., Spurio R., Pon C. L., Gualerzi C. O. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10907–10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 32.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 33.Wijmenga S. S., Mooren M. M. W., Hilbers C. W. (1993) in NMR of Biological Macromolecules: A Practical Approach ( Roberts G. C. K., ed) pp. 216–288, IRL Press, Oxford [Google Scholar]
- 34.Dersch P., Schmidt K., Bremer E. (1993) Mol. Microbiol. 8, 875–889 [DOI] [PubMed] [Google Scholar]
- 35.Shindo H., Iwaki T., Ieda R., Kurumizaka H., Ueguchi C., Mizuno T., Morikawa S., Nakamura H., Kuboniwa H. (1995) FEBS Lett. 360, 125–131 [DOI] [PubMed] [Google Scholar]
- 36.Shindo H., Ohnuki A., Ginba H., Katoh E., Ueguchi C., Mizuno T., Yamazaki T. (1999) FEBS Lett. 455, 63–69 [DOI] [PubMed] [Google Scholar]
- 37.Tippner D., Wagner R. (1995) J. Biol. Chem. 270, 22243–22247 [DOI] [PubMed] [Google Scholar]
- 38.Lingbeck J., Kubinec M. G., Miller J., Reid B. R., Drobny G. P., Kennedy M. A. (1996) Biochemistry 35, 719–734 [DOI] [PubMed] [Google Scholar]
- 39.Bain D. L., Ackers G. K. (1994) Biochemistry 49, 14679–14689 [DOI] [PubMed] [Google Scholar]
- 40.Hardy C. D., Cozzarelli N. R. (2005) Mol. Microbiol. 57, 1636–1652 [DOI] [PubMed] [Google Scholar]
- 41.Madrid C., Nieto J. M., Paytubi S., Falconi M., Gualerzi C. O., Juárez A. (2002) J. Bacteriol. 184, 5058–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leonard P. G., Ono S., Gor J., Perkins S. J., Ladbury J. E. (2009) Mol. Microbiol. 73, 165–179 [DOI] [PubMed] [Google Scholar]
- 43.Bouffartigues E., Buckle M., Badaut C., Travers A., Rimsky S. (2007) Nat. Struct. Mol. Biol. 14, 441–448 [DOI] [PubMed] [Google Scholar]
- 44.Protozanova E., Yakovchuk P., Frank-Kamenetskii M. D. (2004) J. Mol. Biol. 342, 775–785 [DOI] [PubMed] [Google Scholar]