Abstract
Multiple sclerosis (MS) is a disease of the central nervous system with autoimmune etiology. Susceptibility to MS is linked to viral and bacterial infections. Matrix metalloproteinases (MMPs) play a significant role in the fragmentation of myelin basic protein (MBP) and demyelination. The splice variants of the single MBP gene are expressed in the oligodendrocytes of the central nervous system (classic MBP) and in the immune cells (Golli-MBPs). Our data suggest that persistent inflammation caused by environmental risk factors is a step to MS. We have discovered biochemical evidence suggesting the presence of the inflammatory proteolytic pathway leading to MS. The pathway involves the self-activated furin and PC2 proprotein convertases and membrane type-6 MMP (MT6-MMP/MMP-25) that is activated by furin/PC2. These events are followed by MMP-25 proteolysis of the Golli-MBP isoforms in the immune system cells and stimulation of the specific autoimmune T cell clones. It is likely that the passage of these autoimmune T cell clones through the disrupted blood-brain barrier to the brain and the recognition of neuronal, classic MBP causes inflammation leading to the further up-regulation of the activity of the multiple individual MMPs, the massive cleavage of MBP in the brain, demyelination, and MS. In addition to the cleavage of Golli-MBPs, MMP-25 proteolysis readily inactivates crystallin αB that is a suppressor of MS. These data suggest that MMP-25 plays an important role in MS pathology and that MMP-25, especially because of its restricted cell/tissue expression pattern and cell surface/lipid raft localization, is a promising drug target in MS.
MS4 is a chronic, inflammatory, and T cell-mediated autoimmune disease of the central nervous system associated with demyelination, axonal loss, and brain atrophy (1). Activated autoreactive T cells play a central role in MS pathophysiology. Susceptibility to MS is distantly linked to genetic variations and environmental risk factors, including vitamin D deficiency and viral and bacterial infections of the respiratory airways and gastrointestinal or urinary tracts (2, 3).
Experimental autoimmune encephalomyelitis (EAE) is an inducible disease in laboratory animals and a widely accepted model of MS. EAE is induced by autoreactive CD4+ T cells specific for myelin antigens or by immunization with myelin antigens or their peptide fragments. Proteolipid protein, myelin oligodendrocyte glycoprotein, and especially MBP are candidate autoantigens in MS. Immunoreactive MBP fragments appear in the cerebrospinal fluid in MS patients (4, 5).
MBP and its Golli splice variants are transcribed from a single gene in humans and mice (6). There are three transcription start sites in the MBP gene. Transcription from the first site generates Golli-BG21 and -J37. The classic MBP isoforms are transcribed from the two downstream sites (7). Because of the presence of the common exons, the fragmentation of the MBP isoforms can generate similar immunogenic peptides, including the fragment of the 1–15-residue immunogenic region and a source of a dominant T cell clonotype in EAE (8). The expression of the classic MBP transcripts is restricted to myelin-forming cells. BG21 and J37 are expressed in the thymus, spleen, and lymph nodes (7, 9) and in the myeloid lineage cells, including macrophages, dendritic cells, and granulocytes. Golli-MBPs play an incompletely understood but important role in MS (7, 10, 11).
Crystallin αB (CRYAB), a member of the small heat shock protein family (12), also plays an important role in EAE acting as a brake on several inflammatory pathways in both the immune system and central nervous system. As a result, CRYAB−/− mice show worse EAE (13). Antibodies against CRYAB are present in the cerebrospinal fluid of MS patients and in the serum from EAE mice (14).
Furin, PC1/3, PC2, PC4, PC5/6, PC7, and PACE4 proprotein convertases (PCs) selectively cleave the RX(K/R/X)R sequence motif and transform inactive precursors into biologically active proteins and peptides (15, 16). PCs are synthesized as pre-proproteins in which the N-terminal prodomain functions as an autoinhibitor. To become active, PCs autoproteolytically remove the prodomain (16). Self-activated PCs are then capable of activating the downstream proteinases, including MMPs. Because of the overlapping substrate preferences and cell/tissue expression, there is a level of redundancy in the functionality of PCs (17–19).
In MS, MMPs could be responsible for the influx of inflammatory mononuclear cells into the central nervous system, contribute to myelin destruction, and affect the integrity of the blood-brain barrier (20, 21). MMPs comprise a family of 24 enzymes that are expressed by many cell types, especially in malignancy and inflammation. Membrane-tethered MMPs are distinguished from soluble MMPs by the additional transmembrane and cytoplasmic domains (MT1–3-MMP and MT5-MMP). In contrast to these four MMPs, MT6-MMP/MMP-25 is attached to the cell membrane via a glycosylphosphatidylinositol anchor. All MMPs are synthesized as zymogens and require proteolytic removal of the N-terminal prodomain to become active proteinases. Lipid raft-associated MMP-25 with the RX(K/R)R motif in its propeptide is activated by furin (16, 22). Among MMPs, the expression of MMP-25 (initially called leukolysin) is most selectively linked to the leukocyte lineage cells and up-regulated in certain cancer types, including brain tumors (23). Because BG21 relocates to the caveolae-enriched lipid rafts upon phorbol ester stimulation of T cells (24), the presence of both BG21 and MMP-25 in the lipid raft compartment increases the opportunity for the selective MMP-25 proteolysis of the Golli proteins in the stimulated immune system cells.
Earlier, we determined that MMP-25 was superior to all of the other MMPs in cleaving the MBP isoforms (25). We demonstrated that MMP-25 proteolysis of the MBP isoforms generated, with a near quantitative yield, the immunogenic N-terminal 1–15-residue peptide. This peptide selectively stimulated the proliferation of the PGPR7.5 T cell clone isolated from mice with EAE and specific for the 1–15-residue peptide. Consistent with our results, MMP-25 is up-regulated in the spinal cord of mice with EAE induced by adoptive transfer of MBP-reactive T cells (26).
We now hypothesized that persistent inflammation caused by viral and bacterial infections is an integral step to both MMP-25 activation and Golli-MBP degradation and to the onset of autoimmune MS. To test our hypothesis, we demonstrated that LPS caused the up-regulation of PCs, MMP-25, and BG21 in the antigen-presenting macrophages. These events correlated with both the enhanced specific proteolysis of BG21 (a positive regulator of MS) and, simultaneously, the proteolytic inactivation of CRYAB (a negative regulator of MS) and to the presentation of the fragmented MBP sequence by the activated macrophages, thus stimulating the activation and proliferation of the reactive T cell populations resulting in autoimmunity and leading to MS.
Collectively, our experimental results suggest that persistent inflammation caused by viruses and bacteria stimulates the PC/MMP-25/BG21 proteolytic pathway in the activated antigen-presenting cells and that these events represent a step to MS. As a result of our studies and because of its both restricted cell/tissue expression pattern and cell surface localization, we have concluded that MMP-25 is a novel and promising drug target in MS.
MATERIALS AND METHODS
Reagents
All reagents were purchased from Sigma unless indicated otherwise. Human MBP (18.5-kDa isoform, GenBankTM AAH08749) was purchased from Biodesign. The total brain protein samples extracted from the total brain and from the medulla oblongata of healthy and MS patients were purchased from the BioChain Institute (Hayward, CA). The frozen human brain tissue samples obtained from MS patients were kindly provided by the Human Brain and Spinal Fluid Resource Center (UCLA, Los Angeles). The CD4+ T cell clone specific to murine MBP-(1–15) fragment were kindly provided by Dr. Vipin Kumar (Torrey Pines Institute for Molecular Studies, San Diego). An inhibitor of proprotein convertases (decanoyl-Arg-Val-Lys-Arg-chloromethyl ketone; dec-RVKR-cmk) was obtained from Bachem. GM6001 and AG3340 (small molecule hydroxamate inhibitors of MMPs) were purchased from Chemicon and kindly provided by Dr. P. Baciu (Allergan), respectively. A plasmid encoding human CRYAB was a kind gift of Dr. Lawrence Steinman (Stanford University, Palo Alto, CA).
Antibodies
Murine monoclonal antibody (11-5B) against the human 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase; oligodendrocyte marker) and the rabbit polyclonal antibody AB980 against human MBP were from Chemicon. The murine SMI94 and SMI99 monoclonal antibodies against the 70–89- and 131–136-residue fragments of human MBP, respectively, were from Covance. The rabbit polyclonal antibodies to the Golli portion of Golli-MBP and to the human PC2 and MMP-25 were kindly provided by Drs. Celia and Anthony Campagnoni (UCLA) (27), Robert Day (University of Sheerbrooke, Quebec, Canada), and Rafael Fridman (Wayne State University, Detroit, MI), respectively.
Cells
Murine macrophage RAW264.7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Murine mammary carcinoma 4T1 cells were cultured in Dulbecco's modified Eagle's medium, 10% fetal bovine serum supplemented with gentamicin (10 μg/ml). Bone marrow macrophages (BMDM) were derived from murine bone marrow cells by culturing them for 7 days in a 3-fold diluted medium pre-conditioned by mouse L929 cell fibroblasts. Human breast carcinoma MCF-7 cells were stably transfected with the full-length human MMP-25 cDNA gene (GenBankTM AB042328) in the pcDNA3.1-neo vector (MCF-MMP-25 cells). Stable clones were selected using G418 (400 μg/ml) and analyzed using Western blots with the MMP-25 antibody. To avoid clonal effects, the most efficient multiple clones were pooled and used in our studies. As a control, we used MCF-7 cells stably transfected with the original pcDNA3.1-neo plasmid (MCF-mock). Where indicated, cells were treated with 1 μg/ml LPS for 18–48 h.
Recombinant Proteins
Human PC2 (a kind gift of Dr. Robert Day, University of Sheerbrooke, Quebec, Canada) was expressed and purified from the S2 Drosophila expression system (Invitrogen) (28). Soluble human furin was purified from the stably transfected Sf9 insect cell line (29). The murine BG21 and J37 Golli-MBP isoforms and human CRYAB were expressed in Escherichia coli and then isolated from the soluble protein fraction using metal-chelating chromatography (25).
Recombinant MMPs
The individual catalytic domains of MMP-10 and MMP-12 were purchased from Biomol. The individual catalytic domains of MMP-8, MMP-14, MMP-15, MMP-16, MMP-17, MMP-24, and MMP-25 were expressed in E. coli and purified from the inclusion bodies in 8 m urea using metal-chelating chromatography. The purified samples were then refolded to restore their native conformation and proteolytic activity (30). The recombinant pro-forms of the catalytic domains of MMP-2 and MMP-9 were purified from the serum-free medium conditioned by the stably transfected HEK293 cells using the gelatin-column chromatography (31). Pro-MMP-2 and pro-MMP-9 were activated using 4-aminophenylmercuric acetate as described earlier (31). The purity of the isolated MMPs was confirmed by SDS-PAGE followed by Coomassie staining of the gels. The MMP samples (the purity >95%) were used in our subsequent studies. The concentration of the catalytically active MMPs was determined using a fluorescent assay by titration against a standard solution of a hydroxamate inhibitor (GM6001 or AG3340) of known concentration (25, 30). (7-Methoxycoumarin-4-yl)-acetyl-Pro-Leu-Gly-Leu-(3-[2,4-dinitrophenyl]-l-2,3-diaminopropionyl)-Ala-Arg-NH2 (Bachem) was used as a fluorescent substrate.
RT-PCR of PCs and MMPs in Murine Macrophage RAW264.7 Cells
The primer sequence we used for the RT-PCR amplification of furin, PC1/3, PC2, PACE4, PC5/6, and PC7 (the calculated size of the PCR products was as follows: 399, 457, 261, 456, 510, and 471 bp, respectively) was reported earlier (32). The primers used for the amplification of MMP-2, MMP-9, MMP-12, MMP-14, and MMP-25 (the calculated size of the PCR products was as follows: 278, 343, 297, 285, and 299 bp, respectively) are shown in supplemental Table S1. Total RNA was isolated from intact RAW264.7 cells or cells treated for 24 h with 1 μg/ml LPS using a TRIzol reagent and then additionally purified using a mini RNA Isolation II kit column (ZymoResearch). The purified RNA samples (250 ng) were used in the amplification reactions (normally, 30–35 cycles) containing the specific primers (600 nm), Superscript II reverse transcriptase (Invitrogen) and TaqDNA polymerase, or a One-Step RT-PCR system (Qiagen). The resulting PCR products were sequenced to confirm their authenticity. Murine glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 300 bp) was used as a control.
Quantitative RT-PCR (Q-RT-PCR)
Total RNA was isolated from the cells using the RNeasy Maxi kit (Qiagen). RNA (3.0 μg) was primed with oligo(dT) and then reverse-transcribed using the QuantiTech reverse transcription kit (Qiagen). Q-RT-PCR was performed using an Mx3000p instrument (Stratagene). The cDNA (25 ng) and the primers (400 nm) were used in the duplicate 30-μl reactions with the 1× SYBR Green PCR Master Mix (Applied Biosystems). The sequence of the primers is shown in supplemental Table S1. PCRs (40 cycles) were performed using 15 s of denaturing at 95 °C, 1 min of annealing at 58 °C, and 30 s of elongation at 72 °C. PCR product amplification was monitored by SYBR Green fluorescence and normalized relative to the Rox dye standard. A standard curve was generated with β-actin primers for each tested cDNA sample, and it proved to be linear over 4 orders of magnitude. This curve was used to determine the relative differences in cDNA from changes in CT (cycle threshold) values.
Cell Immunostaining
Cells grown in Dulbecco's modified Eagle's medium, 10% fetal bovine serum in LabTek chamber slides were fixed 15 min in 4% paraformaldehyde and blocked using 2% bovine serum albumin. For PC2 staining cells were additionally permeabilized for 4 min using 0.1% Triton X-100. Cells were then incubated for 2 h with the primary polyclonal antibodies (dilution 1:500–1:1000) followed by a 2-h incubation with the goat anti-rabbit secondary antibody conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes). The slides were mounted in the Vectashield medium (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole for the nuclear staining. Images were acquired at a ×400 magnification on an Olympus BX51 fluorescence microscope equipped with an Olympus MagnaFire digital camera and the MagnaFire 2.1C software.
Immunohistochemistry
Frozen brain sections from 10 healthy patients and 10 patients with MS were stained with the SMI94 and SMI99 MBP and the 11-5B CNPase antibodies followed by a diaminobenzidine-based detection method employing the Envision Plus horseradish peroxidase system (DakoCytomation) and an automated Dako immunostainer. The slides were counterstained, dehydrated, and mounted with permanent mounting media. For all tissues examined, the immunostaining procedure was done in parallel using the preimmune serum.
Gene Arrays Analysis and Data Processing
To generate the heat map and the hierarchical clustering of MMPs and their physiological inhibitors (tissue inhibitors of metalloproteinase-1, -2, -3, and -4 and RECK), the neurocrine data set (NCBI GEO code GSE3526) of the genome-wide profiling of 356 samples of 65 normal tissue types was used. The original raw array data were processed using the RMA algorithm and further normalized and analyzed using GeneSpring and the HCE program (33). The processed data were then normalized per every chip and gene using the default procedure in GeneSpring. Per chip normalization was to the 50th percentile. Per gene normalization was to the median. Hierarchical clustering of the array data (log10 data) was computed using the average linkage (Unweighted Pair Group Method with Arithmetic mean) and correlation distance measure options of the HCE program (33). Clustering of the tissue types was done using the whole gene set. The subset of genes was then extracted, and the clustering of genes was performed within the individual tissue types.
Cleavage of Protein Substrates
MBP, BG21, J37, and CRYAB (10 μm each) were co-incubated for 1 h at 37 °C with the individual MMPs (1–100 nm; 1:100–1:10,000 enzyme/substrate molar ratio) in 50 mm HEPES, pH 6.8, supplemented with 10 mm CaCl2 and 50 μm ZnCl2. MBP, BG21, and J37 (∼10 μm each) were also co-incubated for 3 h with furin (1:4 enzyme/substrate molar ratio) and PC2 (3–12 activity units) in 100 mm HEPES, pH 7.5, and 20 mm BisTris, pH 5.6, respectively, both supplemented with 1 mm CaCl2 and 0.005% Brij-35. One activity unit was equal to the amount of the enzyme that was required to cleave 1 pmol/min of the pyroglutamic acid-Arg-Thr-Lys-Arg-methyl-coumaryl-7-amide substrate at 37 °C. Where indicated GM6001 (2.5 μm) and dec-RVKR-cmk (5 μm) were added to the reactions to inhibit MMPs and PCs, respectively. The cleavage reactions were stopped using a 5× SDS sample buffer. The digest samples were analyzed by SDS-gel electrophoresis and MALDI-TOF MS using an Autoflex II MALDI TOF/TOF instrument (Bruker Daltonics). For MS analysis, the reactions were cooled on ice, and equal volumes of a sample and of a sinapic acid (20 mg/ml) in 50% acetonitrile, 0.1% trifluoroacetic acid solution were co-crystallized directly on the MALDI target plate and allowed to dry for 5 min. Mass spectra were processed with FlexAnalysis 2.4. The singly charged cleavage products, which were observed only in the cleavage reactions but not in the controls, were recorded and processed further.
Cleavage of Synthetic Peptides
The synthetic peptide 102VRRRRRYALS (1332 Da) that spans the furin cleavage sites in the human MMP-25 prodomain was synthesized by GenScript. The peptide (1.5 μg) was incubated for 2 h at 37 °C with furin or PC2 (1 activity unit each) in 100 mm HEPES, pH 7.5, and 20 mm BisTris, pH 5.6, respectively, both supplemented with 1 mm CaCl2 and 0.005% Brij-35. The mass of the intact peptide and the 102VRRRRR cleavage product (898 kDa) was determined by MALDI-TOF MS. dec-RVKR-cmk (5 μm) was added to the reactions to inhibit the PC activity. For mass spectrometric analysis, equal volumes of a sample and of an α-cyano-4-hydroxycinnamic acid (20 mg/ml) in 50% acetonitrile, 0.1% trifluoroacetic acid solution were co-crystallized directly on the MALDI target plate and allowed to dry for 5 min.
Prediction and Ranking of the Cleavage Sites
To predict and rank the MMP and furin cleavage sites in the CRYAB, MBP, BG21, and J37 sequence, we used a specialized computer program we have developed and validated earlier (25).
Stimulation of the Murine T Cell Clone PGPR7.5 Specific to the MBP-(1–15) Fragment
Prior to the assay, the E. coli-derived BG21 and J37 samples were purified using an Affinity Pak Detoxi-Gel kit (Thermo Scientific) to remove LPS. MBP, BG21, and J37 (5 μm each) were digested for 1 h at 37 °C by the individual MMPs (an enzyme/substrate molar ratio of 1:100) or for 3 h with furin (an enzyme/substrate molar ratio of 1:4). The irradiated splenocytes from B10.PL mice (1 × 106) were co-incubated for 1 h with the digest reactions. As controls, intact MBP, BG21, and J37 and the murine MBP-(2–18)/Golli-MBP-(135–151)-peptide ASQKRPSQRSKYLATAS (5 μm each) were used. The CD4+ T cells (5 × 105; clone PGPR7.5) specific for murine 1–15-residue peptide presented in the major histocompatibility complex H-2U context were then added to the reactions for 72 h. [3H]Thymidine (1 μCi) was added to the cells for 14 h. The incorporation of the label into the T cells was measured by liquid scintillation counting.
RESULTS
Demyelination and Macrophage Infiltration
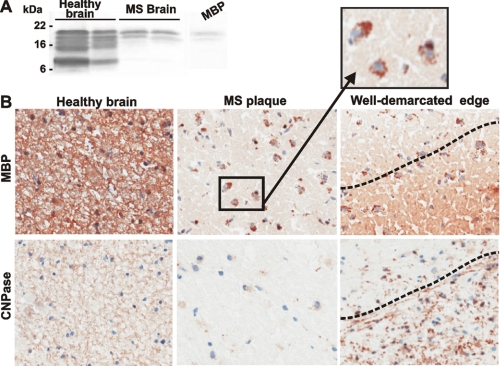
MS is characterized by the active degradation of the myelin sheath resulting in the multiple regions of focal myelin loss (lesions or plaques) in the central nervous system and by infiltration of macrophages and lymphocytes that are reactive against myelin antigens. The results of our Western blotting and immunostaining studies support these conclusions and show the loss of MBP in the chronic active plaques of MS patients. The decrease in MBP correlates both with the presence of the phagocytic macrophages, which co-localize with fragmented MBP, and with a loss of oligodendrocytes in the plaque (Fig. 1).
FIGURE 1.
Representative immunostaining of the healthy and MS brain sections. A, protein extracts (25 μg of total protein each) from a healthy total brain and medulla oblongata (left and right lines, respectively) and from the plaque area of the two MS brain samples were analyzed by Western blotting with the MBP antibody. The extracts were purchased from BioChain Institute (Hayward, CA). The representative samples are shown. Right lane, purified MBP (10 ng). B, immunostaining of the brain sections with the MBP and 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase) antibodies. The staining with the primary antibodies was followed by a diaminobenzidine-based detection method. A well demarcated edge (right) is visible between the plaque and the normal brain area. Magnification, ×160. Inset, the phagocytic macrophages (identified by their characteristic size and morphology) co-localize with fragmented MBP (brown) in the plaque. The identity of the macrophages was additionally confirmed using an antibody against the pan macrophage F4/80 marker (Pharmingen).
MMP-25 Is a Bone Marrow Enzyme
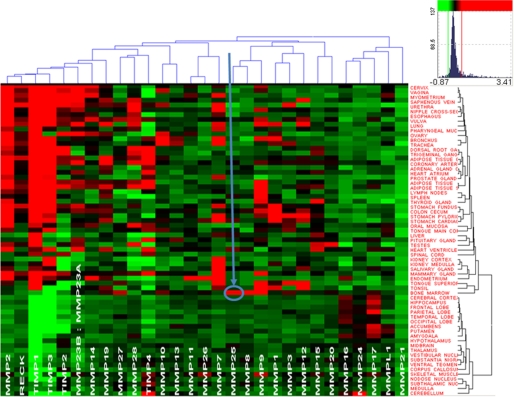
To support the characteristic association of MMP-25 (leukolysin) with the bone marrow-derived leukocyte lineage cells, we analyzed the publicly available data of the genome-wide profiling of 356 samples of 65 normal human tissues each of which included over 47,000 human gene transcripts. A subset of the resulting data that relates to the human MMPs and their physiological inhibitors (TIMPs and RECK) indicates a unique association of MMP-25 with the bone marrow (Fig. 2). Indeed, MMP-25 is the only MMP that is highly expressed in the bone marrow relative to other normal human tissues. This parameter explains the elevated expression of MMP-25 in the bone marrow-derived cells, including the macrophages.
FIGURE 2.
MMP-25 is uniquely enriched in the bone marrow. Heat map and the hierarchical bi-clustering of the expression data (log10 of expression level) of MMPs and their natural inhibitors TIMPs and RECK (Vertical clustering, by the level of expression in the individual tissue types. Right, horizontal clustering, by gene expression levels, on the top). The data were derived from 356 gene arrays of 65 normal tissue types. Red and green correspond to the high and the low expression levels, respectively. Black represents an average level of expression. Color map inset shows the distribution (frequency) of different gene expression levels in the set and related colors. Arrow points to MMP-25 in the bone marrow.
MMP-25 Efficiently Cleaves MBP, BG21, and J37 and Generates the Immunogenic MBP Peptides
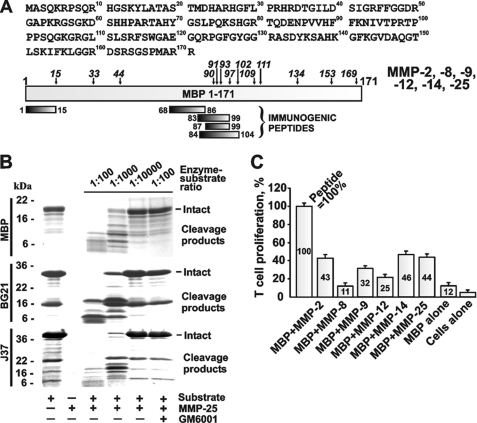
The three purified MBP isoforms (BG21, J37, and the classic neuronal MBP) were readily degraded by MMP-25 in the cleavage reactions in vitro (Fig. 3). The degradation of MBP was detectable at an enzyme/substrate molar ratio of 1:10,000, whereas at a 1:1000 ratio the degradation was largely accomplished. These data support our previous observations that MMP-25 was the most efficient among all of the individual MMPs we tested for cleaving the MBP, BG21, and J37 constructs (25).
FIGURE 3.
MMP proteolysis of MBP and activation of the specific T cell clone. A, 1–171-residue sequence of human MBP (GenBankTM accession number AAH08749). The immunogenic regions are shown at the bottom of the panel using the MBP residue numbering. Following the MBP cleavage by MMPs, the mass and, consecutively, the sequence of the digest fragments were determined by MALDI-TOF MS. The italicized numbers indicate the positions of the cleavage sites. B, MMP-25 proteolysis of MBP, BG21, and J37. Where indicated, an inhibitor (GM6001) was added to the reactions. C, MMPs cleave MBP and generate the N-terminal peptide that stimulates the proliferation of the specific T cell clone. MBP (5 μm) was cleaved by the indicated MMPs (an enzyme/substrate molar ratio of 1:100). The irradiated splenocytes from B10.PL mice were incubated with the cleavage reactions. The PGPR7.5 T cells specific for the murine 1–15-residue MBP peptide and [3H]thymidine were then added to the reactions. The incorporation of the label into the T cells was measured by liquid scintillation counting. For MBP alone, intact MBP (5 μm) was added to the cells. For cells alone, no peptide was added. For peptide, the 1–15-residue ASQKRPSQRSKYLATAS MBP peptide (5 μm) was used as a control (= 100%). The numbers show the percentage relative to the peptide control. The experiments were repeated four times, and the data represent the mean ± S.E.
According to our cleavage data, multiple MMPs, including MMP-25, were capable of readily generating the C-terminal and especially N-terminal digest peptides of the MBP portion of the constructs. These peptides were highly similar to the known immunogenic sequence regions of MBP that were capable of causing EAE in animal models (34–36). Generally, because of the cleavage preference redundancy among MMPs, multiple individual MMPs are capable of generating, albeit with widely varying kinetics, similar cleavage peptide sequences as a result of the proteolysis of the MBP and Golli-MBP isoforms.
To determine which individual MMP most efficiently generates the immunogenic 1–15-residue peptide, we used the digests to stimulate the proliferation of the murine T cell PGPR7.5 clone, which is specific to the 1–15-residue fragment of MBP (37). MBP was cleaved by MMP-2, MMP-8, MMP-9, MMP-12, MMP-14, and MMP-25. The irradiated splenocytes from B10.PL mice were co-incubated with the digests. The PGPR7.5 T cells and [3H]thymidine were then added to the reactions. The incorporation of the label into the T cells was measured by liquid scintillation counting. The digest products were internalized and presented by the splenocytes, and then the presented peptides stimulated the proliferation of the specific PGPR7.5 T cells. As a control, we used the synthetic ASQKRPSQRSKYLATAS peptide (5 μm) that corresponded to the 2–18-residue sequence of murine MBP and the stimulatory effect of which we took as 100%. The MMP-2, MMP-14, and MMP-25 digest of MBP resulted in a similarly high, 43–46%, level of proliferation of PGPR7.5 T cells compared with the equimolar amount of the ASQKRPSQRSKYLATAS peptide, thus suggesting a high yield of the specific immunogenic fragment in the digest (Fig. 3). A comparable level of stimulation of the specific PGPR7.5 T cell clone was observed when the MMP-25 digest of BG21 was used in the T cell proliferation assay (25).
MMP-25, Furin, and PC2, and BG21 Are Up-regulated in the Activated Macrophages
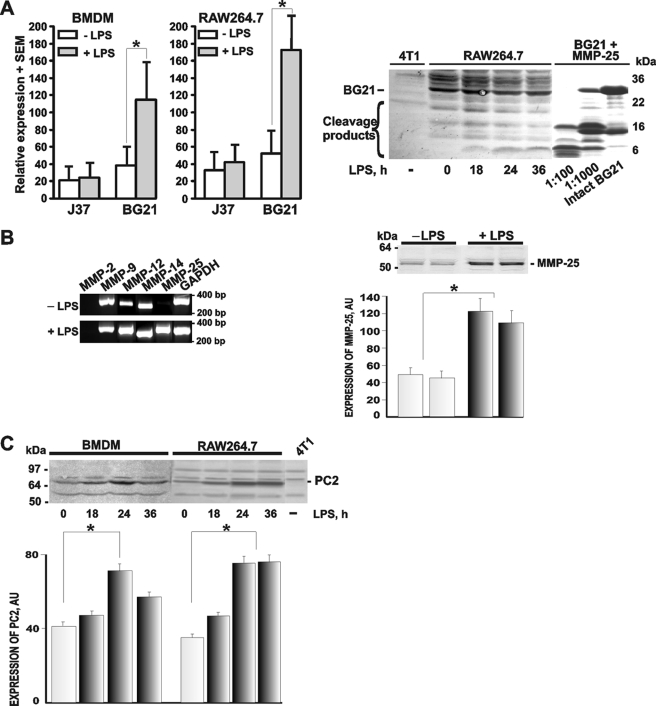
We then determined if the activated macrophages overexpressed PCs, MMPs, and their Golli-MBP substrates. To mimic inflammation, the macrophages were stimulated by LPS. As determined by Q-RT-PCR, LPS caused a several-fold increase of the BG21 mRNA levels both in the bone marrow-derived macrophages and RAW264.7 macrophages cells (Fig. 4). RT-PCR also readily detected a significant increase in the expression levels of MMP-25 in the LPS-stimulated RAW264.7 macrophages, although there was no change in other individual MMPs. The analysis of the RAW264.7 cell lysates using Western blotting and immunostaining with the MMP-25 antibody corroborated the up-regulation of MMP-25 in the LPS-primed macrophage cells (Figs. 4 and 5).
FIGURE 4.
LPS stimulates the proteolytic pathway in the macrophages. A, LPS up-regulates BG21 in the macrophages. Left, BMDM and RAW264.7 macrophage cells were left intact or treated 24 h with LPS. Total RNA extracted from the cells was analyzed by Q-RT-PCR to determine the levels of the BG21 and J37 mRNAs. Right, murine breast carcinoma 4T1 and macrophage RAW264.7 cells were treated with LPS for the indicated time. The cell lysate samples (50 μg of total protein) were analyzed by Western blotting with the Golli-MBP antibody. The three right lanes show the in vitro cleavage of BG21 by MMP-25 at a 1:100–1:1000 enzyme/substrate molar ratio and intact BG21. The experiments were repeated 3–5 times, and the data represent the mean ± S.E. *, p < 0.05. B, LPS up-regulates MMP-25 in the macrophages. Left, RAW264.7 cells were left intact (−LPS) or treated 24 h with LPS (+LPS). Total RNA extracted from the cells was analyzed by RT-PCR to determine the levels of the mRNAs of the individual MMPs. GAPDH was used as a control. The calculated size of the amplified fragments was 278, 343, 297, 285, 299, and 300 bp for MMP-2, -9, -12, -14, -25, and GAPDH, respectively. Right, two independent RAW264.7 cell samples were left intact or treated 24 h with LPS. The cell lysate samples (50 μg of total protein) were analyzed by Western blotting with the MMP-25 antibody (top panel). The images were scanned and digitized using the Fujifilm MultiGauge software. The MMP-25 band density (bottom panel) is shown in arbitrary units (AU). These experiments were repeated 3–5 times with comparable results. *, p < 0.05. C, LPS up-regulates PC2 in the macrophages. BMDM and RAW264.7 cells were left intact or treated with LPS for 18–36 h. The cell lysate samples (50 μg of total protein) were analyzed by Western blotting with the PC2 antibody (top panel). Murine mammary carcinoma 4T1 cells (4T1), which do not produce any significant level of PC2, were used as a negative control. The images were scanned and digitized using the Fujifilm MultiGauge software. The PC2 band density (bottom panel) is shown in arbitrary units (AU). These experiments were repeated 3–5 times with comparable results. *, p < 0.05.
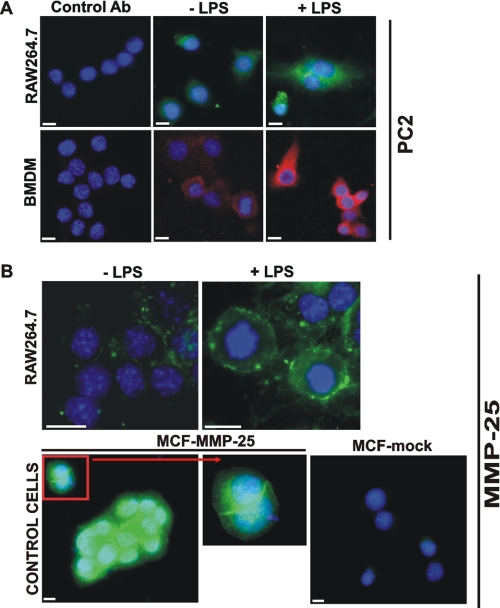
FIGURE 5.
Immunostaining of PC2 and MMP-25. A, macrophage RAW264.7 cells and the murine BMDM were grown in the LabTek chamber slides. The cells were left intact (−LPS) or stimulated 24 h with LPS (1 μg/ml). The cells were fixed, permeabilized, and stained with the PC2 antibody (Ab) followed by the secondary antibody conjugated with Alexa Fluor 488 (green) or Alexa Fluor 594 (red). Staining with the preimmune rabbit serum (control antibody) was negative. The nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). B, macrophage RAW264.7 cells were grown in the LabTek chamber slides. The cells were left intact (−LPS) or stimulated 24 h with LPS (1 μg/ml). The cells were fixed and stained with the MMP-25 antibody. Breast carcinoma MCF7 cells stably transfected with MMP-25 (MCF-MMP-25) or the empty plasmid (MCF-mock) were used as controls. Inset, cell surface staining of MMP-25. The nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). Original magnification ×400; the bar, 10 μm.
The expression of several PCs, including furin, PC2, PACE4, and PC7, was also confirmed in the intact and LPS-stimulated RAW264.7 cells by RT-PCR (supplemental Fig. 1). LPS stimulation of the cells caused an increase in the expression of PC2. The Western blotting and immunostaining analyses confirmed the enhanced expression of PC2 the LPS-stimulated macrophages (Figs. 4 and 5).
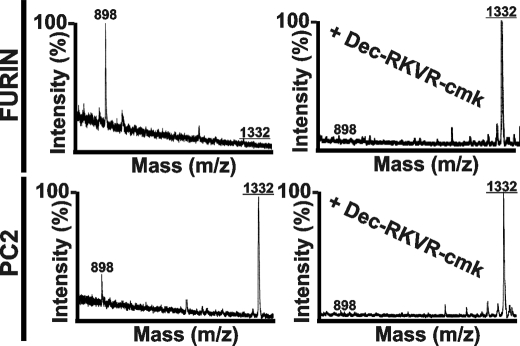
To determine whether PC2 in addition to furin can activate MMP-25, we cleaved the synthetic 102VRRRRRYALS (1332 Da) peptide that spans the furin cleavage site in the human MMP-25 prodomain by using these PCs. The MALDI-TOF MS analysis identified the presence of the 102VRRRRR cleavage fragment (898 Da) in the cleavage reactions (Fig. 6), thus confirming that the MMP-25 zymogen may be activated by both furin and PC2.
FIGURE 6.
MS analysis of the VRRRRRYALS peptide cleavage. The VRRRRRYALS peptide that spans the furin cleavage site in the propeptide of human MMP-25 was incubated for 2 h at 37 °C with furin and PC2 (1 activity unit each). The mass of the cleavage peptides was determined using MALDI-TOF MS. There was no difference between the calculated and the estimated molecular mass of the peptides. Where indicated, the PC inhibitor (dec-RVKR-cmk) was added to the reactions. The mass of the intact peptide was 1332 Da (underlined). The mass of the VRRRRR cleavage product was 898 Da.
Overall, our data suggested that PCs (especially PC2 and ubiquitously expressed furin), MMP-25, and BG21 were specifically up-regulated in the activated macrophages thus providing an opportunity for the activation of the PC/MMP-25/BG21 cleavage pathway in the inflammatory activated macrophages. Furthermore, Western blots confirmed the expression of BG21 in RAW264.7 cells and also detected the presence of the BG21 fragments in the LPS-treated cells. The BG21 fragments we observed in the LPS-stimulated RAW264.7 cells were similar to those that have been generated by MMP-25 proteolysis of the purified BG21 (Fig. 4).
High Levels of Furin Directly Contribute to the MBP Cleavage
In contrast with MMP-25, only exceedingly high levels of furin are capable of directly cleaving the MBP isoforms in the in vitro reactions. At an enzyme/substrate molar ratio of 1:4, there was a noticeable level of furin proteolysis of MBP, BG21, and J37. As a result, the N-terminal immunogenic fragments of MBP were generated. The cleavage map, the sequence, and the mass of the digest peptides are shown in Fig. 7 and supplemental Table S2. High levels of furin, which are required for the direct cleavage of MBP, suggest that in vivo PCs serve to activate macrophage MMP-25 rather than to accomplish themselves the degradation of the MBP isoforms.
FIGURE 7.
Furin proteolysis of MBP and activation of the specific T cell clone. A, furin proteolysis of the MBP isoforms. MBP, J37, and BG21 were incubated for 3 h at 37 °C with furin at an enzyme/substrate molar ratio of 1:4. The digest reactions were analyzed by SDS-gel electrophoresis followed by Coomassie staining. Where indicated, dec-RVKR-cmk was added to the reactions. B, 1–171-residue sequence of human MBP (GenBankTM accession number AAH08749). The immunogenic regions are shown at the bottom of the panel using the MBP residue numbering. Following the MBP cleavage by MMPs, the mass and, consecutively, the sequence of the digest fragments was determined by MALDI-TOF MS. The italicized numbers indicate the positions of the cleavage sites. C, furin proteolysis of MBP generates the N-terminal peptide that stimulates the proliferation of the specific T cell clone. MBP, BG21, and J37 (5 μm each) were co-incubated for 3 h at 37 °C with furin at an enzyme/substrate molar ratio of 1:4. The irradiated splenocytes from B10.PL mice were incubated with the cleavage reactions. The PGPR7.5 T cells specific for the murine MBP-(1–15)-peptide and [3H]thymidine were then added to the reactions. The incorporation of the label into the T cells was measured by liquid scintillation counting. MBP, BG21, and J37 alone indicate intact MBP, BG21, and J37 (5 μm each) added to the cells. Cells alone indicates no peptide. Peptide, the 1–15-residue ASQKRPSQRSKYLATAS MBP peptide (5 μm) was used as a control (= 100%). The numbers show the percentage relative to the peptide control. The experiments were performed in triplicate and repeated twice. The data represent the mean ± S.E.
In Vitro Cleavage of CRYAB by MMPs
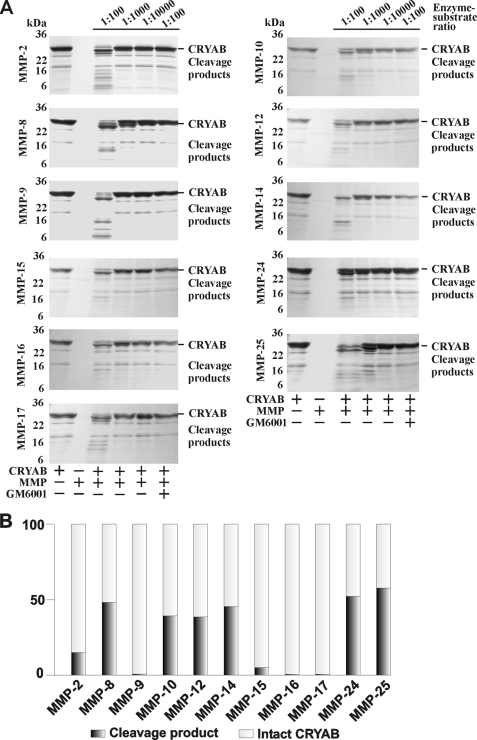
To provide biochemical evidence that the activated MMP-25 and other individual MMPs that cleave the MBP isoforms can simultaneously proteolyze the macrophage CRYAB (a negative regulator of MS) (13), the purified CRYAB construct was co-incubated with MMPs. The digests were then analyzed by gel electrophoresis and also by MALDI-TOF MS to determine the mass and, consecutively, the sequence of the resulting digest fragments (Fig. 8 and supplemental Table S3). CRYAB was sensitive to proteolysis by multiple MMPs. Thus, MMP-8, MMP-10, MMP-12, MMP-14, MMP-24, and especially MMP-25 initiated the digestion of CRYAB at an enzyme/substrate ratio of 1:1000, whereas the proteolytic effect of MMP-9, MMP-15, MMP-16, and MMP-17 were significantly less visible. As a result of MMP proteolysis, including MMP-25 proteolysis, the known immunogenic 1–16-, 153–168-, and 73–83-residue fragments of CRYAB were readily detected in the digest reactions by MALDI-TOF MS (Fig. 9).
FIGURE 8.
Multiple MMPs cleave CRYAB. A, purified 1–175 human crystallin αB was co-incubated with the individual MMPs at the indicated enzyme/substrate molar ratio. Where indicated, an inhibitor (GM6001) was added to the reactions. B, quantification of MMP proteolysis of CRYAB. The images shown in A were scanned and digitized. The ratios of the intact CRYAB (27–28 kDa) to the major digest product (25–26 kDa) are shown for the individual MMPs (the 1:1000 enzyme/substrate ratio except the 1:100 ratio which was used to calculate the data with MMP-24). The experiments were repeated 4–5 times with comparable results. The data from one representative experiment are shown.
FIGURE 9.
MMPs proteolysis of CRYAB generates the immunogenic fragments. A, 1–175-residue sequence of the human crystallin αB construct tagged with the V5 and His6 tags. B, schematic representation of the CRYAB construct and MMP proteolysis. The immunogenic regions are shown at the bottom of the panel. The immunogenicity of these regions was detected using Biozzi ABH, SJL, and 129 mice and Lewis rats (47, 48). Following the CRYAB cleavage by MMPs, the mass and, consecutively, the sequence of the digest fragments were determined by MALDI-TOF MS. The numbers indicate the positions of the cleavage sites.
DISCUSSION
MS is an autoimmune degenerative disease of the central nervous system. Environmental risk factors, including multiple viral and bacterial infections, vitamin D deficiency, and probably the resulting inflammation, appear to play a role in the onset of MS in humans. Several MMP family members contribute to pathology in MS and EAE (38–42). The existing data suggest that there are links among MMP proteolysis of MBP, demyelination, and MS (21). Theses events cause an MBP deficiency, myelin sheath destruction, and axon degeneration leading to MS. When fragmented, MBP generates several peptides that are potent immunogens (1, 43–46). EAE is an animal model of autoimmune MS. Immunization with MBP fragments induces autoimmune demyelinating EAE in animals. Several fragments of MBP are known to generate EAE efficiently (34–36).
There are, however, several forms of MBP. The expression of the classic MBP transcripts is restricted to myelin-forming cells. In turn, the splice variants of MBP called Golli-MBP BG21 and J37 (in mice) are expressed in the thymus, spleen, and lymph nodes and in the myeloid lineage cells. Because of the presence of the common exons in MBP and its Golli variants, their fragmentation can generate similar immunogenic peptides (7, 9).
We have the biochemical evidence that the proteolysis of Golli-MBPs by lipid-raft-associated MMP-25 plays a highly important role in the origin of MS. In contrast to other individual MMPs, MMP-25 is primarily expressed in the normal bone marrow and the bone marrow-derived cells, including the antigen-presenting macrophages. The propeptide of MMP-25 exhibits the furin cleavage motif, and as a result, MMP-25 is activated by furin and related PCs. According to our in vitro cleavage data, MMP-25 is superior to all of the other MMPs in cleaving the MBP and Golli-MBP isoforms. MMP-25 proteolysis of the MBP isoforms efficiently generates the immunogenic peptide sequence that selectively stimulates the proliferation of the T cell clone isolated from mice with EAE. PCs (especially furin and PC2), MMP-25, Golli-MBP, and the fragmentation of the latter are up-regulated in the activated macrophages under the conditions that mimic inflammation in vivo. The fragments of Golli-MBP we observed in the stimulated macrophages were similar to those we generated by MMP-25 proteolysis of the purified Golli-MBP using the in vitro cleavage reactions.
Our data also suggest that, in addition to the proteolysis of Golli-MBP, MMP-25 is effective in fragmenting CRYAB, a suppressor of MS (13). As a result, we now believe that in the course of inflammation caused by environmental risk factors MMP-25 is likely to play a dual role as follows: (i) the proteinase can inactivate CRYAB in the macrophages and decrease the suppressive effect of CRYAB in MS, and (ii) the proteinase can stimulate the degradation of Golli-MBP, thus initiating the autoimmune processes and increasing the predisposition for the development of MS in the affected patients.
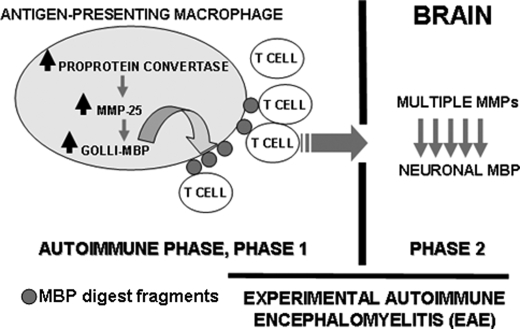
Collectively, our experimental results suggest that there is a proteolytic pathway that is activated in inflammation and that the activation of this pathway in the course of viral and bacterial infections is a step to autoimmune MS (Fig. 10). We now believe that MMP-25 plays an important role in this pathway and that MMP-25, especially because of its restricted cell/tissue expression pattern and its cell surface/lipid raft localization, is a promising drug target in MS.
FIGURE 10.
Inflammatory proteolytic pathway to MS. Phase 1, persistent inflammation caused by viral and bacterial infections and other related factors up-regulates PCs (furin and PC2), MMP-25, and BG21 (up arrows) in the stimulated antigen-presenting macrophages. Autoactivated PCs activate MMP-25, which then cleaves BG21 (the down arrows). The immunogenic fragments of the MBP portion of BG21 are presented in the major histocompatibility complex on the cell surface leading to the activation of the specific populations of the autoimmune T cells. Phase 2, if the brain barrier is not intact, the activated T cells home to the brain, attack the neuronal MBP, and cause inflammation leading to the macrophage infiltration and the follow-up up-regulation of the activity of multiple MMP types, many of which contribute to the further destruction of MBP and to the local demyelination causing plaque formation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA83017 and CA77470 (to A. Y. S.) and RR020843 (to J. W. S. and A. Y. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Fig. 1.
- MS
- multiple sclerosis
- BMDM
- bone marrow-derived macrophage
- CRYAB
- crystallin αB
- EAE
- experimental autoimmune encephalomyelitis
- MBP
- myelin basic protein
- MMP
- matrix metalloproteinase
- PC
- proprotein convertase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- LPS
- lipopolysaccharide
- MALDI-TOF
- matrix- assisted laser desorption ionization time-of-flight
- MS
- mass spectrometry
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- RT
- reverse transcription
- Q-RT-PCR
- quantitative RT-PCR
- CNPase
- 2′,3′-cyclic nucleotide 3′-phosphodiesterase
- dec-RVKR-cmk
- decanoyl-Arg-Val-Lys-Arg-chloromethyl ketone.
REFERENCES
- 1.Steinman L. (2001) Nat. Immunol. 2, 762–764 [DOI] [PubMed] [Google Scholar]
- 2.Goodin D. S. (2009) PLoS ONE 4, e4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fugger L., Friese M. A., Bell J. I. (2009) Nat. Rev. Immunol., in press [DOI] [PubMed] [Google Scholar]
- 4.Hellings N., Barée M., Verhoeven C., D'hooghe M. B., Medaer R., Bernard C. C., Raus J., Stinissen P. (2001) J. Neurosci. Res. 63, 290–302 [DOI] [PubMed] [Google Scholar]
- 5.Weaver A., Goncalves da Silva A., Nuttall R. K., Edwards D. R., Shapiro S. D., Rivest S., Yong V. W. (2005) FASEB J. 19, 1668–1670 [DOI] [PubMed] [Google Scholar]
- 6.Campagnoni A. T., Pribyl T. M., Campagnoni C. W., Kampf K., Amur-Umarjee S., Landry C. F., Handley V. W., Newman S. L., Garbay B., Kitamura K. (1993) J. Biol. Chem. 268, 4930–4938 [PubMed] [Google Scholar]
- 7.Feng J. M. (2007) Neurochem. Res. 32, 273–278 [DOI] [PubMed] [Google Scholar]
- 8.Menezes J. S., van den Elzen P., Thornes J., Huffman D., Droin N. M., Maverakis E., Sercarz E. E. (2007) J. Clin. Invest. 117, 2176–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pribyl T. M., Campagnoni C. W., Kampf K., Kashima T., Handley V. W., McMahon J., Campagnoni A. T. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10695–10699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vos C. M., van Haastert E. S., de Groot C. J., van der Valk P., de Vries H. E. (2003) J. Neuroimmunol. 138, 106–114 [DOI] [PubMed] [Google Scholar]
- 11.Voskuhl R. R., Farris R. W., 2nd, Nagasato K., McFarland H. F., Dalcq M. D. (1996) J. Neuroimmunol. 70, 103–111 [DOI] [PubMed] [Google Scholar]
- 12.van Noort J. M., van Sechel A. C., Bajramovic J. J., el Ouagmiri M., Polman C. H., Lassmann H., Ravid R. (1995) Nature 375, 798–801 [DOI] [PubMed] [Google Scholar]
- 13.Ousman S. S., Tomooka B. H., van Noort J. M., Wawrousek E. F., O'Connor K. C., Hafler D. A., Sobel R. A., Robinson W. H., Steinman L. (2007) Nature 448, 474–479 [DOI] [PubMed] [Google Scholar]
- 14.Stoevring B., Vang O., Christiansen M. (2005) Clin. Chim. Acta 356, 95–101 [DOI] [PubMed] [Google Scholar]
- 15.Seidah N. G., Mayer G., Zaid A., Rousselet E., Nassoury N., Poirier S., Essalmani R., Prat A. (2008) Int. J. Biochem. Cell Biol. 40, 1111–1125 [DOI] [PubMed] [Google Scholar]
- 16.Thomas G. (2002) Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scamuffa N., Calvo F., Chrétien M., Seidah N. G., Khatib A. M. (2006) FASEB J. 20, 1954–1963 [DOI] [PubMed] [Google Scholar]
- 18.Remacle A. G., Shiryaev S. A., Oh E. S., Cieplak P., Srinivasan A., Wei G., Liddington R. C., Ratnikov B. I., Parent A., Desjardins R., Day R., Smith J. W., Lebl M., Strongin A. Y. (2008) J. Biol. Chem. 283, 20897–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiryaev S. A., Remacle A. G., Ratnikov B. I., Nelson N. A., Savinov A. Y., Wei G., Bottini M., Rega M. F., Parent A., Desjardins R., Fugere M., Day R., Sabet M., Pellecchia M., Liddington R. C., Smith J. W., Mustelin T., Guiney D. G., Lebl M., Strongin A. Y. (2007) J. Biol. Chem. 282, 20847–20853 [DOI] [PubMed] [Google Scholar]
- 20.Rosenberg G. A. (2002) Neuroscientist 8, 586–595 [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg G. A. (2009) Lancet Neurol. 8, 205–216 [DOI] [PubMed] [Google Scholar]
- 22.Egeblad M., Werb Z. (2002) Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 23.Sohail A., Sun Q., Zhao H., Bernardo M. M., Cho J. A., Fridman R. (2008) Cancer Metastasis Rev. 27, 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J. M., Fernandes A. O., Campagnoni C. W., Hu Y. H., Campagnoni A. T. (2004) J. Neuroimmunol. 152, 57–66 [DOI] [PubMed] [Google Scholar]
- 25.Shiryaev S. A., Savinov A. Y., Cieplak P., Ratnikov B. I., Motamedchaboki K., Smith J. W., Strongin A. Y. (2009) PLoS ONE 4, e4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toft-Hansen H., Babcock A. A., Millward J. M., Owens T. (2007) J. Neuroinflammation 4, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs E. C., Pribyl T. M., Feng J. M., Kampf K., Spreur V., Campagnoni C., Colwell C. S., Reyes S. D., Martin M., Handley V., Hiltner T. D., Readhead C., Jacobs R. E., Messing A., Fisher R. S., Campagnoni A. T. (2005) J. Neurosci. 25, 7004–7013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fugère M., Limperis P. C., Beaulieu-Audy V., Gagnon F., Lavigne P., Klarskov K., Leduc R., Day R. (2002) J. Biol. Chem. 277, 7648–7656 [DOI] [PubMed] [Google Scholar]
- 29.Gawlik K., Shiryaev S. A., Zhu W., Motamedchaboki K., Desjardins R., Day R., Remacle A. G., Stec B., Strongin A. Y. (2009) PLoS ONE 4, e5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kridel S. J., Sawai H., Ratnikov B. I., Chen E. I., Li W., Godzik A., Strongin A. Y., Smith J. W. (2002) J. Biol. Chem. 277, 23788–23793 [DOI] [PubMed] [Google Scholar]
- 31.Chen E. I., Li W., Godzik A., Howard E. W., Smith J. W. (2003) J. Biol. Chem. 278, 17158–17163 [DOI] [PubMed] [Google Scholar]
- 32.Cheng M., Watson P. H., Paterson J. A., Seidah N., Chrétien M., Shiu R. P. (1997) Int. J. Cancer 71, 966–971 [DOI] [PubMed] [Google Scholar]
- 33.Seo J., Shneiderman B. (2002) Computer 35, 80–86 [Google Scholar]
- 34.Katsara M., Deraos G., Tselios T., Matsoukas J., Apostolopoulos V. (2008) J. Med. Chem. 51, 3971–3978 [DOI] [PubMed] [Google Scholar]
- 35.Matsoukas J., Apostolopoulos V., Kalbacher H., Papini A. M., Tselios T., Chatzantoni K., Biagioli T., Lolli F., Deraos S., Papathanassopoulos P., Troganis A., Mantzourani E., Mavromoustakos T., Mouzaki A. (2005) J. Med. Chem. 48, 1470–1480 [DOI] [PubMed] [Google Scholar]
- 36.Tselios T., Apostolopoulos V., Daliani I., Deraos S., Grdadolnik S., Mavromoustakos T., Melachrinou M., Thymianou S., Probert L., Mouzaki A., Matsoukas J. (2002) J. Med. Chem. 45, 275–283 [DOI] [PubMed] [Google Scholar]
- 37.Acha-Orbea H., Mitchell D. J., Timmermann L., Wraith D. C., Tausch G. S., Waldor M. K., Zamvil S. S., McDevitt H. O., Steinman L. (1988) Cell 54, 263–273 [DOI] [PubMed] [Google Scholar]
- 38.Dasilva A. G., Yong V. W. (2008) J. Neuroimmunol. 199, 24–34 [DOI] [PubMed] [Google Scholar]
- 39.Folgueras A. R., Fueyo A., García-Suárez O., Cox J., Astudillo A., Tortorella P., Campestre C., Gutiérrez-Fernández A., Fanjul-Fernández M., Pennington C. J., Edwards D. R., Overall C. M., López-Otín C. (2008) J. Biol. Chem. 283, 9465–9474 [DOI] [PubMed] [Google Scholar]
- 40.Ulrich R., Baumgärtner W., Gerhauser I., Seeliger F., Haist V., Deschl U., Alldinger S. (2006) J. Neuropathol. Exp. Neurol. 65, 783–793 [DOI] [PubMed] [Google Scholar]
- 41.van Horssen J., Vos C. M., Admiraal L., van Haastert E. S., Montagne L., van der Valk P., de Vries H. E. (2006) Neuropathol. Appl. Neurobiol. 32, 585–593 [DOI] [PubMed] [Google Scholar]
- 42.Werner S. R., Dotzlaf J. E., Smith R. C. (2008) BMC Neurosci. 9, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bar-Or A., Vollmer T., Antel J., Arnold D. L., Bodner C. A., Campagnolo D., Gianettoni J., Jalili F., Kachuck N., Lapierre Y., Niino M., Oger J., Price M., Rhodes S., Robinson W. H., Shi F. D., Utz P. J., Valone F., Weiner L., Steinman L., Garren H. (2007) Arch. Neurol. 64, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 44.Steinman L., Conlon P. (2001) J. Clin. Immunol. 21, 93–98 [DOI] [PubMed] [Google Scholar]
- 45.Steinman L., Waisman A., Altmann D. (1995) Mol. Med. Today 1, 79–83 [DOI] [PubMed] [Google Scholar]
- 46.Wucherpfennig K. W., Catz I., Hausmann S., Strominger J. L., Steinman L., Warren K. G. (1997) J. Clin. Invest. 100, 1114–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starckx S., Van den Steen P. E., Verbeek R., van Noort J. M., Opdenakker G. (2003) J. Neuroimmunol. 141, 47–57 [DOI] [PubMed] [Google Scholar]
- 48.Thoua N. M., van Noort J. M., Baker D., Bose A., van Sechel A. C., van Stipdonk M. J., Travers P. J., Amor S. (2000) J. Neuroimmunol. 104, 47–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.