Abstract
Translocator protein (TSPO) is an 18-kDa cholesterol- and drug-binding protein conserved from bacteria to humans. While surveying for Tspo-like genes, we identified its paralogous gene, Tspo2, encoding an evolutionarily conserved family of proteins that arose by gene duplications before the divergence of avians and mammals. Comparative analysis of Tspo1 and Tspo2 functions suggested that Tspo2 has become subfunctionalized, typical of duplicated genes, characterized by the loss of diagnostic drug ligand-binding but retention of cholesterol-binding properties, hematopoietic tissue- and erythroid cell-specific distribution, and subcellular endoplasmic reticulum and nuclear membrane localization. Expression of Tspo2 in erythroblasts is strongly correlated with the down-regulation of the enzymes involved in cholesterol biosynthesis. Overexpression of TSPO2 in erythroid cells resulted in the redistribution of intracellular free cholesterol, an essential step in nucleus expulsion during erythrocyte maturation. Taken together, these data identify the TSPO2 family of proteins as mediators of cholesterol redistribution-dependent erythroblast maturation during mammalian erythropoiesis.
Translocator protein (TSPO)2 is an 18-kDa protein that was previously known as PBR (peripheral type benzodiazepine receptor) and represents a gene family evolutionarily conserved from bacteria to humans (1). In bacteria, TSPO is the tryptophan-rich sensory protein, an integral membrane protein that acts as a negative regulator of the expression of specific photosynthesis genes in response to oxygen and light (2). It is involved in the efflux of porphyrin intermediates from the cell, and several conserved aromatic residues within TSPO are thought to be involved in binding porphyrin intermediates (2). TSPO of bacterial origin has been shown to have the same ligand binding properties as mammalian TSPO proteins (3). In addition to the binding of porphyrin and heme, mammalian TSPO can replace the activity of its bacterial homologs (2, 4, 5). Rat TSPO was shown to retain its structure within the bacterial outer membrane, to functionally substitute for the bacterial homolog, and to act in a manner similar to TSPO in the outer mitochondrial membrane (6). Therefore, it is conceivable that some conserved functions of the Tspo genes within a cell are maintained from bacteria to plants and to mammals.
In mammals, the biological significance of TSPO has been studied for decades, and TSPO has been shown to be involved in a variety of cellular functions, including cholesterol transport and steroid hormone synthesis, mitochondrial respiration, permeability transition pore opening, apoptosis, and proliferation (7–10). Moreover, its expression correlates with certain pathological conditions such as cancer and endocrine and neurological diseases (8). Although some conserved cellular functions of Tspo are shared from bacteria to mammals, such as cholesterol-binding and transport, their biological significance seems to have adapted to serve specific functions critical for each organism. For instance, cholesterol transport into mitochondria is the rate-determining step in steroidogenesis (8, 11). TSPO serves the similar function in plants (12), insects (13), and mammals (14). However, the appearance of the drug, such as the benzodiazepine diazepam, binding sites on TSPO evolved later than the brain-specific γ-aminobutyric acid A receptor benzodiazepine binding sites (15), although drug binding was observed in both the plant and insect TSPOs (12, 13). Thus, throughout evolution, mammalian Tspo genes have exhibited extraordinary plasticity, a valuable trait to be further exploited.
We sought to reveal the mechanisms controlling the molecular evolution of Tspo and Tspo-like genes and the ligand binding sites in recently sequenced mammalian and other eukaryotic genomes and characterize the relationships and potential functional similarities in cholesterol synthesis, trafficking, and cholesterol-supported steroidogenesis between different Tspo genes. During these studies, we identified a new family of Tspo-like genes involved in cholesterol trafficking and redistribution, which is linked to erythropoiesis and probably to a new mechanism of erythroblast maturation.
EXPERIMENTAL PROCEDURES
Sequence Selection and Phylogenetic Analyses
The TSPO homologous sequences were retrieved from the non-redundant GenBankTM data base through BLAST searches (supplemental Table S1). Neighbor-joining (NJ) trees and maximum parsimony analyses were performed using the MEGA3 software with options of pairwise deletion and Dayhoff PAM matrix model (16). Maximum likelihood analyses were performed using PHYLIP, Version 3.6 (17). Bootstrap support values for NJ and maximum parsimony trees were obtained from 1000 replicates, and support for maximum likelihood trees was from 100 replicates.
Yeast Cell Growth, Transformation, Inducible TSPO Expression, Drug Ligand, and Cholesterol Binding Studies
Saccharomyces cerevisiae (INVSc1; his3 Δ1/his3 Δ1 leu2/leu2 trp1–289/trp1–289 ura3–52/ura3–52) was grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) or on YPD medium plates (containing 2% agar); Minimal medium plates contained 0.67% yeast nitrogen base, 2% agar, auxotrophic supplements, and a carbon source at a concentration of 2%, as follows: glucose for SD medium and galactose for SG medium. Heterologous expression of TSPO genes was performed using transformation of plasmids containing the open reading frame (ORF) of mouse Tspo2 (mTspo2), human TSPO2 (hTSPO2), or mouse Tspo1 (mTspo1) in pYES3/CT vector (Invitrogen) (supplemental Table S2); pYES3/CT/lacZ as a control vector was used to express β-galactosidase in yeast cells under the control of the GAL1 promoter. To induce TSPO overexpression in yeast, INVSc1 yeast host strain was transformed using pYES3/CT constructs containing the ORF of each TSPO, using the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (Invitrogen). Selective plates were incubated with SD medium without tryptophan (SD-W−) plus 2% agar. To induce expression, cells were grown in SG-W− medium at 30 °C for 8 h and collected by centrifugation. Cell pellets were resuspended into lysis buffer (50 mm Tris-HCl, pH 7.5, 1% DMSO, 100 mm NaCl, 1 mm EDTA, 2 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride; 1× proteinase inhibitor mix (Pierce)) containing glass beads. Samples were mixed 3–5 times for 1 min, each time keeping the cells on ice for 1 min. Total cell lysate was kept at −70 °C prior to binding assays.
Binding of [3H]PK11195 (1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide; specific activity, 83.50 Ci/mmol; PerkinElmer Life Sciences) using glutaraldehyde-fixed membranes (30 μg of protein/assay) was performed as we described previously (14). [1,2-3H]Cholesterol (45 Ci/mmol) uptake binding studies were performed on isolated spheroplasts as described previously (18). Specific cholesterol uptake was defined as induced minus uninduced basal values.
To further confirm the increased uptake of cholesterol due to the expression of mTSPO2, we used 22-(N-7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino-23,24-bisnor-5-cholen-3-β-ol (NBD-cholesterol; Molecular Probes) to visualize the cholesterol uptake. The wild-type cells (vector alone; INVSc1-pYES3/CT/lacZ) and TSPO2-overexpressing cells (INVSc1-mTSPO2↑) were incubated with a 1:1 mixture of cholesterol and NBD-cholesterol (20 μg/ml each) in Tween 80 (5 mg/ml). Cells were collected after a 14-h incubation at 24 °C with agitation (shaking; aerobic growth) and/or still (anaerobic) growth, washed with minimal medium containing 1 mg/ml p-phenylenediamine as an anti-bleach compound, and examined by fluorescence microscopy using a UPlanF1 objective (×40, numerical aperture 0.75) on an Olympus BX40 Research Microscope equipped with a DP70 digital camera. Pictures were recorded with equal exposure times, processed to maintain intensity differences, and processed using Adobe Photoshop Version 6.0.
Binding of [3H]cholesterol was performed using NH2 terminus hexahistidine-tagged recombinants and Ni2+-nitrilotriacetic acid-agarose column-based affinity chromatography, as shown previously, with some modifications (19). Briefly, mTSPO2 wild-type (WT) and mutant without CRAC domain (CRAC) recombinant proteins with an NH2-terminal His tag were generated though cloning of the partial ORFs of mTSPO2 into the pET-28c vector (Novagen), using the primers mTSPO2-RnoS(NdeI), mTSPO2-F*(XhoI), and mTSPO-cracFSp(XhoI) (supplemental Table S2). The mature plasmids were transformed into Escherichia coli expression host, BL21(DE3)pLysS (Novagen), and were then induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside to express the recombinant proteins under growth at 30 °C to maximize the solubility of the recombinant proteins. To confirm the solubility, SDS-PAGE and immunoblot analysis were performed, followed by Coomassie Blue staining and probing with His-probe (AD1.1.10) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Then 1 ml of the soluble fractions was loaded onto the Ni2+-nitrilotriacetic acid-agarose column (Qiagen) and washed with 6 ml of 1× binding buffer A (50 mm Tris-chloride at pH 7.4, 150 mm NaCl, 0.1% IGEPAL® (v/v). Proteins bound on the column were incubated with 20 nm [3H]cholesterol (45 Ci/mmol; Amersham Biosciences) at 4 °C for 4 h. Bound cholesterol was eluted with 1 ml of elution buffer (buffer A containing 500 mm imidazole) after two additional washes with 1× buffer A and quantified by scintillation spectrometry.
Mammalian Cell Culture, Plasmid Preparation, and Intracellular Localization of GFP- and/or RFP-tagged TSPOs
MA-10 cells were maintained in Dulbecco's modified Eagle's medium/Ham's F-12 medium supplemented with 5% heat-inactivated fetal bovine serum and 2.5% horse serum (20). NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated newborn calf serum. HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum.
To make GFP-tagged TSPO expression constructs, we cloned the full ORF of both the mouse and human Tspo2 genes into pEGFP-C3 vector (Clontech) to generate GFP-mTSPO2 and GFP-hTSPO2, respectively. The oligonucleotides used are listed in supplemental Table S2. The RFP-mTSPO1 plasmid was constructed from cutting the mTSPO1 insert from mTSPO1-pEGFP-C1 (21) with XhoI/BamHI and then ligating into pDsRed-Monomer-C1 vector (Clontech). For localization of TSPO2, we cultured mouse NIH 3T3 cells, MA-10 cells, and human HeLa cells on a 35-mm FluoroDishTM sterile culture dish (World Precision Instruments). The next day, cells were transfected with GFP-mTSPO2, GFP-hTSPO2, and RFP-mTSPO or co-transfected with either GFP-mTSPO2 or GFP-hTSPO2 and RFP-mTSPO using Lipofectamine 2000 (Invitrogen). At 24 h post-transfection, cells were incubated with ER-TrackerTM Blue-White DPX (Invitrogen) to label the ER. Cells were observed under a multiphoton Zeiss 510 laser-scanning microscope (Zeiss) with the following excitation/emission wavelengths: green (GFP), 498/516 nm; red (RFP), 558/583 nm; blue (ER staining), 748/560 nm.
In Situ Hybridization (ISH) of Tspo2 mRNA in All Stage, Whole Body Sections
Both antisense and sense riboprobes derived from the mTSPO2 cDNA were prepared for ISH. Briefly, the ORF of the mTspo2 cDNA was cloned into pGEM-T vector (Promega) using oligonucleotides listed in supplemental Table S2 and then was linearized with either HindIII or XhoI based on insert direction to make the in vitro transcription run from the T7 promoter. The RNA transcripts were synthesized in vitro according to the manufacturer's specifications (Ambion) and labeled with [35S]UTP (>1000 Ci/mmol; Amersham Biosciences).
ISH was performed using slides containing mouse whole body sections from different developmental stages or tissue sections from each relevant developmental stage (Cytochem). Tissues were frozen, cut into 10-mm sections, mounted on glass microscope slides, fixed in formaldehyde, and hybridized with 35S-labeled RNA probes, antisense and sense, respectively, overnight at 55 °C in 50% deionized formamide, 0.3 m NaCl, 20 mm Tris-HCl, pH 7.4, 5 mm EDTA, 10 nm NaPO4, 10% dextran sulfate, 1× Denhardt's solution, 50 mg/ml total yeast RNA, and 50–80,000 cpm/ml 35S-labeled RNA probe. The tissue was subjected to stringent washing at 65 °C in 50% formamide, 2× SSC, and 10 mm dithiothreitol, followed by washing in PBS before treatment with 20 mg/ml RNase A at 37 °C for 30 min. After washes in 2× SSC and 0.1× SSC for 10 min at 37 °C, the slides were dehydrated, exposed to x-ray film for 5 days, and then dipped in Kodak NTB-2 nuclear track emulsion and exposed for 18 days in light-tight boxes with desiccant at 4 °C. Tissues were validated by ISH with riboprobes to LDL receptor mRNA (data not shown).
After ISH, gene expression patterns were analyzed by both x-ray film autoradiography (5 days exposure time) and emulsion autoradiography (19 days exposure time). Lightly cresyl violet-stained sections were viewed under either dark field or bright field illumination. Selected slides were then scanned into Photoshop and labeled, as indicated. Anatomical data are shown at low magnification as bright labeling under dark field illumination. Cellular level results are revealed at higher microscopic magnification as black labeling by silver grains on a cresyl violet-stained background.
RT-PCR and Sequencing
The total RNA of various cell lines used was isolated with TRIzol reagent (Invitrogen). In addition, Mouse Total RNA Master Panel and human total RNA were obtained from Clontech and Ambion, respectively. First strand cDNA was synthesized by using the SuperScriptTM first strand synthesis system for RT-PCR (Invitrogen). Amplification was performed with 30 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 30 s and used 2 μl of cDNA and gene-specific primer pairs (supplemental Table S2) for mTSPO2, mTSPO1, mTubulin, hTSPO2, hTSPO1, and hHPRT. PCR products were sequenced using the ABI 3700 sequencer (Applied Biosystems).
Cytometry of Mouse Bone Marrow and Spleen
Bone marrow cells were collected from femurs by flushing the shaft with wash buffer (PBS plus 0.5% bovine serum albumin) using a 26-gauge needle. Spleens were mechanically dissociated briefly with a tissue homogenizer in the presence of PBS and then pushed through a 70-μm cell strainer with wash buffer. Freshly isolated bone marrow or spleen cells were immunostained at 4 °C in staining buffer (PBS, 2% serum in the presence of purified mouse anti-rat CD32 (IgG; 0.5 mg/ml; BD Pharmingen)) to block Fc receptors. Cells were incubated with phycoerythrin-conjugated anti-Ter119 (0.2 mg/ml; BD Pharmingen) and fluorescein isothiocyanate-conjugated anti-CD71 monoclonal antibodies (0.5 mg/ml; BD Pharmingen) in the dark for 40 min. Cells were also stained with propidium iodide solution (50 μg/ml; BD Pharmingen) to exclude dead cells from analysis. Following staining, the cells were washed with sorting buffer (PBS, 2% serum, and 3 μg/ml propidium iodide) three times and then resuspended into sorting buffer to a final concentration of 30 × 106 cells/ml. Four cell populations (Ter119medCD71high, proerythroblasts; Ter119highCD71high, basophilic erythroblasts; Ter119highCD71med, late basophilic and polychromatophilic erythroblasts; Ter119highCD71low, orthochromatophilic erythroblasts or normoblasts) were sorted simultaneously according to a previous report (22, 23). The different cell populations are indicated in the histogram, and the sorted cells were deposited into 15-ml conical tubes on a FACSAria sorter (McGill University Flow Cytometry Facility). Expression of Tspo2 in each subset of the cell populations was evaluated by RT-PCR using the oligonucleotides listed in supplemental Table S2.
Establishment of Stable Cell Lines of Overexpression of hTSPO2
Human erythroid K562 cells were maintained in Iscove's modified Dulbecco's medium (Invitrogen), supplemented with 10% heat-inactivated fetal bovine serum, at 37 °C, in a humidified 95% air, 5% CO2 incubator. To prepare stable cell lines overexpressing TSPO2, a cDNA encoding hTSPO2 was subcloned into the mammalian expression vector pcDNA3.1/ Hygro(+) via flanking NheI and XhoI restriction enzyme sites using the specific primer set (supplemental Table S2). This vector directed the constitutive expression of TSPO2 under the cytomegalovirus immediate early gene promoter control. The resultant plasmid (pCMV-hTSPO2) was introduced into K562 cells using electroporation (30 μg of DNA for 0.4 ml of cells). Stably transfected cells were selected for 3–4 weeks in medium containing 100–200 μg/ml hygromycin. Hygromycin-resistant clones were isolated and expanded in the same medium. Stable clones incorporating the parental pcDNA3.1/Hygro(+) vector alone were isolated in an identical fashion and used as a negative control cell line.
Cholesterol Staining and Laser-scanning Confocal Microscopy
K562 cells and stable cell lines (K562-hTSPO2, which stably expresses hTSPO2, and K562-vec, which was transfected with vector alone) were grown in 35-mm culture dishes. The next day, 2 ml of medium containing 1 μg/ml NBD-cholesterol in ethanol (final concentration 0.1%) was added, and cells were incubated for 10 min at 37 °C. For microscopy, cells were washed with PBS, stained with Hoechst 33342 (Invitrogen) for 15 min, and then washed with PBS at room temperature. Cells were then resuspended in culture medium without serum and transferred into a 35-mm FluoroDishTM sterile culture dish (World Precision Instruments). To investigate whether the COOH-terminal CRAC domain of TSPO2 is responsible for cholesterol trafficking in vivo, we generated two DsRed fusion proteins (a mutant protein without its COOH-terminal CRAC domain, DsRed-hTSPO2-crac, and a wild-type protein, DsRed-hTSPO2-wt), using pDsRed-Monomer-Hyg-C1 vector (Invitrogen) and the oligonucleotides listed in supplemental Table S2. Vector alone was used as control. HeLa cells grown in a 35-mm FluoroDishTM sterile culture dish (World Precision Instruments) were transfected with the above described vectors. Transfected cells were incubated with NBD-cholesterol and Hoechst 33342 as indicated above. Cells were examined using a laser-scanning confocal microscope (Olympus FluoViewTM FV1000) with an oil immersion objective (UPLSAP, ×100). NBD-cholesterol was excited with the 488-nm line of an argon-ion laser, and Hoechst 33342 was excited with the 405-nm line of a diode laser. To stain the intracellular free unesterified cholesterol, fluorescent filipin staining was performed by using a cholesterol cell-based detection assay kit (Cayman Chemical). Briefly, 6 × 104 cells were seeded onto a 24-well plate overnight. Control cells were treated with a 0.1 μg/ml concentration of an inhibitor of desmosterol Δ24-reductase (3-β(2-diethylaminoethoxy)androst-5-en-17-one hydrochloride (U18666A)). The next day, cells were fixed with 10% formaldehyde in Tris-buffered saline (pH 7.4) and stained with Filipin III for 60 min. Stained cells were examined using a laser-scanning confocal microscope (Olympus FluoViewTM FV1000) with an oil immersion objective (UPLSAP, ×100) with a UV diode laser providing excitation at 405 nm. Images were captured with FluoView software (Version 1.70.16; Olympus) and processed using Adobe Photoshop and Image-Pro Plus (Version 6.3).
RESULTS
Tspo2 Genes Arose by an Ancient Gene Duplication Event before the Divergence of Bird and Mammal Lineages
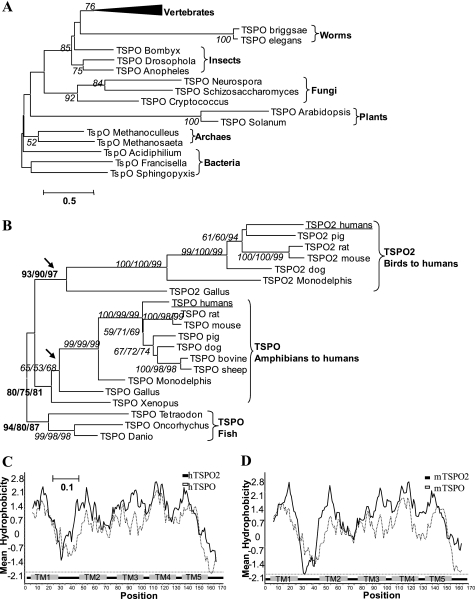
To distinguish the ortholog/paralog relationships within the TSPO family, we performed a phylogenetic analysis using 35 representative sequences (supplemental Fig. S1 and Table S1). The alignments of the deduced amino acid sequences were made based on the entire TSPO protein, and then only the best aligned sequences were used. Fig. 1A shows an NJ tree depicting the relationships among the TSPO proteins. Support for the topology was estimated from 1000 bootstrap replicates, and nodes occurring in more than 50% of the replicates are indicated. The phylogenetic relationships of all Tspo genes are in agreement with organism evolution, which is consistent with the notion that Tspo1 is a housekeeping gene conserved from bacteria to humans (1). However, Tspo2 appears only in the avian and mammalian species (Fig. 1, A and B). The bootstrap values for the branch point between Tspo2 and Tspo1 subgroups are 90/80/87, strongly indicating that the split is bona fide and suggesting that the Tspo2 arose from Tspo1 through ancient gene duplication before the divergence of avians and mammals. In addition, these two genes are involved in different gene networks, suggesting that their different functions evolved during the evolution of the TSPO gene family (supplemental Fig. S2 and Tables S4 and S5).
FIGURE 1.
TSPO2 is a new subfamily of TSPO proteins. A, NJ trees of TSPO proteins from three domains of life organisms, rooted with TSPO proteins from bacteria. B, NJ trees of TSPO and TSPO2 proteins from vertebrates rooted with TSPO from fish. TSPO2 proteins from avians to mammals are in a separate group from the rest of the TSPO proteins with a strong bootstrap support (930/900/970 in 1000 replicates), whereas TSPO proteins are widely distributed within three domains of life. The numbers at the nodes to assess the robustness of the trees represent the NJ bootstrap value, the maximum likelihood bootstrap value, and the maximum parsimony bootstrap value, respectively. Only bootstrap values larger than 50% are shown, and the boldface letters indicate the main branches. The estimated genetic distance indicated as the number of substitutions per amino acid site is proportional to the horizontal length of each branch. GenBankTM accession numbers and other abbreviations used are listed in supplemental Table S1. C and D, comparison of the hydropathy plots of the predicted amino acid sequences of human and mouse TSPO/TSPO2. The hydropathicity indices were determined as described (53). The shaded bars below the plots indicate transmembrane domains predicted by the TMpred algorithm (24). Hydrophobic residues are positive.
TSPO2 Is Localized at Nuclear and ER Membrane
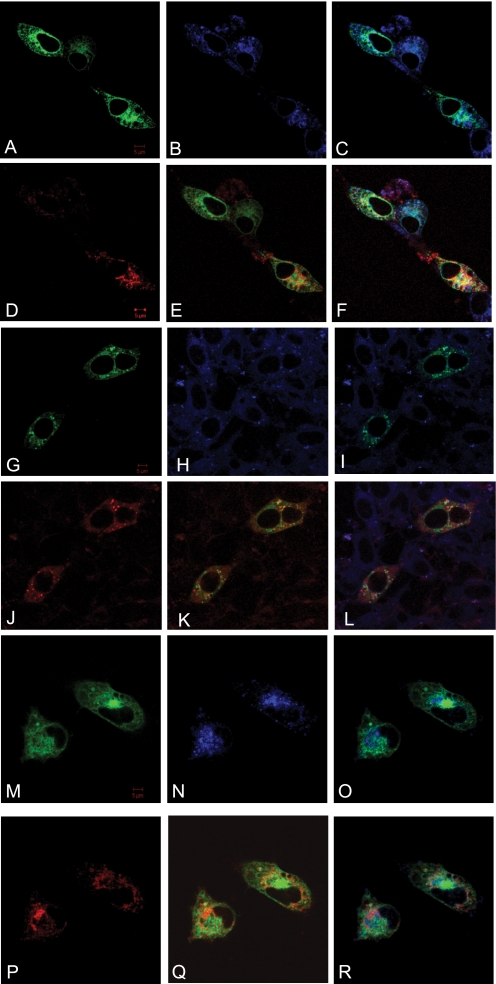
A comparison of the hydropathy plots of the predicted amino acid sequences of human and mouse TSPO/TSPO2 is shown, demonstrating the degree of similarity between members of the TSPO family in both hydrophobic and hydrophilic regions (Fig. 1, C and D). The plots also indicated extensive transmembrane domains predicted by the TMpred algorithm (24). Indeed, TSPO1 is located on the outer mitochondria membrane (OMM), although occasionally (i.e. in metastatic breast cancer cells) it is also found in and around the nucleus (25). To examine the localization of TSPO2, we transfected the mouse and human cell lines, NIH 3T3 (mouse fibroblast), MA-10 (mouse tumor Leydig), and HeLa (human cervical cancer) cells with GFP-mTSPO2 and GFP-hTSPO2, respectively. Cells were co-transfected with RFP-mTSPO1 to determine if these two proteins co-localize. Confocal laser-scanning microscopy indicated that GFP-TSPO2 co-localized with the ER marker Blue-White DPX but not with RFP-mTSPO1, which was seen in mitochondria (21) (Fig. 2 and supplemental Fig. S3). Furthermore, co-expression of an ER marker (red fluorescent protein with ER retention sequence, KDEL, using pDsRed2-ER vector, Clontech) with GFP-mTSPO2 and GFP-hTSPO2, respectively, confirmed our observation (supplemental Fig. S4). In addition, using single transfection with each plasmid, we confirmed that the co-transfection performed did not influence the localization of TSPO2 (supplemental Fig. S5). Thus, our results show that TSPO2 is localized primarily to the ER and nuclear membranes.
FIGURE 2.
Subcellular localization of mouse and human TSPO2 proteins in cultured mouse and human cells. GFP-tagged mTSPO2 and hTSPO2 were cotransfected with RFP-tagged mTSPO1 into NIH 3T3 (A–F), MA-10 (G–L), or HeLa (M–R) cells. The ER membrane was labeled with ER-Tracker Blue-White DPX. A, G, and M, GFP; B, H, and N, ER-Tracker Blue-White DPX; C, I, and O, GFP and ER-Tracker merged; D, J, and P, RFP; E, K, and Q, GFP and RFP merged; F, L, and R, GFP, RFP, and ER-Tracker merged images. Bars, 5 μm.
TSPO2 Has Lost TSPO1-specific Drug Ligand Binding Properties
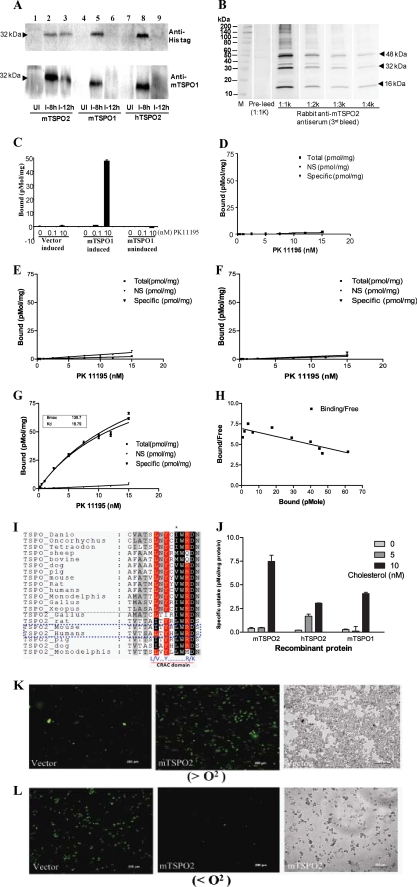
Based on the sequence analysis, we showed that TSPO2 functional properties have diverged from those of TSPO1, and it probably has lost the TSPO1-specific ligand binding properties. To test this hypothesis, we cloned and expressed the TSPO2s as well as TSPO1 in yeast. This has proven to be an ideal system to study the TSPO1 ligand binding properties, because yeast has lost the TSPO1 orthologous gene (26). Heteroexpression of His-tagged recombinant proteins were confirmed using anti-His tag-, anti-TSPO1-, and anti-TSPO2 peptide-specific antibodies (Fig. 3, A and B). The optimized induction conditions for TSPO2 were 8 h in the present of SG medium. The generated anti-TSPO2 peptide antibody recognized the TSPO2, which was found to form homo-oligomers under even denaturing conditions, as previously reported for TSPO1 (27). The specificity of TSPO2 detection was assessed using vector alone and uninduced mTspo1 gene-containing samples as negative controls and induced mTspo1 expression as a positive control. Recombinant mTSPO1 bound the high affinity diagnostic PK 11195 drug ligand, demonstrating that the system produces functional protein (Fig. 3, C, G, and H). In contrast, TSPO2 failed to exhibit specific binding for PK11195 (Fig. 3, D–F), indicating that TSPO2 lost the drug binding function. These results confirm the earlier sequence analysis, which suggested a loss of drug binding due to the substitutions of residues Trp42 and Trp107 (two of the main amino acids responsible for PK 11195 binding) to L/M (supplemental Figs. S1 and S6).
FIGURE 3.
Characterization of [3H]PK 11195- and cholesterol-binding properties of recombinant TSPO proteins in yeast cells. A, expression of human and mTSPO2 and mTSPO1 proteins in S. cerevisiae INVSc1. Shown is Western blot analysis of total yeast proteins from strains transformed with plasmid pYES3/CT-mTSPO2, pYES3/CT-hTSPO2, and pYES3/CT-mTSPO1 and induced with 2% galactose under different time points: 0 h (uninduced control), 8 h, and 12 h. Anti-His tag antibody and anti-mTSPO1 were used to monitor the induced heterologous expression of the recombinant proteins. B, SDS resistance oligomerization of recombinant mTSPO2 in S. cerevisiae. The same amount (6 μg) of mTSPO2 recombinant protein was loaded on each well, transferred onto polyvinylidene difluoride membrane, and then incubated with a serially diluted antiserum (third bleed) from rabbit 4483 immunized with mTSPO2 peptide (IHQPSSRCEDERKLPWC), and different sample preparations were carried out to check the oligomerization of mTSPO2, which is one of the features of TSPO proteins and might be related to the formation of dityrosines as the covalent cross-linker between each monomer (27). C, system test of PK11195 binding of mTSPO1 protein using strains containing vector alone and uninduced mTSPO1 as controls. In the assay, 20 μg of each yeast lysate and different concentrations of PK11195 were used as indicated. D–H, [3H]PK11195-binding properties of vector alone (D), mTSPO2 (E), hTSPO2 (F), and mTSPO1 (G). H, Scatchard plot of [3H]PK11195 binding in mTSPO1 is shown where specific binding of [3H]PK11195 was calculated as total binding (in the absence of competitor) minus nonspecific binding (determined in the presence of 10 μm PK11195). Radioligand concentrations were in the range of 0.1–15 nm; each point represents the average of triplicate determinations at each concentration. D–H, 15 μg of total yeast lysate was used in the binding assay. I, comparison of conserved signature cholesterol binding motif (CRAC) between TSPO2 and TSPO1. J, cholesterol uptake of S. cerevisiae spheroplasts with induced overexpression of mTSPO2, hTSPO2, and mTSPO1 and different concentrations of cholesterol, as indicated. The specific cholesterol uptake is defined as recombinant proteins induced minus vector alone basal values. K, NBD-cholesterol uptake assay. S. cerevisiae strains were induced with 2% galactose to overexpress mTSPO2 in the presence of NBD-cholesterol and under aerobic conditions (>O2). Vector alone cells were used as control. L, NBD-cholesterol uptake assay. S. cerevisiae strains were induced with 2% galactose to overexpress mTSPO2 in the presence of NBD-cholesterol and under anaerobic conditions (<O2). Vector alone was used as the control. Differential interference contrast images of the cells with less cholesterol uptake are shown, respectively, and another two differential interference contrast images are shown in supplemental Fig. S7.
TSPO2 Is Involved in Cholesterol Uptake and Trafficking
Since TSPO2 retained the conserved cholesterol binding domain (previously known as the cholesterol-binding cholesterol recognition/interaction amino acid consensus sequence, CRAC domain, in TSPO1 proteins) at its carboxyl terminus (28) (Fig. 3I and supplemental Fig. S6), we hypothesized that TSPO2 probably functions in the intracellular cholesterol transport pathway. Under aerobic growth conditions, yeast does not take up exogenous cholesterol, because the cell wall acts as a barrier (29). For that reason, we made yeast protoplasts and incubated them with 20 μm cold cholesterol and three concentrations of radiolabeled cholesterol. As shown in Fig. 3J, the TSPO2-expressing yeast strains specifically imported more cholesterol than yeast transformed with vector alone. To further confirm the involvement of TSPO2 in cholesterol uptake, we used NBD-cholesterol as a fluorescent probe to monitor its processing. Yeast cells transfected with the control vector alone did not take up cholesterol under aerobic conditions (Fig. 3K and supplemental Fig. S7). As we expected under anaerobic conditions, empty vector-transfected cells imported NBD-cholesterol to meet the requirements of anaerobic growth (Fig. 3L and supplemental Fig. S7). However, strains overexpressing mTSPO2 took up exogenous NBD-cholesterol under aerobic conditions, suggesting that the overexpressed mTSPO2 protein may alter the cholesterol import and/or synthesis pathways. Under anaerobic conditions with overexpression of TSPO2, yeast cells were unable to import exogenous cholesterol, suggesting that overexpressed TSPO2 may complement the cholesterol synthesis deficiency (Fig. 3L and supplemental Fig. S7). Although the exact mechanism of TSPO2 function in this pathway remains unclear, these data indicate that TSPO2 is involved in cholesterol synthesis, import, and/or trafficking.
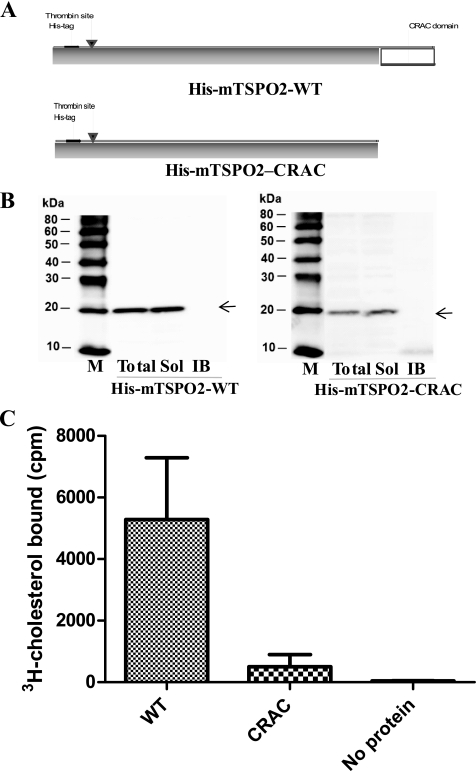
To further confirm that TSPO2 has a functional cholesterol binding domain at its carboxyl terminus, we cloned the ORF and partial ORF without the CRAC domain of mTSPO2 into pET28c vector to generate NH2 terminus hexahistidine-tagged recombinant proteins, His-mTSPO2-WT and His-mTSPO2-CRAC (Fig. 4A). Solubility of the recombinant proteins was confirmed by SDS-PAGE and under our growth conditions. Both recombinant proteins were seen in the soluble fractions of the total cell lysates but not in the purified inclusion body fraction (IB in Fig. 4B). Taking advantage of the soluble recombinant proteins, we performed column-based affinity chromatography to test whether the recombinant proteins bind cholesterol (19). The results obtained showed that recombinant wild-type mTSPO2 specifically bound cholesterol. Removal of the CRAC domain from the carboxyl terminus of the protein dramatically reduced the ability of the protein to bind cholesterol, suggesting that cholesterol bound on the column is mainly due to the presence of the CRAC domain (Fig. 4C). Our observation is consistent with our previous reports demonstrating the CRAC domain of TSPO1 as the site of cholesterol binding to this protein (27, 28, 30).
FIGURE 4.
Cholesterol binding of mTSPO2. A, the schematic structure of NH2 terminus hexahistidine-tagged recombinant proteins, His-mTSPO2-WT and His-mTSPO2-CRAC. B, solubility of the recombinant mTSPO2 proteins. Western blot analysis was performed to check different fractions of cell lysates. M, protein standards; Total, total cell lysate; Sol, soluble fraction; IB, purified inclusion bodies. C, [3H]cholesterol binding. Column-based affinity chromatography was used to test the [3H]cholesterol binding properties. WT, His-mTSPO2-WT; CRAC, His-mTSPO2-CRAC. Results shown are means ± S.E. form three independent experiments.
Temporal and Spatial Distribution of Tspo2 in Hematopoietic Tissues
We analyzed the expression of Tspo2 mRNA by ISH in embryonic mouse sections throughout development. A restricted pattern of Tspo2 mRNA distribution was observed in embryonic, postnatal, and adult mice (Fig. 5) and changed during developmental stages. Tspo2 mRNA was not detected at the e10.5 stage but was detected in the midgestation e12.5 stage primordial liver, when the hematopoietic producing sites switch from the yolk sac to the fetal liver to produce definitive erythrocytes (31). Increased levels of hepatic Tspo2 expression were noted on day 15.5, followed by a decline throughout newborn stage p1 and postnatal stages p5 and p10, concomitant to the process of cell mass reduction of the liver erythroid centers. Tspo2 expression seemed totally silent in the adult liver, because the hematopoietic cells in the liver had receded in hematopoietic function with hepatocytes gradually, and the hematopoietic cells migrate from the liver to the bone marrow and spleen, where erythrocytes develop (32) (Fig. 5E). These results suggest that Tspo2 mRNA expression is restricted to hematopoietic cells in the fetal liver during midgestation embryos until postnatal stages.
FIGURE 5.
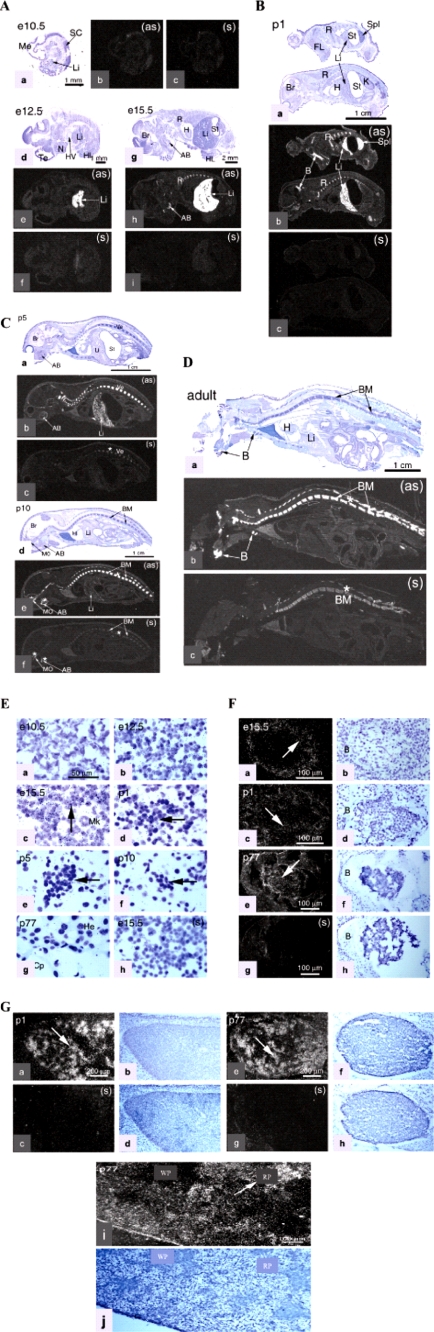
Tspo2 expression in mice. A, Tspo2 expression in the embryonic mouse (e10.5, e12.5, and e15.5). a, d, and g, anatomical view of an embryonic mouse after staining with cresyl violet. b, e, and h, x-ray film autoradiography following hybridization with antisense riboprobe after a 5-day exposure. There is no labeling evident at this e10.5 stage, but a pattern of Tspo2 mRNA distribution is shown to be restricted to hepatic primordium at the e12.5 stage and in the hepatic primordium and bone, including the ribs and alveolar bone in the mandibles at stage 15.5, seen as bright labeling under dark field illumination at the e12.5 stage. c, f, and i, control sense (s) hybridization of an adjacent section comparable with b, e, and h, respectively. B, Tspo2 mRNA in the newborn (p1). a, anatomical view of the p1 mouse after staining with cresyl violet. b, x-ray film autoradiography following hybridization with antisense riboprobe showing a pattern of Tspo2 mRNA labeling seen as bright labeling under dark field illumination. Tspo2 mRNA occurs in the liver, bone, and spleen. c, control sense (s) hybridization of an adjacent section comparable with b. C, Tspo2 mRNA in the postnatal (p5 and p10). a and d, anatomical view of the postnatal (p5 and p10) after staining with cresyl violet. b and e, x-ray film autoradiography following hybridization with antisense riboprobe showing a pattern of Tspo2 mRNA labeling seen as bright labeling under dark field illumination. Tspo2 mRNA occurs in the liver, in vertebrae bone marrow, and in alveolar bone at stage p5; however, Tspo2 mRNA apparently can only be seen in the vertebrae bone marrow and alveolar bone at stage p10. In comparison with the p5 stage, a dramatic decrease in gene 1 labeling is observed in the liver. c and f, control sense (s) hybridization of an adjacent section comparable with b and e, respectively. D, Tspo2 mRNA in the adult mouse (p77). a, anatomical view of the adult mouse, after staining with cresyl violet. b, Tspo2 mRNA labeling mainly in the vertebrae bone marrow and bone tissue, no labeling in the liver. c, control sense (s) hybridization of an adjacent section comparable with b. Note the relatively high sense labeling in the vertebrae bone marrow, molars, and alveolar bone (asterisks) in p10 and adult. E, Tspo2 expression in the liver development. a–g, emulsion autoradiography showing Tspo2 expression in the liver seen as silver grains under light field illumination at high magnification (×540) of tissue stained with cresyl violet. Developmental stages are indicated. The arrows point to the groups of labeled cells. h, control (sense) hybridization shown in the liver primordium on day 15.5. F, Tspo2 expression in the bone marrow development. a–f, emulsion autoradiography showing Tspo2 mRNA labeling (arrows) in the bone marrow under light field illumination at medium magnification (×220 and ×130), tissue stained with cresyl violet. Developmental stages are indicated. The arrows point to the groups of labeled cells. h, control (sense) hybridization shown in the adult bone marrow (p77). G, Tspo2 expression in the newborn (p1) and adult (p77) mouse spleen. a, e, and i, emulsion autoradiography showing Tspo2 mRNA labeling (arrow) in the newborn and adult mouse spleen at medium (×61) and high (×110) magnification, showing the white pulplike labeling pattern (arrow). b, f, and j, the same region as in a, e, and i seen with cresyl violet staining under bright field illumination. c and g, control (sense) hybridization in the adjacent section comparable with a and e, respectively. d and h, the same region as in c and g seen with cresyl violet staining under bright field illumination. AB, alveolar bone in the mandible; B, bone; BM, bone marrow; Br, brain; C, capsule; Cp, capillary; FL, forelimb; H, heart; He, hepatocyte; HL, hind limb; HV, heart ventricle; Li, liver primordium; Me, mesencephalon; Mk, megakaryocyte; N, nose; R, ribs; RP, red pulp; SC, spinal cord; Spl, spleen; St, stomach; Te, telencephalon; Ve, vertebrae; WP, white pulp; as, antisense; s, sense.
Bone marrow is the major hematopoietic organ mainly found in bones, such as vertebrae, ribs, and alveolar bone (Fig. 5, A–D and F). Dissimilar to transient expression in the liver, Tspo2 expression in the bone marrow spanned the late gestation stages and remained elevated until adulthood. Thus, in the rudimental states of the bone, a discrete band of tissue displayed Tspo2 mRNA-labeled cells, where the undifferentiated bone marrow cells are located (Fig. 5F). At postnatal stages p1, p5, and p10 and during adulthood, Tspo2 mRNA labeling was seen in the red marrow cells (Fig. 5C). Notably, sense hybridization produced weak detectable labeling in the bone marrow tissue, which was not seen in other unrelated genes in the bone marrow.
In newborn and adult mice, Tspo2 mRNA labeling was observed in the spleen (Fig. 5, B and G), showing regional differences. The emerging red pulp sites, which are centers of erythropoiesis and megakaryocyte production (33), were highly labeled, but no mRNA was detected in the white pulp regions, which consist of germinal centers for lymphocytes, plasma cells, and macrophages. These results indicate that Tspo2 mRNA is specifically expressed in the erythroid cells in postnatal hematopoietic spleen. This hematopoietic tissue-specific expression of the Tspo2 gene in mice as well as in humans was independently confirmed by RT-PCR (supplemental Fig. S8).
Tspo2 mRNA Is Expressed in an Erythroid Cell Type-specific Manner
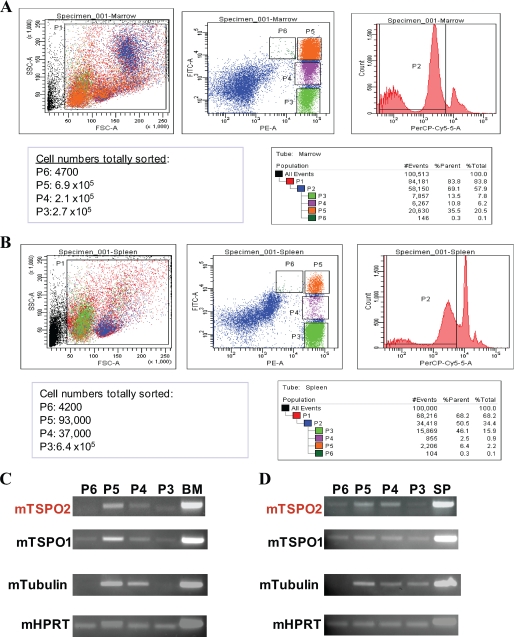
To further confirm the erythroid-specific expression of Tspo2, we isolated four mature erythroid cell types from mice. During erythroid maturation, the last four developmental stages before enucleation into reticulates are (i) proerythroblasts, (ii) basophilic erythroblasts, (iii) late basophilic and polychromatophilic erythroblasts, and (iv) orthochromatophilic erythroblasts (Fig. 6, A and B). Tspo2 expression was weakly detected in the proerythroblasts and orthochromatophilic erythroblasts. However, Tspo2 was highly expressed in the basophilic erythroblasts and late basophilic and polychromatophilic erythroblasts during erythroid maturation (Fig. 6, C and D). Tubulin mRNA was not detected in either proerythroblasts or orthochromatophilic erythroblasts in bone marrow-isolated cells. Also, tubulin mRNA was not detected in proerythroblasts from spleen cells. These features may also serve as a cell subset-specific marker, since erythroblasts express some variants of β-tubulin during erythroid maturation (34). Both Tspo1 and HPRT mRNAs served as controls to show the presence of mRNAs extracted from each sorted subset of cell types.
FIGURE 6.
Tspo2 expression in bone marrow and spleen cells purified from flow cytometry at differentiation stages of erythroblast maturation. A and B, fluorescence-activated cell sorting for progenitor cells. Freshly dissociated wild-type mouse bone marrow and spleen cells were labeled with a fluorescein isothiocyanate-conjugated monoclonal antibody to CD71 and a phycoerythrin-conjugated anti-Ter119 monoclonal antibody. Dead cells (staining positive with propidium iodide, shown in the PerPC-Cy5-A channel) and anucleated red cells (with low forward scatter (FSC)) were excluded from analysis. The upper panels illustrate a density plot of all viable bone marrow and spleen cells; the axes indicate relative fluorescence units for phycoerythrin (x axis) and fluorescein isothiocyanate (y axis). The lower panel shows that regions SP1–SP6 were selected as indicated. These are predominantly proerythroblasts in region SP6, basophilic erythroblasts in region SP5, late basophilic and chromatophilic erythroblasts in region SP4, and orthochromatophilic erythroblasts (possibly containing some enucleated reticulocytes) in region SP3. SSC, side scatter. PE, phycoerythrin; FITC, fluorescein isothiocyanate; PI, propidium iodide (shown in the PerPC-Cy5-A channel). C and D, RT-PCR of mTSPO2 gene expression in sorted cells from each of regions SP6 to SP3 were performed using the total RNAs prepared from each purified cell subset of bone marrow (C) and spleen (D). Mouse tubulin and HPRT gene expression and unseparated cells from bone marrow or spleen were used as controls for RNA content per sample and positive gene transcripts, respectively. The Tspo1 was also presented in comparison with Tspo2.
hTSPO2-mediated Cholesterol Redistribution in Human Erythroleukemia Cells
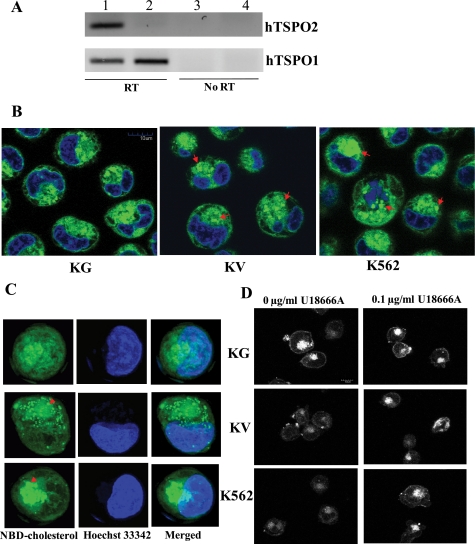
Cholesterol metabolism can profoundly influence the maturation of mammalian erythrocytes. To determine whether hTSPO2 influences the cholesterol distribution of committed erythroid cells, we used the human erythroid progenitor cell line K562 as a model and established two stable cell lines, k562-hTSPO2 and k562-vec, which were stably transfected with a hTSPO2 expression vector or the parent pcDNA3.1/Hygro(+) vector alone as a control. The expression of hTSPO2 in the stable cell lines was determined by RT-PCR (Fig. 7A). The stable cell lines and parental K562 cells were stained with NBD-cholesterol, and the fluorescent signal that localized to the nonpolar core of cytoplasmic lipid droplets was smeared out in the hTSPO2 overexpression cell lines. However, the clear outline of the droplets was observed in the K562-vec control cell line as well as in the parental K562 cells (Fig. 7, B and C). These data suggest that hTSPO2 plays a role in the intracellular trafficking of cholesterol and in the maintenance and equilibrium of free cholesterol storage and cell membrane requirements for integrity and fluidity.
FIGURE 7.
Overexpression of hTSPO2 in human erythroid leukemia cell line K562 and redistribution of intracellular cholesterol. A, confirmation of the stably overexpressed hTSPO2 in cell line K562 by RT-PCR. Lanes 1 and 3, overexpression of hTSPO in stable cell line K562-hTSPO2; lanes 2 and 4, vector alone in stable cell line K562-vec. Detection of the human TSPO1 gene was used as positive control and the no-reverse transcription PCRs were used as negative control. Scale bar, 10 μm (left) and 50 pixels (right). B, redistribution of NBD-cholesterol uptake. KG, overexpression of hTSPO in the stable cell line K562-hTSPO2; KV, vector alone in stable cell line K562-vec; K562, wild-type parental cell line without transfection. Arrow, accumulated endosomal lipid droplets. C, three-dimensional graph to highlight the redistribution of free cholesterol from the cells with overexpression of hTSPO2, vector alone, and wild type. D, redistribution of intracellular free cholesterol as measured by filipin staining after the cells were treated with or without an inhibitor of desmosterol Δ24-reductase (U18666A). Scale bar, 10 μm.
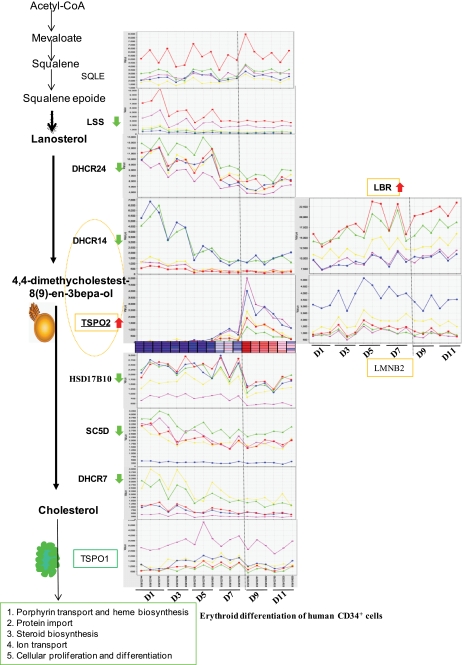
In addition, excess cellular cholesterol has been shown to inhibit erythroid maturation (35). To examine whether overexpression of the hTSPO2 gene affects intracellular free cholesterol accumulation and/or redistribution, we measured unesterified cholesterol by filipin staining followed by confocal microscopy. The results suggest that hTSPO2 overexpression increases and/or redistributes unesterified cholesterol in the cells. The altered cholesterol staining in TSPO2-overexpressing cells was similar to the observed effects of a well known class 2 amphiphilic pharmacological agent, U18666A, which inhibits cholesterol transport (36) (Fig. 7D). The COOH-terminal CRAC domain of TSPO2 was shown earlier to be responsible for cholesterol binding in vitro (Fig. 4). The function of the CRAC domain in cholesterol trafficking/redistribution was investigated in HeLa cells transiently transfected with plasmids containing wild-type and mutant hTSPO2, missing the CRAC domain, fused to pDsRed monomer C1 (supplemental Fig. S9). The hTSPO2 mutant protein missing the CRAC domain showed a subcellular distribution/localization similar to that of the wild-type protein, although the expressed protein formed aggregates (supplemental Fig. S9, A–C). We recently reported the formation of such aggregates in cells transfected with a mutant TSPO1 carrying deletions in the COOH terminus (37). Remarkably, the localization and aggregate formation of mutant TSPO2 did not affect the distribution of intracellular NBD-cholesterol (supplemental Fig. S9, C and L), also evidenced by the unevenly dispersed cholesterol droplets (supplemental Fig. S9K). In contrast, wild-type TSPO2 localized in the ER and nuclear membranes (Fig. 2 and supplemental Fig. S9, D–F) and co-localized with NBD-cholesterol (supplemental Fig. S9, M–O). These data indicate that the cholesterol binding CRAC domain of TSPO2 plays an essential role in cholesterol trafficking/redistribution as well as the proper intracellular localization of TSPO2. Moreover, the appearance of Tspo2 in erythroblasts strongly correlates with the down-regulation of all other main enzymes in the cholesterol biosynthesis pathway, suggesting that TSPO2 mediates cholesterol trafficking and redistribution but not biosynthesis during hematopoiesis (Fig. 8).
FIGURE 8.
Cholesterol biosynthetic pathway in erythroblasts. Main enzymes related to cholesterol synthesis and putative TSPO2-nuclear complex are listed with the ID number in the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array, as follows: squalene epoxidase (SQLE) (209218_at); lanosterol synthase (LSS) (202245_at); 3β-hydroxysteroid-Δ24-reductase (DHCR24) (200862_at); 3β-hydroxysterol Δ-(14)-reductase (C14SR, TM7SF2) (DHCR14A) (210130_s_at); hydroxysteroid (17β) dehydrogenase 10 (HSD17B10) (202282_at); 3β-hydroxysteroid-Δ5-desaturase (lathosterol dehydrogenase) (SC5D) (211423_s_at); 3β-hydroxysteroid-Δ7-reductase (DHCR7) (201791_s_at); lamin B receptor (LBR) (201795_at); lamin B2 (LBR) (201795_at); translocator protein 2 (TSPO2) (215449_at); translocator protein 1 (TSPO1) (202096_s_at). A heat map view of TSPO2 mRNA is shown below the profiling plot, where the largest values are displayed as the deepest red (hot), the smallest values are displayed as the deepest blue (cool), and intermediate values are a lighter shade of either blue or red. The red line represents the gene indicated at the left side of each profiling plot, and the remaining colors indicate the top four genes with the closest similar patterns of gene expression during the erythroid differentiation. These were predicted using Class Neighbors in GenePattern 3.11. D1–D11, the days in the time course of in vitro erythroid differentiation of human adult-derived peripheral blood CD34+ cells (GEO accession number GSE4655) (43).
DISCUSSION
Gene expansion of the TSPO family before the divergence of birds and mammals (about 300 million years ago) must have attributed some specific biological function to the newly discovered TSPO2 proteins. The Tspo gene is evolutionarily conserved and distributed from bacteria to plants and humans (7, 8). Mammalian TSPO is ubiquitously expressed in the body and primarily localized to the mitochondria, where it is involved in many cellular functions. However, the TSPO protein has also been shown to be present in the plasma membrane of red blood cell, a cell that has lost both the nucleus and mitochondria (38), as well as in the plasma membrane of neutrophils detected by immunoprecipitation and immunocytochemistry, where it was shown to stimulate NADPH-oxidase activation of these cells (39). The plasma membrane and mitochondrial forms of TSPO must have some implications for its putative function, including steroidogenesis, mitochondrial respiration, heme metabolism, calcium channel modulation, cell growth, and immunomodulation (40).
The discovery of the TSPO2 subfamily of proteins in hematopoietic tissues has made this scenario more complicated. Considering that birds and mammals are warm blooded animals, the Tspo gene expansion in these two classes may have an unknown yet significant biological importance. The hydropathy plots of the predicted amino acid sequences of human and mouse TSPO/TSPO2 demonstrated a high degree of similarity between members of the TSPO family in both hydrophobic and hydrophilic regions as well in the identified transmembrane domains. Moreover, the carboxyl-terminal domains of both proteins retained the ability to bind cholesterol. Furthermore, the finding that anti-TSPO2 specific anti-peptide polyclonal antiserum cross-reacts with TSPO1 indicated that these proteins share epitopes in the cytoplasmic loop 1 (C1), the most variable region between TSPO2 and TSPO1. Thus, experiments performed with hematopoietic tissues using anti-TSPO antiserum must be interpreted with caution. Despite these similarities, the subfunctionalization of TSPO2 introduced a number of unique features, which include the intracellular localization on ER and nuclear membranes, loss of TSPO1-specific drug ligand binding properties (a functional feature of this protein), and the erythrocyte-specific distribution of TSPO2 in hematopoietic tissues.
The erythroid cell type-specific expression of Tspo2 further validated previous reports of DNA microarray data, which identified an expressed sequence tag resembling Tspo in mouse nucleated erythrocytes from whole bone marrow (WBM; Ter-119+, CD3−, CD4−, CD8−, Mac-1−, Gr-1−, and B220−) as a marker or lineage “fingerprint” for erythrocyte progenitor differentiation and self-renewal (41) and serial oligonucleotide microarray analysis of human normal CD34+ bone marrow cells during lineage-specific differentiation where the same gene was found to be differentially expressed in erythropoiesis (42, 43). Additionally, it seems that Tspo2 expression, restricted to the earlier erythroid blasts stage, could be used as a marker for committed erythroid precursors, since a few genes previously claimed to be erythroid markers are also expressed in other tissues, such as erythroid Kruppel-like factor 1 and GATA1 in testis (44–46). Therefore, TSPO2 could serve as marker in blood transfusion and/or hemopoietic stem cell transplantation studies, in combination with other genes, such as Kell blood group gene, hemoglobin, and erythroid Kruppel-like factor. Furthermore, erythroid cell type-specific expression of Tspo2 may have clinical significance, especially in highly active erythropoiesis due to hypoxia and/or anemia.
Erythropoiesis is the process of red blood cell maturation, in which progressive hematopoietic differentiation starts from hemopoietic stem cells and moves through a series of steps that include the formation of early proerythroblasts, late basophilic erythroblasts, polychromatophilic erythroblasts, and finally morphologically recognizable normoblasts (orthochromatic erythroblasts) and mature erythrocytes (47, 48). In the erythropoietic process, organelles, such as mitochondria, endoplasmic reticulum, ribosomes, and Golgi apparatus, are lost. The most prominent phenomenon during this process is that the nucleus is extruded from the maturing cell to form enucleated erythrocytes that give rise to the mature erythrocytes. Nucleus expulsion is inhibited by excess cellular cholesterol (35), and the high requirement of cholesterol during this process has been linked to hypocholesterolemia (49). Redistribution of intracellular cholesterol to change the nuclear membrane rigidity prior to erythrocyte maturation is a necessary step. Previous reports have shown that membrane cholesterol levels substantially decrease during maturation of erythroblasts to erythrocytes (50). Indeed, in a detailed data analysis of the proteins involved in cholesterol biosynthesis, we observed that the entire cholesterol biosynthetic pathway slows down during the erythropoietic process (Fig. 8). Also during this process, the hematopoietic progenitor cells, characterized by the expression of CD34, evolve from early erythroid progenitors (burst-forming units, erythroid) to colony-forming units, erythroid, which then divide to give rise to proerythroblasts and orthochromatophilic erythroblasts (normoblasts) and finally expel the nucleus to form mature erythrocytes (Fig. 8). However, the mechanism behind this is not fully understood. Here, we provide the first evidence that TSPO2 redistributes free cholesterol, coming either from extracellular or internal sources, suggesting that TSPO2 may play an important role during erythrocyte maturation. During the preparation of our manuscript, a short report about the orthologous gene PBRL (peripheral benzodiazepine receptor-like), which corresponds to Tspo2, in chicken erythropoiesis was published (51). Although it is interesting to see the PBRL/Tspo2 sequence in the genome of lizard, no trace of expression and functional data in any species was shown. Moreover, the localization data for the chicken PBRL/TSPO2 presented in that paper indicated a mitochondrial localization, similar to that of TSPO1. These findings are in contrast to the observations reported herein as well as to a previous study that identified an 18-kDa protein (p18) sharing amino acid sequence similarities to mitochondrial TSPO1 in the chicken erythrocyte nuclear envelope forming a complex with nuclear lamins and the lamin B receptor (52). Based on our findings, the data presented by Nakazawa et al. (51) would need to be reinterpreted.
Supplementary Material
Acknowledgments
We thank Jacynthe Laliberte for assistance with confocal microscopy, Martin Marcinkiewicz (Cytochem Inc., Montreal, Canada) for the mRNA mapping, and Ken McDonald for help with flow cytometry.
This work was supported, in whole or in part, by National Institutes of Health Grants HD37031 and ES07747. This work was also supported by a Canada Research Chair in Biochemical Pharmacology (to V. P.). The Research Institute of the McGill University Health Center was supported by a center grant from Le Fonds de la Recherche en Santé du Québec.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Figs. S1–S9.
- TSPO
- translocator protein
- NJ
- neighbor-joining
- ORF
- open reading frame
- NBD-cholesterol
- 22-(N-7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino-23,24-bisnor-5-cholen-3-β-ol
- WT
- wild-type
- GFP
- green fluorescent protein
- RFP
- red fluorescent protein
- ER
- endoplasmic reticulum
- ISH
- in situ hybridization
- PBS
- phosphate-buffered saline
- RT
- reverse transcription
- OMM
- outer mitochondria membrane.
REFERENCES
- 1.Papadopoulos V., Baraldi M., Guilarte T. R., Knudsen T. B., Lacapere J. J., Lindemann P., Norenberg M. D., Nutt D., Weizman A., Zhang M. R., Gavish M. (2006) Trends Pharmacol. Sci. 27, 402–409 [DOI] [PubMed] [Google Scholar]
- 2.Yeliseev A. A., Kaplan S. (1995) J. Biol. Chem. 270, 21167–21175 [DOI] [PubMed] [Google Scholar]
- 3.Donohue T. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4821–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma A., Nye J. S., Snyder S. H. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2256–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taketani S., Kohno H., Okuda M., Furukawa T., Tokunaga R. (1994) J. Biol. Chem. 269, 7527–7531 [PubMed] [Google Scholar]
- 6.Yeliseev A. A., Krueger K. E., Kaplan S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5101–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavish M., Bachman I., Shoukrun R., Katz Y., Veenman L., Weisinger G., Weizman A. (1999) Pharmacol. Rev. 51, 629–650 [PubMed] [Google Scholar]
- 8.Papadopoulos V., Lecanu L., Brown R. C., Han Z., Yao Z. X. (2006) Neuroscience 138, 749–756 [DOI] [PubMed] [Google Scholar]
- 9.Azarashvili T., Grachev D., Krestinina O., Evtodienko Y., Yurkov I., Papadopoulos V., Reiser G. (2007) Cell Calcium 42, 27–39 [DOI] [PubMed] [Google Scholar]
- 10.Li W., Hardwick M. J., Rosenthal D., Culty M., Papadopoulos V. (2007) Biochem. Pharmacol. 73, 491–503 [DOI] [PubMed] [Google Scholar]
- 11.Rone M. B., Fan J., Papadopoulos V. (2009) BBA-Molecular and Cell Biology of Lipids 1791, 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindemann P., Koch A., Degenhardt B., Hause G., Grimm B., Papadopoulos V. (2004) Plant Cell Physiol. 45, 723–733 [DOI] [PubMed] [Google Scholar]
- 13.Snyder M. J., Van Antwerpen R. (1998) Cell Tissue Res. 294, 161–168 [DOI] [PubMed] [Google Scholar]
- 14.Papadopoulos V., Mukhin A. G., Costa E., Krueger K. E. (1990) J. Biol. Chem. 265, 3772–3779 [PubMed] [Google Scholar]
- 15.Bolger G. T., Weissman B. A., Lueddens H., Basile A. S., Mantione C. R., Barrett J. E., Witkin J. M., Paul S. M., Skolnick P. (1985) Brain Res. 338, 366–370 [DOI] [PubMed] [Google Scholar]
- 16.Kumar S., Tamura K., Nei M. (2004) Brief. Bioinform. 5, 150–163 [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. (2004) PHYLIP (Phylogeny Inference Package), Version 3.6, University of Washington, Seattle, WA [Google Scholar]
- 18.Li H., Papadopoulos V. (1998) Endocrinology 139, 4991–4997 [DOI] [PubMed] [Google Scholar]
- 19.Infante R. E., Radhakrishnan A., bi-Mosleh L., Kinch L. N., Wang M. L., Grishin N. V., Goldstein J. L., Brown M. S. (2008) J. Biol. Chem. 283, 1064–1075 [DOI] [PubMed] [Google Scholar]
- 20.Hauet T., Yao Z. X., Bose H. S., Wall C. T., Han Z., Li W., Hales D. B., Miller W. L., Culty M., Papadopoulos V. (2005) Mol. Endocrinol. 19, 540–554 [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Rone M. B., Papadopoulos V. (2006) J. Biol. Chem. 281, 38879–38893 [DOI] [PubMed] [Google Scholar]
- 22.Socolovsky M., Nam H., Fleming M. D., Haase V. H., Brugnara C., Lodish H. F. (2001) Blood 98, 3261–3273 [DOI] [PubMed] [Google Scholar]
- 23.Meurs I., Hoekstra M., van Wanrooij E. J. A., Hildebrand R. B., Kuiper J., Kuipers F., Hardeman M. R., Van Berkel T. J., Van Eck M. (2005) Exp. Hematol. 33, 1309–1319 [DOI] [PubMed] [Google Scholar]
- 24.Hofmann K., Stoffel W. (1993) Biol. Chem. Hoppe-Seyler 347, 166. [DOI] [PubMed] [Google Scholar]
- 25.Hardwick M., Fertikh D., Culty M., Li H., Vidic B., Papadopoulos V. (1999) Cancer Res. 59, 831–842 [PubMed] [Google Scholar]
- 26.Joseph-Liauzun E., Farges R., Delmas P., Ferrara P., Loison G. (1997) J. Biol. Chem. 272, 28102–28106 [DOI] [PubMed] [Google Scholar]
- 27.Delavoie F., Li H., Hardwick M., Robert J. C., Giatzakis C., Peranzi G., Yao Z. X., Maccario J., Lacapere J. J., Papadopoulos V. (2003) Biochemistry 42, 4506–4519 [DOI] [PubMed] [Google Scholar]
- 28.Li H., Yao Z., Degenhardt B., Teper G., Papadopoulos V. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1267–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiner S., Micolod D., Schneiter R. (2005) Biochem. Soc. Trans. 33, 1186–1188 [DOI] [PubMed] [Google Scholar]
- 30.Lacapere J. J., Delavoie F., Li H., Peranzi G., Maccario J., Papadopoulos V., Vidic B. (2001) Biochem. Biophys. Res. Commun. 284, 536–541 [DOI] [PubMed] [Google Scholar]
- 31.Whitelaw E., Tsai S. F., Hogben P., Orkin S. H. (1990) Mol. Cell Biol. 10, 6596–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodsky I., Dennis L. H., Kahn S. B., Brady L. W. (1966) Cancer Res. 26, 198–201 [PubMed] [Google Scholar]
- 33.Sasaki K., Matsumura G., Ito T. (1982) Arch. Histol. Cytol. 45, 247–255 [DOI] [PubMed] [Google Scholar]
- 34.Murphy D. B., Grasser W. A., Wallis K. T. (1986) J. Cell Biol. 102, 628–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holm T. M., Braun A., Trigatti B. L., Brugnara C., Sakamoto M., Krieger M., Andrews N. C. (2002) Blood 99, 1817–1824 [DOI] [PubMed] [Google Scholar]
- 36.Koh C. H., Cheung N. S. (2006) Cell. Signal. 18, 1844–1853 [DOI] [PubMed] [Google Scholar]
- 37.Rone M. B., Liu J., Blonder J., Ye X., Veenstra T. D., Young J. C., Papadopoulos V. (2009) Biochemistry 48, 6909–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olson J. M., Ciliax B. J., Mancini W. R., Young A. B. (1988) Eur. J. Pharmacol. 152, 47–53 [DOI] [PubMed] [Google Scholar]
- 39.Zavala F., Masson A., Brys L., de Baetselier P., Scamps-Latscha B. (1991) Biochem. Biophys. Res. Commun. 176, 1577–1583 [DOI] [PubMed] [Google Scholar]
- 40.Woods M. J., Williams D. C. (1996) Biochem. Pharmacol. 52, 1805–1814 [DOI] [PubMed] [Google Scholar]
- 41.Chambers S. M., Boles N. C., Lin K.-Y., Tierney M. P., Bowman T. V., Bradfute S. B., Chen A. J., Merchant A. A., Sirin O., Weksberg D. C., Merchant M. G., Fisk C. J., Shaw C. A., Goodell M. A. (2007) Cell Stem Cell 1, 578–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komor M., Guller S., Baldus C. D., de V. S., Hoelzer D., Ottmann O. G., Hofmann W. K. (2005) Cell Stem Cell 23, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 43.Keller M. A., Addya S., Vadigepalli R., Banini B., Delgrosso K., Huang H., Surrey S. (2006) Physiol. Genomics 28, 114. [DOI] [PubMed] [Google Scholar]
- 44.Ito E., Toki T., Ishihara H., Ohtani H., Gu L., Yokoyama M., Douglas Engel J., Yamamoto M. (1993) Nature 362, 466–468 [DOI] [PubMed] [Google Scholar]
- 45.Anderson K. P., Kern C. B., Crable S. C., Lingrel J. B. (1995) Mol. Cell. Biol. 15, 5957–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakabayashi J., Yomogida K., Nakajima O., Yoh K., Takahashi S., Engel J. D., Ohneda K., Yamamoto M. (2003) Genes Cells 8, 619–630 [DOI] [PubMed] [Google Scholar]
- 47.Molineux G., Foote M. A., Elliott S. G. (2006) Erythropoietins and Erythropoiesis: Molecular, Cellular, Preclinical, and Clinical Biology, Springer-Verlag New York Inc., New York [Google Scholar]
- 48.Moritz K. M., Lim G. B., Wintour E. M. (1997) Am. J. Physiol. 273, 1829–1844 [DOI] [PubMed] [Google Scholar]
- 49.Shalev H., Kapelushnik J., Moser A., Knobler H., Tamary H. (2007) Am. J. Hematol. 82, 199–202 [DOI] [PubMed] [Google Scholar]
- 50.Montfoort A., Boere W. A. (1978) Lipids 13, 580–587 [DOI] [PubMed] [Google Scholar]
- 51.Nakazawa F., Alev C., Shin M., Nakaya Y., Jakt L. M., Sheng G. (2009) Gene Expr. Patterns 9, 114–121 [DOI] [PubMed] [Google Scholar]
- 52.Simos G., Maison C., Georgatos S. D. (1996) J. Biol. Chem. 271, 12617–12625 [DOI] [PubMed] [Google Scholar]
- 53.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.