Abstract
p63, a p53 family member, plays an essential role in epidermal development by regulating its transcriptional program. Here we report a previously uncovered role of p63 in controlling bone morphogenetic protein (BMP) signaling, which is required for maintaining low expression levels of several non-epidermal genes. p63 represses transcription of the inhibitory Smad7 and activates Bmp7, thereby sustaining BMP signaling. In the absence of p63, compromised BMP signaling leads to inappropriate non-epidermal gene expression in postnatal mouse keratinocytes and in embryonic epidermis. Reactivation of BMP signaling by Smad7 knockdown and/or, to a lesser extent, by BMP treatment suppresses expression of non-epidermal genes in the absence of p63. Canonical BMP/Smad signaling is essential for control of non-epidermal genes as use of a specific inhibitor, or simultaneous knockdown of Smad1 and Smad5 counteract suppression of non-epidermal genes. Our data indicate that p63 prevents ectopic expression of non-epidermal genes by a mechanism involving Smad7 repression and, to a lesser extent, Bmp7 induction, with consequent enhancement of BMP/Smad signaling.
Mouse embryonic skin develops from an initial undifferentiated monolayer of epithelial cells that subsequently undergoes stratification beginning at embryonic day 14.5 (E14.5) (recently reviewed in Ref. 1). During embryogenesis, the surface ectoderm initiates the expression of gene products characteristic of the basal cells of the epidermis, including keratins Krt5 and Krt14. When stratification begins, suprabasal keratins Krt1 and Krt10 start to be expressed in the upper layers, followed by expression of late differentiation markers, including late cornified envelope proteins (Lce). p63 is one of the first genes to be specifically expressed in the surface ectoderm prior to Krt5 and Krt14 expression at E7.5–E8, and continues to be expressed during embryonic skin development and in the basal proliferative layer in postnatal life (2–4). The p63 gene encodes a tetrameric transcription factor that can be expressed in at least six isoforms with widely different transactivation potential that share an identical DNA binding domain (4). Alternative transcription start sites give rise to transactivation (TA)6 isoforms, encoding proteins with a canonical transactivation domain similar to p53, and ΔN isoforms containing an alternative transactivation domain (4, 5). ΔNp63α, the most abundant isoform in isolated keratinocytes and in the epidermis (4), contains both a transactivation domain and an inhibitory domain, and has been shown to activate or repress gene transcription depending on the promoter context (reviewed in Ref. 6). Mice lacking the p63 gene die from dehydration shortly after birth and display cleft palate, limb truncation, and absence of all stratified epithelia, including the epidermis (7, 8), suggesting that p63 plays a non-redundant role in these tissues. Defects in the surface epithelium of p63-null mice have been ascribed to loss of proliferative potential of keratinocyte stem cells (8, 9), and/or altered epidermal stratification and cell differentiation associated with reduced expression levels of Krt5/Krt14 and Krt1/Krt10 (2, 7, 10). In parallel with suppression of epidermal keratins, loss of p63 results in aberrant expression of the simple epithelial keratins Krt8 and Krt18 both in vivo and in vitro (2, 11), suggesting that p63 may be involved in maintaining an epithelial gene expression program in mammalian cells.
In non-mammalian vertebrates BMP signaling is an important determinant of epidermal fate specification, acting as an epidermal inducer and suppressing neural fate in early development (reviewed in Ref. 12). In mammals a putative BMP function in regulating epidermal fate or specific gene expression has not been demonstrated consistent with a possible redundant function among BMP family members or with other signaling pathways. The BMP family consists of more than 20 secreted proteins that belong to the transforming growth factor-β (TGF-β) family. Members of the BMP family are differentially expressed in embryonic skin. Expression of Bmp2 and Bmp4 transcripts in developing murine skin is restricted to the hair follicle epithelium and mesenchyme, respectively (13, 14). Strong expression of Bmp7 mRNA is seen in mouse and rat epidermis during embryonic development from early stages (3, 15), whereas Bmp6 is expressed in suprabasal layers of the embryonic murine epidermis at E15.5 (14). Interestingly, Bmp7 transcripts are strongly down-regulated in the ectoderm of p63 -null mice (3), although the functional significance of this finding has not been addressed.
An important role in BMP/TGF-β signaling is played by the inhibitory Smads, Smad6 and Smad7, which block signaling by several mechanisms including competition with R-Smads for interaction with the activated receptors, ubiquitination, and degradation of the receptors (for a review, see Ref. 16). In addition Smad7 also plays noncanonical functions by regulating several other signaling proteins, including β-catenin (16, 17).
Here we report that p63, and more specifically the ΔNp63α isoform, activates BMP signaling both in vitro and in vivo. p63 directly binds to an evolutionary conserved regulatory region on the Smad7 promoter thereby repressing its expression. At the same time, p63 sustains Bmp7 expression in the epidermis and indirectly controls Bmp4 expression in the dermis. Induction of BMP signaling maintains physiological levels of non-epidermal genes downstream of p63 in a Smad1/5-dependent manner. Taken together, these findings reveal a previously uncovered role of BMP/Smad signaling downstream of p63 in suppressing non-epidermal gene expression in keratinocytes.
EXPERIMENTAL PROCEDURES
Cell Cultures, Constructs, Transfections, Reporter Assays, and Retroviral Infections
Primary mouse keratinocytes were isolated from 2-day-old Swiss CD1 mice and cultured as previously described (18). Transfections were performed 5 days after plating using Lipofectamine 2000 (Invitrogen). Reporter plasmids (250 ng) were co-transfected with pCMV2-FLAG-ΔNp63α or pCMV2-FLAG control (18). A 4.3-kb Smad7 promoter (3.6-kb promoter region and 0.7-kb 5′-untranslated region upstream) (19) was cloned into the XhoI-HindIII sites in the pGL3 reporter plasmid (Promega). The Smad7 fragment (−3.0/−2.6) was generated by deletion of the Smad7 promoter using NheI-PstI and cloned into a pGL3-TKLuc reporter plasmid (20). Mutations in the p63 binding sites were generated using the QuikChange Site-directed mutagenesis kit (Stratagene). Luciferase activity was determined 48 h after transfection with the dual-luciferase reporter assay kit (Promega). pCMV-Renilla reporter (20 ng; Promega) was used to normalize transfection efficiency. A total amount of 200 nm siRNA (Stealth siRNA, Invitrogen) for pan-p63, for specific p63 isoforms (18), and/or for Smad7, Smad1, Smad5, Bmp7, or control medium GC-rich siRNA (Stealth siRNA, Invitrogen) were transfected by Lipofectamine 2000. In some experiments cells were treated with BMP7 (20 ng/ml) (R&D Systems) 24 h after transfection. The BMP type I receptor inhibitor LDN-193189 (200 nm) was given to the cells 30 min before BMP7 addition. High titer retrovirus production was obtained in human embryonic kidney-293T cells by transient transfection of the pBABE-Smad7 (21) using Lipofectamine 2000 as previously described (20). Primary keratinocytes were infected in the presence of 8 μg/ml Polybrene (Sigma), selected with 2 μg/ml puromycin for 48 h, and grown after selection for an additional 24 h in the absence of puromycin.
Analysis of Gene Expression, Real Time RT-PCR, and ChIP
Gene expression profiling was obtained in p63 knockdown versus control keratinocytes 48 h after transfection using Affymetrix Mouse Genome 430A 2.0 chips and analyzed as reported (22). Among the genes affected by loss of p63, 106 genes were up-regulated more than 5-fold by p63 knockdown (supplemental Table S1). Tissue expression profiling of up-regulated genes was obtained from a custom made mouse GNF1M (MAS5) GNF gene expression data base (23). Among 72 genes that were up-regulated more than 5-fold by p63 knockdown (false discovery rate < 0.25), 48 (67%) were not expressed in normal epidermis, but rather in other tissues. Total RNA was extracted 48 h after transfection from primary keratinocytes using TRIzol reagent (Invitrogen), and from mouse embryonic skin (E14.5) using the RNAspin Mini RNA isolation kit (GE Healthcare) according to the manufacturer's instruction. RNA samples were treated with RNase-free DNase I (Promega), and cDNA was synthesized using SuperScript Vilo (Invitrogen). Two-step real time RT-PCR was performed using the SYBR Green PCR master mixture in an ABI PRISM 7500 (Applied Biosystems). Levels of the target genes were quantified using specific oligonucleotide primers and normalized for glyceraldehyde-3-phosphate dehydrogenase or anti-β-actin expression as indicated in the figure legends. Approximately 3 × 106 primary keratinocytes were fixed with 1% formaldehyde and ChIP was performed using anti-p63 antibodies (H-137, Santa Cruz Biotechnology), and anti-ERK1 antibodies as negative control. ChIP and real time PCR were performed as previously described (18). For oligonucleotide sequences, see supplemental Table S2.
Immunostaining, Immunoblotting, and in Situ Hybridization
Embryos were fixed in 4% paraformaldehyde and either embedded in OCT (Sakura) or paraffin. Fluorescent signals were monitored under a Zeiss confocal microscope LSM510meta using a Zeiss EC Plan-Neofluar ×40/1.3 oil immersion objective. For immunoblotting cells were lysed in sample buffer or in 1% Triton X-100 lysis buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA) for Smad7, and protein extracts were run on SDS-PAGE gels, transferred on Immobilon-P transfer membranes (Millipore), probed with the indicated antibodies, and detected by chemiluminescence (ECL, GE Healthcare). For a full list of antibodies, see the supplemental data. In situ hybridization was performed on frozen sections of P1 skin and E14.5 embryos as previously described (24). Digoxigenin-labeled antisense and sense RNA probes were transcribed from the SP6 and T7 promoters using a digoxigenin labeling kit (Roche Applied Science) according to manufacturer's instructions. The Smad7 probe corresponding to a 485-bp cDNA fragment was kindly provided by Xiao-Jing Wang (17). The Bmp6 probe was synthesized from a 893-bp SacI-EcoRI cDNA fragment kindly provided by Dr. M. Mikkola. The Bmp7 probe corresponds to a 440-bp long cDNA fragment generated by PCR and inserted in the pCR®II vector (Invitrogen). For PCR oligonucleotide sequences, see supplemental Table S2. Digoxigenin labeling was monitored under a Zeiss Axioskop2 plus microscope using a Zeiss Plan-Neofluar ×20/0.50 objective.
Mice and Skin Explants
All experiments performed with mice were conducted under IACUC approval. p63-null mice (B6.129S7-Trp63tm1Brd/J) were obtained from the Jackson Laboratory. For skin explants, dorsal skins from embryos at E14.5 were laid on a culture plate insert (Millipore) and cultured in the presence or absence of BMP7 in Dulbecco's modified Eagle's medium with or without 5% fetal bovine serum, overnight at 37 °C and 5% CO2.
RESULTS
p63 Positively Controls BMP Signaling
To gain insight into the role of p63 in controlling gene expression in epidermal cells, we recently identified a large number of putative p63 target genes in primary mouse keratinocytes (22). A subset of genes encoding for members of the BMP family and their regulators were differentially expressed in p63 knockdown versus control keratinocytes.
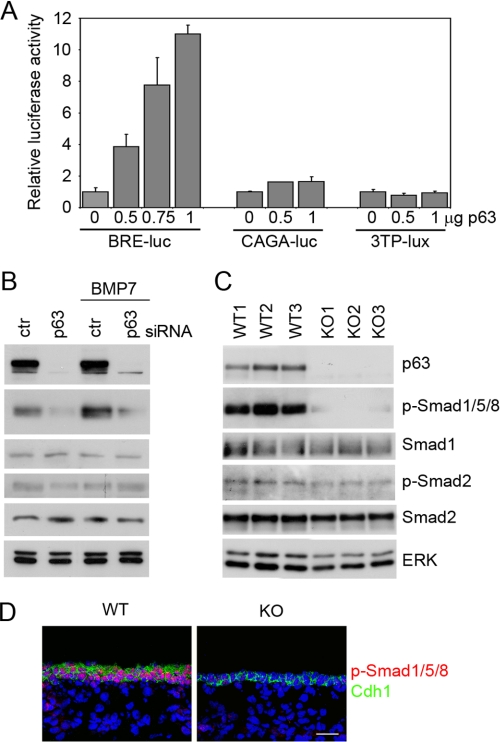
To assess whether p63 affected BMP or TGF-β signaling, we first co-transfected varying amounts of a ΔNp63α-expressing plasmid together with a luciferase reporter gene under the control of either BMP (BRE) or TGF-β (CAGA and 3TP) responsive elements in primary mouse keratinocytes. As shown in Fig. 1A, expression of p63 resulted in a dose-dependent induction of the BMP-responsive element, without affecting the TGF-β ones. In parallel, p63 knockdown resulted in a significant reduction in Smad1/5/8 phosphorylation under basal conditions and upon BMP7 treatment (Fig. 1B). In contrast, p63 knockdown elicited no effect on Smad2 phosphorylation, a direct TGF-β effector, either under basal conditions or upon TGF-β1 stimulation (Fig. 1B and data not shown).
FIGURE 1.
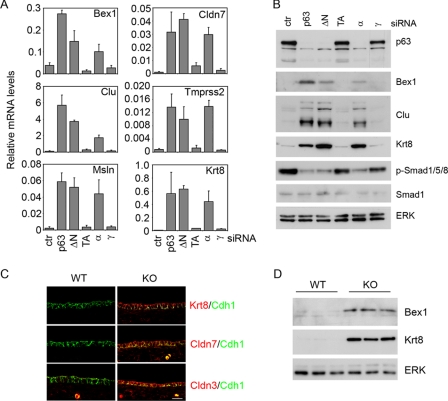
p63 positively regulates BMP signaling. A, different amounts of the ΔNp63α expression vector were co-transfected in primary mouse keratinocytes as indicated with the BMP-responsive reporter BRE-luc (50), or with TGF-β responsive reporters CAGA-luc (51) or 3TP-lux (52), and examined 48 h after transfection. Values are normalized to Renilla luciferase activity and are expressed as fold-changes over the promoter activity in the absence of ΔNp63α, and represent mean ± S.E. of three independent experiments. B, immunoblotting of primary keratinocytes transfected with p63 siRNA or control siRNA for 48 h, and either treated for 24 h with BMP7 (20 ng/ml) or left untreated, and probed with the indicated antibodies. Activation of BMP and TGF-β signaling was measured using phosphorylation-specific antibodies for Smad1/5/8 (p-Smad1/5/8) and Smad2 (p-Smad2), respectively. Total Smad1 and Smad2 expression is shown. ERK was used as loading control. C, immunoblotting of total skin extracts isolated from wild-type (WT) and p63-null embryos (KO) at E14.5, probed with the indicated antibodies. D, immunofluorescence staining was performed with anti-p-Smad1/5/8 (red) and anti-E-cadherin antibodies (Cdh1, green) in WT and p63 KO at E15.5, and detected by confocal microscopy. Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). Scale bar is 20 μm.
To assess whether p63 regulates BMP signaling during embryonic skin development, we measured Smad1/5/8 phosphorylation in p63-null and in wild-type skin. Immunoblotting analysis and immunofluorescence staining using anti-phospho-Smad1/5/8 revealed strong activation of BMP signaling in the developing wild-type epidermis, whereas the Smad1/5/8 signal was dramatically reduced in the p63-null epidermis (Fig. 1, C and D). In contrast Smad2 phosphorylation was similar in wild-type and p63-null skin at this embryonic stage (Fig. 1C). Taken together, these data indicate that p63 positively regulates BMP signaling in primary mouse keratinocytes and in embryonic epidermis, without significantly affecting TGF-β signaling.
p63 Transcriptionally Controls Smad7 and Bmp7 Gene Expression
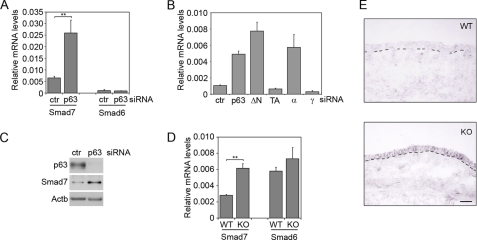
Our previous gene expression profiling indicated that p63 knockdown might affect Smad7 expression (22). Given the key role of Smad7 in regulating BMP signaling, we sought to confirm and expand this finding. p63 knockdown in keratinocytes caused a significant induction of Smad7 both at the RNA and at the protein levels, without affecting expression of the related gene Smad6 (Fig. 2, A and C). In addition, specific knockdown of ΔNp63 and p63α isoforms induced Smad7 expression, whereas knockdown of TAp63 and p63γ isoforms had no effect (Fig. 2B), indicating that the ΔNp63α isoform is required for Smad7 repression. In parallel with these findings, Smad7 was expressed at low levels in wild-type embryonic and newborn epidermis (Fig. 2, D and E, and supplemental Fig. S1A), whereas its expression was significantly increased in the p63-null epidermis (Fig. 2, D and E).
FIGURE 2.
Smad7 is repressed by p63. A, Smad7 and Smad6 mRNA levels were measured by real time RT-PCR in primary keratinocytes transfected with p63 (p63) or control siRNA (ctr). Cells were collected 48 h after transfection. Values are expressed as glyceraldehyde-3-phosphate dehydrogenase-normalized mRNA levels, and represent the mean of independent experiments ± S.E. (**, p < 0.005, n = 8). B, Smad7 expression was measured in primary mouse keratinocytes transfected with siRNA specific for pan-p63, ΔNp63, TAp63, p63α, and p63γ isoforms, or control. RNA levels were expressed as in A. Isoform-specific siRNA specifically inhibited the corresponding isoforms (data not shown). C, Smad7 protein levels were measured in p63 knockdown and control keratinocytes 48 h after transfection. Protein extracts were normalized using anti-β-actin (Actb) antibodies. D, Smad7 and Smad6 mRNA levels were measured by real time RT-PCR in p63 KO and wild type (WT) skin at E14.5 (**p < 0.005, n = 5 embryos). E, RNA in situ hybridization of mouse embryonic skin sections at E14.5 from WT and p63 knock-out (KO) using a digoxigenin-labeled antisense probe for mouse Smad7. The dashed line indicates the dermal-epidermal junction. A Smad7 sense probe gave no detectable signal under the same conditions (data not shown). Scale bar is 50 μm.
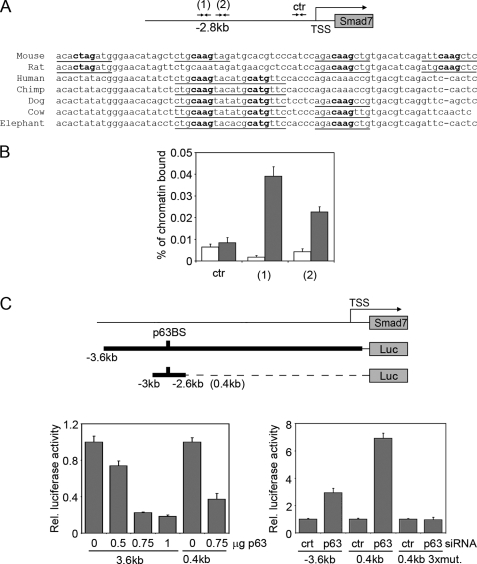
To test the possibility that p63 might directly regulate Smad7, we tested a putative high affinity p63-binding region located at −2.8 kb from the transcription start site, which emerged from our previous ChIP-on-ChIP experiment in a 48.5-kb genomic sequence spanning the Smad7 locus (22). The p63-binding region was centered on an evolutionary conserved genomic sequence containing four canonical p63 binding hemisites (Fig. 3A). ChIP followed by real time PCR with two independent sets of oligonucleotides confirmed that p63 specifically bound this genomic region (Fig. 3B). In addition, p63 overexpression significantly inhibited the activity of a 3.6-kb Smad7 promoter, as well as the activity of a 0.4-kb fragment containing the p63 binding sites (Fig. 3C, left panel). Conversely, p63 knockdown resulted in enhancement of the activity both of the Smad7 promoter and the fragment containing the p63 binding sites (Fig. 3C, right panel). Mutations in three canonical p63 binding hemisites in the promoter fragment abolished the ability of p63 knockdown to enhance promoter activity. Taken together, these data strongly suggest that p63 directly represses Smad7 in primary mouse keratinocytes and in embryonic skin.
FIGURE 3.
Smad7 is a transcriptional target of p63. A, a conserved p63-binding site is located at −2.8 kb from the Smad7 transcription start site. The predicted p63-binding hemisites are indicated with their nucleotide sequence and phylogenetic conservation in multiple species. Bold nucleotides correspond to the core nucleotide sequence required for p63 binding, whereas underlined nucleotides are matches in the consensus (22, 53). B, specific binding of endogenous p63 to the mouse Smad7 promoter. Primary keratinocytes were processed for ChIP with antibodies specific for p63 (gray bars), or unrelated anti-ERK antibodies as control (white bars), followed by real-time PCR amplification using oligonucleotide primers designed at the indicated position from the transcription start site. The amount of precipitated DNA was calculated relative to the total input chromatin, and expressed as the percentage of the total DNA, as previously described (54). C, the activity of a 3.6-kb Smad7 promoter region and its 0.4-kb fragment spanning the p63-binding sites (p63BS) were measured in the presence of the indicated amounts of a ΔNp63α-expressing construct (p63) (left panel), or p63 or control (ctr) siRNA (right panel). A 0.4-kb fragment containing 2-bp mutations in each of the first three binding hemisites indicated in A (0.4kb 3xmut) was also co-transfected with p63 or control siRNA. Values are expressed as described in Fig. 1A.
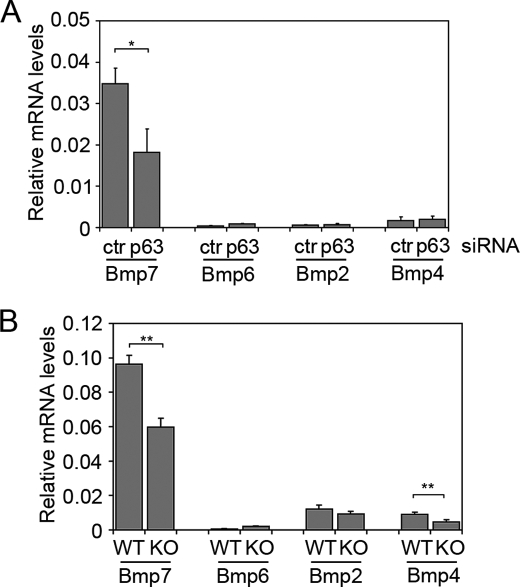
Next we analyzed the expression of selected BMP family members known to be expressed in skin, in the presence or absence of p63. Bmp2 and Bmp4 were poorly expressed in primary mouse keratinocytes and their expression was unaffected by p63 knockdown (Fig. 4A). Bmp6 was modestly induced by p63 knockdown, however, its expression was very low under non-differentiating conditions, consistent with its suprabasal expression in newborn skin (supplemental Fig. S1A) and in the developing murine epidermis (14, 25). In contrast Bmp7 was by far the most highly expressed BMP family member in keratinocytes, and its expression was inhibited ∼50% by p63 knockdown. Consistent with this observations, low levels of Bmp6 were modestly up-regulated in p63-null embryonic skin at E14.5, whereas Bmp2 was not significantly altered (Fig. 4B). Interestingly, Bmp4, which is mainly expressed in the mesenchymal component of the skin, was reduced in p63-null skin, suggesting that p63 may indirectly control Bmp4 expression in the adjacent dermis, thus possibly contributing to the reduction in BMP signaling observed in the p63-null epidermis. Strong signaling for Bmp7 mRNA was detected by in situ hybridization both in the basal layer of newborn epidermis and in embryonic epidermis (supplemental Fig. S1, A and B). Bmp7 was previously reported to be absent in p63-null skin at E14 by in situ hybridization, although no quantitative data were provided (3). We observed a similar, although more modest, reduction of Bmp7 transcript in p63-null versus wild-type skin at E14.5 as assessed by real time RT-PCR and in situ hybridization (Fig. 4B and supplemental Fig. S1B), in agreement with the reduction observed in p63 knockdown keratinocytes. ChIP analysis revealed that p63 bound to an evolutionary conserved genomic region in intron 1 (supplemental Fig. S1C), previously shown to bind all p53 family members in a breast cancer cell line (26). Taken together these data demonstrate that p63 regulates Smad7 and Bmp7 in epidermal cells, inducing one and repressing the other, by direct binding to specific consensus sequences in highly conserved genomic regions proximal to their transcription start sites.
FIGURE 4.
Bmp7 is the most abundant BMP family member in keratinocytes and embryonic skin and is down-regulated in the absence of p63. A, expression levels of the indicated BMPs were measured by real time RT-PCR in primary keratinocytes transfected with p63 or control (ctr) siRNA as described in the legend to Fig. 2A. Values represent mean of independent experiments ± S.E. (*, p < 0.05, n = 4). B, BMP mRNA levels were measured in p63 knockout (KO) and wild-type (WT) skin at E14.5. Bmp7 and Bmp4 are significantly down-regulated in p63 KO skin (**, p < 0.005, n = 4 embryos).
Loss of p63 Induces Expression of Non-epidermal Genes in Primary Keratinocytes and in Embryonic Epidermis
Global gene expression profiling of p63 knockdown versus control keratinocytes (22) revealed that p63 negatively regulates a set of genes that are preferentially expressed in other tissues (non-epidermal genes), including the previously reported Krt8 and Krt18 (11, 27). 67% of the genes up-regulated more than 5 times in the absence of p63 were expressed at very low levels in wild-type keratinocytes, and were enriched in genes expressed in early embryonic development, in simple epithelia, or in neural tissues (supplemental Table S1).
Aberrant expression of non-epidermal genes in p63 knockdown keratinocytes was confirmed at the RNA and protein levels for some relevant genes (Fig. 5, A and B, and data not shown). Knockdown of the ΔNp63 and the p63α isoforms strongly induced expression of non-epidermal genes and their gene products and at the same time inhibited Smad1/5/8 phosphorylation, whereas knockdown of TAp63 and p63γ isoforms had no effect. To test whether the effect elicited by p63 knockdown in primary keratinocytes correlated with p63 function in vivo, we analyzed expression of non-epidermal genes in total embryonic skin of p63-null embryos and their wild-type counterparts at E13.5 and E14.5. Importantly, non-epidermal gene products were strongly expressed in the p63-null epidermis but not in wild-type epidermis (Fig. 5, C and D). Taken together these data indicate that p63 is required to suppress several non-epidermal genes during embryonic skin development and in postnatal keratinocytes, and that this regulation is unlikely to occur directly.
FIGURE 5.
Aberrant expression of non-epidermal genes in the absence of p63. A, expression of the indicated non-epidermal genes was measured in primary mouse keratinocytes transfected with siRNA specific for pan-p63, ΔNp63, TAp63, p63α, and p63γ isoforms, or control (ctr). RNA levels were expressed as described in the legend to Fig. 2A. Efficient knockdown for the specific isoforms was demonstrated at the mRNA level (data not shown). B, immunoblotting of total cell lysates prepared from primary keratinocytes transfected with siRNA as in A. C, immunofluorescence analysis of the indicated non-epidermal proteins (in red) and E-cadherin (Cdh1, in green) in dorsal skin of p63 knockout (KO) and wild-type (WT) embryos at E13.5. Nonspecific staining in the dermis is due to some autofluorescent cells. Scale bar is 30 μm. D, immunoblotting of total cell lysates prepared from the dorsal skin of p63 KO and WT embryos at E14.5 and probed with the indicated antibodies.
p63 Represses Non-epidermal Genes through a Canonical BMP/sSmad-dependent Mechanism
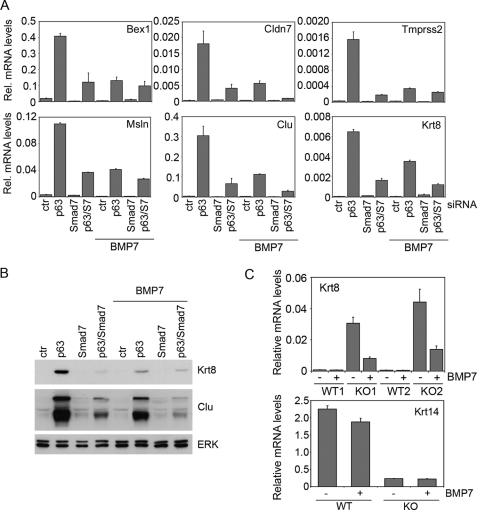
Non-epidermal genes were not identified as early target genes upon p63 activation (22), and their induction occurred at a late interval upon p63 knockdown (data not shown), suggesting that they are not directly regulated by p63. Because in lower vertebrates BMP signaling has been shown to be a crucial determinant of the epidermal cell fate during ectodermal development by suppressing alternative fates, we hypothesized that p63 could maintain low levels of non-epidermal genes via a BMP-dependent mechanism. To this end we examined whether re-activation of BMP signaling in p63 knockdown keratinocytes by either Smad7 knockdown and/or BMP treatment could restore physiological levels of non-epidermal genes. Smad7 knockdown resulted in a significant re-activation of Smad1/5/8 signaling in the absence of p63, without affecting Smad2 activation (supplemental Fig. S2A). Conversely, Smad7 overexpression inhibited BRE-Luc activity without affecting CAGA-Luc (supplemental Fig. S2B). In parallel, Smad7 knockdown resulted in strong down-regulation of non-epidermal gene expression in p63 knockdown keratinocytes both at the RNA and protein levels (Fig. 6, A and B), suggesting that Smad7 depletion counteracted the effect of p63 knockdown. A similar counteracting effect was also observed with a second distinct siRNA oligonucleotide against Smad7 (supplemental Fig. S2C), confirming the specificity of this effect. Inhibition of non-epidermal genes was also observed by treating p63 knockdown keratinocytes with BMP7, BMP4, or BMP6 (Fig. 6, A and B, and data not shown). Concomitant Smad7 knockdown and BMP treatment had little additional effect on the expression of non-epidermal markers as compared with each treatment alone, suggesting that these treatments repressed non-epidermal genes through overlapping mechanisms. Neither Smad7 knockdown nor BMP treatment affected p63 expression (data not shown), excluding a feedback loop mechanism. TGF-β treatment under the same experimental conditions did not suppress non-epidermal genes (data not shown).
FIGURE 6.
Re-activation of BMP signaling restores low levels of non-epidermal genes in p63 knockdown keratinocytes and p63 KO skin explants. A, expression of the indicated non-epidermal genes was measured in primary keratinocytes transfected with p63, Smad7, p63 and Smad7 (p63/S7), or control (ctr) siRNA, and cultured for 48 h. Cells were either treated with BMP7 for the last 24 h or left untreated. Relative mRNA levels was measured as described in the legend to Fig. 2A. B, total protein extracts from primary keratinocytes treated as in A were subjected to immunoblotting with the indicated specific antibodies. Protein extracts were normalized using anti-ERK polyclonal antibodies. C, skin explants at E14.5 were cultured overnight and then treated with BMP7 (80 ng/ml) or left untreated for an additional 24 h in the absence (WT1 and KO1) or presence of fetal bovine serum (WT2 and KO2). Krt8 and Krt14 mRNA levels were evaluated by real time RT-PCR and normalized as in A. Data presented are the average of two independent experiments and the S.E. is indicated. KO, knockout.
To test whether BMP signaling functions specifically downstream of p63 to regulate non-epidermal genes, or has a broader compensatory role on p63 downstream targets, we measured expression of the Lce genes (or Sprrl), which encode epidermal markers involved in late differentiation and are up-regulated by p63 knockdown (supplemental Table S1). Neither BMP treatment nor Smad7 knockdown rescued the effect of p63 knockdown on Lce genes (supplemental Fig. S2D), indicating that BMP signaling selectively restored low levels of non-epidermal genes.
To demonstrate that loss of BMP signaling contributes to the expression of non-epidermal genes in p63-null epidermis, skin explants were isolated at E14.5 from p63-null and wild-type mice and cultured with or without BMP7. BMP7 treatment significantly down-regulated Krt8 expression in p63-null skin (Fig. 6C), consistent with a role of BMP signaling in repressing non-epidermal genes in the embryonic epidermis. In contrast, BMP7 treatment was insufficient to rescue loss of Krt14 expression in p63-null skin explants, indicating that BMP7 elicited a selective effect on non-epidermal genes in embryonic epidermis.
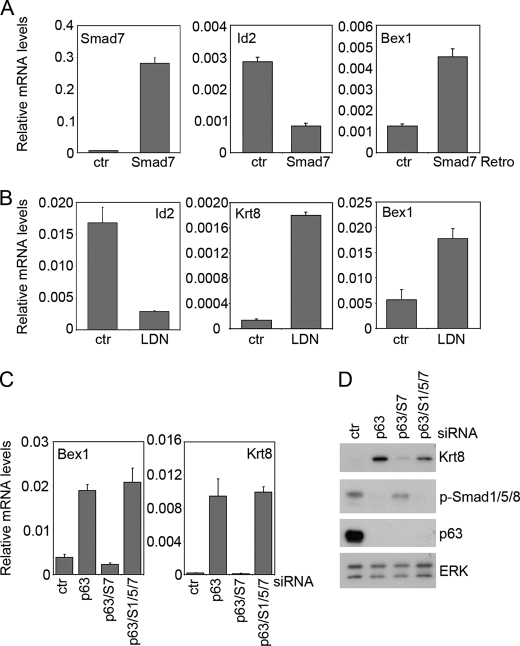
We then explored the possibility that loss of BMP signaling may be sufficient to cause aberrant expression of non-epidermal genes. Smad7 overexpression up-regulated non-epidermal genes in primary keratinocytes (Fig. 7A and data not shown), although to a lesser extent than p63 knockdown, while inhibiting the expression of the BMP direct target gene Id2 as expected. Treatment with LDN-193189, a selective small molecule inhibitor of the BMP type I receptor kinases (28), similarly induced expression of non-epidermal genes and repressed Id2 (Fig. 7B), whereas SB431542, a selective inhibitor of the TGF-β type I receptor did not (data not shown). However, in the embryonic epidermis Smad7 overexpression (17) or homozygous deletion of Bmp7 (29) were insufficient to induce non-epidermal genes (data not shown). Taken together, these data indicate that p63 represses non-epidermal genes in a BMP-dependent manner, and that, at least in isolated keratinocytes, loss of BMP signaling by itself leads to induction of non-epidermal genes.
FIGURE 7.
Canonical BMP/Smad pathway regulates the expression of non-epidermal genes. A, expression of the indicated genes was measured by real time RT-PCR in primary keratinocytes infected with a Smad7 expressing or a control retrovirus. B, expression of the indicated genes was measured by real time RT-PCR in primary keratinocytes treated with 200 nm LDN-193189 for 24 h. C, expression of the indicated non-epidermal genes was measured by real time RT-PCR in p63 knockdown primary keratinocytes in the presence or absence of siRNA targeting Smad7 (S7), Smad1 (S1), and Smad5 (S5) in the indicated combinations. D, expression of the indicate proteins was measured by immunoblotting in primary mouse keratinocytes co-transfected with the indicated combinations of siRNA. Protein extracts were normalized using ERK antibodies.
BMP-mediated receptor activation leads to induction of a canonical signaling pathway mediated by Smad1/5/8 and to activation of other signaling molecules, including p38 mitogen-activated protein kinase (MAPK). Similarly, Smad7 has also been shown to regulate R-Smad independent pathways as discussed above. Among the BMP-responsive Smads, Smad1 and Smad5 were highly expressed in primary mouse keratinocytes (supplemental Fig. S3, A and B). Concomitant Smad1/5 knockdown inhibited Id1 and Id2 expression, consistent with decreased BMP/Smad signaling, whereas knockdown of either Smad1 or Smad5 alone was insufficient to elicit any effect (supplemental Fig. S3C and data not shown). We then tested whether activation of Smad1/5 is required for suppression of non-epidermal genes. Concomitant Smad1/5 depletion completely restored high levels of non-epidermal genes in the absence of p63 and Smad7 (Fig. 7, C and D). A requirement for Smad1/5 was similarly obtained upon treatment with BMP7 (data not shown). Thus the canonical BMP/Smad signaling pathway is required for suppression of non-epidermal genes downstream of p63.
DISCUSSION
p63 acts as a crucial regulator of gene expression in the epidermis and is essential for epidermal development, however, its interaction with signaling pathways involved in epidermal development is still poorly understood. We previously demonstrated a cross-talk between p63 and Notch, in which p63 transcriptionally represses the Notch effector Hes1, thus regulating keratinocyte differentiation (30). Here we establish a novel function of p63 in repressing non-epidermal genes through a mechanism that involves BMP/Smad signaling. We show that besides the previously reported Krt8 and Krt18, many other non-epidermal genes are induced by loss of p63 in epidermal cells both in vitro and in vivo. Interestingly, loss of ΔNp63α expression in human keratinocytes also induces the expression of a subset of non-epidermal genes (31), further reinforcing the notion that p63 may participate in maintaining an epithelial gene expression program in mammalian cells.
BMP7 was recently described as a novel direct target of the p53 family in breast cancer cells (26). BMP7 is induced by p53, p63, and p73, and all three members of the family directly bind to the p53 responsive element located in the BMP7 intron 1. In keratinocytes, we find that Bmp7 is inhibited in the absence of p63, although the remaining levels of Bmp7 are still fairly high, possibly suggesting that other p53 family members are responsible for its basal expression in keratinocytes. BMP7 promotes cell survival in p53-deficient breast carcinoma cells, at least in part, through Id2 (26), and has been demonstrated to promote cell survival in other contexts and tissues (32–35). Survival of epidermal cells is compromised in the absence of p63 (36). Thus, besides playing a role in maintaining epidermal identity, an interesting possibility that will require further studies is that Bmp7 may cooperate with other p63 downstream target genes to promote epidermal cell survival.
We demonstrate that p63 positively regulates BMP signaling both in isolated keratinocytes as well as in the embryonic epidermis. We provide entirely novel and compelling evidence that p63 directly suppresses the inhibitory Smad7 by binding to a highly conserved genomic region in the Smad7 promoter. Consistent with this notion, Smad7 is poorly expressed in the developing epidermis and is induced in the p63-null epidermis. It has been previously reported that Smad7 induces expression of Cripto-1, and that both Smad7 and Cripto-1 induce Krt8 expression in mouse keratinocytes (21, 37). Loss of p63 does not affect Cripto-1 expression,7 suggesting that other molecules downstream of Smad7 are required for repressing non-epidermal genes. Accordingly, we show that aberrant expression of non-epidermal genes seen in p63-deficient cells can be reversed by Smad7 knockdown or, to a lesser extent, by BMP7 treatment and that this effect is dependent on the Smad1/5 canonical pathway. The lack of synergy between BMP7 treatment and Smad7 knockdown suggests a threshold response, for which a critical level of BMP signaling is necessary, but additional signaling does not further increase the response.
Loss of BMP signaling by itself induces aberrant expression of non-epidermal genes in cultured keratinocytes, whereas Bmp7 deletion or Smad7 overexpression are insufficient to induce non-epidermal genes in embryonic epidermis. A role for BMP signaling in epidermal development has remained elusive despite the strong Smad1/5/8 phosphorylation observed in the interfollicular epidermis during embryogenesis (present work and Ref. 38). Lack of an epidermal phenotype in mice carrying a deletion of single components of the pathway may be due at least in part to functional redundancy within the pathway. Among the BMP receptors, Bmpr1A/ALK3 plays a crucial role in hair follicle development with little phenotype in the epidermis (39–41). However, Bmpr1B/ALK6 and Acvr1/ALK2 are also expressed in the embryonic epidermis (42–44), and Acvr1/ALK2 is the main receptor for BMP7 (45–47), suggesting that depletion of multiple receptors may be required to observe an epidermal phenotype.
In addition to decreased Bmp7 and to the concomitant induction of Smad7, we observed that Bmp4 is down-regulated in the p63-null skin at E14.5. Bmp4 is poorly expressed in keratinocytes and is unaffected by p63 knockdown, indicating that as yet unidentified indirect signals derived from the p63-null epidermis inhibit Bmp4 expression in the dermis. BMP7 and BMP4 are capable of forming heterodimers 3–10 times more active than either BMP4 or BMP7 homodimers (48), suggesting that the concomitant reduction in both molecules observed in the p63-null skin could have more severe consequences on BMP signaling in the epidermis than loss of BMP7 alone. Interestingly, Bmp7 deletion in the urethra is by itself sufficient to induce Krt8 expression (49), whereas in the developing epidermis multiple signaling molecules may be involved in this function. Thus our data indicate that BMP signaling plays a crucial and selective role in suppressing the expression of non-epidermal genes downstream of p63 in embryonic and postnatal epidermal cells, possibly in conjunction with other as yet unidentified mechanisms. Further assessment of the role of BMP signaling in the embryonic epidermis will be a key subject for subsequent investigations.
Supplementary Material
Acknowledgments
We thank Tommaso Russo and Domenico Salvatore for critical reading of the manuscript, and Xiao-Jing Wang for providing the Smad7 riboprobe and tissue sections from gene-switch Smad7 transgenic mice. We are grateful to Leif Oxburg for providing Bmp7-null embryos, Marja L. Mikkola for sharing the skin explant protocol and the Bmp6 riboprobe, Adam Glick for providing the pBABE-Smad7 construct, Frank Margolis for Bex1 polyclonal antibodies, and Yan Chen for providing the Smad7-βGal construct. We also thank the CEINGE Dynamic Imaging Microscopy and the histology core facility.
This work was supported in part by Italian Telethon Foundation Grant GGP06243 and the National Foundation for Ectodermal Dysplasia.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
L. De Rosa, D. Antonini, G. Ferone, M. T. Russo, P. B. Yu, R. Han, and C. Missero, unpublished observations.
- TA
- transactivation
- TGF-β
- transforming growth factor-β
- BMP
- bone morphogenetic protein
- siRNA
- small interfering RNA
- RT
- reverse transcriptase
- ChIP
- chromatin immunoprecipitation
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Fuchs E. (2007) Nature 445, 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koster M. I., Kim S., Mills A. A., DeMayo F. J., Roop D. R. (2004) Genes Dev. 18, 126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laurikkala J., Mikkola M. L., James M., Tummers M., Mills A. A., Thesleff I. (2006) Development 133, 1553–1563 [DOI] [PubMed] [Google Scholar]
- 4.Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 5.Helton E. S., Zhu J., Chen X. (2006) J. Biol. Chem. 281, 2533–2542 [DOI] [PubMed] [Google Scholar]
- 6.Perez C. A., Pietenpol J. A. (2007) Cell Cycle 6, 246–254 [DOI] [PubMed] [Google Scholar]
- 7.Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999) Nature 398, 708–713 [DOI] [PubMed] [Google Scholar]
- 8.Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) Nature 398, 714–718 [DOI] [PubMed] [Google Scholar]
- 9.Senoo M., Pinto F., Crum C. P., McKeon F. (2007) Cell 129, 523–536 [DOI] [PubMed] [Google Scholar]
- 10.Romano R. A., Ortt K., Birkaya B., Smalley K., Sinha S. (2009) PLoS ONE 4, e5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truong A. B., Kretz M., Ridky T. W., Kimmel R., Khavari P. A. (2006) Genes Dev. 20, 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stern C. D. (2006) Curr. Opin. Cell Biol. 18, 692–697 [DOI] [PubMed] [Google Scholar]
- 13.Bitgood M. J., McMahon A. P. (1995) Dev. Biol. 172, 126–138 [DOI] [PubMed] [Google Scholar]
- 14.Lyons K. M., Pelton R. W., Hogan B. L. (1989) Genes Dev. 3, 1657–1668 [DOI] [PubMed] [Google Scholar]
- 15.Takahashi H., Ikeda T. (1996) Dev. Dyn. 207, 439–449 [DOI] [PubMed] [Google Scholar]
- 16.Hoover L. L., Kubalak S. W. (2008) Sci. Signal. 1, pe48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han G., Li A. G., Liang Y. Y., Owens P., He W., Lu S., Yoshimatsu Y., Wang D., Ten Dijke P., Lin X., Wang X. J. (2006) Dev. Cell 11, 301–312 [DOI] [PubMed] [Google Scholar]
- 18.Antonini D., Dentice M., Mahtani P., De Rosa L., Gatta G. D., Mandinova A., Salvatore D., Stupka E., Missero C. (2008) J. Invest. Dermatol. 128, 1676–1685 [DOI] [PubMed] [Google Scholar]
- 19.Liu X., Chen Q., Kuang C., Zhang M., Ruan Y., Xu Z. C., Wang Z., Chen Y. (2007) Biochim. Biophys. Acta 1769, 149–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonini D., Rossi B., Han R., Minichiello A., Di Palma T., Corrado M., Banfi S., Zannini M., Brissette J. L., Missero C. (2006) Mol. Cell. Biol. 26, 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Lee J., Cooley M., Bhogte E., Hartley S., Glick A. (2003) Cancer Res. 63, 7760–7768 [PubMed] [Google Scholar]
- 22.Della Gatta G., Bansal M., Ambesi-Impiombato A., Antonini D., Missero C., di Bernardo D. (2008) Genome Res. 18, 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brancaccio A., Minichiello A., Grachtchouk M., Antonini D., Sheng H., Parlato R., Dathan N., Dlugosz A. A., Missero C. (2004) Hum. Mol. Genet. 13, 2595–2606 [DOI] [PubMed] [Google Scholar]
- 25.Wall N. A., Blessing M., Wright C. V., Hogan B. L. (1993) J. Cell Biol. 120, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan W., Chen X. (2007) Cancer Res. 67, 9117–9124 [DOI] [PubMed] [Google Scholar]
- 27.Koster M. I., Dai D., Marinari B., Sano Y., Costanzo A., Karin M., Roop D. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3255–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu P. B., Deng D. Y., Lai C. S., Hong C. C., Cuny G. D., Bouxsein M. L., Hong D. W., McManus P. M., Katagiri T., Sachidanandan C., Kamiya N., Fukuda T., Mishina Y., Peterson R. T., Bloch K. D. (2008) Nat. Med. 14, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudley A. T., Lyons K. M., Robertson E. J. (1995) Genes Dev. 9, 2795–2807 [DOI] [PubMed] [Google Scholar]
- 30.Nguyen B. C., Lefort K., Mandinova A., Antonini D., Devgan V., Della Gatta G., Koster M. I., Zhang Z., Wang J., Tommasi di Vignano A., Kitajewski J., Chiorino G., Roop D. R., Missero C., Dotto G. P. (2006) Genes Dev. 20, 1028–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbieri C. E., Tang L. J., Brown K. A., Pietenpol J. A. (2006) Cancer Res. 66, 7589–7597 [DOI] [PubMed] [Google Scholar]
- 32.Mitu G. M., Wang S., Hirschberg R. (2007) Am. J. Physiol. Renal Physiol. 293, F1641–F1648 [DOI] [PubMed] [Google Scholar]
- 33.Vukicevic S., Basic V., Rogic D., Basic N., Shih M. S., Shepard A., Jin D., Dattatreyamurty B., Jones W., Dorai H., Ryan S., Griffiths D., Maliakal J., Jelic M., Pastorcic M., Stavljenic A., Sampath T. K. (1998) J. Clin. Invest. 102, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yabe T., Samuels I., Schwartz J. P. (2002) J. Neurosci. Res. 68, 161–168 [DOI] [PubMed] [Google Scholar]
- 35.Yang S., Pham L. K., Liao C. P., Frenkel B., Reddi A. H., Roy-Burman P. (2008) Cancer Res. 68, 198–205 [DOI] [PubMed] [Google Scholar]
- 36.Keyes W. M., Wu Y., Vogel H., Guo X., Lowe S. W., Mills A. A. (2005) Genes Dev. 19, 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shukla A., Ho Y., Liu X., Ryscavage A., Glick A. B. (2008) Mol. Cancer Res. 6, 509–516 [DOI] [PubMed] [Google Scholar]
- 38.Mou C., Jackson B., Schneider P., Overbeek P. A., Headon D. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9075–9080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andl T., Ahn K., Kairo A., Chu E. Y., Wine-Lee L., Reddy S. T., Croft N. J., Cebra-Thomas J. A., Metzger D., Chambon P., Lyons K. M., Mishina Y., Seykora J. T., Crenshaw E. B., 3rd, Millar S. E. (2004) Development 131, 2257–2268 [DOI] [PubMed] [Google Scholar]
- 40.Kobielak K., Pasolli H. A., Alonso L., Polak L., Fuchs E. (2003) J. Cell Biol. 163, 609–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobielak K., Stokes N., de la Cruz J., Polak L., Fuchs E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10063–10068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dewulf N., Verschueren K., Lonnoy O., Morén A., Grimsby S., Vande Spiegle K., Miyazono K., Huylebroeck D., Ten Dijke P. (1995) Endocrinology 136, 2652–2663 [DOI] [PubMed] [Google Scholar]
- 43.He W., Li A. G., Wang D., Han S., Zheng B., Goumans M. J., Ten Dijke P., Wang X. J. (2002) EMBO J. 21, 2580–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verschueren K., Dewulf N., Goumans M. J., Lonnoy O., Feijen A., Grimsby S., Vandi Spiegle K., ten Dijke P., Morén A., Vanscheeuwijck P., Heldin C. H., Miyazono K., Mummery C., Van Den Eijnden-Van Raaij J., Huylebroeck D. (1995) Mech. Dev. 52, 109–123 [DOI] [PubMed] [Google Scholar]
- 45.Macías-Silva M., Hoodless P. A., Tang S. J., Buchwald M., Wrana J. L. (1998) J. Biol. Chem. 273, 25628–25636 [DOI] [PubMed] [Google Scholar]
- 46.ten Dijke P., Yamashita H., Sampath T. K., Reddi A. H., Estevez M., Riddle D. L., Ichijo H., Heldin C. H., Miyazono K. (1994) J. Biol. Chem. 269, 16985–16988 [PubMed] [Google Scholar]
- 47.Yamashita H., ten Dijke P., Huylebroeck D., Sampath T. K., Andries M., Smith J. C., Heldin C. H., Miyazono K. (1995) J. Cell Biol. 130, 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aono A., Hazama M., Notoya K., Taketomi S., Yamasaki H., Tsukuda R., Sasaki S., Fujisawa Y. (1995) Biochem. Biophys. Res. Commun. 210, 670–677 [DOI] [PubMed] [Google Scholar]
- 49.Suzuki K., Haraguchi R., Ogata T., Barbieri O., Alegria O., Vieux-Rochas M., Nakagata N., Ito M., Mills A. A., Kurita T., Levi G., Yamada G. (2008) Eur. J. Hum. Genet. 16, 36–44 [DOI] [PubMed] [Google Scholar]
- 50.Korchynskyi O., ten Dijke P. (2002) J. Biol. Chem. 277, 4883–4891 [DOI] [PubMed] [Google Scholar]
- 51.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wrana J. L., Attisano L., Cárcamo J., Zentella A., Doody J., Laiho M., Wang X. F., Massagué J. (1992) Cell 71, 1003–1014 [DOI] [PubMed] [Google Scholar]
- 53.Yang A., Zhu Z., Kapranov P., McKeon F., Church G. M., Gingeras T. R., Struhl K. (2006) Mol. Cell 24, 593–602 [DOI] [PubMed] [Google Scholar]
- 54.Frank S. R., Schroeder M., Fernandez P., Taubert S., Amati B. (2001) Genes Dev. 15, 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.