Abstract
Histone deacetylases (HDACs) are globally implicated in the growth and differentiation of mammalian cells; however, relatively little is known about their specific roles in hematopoiesis. In this study, we investigated the expression of HDACs in human hematopoietic cells and their functions during hematopoiesis. The expression of HDACs was very low in hematopoietic progenitor cells, which was accompanied by histone hyperacetylation. HDACs were detectable in more differentiated progenitors and erythroid precursors but down-regulated in mature myeloid cells especially granulocytes. In contrast, acute myeloid leukemias showed HDAC overexpression and histone hypoacetylation. Transcription of the HDAC1 gene was repressed by CCAAT/enhancer binding proteins during myeloid differentiation, and activated by GATA-1 during erythro-megakaryocytic differentiation. Small interfering RNA-mediated knockdown of HDAC1 enhanced myeloid differentiation in immature hematopoietic cell lines and perturbed erythroid differentiation in progenitor cells. Myeloid but not erythro-megakaryocytic differentiation was blocked in mice transplanted with HDAC1-overexpressing hematopoietic progenitor cells. These findings suggest that HDAC is not merely an auxiliary factor of genetic elements but plays a direct role in the cell fate decision of hematopoietic progenitors.
Hematopoiesis is an ordered process involving self-renewal of stem cells, expansion of the lineage-committed progenitor population, and maturation into terminal elements (1). Each step is tightly regulated by several transcription factors, which confer proper expression of lineage-specific genes and/or cell cycle control genes in hematopoietic stem and progenitor cells (2). For instance, extensive biological and genetic studies indicate that GATA-1, whose expression is confined to erythroblasts, megakaryocytes, eosinophils, and mast cells, is a master regulator of erythro-megakaryocytic differentiation (3, 4), whereas GATA-2 is mainly expressed in hematopoietic stem and early progenitor cells and plays a pivotal role in self-renewal (5, 6). During myeloid differentiation, CCAAT/enhancer-binding protein (C/EBP)3 family proteins bind to cognate sequences and transactivate a variety of myeloid-specific genes (7, 8). Although there may be some functional redundancy and overlap among C/EBPs, genetic studies reveal that Cebpa and Cebpb are essential for granulocytic differentiation and macrophage functions, respectively (8, 9).
Acute myeloblastic leukemia (AML) is characterized by deregulated proliferation and impaired differentiation of hematopoietic stem cells or immature progenitor cells (10). Recent investigations have greatly increased our understanding of the molecular basis of the biological properties of AML. Deregulated proliferation is mostly caused by aberrant activation of signal transduction pathways downstream of hematopoietic growth factor receptors. Prototype abnormalities of this type include mutations in growth factor receptors, such as FLT3 (Fms-like tyrosine kinase 3) and c-KIT. On the other hand, impaired differentiation is attributable to alterations of transcription factors, which result in the disruption of normal functions governing hematopoiesis. They are exemplified by fusion gene formation associated with chromosomal translocations, such as PML/RARα and AML1/ETO, and loss-of-function mutations of CEBPA and AML1. These two functionally distinct groups are termed class I and class II abnormalities, respectively (11). Accumulating genetic evidence strongly suggests that AML develops when a class I mutation confers a growth advantage to hematopoietic stem/progenitor cells in which differentiation is blocked by a class II mutation.
As described above, several lines of evidence underscore the importance of genetic elements in normal hematopoiesis and their disruptions in AML. In contrast, relatively little is known about the role of epigenetics in hematopoiesis as well as leukemogenesis. However, recent analyses of leukemic fusion proteins point to the involvement of histone deacetylases (HDACs) and the therapeutic implications. HDACs are a family of enzymes that catalyze the removal of acetyl groups from core histones, which results in chromatin compaction and transcriptional repression (12). Mammalian HDACs are divided into four groups: class I (HDAC1, -2, -3, and -8), IIa (HDAC4, -5, -7, and -9), IIb (HDAC6 and -10), and IV (HDAC11). Class I HDACs are ubiquitously expressed and are generally involved in cell growth and differentiation (13), whereas class II HDACs have a more restricted pattern of expression (skeletal muscle, heart, and brain) and act in association with tissue-specific transcription factors. Leukemic fusion proteins, such as PML/RARα and AML-1/ETO, form a complex with HDACs with higher affinities than their normal counterparts and aberrantly suppress the expression of genes required for cell differentiation and growth control, leading to the transformation of hematopoietic progenitor cells (14, 15). Therefore, HDACs are considered direct targets of treatment in these cases. Indeed, a variety of small compounds that inhibit HDAC activity have been developed and tested as therapeutic agents for hematologic malignancies, including AML with fusion gene products, and solid tumors (16).
HDAC inhibitors can induce differentiation, cell cycle arrest, and apoptosis in AML cells irrespective of the presence of leukemic fusion proteins, suggesting that HDACs are generally involved in leukemogenesis via multiple mechanisms (16). These effects provide a rational backbone for the clinical application of HDAC inhibitors to AML. Intriguingly, recent clinical trials have revealed that HDAC inhibitors have only moderate hematologic toxicity (17, 18), but the underlying mechanisms are to be determined. Since these observations are biologically interesting and clinically important, their molecular basis is worth investigating. In this study, we therefore examined the expression of HDACs in human hematopoietic cells and their functions during hematopoiesis and found that the expression levels of HDACs determine the fate of hematopoietic progenitor cells.
EXPERIMENTAL PROCEDURES
Cells
Human bone marrow mononuclear cells (BMMNCs) were purchased from Cambrex BioScience (Walkersville, MD). CD34+ cells were purified by positive selection with CD34 MicroBeads and MACS separation columns (Miltenyi Biotec, Gladbach, Germany). More than 95% of enriched cells were shown to be positive for CD34 and negative for lineage markers (19). CD34+ BMMNCs represent early hematopoietic progenitors, since most express CD38 (data not shown). The remaining cells were used as CD34− BMMNCs after depleting lineage marker-expressing cells with a lineage cell depletion kit (Miltenyi Biotec). This fraction mainly consists of committed progenitors of multiple lineages and does not contain terminally differentiated elements, such as mature myeloid cells, erythroblasts, and lymphocytes.
Human AML cell lines, HL60, U937, and K562, were differentiated in serum-free GIT medium (Nihon Pharmaceutical, Tokyo, Japan) containing appropriate chemicals, as described previously (20, 21). Primary AML cells were obtained from patients at diagnosis by sedimentation on Ficoll-Hypaque density gradients. Informed consent was obtained from all subjects in accord with the requirements of the institutional review board. Samples were selected for the study only when they contained more than 90% leukemic cells and did not carry chromosomal translocations.
Cell Culture
For clonogenic growth assays, human CD34+ BMMNCs and primary AML cells were plated at 0.5–1 × 103 cells/ml in methylcellulose medium supplemented with full cytokines (H4435, Stem Cell Technologies, Vancouver, Canada) (19). AML cell lines were cultured in methylcellulose medium supplemented with 10% fetal calf serum for 7 days. To form colony-forming unit-granulocyte/macrophage (CFU-GM) and colony-forming unit-erythroid (CFU-E), murine bone marrow cells were plated at 2.5 × 103 cells/ml in methylcellulose medium supplemented with a combination of stem cell factor, interleukin-3, and interleukin-6 (M3534) and erythropoietin alone (M3334), respectively.
Semiquantitative and Real Time Quantitative RT-PCR
Total cellular RNA was isolated from 1 × 104 cells and reverse-transcribed into cDNA using SuperScript reverse transcriptase and oligo(dT) primers (Invitrogen). We performed subsequent semiquantitative PCR, as described previously (2), and real time quantitative RT-PCR using Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK). Detailed information on primers, including sequences, corresponding nucleotide positions, and PCR product sizes, is shown in supplemental Table S1.
RNA Blotting
An equal amount (15 μg) of total cellular RNA was electrophoresed in 1% agarose gels containing formaldehyde and blotted onto Hybond N synthetic nylon membranes (Amersham Biosciences). The membranes were hybridized with 32P-labeled probes in Rapid-hyb buffer (Amersham Biosciences).
Immunoblotting
Immunoblotting was carried out according to the standard method using the following antibodies: anti-HDAC1 (Sigma), anti-HDAC2 (MBL International, Woburn, MA), anti-HDAC3 (BD Pharmingen, San Jose, CA), and anti-β-actin (Ab-1; Oncogene Science, Uniondale, NY). We purchased site-specific antibodies against acetylated histones (H3-Lys9, H3-Lys18, H4-Lys12, and H4-Lys16) from Cell Signaling Technology (Beverly, MA).
Confocal Laser Microscopy
Confocal microscopic analysis was performed using anti-HDAC1 polyclonal (Sigma) and anti-CD34 monoclonal (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies. We used Alexa 488-conjugated goat antibody to mouse immunoglobulin (Molecular Probes, Inc., Eugene, OR) and Cy3-conjugated goat antibody to rabbit immunoglobulin (Amersham Biosciences) as secondary antibodies.
Flow Cytometry Analysis and Fluorescence-activated Cell Sorting
Flow cytometry analysis and cell sorting were carried out with BD FACSaria (BD Biosciences) as described previously (19).
Plasmids and Transfection
Retroviral expression vector for HDAC1 was constructed by inserting full-length cDNA (provided by Dr. Stuart Schreiber, Harvard University, Boston, MA) upstream of the internal ribosome entry site-enhanced green fluorescent protein cassette of pMYs plasmid, as described previously (22). Retrovirus production was carried out by transfecting the plasmids into Plat-E packaging cells. Expression vectors for MZF-1, C/EBPα, C/EBPβ, GATA-1, GATA-2, and Sp1 were kindly provided by Drs. Robert Hromas (University of New Mexico, Albuquerque, NM), Atsushi Iwama (Department of Cellular and Molecular Medicine, Chiba University, Chiba, Japan), and Mitsuru Nakamura (National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan).
We used a lentiviral shRNA/siRNA expression vector pLentiLox3.7 for knockdown of HDAC1. Target sequences were designed to be homologous to wild-type cDNA sequences: HDAC1 (forward), TggcaaaggcaagtattatgTTCAAGAGAcataatacttgcctttgccTTTTTTC; HDAC1 (reverse), TCGAGAAAAAAggcaaaggcaagtattatgTCTCTTGAAcataatacttgcctttgcc. Scrambled sequences were used as controls. Lentiviruses were then added to cell suspensions in the presence of 8 μg/ml Polybrene and transduced for 24 h, as described previously (19).
Reporter Assays
We amplified the promoter regions of the HDAC1 gene (−1170 to +397 and −73 to +397) by PCR and inserted them into pGL4.10 firefly luciferase vector (Promega, Madison, WI) to generate reporter plasmids. HEK293 cells were transfected with reporter plasmids along with pGL4.73 Renilla luciferase vector (Promega), which served as a positive control to determine transfection efficiencies, in the presence of test plasmids encoding Sp1, GATA-1, GATA-2, MZF-1, CEBPA, and CEBPB or empty vectors. After 48 h, firefly and Renilla luciferase activities were discriminately measured using the Dual-Luciferase reporter assay system (Promega).
Chromatin Immunoprecipitation Assays
We used the ChIP-IT chromatin immunoprecipitation kit (Active Motif, Carlsbad, CA) to perform chromatin immunoprecipitation assays. In brief, cells were fixed with 1% formaldehyde and sonicated to obtain chromatin suspensions. After centrifugation, supernatants were incubated with antibodies of interest in the presence of protein A-agarose beads. DNA fragments bound to the beads were purified with washing and subjected to PCR using primer pairs spanning −377 to −77 of the HDAC1 gene.
Stem Cell Transplantation in Syngeneic Mice
Bone marrow mononuclear cells were isolated from C57BL/6 (Ly-5.1) donor mice (8–12 weeks of age). c-KIT-positive cells were isolated by CD117 MicroBeads in MACS separation columns (Miltenyi Biotec), and cultured overnight in Iscove's modified Dulbecco's medium supplemented with BIT 9500 and 50 ng/ml each of stem cell factor, FLT3 ligand, interleukin-3, and thrombopoietin. Prestimulated cells were infected with retroviruses harboring either pMYs-HDAC1 or an empty vector (mock) in 6-well dishes for 24 h (22, 23). Then 1–6 × 105 cells were injected through the tail vein into lethally irradiated (9.5 grays) C57BL/6 (Ly-5.2) recipient mice (8–12 weeks of age). Engraftment of transplanted cells was confirmed by measuring the percentages of GFP+ and/or Ly-5.1+ cells in the peripheral blood of recipients. All animal studies were approved by the Institutional Animal Ethics Committee and performed in accordance with the Guide for the Care and Use of Laboratory Animals formulated by the National Academy of Sciences.
RESULTS
Relative Resistance of Normal Human Hematopoietic Progenitors to HDAC Inhibitors
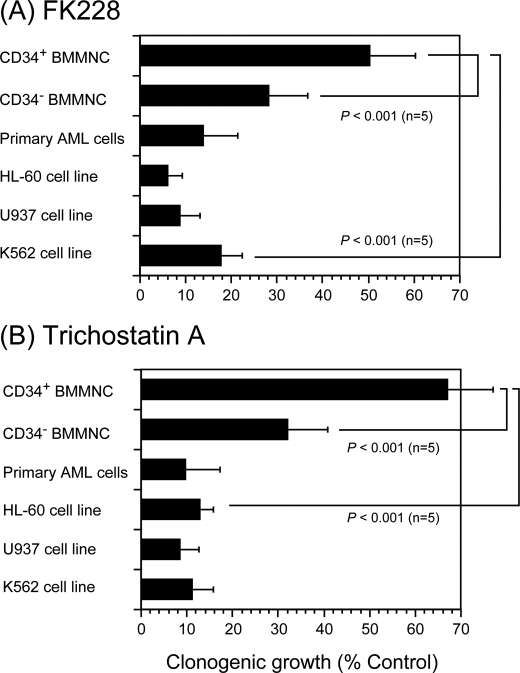
Given the relatively weak hematological toxicity reported in clinical trials (17, 18), we reasoned that normal human hematopoietic stem/progenitor cells are resistant to HDAC inhibitors. Indeed, normal human hematopoietic progenitors (CD34-positive/CD38-positive/lineage marker-negative bone marrow mononuclear cells (CD34+ BMMNC)) generated more colonies in the presence of HDAC inhibitors than primary blasts from patients with AML and myeloid leukemia cell lines in clonogenic growth assays (Fig. 1). Interestingly, normal hematopoietic progenitors were more resistant to HDAC inhibitors than their differentiated offspring (CD34-negative and lineage marker-negative bone marrow mononuclear cells (CD34− BMMNC), which correspond to committed progenitors of multiple lineages).
FIGURE 1.
Relative resistance of normal hematopoietic progenitor cells to HDAC inhibitors. We seeded normal human CD34+ and CD34− BMMNCs and primary AML cells at 1 × 103 cells/ml in methylcellulose medium supplemented with stem cell factor (50 ng/ml), interleukin-3 (10 ng/ml), interleukin-6 (10 ng/ml), granulocyte/macrophage colony-stimulating factor (10 ng/ml), granulocyte colony-stimulating factor (10 ng/ml), and erythropoietin (3 units/ml) and cultured in the absence or presence of either 2 nm FK228 (A) or 10 ng/ml trichostatin A (B) for 14 days. Three AML cell lines (HL-60, U937, and K562) were cultured in methylcellulose medium supplemented with 10% fetal calf serum for 7 days. Each column indicates the relative colony numbers setting untreated controls at 100%. The means ± S.D. (bars) of five independent experiments are shown. p values were calculated by one-way analysis of variance with the Bonferroni post hoc test.
Differential Expression of HDACs in Normal Hematopoietic Cells and AML Blasts
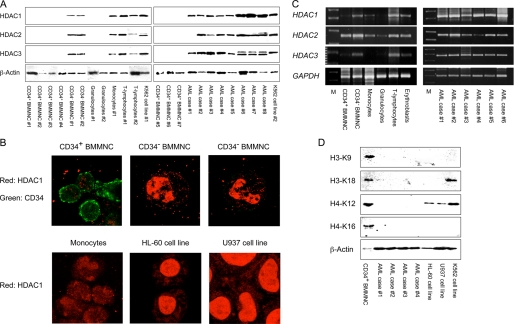
To understand the mechanisms underlying the relative resistance of normal hematopoietic progenitor cells to HDAC inhibitors, we screened for the expression of three major class I HDACs (HDAC1, -2, and -3) by immunoblotting using specific antibodies. The selection was based on the fact that these HDACs, especially HDAC1, represent the vast majority of cellular HDAC activities (13, 24) and major class II HDACs (HDAC4, -5, and -7) were scarcely expressed in hematopoietic cells in our pilot experiments (data not shown). Moreover, class I HDACs, especially HDAC1, are known to be implicated in leukemogenesis and carcinogenesis and thus are pharmacological targets of most HDAC inhibitors (25, 26). As shown in Fig. 2A, the expression of HDAC proteins was below the detection limit in immature progenitor cells from normal human bone marrow (CD34+ BMMNC). Immunoblotting with different antibodies yielded the same results, negating the possibility of the low sensitivity of the antibodies used for detection (data not shown). HDAC proteins were readily detectable in committed progenitors of multiple lineages (Lin−/CD34− BMMNC) and T-lymphocytes but were weakly expressed in monocytes and nearly absent in mature granulocytes from the peripheral blood of healthy volunteers (see quantified data in supplemental Fig. S1). Primary leukemic cells from AML patients showed higher expression levels of HDACs than their normal counterparts (immature progenitor cells), although there was a minor case-to-case variation. In particular, HDAC1 was more abundantly expressed than HDAC2 and HDAC3 in approximately half of the cases with AML (Fig. 2A). We further confirmed the expression of HDAC1 using immunocytochemistry. As shown in Fig. 2B, HDAC1 was not detected in CD34+ BMMNC, moderately expressed in the nuclei of CD34− BMMNC and monocytes, and apparently overexpressed in AML cells. The differential expression of HDACs does not simply reflect the proliferative states of these cells, because the abundance of HDACs was only modestly increased along with cell cycle entry of mitogen-stimulated T-lymphocytes (supplemental Fig. S2).
FIGURE 2.
Differential expression of HDACs in normal hematopoietic cells and AML blasts. A, whole cell lysates were prepared from the indicated cell types isolated as described under “Experimental Procedures” and subjected to immunoblot analysis for the expression of HDAC1, HDAC2, HDAC3, and β-actin (loading control). Signals were obtained with the ChemiDoc XRS Molecular Imager (Bio-Rad) and used without digital manipulation except for the conversion to TIF files. See supplemental Fig. S1 for data quantification and statistical analysis. B, CD34+ and CD34− BMNNC were stained with anti-HDAC1 (red) and anti-CD34 (green) antibodies and analyzed under confocal microscopy. Original magnification is ×600 for all panels. C, total cellular RNA was isolated from the indicated cell types, and 2.5 μg (normal hematopoietic cells) or 1.0 μg (primary AML cells) was subjected to semiquantitative RT-PCR analysis for the expression of HDAC1, HDAC2, HDAC3, and GAPDH (internal control). Five μl of the amplified products were visualized by ethidium bromide staining after 2% agarose gel electrophoresis. The results of suboptimal amplification cycles are shown: 35 and 30 cycles for HDAC genes and GAPDH, respectively. M, a molecular size marker (BioMarker Low; BioVentures, Inc., Murfreesboro, TN). See supplemental Fig. S3 for data quantification and statistical analysis. D, protein samples were analyzed by immunoblotting using specific antibodies against histone H3 acetylated at lysine 9 (H3-K9), histone H3 acetylated at lysine 18 (H3-K18), histone H4 acetylated at lysine 12 (H4-K12), histone H4 acetylated at lysine 16 (H4-K16), and β-actin (loading control). Data shown are the representative results of multiple independent experiments.
Next, we carried out similar analyses with semiquantitative RT-PCR. The expression of HDAC mRNAs was very weak in normal hematopoietic progenitors (CD34+ BMMNC) as well as in mature myeloid cells (monocytes and granulocytes), except for HDAC2 in CD34+ BMMNC (Fig. 2C). HDAC transcripts were moderately expressed in committed progenitors (Lin−/CD34− BMMNC), purified erythroblasts from bone marrow, and peripheral blood T-lymphocytes. In contrast, primary AML cells strongly expressed HDAC genes, especially HDAC1. The virtually identical pattern was obtained with real time quantitative RT-PCR (supplemental Fig. S3). Overall, the expression pattern of HDAC transcripts is nearly equal to that of HDAC proteins in normal and malignant hematopoietic cells. These results suggest that the expression of HDACs is regulated primarily at mRNA levels, but post-translational modification is also involved especially in HDAC2, as suggested by previous studies (27).
Finally, we investigated whether the acetylation status of intracellular histones reflects the distinct expression levels of HDACs in hematopoietic cells. To this end, we performed immunoblotting using site-specific antibodies against acetylated histones. As shown in Fig. 2D, histones H3 and H4 were heavily acetylated at Lys9/Lys18 and Lys12/Lys16, respectively, in normal hematopoietic progenitors but not in primary AML cells. AML cell lines also lacked the acetylation of H3-Lys9 and H4-Lys16, but H4-Lys12 and H3-Lys18 were acetylated in all three cell lines examined and in K562, respectively.
Lineage-specific Regulation of HDAC Expression during Hematopoietic Differentiation
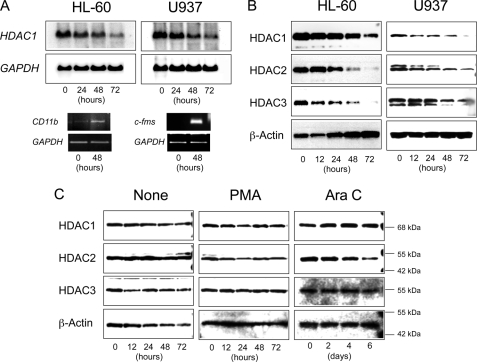
To further delineate the differential expression of HDACs in normal and malignant hematopoietic cells, we took advantage of the cell line model system. To recapitulate myeloid differentiation in vitro, we cultured HL-60 and U937 cells with chemical inducers of differentiation: phorbol ester, dimethyl sulfoxide, and retinoic acid (20). Successful induction of differentiation was verified by the appearance of mature myelomonocytic markers, such as CD11b and c-Fms (Fig. 3A, bottom). During myeloid differentiation, HDACs were down-regulated at both mRNA (Fig. 3A, top) and protein levels (Fig. 3B). We then performed similar experiments with the K562 cell line, which can be differentiated into megakaryocytic and erythroid lineages by phorbol ester and cytosine arabinoside, respectively (supplemental Fig. S4) (21). Unlike myeloid differentiation, the abundance of HDACs, especially HDAC1 and HDAC3, was unchanged during megakaryocytic and erythroid differentiation (Fig. 3C). Since HDAC1 seemed to be slightly increased in cytosine arabinoside-treated K562 cells, we monitored the expression of HDAC1 during in vitro differentiation of primary CD34+ BMMNC using real time RT-PCR. When purified CD34+ BMMNCs were cultured with a combination of cytokines directing erythropoiesis (2), up-regulation of HDAC1 expression coincided with morphological differentiation of immature progenitor cells into erythroid precursors (supplemental Fig. S5). In addition, this culture system yielded the confirmation of HDAC1 down-regulation during myeloid differentiation of normal progenitors (data not shown).
FIGURE 3.
Lineage-specific regulation of HDAC expression during hematopoietic differentiation. HL-60 and U937 cells were cultured with all-trans-retinoic acid and phorbol-12-myristate-13-acetate (PMA) to induce granulocytic and monocytic differentiation, respectively. A, total cellular RNA was isolated at the indicated time points and subjected to semiquantitative RT-PCR analysis for CD11b, c-fms, and GAPDH (internal control) expression and Northern blot analysis for HDAC1 and GAPDH expression. B, whole cell lysates were isolated at the indicated time points and subjected to immunoblot analysis for the expression of HDACs and β-actin (loading control). C, K562 cells were cultured with PMA and cytosine arabinoside (AraC) to induce megakaryocytic and erythroid differentiation, respectively. See supplemental Fig. S4 for the proper achievement of differentiation induction. Whole cell lysates were isolated at the indicated time points and subjected to immunoblot analysis for the expression of HDACs and β-actin (loading control). Signals were obtained with the ChemiDoc XRS Molecular Imager (Bio-Rad) and used without digital manipulation except for the conversion to TIF files. Data shown are representative results of multiple independent experiments.
Regulation of HDAC1 Promoter by Hematopoietic Transcription Factors
Our analyses demonstrated that HDAC expression was very low in immature hematopoietic progenitors, induced in more differentiated progenitor cells, and down-regulated during myeloid differentiation with nearly complete disappearance in mature granulocytes, whereas it was retained during erythroid and megakaryocytic differentiation. In contrast, HDACs, especially HDAC1, were overexpressed in virtually all AML cases and cell lines. To corroborate the regulatory mechanisms underlying this unique expression pattern, we isolated putative promoter regions of the HDAC1 gene and subjected them to functional analysis. The HDAC1 promoter contains canonical binding sites of Sp1 (GC box), MZF-1, C/EBPs (CCAAT box), and GATA transcription factors (supplemental Fig. S6). We subcloned two promoter fragments, −1179 to +397 and −73 to +397, into pGL4 luciferase vector to generate reporter plasmids as illustrated in Fig. 4A. The selection of analyzed regions was based on previous studies on the murine Hdac1 promoter (28) and the results of our pilot experiments in which the segment between −1179 and +397 confers full promoter activity (data not shown). We carried out reporter assays with HEK293, because this cell line lacks the expression of transcription factors to be tested except for Sp1 (data not shown). When the −1170 construct was used, reporter assays revealed that HDAC1 transactivation was driven by Sp1 and GATA-1 (Fig. 4A, middle). In contrast, members of the C/EBP family transcription factors, C/EBPα and C/EBPβ, negatively regulated transcription of the HDAC1 gene. GATA-2 and MZF-1 showed only a modest effect on promoter activity. Sp1 was able to activate the −73 construct to a similar extent as the −1170 construct, suggesting that GC boxes surrounding the transcription start site are responsible for Sp1-mediated transactivation of HDAC1 (Fig. 4A, bottom). However, the activity of GATA-1 was significantly diminished when the −73 construct was used, implying that consensus sequences at the positions −973 and −91 are necessary for GATA-1 to fully activate the HDAC1 promoter. In contrast, C/EBPα and C/EBPβ were still able to repress the HDAC1 promoter in the −73 construct. Since the −73 construct lacks CCAAT boxes, the suppressor function of C/EBPs may be mediated via interaction with activator proteins, most likely GATA-1 (7, 29). To verify this hypothesis, we examined the effects of C/EBPα and C/EBPβ on GATA-1-mediated transactivation of the HDAC1 gene using co-transfection. As anticipated, both C/EBPα and C/EBPβ significantly suppressed HDAC1 promoter activation by GATA-1. In particular, C/EBPα suppressed the activity of GATA-1 to about one-tenth (Fig. 4B, bottom). More importantly, GATA-2 and MZF-1 individually antagonized GATA-1-mediated activation of the HDAC1 promoter in a dose-dependent manner (Fig. 4B, top), and the two factors synergistically suppressed it (Fig. 4B, bottom).
FIGURE 4.
Regulation of HDAC1 promoter by hematopoietic transcription factors. A, top, schematic representation of HDAC1 promoter constructs used in this study. Promoter regions of the HDAC1 gene (−1170 to +397 and −73 to +397) were linked to the luciferase gene in pGL4.10 vector as indicated. Relative locations of the putative binding sites of hematopoietic transcription factors are approximated by the symbols shown in the box. See supplemental Fig. S6 for the nucleotide sequence. Bottom, we transfected pGL4.10 plasmid containing HDAC1 promoter sequences between −1170 and +397 (−1170) or between −73 and +397 (−73) into HEK293 cells along with expression vectors encoding Sp1, GATA-1, GATA-2, MZF-1, CEBPA, and CEBPB at various doses (1, 2, and 4 μg) and measured luciferase activities after 48 h. HDAC1 promoter activity (y axis) was calculated as firefly luciferase activities of cells transfected with either pGL4.10−1170 or pGL4.10−73 and an empty expression vector setting at 1.0 after normalization of transfection efficiencies using Renilla luciferase activity. Data shown are the means ± S.D. of three independent experiments. B, top, we transfected pGL4.10−1170 reporter plasmid into HEK293 cells along with expression vectors encoding GATA-1, GATA-2, and MZF-1 at the indicated doses (μg) and measured luciferase activity after 48 h. HDAC1 promoter activity (y axis) was calculated as firefly luciferase activities of cells transfected with pGL4.10−1170 and an empty expression vector set at 1.0 after normalization of transfection efficiencies using Renilla luciferase activities. Bottom, we transfected pGL4.10−1170 reporter plasmid and 1 μg of GATA-1 expression vector into HEK293 cells in the absence or presence of 1 μg of expression plasmids encoding GATA-2, MZF-1, CEBPα, and CEBPβ and measured luciferase activities after 48 h. Relative promoter activity (y axis) was calculated as firefly luciferase activities of cells transfected with pGL4.10−1170 and GATA-1 expression vector setting at 100% after normalization of transfection efficiencies using Renilla luciferase activities. Data shown are the means ± S.D. of three independent experiments. p values were calculated by one-way analysis of variance with the Student-Newman-Keuls multiple comparisons test. C, HL60 and K562 were cultured in the presence of PMA for 4 days and harvested before and after differentiation for chromatin immunoprecipitation assays. Chromatin suspensions were immunoprecipitated with the indicated antibodies and corresponding control antibodies. The resulting precipitants were subjected to PCR to amplify the promoter region (−377 to −77) of the HDAC1 gene. The amplified products were visualized by ethidium bromide staining after 2% agarose gel electrophoresis. Representative data of 50 cycles are shown. Input, PCR was performed with genomic DNA.
Next, we investigated the binding of these factors to HDAC1 promoter in vivo and its changes during hematopoietic differentiation using chromatin immunoprecipitation assays. In an undifferentiated HL-60 cell line, Sp1 and GATA-1 bound to the sequence between −377 and −77, which was shown to be the active regulatory region by reporter assays (Fig. 4C, day 0). This suggests that these two factors confer the base-line expression of HDAC1 in the myeloid-committed cell line HL-60. Upon differentiation, both Sp1 and GATA-1 dissociated from the HDAC1 promoter, and GATA-2 and a small amount of C/EBPα became detectable (Fig. 4C, day 4). These results suggest that the exchange of positive to negative regulators on the promoter contributes to the silencing of HDAC1 during myeloid differentiation. On the other hand, the binding of Sp1, MZF-1, and GATA-2 was demonstrated in an untreated K562 cell line, whereas GATA-1 binding was not observed (Fig. 4C, day 0). The difference of binding factors may be attributed to the fact that K562 is more immature than HL-60 and possesses pluripotency (21). Sp1 and GATA-2 may render the base-line expression of HDAC1 in K562 cells, because GATA-2 appears to be a weak activator in the absence of GATA-1 in reporter assays. Upon megakaryocytic differentiation, the strongest activator GATA-1 was recruited to the promoter along with the dissociation of its inhibitors MZF-1 and GATA-2 in K562 cells. In addition, there was a slight increase in the binding of Sp1 (Fig. 4C, day 4). These changes may underlie the sustained expression of HDAC1 during megakaryocytic differentiation.
Expression Levels of HDAC1 Affect the Differentiation Program of Hematopoietic Progenitor Cells
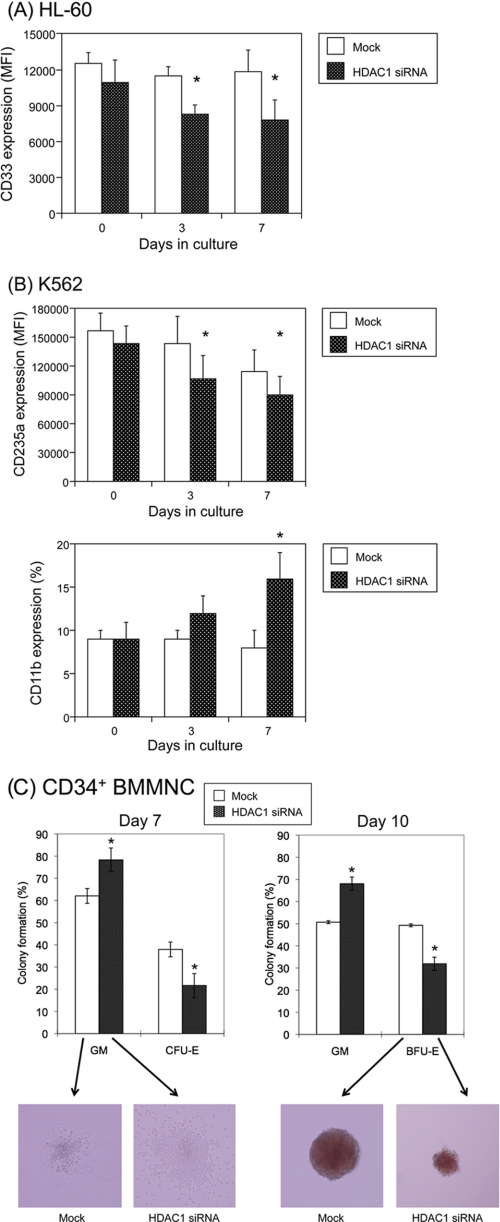
One important question regarding the expression pattern of HDACs during differentiation is whether the change is a simple consequence of differentiation or if it has a functional meaning. To address this question, we carried out loss-of-function studies using siRNA against HDAC1, which represents more than half of HDAC activities in mammalian cells and cannot be compensated by other class I HDACs (13, 24). In a myeloid-committed cell line HL-60, siRNA-mediated knockdown of HDAC1 promoted myeloid differentiation, as revealed by a decrease in the expression of CD33, a marker of immature myeloid cells (30) (Fig. 5A). This suggests that HDAC1 is indispensable for the maintenance of an immature state in HL-60 cells. In multipotent K562 cells, the suppression of HDAC1 caused a decrease of an erythroid marker CD235a and a reciprocal increase of a mature myeloid marker CD11b (Fig. 5B). This implies that overexpressed HDAC1 maintains the erythroid properties of K562 cells, and its reduction results in retrograde differentiation into a myeloid lineage. This is compatible with the expression pattern of HDACs in hematopoietic cells. In addition, we examined the effects of HDAC1 knockdown on normal progenitor cells in clonogenic assays. CD34+ human BMMNCs were transduced with siRNA expression vectors and cultured in methylcellulose medium with full cytokines to induce both myeloid and erythroid differentiation. HDAC1 knockdown reduced the numbers and sizes of erythroid colonies, CFU-E and burst-forming unit-erythroid (BFU-E), with a reciprocal increase in granulocyte/macrophage colonies (Fig. 5C). Taken together, these results point to an instructive role of HDACs in lineage specification during hematopoiesis.
FIGURE 5.
siRNA-mediated knockdown of HDAC1 affects the differentiation program of hematopoietic progenitor cells. A, HL-60 cells were transfected with lentivirus vectors carrying shRNA/siRNA against scrambled sequences (Mock) or HDAC1 (HDAC1 siRNA) and subjected to flow cytometric analysis at the indicated time points. Data shown are the means ± S.D. (bars) of the mean fluorescence intensity (MFI) of CD33 in GFP-positive fractions (n = 3). p values were calculated by paired Student's t test (*, p < 0.05). B, K562 cells were transfected with lentiviral vectors carrying shRNA/siRNA against scrambled sequences or HDAC1 and subjected to flow cytometric analysis at the indicated time points. Data shown are the means ± S.D. (bars) of the mean fluorescence intensity of CD235a and a percentage of CD11b-positive cells in GFP-positive fractions (n = 3). p values were calculated by paired Student's t test (*, p < 0.05). C, CD34+ human bone marrow mononuclear cells were transfected with lentivirus vectors carrying shRNA/siRNA against scrambled sequences or HDAC1. GFP-positive cells were sorted and seeded at 0.5–1 × 103 cells/ml in methylcellulose medium supplemented with full cytokines as described in the legend to Fig. 1. The numbers of CFU-GM/CFU-E and CFU-GM/BFU-E were counted after 7 and 10 days of culture, respectively, and expressed as relative percentages. Data shown are the means ± S.D. of three independent experiments. p values were calculated by paired Student's t test (*, p < 0.05). Representative photographs are shown below (original magnification, ×100).
HDAC1 Overexpression Blocks Myeloid Differentiation in a Murine Stem Cell Transplantation Model
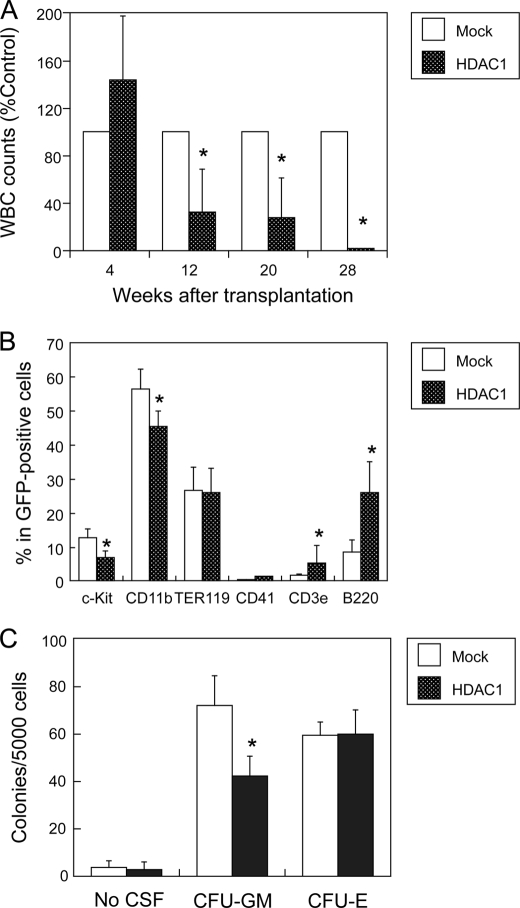
The data so far suggest that HDAC expression should be repressed in normal hematopoietic progenitors, and HDAC overexpression alters the differentiation program, leading to myeloid leukemogenesis. To investigate whether HDAC overexpression actually results in the deregulation of hematopoietic differentiation, we performed stem cell transplantation studies in syngeneic mice (22, 23). We isolated c-KIT-positive bone marrow mononuclear cells from C57BL/6 (Ly-5.1) mice and infected them with retroviruses carrying either HDAC1 cDNA with green fluorescent protein (GFP) or GFP alone (mock). The validity of the expression vector was confirmed by checking HDAC1 overexpression and target gene silencing in transfected cells (supplemental Fig. S7). We transplanted HDAC1- or mock-transfected cells into a lethally irradiated C57BL/6 (Ly-5.2) strain and monitored engraftment by measuring the percentages of GFP-positive and/or Ly-5.1- positive cells in peripheral blood serially. As shown in Fig. 6A, both HDAC1- and mock-transfected progenitor cells successfully reconstructed hematopoiesis in recipient mice 4 weeks after transplantation. However, donor-derived leukocytes gradually declined in the peripheral blood of mice that received HDAC1-transduced cells after 12 weeks and became nearly undetectable after 28 weeks (Fig. 6A). In contrast, there were no significant changes in the numbers of red blood cells and platelets between HDAC1- and mock-transplanted mice (data not shown). This is in line with the results of our in vitro studies indicating that HDAC expression is required for erythro-megakaryocytic differentiation. Leukemic transformation of transplanted cells was not observed up to 60 weeks after transplantation.
FIGURE 6.
Overexpression of HDAC1 blocks myeloid differentiation in a murine stem cell transplantation model. Purified c-KIT-positive bone marrow cells from C57BL/6 (Ly-5.1) mice were transfected with retroviruses carrying either GFP alone (Mock) or HDAC1/GFP (HDAC1) and transplanted into lethally irradiated C57BL/6 (Ly-5.2) mice. A, peripheral blood was drawn at the indicated time points and subjected to blood counting. The numbers of white blood cells are shown as percentages of those of mock-transplanted mice. B, bone marrow mononuclear cells were subjected to flow cytometric analysis of surface marker expression. Data shown are the means ± S.D. (bars) of percentages of each surface marker in GFP-positive fractions (n = 3). C, GFP-positive bone marrow mononuclear cells were cultured in semisolid culture medium supplemented with appropriate cytokines as described under “Experimental Procedures.” Colony numbers were counted after 14 days. Data shown are the means ± S.D. of three independent experiments. p values were calculated by paired Student's t test (*, p < 0.05).
Next, we analyzed the components of bone marrow 10 weeks after transplantation. In HDAC1-transduced mice, there was a significant reduction of c-KIT-positive progenitors and CD11b-positive myeloid cells, whereas CD3- and B220-positive lymphoid cells were relatively increased compared with mock-transfected mice (Fig. 6B). Consistent with normal numbers of red blood cells and platelets in peripheral blood, immature cells of both lineages were normal or slightly increased in the bone marrow of HDAC1-transduced mice. These results indicate that HDAC1 exclusively perturbs myelopoiesis when overexpressed in hematopoietic progenitor cells. To corroborate this notion, we isolated c-KIT/GFP double-positive cells from bone marrow of mice transplanted with either HDAC1- or mock-transduced progenitors and subjected them to in vitro clonogenic growth assays. As shown in Fig. 6C, HDAC1 overexpression reduced the formation of CFU-GM, which corresponds to committed progenitors of myeloid lineage, without affecting mature erythroid progenitors CFU-E. HDAC1-transduced progenitors did not produce spontaneous colonies in this assay (Fig. 6C, No CSF). Furthermore, replating experiments yielded few or no secondary and tertiary colonies in semisolid medium even in the presence of growth factors (data not shown). These results suggest that HDAC1 overexpression alone cannot immortalize hematopoietic stem/progenitor cells, which is in line with the observation of transplantation studies.
DISCUSSION
Class I histone deacetylases are globally implicated in the growth and differentiation of mammalian cells by modifying chromatin structures and gene expression (12, 13). However, relatively little is known about their specific roles in normal hematopoiesis. In this study, we investigated the expression and function of major class I HDACs in normal human hematopoietic cells and obtained the following novel findings. First, the expression of HDACs is very low in CD34+ hematopoietic progenitor cells. Second, HDAC1 plays an instructive role in lineage specification during hematopoiesis. Finally, HDAC1 is overexpressed in AML cells and contributes to leukemogenesis by perturbing myeloid differentiation. Overall, these findings indicate that HDAC is not merely an auxiliary factor of genetic elements but plays a direct role in the cell fate decision of hematopoietic progenitors.
The biological significance of the low level expression of HDACs in hematopoietic progenitor cells has yet to be determined; however, this phenomenon explains why hematological toxicity is relatively weak in patients treated with HDAC inhibitors (17, 18). Inoue et al. (31) have also reported that the expression of Hdac1 mRNA is extremely low in purified murine hematopoietic stem cells compared with cumulus and other somatic cells. They speculate that low HDAC activity underlies insufficient reprogramming of the genome from hematopoietic stem cells after nuclear transfer, which is consistent with the general consensus that hematopoiesis is difficult to reconstruct in vitro by genetic manipulation (32). Our findings fully support their view and further suggest that hematopoietic stem/progenitor cells may possess a unique genetic program, which characterizes the hematopoietic system.
Accumulating evidence indicates that hematopoietic stem/progenitor cells express a wide variety of genes required for multiple differentiation programs, most of which become repressed as lineage choices are restricted during terminal differentiation (33–35). This “priming” of multiple genes is considered to be essential for hematopoietic stem/progenitor cells to maintain multipotency. To accomplish this unique property, hematopoietic stem/progenitor cells have an open chromatin state, which is achieved by hyperacetylation of promoter histones. The low abundance of HDACs should contribute to histone hyperacetylation of “primed” genes in hematopoietic stem/progenitor cells. Indeed, multiple sites of core histones were hyperacetylated in CD34+ progenitor cells compared with AML cells in our study. Moreover, up-regulation of HDACs in more differentiated/committed progenitors is consistent with this notion. If this scenario is true, exogenous interference with the up-regulation of HDACs may inhibit the differentiation of progenitor cells, leading to their expansion. Recent investigations suggest that this is the case: pharmacological down-regulation of HDACs effectively expands hematopoietic progenitor cells ex vivo (36, 37). Similarly, Haumaitre et al. (38) succeeded in amplifying endocrine progenitor cells using HDAC inhibitors.
In this study, we also found that HDAC1 has an instructive role in lineage specification of human hematopoietic cells. HDAC1 is up-regulated in differentiating progenitor cells of multiple lineages by down-regulation of GATA-2 and MZF-1, which are abundantly expressed in early progenitor cells (39, 40). Common myeloid progenitors differentiate into erythro-megakaryocytic lineages when HDAC1 expression is sustained by GATA-1. In contrast, they differentiate into myeloid cells, especially granulocytes, when HDAC1 is down-regulated by C/EBPs. This view is compatible with the recent report by Yamamura et al. (41), in which pharmacological inhibition of HDAC activities enhances interleukin-3 and stem cell factor-mediated generation of committed progenitors from human peripheral blood-derived CD34+ cells and inhibits erythropoietin-induced erythroid differentiation of committed progenitors. Furthermore, experiments using HDAC inhibitors have revealed the roles of HDACs in the differentiation of several cell types, including neurons, oligodendrocytes, osteoblasts, intestinal epithelial cells, adipocytes, and regulatory T cells (42–44). Our results provide the molecular basis of these observations and the rationale for medical application of HDAC inhibitors to tissue regeneration.
Finally, we found that HDACs are overexpressed in nearly all primary AML cells and cell lines without fusion gene products caused by chromosomal translocations. The involvement of HDACs in leukemogenesis in AML cells carrying fusion gene products has been well characterized and provides a theoretical basis for the efficacy of HDAC inhibitors for acute leukemias with chromosomal translocations (14, 15). Our findings extend this view and propose more generalized roles of HDACs in leukemogenesis and the validity of class I HDACs as therapeutic targets in various hematological malignancies. As a result of HDAC overexpression, histone acetylation was diminished at multiple residues, including histone H4-lysine 16. Loss of acetylation at histone H4-lysine 16 was shown to be an epigenetic hallmark of human cancer, including AML, by Fraga et al. (45). Therefore, overexpression of HDACs may be a common feature of malignant cells, playing a fundamental role in oncogenesis. Indeed, overexpression of HDACs, mostly HDAC1, has been described in gastric, prostate, breast, cervical, and colon cancers (46, 47). As for the mechanisms of HDAC overexpression in AML, we speculate that loss-of-function mutations of C/EBP or abnormalities in the expression of MZF-1 and GATA2 cause aberrant expression of HDAC1 in hematopoietic stem/progenitor cells. Although further investigation is required to elucidate the underlying mechanisms, HDAC overexpression justifies the application of HDAC inhibitors for cancer treatment.
According to the two-hit theory, two independent genetic abnormalities are required for the development of AML (11). Class I mutations confer a growth advantage on hematopoietic stem cells, and class II mutations block differentiation. The results of our transplantation studies suggest that HDAC1 acts as a novel class II transforming gene upon overexpression in hematopoietic stem/progenitor cells. From a mechanistic standpoint, two questions emerge immediately. What is the identity of the downstream effectors? What are the accompanying class I mutations? To address the first question, we performed DNA microarray analysis of HDAC1-transduced c-KIT-positive bone marrow cells from recipient mice. Among 22,201 genes screened, the expressions of 153 genes (0.69%) were up-regulated more than 2-fold in HDAC1-transduced c-KIT+ progenitor cells compared with mock-transfected control (supplemental Fig. S8). Similarly, down-regulation was observed in 63 genes (0.28%). Supplemental Table S2 shows part of the HDAC1-regulated genes, in which the alteration of the expression levels was confirmed by real time quantitative RT-PCR. Among them, up-regulation of c-Mpl is most interesting, because it is essential for megakaryocyte development (48) and for maintaining hematopoietic stem cells in the G0 phase of the cell cycle (49). It is possible that up-regulated c-MPL directs HDAC1-expressing progenitors into quiescence and also skews their differentiation program into erythro- megakaryocytic lineage. Regarding the second question, we have found a positive correlation of the presence of internal tandem duplication of FLT3 (FLT3-ITD), the most frequent class I abnormality in de novo AML (50), with the expression levels of HDAC1 (supplemental Fig. S9). This strongly suggests that HDAC1 overexpression and FLT3 mutations collaborate to transform progenitor cells into leukemic clones. Extensive investigation is currently under way in our laboratory to address the functional interplay between HDAC1 overexpression and c-MPL up-regulation and/or FLT3 mutations in leukemogenesis.
In summary, our findings add new insight into the epigenetic regulation of normal and malignant hematopoiesis and ultimately contribute to the development of better treatment strategies in diseases of the hematopoietic system.
Supplementary Material
Acknowledgments
We are grateful to Dr. Tsukasa Ohmori (Jichi Medical School) for helpful discussions and technical advice and Drs. Stuart Schreiber, Robert Hromas, Atsushi Iwama, and Mitsuru Nakamura for materials.
This work was supported in part by the High-Tech Research Center Project for Private Universities: Matching Fund Subsidy from MEXT 2002–2006 and grants from the Vehicle Racing Commemorative Foundation and the Sankyo Foundation of Life Science (to Y. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S7.
- C/EBP
- CCAAT/enhancer-binding protein
- HDAC
- histone deacetylase
- AML
- acute myeloblastic leukemia
- BMMNC
- bone marrow mononuclear cells
- CFU-GM
- colony-forming unit-granulocyte/macrophage
- CFU-E
- colony-forming unit-erythroid
- BFU-E
- burst-forming unit-erythroid
- siRNA
- small interfering RNA
- RT
- reverse transcription
- GFP
- green fluorescent protein
- shRNA
- short hairpin RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Orkin S. H., Zon L. I. (2008) Cell 132, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furukawa Y., Kikuchi J., Nakamura M., Iwase S., Yamada H., Matsuda M. (2000) Br. J. Haematol. 110, 663–673 [DOI] [PubMed] [Google Scholar]
- 3.Martin D. I., Zon L. I., Mutter G., Orkin S. H. (1990) Nature 344, 444–447 [DOI] [PubMed] [Google Scholar]
- 4.Orkin S. H., Shivdasani R. A., Fujiwara Y., Mcdevitt M. A. (1998) Stem Cells 16, 79–83 [DOI] [PubMed] [Google Scholar]
- 5.Heyworth C., Gale K., Dexter M., May G., Enver T. (1999) Genes Dev. 13, 1847–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling K. W., Ottersbach K., van Hamburg J. P., Oziemlak A., Tsai F. Y., Orkin S. H., Ploemacher R., Hendriks R. W., Dzierzak E. (2004) J. Exp. Med. 200, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasaki H., Mizuno S., Arinobu Y., Ozawa H., Mori Y., Shigematsu H., Takatsu K., Tenen D. G., Akashi K. (2006) Genes Dev. 20, 3010–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang D. E., Zhang P., Wang N. D., Hetherington C. J., Darlington G. J., Tenen D. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka T., Akira S., Yoshida K., Umemoto M., Yoneda Y., Shirafuji N., Fujiwara H., Suematsu S., Yoshida N., Kishimoto T. (1995) Cell 27, 353–361 [DOI] [PubMed] [Google Scholar]
- 10.Fröhling S., Scholl C., Gilliland D. G., Levine R. L. (2005) J. Clin. Oncol. 23, 6285–6295 [DOI] [PubMed] [Google Scholar]
- 11.Gilliland D. G. (2002) Semin. Hematol. 39, 6–11 [DOI] [PubMed] [Google Scholar]
- 12.Yang X. J., Seto E. (2008) Nat. Rev. Mol. Cell Biol. 9, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagger G., O'Carroll D., Rembold M., Khier H., Tischler J., Weitzer G., Schuettengruber B., Hauser C., Brunmeir R., Jenuwein T., Seiser C. (2002) EMBO J. 21, 2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin R. J., Nagy L., Inoue S., Shao W., Miller W. H., Jr., Evans R. M. (1998) Nature 391, 811–814 [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Hoshino T., Redner R. L., Kajigaya S., Liu J. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10860–10865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu W. S., Parmigiani R. B., Marks P. A. (2007) Oncogene 26, 5541–5552 [DOI] [PubMed] [Google Scholar]
- 17.Byrd J. C., Marcucci G., Parthun M. R., Xiao J. J., Klisovic R. B., Moran M., Lin T. S., Liu S., Sklenar A. R., Davis M. E., Lucas D. M., Fischer B., Shank R., Tejaswi S. L., Binkley P., Wright J., Chan K. K., Grever M. R. (2005) Blood 105, 959–967 [DOI] [PubMed] [Google Scholar]
- 18.O'Connor O. A., Heaney M. L., Schwartz L., Richardson S., Willim R., MacGregor-Cortelli B., Curly T., Moskowitz C., Portlock C., Horwitz S., Zelenetz A. D., Frankel S., Richon V., Marks P., Kelly W. K. (2006) J. Clin. Oncol. 24, 166–173 [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi J., Shimizu R., Wada T., Ando H., Nakamura M., Ozawa K., Furukawa Y. (2007) Stem Cells 25, 2439–2447 [DOI] [PubMed] [Google Scholar]
- 20.Zhao K. W., Li X., Zhao Q., Huang Y., Li D., Peng Z. G., Shen W. Z., Zhao J., Zhou Q., Chen Z., Sims P. J., Wiedmer T., Chen G. Q. (2004) Blood 104, 3731–3738 [DOI] [PubMed] [Google Scholar]
- 21.Terui Y., Furukawa Y., Kikuchi J., Iwase S., Hatake K., Miura Y. (1998) Exp. Hematol. 26, 236–244 [PubMed] [Google Scholar]
- 22.Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. (2003) Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 23.Ono R., Nakajima H., Ozaki K., Kumagai H., Kawashima T., Taki T., Kitamura T., Hayashi Y., Nosaka T. (2005) J. Clin. Invest. 115, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Lin Q., Wang W., Wade P., Wong J. (2002) Genes Dev. 16, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senese S., Zaragoza K., Minardi S., Muradore I., Ronzoni S., Passafaro A., Bernard L., Draetta G. F., Alcalay M., Seiser C., Chiocca S. (2007) Mol. Cell Biol. 27, 4784–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glozak M. A., Seto E. (2007) Oncogene 26, 5420–5432 [DOI] [PubMed] [Google Scholar]
- 27.Krämer O. H., Zhu P., Ostendorff H. P., Golebiewski M., Tiefenbach J., Peters M. A., Brill B., Groner B., Bach I., Heinzel T., Göttlicher M. (2003) EMBO J. 22, 3411–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuettengruber B., Simboeck E., Khier H., Seiser C. (2003) Mol. Cell Biol. 23, 6993–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong Q., Tsai J., Tan G., Dalgin G., Hotamisligil G. S. (2005) Mol. Cell Biol. 25, 706–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stelzer G. T., Shults K. E., Loken M. R. (1993) Ann. N.Y. Acad. Sci. 677, 265–280 [DOI] [PubMed] [Google Scholar]
- 31.Inoue K., Ogonuki N., Miki H., Hirose M., Noda S., Kim J. M., Aoki F., Miyoshi H., Ogura A. (2006) J. Cell Sci. 119, 1985–1991 [DOI] [PubMed] [Google Scholar]
- 32.Ledran M. H., Krassowska A., Armstrong L., Dimmick I., Renström J., Lang R., Yung S., Santibanez-Coref M., Dzierzak E., Stojkovic M., Oostendorp R. A., Forrester L., Lako M. (2008) Cell Stem Cell 3, 85–98 [DOI] [PubMed] [Google Scholar]
- 33.Hu M., Krause D., Greaves M., Sharkis S., Dexter M., Heyworth C., Enver T. (1997) Genes Dev. 11, 774–785 [DOI] [PubMed] [Google Scholar]
- 34.Warren L., Bryder D., Weissman I. L., Quake S. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17807–17812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H. F., John R. M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A. G. (2006) Nat. Cell Biol. 8, 532–538 [DOI] [PubMed] [Google Scholar]
- 36.De Felice L., Tatarelli C., Mascolo M. G., Gregorj C., Agostini F., Fiorini R., Gelmetti V., Pascale S., Padula F., Petrucci M. T., Arcese W., Nervi C. (2005) Cancer Res. 65, 1505–1513 [DOI] [PubMed] [Google Scholar]
- 37.Bug G., Gül H., Schwarz K., Pfeifer H., Kampfmann M., Zheng X., Beissert T., Boehrer S., Hoelzer D., Ottmann O. G., Ruthardt M. (2005) Cancer Res. 65, 2537–2541 [DOI] [PubMed] [Google Scholar]
- 38.Haumaitre C., Lenoir O., Scharfmann R. (2008) Mol. Cell Biol. 28, 6373–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagai T., Harigae H., Ishihara H., Motohashi H., Minegishi N., Tsuchiya S., Hayashi N., Gu L., Andres B., Engel J. D., Yamamoto M. (1994) Blood 84, 1074–1084 [PubMed] [Google Scholar]
- 40.Gaboli M., Kotsi P. A., Gurrieri C., Cattoretti G., Ronchetti S., Cordon-Cardo C., Broxmeyer H. E., Hromas R., Pandolfi P. P. (2001) Genes Dev. 15, 1625–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamura K., Ohishi K., Katayama N., Yu Z., Kato K., Masuya M., Fujieda A., Sugimoto Y., Miyata E., Shibasaki T., Heike Y., Takaue Y., Shiku H. (2006) Br. J. Haematol. 135, 242–253 [DOI] [PubMed] [Google Scholar]
- 42.Hsieh J., Nakashima K., Kuwabara T., Mejia E., Gage F. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16659–16664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen S., Sandoval J., Swiss V. A., Li J., Dupree J., Franklin R. J., Casaccia-Bonnefil P. (2008) Nat. Neurosci. 11, 1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao R., de Zoeten E. F., Ozkaynak E., Chen C., Wang L., Porrett P. M., Li B., Turka L. A., Olson E. N., Greene M. I., Wells A. D., Hancock W. W. (2007) Nat. Med. 13, 1299–1307 [DOI] [PubMed] [Google Scholar]
- 45.Fraga M. F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Iyer N. G., Pérez-Rosado A., Calvo E., Lopez J. A., Cano A., Calasanz M. J., Colomer D., Piris M. A., Ahn N., Imhof A., Caldas C., Jenuwein T., Esteller M. (2005) Nat. Genet. 37, 391–400 [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z., Yamashita H., Toyama T., Sugiura H., Ando Y., Mita K., Hamaguchi M., Hara Y., Kobayashi S., Iwase H. (2005) Breast Cancer Res. Treat. 94, 11–16 [DOI] [PubMed] [Google Scholar]
- 47.Zhu P., Martin E., Mengwasser J., Schlag P., Janssen K. P., Göttlicher M. (2004) Cancer Cell 5, 455–463 [DOI] [PubMed] [Google Scholar]
- 48.Gurney A. L., Carver-Moore K., de Sauvage F. J., Moore M. W. (1994) Science 265, 1445–1447 [DOI] [PubMed] [Google Scholar]
- 49.Yoshihara H., Arai F., Hosokawa K., Hagiwara T., Takubo K., Nakamura Y., Gomei Y., Iwasaki H., Matsuoka S., Miyamoto K., Miyazaki H., Takahashi T., Suda T. (2007) Cell Stem Cell 1, 685–697 [DOI] [PubMed] [Google Scholar]
- 50.Parcells B. W., Ikeda A. K., Simms-Waldrip T., Moore T. B., Sakamoto K. M. (2006) Stem Cells 24, 1174–1184 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.