Abstract
Kelch-related protein 1 (Krp1) is up-regulated in oncogene-transformed fibroblasts. The Kelch repeats interact directly with the actin-binding protein Lasp-1 in membrane ruffles at the tips of pseudopodia, where both proteins are necessary for pseudopodial elongation. Herein, we investigate the molecular basis for this interaction. Probing an array of overlapping decapeptides of Rattus norvegicus (Rat) Krp1 with recombinant Lasp-1 revealed two binding sites; one (317YDPMENECYLT327) precedes the first of five Kelch repeats, and the other (563TEVNDIWKYEDD574) is in the last of the five Kelch repeats. Mutational analysis established that both binding sites are necessary for Krp1-Lasp-1 interaction in vitro and function in vivo. The crystal structure of the C-terminal domain of rat Krp1 (amino acids 289–606) reveals that both binding sites are brought into close proximity by the formation of a novel six-bladed β-propeller, where the first blade is not formed by a Kelch repeat.
Krp1,2 a member of the BTB-BACK-5Kelch repeat subfamily (1, 2), was identified as a muscle specific protein sarcosin (3) that plays an important role in the assembly of myofibrils through interactions with actin and N-RAP (4, 5). Krp1 is up-regulated in oncogene-transformed rat fibroblasts, where it is important for pseudopodial elongation and cell migration through the regulated direct interaction with Lasp-1 (6, 7) at sites of dynamic membrane associated actin cytoskeleton rearrangements.
The Kelch repeat superfamily consists of several distinct subfamilies, which display a wide variety of functions. Kelch motifs range from 44 to 56 amino acids in length, usually arranged in a series of 5–7 repeats near the C terminus of the protein (1, 2). The signature motifs in each Kelch repeat are a series of 4 hydrophobic amino acids followed by a glycine doublet, a conserved tyrosine, and a conserved tryptophan (1). Each Kelch repeat folds into four twisted antiparallel β-strands (A–D) connected by intrablade loops to form a single blade of a β-propeller (8–10). A C-terminal strand closure mechanism links the first and last blades to complete the propeller. There is little sequence identity between one Kelch repeat and another, suggesting that individual Kelch proteins may interact with multiple partners (11–14). Kelch β-propellers primarily function as scaffolds for protein-protein interactions (10–12, 15). The largest subfamily of Kelch proteins has an N-terminal BTB domain separated from either five or six Kelch repeats by a BACK domain (see Fig. 2a). Some members of the BTB-Kelch domain subfamily are known to interact directly with actin (1, 2, 16) or to regulate the actin cytoskeleton, thereby affecting cell-cell interactions, cell-substrate interactions, and cell migration (6, 7, 17).
FIGURE 2.
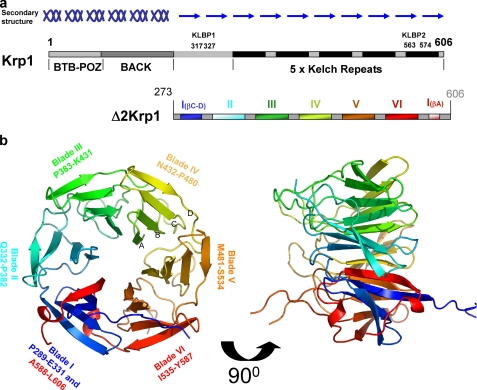
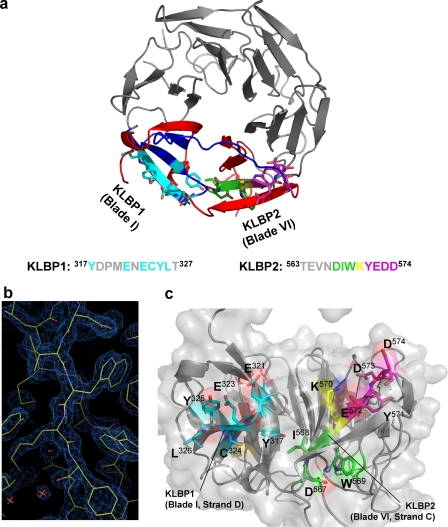
The structure of the β-propeller of Krp1. a, Krp1 is 606-amino acid protein with an N-terminal BTB/POZ (pox viruses and zinc fingers) domain, a central BACK domain, and a C-terminal domain that includes five Kelch repeats. Secondary structure predictions suggest that the protein is mainly α-helical from 1 to 270 and then β-stranded to the C terminus. KLBP1 is located in a β-stranded region outside the Kelch domain, whereas KLBP2 is located in the fifth Kelch repeat. The recombinant protein used for crystallization, Δ2Krp1-(273–606), includes all Kelch repeats and this additional β-stranded region. The observable crystal structure of the C-terminal domain spans residues 289–606 and is a six-bladed β-propeller arising from the five Kelch repeats, with blade I provided by the additional β-stranded region. b, the crystal structure revealed six blades of ∼50 amino acids assembling on a six-fold axis of symmetry around a central pore. Each Kelch blade is composed of four β-strands (A–D as noted on blade IV), forming an antiparallel β-sheet. Strands of blade I are not antiparallel as strands A and B are extended and twist 90° to cross the face of the blade. The propeller is completed by a C-terminal strand closure mechanism where the strand A of blade I is provided by a C-terminal strand that follows blade VI. The upper face of the propeller is formed by the β-strands, whereas the lower face is decorated by the extensive loops that link the strands in each blade.
Despite the ubiquity of Kelch-containing molecules, only four structures exist in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB), and of those, only two, galactose oxidase (9) and Keap1 (10, 13, 18), have been characterized biologically. The galactose oxidase of Dactylium dendroides was the first to be solved and demonstrates the Kelch domain of the molecule to be a highly symmetrical seven-bladed β-propeller that coordinates a central cupric ion that is key to the catalysis. The mammalian Keap1 protein, a substrate adaptor protein for a ubiquitin ligase complex (15, 19), has a Kelch repeat domain with six blades that form the highly symmetrical β-propeller. Keap1 has been used to illustrate a role for the conserved residues of the Kelch motif in maintaining the structural integrity of the β-propeller, and the structure has been posited as the archetypal Kelch propeller (10). A number of interactions have been demonstrated for this protein (13, 14, 18, 20). The crystal structures of the Keap1 Kelch domain in complex with peptides from Nrf2 (a transcription factor regulating cryoprotective genes (18)) and prothymosin (a nuclear oncoprotein (20)) have been solved and demonstrate that Keap1 binds both partners at sites on the variable loops that extend from the blades to the underside of the propeller structure, with both peptides binding to the same general location (13, 14). The remaining two structures available in the PDB, but as yet unpublished, are YJHT (PDB code 2UVK) and KLHL12 (PDB code 2VPJ). These are similar to Keap1, with standard six-bladed propellers. To better understand the molecular basis for the interaction between Krp1 and Lasp-1 a combined strategy of oligopeptide array analysis, cell-based assays, in vitro binding assays, and x-ray crystallography was adopted to define the amino acids that mediate the association.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A cDNA encoding residues 273–606 of Rattus norvegicus Krp1 (Δ2Krp1) (6) (see Fig. 2a) (National Center for Biotechnology Information (NCBI) reference sequence NP_476539.1) was cloned into the pGEX6-P-1 vector (GE Healthcare) and expressed in Escherichia coli BL21 (DE3) pLysS (Stratagene). GST-Δ2Krp1 was purified by glutathione-Sepharose affinity chromatography (equilibrated and washed in 50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 3 mm DTT, 1 mm EDTA and eluted in this buffer supplemented with 20 mm reduced glutathione) and used in screening peptide arrays and protein pulldown assays (7). Protein for crystallization was further purified by affinity tag cleavage followed by gel filtration chromatography on Superdex 75 and Superdex 200 columns in a buffer of 10 mm Tris-HCl, pH 7.5, 300 mm NaCl, 3 mm DTT, 1 mm EDTA. Pure Δ2Krp1 was concentrated to 8 mg/ml (216.8 μm). Selenomethionine (Se-Met)-substituted protein was prepared in a similar way, in E. coli B834 DE3 pLysS cells using a minimal medium supplemented with Se-Met. Rat His6-Lasp-1 (21) was expressed in E. coli BL21 DE3 pLysS and purified by nickel-nitrilotriacetic acid metal affinity chromatography followed by gel filtration on Superdex S75. For Se-Met protein purification, the buffers were those used in the wild-type protein purification but containing 10 mm DTT.
Screening of Peptide Arrays
All peptide arrays of Krp1(6) and Lasp-1 (21) sequences were synthesized by the Cancer Research UK (CRUK) peptide synthesis facility. Each decapeptide shifted by +1 amino acid throughout the full sequence. Alanine substitution peptide arrays were based on the amino acid sequences of KLBP1 and KLBP2. The peptide arrays were essentially treated as Western blots (7). After incubation with blocking buffer, each membrane was incubated overnight with 20 ml of 32.5 nm His6-Lasp-1 in blocking buffer. Bound His6-Lasp-1 was detected using a rabbit anti-Lasp-1 primary antibody (Beatson Institute Antibody Production Service) and an anti-rabbit-horseradish peroxidase conjugate secondary antibody (Sigma).
Mammalian Expression
All mammalian expression was performed with pcDNA3.1Myc/His(−)B (Invitrogen) with inserts of Krp1-Myc, Δ2Krp1-Myc, or mutations of Δ2Krp1-Myc (created by site-directed mutagenesis (Stratagene)) or HA-Lasp-1. Transfection of rat FBR cells or COS7 was performed using FuGENE 6 (Roche Diagnostics) according to the manufacturer's instructions. HA-Lasp-1 in COS7 cells was prepared for use in pulldown experiments (7), and Krp1-Myc in FBRs were prepared for confocal microscopy (7).
Streptavidin Pulldowns
Streptavidin HP spin trap (GE Healthcare) columns were equilibrated in GST-FISH buffer (10% glycerol, 50 mm Tris, pH 7.4, 100 mm NaCl, 1% Nonidet P-40, 2 mm MgCl2 plus protease inhibitors (protease inhibitor cocktail Set I, Calbiochem) and phosphatase inhibitors (5 mm sodium pyrophosphate, 50 mm NaF)), which were used in all subsequent washes. 0.4 μg/μl biotinylated peptides (CRUK, Protein and Peptide Chemistry) were added to the columns and incubated at room temperature for 30 min. Columns were washed once and then blocked in 300 μl of 5% bovine serum albumin (Sigma, fraction V) in GST-FISH buffer plus 0.5 mm DTT for 30 min. After three washes with FISH buffer plus 0.5 mm DTT, columns were incubated with Lasp-1 for 1 h at room temperature. For HA-Lasp-1, 200 μl of total cell lysate plus 200 μl of FISH buffer were used. For purified bacterially expressed His6-Lasp-1, 4.45 nm in 400 μl of FISH buffer was used. Columns were washed five times. Beads were removed from each column and resuspended in 200 μl of 1× SDS-PAGE sample buffer and separated on a 4–20% gradient SDS-PAGE gel according to standard techniques.
GST Pulldowns
GST pulldowns were carried out essentially as described previously (7). For peptide competitions, a 4 m excess of peptide with respect to GST-Δ2Krp1 bound to beads was added simultaneously with 200 μl of HA-Lasp-1-expressing COS7 lysate.
Crystallization, Data Collection, and Data Processing
Crystals were grown in 22% polyethylene glycol 4000, 150 mm sodium acetate, 100 mm Tris-HCl, pH 7.5, and 3 mm DTT by mixing 1 μl of protein stock with 1 μl of crystallizing agent for hanging drop vapor diffusion at 18 °C. Crystals were flash-cooled using 10 mm Tris-HCl, pH 7.0, 300 mm NaCl, 20% polyethylene glycol 8000, 20% Glycerol, 1 mm EDTA, 3 mm DTT as a cryoprotectant. Se-Met-substituted protein crystallized in the same conditions in the presence of 10 mm DTT. Native data sets were collected at European Synchrotron Radiation Facility (ESRF) ID14.4, and Se-Met data were collected at ESRF ID29 in a cryostream at 93 K. Reflection data were processed using MOSFLM and SCALA (Collaborative Computational Project, Number 4 (CCP4) ((22)), and the structure was solved by single wavelength anomalous dispersion using SHARP (Global Phasing). Building was done manually in COOT (23). Refinement and validation were performed using REFMAC (22) and PROCHECK (CCP4), and additional analyses were performed using CONTACT (22) (CCP4) and AREAIMOL (22) (CCP4). Structure images were generated using PyMOL (24).
RESULTS
A Krp1 Oligopeptide Array Screened with Lasp-1 Identifies Two C-terminal Binding Sites
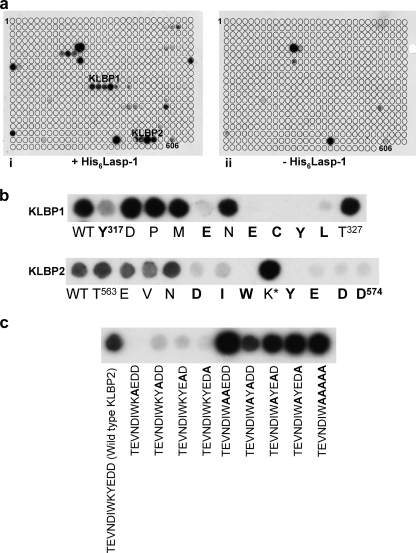
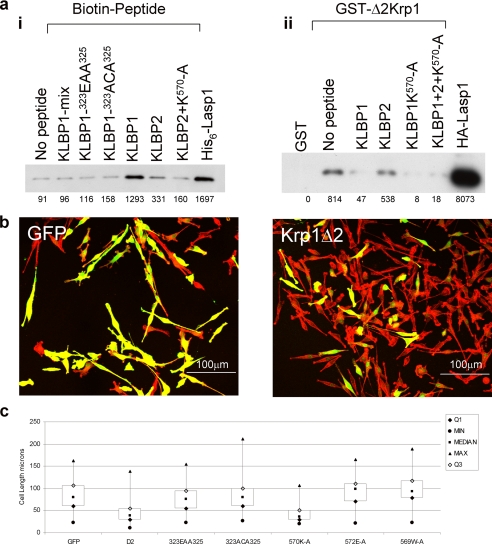
Three series of Krp1 oligopeptides bound Lasp-1 in oligopeptide array screens (Fig. 1a). The first is in the BACK domain, outside the Kelch repeat that is the focus of this study, and was not investigated further. Of the two remaining C-terminal sequences, the first, Kelch-Lasp-1 binding peptide 1 (KLBP1), Tyr317–Thr327, appears to be located outside the Kelch domain in the region following the C-terminal end of the BACK domain. The second, Kelch-Lasp-1 binding peptide 2 (KLBP2), Thr563–Asp574, mapped to the fifth and final Kelch repeat. Amino acids interacting with LASP-1 were identified by scanning through the oligopeptides with alanine mutations (Fig. 1b). In KLBP1, only replacement of Tyr317, Glu321, Glu323, Cys324, Tyr325, and Leu326 with alanine decreased the binding for Lasp-1. In KLBP2, replacement of Asp567, Ile568, Trp569, Tyr571, Glu572, Asp573, and Asp574 with alanine resulted in decreased binding of Lasp-1. Interestingly, replacement of Lys570 with alanine resulted in a reproducible increase in Lasp-1 binding signal. Surprisingly, once Lys570 was changed to alanine, the remaining Tyr571–Glu572–Asp573–Asp574 were no longer required for binding (Fig. 1c). To understand the organization of the KLBP1 and KLBP2 binding sites in Krp1, the crystal structure of the C-terminal domain was determined.
FIGURE 1.
Probing oligopeptide arrays revealed Lasp-1 binding motifs in Krp1. a, oligopeptide membranes that were arrays of decamer oligopeptides spanning the entire length of Krp1, modulating by +1 amino acid at a time, were probed with recombinant His6-Lasp-1. Two binding sites in the region of the C-terminal domain were identified (panel i), and these oligopeptides did not give a positive result on control membranes where no His6-Lasp-1 had been applied (panel ii). These two motifs corresponded to Tyr317–Thr327 (KLBP1) and Thr563–Asp574 (KLBP2). b, each motif was scanned by mutating each residue in turn to alanine. This demonstrated that the residues involved in Lasp-1 binding were Tyr317, Glu321, Glu323, Cys324, Tyr325, and Leu326 in KLBP1 and Asp567, Ile568, Trp569, Tyr571, Glu572, Asp573, and Asp574 in KLBP2. Mutation of Lys570 to alanine in KLBP2 consistently gave an increase in signal from Lasp-1 binding in replicate blots. WT, wild type. c, mutation of any of the 4 residues following Lys570 in KLBP2 resulted in an almost complete loss of Lasp-1 binding (all mutations are noted in bold). However, any or all of residues 571–574 could be removed without effect if Lys570 was mutated to alanine.
Structure of the Krp1 β-Propeller and the Component Blades
Secondary structure predictions (Jpred) suggest that the molecule is α-helical in the N-terminal 273 amino acids and then mainly β-stranded to the C terminus. The change in character of secondary structure occurs prior to the Kelch domain and following the C-terminal boundary of the BACK domain (Fig. 2a). Crystals of a 36.9-kDa truncate (Δ2KRP; residues 237–606) comprising the entire predicted β-stranded region of Krp1 (Fig. 2a). Crystallized in space group P21212 and was solved by Se-Met single wavelength anomalous diffraction and refined to 2.0 Å resolution (Table 1). The observable structure for the C-terminal domain of Krp1 spans from Pro289 to Leu606, whereas the N-terminal 15 residues of Δ2Krp1 and the loops at 356–362 and 555–558 were not observable and therefore considered to be disordered.
TABLE 1.
Data collection, phasing, and refinement statistics
| Native | Selenomethionine | |
|---|---|---|
| Data collection | ||
| Space group | P21212 | P21212 |
| a, b, c (Å) | 66.11, 98.53, 46.97 | 65.86, 98.52, 46.78 |
| Molecules per asymmetric unit | 1 | 1 |
| Resolution (Å) | 2.0 | 2.5 |
| Measured/unique reflections | 84247/20693 | 62858/16543 |
| Completeness (%)a | 97.1 (99.6) | 99.8 (100) |
| Multiplicity | 4.1 | 3.8 |
| Rmergeb | 5.9 (40.2) | 7.2 (37.8) |
| Mean I/σI | 16.4 (3.6) | 17.2 (4.2) |
| Selenium atoms per asymmetric unit | 6 expected/5 observed | |
| Mean FOMc | 0.86 | |
| Refinement | ||
| Rcryst/Rfreed | 23.7/27.9 | |
| r.m.s.d.e bond lengths | 0.015 | |
| r.m.s.d.e bond angles | 2.062 | |
| Mean B-factor (Å) | 28.95 | |
| No. of atoms (protein/water) | 2388/64 | |
| Ramachandran analysis | ||
| Favored regions | 90.4% | |
| Additionally allowed regions | 10.6% | |
| Generously allowed regions | 1.9% | |
| Disallowed regions | 1.5% | |
| Protein Data Bank code | 2woz | |
Values in parentheses refer to highest resolution shell.
Rmerge = Σ‖I − 〈I〉|/|Σ〈I〉|.
Figure of merit following phasing and density modification with SHARP/SOLOMON.
Rcryst = (Σ‖Fo − Fo‖)/(Σ‖Fo‖).
r.m.s.d., root mean square deviation.
The crystal structure reveals that Krp1 has a six-bladed propeller. Blades II to VI are formed by the five Kelch repeats, but the first blade is formed by a non-Kelch amino acid sequence (Fig. 2b). The β-propeller is ∼43 Å diameter and 40 Å in height with a central 9 Å pore containing a number of ordered water molecules. Each Kelch blade contains ∼50 residues forming a twisted β-sheet formed by four β-strands. In each Kelch blade, strand A lines the central pore and lies roughly on the six-fold axis of symmetry of the propeller (10). The Kelch blade radiates outwards from the pore, twisting around 90° until the outer strand orients perpendicular to the inner strand, forming the outer circumference of the circular propeller. Blade I is composed of three strands from the N-terminal region of the crystallized protein, whereas the fourth strand is provided by a common C-terminal closure mechanism involving the sequence immediately after the final strand of blade VI (1). Blade I arises from a non-Kelch sequence and adopts a novel conformation that is distinct from a typical Kelch blade.
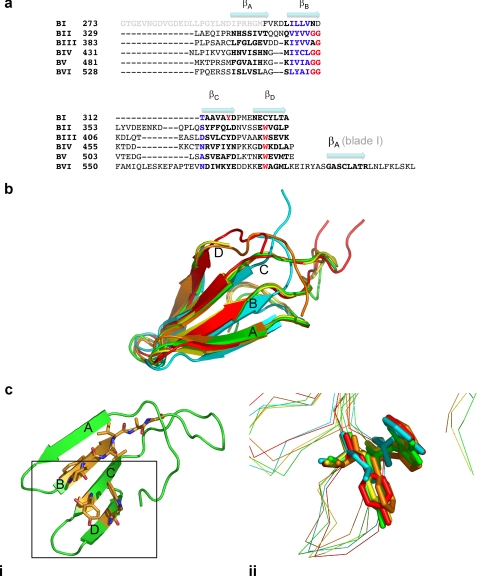
A high degree of symmetry between the five Kelch blades is observed (Fig. 3, a and b) with the largest variation arising from the large loop between strand B and C. These extended loops at the bottom of the propeller are largely equal in size except for the loop extending from blade VI that is significantly extended, and the outer reaches of the loop are disordered in the structure. A Kelch repeat signature consists of several highly conserved features, namely a hydrophobic motif followed by a glycine doublet in strand B and a tyrosine in strand C that is positioned to interact with a tryptophan in strand D (Fig. 3c, panel i). Only some of these key features are present in the five Kelch repeat blades of Krp1. All six of the Kelch blades display a series of hydrophobic residues that form strand B. However, the characteristic double glycine motif at the beginning of the strand B–strand C loop is present in only four of the five Kelch blades with the Gly-Gly motif altered to Ala-Gly in blade III. This substitution does not seem to alter the conformation of the loop immediately following strand B of this blade. The glycine doublet does form hydrogen bonds with an arginine positioned at the beginning of strand A in four of the blades, but it does not occur in blade IV, where the arginine is replaced by a valine, and this smaller side chain does not extend sufficiently to make contacts with the GG motif (data not shown). The signature tryptophan residue in strand D is present in all of the Kelch blades. A significant hydrophobic interaction occurs at this tryptophan in the Kelch blade, but the typical tyrosine is not conserved and is replaced by a leucine in blade II and a phenylalanine in blade V (Fig. 3c, panel ii).
FIGURE 3.
The structure of the five Kelch blades. a, a structural alignment of each of the blades of the Krp1 β-propeller illustrates that blade I is significantly different from the five Kelch repeats. Residues that are considered to be hallmarks of the Kelch repeat are noted, but these residues are not totally conserved in Krp1 Kelch repeats. b, the degree of structural identity in the Kelch repeats is revealed when all five blades are superimposed (propeller blade II, cyan; propeller blade III, green; propeller blade IV, yellow; propeller blade V, orange; propeller blade VI, red). The highest degree of variation in the structure is in the interstrand loops, particularly the loop linking strand B to strand C. c, a standard Kelch arrangement is found in blade III of Krp1 (panel i). The current definition of a Kelch repeat requires 4 hydrophobic amino acids followed by a glycine doublet to form strand B and a tyrosine toward the end of strand C that interacts with a tryptophan on strand D. It has been proposed that the hydrophobic interactions between the aromatic rings of the conserved tyrosine and tryptophan are critical to blade folding and that additional hydrogen bonding from the tyrosine hydroxyl group is also important (31). However, the tyrosine is not completely conserved in Krp1 as this position is occupied by a phenylalanine in blade V, eliminating the availability of a hydroxyl group from hydrogen bonding, and a leucine in blade II, demonstrating that the hydrophobic residue at this position need not be an aromatic (panel ii).
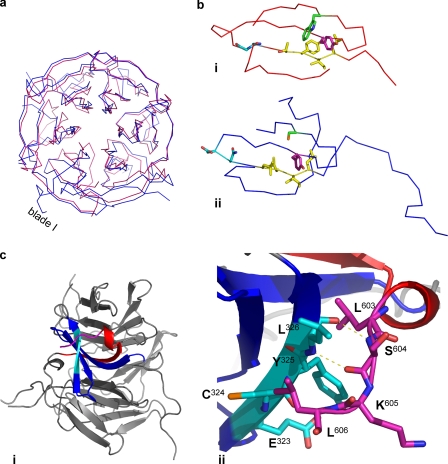
Blade I of the Krp1 propeller differs significantly from blade I of Keap1 (Fig. 4a) and from other Kelch blades (9, 10). A structural alignment demonstrates that all Krp1 blades have the typical four β-strands, but the key motifs that define a Kelch motif are not present in blade I (Fig. 4b). Although strand B has a run of 4 hydrophobic amino acids, the glycine doublet that precedes the usually extensive strand B to strand C loop is replaced by an asparagine and aspartic acid. In blade I, there is no extensive loop here, and the structure turns quickly from strand B to C. Strand C does possess a tyrosine in the same position as the Kelch blade, but the tryptophan of strand D is absent and is replaced by a cysteine (Cys324). When compared with a typical Kelch blade, there is a significant alteration in the topology of strands A and B of blade I that are involved in the C-terminal closure mechanism. Rather than the usual antiparallel arrangement, these extended strands twist through 90° and cross the upper face of the blade (Fig. 4, a and b). On twisting, the β-strands extend far enough away from the blade so as to make no major contacts with strands C or D of blade I. Strand B has a significant N-terminal extension, and this seems to contact only with the C-terminal closure strand A of blade I, where it serves to stabilize the twisted conformation of strand A as it crosses the upper face of the blade (Fig. 4c, panel i). The remainder of this long extension suggests that there is a significant linker region between the β-propeller and the BACK domain of Krp1. There is a 9-amino acid C-terminal extension beyond strand A, and the final 4 residues (603–606) make significant contacts with residues on strand D of blade I, especially Tyr325 and Leu326 (Fig. 4c, panel ii). This C-terminal extension is anchored here by two polar contacts between the respective main chain nitrogens and carbonyl oxygens of Ser604 and Leu326. In addition, analysis by CONTACT (CCP4) (22) reveals a substantial number of van der Waals contacts between these two motifs.
FIGURE 4.
The structure of blade I is different to that of a Kelch blade. a, the structure of the Krp1 β-propeller (blue) is superimposed on the Kelch domain of Keap1 (1U6D) (red). The structure of the Kelch blades is conserved, but this homology is not evident in blade I as strands A and B do not conform to the usual antiparallel arrangement. b, other differences are observed when comparing blade I of Keap1 (panel i) and Krp1 (panel ii). Consistent with a Kelch repeat, there are 4 hydrophobic residues on strand B (yellow) and a tyrosine in the correct position in strand C (magenta) of the Krp1 blade I. However, there is no glycine doublet at the end of strand B, and the glycines are replaced by asparagine and aspartic acid (cyan). Unlike all the Kelch repeats of Krp1, there is no tryptophan on strand D, and this location is occupied by a cysteine (green). c, panel i, strand A of blade I facilitates the C-terminal strand closure mechanism. There is a significant C-terminal extension following the strand, and the C-terminal residues of Krp1 (magenta) interact with residues forming strand D (cyan). Panel ii, the interaction is achieved by a number of van der Waals contacts and two polar contacts between the amide nitrogens and carbonyl oxygens of Ser604 and Leu326. Interestingly, the residues of strand D that interact with the C-terminal extension are key Lasp-1-interacting residues of KLBP1(Glu323–Leu326).
The Structure of the Putative Lasp-1 Binding Site
Peptide mapping experiments identified two distinct binding motifs in the β-propeller domain of Krp1. The two motifs were distant in the primary amino acid sequence, but due to the cyclical nature of the propeller, they proved to be very close in the three-dimensional structure (Fig. 5a). The N-terminal binding site KLBP1 (Tyr317–Thr327) maps to strand D of blade I, whereas KLBP2 (Thr563–Asp574) maps to strand C and loop of blade VI. An alanine scanning experiment identified which residues in these motifs were key to Lasp-1 binding. In KLBP1, Tyr317 and Glu321 are implicated in Lasp-1 binding, whereas the residues from Glu323 to Leu326 are critical. The Glu323–Leu326 motif forms the outer strand D of blade I. Inspection of the structure and analysis by AREAIMOL (CCP4) (22) demonstrated that, with the exception of Tyr317, these are all largely surface-exposed residues (Fig. 5c). Glu321 is the outermost residue on the loop between stands C and D and is also fully exposed to the solvent. As the main components of the outer strand D, Glu323–Leu326 are also exposed to the solvent despite interacting with the C-terminal extension of strand A in this location. The critical residues of the KLBP2 peptide appear to be Asp567, Ile568, and Trp569. The side chains of both the Trp569 and the Asp567 are not as exposed as the residues of the KLBP1 binding site but do contribute to a pocket formed by these residues plus the side chain of Lys583 (Fig. 5b). The base of this pocket is provided by the carbonyl oxygen of Ile568 with the side chain firmly buried in the core of the structure. Strand D of blade VI crosses strand C after Trp569. The sequence on the other side of this dissection also proved important for Lasp-1 binding in peptide mapping experiments. A consistent increase in the signal for Lasp-1 binding was observed when Lys570 was replaced by alanine, but the replacement of any of the 4 following residues (Tyr571 or the 3 acidic residues Glu572, Asp573, and Asp574) with alanine eliminated Lasp-1 binding (Fig. 1c). The influence of residues Tyr571–Asp574 is only important in the context of a peptide containing an intact Lys570. Tyr571 is largely buried, whereas the 3 acidic residues are all surface-exposed. However, as shown above, these residues do not appear to interact directly with Lasp-1.
FIGURE 5.
KLBP1 and KLBP2 converge in the Krp1 structure to form a binding site for Lasp-1. a, KLBP1 (cyan) is located on the strand C–D loop and strand D of blade I. KLBP2 (green, yellow, and magenta) is located on strand C and the strand C–D loop of blade VI. The cyclical structure of the domain means that these two motifs are in close proximity. b, a typical example of the 2Fo − Fc electron density contoured at 1.8 σ in COOT (23). The figure illustrates the region around KLBP2. c, the binding site(s) are accessible from the side face of the propeller. The majority of interacting residues on KLBP1 (cyan) are solvent-exposed. The Asp567–Ile568–Trp569 of KLBP2 are required for Lasp-1 binding but are less surface-exposed than the residues of KLBP1. It is possible that Lasp-1 binding may induce a conformational change that increases the accessibility of these residues. Lys570 (yellow) is not essential for Lasp-1 interaction, but an increased binding signal is observed when this residue is mutated to alanine. The 4 residues following Lys570 (Tyr571–Glu572–Asp573–Asp574) (magenta) are located on the exposed strand C–D loop of blade VI and are also essential for Lasp-1 binding but only in oligopeptides containing Lys570.
The Lasp-1 Binding Sites Are Functional
To confirm that the peptide binding sites interact directly with Lasp-1, biotinylated peptides bound to streptavidin beads were incubated with recombinant His6-Lasp-1 (Fig. 6a, panel i). KLBP1 and, to a lesser extent, KLBP2 captured Lasp-1, but a peptide with a scrambled sequence did not. When the amino acids implicated in binding were altered (Glu323–Cys324–Tyr325 to Glu-Ala-Ala or Ala-Cys-Ala), binding was also inhibited. The KLBP2 K570A mutant that showed increased binding on the oligopeptide array did not significantly or consistently interact with Lasp-1 in these assays.
FIGURE 6.
KLBP1 and KLBP2 affect the interaction between Krp1 and Lasp-1 in vitro and in vivo. a, in vitro, panel i, biotinylated peptides representing either wild-type, mutant, or scrambled (mix) sequences of KLBP1 and KLBP2 captured recombinant Lasp-1 on streptavidin beads. Bound Lasp-1 was identified by Western analysis. Panel ii, GST-Δ2Krp1 glutathione beads specifically captured HA-Lasp-1 from COS7 cell lysates, and this binding was inhibited by a 4-fold molar excess of KLBP1 and KLBP2 K570A peptides. KLBP2 peptide marginally reduced this interaction. A pool of the three peptides inhibited binding. b, in vivo bipolar FBR cells were nucleofected with vectors expressing enhanced GFP (control), Myc-Δ2Krp1, or mutants in KLBP1 or KLBP2. After 24 h, the cells were fixed and stained with phalloidin-TRITC, Myc tag monoclonal antibody 9E10, and anti-mouse IgG-Fl. Confocal microscopy identified the expressing cells. The GFP panel depicts control cells, whereas the Krp1Δ2 panel represents cells expressing Δ2Krp1, which display truncated pseudopodia. c, to determine the effect of Δ2Krp1 and its mutants on cell length, images of cells were collected using a ×20 objective, and the length of at least 80 cells for each expression vector was measured using the ImageJ software. Length values of minimum (min), maximum (max), median, and first and third quartiles were calculated and plotted in Excel as box plots. Unpaired t tests were carried out between each data set to confirm statistical significance. Δ2Krp1 and Δ2Krp1–570K-A expression results in significantly truncated cells, whereas cells expressing mutants in either of the two binding sites remain equivalent to the control enhanced GFP cells. (Unpaired t test results are as follows: GFP control 1.65 × 10−49; 323EAA325 1.32 × 10−40; 323ACA325 3.77 × 10−56; 572EA573 1.63 × 10−44; 569WA570 2.65 × 10−40; 570KA571 0.683248).
Previously, the Kelch domain of Krp1 was shown to bind to Lasp-1 (6, 7). To determine whether KLBP1 and KLBP2 were involved in this binding, recombinant GST-Δ2Krp1 was used to capture HA-Lasp-1 from COS7 cell lysates in the absence or presence of peptides with wild-type or mutant sequences of the binding sites (Fig. 6a, panel ii). This demonstrated that GST-Δ2Krp1 bound Lasp-1 but GST alone did not. The capture of Lasp-1 by GST-Δ2Krp1 was inhibited by the addition of a 4 m excess of KLBP1 but not KLBP2. In this format, the KLBP2 K570A mutant did prevent binding of Lasp-1 to GST-Δ2Krp1. This demonstrated that KLBP1 and KLBP2 K570A blocked the binding of GST-Δ2Krp1 to Lasp-1, indicating that KLBP1 and KLBP2 are functional binding sites on GST-Δ2Krp1 for Lasp-1.
FBR cells normally exist as bipolar cells with long pseudopodia (25). The Kelch repeats of Krp1 when expressed in FBR cells function as a dominant negative mutant resulting in truncated pseudopodia (6, 7). To determine whether the binding site on Δ2Krp1 was functional in vivo, wild-type and mutant forms of Myc-Δ2Krp1 were expressed in FBR cells. The expression of Δ2Krp1-Myc in FBR cells resulted in truncated pseudopodia (Fig. 6, b and c). Expression of Δ2Krp1-Myc KLBP1 mutants C324A,Y325A and E323A,Y325A did not result in truncated pseudopodia. Expression of Δ2Krp1-Myc KLBP2 mutant K570A did result in truncated pseudopodia; however, KLBP2 mutants E572A or W569A did not result in truncated pseudopodia. These results indicate that both binding sites are required for a functional Krp1 Kelch domain in vivo.
DISCUSSION
We have previously demonstrated that an interaction occurs between Lasp-1 and the C-terminal domain of Krp1 that features several Kelch repeats (7). Two potential binding sites in this C-terminal region of Krp1 were identified, but it was not clear whether these binding sites work cooperatively or separately or interact to contribute to a single interface for Lasp-1 binding. To understand the organization, we determined to solve the x-ray crystal structure of the C-terminal domain of Krp1.
There is relatively little structural information available for Kelch molecules, and the first to be solved was the seven-bladed propeller of galactose oxidase from D. dendroides (PDB ID: 1GOF) (9). There are three crystal structures of mammalian Kelch domains in the Protein Data Bank: Keap1 (PDB ID: 1ZGK and 1U6D) (10), KLHL12 (PDB ID: 2VPJ), and YJHT (PDB ID: ID 2UVK). All three of these proteins exhibit highly symmetrical six-bladed β-propellers arising from six clearly defined Kelch motifs. Of these Kelch domains, only the Keap1 structure has been evaluated in a biological context (13, 14, 18, 19).
Based on the presence of five Kelch repeats, Krp1 was proposed to be a five-bladed β-propeller. However, any recombinant protein comprised only of the five Kelch repeats would not express and/or crystallize. The only recombinant protein that would crystallize, Δ2Krp1-(273–606), contained additional N-terminal residues prior to the five Kelch repeats and included the β-strands that had previously been considered outside the Kelch domain, and these residues provide an additional unanticipated blade.
At first glance, the Krp1 propeller appears very similar to the other mammalian Kelch domain structures, with a six-fold axis of symmetry built around repeating blades of ∼50 amino acids. However, there are several notable differences including some alterations that have consequences for partner binding. A detailed examination of the five Krp1 Kelch repeats shows that only two display the full repertoire of the signature motifs. The structure of Keap1 provided the opportunity to assign structural roles to the signature motifs of the Kelch repeat (10, 13, 14, 31), an ∼50-amino acid polypeptide that folds in a four-stranded antiparallel β-sheet with 4 hydrophobic amino acids constituting strand B followed by a glycine doublet and a tyrosine on strand C in position to form a C–H … π hydrogen bond with a tryptophan on strand D. The structure of Krp1 demonstrates that the tertiary structure of the blade can persist in the absence of one or more of these signature motifs and questions the accepted role that each motif plays in maintaining the structure of the blades of the β-propeller. In the five Krp1 Kelch blades, the hydrophobic motif on strand B was conserved, as was the tryptophan on strand D. However, in one of the glycine doublets, there was a conservative alteration to alanine, illustrating that the sequence here is not invariably a Gly-Gly. Most notably, the tyrosine of strand C is not conserved. It was previously suggested that a tyrosine at this position is essential for C–H … π hydrogen bonding with the strand D tryptophan and that the hydroxyl group of the tyrosine establishes hydrogen bonds that further underpin the blade. However, the residue at this position in Krp1 blade V is substituted to phenylalanine, demonstrating that the hydrogen bonding of the hydroxyl group is not essential. Indeed the residue at this position on blade II is substituted to isoleucine, illustrating that diverse hydrophobic residues are sufficient in this position. The signature motifs of Caenorhabditis elegans SPE-26 are even more degenerate (26). In this case, the tryptophan of strand D appears to be absent in blades IV and V and the interacting aromatic residue of strand C is never a tyrosine. Our findings suggest that it is difficult to construct a specific definition that would allow for the identification of a Kelch repeat from the primary sequence alone as the key hydrophobic interactions that are central to the integrity of the Kelch blade can be established by a broad range of hydrophobic residues.
Blade I of the propeller is formed by residues previously considered to be outside the Kelch domain. Overall, blade I does retain some of the character of a Kelch repeat in terms of general structure (a four-stranded blade) and the conservation of hydrophobic residues on strand B and the tyrosine on strand C. However, the hydrophobic residues are not followed by the double glycine, and there is no tryptophan on strand D to interact with the tyrosine on strand C. Strands A and B are extended and twist across the upper face of the blade in a manner not observed in Kelch repeat blades. It is possible that blade I originated as a Kelch repeat but has altered to allow partner protein binding. Alternatively, the Kelch-like elements of blade I may be a product of convergent evolution where the hydrophobic elements in question are simply required to maintain the structural integrity in the core of a blade. These differences may have a significant impact on partner protein binding.
Prior to this study, little was known about the mode of interaction of Krp1 with its binding partners. Our previous investigation demonstrated that Lasp-1 was expressed in both normal and transformed fibroblasts, and it physically associated with Krp1 at sites of dynamic membrane-actin reorganization in the latter (7). Both proteins are required for pseudopodial extension in transformed fibroblasts, whereas Lasp-1 enhances the migration of other cancer-derived cell lines and is up-regulated in some breast cancers (27–30). Thus, an understanding of the structural basis for the interaction would be advantageous. As Krp1 proved to be a six-bladed propeller, the two binding sites identified in the peptide mapping experiments are brought into close proximity. Two structures of Keap1 interacting with peptides from binding partners have been solved (13, 14). In both cases, the partner binds to the loops on the underside of the propeller, at the most distant point from the β-propeller. These locations are the most divergent in terms of sequence and structure. Lasp-1 appears to bind directly to the β-propeller of Krp1 but also at a site of significant structural and sequence divergence. KLBP1 is perhaps the most interesting of the two binding motifs as it arises from the non-Kelch sequence of blade I and performed consistently well in pulldown assays that validate the interaction.
Strand D of the Kelch repeats of Krp1 always contains a tryptophan, but there is no large hydrophobic side chain in this position of strand D in blade I as this strand is formed by KLBP1 and the equivalent position is occupied by Cys324. Thus, the neutral character of the side chain at this position is conserved, and the Cα and side chain orientation of the cysteine is similar to that of the tryptophan, but there is no C–H … π hydrogen bonding between the much smaller aliphatic cysteine and the tyrosine to underpin the core of the blade. The absence of a large hydrophobic side chain in strand D may be required for Lasp-1 binding, but it does weaken the usual strong hydrophobic interaction that promotes the structural integrity of the blade. There is a significant interaction between the C-terminal extensions of strand A and the residues of KLBP1. It is possible that the twisted torsion of strands A and B is required to provide the additional interaction with strand D, which acts as a “cross-brace” to stabilize the blade structure in the absence of the C–H … π hydrogen bonding normally provided by the tryptophan. The strand A extension may also provide a regulatory mechanism that could control access of Lasp-1 to KLBP1. Interestingly, the main polar interactions on the extension arise from a surface-exposed serine that may be phosphorylated. Indeed the residues of KLBP1 also prompt the notion of regulation with a surface-exposed Tyr325 that may be phosphorylated, and Cys324 is available for redox modification. KLBP2 did interact with Lasp-1 in pulldown experiments but did not perform as well as KLBP1. It is possible that KLBP2 acts as a secondary binding site to stabilize the interaction with Lasp-1 at KLBP1. In addition, the Asp567, Ile568, and Trp569 that are critical for the interaction arise from strand C of blade VI and are not as surface-exposed as the key residues of KLBP1. However, the accessibility of these residues may increase in the event of a conformational change on Lasp-1 binding. The efficiency of KLBP2 increased when Lys570 was mutated to alanine, and this was consistent with the increased binding signal observed on oligopeptide membranes.
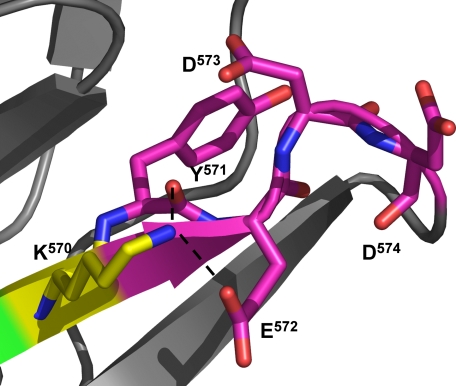
Residues Tyr571–Asp574 also appeared critical for Lasp-1 binding to KLBP2, but their importance was completely negated if Lys570 was removed. Lys570 is surface-exposed in the crystal structure but remains very well ordered with an average B-factor for the residue of 27.40 Å2 (averaged B-factor for the molecule is 28.96 Å2). The structure shows that the lysine side chain is tethered in position by hydrogen bonds to the carbonyl oxygen of Tyr571 and a side chain carboxyl oxygen of Glu572 (Fig. 7). It is possible that all 4 resides are required to provide a structure that facilitates the tethering of the lysine, which may otherwise interfere with Lasp-1 binding to KLBP2 at the Asp567–Ile568–Trp569. When Lys570 is removed, this strategy is no longer required, so residues 571–574 lose their role in the binding mechanism. Admittedly, it is surprising that this hypothesis should hold true in the context of KLBP2 oligopeptides both on membranes and in solution.
FIGURE 7.
Residues 571–574 reduce the inhibition of Lasp-1 binding to the Asp567–Ile568–Trp569 motif by Lys570. In the crystal structure, the side chain of Lys570 is exposed to the solvent but is surprisingly well ordered (averaged B-factor is 27.40 Å2 for Lys570, 28.96 Å2 for the Krp1 molecule). It appears to be held in place by a number of interactions including hydrogen bonds with a carboxyl oxygen of the Glu572 side chain and the main chain carbonyl oxygen of Tyr571. The positioning of residues 571–574 seems to be critical in a mechanism that tethers the Lys570 side chain in position.
This study that demonstrates the interactions between Kelch β-propellers and partners can occur in locations other than the extensive loops protruding from the underside of the propeller. It is interesting to propose that the structure of a β-propeller, with a large surface area to volume ratio, is ideal for a domain that may act as a binding hub for several partners. The structures of Keap1 with Nrf2 and prothymosin peptides illustrate the potential for the loops in this regard. This study implicates direct interactions with the core of the β-propeller. Other interactions may be possible. For example, the Krp1 β-propeller has a canonical SH3 binding motif (Pro382–Pro383–Leu384–Pro385) on the loop that connects strand D of blade II to strand A of blade III.
In summary, we had initially assumed that the Krp1 C-terminal domain would be a five-bladed Kelch β-propeller and that solving the structure may provide us with an understanding of the organization of the Lasp-1 binding sites. The resulting structure proved more interesting than anticipated as it revealed a six-bladed propeller with the BACK domain binding site KLBP1 presented on a non-Kelch blade I that acts with the Kelch blade VI to form a Lasp-1 binding site. Also, an examination of the structure of the Kelch blades questions the accepted definition of the Kelch repeat as we clearly illustrate that the typical structure of a blade can be achieved in the absence of many of the Kelch signature motifs that have previously been considered essential.
Supplementary Material
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and for assistance in using beamlines ID14-4 and ID29. We thank Jim Norman, David Gillespie, Neil Issacs, and Frank Kozielski for assistance.
This work was funded by Cancer Research UK (to B. W. O.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 2woz) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Krp1
- Kelch-related protein 1
- BTB
- bric-à-brac tramtrack broad complex
- GST
- glutathione S-transferase
- DTT
- dithiothreitol
- Se-Met
- selenomethionine
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Adams J., Kelso R., Cooley L. (2000) Trends Cell Biol. 10, 17–24 [DOI] [PubMed] [Google Scholar]
- 2.Prag S., Adams J. C. (2003) BMC Bioinformatics 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor A., Obholz K., Linden G., Sadiev S., Klaus S., Carlson K. D. (1998) Mol. Cell. Biochem. 183, 105–112 [DOI] [PubMed] [Google Scholar]
- 4.Greenberg C. C., Connelly P. S., Daniels M. P., Horowits R. (2008) Exp. Cell Res. 314, 1177–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu S., Carroll S. L., Herrera A. H., Ozanne B., Horowits R. (2003) J. Cell Sci. 116, 2169–2178 [DOI] [PubMed] [Google Scholar]
- 6.Spence H. J., Johnston I., Ewart K., Buchanan S. J., Fitzgerald U., Ozanne B. W. (2000) Oncogene 19, 1266–1276 [DOI] [PubMed] [Google Scholar]
- 7.Spence H. J., McGarry L., Chew C. S., Carragher N. O., Scott-Carragher L. A., Yuan Z., Croft D. R., Olson M. F., Frame M., Ozanne B. W. (2006) Mol. Cell Biol. 26, 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bork P., Doolittle R. F. (1994) J. Mol. Biol. 236, 1277–1282 [DOI] [PubMed] [Google Scholar]
- 9.Ito N., Phillips S. E., Yadav K. D., Knowles P. F. (1994) J. Mol. Biol. 238, 794–814 [DOI] [PubMed] [Google Scholar]
- 10.Li X., Zhang D., Hannink M., Beamer L. J. (2004) J. Biol. Chem. 279, 54750–54758 [DOI] [PubMed] [Google Scholar]
- 11.Angers S., Thorpe C. J., Biechele T. L., Goldenberg S. J., Zheng N., MacCoss M. J., Moon R. T. (2006) Nat. Cell Biol. 8, 348–357 [DOI] [PubMed] [Google Scholar]
- 12.Rondou P., Haegeman G., Vanhoenacker P., Van Craenenbroeck K. (2008) J. Biol. Chem. 283, 11083–11096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo S. C., Li X., Henzl M. T., Beamer L. J., Hannink M. (2006) EMBO J. 25, 3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmanabhan B., Nakamura Y., Yokoyama S. (2008) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa M., Xiong Y. (2005) Mol. Cell Biol. 25, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salinas G. D., Blair L. A., Needleman L. A., Gonzales J. D., Chen Y., Li M., Singer J. D., Marshall J. (2006) J. Biol. Chem. 281, 40164–40173 [DOI] [PubMed] [Google Scholar]
- 17.Williams S. K., Spence H. J., Rodgers R. R., Ozanne B. W., Fitzgerald U., Barnett S. C. (2005) J. Neurosci. Res. 81, 622–631 [DOI] [PubMed] [Google Scholar]
- 18.Copple I. M., Goldring C. E., Kitteringham N. R., Park B. K. (2008) Toxicology 246, 24–33 [DOI] [PubMed] [Google Scholar]
- 19.Zhang D. D., Lo S. C., Sun Z., Habib G. M., Lieberman M. W., Hannink M. (2005) J. Biol. Chem. 280, 30091–30099 [DOI] [PubMed] [Google Scholar]
- 20.Karapetian R. N., Evstafieva A. G., Abaeva I. S., Chichkova N. V., Filonov G. S., Rubtsov Y. P., Sukhacheva E. A., Melnikov S. V., Schneider U., Wanker E. E., Vartapetian A. B. (2005) Mol. Cell Biol. 25, 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chew C. S., Parente J. A., Jr., Zhou C., Baranco E., Chen X. (1998) Am. J. Physiol. 275, C56–67 [DOI] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.DeLano W. L. (2002) Curr. Opin. Struct. Biol. 12, 14–20 [DOI] [PubMed] [Google Scholar]
- 25.Hennigan R. F., Hawker K. L., Ozanne B. W. (1994) Oncogene 9, 3591–3600 [PubMed] [Google Scholar]
- 26.Varkey J. P., Muhlrad P. J., Minniti A. N., Do B., Ward S. (1995) Genes Dev. 9, 1074–1086 [DOI] [PubMed] [Google Scholar]
- 27.Grunewald T. G., Butt E. (2008) Mol. Cancer 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grunewald T. G., Kammerer U., Winkler C., Schindler D., Sickmann A., Honig A., Butt E. (2007) Br. J. Cancer 96, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y. H., Park Z. Y., Lin D., Brahmbhatt A. A., Rio M. C., Yates J. R., 3rd, Klemke R. L. (2004) J. Cell Biol. 165, 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomasetto C., Moog-Lutz C., Régnier C. H., Schreiber V., Basset P., Rio M. C. (1995) FEBS Lett. 373, 245–249 [DOI] [PubMed] [Google Scholar]
- 31.Beamer L. J., Li X., Bottoms C. A., Hannink M. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 1335–1342 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.