Abstract
Glial cell line-derived neurotrophic factor (Gdnf) promotes neurite outgrowth and survival of neuronal cells, but its transcriptional regulation is poorly understood. Here, we sought to investigate the mechanism underlying fibroblast growth factor-2 (FGF2) induction of Gdnf expression in astrocytes. We found that FGF2 stimulation of rat astrocytes induced expression of Egr-1 at a high level. Sequence analysis of the rat Gdnf gene identified three overlapping Egr-1-binding sites between positions −185 and −163 of the rat Gdnf promoter. Transfection studies using a series of deleted Gdnf promoters revealed that these Egr-1-binding sites are required for maximal activation of the Gdnf promoter by FGF2. Chromatin immunoprecipitation analysis indicated that Egr-1 binds to the Gdnf promoter. Furthermore, the induction of Gdnf expression by FGF2 is strongly attenuated both in C6 glioma cells stably expressing Egr-1-specific small interfering RNA and in primary cultured astrocytes from the Egr-1 knock-out mouse. Additionally, we found that stimulation of the ERK and JNK pathways by FGF2 is functionally linked to Gdnf expression through the induction of Egr-1. These data demonstrate that FGF2-induced Gdnf expression is mediated by the induction of Egr-1 through activation of the ERK and JNK/Elk-1 signaling pathways.
Astrocytes play important roles in the regulation of neurogenesis and synaptogenesis of the central nervous system, as well as in the regulation of its ionic and neurotransmitter environments (1). By releasing various growth factors, such as fibroblast growth factor-2 (FGF2),2 glial cell line-derived neurotrophic factor (Gdnf), nerve growth factor, and ciliary neurotrophic factor, astrocytes also provide neuroprotection against various types of neuronal damage (2).
Gdnf is a neurotrophic factor that promotes neurite outgrowth and the survival and differentiation of distinct populations of neuronal cells, as well as astrocyte proliferation (3, 4). Targeted disruption of the mouse Gdnf gene causes various defects in sensory and enteric neuron subpopulations, renal abnormalities, and a failure of spermatogenesis (5–8). Gdnf gene expression is elevated during embryogenesis and continues at a low level in adults (9). Gdnf is rapidly up-regulated in response to brain injury (10) and specific pharmacological treatments. However, the details of the mechanism underlying the transcriptional regulation of Gdnf expression are not clearly understood.
Egr-1 (also known as nerve growth factor-induced-A (NGFI-A), zinc-finger clone 28 (zif268), Krüppel box 24 (krox24), and TPA-induced sequence 8 (Tis8)) is a transcription factor encoded by an immediate-early response gene. Egr-1 is present at high concentrations in neurons of the cerebral cortex, hippocampus, thalamus, amygdaloid nuclei, and striatum (11, 12) and is maintained by synaptic plasticity occurring in response to physiological stimuli (13), suggesting that Egr-1 may play a role in the nervous system.
The regulatory regions of the murine and human Gdnf genes contain putative consensus sequences for Egr-1 binding. Because Egr-1 (14) and Gdnf (15) can be induced by FGF2, we investigated whether Egr-1 is involved in Gdnf expression. Here, we show that Egr-1 directly binds to the Gdnf promoter and promotes transcriptional activation of the Gdnf gene. FGF2 induces Egr-1 expression through the ERK or JNK pathways. Inhibition of either of these pathways abrogates FGF2-induced expression of Egr-1 and Gdnf. An important role for Egr-1 in Gdnf expression is supported by the demonstration that FGF2-induced GDNF expression is attenuated both in C6 glioma cells stably expressing Egr-1-specific small interfering RNA (siRNA) and in primary cultured astrocytes derived from the Egr-1 knock-out mouse. These findings suggest a novel role for Egr-1 in the regulation of Gdnf transcription by FGF2 in astrocytes.
EXPERIMENTAL PROCEDURES
Materials
Egr-1 knock-out mice developed on a C57BL6 background were from Dr. Jeffrey Milbrandt (Washington University, St. Louis, MO) and have been described elsewhere (16). Mouse recombinant FGF2 was purchased from Calbiochem (San Diego, CA). The firefly and Renilla Dual-GloTM Luciferase Assay System was purchased from Promega (Madison, WI).
Plasmids
The plasmid pRL-null, which encodes Renilla luciferase, was purchased from Promega. Plasmid pFA2-Elk1, which encodes a fusion protein consisting of the yeast Gal4 DNA-binding domain (amino acids 1–147) and the activation domain of Elk-1 (amino acids 307–427), and plasmid pFR-Luc, which contains five Gal4-binding element repeats upstream of the luciferase gene, were purchased from Stratagene (La Jolla, CA). Plasmids expressing dominant negative (DN)-MEK1 (pCGN1/MEK DN), DN-ERK2 (pHA-ERK2 K52R), DN-JNK1 (pSRα/HA-JNK T183A/Y185F), and kinase-dead p38 kinase (pCDNA3/FLAG-p38 T180A/Y182F) were kindly provided by Dr. D. S. Min (Department of Molecular Biology, College of Natural Science, Pusan National University, Korea). The pCDNA3.1/Egr-1(I293F) plasmid, which expresses dominant-active (DA)-Egr-1, was generated as previously described (17).
Cell Culture
The rat glioma cell line C6 was obtained from the American Type Culture Collection (Manassas, VA). C6 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (HyClone, Logan, UT). Primary rat astrocytes and astrocytes from Egr-1 knock-out (Egr-1−/−) and heterozygous (Egr-1+/−) mice were prepared from cerebral cortices of newborn pup littermates as previously described (18) and cultured in modified Eagle's medium supplemented with 10% fetal bovine serum (HyClone). The Egr-1 genotype was determined by PCR amplification from genomic DNA using the forward primer 5′-aaccggcccagcaagacacc-3′ and the reverse primer 5′-ctcgtgctttacggtatcgc-3′. Astrocyte purity was determined by immunostaining with anti-glial fibrillary acidic protein antibody (1:500, Dako Corp., Carpinteria, CA).
Quantitative Real Time PCR and Reverse Transcription-PCR (RT-PCR)
Total RNA was extracted using a TRIzol RNA extraction kit (Invitrogen) from cells stimulated with FGF2 (10 ng/ml) for various lengths of time. The first-strand cDNA was synthesized from 500 ng of total RNA by using an iScript cDNA synthesis kit (Bio-Rad). Quantitative real time PCR was performed an icycler iQTM system (Bio-Rad), using the TaqMan-iQTM supermix kit (Bio-Rad) according to the manufacturer's recommendation. The TaqManTM fluogenic probes and PCR primers for rat Gdnf and glyceraldehyde-3-phosphate dehydrogenase (gapdh) were designed by Metabion Int. (Martinsreid, Deutschland). The sequences of primers were 5′-CTGCCCGCCGGTAAGAGG-3′ (forward for Gdnf) and 5′-CGTCATCAAACTGGTCAGGATAATC-3′ (reverse for Gdnf), 5′-GTGATGCTGGTGCTGAGTATGTC-3′ (forward for gapdh), and 5′-GCGGAAGGGGCGGAGATG-3′ (reverse for gapdh). The TaqMan fluorogenic probes used were 5′-FAM-CGCCCGCCGAAGACCACTCCCTC-BHQ-3′ (for Gdnf) and 5′-Yakima Yellow TM-ACCCTTCAGGTGAGCCCCAGCCTT-BHQ-1-3′ (for gapdh). Threshold cycle, Ct, which correlates inversely with the target mRNA levels, was measured as the cycle number at which the reporter fluorescent emission increases above a threshold level. The relative changes in Gdnf mRNA levels were normalized for gapdh mRNA in the same samples.
For RT-PCR analysis, gene-specific primers for murine Gdnf (forward, 5′-GGTCTACGGAGAGACCGATCCGAGGTGC-3′; reverse, 5′-TCTCTGGAGCCAGGGTCAGATACATC-3′) and actin (forward, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′; reverse, 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′) were used. The PCR conditions for all primers were as follows: hold for 5 min at 94 °C, followed by 30 cycles consisting of denaturation at 94 °C (30 s), annealing at 55 °C (30 s), and elongation at 72 °C (1 min). The amplified products were subjected to electrophoresis on a 1% agarose gel.
Construction of Full-length and Deletion Mutant Mouse Gdnf Promoter-Reporter Constructs
A Gdnf promoter fragment spanning nucleotides −983 to +3 was synthesized from rat genomic DNA (Promega) by PCR using primers 5′-ccggtaccTCCATCAAAGTCCAACTTGGCAAATA-3′ (forward) and 5′-gaagatctTCTCAGCTCTTAGGTCTCCTCTGCC-3′ (reverse). (KpnI and BglII restriction sites are indicated by lowercase letters.) The amplified PCR product was ligated into the KpnI-BglII site of the pGL3-basic vector (Promega) to yield pGDNF-Luc(−983/+3).
A series of deletion constructs of rat Gdnf promoter fragments were synthesized by PCR using the pGDNF-Luc(−983/+3) plasmid as a template. The forward and reverse primers used in the PCR to generate deletion mutant promoters contained KpnI and BglII restriction enzyme sites, respectively, to facilitate subsequent ligation into the KpnI-BglII site of pGL3-basic. The forward primer sequences were 5′-ccggtaccCGCCCTCATGTCTTCAAGGG-3′ (−493 to +3) ccggtaccGAGGAGGTGCAGAGTGAGGC (−374 to +3) and 5′-ccggtaccCCGGTACCTGGATTGCGTGCTCTTGCTC-3′ (−114 to +3). One reverse primer (5′-gaagatctTCTCAGCTCTTAGGTCTCCTCTGCC-3′) was used to generate all deletion mutant promoters (lowercase letters in the forward and reverse primers indicate KpnI and BglII restriction sites, respectively). The PCR products were digested with KpnI and BglII and then ligated into the pGL3-basic vector.
An internal deletion construct lacking Egr-1-binding motifs (−185 to −163) in pGDNF-Luc(−493/+3) was generated using two-step PCR yielding pGDNF-Luc(−493/+3ΔEgr1). The primers used for the first step were 5′-ccggtaccCGCCCTCATGTCTTCAAGGG-3′ (forward) and 5′-tctcgagAGGAGGGCGAAGGC-3′ (−493 to −185; reverse); 5′-actcgagACACGGTGGCCGC-3′ (forward) and 5′-gaagatctTCTCAGCTCTTAGGTCTCCTCTGCC-3′ (−163 to +3; reverse). (Lowercase letters indicate XhoI restriction sites.) The PCR products were digested with XhoI and ligated. In the second step, the primers were 5′-ccggtaccCGCCCTCATGTCTTCAAGGG-3′ (forward) and 5′-gaagatctTCTCAGCTCTTAGGTCTCCTCTGCC-3′ (reverse). All constructs were verified by DNA sequencing and restriction analysis.
Luciferase Reporter Assay
C6 cells or primary cultured astrocytes were seeded into 12-well plates and transfected with 0.5 μg of promoter-reporter construct using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions. For Elk-1 trans-acting activity, cells were co-transfected with 50 ng of trans-activator plasmid (pFA2-Elk1) and 0.5 μg of reporter plasmid (pFR-Luc). To monitor transfection efficiency, 50 ng of the pRL-null plasmid encoding Renilla luciferase was included in all samples. Where indicated, 0.2 μg of mammalian expression vector was also included. At 24-h post-transfection, the levels of firefly and Renilla luciferase activity were measured sequentially from a single sample using the Dual-Glo Luciferase assay system (Promega) with a luminometer (Centro LB960; Berthold Tech, Bad Wildbad, Germany). Firefly luciferase activity was normalized to Renilla luciferase activity, and the relative amount of luciferase activity in the untreated cells was designated as 1.
Western Blot Analysis
Cells were lysed in 20 mm HEPES (pH 7.2) containing 1% Triton X-100, 10% glycerol, 150 mm NaCl, 10 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride. The protein samples (20 μg/lane) were then separated by 10% SDS-PAGE and transferred onto nitrocellulose filters. Western blotting was performed according to standard procedures using antibodies specific for Egr-1 (1:1000 dilution), ERK2 (1:5000), or GAPDH (1:2000) (all from Santa Cruz Biotechnology, Santa Cruz, CA) or for phospho-Raf1 (1:1000), phospho-(Thr202/Tyr204)-ERK1/2 (1:1000), phospho-(Thr180/Tyr182)-p38 (1:1000), or phospho-(Thr183/Tyr185)-JNK (1:1000) (all from Cell Signaling Technology, Danvers, MA). The signals were developed using an enhanced chemiluminescence detection system (Amersham Biosciences Inc.).
Expression of Egr-1 siRNA
The small hairpin RNA (shRNA) plasmid targeting mouse Egr-1 mRNA (pSilencer/siEgr1) and the generation of C6 cells expressing Egr-1 siRNA (C6/SiEgr-1) or control siRNA (C6/Scramble) have been described previously (19).
Confocal Microscopy
C6 cells or primary cultured astrocytes plated on coverslips were treated with 0 or 10 ng/ml of FGF2 for 24 h, fixed with 4% paraformaldehyde, and permeabilized in 0.1% Triton X-100 and 2% bovine serum albumin as described previously (20). The coverslips were co-stained with rabbit anti-Gdnf (1:100) and mouse anti-S100 (1:200) antibodies for the mouse astrocyte marker, for 90 min at room temperature and then incubated for 30 min with Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 555-conjugated anti-rabbit secondary antibodies, yielding green and red signals, respectively. The labeled cells were examined under an Olympus FV-1000 Spectral confocal laser-scanning microscope with excitation at 488 nm and emission at 550 nm.
Chromatin Immunoprecipitation (ChIP) Assay
Primary cultured astrocytes treated with FGF2 (10 ng/ml) were treated with 1% formaldehyde to cross-link the DNA, lysed, and chromatin immunoprecipitated using rabbit anti-Egr-1 antibody (Santa Cruz Biotechnology) or normal rabbit IgG as described previously (21). The precipitated DNA was analyzed by standard RT-PCR methods.
Immunohistochemistry
All procedures were carried out with the approval of the Konkuk University Institutional Animal Care and Use Committee. For preparation of samples, mice were anesthetized with a mixture of Zoletil (22.5 mg/kg; Virbac, Carros, France) and Xylazine (7.5 mg/kg; Bayer, KS) and perfused transcardially with a fixative solution containing 4% paraformaldehyde in 0.1 m phosphate-buffered saline (pH 7.4). Brains were post-fixed for 24 h in the same fixative solution and bisected sagitally at the midline. The samples were embedded in paraffin, cut into serial sections 4-μm thick, and stained with hematoxylin and eosin. Immunohistochemical analysis was carried out using a Vectastain Elite ABC kit (Vector Labs, Burlingame, CA). The paraffin sections were deparaffinized, hydrated, heated in 0.01 m sodium citrate for 10 min in a microwave to retrieve the antigen, and treated with 3% hydrogen peroxide in methanol. They were then incubated with blocking solution containing 1.5% anti-horse serum and 5% bovine serum albumin. The sections were incubated with anti-Gdnf antibody (1:50; Santa Cruz Biotechnology) overnight at 4 °C. After washing in phosphate-buffered saline, the sections were incubated with biotinylated secondary antibody for 60 min and then with a avidin-biotin peroxidase complex for 30 min at room temperature. Sections were treated with peroxidase substrate kits (Vector Labs) to visualize immunoreactivity, counterstained with hematoxylin, and dehydrated in ethanol. After clearing in xylene, sections were coverslipped and examined under a Olympus BX51 microscope (Olympus Optical, Tokyo, Japan). Immunoreactive cells were quantitatively analyzed by a computerized image analyzer MetaMorph 7.5 (Molecular Devices, Downington, PA). The immunopositive cells in three nonconsecutive sections per mouse were counted by two blinded independent investigators using the image analyzer at a magnification of ×200. Results are expressed as the mean number of cells per mm2 ± S.D. Statistical significance was evaluated by one-way analysis of variance using SPSS for Windows (version 14.0, SPSS Inc., Chicago, IL).
GDNF Enzyme-linked Immunosorbent Assay
For the assay of Gdnf release, Egr-1+/− or Egr-1−/− astrocytes were cultured with or without 10 ng/ml of FGF2 at a density of 13 × 104/cm2 on a 12-well plate. After 48 h treatment, conditioned medium was collected and stored at −80 °C until assayed. GDNF protein levels in cell-conditioned media were determined using a Gdnf enzyme-linked immunosorbent assay according to the manufacturer's instructions (Promega).
RESULTS
FGF2 Induces Gdnf Expression at the Transcriptional Level in Rat Astrocytes
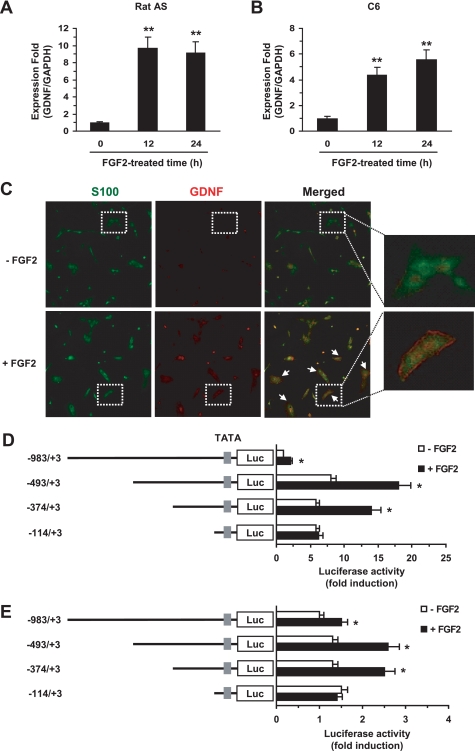
To investigate the molecular mechanism of FGF2 regulation of Gdnf expression, we first determined the ability of FGF2 to induce the expression of Gdnf by quantitative real time-PCR using the TaqMan fluogenic probes in rat astrocytes. After 12 and 24 h of exposure to FGF2, Gdnf mRNA was increased 9.7- and 9.2-fold, respectively, as compared with control cells in a dissociated culture of rat astrocytes (Fig. 1A), and increased 4.4- and 5.6-fold, respectively, as compared with control cells in C6 glioma cells (Fig. 1B). All of the dissociated cultures of rat astrocytes were shown to express S-100, an astrocyte marker, by confocal microscopy. Notably, FGF2 treatment enhanced Gdnf immunoreactivity in the dissociated rat astrocyte culture (Fig. 1C). These data clearly demonstrate that FGF2 induces up-regulation of Gdnf expression in rat astrocytes.
FIGURE 1.
FGF2 activates transcription of Gdnf expression in rat astrocytes. A and B, primary cultured rat astrocytes (A) or C6 rat glioma cells (B) were treated with 0 or 10 ng/ml of FGF2 for 12 h. Total RNA was extracted and then subjected to RT-PCR to quantify Gdnf expression. Actin expression was used as an internal control. **, p < 0.01 compared with basal luciferase activity. C, immunodetection of Gdnf. Primary rat astrocytes cultured on coverslips were treated with 0 or 10 ng/ml of FGF2 for 18 h. After incubation with anti-Gdnf (1:100) and anti-S100 (1:200) antibodies, the cells were observed under a confocal fluorescence microscope; green, S-100; red, Gdnf. Arrows indicate immunoreactive cells for Gdnf. D and E, Gdnf promoter constructs (0.2 μg each) were transiently co-transfected with the pRL-null vector (50 ng) into primary rat astrocytes (D) or C6 glioma cells (E). At 24-h post-transfection, cells were treated with 0 or 10 ng/ml of FGF2 for 8 h, and luciferase activity was measured. The firefly luciferase activity was normalized to Renilla luciferase activity. The data represent the mean ± S.D. (error bars) of three independent experiments, each performed in triplicate. *, p < 0.05 compared with basal luciferase activity (Student's t test).
To identify the cis-acting region that mediates the responsiveness of the rat Gdnf gene to FGF2, progressively deleted Gdnf promoter-reporter constructs were made and transfected into rat primary astrocytes. The basal promoter activity of the −493/+3 and −114/+3 constructs was 10 and 5 times higher, respectively, than that of the −983/+3 construct (Fig. 1D), suggesting negative regulatory elements between nucleotides −983 and −492 control basal transcription. In contrast, FGF2-induced promoter activity approximately doubled in the −983/+3, −493/+3, and −374/+3 constructs but remained the same in the −114/+3 construct. Similar results were obtained in C6 glioma cells, although the observed increases in basal activity were much smaller than those seen in primary cultured astrocytes (Fig. 1E). These data suggest that a proximal region between −374 and −114 of the Gdnf promoter contains a cis-acting regulatory element responsive to FGF2.
Egr-1-binding Sequences Are Required for FGF2-mediated Activation of the Gdnf Promoter
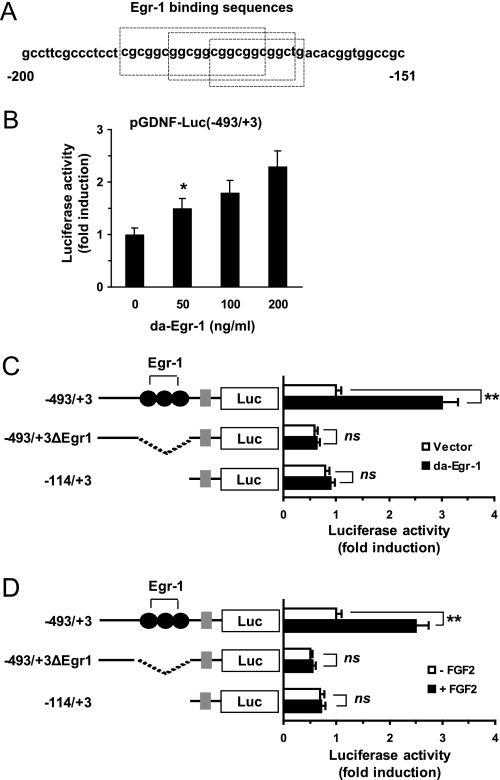
An analysis of the Gdnf promoter sequences spanning −374 to −114 performed using the program MatInspector revealed the presence of three putative binding motifs for Egr-1 proximal to the transcription start site at positions −185 to −163 (Fig. 2A). Egr-1 is a Cys2/His2-type zinc finger transcription factor that regulates cell growth, differentiation, and development (22). To assess whether Egr-1 activates the Gdnf promoter, we co-transfected the −493/+3 Gdnf promoter construct and the expression plasmid for DA-Egr-1, pcDNA3.1/Egr1(I293F), into rat primary cultured astrocytes. In the absence of FGF2 treatment, the forced expression of DA-Egr-1 caused basal reporter activity to increase as the amount of transfected DNA increased (Fig. 2B).
FIGURE 2.
Functional role of Egr-1 and Egr-1-binding sequences in Gdnf promoter activation. A, sequence of the promoter region in the rat Gdnf gene (−185 to −163). The three putative Egr-1-binding sites are boxed, B, primary rat astrocytes were co-transfected with pGDNF-Luc(−493/+3) (0.2 μg), various concentrations of the DA-Egr-1 expression plasmid pCDNA3.1/Egr1(I293F), and the pRL-null vector (50 ng). At 24 h post-transfection, cells were collected, and luciferase activity was measured. *, p < 0.05 compared with basal luciferase activity (Student's t test). C, primary rat astrocytes were co-transfected with the DA-Egr-1 expression plasmid pCDNA3.1/Egr1(I293F) and 0.2 μg of a Gdnf promoter construct (pGDNF-Luc(−493/+3), pGDNF-Luc(−493/+3ΔEgr1), or pGDNF-Luc(−114/+3)). At 24 h post-transfection, cells were collected, and luciferase activity was measured. **, p < 0.01 compared with empty vector-transfected basal luciferase activity; ns, not significant (Student's t test). D, primary rat astrocytes were transfected as in C. At 24 h post-transfection, they were treated with 0 or 10 ng/ml of FGF2 for 8 h, and luciferase activity was measured. The firefly luciferase activity was normalized to the Renilla luciferase activity. The data shown represent the mean ± S.D. (error bars) of three independent experiments, each performed in triplicate. **, p < 0.01 compared with empty vector-transfected basal luciferase activity; ns, not significant (Student's t test).
To characterize whether the Egr-1-binding motifs were responsible for stimulation of the GDNF gene promoter by Egr-1, we generated an internal deletion construct lacking all three Egr-1-binding motifs (−493/+3ΔEgr1). As shown in Fig. 2C, transfection of DA-Egr-1 into rat primary astrocytes had no appreciable effect on the reporter activity of the −493/+3ΔEgr1 or −114/+3 constructs, which lack apparent Egr-1-binding sequences. To assess the contribution of the Egr-1-binding sequences to the activation of the GDNF promoter by FGF2, the transcriptional activity of these deletion constructs in response to FGF2 was evaluated in rat primary astrocytes. As shown in Fig. 2D, FGF2 approximately doubled the reporter activity in the −493/+3 construct, but not in the −493/+3ΔEgr1 or −114/+3 constructs. Similar results were obtained in C6 glioma cells (data not shown). These observations indicate that the Egr-1-binding motifs are cis-acting elements necessary for maximal activation of the GDNF promoter by FGF2.
Egr-1 Is Induced by FGF2
Egr-1 expression is induced by FGF2 in several cell lines (23–25). Consistent with previous findings, Egr-1 expression was clearly up-regulated by FGF2 as a function of time in primary rat astrocytes (Fig. 3A) or in C6 glioma cells (Fig. 3B). In addition, FGF2 increased Egr-1 promoter activity in a concentration-dependent manner in primary rat astrocytes (Fig. 3C) and C6 cells (Fig. 3D).
FIGURE 3.
Induction of Egr-1 expression by FGF2. A and B, primary rat astrocytes (A) or C6 cells (B) were cultured in the presence of 0.5% serum for 24 h and then treated with 10 ng/ml of FGF2 for various lengths of time (15–240 min). The amount of Egr-1 in whole cell lysates was measured by Western blotting (15 μg of protein/lane) with rabbit anti-Egr-1 antibody. In the lower panel, the blot was re-probed with anti-GAPDH antibody as a loading control. C and D, primary rat astrocytes (C) or C6 cells (D) cells grown in 12-well plates were co-transfected with 0.5 μg of the Egr-1 promoter construct pEgr1-Luc(−780/+1) and 50 ng of pRL-null vector. Twenty-four hours after transfection, the cells were treated with varying concentrations of FGF2 (0–20 ng/ml) for 8 h. The firefly luciferase activity was normalized to the Renilla luciferase activity. The data shown represent the mean ± S.D. (error bars) of three independent experiments performed in triplicate. E, in ChIP experiments, rat primary astrocytes were treated with 0 or 10 ng/ml of FGF2 for 1 h, cross-linked, lysed, and immunoprecipitated with rabbit anti-Egr-1 antibody or with normal rabbit IgG (negative control). The precipitated DNA was subjected to PCR with primers specific to the Gdnf promoter. An aliquot of input DNA was used as a positive control. The schematic representation at the right shows the locations of the Egr-1-binding sites and PCR primers in the Gdnf promoter.
Egr-1 Is Associated with the Gdnf Promoter
To determine whether Egr-1 protein binds to the Egr-1-binding sequences in vivo, we performed a ChIP assay. Cross-linked rat primary astrocytes were immunoprecipitated with rabbit anti-Egr-1 antibody or normal rabbit IgG. The resulting immunoprecipitates were analyzed by PCR assays using primers flanking the Egr-1-binding sequences (−324 to −88) of the Gdnf promoter. As shown in Fig. 3E, a noticeable increase in the intensity of the DNA band was observed for rabbit anti-Egr-1 antibody but not for the normal rabbit IgG. The off-target region (from −443 to −243) was not amplified, although positive results were obtained from input DNA. This result indicates that a physical interaction between Egr-1 and the Gdnf promoter occurs in vivo upon FGF2 treatment.
ERK and JNK Mitogen-activated Protein Kinases (MAPKs) Are Involved in FGF2-induced Egr-1 Expression
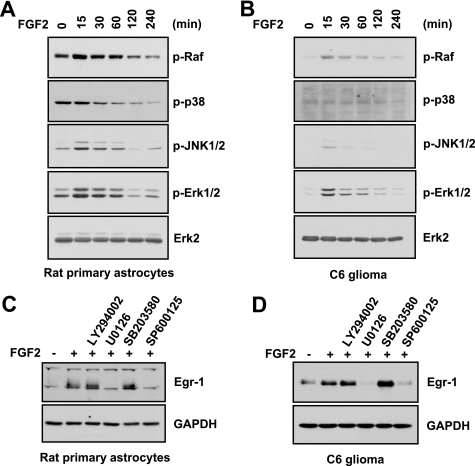
In many cell systems, activation of MAPKs, including the ERK, JNK, and p38 MAPKs, induces Egr-1 promoter activity (26–31). To identify the signal transduction pathway that mediates FGF2-induced expression of Egr-1 in primary rat astrocytes, we examined the ability of FGF2 to stimulate MAPK phosphorylation (Fig. 4A). After serum-starved rat astrocytes were treated with FGF2 for various lengths of time, the activation status of MAPKs was assessed using phospho-specific antibodies. The levels of phosphorylated Raf, p38 kinase, JNK1/2, and ERK1/2 increased in a time-dependent manner upon FGF2 treatment. These increases in the phosphorylated levels were evident within 15 min after FGF2 treatment. Similar results were observed in C6 cells (Fig. 4B).
FIGURE 4.
Role of ERK and JNK in FGF2-induced expression of Egr-1. A and B, primary rat astrocytes (A) and C6 cells (B) were cultured in the presence of 0.5% serum for 24 h and then treated with 10 ng/ml of FGF2 for various lengths of time. Total cell lysates were prepared, and Western blotting was performed using total protein extracts and antibodies against phospho-Raf1 (Ser259), phospho-(Thr180/Tyr182)-p38 kinase, phospho-(Thr183/Tyr185)-JNK1/2, phospho-(Thr202/Tyr204)-ERK1/2, and ERK2. Each blot is representative of at least three separate experiments. C and D, serum-starved primary rat astrocytes (C) and C6 cells (D) were treated with LY294002 (20 μm), U0126 (10 μm), SB203580 (20 μm), or SP600125 (20 μm) for 30 min and then treated with 0 or 10 ng/ml of FGF2 for 1 h. Total cell lysates were prepared and subjected to Western blotting with anti-Egr-1 antibody. The same blot was re-probed with anti-GAPDH antibody as an internal control. Each blot shown is representative of at least three separate experiments.
To determine whether activation of these MAPKs was associated with FGF2-induced Egr-1 expression, the effects of chemical inhibitors were assessed. Pretreatment of primary rat astrocytes with the MEK inhibitor U0126 or the JNK inhibitor SP600125 completely inhibited FGF2-stimulated Egr-1 expression, whereas neither the phosphatidylinositol 3-kinase inhibitor LY294002 nor the p38 MAPK inhibitor SB203580 had any effect (Fig. 4C). Similar results were obtained in C6 cells (Fig. 4D).
ERK and JNK MAPKs Are Involved in FGF2 Activation of the Gdnf Promoter
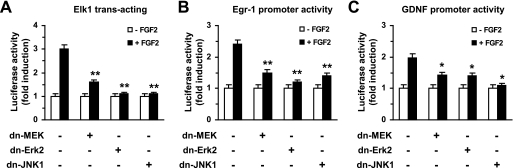
As a ternary complex factor, Elk-1 forms a complex with serum response factor on the serum response element (32). Elk-1 is activated by ERK, and JNK (33), and its transactivation plays a crucial role in extracellular signaling-induced transcription of Egr-1 (34, 35). To determine whether the ERK and JNK pathways induce Egr-1 expression via Elk-1 transactivation upon FGF2 stimulation in primary astrocytes, Elk-1 trans-acting activity was examined using a reporter assay system. Primary rat astrocytes were co-transfected with the trans-acting plasmid Gal4-Elk-1 and the pFR-Luc reporter plasmid containing Gal4-binding sequences. Treatment with FGF2 tripled the trans-acting activity of Gal4-Elk-1 (Fig. 5A). However, this activity was strongly inhibited in the presence of DN-MEK1, DN-ERK2, or DN-JNK1. Similarly, FGF2-induced Egr-1 promoter activity (Fig. 5B) and Gdnf promoter activity (Fig. 5C) were strongly attenuated by the expression of DN-MEK, DN-ERK2, or DN-JNK1. These data demonstrate that both ERK and JNK pathways are involved in FGF2-induced expression of Egr-1 and Gdnf.
FIGURE 5.
Role of ERK and JNK in FGF2 activation of the Gdnf promoter. Primary rat astrocytes were co-transfected with 50 ng of Elk-1 trans-activator plasmid (pFA2/Gal4-Elk-1), 50 ng of pRL-null vector, and 0.5 μg of the reporter plasmid pFR-Luc (A), the Egr-1 promoter-reporter plasmid pEgr1-Luc(−780/+1) (B), or the Gdnf promoter-reporter plasmid pGDNF Luc(−493/+3) (C), together with or without 0.2 μg of a plasmid expressing DN-MEK1 (pCGN1/MEK DN), DN-ERK2 (pHA-ERK2 K52R), or DN-JNK1 (pSRα/HA-JNK T183A/Y185F), as indicated. Twenty-four hours after transfection, the cells were treated with 0 or 10 ng/ml of FGF2 for 8 h. The resulting firefly luciferase activity was normalized to the Renilla luciferase activity. The data represent the mean ± S.D. (error bars) of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01, compared with FGF2-treated control cells transfected with empty vector (Student's t test).
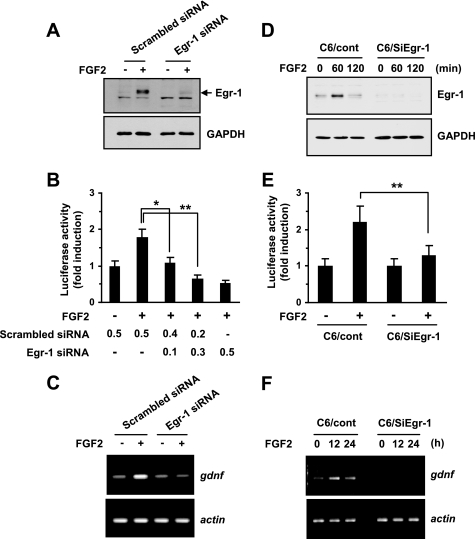
Silencing of Egr-1 Reduces FGF2-induced Gdnf Expression
To determine whether Egr-1 plays a functional role in FGF2 regulation of Gdnf transcription, we used RNA interference. First, the knockdown of Egr-1 expression in primary rat astrocytes by transient transfection of shRNA plasmids generating Egr-1-specific siRNA (pSilencer/siEgr1) was verified (Fig. 6A). Then, FGF2-induced Gdnf expression was measured under these conditions using Western blotting. The transient expression of Egr-1 siRNA clearly attenuated activation of the Gdnf promoter (Fig. 6B) and accumulation of Gdnf mRNAs (Fig. 6C) by FGF2.
FIGURE 6.
Effect of Egr-1 knockdown on FGF2-induced expression of Gdnf. A, primary rat astrocytes were transiently transfected with an shRNA plasmid expressing scrambled (pSilencer/scrambled) or Egr-1 (pSilencer/siEgr1) siRNA. After 48 h, cells were either untreated or treated with 10 ng/ml of FGF2 for 2 h, and the Egr-1 expression level was determined by Western blot analysis. The same blot was re-probed with anti-GAPDH antibody as a loading control. Each blot is representative of at least three separate experiments. B, primary rat astrocytes were transiently co-transfected with a Gdnf promoter-reporter plasmid (pGDNF-Luc(−493/+3)) and an shRNA plasmid expressing scrambled siRNA (pSilencer/scrambled) or Egr-1 siRNA (pSilencer/siEgr1), or with both shRNA plasmids, as indicated. After 48 h, cells were treated with 10 ng/ml of FGF2 for 8 h, and luciferase activity was measured. The firefly luciferase activity was normalized to the Renilla luciferase activity. The data represent the mean ± S.D. (error bars) of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01 (Student's t test). C, primary rat astrocytes were transiently transfected with a scrambled siRNA (pSilencer/scrambled) or Egr-1 siRNA (pSilencer/siEgr1). After 48 h, cells were treated with 10 ng/ml of FGF2 for 12 h. Total RNA was extracted and assessed for Gdnf mRNA expression by RT-PCR. Similar results were obtained from three independent experiments. D, serum-starved C6 transfectants stably expressing scrambled siRNA (C6/cont) or Egr-1 siRNA (C6/siEgr1) were treated with 10 ng/ml of FGF2 for various lengths of time. At the indicated time points, the cells were collected and analyzed for Egr-1 expression using Western blotting. The same blot was re-probed with anti-GAPDH antibody as a loading control. Each blot shown is representative of at least three separate experiments. E, C6/cont and C6/siEgr1 cells were transiently transfected with a Gdnf promoter-reporter plasmid (pGDNF-Luc(−493/+3)). After 48 h, cells were treated with 10 ng/ml of FGF2 for 8 h, and luciferase activity was measured. The firefly luciferase activity was normalized to the Renilla luciferase activity. The data represent the mean ± S.D. (error bars) of three independent experiments performed in triplicate. **, p < 0.01 (Student's t test). F, C6/cont or C6/siEgr1 cells were treated with FGF2 for 12 or 24 h. Total RNA was extracted and assessed for Gdnf mRNA expression by RT-PCR. Similar results were obtained from three independent experiments.
To further evaluate the importance of Egr-1 expression in FGF2-mediated transcriptional activation of the Gdnf gene, we used C6 cell lines stably expressing Egr-1 siRNA (19). The effect of Egr-1 siRNA-mediated knockdown of the Egr-1 protein was evaluated after FGF2 treatment in serum-starved cells (Fig. 6D). Stable expression of Egr-1 siRNA strongly inhibited FGF2-induced Gdnf promoter activity (Fig. 6E) and mRNA expression (Fig. 6F) in C6 cells, demonstrating that Egr-1 induction is important for Gdnf gene activation by FGF2.
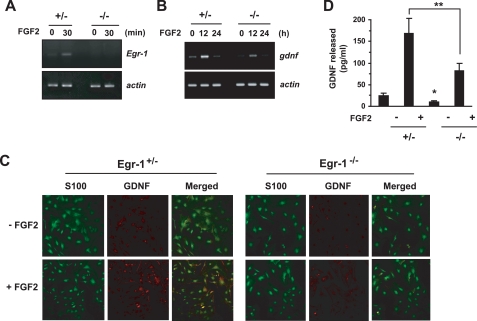
To confirm the functional role of Egr-1 in Gdnf expression, we prepared primary astrocytes from Egr-1 heterozygous (+/−) and knock-out (−/−) mice. In astrocytes from Egr-1−/− mice, no FGF2-induced expression of specific Egr-1 transcripts was detectable by RT-PCR (Fig. 7A). When we examined Gdnf mRNA levels in Egr-1−/− astrocytes treated or not treated with FGF2, we found that both the basal and FGF2-induced levels of Gdnf mRNA were substantially lower than in the Egr-1+/− astrocytes (Fig. 7B). However, Gdnf mRNA was still detectable in the Egr-1−/− astrocytes, suggesting that other regulatory factors are also involved in FGF2-induced Gdnf expression. Double immunofluorescence staining was performed to assess the expression of the Gdnf protein in primary cultured astrocytes. Confocal microscopy revealed that after FGF2 treatment, the amount of immunoreactivity to anti-Gdnf antibody was far lower in Egr-1−/− astrocytes than in Egr-1+/− astrocytes (Fig. 7C). To further ensure the role of Egr-1 in the production of Gdnf proteins, enzyme-linked immunosorbent assay was used to detect proteins present in the conditioned medium. The amount of secreted Gdnf protein was significantly decreased after FGF2 treatment in the Egr-1−/− astrocytes as compared with Egr-1+/− astrocytes (Fig. 7D). Thus, it is likely that Egr-1 is important for basal expression of Gdnf and is necessary, but not sufficient, for FGF2-induced activation of Gdnf transcription in astrocytes.
FIGURE 7.
FGF2-induced Gdnf expression is suppressed in Egr-1−/− astrocytes. A and B, Egr-1+/− or Egr-1−/− astrocytes were treated with 10 ng/ml of FGF2 for various lengths of time, and total RNA was extracted. Egr-1 (A) or Gdnf (B) mRNA expressions were determined by RT-PCR. Similar results were obtained from three independent experiments. C, Egr-1+/− or Egr-1−/− astrocytes cultured on coverslips were treated with 10 ng/ml of FGF2 for 18 h. After incubation with anti-Gdnf (1:100) and anti-S100 (1:200) antibodies, the cells were examined under a confocal fluorescence microscope; red, Gdnf; green, S100. D, Egr-1+/− or Egr-1−/− astrocytes cultured in 24-well plates were incubated with or without 10 ng/ml of FGF2. After 48 h, the Gdnf concentration in the conditioned medium was measured using an enzyme-linked immunosorbent assay. The data represent the mean ± S.D. (error bars) of three independent experiments performed in triplicate. *, p < 0.05 compared with Egr-1+/− conditioned medium in the absence of FGF2; **, p < 0.01 (Student's t test).
Gdnf Expression Is Impaired in the Egr-1−/− Mouse Brain
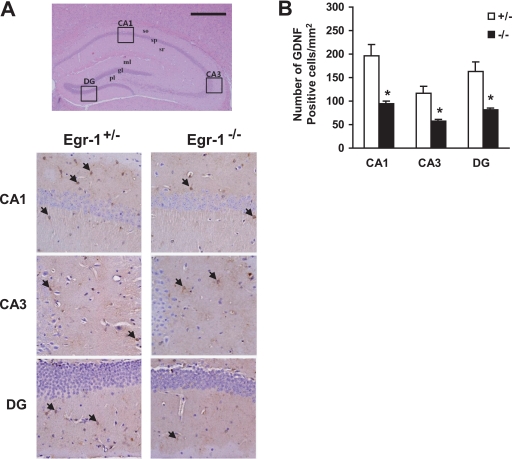
To investigate whether a lack of Egr-1 expression causes a defect in Gdnf expression in vivo, brain sections were prepared from Egr-1+/− and Egr-1−/− mice. Gdnf expression in Cornu Ammonis 1 (CA1), CA3, and dentate gyrus regions in the hippocampus was detected using immunohistochemistry (Fig. 8A). The number of Gdnf-immunopositive cells in the hippocampal regions of the Egr-1−/− mouse was significantly lower than in the Egr-1+/− mouse (Fig. 8B). These data further support the hypothesis that Egr-1 is required for Gdnf expression in vivo.
FIGURE 8.
Impairment of GdnF expression in the Egr-1−/− mouse brain. A, brain sections were prepared, stained with hematoxylin and eosin (upper panel), and immunostained with anti-Gdnf antibody (lower panels). Gdnf immunoreactive cells in the CA1, CA3, and dentate gyrus (DG) regions of the hippocampus (boxes in upper panel) are shown in the lower panels. Arrows indicate immunopositive cells. Scale bar, 500 μm. B, quantification of Gdnf-immunoreactive cells in the CA1, CA3, and dentate gyrus (DG) regions of an 8-month-old mouse brain. Values represent the mean ± S.D. (error bars). *, p < 0.05 (n = 6) for Egr-1+/− versus Egr-1−/− mice (one-way analysis of variance analysis).
DISCUSSION
Astrocyte-derived Gdnf acts as a synaptogenic and neurotrophic factor (4, 36). The neurotrophic activity of Gdnf may therefore have therapeutic implications for neurodegenerative disorders such as Parkinson disease (3). Therefore, an understanding of the molecular mechanism of Gdnf gene expression regulation might aid the development of effective neuroprotective agents.
Egr-1, a transcription factor encoded by an immediate-early response gene, has emerged as a major neuroregulatory factor. It is essential for stabilizing synaptic plasticity in the hippocampus and establishing late long-term potentiation and long-term memories (13, 37, 38). In the present study, we demonstrated the important role of Egr-1 in FGF2-induced Gdnf expression and identified the signaling pathway leading to Egr-1 expression.
To functionally analyze the molecular pathways mediating the induction of Gdnf gene expression by FGF2, we used 5′-deletion analysis of the rat Gdnf promoter to identify the participating cis-acting response elements. We found that the promoter region spanning positions −493 to −114 is indispensable for FGF2-triggered Gdnf promoter activity. This promoter region contains the three putative Egr-1-binding sites overlapped in the region from −185 to −163. The Egr-1-binding sequence, 5′-GCGGCGGC-3′, is highly conserved in the human and murine Gdnf promoters (39), which may reflect its importance as a regulatory element.
In previous studies, Gdnf expression was shown to be enhanced by diverse biological factors and pharmacological agents, including FGF2 (15), inflammatory cytokines (40), sphingosine 1-phosphate (41), endothelin-1 (42), adenosine (43), serotonin (44), and antidepressants (44–46). FGF2 (47) and antidepressants (46) induce Egr-1 expression in primary astrocytes and C6 glioma cells. However, no information was available about the involvement of Egr-1 in Gdnf expression, although whether Egr-1 participates in Gdnf production by astrocytes is an issue of considerable interest.
To test the role of Egr-1 in Gdnf expression, we generated internal deletion mutants of the Gdnf promoter lacking Egr-1-binding sequences. Deletion of the overlapping Egr-1-binding elements in the −185/−163 region completely abrogated FGF2-induced Gdnf promoter-reporter activity in both rat primary astrocytes and C6 cells. Using ChIP experiments, we found that Egr-1 binding to the Gdnf promoter was markedly increased in response to FGF2. We also found that the forced expression of DA-Egr-1 by itself enhanced Gdnf promoter activity, and the introduction of Egr-1-specific siRNA into rat primary astrocytes or C6 cells strongly attenuated FGF2-induced Gdnf promoter activity. More importantly, compared with Egr-1+/− astrocytes, Egr-1−/− astrocytes were drastically impaired in their ability to express Gdnf mRNA and protein in response to FGF2. These results provide strong evidence that Egr-1 contributes to activation of the Gdnf gene promoter, at least upon FGF2 stimulation.
Although these results have greatly increased our understanding of Egr-1 action in Gdnf expression, some issues remain to be elucidated. For example, we saw that even Egr-1−/− astrocytes were susceptible to FGF2 stimulation, although their response to FGF2 was markedly reduced compared with that of Egr-1+/− astrocytes. The ability of Egr-1−/− astrocytes to respond to FGF2 might reflect the participation of other factors in Gdnf induction by FGF2. The cAMP response element (CRE)-binding protein (CREB) might be one such factor. CREB is required for long-term memory and for the development and plasticity of the nervous system (48). Previous studies have demonstrated that CREB is activated by FGF2 in astrocytes (49) and that CREB phosphorylation is associated with Gdnf expression (50–52). Thus, we cannot rule out the possibility that CREB might play a role in the modulation of Gdnf expression in response to various signals. The CRE of the mouse Gdnf gene is located at +43/+50 in exon I (53). Alternatively, other members of the Egr family, such as Egr-2, Egr-3, and Egr-4, might functionally compensate for the absence of Egr-1 in vivo; they recognize the same DNA binding site as Egr-1, and their DNA-binding domains are highly homologous to that of Egr-1 (13). Nonetheless, our results clearly implicate Egr-1 as the regulator of Gdnf transcription in FGF2-stimulated astrocytes, regardless of other possible contributions to Gdnf promoter activation in the absence of Egr-1.
We also investigated the intracellular signaling pathways that mediate FGF2-induced expression of Egr-1 and Gdnf. In many cell types, activation of ERK, JNK, or p38 MAPK leads to an increase in Elk-1 transactivation (32). Elk-1, a member of the Ets family of transcription factors (32), plays a crucial role in growth factor-induced Egr-1 transcription by forming a complex with the serum response factor on the serum response element of the Egr-1 promoter (35). Elk-1 is phosphorylated and trans-activated by the ERK, JNK, and p38 MAPKs (33). Therefore, we also examined whether these MAPKs contribute to FGF2-induced Egr-1 and Gdnf expression. We found that FGF2 increased the level of phosphorylation of all three major MAPKs in both C6 and primary rat astrocytes. Moreover, inhibition of ERK or JNK, but not p38 kinase, suppressed FGF2-induced Elk-1 trans-acting activity, Egr-1 promoter activity, and Gdnf promoter activity. A previous report (54) demonstrated a similar involvement of Elk-1, which is activated by ERK1/2 but not by JNK or p38, in the stimulation of Gdnf expression in C6 glioma cells by the antidepressant amitriptyline. In addition, serotonin (5-hydroxytryptamine) induces Gdnf mRNA expression in C6 cells via ERK phosphorylation mediated by transactivation of the FGF receptor (44). Taking these observations into consideration, we propose that different MAPK pathways might converge on Egr-1 expression via Elk-1 transactivation, which in turn may lead to Gdnf promoter activation in astrocytes.
In summary, our data provide evidence that Egr-1 mediates FGF2-inducible expression of the Gdnf gene in murine astrocytes. The importance of Egr-1 in Gdnf gene expression is confirmed by several pieces of supporting data: (i) Egr-1 is rapidly induced by FGF2, (ii) the Egr-1-binding motif in the proximal region of the Gdnf promoter is necessary for maximal promoter activation by FGF2, (iii) exogenous Egr-1 transactivates the Gdnf promoter, and (iv) Egr-1 deficiency suppresses FGF2-inducible Gdnf expression. Our study therefore supports the view that Egr-1 is an important transcription factor with diverse neuronal functions.
This work was supported by Disease Network Research Program Grant 20090084181 from the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology, Republic of Korea.
- FGF2
- fibroblast growth factor-2
- Gdnf
- glial cell line-derived neurotrophic factor
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun NH2-terminal kinase
- siRNA
- small interfering RNA
- DN
- dominant negative
- DA
- dominant active
- RT
- reverse transcriptase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- shRNA
- small hairpin RNA
- ChIP
- chromatin immunoprecipitation
- MAPK
- mitogen-activated protein kinase
- CA
- cornu ammonis
- CRE
- cAMP-response element
- CREB
- cAMP response element-binding protein.
REFERENCES
- 1.Ullian E. M., Christopherson K. S., Barres B. A. (2004) Glia 47, 209–216 [DOI] [PubMed] [Google Scholar]
- 2.Darlington C. L. (2005) Curr. Opin. Investig. Drug 6, 700–703 [PubMed] [Google Scholar]
- 3.Lin L. F., Doherty D. H., Lile J. D., Bektesh S., Collins F. (1993) Science 260, 1130–1132 [DOI] [PubMed] [Google Scholar]
- 4.Pozas E., Ibáñez C. F. (2005) Neuron 45, 701–713 [DOI] [PubMed] [Google Scholar]
- 5.Moore M. W., Klein R. D., Fariñas I., Sauer H., Armanini M., Phillips H., Reichardt L. F., Ryan A. M., Carver-Moore K., Rosenthal A. (1996) Nature 382, 76–79 [DOI] [PubMed] [Google Scholar]
- 6.Pichel J. G., Shen L., Sheng H. Z., Granholm A. C., Drago J., Grinberg A., Lee E. J., Huang S. P., Saarma M., Hoffer B. J., Sariola H., Westphal H. (1996) Nature 382, 73–76 [DOI] [PubMed] [Google Scholar]
- 7.Sánchez M. P., Silos-Santiago I., Frisén J., He B., Lira S. A., Barbacid M. (1996) Nature 382, 70–73 [DOI] [PubMed] [Google Scholar]
- 8.Naughton C. K., Jain S., Strickland A. M., Gupta A., Milbrandt J. (2006) Biol. Reprod. 74, 314–321 [DOI] [PubMed] [Google Scholar]
- 9.Strömberg I., Björklund L., Johansson M., Tomac A., Collins F., Olson L., Hoffer B., Humpel C. (1993) Exp. Neurol. 124, 401–412 [DOI] [PubMed] [Google Scholar]
- 10.Kitagawa H., Hayashi T., Mitsumoto Y., Koga N., Itoyama Y., Abe K. (1998) Stroke 29, 1417–1422 [DOI] [PubMed] [Google Scholar]
- 11.Mack K., Day M., Milbrandt J., Gottlieb D. I. (1990) Brain Res. Mol. Brain Res. 8, 177–180 [DOI] [PubMed] [Google Scholar]
- 12.Schlingensiepen K. H., Lüno K., Brysch W. (1991) Neurosci. Lett. 122, 67–70 [DOI] [PubMed] [Google Scholar]
- 13.Beckmann A. M., Wilce P. A. (1997) Neurochem. Int. 31, 477–510 [DOI] [PubMed] [Google Scholar]
- 14.Gentilella A., Passiatore G., Deshmane S., Turco M. C., Khalili K. (2008) Oncogene 27, 5011–5018 [DOI] [PubMed] [Google Scholar]
- 15.Suter-Crazzolara C., Unsicker K. (1996) Brain Res. Mol. Brain Res. 41, 175–182 [DOI] [PubMed] [Google Scholar]
- 16.Lee S. L., Wang Y., Milbrandt J. (1996) Mol. Cell Biol. 16, 4566–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo M. W., Matheny C., Milbrandt J. (1993) Mol. Cell Biol. 13, 6858–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J. M., Calkins M. J., Chan K., Kan Y. W., Johnson J. A. (2003) J. Biol. Chem. 278, 12029–12038 [DOI] [PubMed] [Google Scholar]
- 19.Choi B. H., Kim C. G., Bae Y. S., Lim Y., Lee Y. H., Shin S. Y. (2008) Cancer Res. 68, 1369–1377 [DOI] [PubMed] [Google Scholar]
- 20.Shin S. Y., Bahk Y. Y., Ko J., Chung I. Y., Lee Y. S., Downward J., Eibel H., Sharma P. M., Olefsky J. M., Kim Y. H., Lee B., Lee Y. H. (2006) EMBO J. 25, 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S. H., Sharrocks A. D. (2005) EMBO J. 24, 2161–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiel G., Cibelli G. (2002) J. Cell. Physiol. 193, 287–292 [DOI] [PubMed] [Google Scholar]
- 23.Jin Y., Sheikh F., Detillieux K. A., Cattini P. A. (2000) Mol. Pharmacol. 57, 984–990 [PubMed] [Google Scholar]
- 24.Santiago F. S., Lowe H. C., Day F. L., Chesterman C. N., Khachigian L. M. (1999) Am. J. Pathol. 154, 937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fahmy R. G., Dass C. R., Sun L. Q., Chesterman C. N., Khachigian L. M. (2003) Nat. Med. 9, 1026–1032 [DOI] [PubMed] [Google Scholar]
- 26.Kim C. G., Choi B. H., Son S. W., Yi S. J., Shin S. Y., Lee Y. H. (2007) Cell Signal. 19, 1290–1300 [DOI] [PubMed] [Google Scholar]
- 27.Guha M., O'Connell M. A., Pawlinski R., Hollis A., McGovern P., Yan S. F., Stern D., Mackman N. (2001) Blood 98, 1429–1439 [DOI] [PubMed] [Google Scholar]
- 28.Tsai J. C., Liu L., Guan J., Aird W. C. (2000) Am. J. Physiol. Cell Physiol. 279, C1414–C1424 [DOI] [PubMed] [Google Scholar]
- 29.Lim C. P., Jain N., Cao X. (1998) Oncogene 16, 2915–2926 [DOI] [PubMed] [Google Scholar]
- 30.Cohen D. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11242–11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hipskind R. A., Baccarini M., Nordheim A. (1994) Mol. Cell. Biol. 14, 6219–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling Y., Lakey J. H., Roberts C. E., Sharrocks A. D. (1997) EMBO J. 16, 2431–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S. H., Sharrocks A. D. (2006) Biochem. Soc. Symp. 73, 121–129 [DOI] [PubMed] [Google Scholar]
- 34.Schratt G., Weinhold B., Lundberg A. S., Schuck S., Berger J., Schwarz H., Weinberg R. A., Rüther U., Nordheim A. (2001) Mol. Cell. Biol. 21, 2933–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson D. K., Robinson L., Hodge D. R., Kola I., Papas T. S., Seth A. (1997) Oncogene 14, 213–221 [DOI] [PubMed] [Google Scholar]
- 36.Ledda F., Paratcha G., Sandoval-Guzmán T., Ibáñez C. F. (2007) Nat. Neurosci. 10, 293–300 [DOI] [PubMed] [Google Scholar]
- 37.Jones M. W., Errington M. L., French P. J., Fine A., Bliss T. V., Garel S., Charnay P., Bozon B., Laroche S., Davis S. (2001) Nat. Neurosci. 4, 289–296 [DOI] [PubMed] [Google Scholar]
- 38.Bozon B., Davis S., Laroche S. (2002) Hippocampus 12, 570–577 [DOI] [PubMed] [Google Scholar]
- 39.Woodbury D., Schaar D. G., Ramakrishnan L., Black I. B. (1998) Brain Res. 803, 95–104 [DOI] [PubMed] [Google Scholar]
- 40.Verity A. N., Wyatt T. L., Hajos B., Eglen R. M., Baecker P. A., Johnson R. M. (1998) J. Neurochem. 70, 531–539 [DOI] [PubMed] [Google Scholar]
- 41.Yamagata K., Tagami M., Torii Y., Takenaga F., Tsumagari S., Itoh S., Yamori Y., Nara Y. (2003) Glia 41, 199–206 [DOI] [PubMed] [Google Scholar]
- 42.Koyama Y., Tsujikawa K., Matsuda T., Baba A. (2003) Biochem. Biophys. Res. Commun. 303, 1101–1105 [DOI] [PubMed] [Google Scholar]
- 43.Yamagata K., Hakata K., Maeda A., Mochizuki C., Matsufuji H., Chino M., Yamori Y. (2007) Neurosci. Res. 59, 467–474 [DOI] [PubMed] [Google Scholar]
- 44.Tsuchioka M., Takebayashi M., Hisaoka K., Maeda N., Nakata Y. (2008) J. Neurochem. 106, 244–257 [DOI] [PubMed] [Google Scholar]
- 45.Shin S. Y., Kim S. Y., Kim J. H., Min D. S., Ko J., Kang U. G., Kim Y. S., Kwon T. K., Han M. Y., Kim Y. H., Lee Y. H. (2001) J. Biol. Chem. 276, 7797–7805 [DOI] [PubMed] [Google Scholar]
- 46.Chung E. Y., Shin S. Y., Lee Y. H. (2007) Neurosci. Lett. 422, 43–48 [DOI] [PubMed] [Google Scholar]
- 47.Biesiada E., Razandi M., Levin E. R. (1996) J. Biol. Chem. 271, 18576–18581 [DOI] [PubMed] [Google Scholar]
- 48.Carlezon W. A., Jr., Duman R. S., Nestler E. J. (2005) Trends Neurosci. 28, 436–445 [DOI] [PubMed] [Google Scholar]
- 49.Bayatti N., Engele J. (2001) J. Neurochem. 78, 972–980 [DOI] [PubMed] [Google Scholar]
- 50.Koyama Y., Egawa H., Osakada M., Baba A., Matsuda T. (2004) Biochem. Pharmacol. 68, 275–282 [DOI] [PubMed] [Google Scholar]
- 51.Cen X., Nitta A., Ohya S., Zhao Y., Ozawa N., Mouri A., Ibi D., Wang L., Suzuki M., Saito K., Ito Y., Kawagoe T., Noda Y., Ito Y., Furukawa S., Nabeshima T. (2006) J. Neurosci. 26, 3335–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hisaoka K., Maeda N., Tsuchioka M., Takebayashi M. (2008) Brain Res. 1196, 53–58 [DOI] [PubMed] [Google Scholar]
- 53.Tanaka M., Ito S., Matsushita N., Mori N., Kiuchi K. (2000) Brain Res. Mol. Brain Res. 85, 91–102 [DOI] [PubMed] [Google Scholar]
- 54.Hisaoka K., Takebayashi M., Tsuchioka M., Maeda N., Nakata Y., Yamawaki S. (2007) J. Pharmacol. Exp. Ther. 321, 148–157 [DOI] [PubMed] [Google Scholar]